Abstract
Relative gene expression analysis through RT-qPCR is an important molecular technique that helps understanding different molecular mechanisms, such as the plant defense response to insect pests. However, the use of RT-qPCR for gene expression analysis can be affected by factors that directly affect the reliability of the results. Among these factors, the appropriate choice of reference genes is crucial and can strongly impact RT-qPCR relative gene expression analyses, highlighting the importance in correctly choosing the most suitable genes for the success of the analysis. Thus, this study aimed to select and validate reference genes for relative gene expression studies through RT-qPCR in hybrids of Eucalyptus tereticornis × Eucalyptus camaldulensis (drought tolerant and susceptible to Leptocybe invasa) under conditions of inoculation by the Beauveria bassiana fungus and subsequent infestation by L. invasa. The expression level and stability of eleven candidate genes were evaluated. Stability was analyzed using the RefFinder tool, which integrates the geNorm, NormFinder, BestKeeper, and Delta-Ct algorithms. The selected reference genes were validated through the expression analysis of the transcriptional factor EcDREB2 (dehydration-responsive element-binding protein 2). For all treatments evaluated, EcPTB, EcPP2A-1, and EcEUC12 were the best reference genes. The triplets EcPTB/EcEUC12/EcUBP6, EcPP2A-1/EcEUC12/EcPTB, EcIDH/EcSAND/Ecα-TUB, EcPP2A-1/Ecα-TUB/EcPTB, and EcPP2A-1/EcUPL7/EcSAND were the best reference genes for the control plants, mother plants, plants inoculated with B. bassiana, plants infested with L. invasa, and plants inoculated with B. bassiana and subsequently infested with L. invasa, respectively. The best determined reference genes were used to normalize the RT-qPCR expression data for each experimental condition evaluated. The results emphasize the importance of this type of study to ensure the reliability of relative gene expression analyses. Furthermore, the findings of this study can be used as a basis for future research, comprising gene expression analysis of different eucalyptus metabolic pathways.
Similar content being viewed by others
Introduction
The Eucalyptus genus belongs to the Myrtaceae family and is composed by approximately 600 species and subspecies, being one of the main global sources of wood and widely cultivated for industrial use1,2. Worldwide, raw materials derived from the forestry sector are used in the production of various products, such as civil construction structures, furniture, paper, pharmaceutical, and cosmetic products, being also used for energy generation3. Globally, Eucalyptus spp. is the most extensively cultivated forest genus, with a planted area of around 25 million hectares4. Brazil stands out as the world's largest eucalyptus producer, with a cultivation area of more than 7 million hectares, which corresponds to 75.8% of the total area planted with trees, playing an important role in its economy, and generating a total revenue of more than 47 billion of dollars and 2 million jobs5. Thus, the economic and socioeconomic importance of forest resources has driven the growth of the sector, although the incidence of biotic factors, such as pests, still represents a challenge to be overcome6,7.
The eucalyptus gall wasp (Leptocybe invasa) is a biotic factor that affects the sustainability of eucalyptus plantations, as it causes serious damage through the induction of galls along the entire length of veins and on petioles of young leaves, and also in internodes of branch apices. Infestation by this pest can lead to devastating outcomes, such as rough appearance of plants, stunted growth, and in extreme cases, plant death8,9,10. To meet this challenge, it is essential to implement the best combination of integrated tools and techniques. Thus, studies related to plant-microorganism association, especially with endophytic organisms, are essential considering the great damage caused by the attack of insect pests, such as the eucalyptus gall wasp11,12. Endophytic fungi act symbiotically, triggering, for instance, local and/or systemic defenses13. The species Beauveria bassiana (Bals.) Vuill is an entomopathogenic fungus often used as a biopesticide in agricultural crops due to its environmental safety, not causing any risk to human health and displaying only minimal adverse effects on non-target organisms12,14. In Eucalyptus, the use of B. bassiana as an alternative biological agent to control the galling insect L. invasa has shown great potential for plant resistance to attack by this pest12.
The use of molecular techniques to study eucalyptus species, as well as studies aimed at biological control, has gained progressive attention due to the diverse possibilities they offer for a better comprehension of plant metabolism. The sequencing of the Eucalyptus grandis genome has enabled significant advances in molecular studies by providing information on genes related to metabolic pathways of great economic interest15. In this context, gene expression analysis is an important tool to better understand the molecular mechanisms behind different biological processes16 such as the defense response to L. invasa infestation under the presence of B. bassiana. Currently, several methods of gene expression analysis in plants are used17, with the quantitative Real-Time Polymerase Chain Reaction with reverse transcription (RT-qPCR) being one of the most common and widely used method due to its speed, high sensitivity, reproducibility, and precision in determining gene expression levels18,19,20. However, it is important to consider that the use of RT-qPCR for RNA quantification is prone to several factors that can directly affect the reliability of the results obtained21.
RNA (ribonucleic acid) integrity and quality, cDNA (complementary DNA) synthesis and amplification efficiencies, and the choice of reference genes are crucial factors that can strongly impact the results of RT-qPCR studies20,22. Therefore, to ensure reproducibility and minimize the variability of RT-qPCR assays, it is crucial to evaluate these parameters21. In particular, among the factors previously mentioned, the determination of reference genes is one of the most important aspects when using gene expression analysis by RT-qPCR1, since the efficiency of the technique depends on the validation of appropriate reference genes for accurate assay normalization and correction of nonspecific variations23,24,25. For this reason, the use of validated reference genes, based on their expression stability in different tissues and experimental conditions, is essential to guarantee the quality and reliability of the results26.
The use of reference genes for the normalization of RT-qPCR gene expression data is based on the fact that, in theory, their expression levels are constant, regardless of the tissues, developmental stages or physiological conditions of the evaluated species17,27. Thus, different genes involved in the maintenance of basic cellular functions such as cell division, growth and development, apoptosis, and other physiological processes are often used as reference genes due to their ability to be expressed in basically every cell of the organism26. Plants genes such as ACT, TUB, ribosomal RNA genes (18S, rRNA, 26S rRNA), GAPDH, UBQ, EF-1α, UBC, and PP2A are commonly employed as reference genes17,25 However, different studies have shown that the expression of these genes may vary, leading to incorrect normalization of target genes28,29 and confirming that there is no universal reference gene30,31,32,33.
Different studies aiming to determine the best eucalyptus reference genes have been performed1,2,34,35,36,37,38,39. However, such studies are still necessary when specific genotypes, tissues or stress conditions are under study40,41. In this context, studies aimed to determine the best reference genes in eucalyptus under L. invasa infestation and inoculation of the B. bassiana fungus have not been carried out yet. Therefore, considering the importance of the RT-qPCR technique for gene expression studies and the need to validate suitable reference genes for specific conditions, the objective of this study was to select and validate reference genes for RT-qPCR studies in the Eucalyptus. tereticornis × Eucalyptus. camaldulensis hybrid (drought tolerant and susceptible to L. invasa) under the inoculation of B. bassiana and subsequently infestation by L. invasa.
Results
Identification and selection of target and reference genes
The literature review returned seven articles related to the selection of reference genes in Eucalyptus spp., from which four were included in this analysis. The inclusion criteria were based on the possibility of identifying the gene sequences described in each study. After selecting the articles, 12 reference genes were chosen, following the recommendation criteria of the study in which they were analyzed. In other words, the genes that showed the greatest expression stability in the samples evaluated in each study were selected. Thus, the following genes were chosen: PP2A-1 (Protein phosphatase 2A-2), SAND (SAND family, trafficking protein Mon1), UPL7 (Ubiquitin-protein ligase 7), PTB (Polypyrimidine tract-binding protein 1)38; Actin-2 (Actin 2), 18srRNA (RNA ribosomal 18S), IDH (NADP-isocitrate dehydrogenase)34, α-TUB (α-Tubulin), UBC (Ubiquitin C), EF-1α (Elongation factor 1-α)37, EUC12 (Putative RNA binding protein), and H2B (Histone H2B)36.
In order to identify and select a target gene, gene expression levels of 155 genes from the AP2/EREBP superfamily (APETALA2/Ethylene Responsive Element Binding Protein) were analyzed during L. invasa infestation, using the gene expression graph generated by the ExHeatmap option, as described by Sundell et al.42. The transcriptional factor DREB2 (dehydration-responsive element-binding protein) stood out from the analyzed genes, displaying high expression levels in E. grandis tissues subjected to L. invasa attack, being selected for the further analyses.
Expression levels of the candidate reference genes
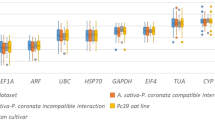
The expression analyses of the candidate reference genes in all treatments (Fig. 1A) indicated a wide variation in the Cqs average values, with the minimum and maximum values of 19.33 and 25.87, respectively. When each treatment was separately analyzed, it was possible to observe a similar expression pattern to the one observed when all treatments are analyzed together (Fig. 1A). The Cq average values in the control treatment (Fig. 1B), in mother plants (Fig. 1C), in plants inoculated with B. bassiana (Fig. 1D), in plants infested with L. invasa (Fig. 1E), and in plants inoculated with B. bassiana and infested with L. invasa (Fig. 1F) ranged from 18.77, 18.79, 18.64, 20.10, and 20.55 (lowest value) to 25.58, 25.88, 24.99, 26.06, and 26.94 (highest value), respectively.
Candidate reference gene expression levels based on the Cq (Cycle of Quantification) data obtained from stem, leaves and leaf apex of eucalyptus plants under different treatments conditions (a), eucalyptus control plants (b), eucalyptus mother plants (c), eucalyptus plants inoculated with B. bassiana (d), eucalyptus plants infested with L. invasa (e), and eucalyptus plants inoculated with B. bassiana and infested with L. invasa (f). Vertical bars represent the standard deviation, and the black dots represent the mean Cq values.
The Cq average values for all genes revealed that, regardless of the evaluated condition, the three genes with the highest and lowest expression levels were the same. When comparing the average Cq values of each treatment (Fig. 1), EcACT, EcH2B, and EcIDH showed the highest expression levels, with values varying from 18.64 to 21.27. In contrast, EcUPL7, EcEF1-α, and EcPTB displayed the lowest expression levels, with Cq average values ranking from 23.91 to 26.94.
Expression stability of the candidate reference genes
Tables 1, 2, 3, 4 and 5, along with Fig. 2 describe the expression stability analysis of the candidate reference genes using the geNorm, NormFinder, BestKeeper, Delta-Ct, and RefFinder algorithms. The data presented in the previously mentioned tables comprise the analysis of gene stability in 5 sample sets, and the Cqs of all genes and conditions evaluated can be found in Tables (Supplementary S1), making it possible to combine several other sample sets.
Ranking of the candidate reference genes according to their stability value calculated by the geNorm (a), NormFinder (b), BestKeeper (c), Delta-Ct (d), and RefFinder (e) algorithms using the Cq data (Cycle of Quantification) obtained from all five treatments (Control plants; Mother plants; Plants infested with L. invasa; Plants inoculated with B. bassiana; Plants infested with L. invasa and inoculated with B. bassiana).
Control plants
In control plants (Table 1), the geNorm, NormFinder, and Delta-Ct algorithms indicated that EcPTB, EcEUC12, and EcPP2A-1 were the most stable genes. The BestKeeper algorithm, on the other hand, found that EcUBP6, EcPTB, and EcEUC12 were most stable in this treatment. In terms of the general classification provided by the RefFinder algorithm, EcPTB, EcEUC12, and EcUBP6 were the best reference genes. In relation to the least stable genes, EcH2B, EcEF1-α, and EcIDH were identified by the geNorm, NormFinder, and Delta-Ct algorithms as the least stable genes. BestKeeper classified EcEF1-α, EcACT, and EcIDH as the most variable genes and RefFinder considered EcEF1-α, EcH2B, and EcIDH as the genes with the lower stability values.
Mother plants
The stability values of the candidate reference genes for the mother plants (Table 2) presented different results among the algorithms analyzed. EcPP2-A, EcEUC12, and EcPTB were indicated as the most stable genes by geNorm and Delta-Ct algorithms, while EcEUC12, EcPP2-A, and Ecα-TUB were classified as the most stable by NormFinder. On the other hand, according to BestKeeper, EcUBP6, EcIDH, and EcSAND are most stable genes for mother plants and for RefFinder EcPP2-A, EcEUC12, and EcPTB displayed the higher expression stability. About the least stable genes, all algorithms identified EcH2B, EcACT, and EcEF1-α as the worst (least stable) genes, except for NormFinder that, instead of EcEF1-α, classified EcUBP6 among the three least stable genes.
Plants inoculated with B. bassiana
For the plants inoculated with the B. bassiana fungus, the expression stability analysis revealed significant differences among the five algorithms used (Table 3). The geNorm listed EcIDH, Ecα-TUB, and EcUPL7 as the most stable genes, while NormFinder considered EcIDH, EcSAND, and EcUPL7. BestKeeper classified EcEUC12, EcPTB, and EcUBP6 as the most stable genes and for Delta-Ct, on the other hand, EcSAND, EcIDH, and EcUPL7 showed the best stability levels. Finally, RefFinder, which takes into account the results of all algorithms, classified EcIDH, EcSAND, and Ecα-TUB as the most stable genes. In relation to the genes with relatively lower stability, geNorm, NormFinder, and Delta-Ct classified EcEF1-α, EcACT, and EcEUC12 as the least stable genes. Similarly, the BestKeeper algorithm also identified EcEF1-α and EcACT as the genes with lower stability levels and differed only in the inclusion of Ecα-TUB, instead of EcEUC12, when compared to the previously mentioned algorithms. For the overall ranking defined by RefFinder, EcEF1-α, EcACT, and EcUBP6 were found to be the least stable genes.
Plants infested with L. invasa
When eucalyptus plants were under L. Invasa infestation, the four algorithms used consistently indicated the same set of genes as the most stable ones: EcPP2A-1, Ecα-TUB, and EcPTB (Table 4). Similarly, these algorithms also identified the same genes as the least stable ones: EcACT, EcH2B, and EcIDH, except for BestKeeper, which indicated the EcEF1-α instead of EcIDH. The only difference, in both cases, was the position in which the genes were classified (Table 4).
Plants inoculated with B. bassiana and infested with L. invasa
Similarities and divergences could be found for eucalyptus plants under B. bassiana inoculation and L. invasa infestation (Table 5). For the most stable genes, geNorm pointed out the genes EcPP2A-1, EcSAND, and EcEUC12 as the best ones. NormFinder identified EcUPL7, EcPP2A-1 and, EcEUC12 as the most stable genes, while BestKeeper presented a slightly different classification, classifying EcUBP6, EcIDH, and EcSAND as the genes with higher expression stability. The Delta-Ct and the RefFinder considered the same set of genes, EcPP2A-1, EcUPL7, and EcSAND, as the most stable in terms of expression, although in different positions (Table 5). For the least stable genes, the geNorm, NormFinder, and DeltaCT algorithms indicated EcUBP6, EcH2B, and ECEF1-α as the genes displaying higher expression variability. On the other hand, BestKeeper ranked EcH2B, EcEF1-α, and EcACT as the least stable genes, just like RefFinder (Table 5).
Overall treatment analysis
Cq values from the five evaluated treatments were combined to analyze the overall expression stability of the candidate reference genes (Fig. 2). EcPP2A-1, EcPTB, and Ecα-TUB were defined by NormFinder and DeltaCt algorithms as the most stable genes. geNorm indicated EcPTB, EcEUC12, and EcPP2A-1 as the genes with higher expression stability, while BestKepper listed EcUBP6, EcPTB, and EcEUC12. RefFinder classified EcPTB, EcPP2A-1, and EcEUC12 as the most stable reference genes when every treatment is analyzed at the same time. Regarding the least stable genes, the geNorm, BestKepper, DeltaCt, and RefFinder algorithms indicated the same set of genes, EcH2B, EcACT, and EcEF1-α, as those with higher expression variability, while for NormFinder, EcH2B, EcACT, and EcUBP6 were classified as least stable genes.
Reference gene validation
The impact of choosing different reference genes was observed by analyzing the relative expression of EcDREB2 (Fig. 3). The expression profile of the EcDREB2 obtained from plants inoculated with the fungus B. bassiana and infested with the wasp L. invasa showed that significant differences can occur depending on the reference genes chosen for the normalization process.
EcDREB2 expression pattern, normalized with the most (EcPP2A-1, EcULP7 and EcSAND) and least (EcH2B, EcEF1-α e EcACT) stable reference genes, according to the RefFinder algorithm, in stem and leaf Apex from eucalyptus plants submitted to L. invasa infestation and B. Bassiana inoculation. Columns represent the fold different in gene expression in relation to a calibrator sample (stem). Expression levels were obtained from three biological replicates and the error bars represent the standard error among these replicates.
When EcDREB2 was normalized by using the most stable reference genes according to RefFinder (EcPP2A-1, EcULP7, and EcSAND) its expression was more than two times higher in leaf apex when compared to stem tissues (Fig. 3). However, when normalized by the least stable reference genes (EcH2B, EcEF1-α, and EcACT) there was no difference in EcDREB2 expression between these tissues.
Discussion
RT-qPCR analysis is universally accepted as a robust tool for quantifying gene expression levels33,43. However, it is important to highlight that the calculation used to measure the gene expression levels requires an essential step that is data normalization44. Normalization is necessary to correct experimental variations, such as differences in sample collection, total RNA extraction, cDNA synthesis, and procedures inherent to the RT-qPCR technique itself, guaranteeing the precision, reproducibility, and reliability of the results45,46. In this context, a common strategy employed is the use of reference genes as internal normalization standards. This approach is widely recognized as one of the most efficient for correcting nonspecific variations and achieving accurate normalization of gene expression data by RT-qPCR25,47.
The formulas used to calculate gene expression levels by RT-qPCR, such as those described by Livak and Schmittgen48 and Pfaffl49, necessarily require the use of reference genes. Therefore, their appropriate selection, as performed in this research, is a fundamental preliminary step in gene expression studies, since the use of inappropriate reference genes can significantly compromise the accuracy and interpretation of the results33,50. In this sense, several studies have been carried out, in different organisms and experimental conditions, with the aim of selecting and validating the best reference genes for RT-qPCR gene expression analyses51,52,53,54.
The results obtained in the present study provides the most suitable reference genes for the normalization of RT-qPCR data in eucalyptus plants under the inoculation by B. bassiana and L. invasa infestation. Although several studies related to the selection of reference genes in eucalyptus have been performed so far1,2,34,35,36,37, this is the first time that such analysis is conducted in Eucalyptus (E. tereticornis × E. camaldulensis hybrid) plants under B. bassiana inoculation and L. invasa infestation.
Expression levels of all candidate reference genes
Selection of suitable reference gene requires meeting several criteria, including: (1) to be expressed at different developmental stages, physiological conditions, and tissues; (2) to display stable expression levels among different tissues and developmental, physiological and environmental conditions; (3) to display moderate to high expression levels (Cq value between 15 and 30); (4) to display the ability to reflect variations in the quantity and quality of RNA47,55. These criteria are essential to ensure accurate and reliable choice of a reference gene.
The results found here demonstrated that the maximum and minimum Cq values from all tissues and experimental conditions were within the range recommended in the literature (minimum of 19.33 and maximum of 25.87)55. Furthermore, when comparing the data obtained in this study with other studies carried out in eucalyptus that used the same analysis approach, a similar Cq variation was observed among the candidate reference genes. In the study conducted by Boava et al.35, the maximum and minimum Cq values of the 13 reference genes varied from 16 to 27. De Almeida et al.36 analyzed the expression of 11 candidate reference genes and obtained minimum and maximum Cq values of 12 and 25, a difference of 13 cycles. In the study conducted by Fernández et al.37, 10 genes were evaluated and the Cq variation among them was of approximately 7 cycles (Cqs from 21 to 28). In our data, the minimum and maximum Cq values among all genes evaluated varied from 18 to 27.
These results are in accordance with the variations observed in the literature and also with two of the essential criteria highlighted by Ling et al.55 and Mughal et al.47 for the selection of adequate reference genes: (1) to be expressed at different developmental stages, physiological conditions, and tissues; (2) to display moderate to high expression levels (Cq value between 15 and 30). This contributes to the reliability of the analyses performed in this study and reinforces the importance of carefully evaluating gene expression levels when aiming to select the most appropriate reference genes for normalizing RT-qPCR gene expression data. Furthermore, this analysis has been widely explored in several studies with similar objectives51,54,56,57,58.
Reference gene expression stability
Reference genes have been extensively used for the normalization of RT-qPCR gene expression data, as they are assumed to be expressed at constant levels, regardless of the tissues under analysis, experimental treatment, developmental stages or physiological conditions of the species evaluated28,56. Thus, the use of genes involved in basal metabolism and structural integrity of the cell as reference genes is based on the general assumption that their expression levels are stable, independent of environmental/experimental conditions and cell type59. Therefore, genes that play essential roles in maintaining basic cellular functions, such as growth and development processes, apoptosis, cell division, and other physiological processes, are commonly used as reference genes, due to their ability to be expressed in all cells of an organism26.
ACT, GAPDH, EF1-α, TUB, UBC, and PP2A are often used as reference genes for the normalization of RT-qPCR studies25. However, several studies have demonstrated that the expression of many commonly used reference genes can significantly vary their expression pattern, and this may lead to inadequate normalization of target genes19,28,29,44,60. This reinforces that no universal reference gene has been identified, since several studies have shown that none of the reference genes tested so far can be used across different species, environmental conditions, developmental stages, or different tissues30,31,32,61. Therefore, evaluating reference gene stability in different tissues, developmental, physiological, and environmental conditions is one of the fundamental criteria during the process of selecting the most suitable reference genes47,55.
Genes commonly used and described in literature as adequate reference genes, such as ACT, TUB, PP2A, UBC, and EF1-α, were evaluated in this study and the results confirmed that the selection of more stable reference genes may vary depending on the experimental conditions. This pattern has also been observed in other eucalyptus reference genes studies1,2,35. Thus, since the ideal reference gene must present relatively stable expression levels in different cultivars, tissues and conditions47,55, the results described in the literature highlight the challenge of identifying suitable genes for different conditions and the importance of validating reference genes.
The algorithms used in this study, geNorm59, NormFinder62, BestKeeper63, Delta-Ct64, and RefFinder65,66 have been commonly used to evaluate the stability of reference genes in plants19,25,54, animals67,68,69 and microorganisms70,71,72. They are used to calculate the stability of gene expression using different mathematical models. Consequently, when analyzing the results among these algorithms, divergences can be observed in the establishment of the most stable genes60. In order to increase the reliability of the reference gene selection process, at least two different algorithms should be used to select the most stable reference genes73, since there may be some variation in the selected genes among them1,28,44,60,74. For this reason, RefFinder stands as an interesting alternative, instead of using these algorithms individually, since it uses the individual results obtained by each algorithm to perform a reanalysis and generate an overall classification of the most stable genes66. The RefFinder tool has been widely used in studies aimed at determining the best reference genes51,54,56,74.
In the present study, the Cq data of all candidate reference genes were submitted to a stability analysis using RefFinder, which allowed the identification of the genes that showed the greater stability in each treatment and enabled the observation of some similarities with other studies conducted eucalyptus plants. In our results, SAND gene was one of the most stable genes in plants inoculated with B. bassiana (Table 3) and plants inoculated with B. bassiana and infested with L. invasa (Table 5). Moura et al.1, for example, showed that SAND was one of the most stable genes under different conditions and different eucalyptus species. Similarly, De Almeida et al.36 defined the SAND gene as the most stable genes according to the geNorm algorithm during in vitro adventitious rooting of Eucalyptus globulus Labill. The TUB gene expression pattern in plants inoculated with B. bassiana (Table 3) and plants infested with L. invasa (Table 4) was one of the most stable when compared to the other analyzed genes, similar to the study conducted by Fernández et al.37, where TUB was also one of the best reference genes across different acclimation and de-acclimation treatments of E. globulus. Considering all treatments evaluated (Fig. 2), control plants (Table 1), and mother plants (Table 2), one of the best reference genes analyzed was EUC12. De Almeida et al.36 analyzed in vitro adventitious rooting in E. globulus and also found that EUC12 displayed one of the most stable expression patterns among the genes analyzed according to the geNorm algorithm. It is important to highlight that the comparison between the best genes defined in other studies must consider the species, tissues and treatments analyzed. However, the fact that, in some cases, the same genes from different studies are defined as most stable ones suggests a high expression stability of them in different eucalyptus species, conditions and tissues.
Reference gene validation
Plants are organisms capable of sensing environmental changes and adjusting their physiological state to adapt to new conditions. Successful plant defense responses depend on the timely accumulation of defense compounds, which are activated according to the nature of the pathogen. In this context, pathogen attack can lead to changes in gene expression and metabolic modifications that allow the establishment of an efficient defense response. Transcription factors play a crucial role in the plant innate immunity. Specifically, ethylene response factors (ERF) are important integrators of hormonal pathways and play a direct role in the transcriptional regulation of several defense genes activated by jasmonate/ethylene75.
The AP2/EREBP superfamily is one of the largest groups of plant-specific transcription factors76. Genes belonging to this superfamily play a crucial role in plant development, as well as in their ability to tolerate biotic, and abiotic stresses77. These genes are essential for plant growth and the development and response to various stresses, such as extreme temperatures, drought, high salinity, and pathogen infection. Furthermore, they are involved in several hormonal signaling pathways, including abscisic acid, ethylene, cytokinins, and jasmonates76,78.
The DREB (drought-responsive binding elements) genes belong to the AP2/EREBP superfamily and are important regulators of the response to abiotic stress79. Particularly, DREB2 is mainly involved in dehydration/heat tolerance80. Although its main function is associated with the response to these stresses, DREB2 also displays high expression levels in tissues under insect attack42. However, studies investigating the transcriptional expression of these genes in response to galling insect attacks are still scarce. Therefore, in the present work, the relative expression of EcDREB2 was analyzed, allowing us to observe its expression in tissues subjected to B. bassiana inoculation and gall wasp attack. In comparison with the data available in the tool developed by Sundell et al.42, it could be observed that the EcDREB2 expression pattern, when the RT-qPCR data were normalized with the best reference genes (Fig. 3), is similar, showing high expression levels in plants infested with L. invasa. Relatively higher expression levels of EcDREB2 in leaf apex was expected, considering that L. invasa infestation primarily occurs in leaf tissues12,81. However, when the data was normalized with the least stable reference genes (Fig. 3), the expression pattern was modified and EcDREB2 expression in leaf apex and stem showed no difference, thus confirming the influence of the reference genes used in the final result of the RT-qPCR gene expression analysis.
The results obtained in the study shows that the expression pattern of a target gene is closely associated with the reference genes used for data normalization. When employing reference genes with lower stability to calculate EcDREB2 expression, a reduction in the relative expression level was observed, which can lead to erroneous interpretations and conclusions. These data highlight the relevance of carefully validating reference genes before applying them in gene expression studies.
Conclusion
In the present study, for the first time, the analysis and selection of the best reference genes for gene expression normalization in Eucalyptus (E. tereticornis × E. camaldulensis hybrid) plants subjected to B. bassiana inoculation and L. invasa infestation has been conducted. EcPTB, EcPP2A-1, and EcEUC12 were the best reference genes when all treatments were evaluated. The triplets EcPTB/EcEUC12/EcUBP6, EcPP2A-1/EcEUC12/EcPTB, EcPP2A-1/Ecα-TUB/EcPTB, EcIDH/EcSAND/Ecα-TUB, and EcPP2A-1/EcUPL7/EcSAND were the best reference genes for control plants, mother plants, plants infested with L. invasa, plants inoculated with B. bassiana, and plants inoculated with B. bassiana and infested with L. invasa, respectively. In relation to the worst reference genes, EcPTB, EcPP2A-1, and EcEUC12 were defined as the least suitable genes when all treatments are evaluated. The data obtained highlight the importance of developing this type of study to increase the reliability of relative expression analyses. Furthermore, the reported findings can serve as a basis for the development of studies aimed at analyzing the relative expression of genes related to different metabolic pathways of interest, such as the defensive responses of eucalyptus plants inoculated with the entomopathogenic fungus B. bassiana and infested with the gall wasp L. invasa.
Methods
Experiment design
Study area
The experiment was carried out under greenhouse conditions at the Experimental Research Station of the Federal University of Tocantins – UFT, Gurupi, Tocantins, Brazil (11°43’ S and 49°04’ W, 284 m altitude). The region's climate is Aw type (tropical climate, with dry winter), with an average annual temperature and rainfall of 26.1 ºC and 1776.4 mm, respectively.
Eucalyptus seedling production
Rooted cuttings of the hybrid clone of E. tereticornis × E. camaldulensis were produced under greenhouse conditions. After 120 days, the cuttings were transplanted from tubes into 3.8-L pots containing Bioplant® (Bioplant, Ponte Nova, MG, Brazil) commercial substrate based on pine bark, carbonized rice husk, vermiculite, macronutrients, and micronutrients. After transplantation of the seedlings, plants were acclimatized in the greenhouse for 30 days and then taken to an area in full sun. The plant material used in this study complies with international, national and/or institutional guidelines.
B. bassiana inoculation
The fungus B. bassiana (strain PL 63) was obtained from the mycological collection of the Insect-Microorganism Symbioses Laboratory of UFT (Gurupi Campus). Plates from the collection were re-plated into new plates containing P.D.A. medium (Potato, Dextrose (Labsybth, Diadema, SP, Brazil), Agar (Labsybth, Diadema, SP, Brazil)), supplemented with amoxicillin (500 μg mL−1) (EMS Pharma Hortolândia, SP, Brazil), and maintained at B.O.D. (Biochemical Oxygen Demand) for a period of 12 days at 25 ºC ± 2 °C and a 12-h photoperiod.
After this period, plates were opened in a laminar flow chamber and, under aseptic conditions, spores were removed from the colonies. This step was performed by adding 10 ml of distilled water and Tween 80® 0.02% (v/v) (Labsybth, Diadema, SP, Brazil), previously autoclaved (121.0 °C for 15 min), with the spores being gently scraped with a sterilized spatula. These spores were transferred to a sterile beaker containing 100 ml of sterilized distilled water. This solution was stirred in an incubator with orbital shaking at 6 × g (RCF) for 10 min at room temperature. After this process, the suspension was filtered into an autoclaved (121.0 °C for 15 min) beaker through a double layer of sterilized gauze to retain the mycelium fragments and remains of the culture medium. An aliquot of this solution was placed in a Neubauer chamber to count spores using an optical microscope. Then, the concentration of the solution was adjusted to 108 spores/ml with distilled water containing 0.02% (v/v) Tween 80®. The inoculation with this solution was performed immediately after its preparation. For control plants, an autoclaved (121.0 °C for 15 min) solution of distilled water solution containing 0.02% (v/v) Tween 80® was used. From the same solution, a 15 ml aliquot was used to perform the viability test with the aid of the Neubauer chamber. After a period of 48 h, the viability test was carried out, where the conidia evaluated displayed a viability of 80%. The adaxial epidermis of the fourth, fifth and sixth fully expanded leaves from each plant were slightly injured with a soft sponge. Spraying was carried out, especially on injured leaves. After spraying, six branches from the upper third of each plant were covered with transparent plastic bags for 36 h82.
L. invasa breeding and infestation
The L. invasa individuals used in this work were obtained from a breeding cage (2.8 m × 5.2 m × 3.0 m—height × length × width, and branches with galls close to emergence were cut from eucalyptus plants (hybrid E. tereticornis × E. camaldulensis) in the breeding cage. After cutting the branches, they were immediately taken to the laboratory and placed in a beaker with distilled water inside a bench cage covered with organza. A container with a solution of water and honey (3:2 ratio) was also placed inside the cage to feed the wasp individuals that emerged from the galls. The cage was kept in the laboratory, at room temperature, for a period of 24 h. After this period, the individuals that emerged were collected with manual suckers and placed in microcentrifuge tubes.
At 45 days after inoculation of B. bassiana in eucalyptus plants, all plants (inoculated and non-inoculated), except control and mother plants, were infested with L. invasa. For this, six branches of each plant were wrapped in an organza bag containing a closed 2.0 ml microcentrifuge tube with two individuals of L. invasa10. Then, the microtubes were opened so that the wasps could come out to oviposit. After 48 h of infestation, the bags and wasps were removed, and after 48 h from this, samples were collected.
Plant material and treatments
Samples consisted of roots, stems, fully expanded leaves, and leaf apices from mother plants maintained under field conditions and the plants produced as previously described. Each of these materials were collected from plants of the following treatments: mother plants, control plants (without inoculation and without infestation), plants infested with L. invasa, seedlings inoculated with B. bassiana, and plants inoculated with B. bassiana and also infested with L. invasa. Three biological replicates were used for each sample type, with each biological replicate consisting of four plants. Samples were collected and immediately frozen in liquid nitrogen and subsequently stored at − 80 °C until RNA extraction. Molecular analyses were carried out at the Molecular Analysis Laboratory (LAM) of the Federal University of Tocantins (UFT), Palmas campus.
RNA extraction
RNA extraction was performed using the CTAB (cetyltrimethylammonium bromide) method83 modified by Gonçalves et al.84 and with minor alterations (Supplementary S2). After extraction, the RNA quantity and purity (A260/A280 and A260/A230 ratios) were determined through a spectrophotometer (Nanodrop® One Spectrophotometer, Thermo Fisher Scientific, Wilmington, DE), while RNA integrity was verified using the agarose gel (0.8%).
DNase treatment and cDNA synthesis
RNA samples (5 μg) were treated with DNase I, using the Turbo DNA-free kit (Applied Biosystems, Thermo Fisher Scientific, Vilnius, Lithuania) and following its instructions, to eliminate residual DNA contamination. Subsequently, RNA was evaluated for its quantity and purity (A260/A280 and A260/A230 ratios) through spectrophotometry (Nanodrop® One Spectrophotometer) and its integrity was analyzed through agarose gels (0.8%). cDNA was synthesized from 1.0 μg of RNA using the High-Capacity cDNA Reverse Transcription kit (Invitrogen, Thermo Fisher Scientific, Vilnius, Lithuania) and following the manufacturer's protocol. cDNA samples were then stored at − 20 °C.
Identification and selection of target and reference genes
The reference genes analyzed in this study were chosen from a search for eucalyptus reference gene studies on the Web of Science database (www.webofknowledge.com), using the following keywords: housekeeping gene, endogenous gene, reference gene, and Eucalyptus. The Boolean interpolator “and” was used. From this search, the selection of the reference genes followed the recommendation criteria of the article in which they were analyzed, that is, the genes that had the best results (greater expression stability in the sample set used) were prioritized. Following this approach, 12 different genes were selected: PP2A-1, SAND, UPL7, PTB38; Actin-2, 18srRNA, IDH34; α-TUB, UBC, EF-1α37; EUC12, and H2B36.
The selection of the target gene used to validate the selected reference genes was based on the gene expression data generated by the tool (https://plantgenie.org/) described by Sundell et al.42. For this, the PFAM number (PF00847)85 corresponding to the AP2/EREBP superfamily was used to search on the available E. grandis database of the tool, all gene sequences referring to this superfamily. After identifying the sequences, they were selected and used to generate a gene expression graph using the ExHeatmap option. After analyzing the graph, the gene DREB2 (Eucgr.F02440.1) showed a high expression level in E. grandis tissues subjected to L. invasa infestation, being selected for validation analysis.
Gene sequence identification
The reference and target gene sequences were obtained using the BLAST tool (Basic Local Alignment Search)86 through the comparison of their nucleotide sequences from E. grandis, obtained on the Phytozome database (https://phytozome-next.jgi.doe.gov/), with the Eucalyptus camaldulensis Genome Database (https://www.kazusa.or.jp/eucaly/index.html).
Primer design
RT-qPCR primers for RT-qPCR (Table 6) were designed by using the reference and target gene sequences obtained from the Eucalyptus camaldulensis Genome Database and the OligoPerfect program (apps.thermofisher.com/apps/oligoperfect/). Quality assessment of the designed primers was evaluated by the OligoAnalyzer tool (http://www.idtdna.com/calc/analyzer), and the melting curves of all genes analyzed here are described in Supplementary S3. Corrections of primer efficiency differences were performed by applying the mathematical model proposed by Pfaffl49.
RT-qPCR analysis
RT-qPCR analyses were carried out on an ABI PRISM 7500 Real-Time PCR thermocycler (Applied Biosystems, Thermo Fisher Scientific, Singapore), using the PowerUp™ SYBR™ Green Master Mix (Applied Biosystems, Thermo Fisher Scientific, Vilnius, Lithuania). Reactions were performed in 10 μL final volume: 1.0 μL of cDNA (diluted 1:5), 0.2 μL of each primer at 10 μM, and 5.0 μL of PowerUp™ SYBR™ Green Master Mix, and 3.6 μL of RNase-DNase-free water. Three biological replicates were used, and reactions were run in triplicates as technical repetitions. Amplification reactions were carried out with the following conditions: 2 min at 50 °C, 2 min at 95 °C, followed by 40 cycles of 15 s at 95 °C and 1 min at 60 °C. To confirm the specificity of the primers, melting curves were generated after 40 amplification cycles for each primer pair by raising the temperature from 60 to 95 °C, with 1 °C increase in temperature every 5 s. Cq was determined by the number of cycles in which the fluorescence generated within a reaction crosses the threshold line.
Expression stability and validation of candidate reference genes
The algorithms geNorm59, NormFinder62, BestKeeper63, and Delta-Ct64 were used to calculate the stability values. The algorithms generate a classification based on the stability value of each evaluated gene. Subsequently, the data generated by the algorithms were analyzed by RefFinder, which integrates the four algorithms and provides a general classification of all candidate reference gene tested65,66. The following sample sets were used by the RefFinder tool (www.ciidirsinaloa.com.mx/RefFinder-master/) to evaluate the stability of the evaluated genes: root, stem, leaf and leaf apex in all treatments; and root, stem, leaf, and leaf apex in each treatment. Box plots were plotted to illustrate the expression levels and variations of the tested genes by the SigmaPlot program.
Reference gene validation was performed through expression analyzes of the target gene, EcDREB2. The mathematical model proposed by Pfaffl49, which is based in primer amplification efficiencies, was used to calculate the relative expression values. Expression data was normalized by using more than one reference gene, in accordance with Bustin et al.46 EcDREB2 expression pattern was normalized with the most (EcPP2A-1, EcULP7 and EcSAND) and least (EcH2B, EcEF1-α e EcACT) stable reference genes, according to the RefFinder algorithm, and analyzed in stem and leaf apex from eucalyptus plants submitted to L. invasa infestation and B. Bassiana inoculation. Relative expression graphs were plotted using the SigmaPlot program (version 12.0, Systat Software Inc, San Jose, CA, USA).
Data availability
All data generated and analyzed for this study are included in this published article and its Supplementary Information files. All programs used to analyze the data are publicly available.
References
Moura, J. C. M. S. et al. Validation of reference genes from Eucalyptus spp. under different stress conditions. BMC Res. Notes 5, 1–10 (2012).
De Oliveira, L. A. et al. Reference genes for the normalization of gene expression in Eucalyptus species. Plant Cell Physiol. 53, 405–422 (2012).
Sistema Nacional de Informações Florestais (SNIF). Cadeia Produtiva. http://snif.florestal.gov.br/pt-br/cadeia-produtiva (2020).
Florêncio, G. W. L., Martins, F. B. & Fagundes, F. F. A. Climate change on Eucalyptus plantations and adaptive measures for sustainable forestry development across Brazil. Ind. Crops Prod. 188, (2022).
Indústria Brasileira de Árvores (IBÁ). Relatório Anual. https://www.iba.org/datafiles/publicacoes/relatorios/relatorio-anual-iba2022-compactado.pdf (2022).
Booth, T. H. Eucalypt plantations and climate change. For. Ecol. Manag. 301, 28–34 (2013).
de Moraes Goncalves, J. L. et al. Integrating genetic and silvicultural strategies to minimize abiotic and biotic constraints in Brazilian eucalypt plantations. For. Ecol. Manag. 301, 6–27 (2013).
Mendel, Z., Protasov, A., Fisher, N. & La Salle, J. Taxonomy and biology of Leptocybe invasa gen. & sp. N. (Hymenoptera: Eulophidae), an invasive gall inducer on Eucalyptus. Aust. J. Entomol. 43, 101–113 (2004).
Kumari, N. K., Harish, K., Vastrad, A. S. & Goud, K. B. Biology of eucalyptus gall wasp, Leptocybe invasa Fisher and La Salle (Hymenoptera: Eulophidae). Karnataka J. Agric. Sci. 23, 211–212 (2010).
Sarmento, M. I. et al. Differential development times of galls induced by Leptocybe invasa (Hymenoptera: Eulophidae) reveal differences in susceptibility between two Eucalyptus clones. Pest Manag. Sci. 77, 1042–1051 (2021).
Baron, N. C. et al. Filamentous fungi in biological control: current status and future perspectives. Chil. J. Agric. Res. 79, 307–315 (2019).
Rocha, J. P. L. et al. Morphophysiological responses in Eucalyptus demonstrate the potential of the entomopathogenic fungus Beauveria bassiana to promote resistance against the galling wasp Leptocybe invasa. Forests 14, 1349 (2023).
Bacon, C. W. Biotechnology of endophytic fungi of grasses. (ed. Bacon, C. W & White, J. F), 1–213 (CRC Press, 2018).
Zimmermann, G. Review on safety of the entomopathogenic fungi Beauveria bassiana and Beauveria brongniartii. Biocontrol Sci. Technol. 17, 553–596 (2007).
Myburg, A. A. et al. The genome of Eucalyptus grandis. Nature 510, 356–362 (2014).
Brazma, A. & Vilo, J. Gene expression data analysis. FEBS Lett. 480, 17–24 (2000).
Yang, Z., Zhang, R. & Zhou, Z. Identification and validation of reference genes for gene expression analysis in Schima superba. Genes (Basel) 12, 732 (2021).
Gachon, C., Mingam, A. & Charrier, B. Real-time PCR: What relevance to plant studies?. J. Exp. Bot. 55, 1445–1454 (2004).
Fernandes-Brum, C. N. et al. A panel of the most suitable reference genes for RT-qPCR expression studies of coffee: Screening their stability under different conditions. Tree Genet. Genomes 13, 131 (2017).
Lucho, S. R. et al. Validation of reference genes for RT-qPCR studies in Stevia rebaudiana in response to elicitor agents. Physiol. Mol. Biol. Plants 24, 767–779 (2018).
Nolan, T., Hands, R. E. & Bustin, S. A. Quantification of mRNA using real-time RT-PCR. Nat. Protoc. 1, 1559–1582 (2006).
Pfaffl, M. W. Quantification strategies in real-time PCR. AZ Quant. PCR 1, 89–113 (2004).
Yang, Q. et al. Reference gene selection for qRT-PCR in Caragana korshinskii Kom. under different stress conditions. Mol. Biol. Rep. 41, 2325–2334 (2014).
Zhu, J. et al. Reference gene selection for quantitative real-time PCR normalization in Caragana intermedia under different abiotic stress conditions. PLoS One 8, e53196 (2013).
Lin, Y. et al. Identification and validation of reference genes for qRT-PCR analyses under different experimental conditions in Allium wallichii. J. Plant Physiol. 281 (2023).
Joseph, J. T., Poolakkalody, N. J. & Shah, J. M. Plant reference genes for development and stress response studies. J. Biosci. 43, 173–187 (2018).
Nguyen, D. Q., Eamens, A. L. & Grof, C. P. L. Reference gene identification for reliable normalisation of quantitative RT-PCR data in Setaria viridis. Plant Methods 14 (2018).
Li, G. et al. Identification of reference genes for reverse transcription-quantitative PCR analysis of ginger under abiotic stress and for postharvest biology studies. Front. Plant Sci. 13 (2022).
Reddy, D. S. et al. Identification and validation of reference genes and their impact on normalized gene expression studies across cultivated and wild Cicer species. PLoS One 11, (2016).
Zhong, Y. et al. Selection and validation of reference genes for quantitative real-time PCR normalization in Psoralea corylifolia (Babchi) under various abiotic stress. J. Plant Physiol. 274, 153722 (2022).
de Oliveira, L. F. et al. Selection and validation of reference genes for measuring gene expression in Piper species at different life stages using RT-qPCR analysis. Plant Physiol. Biochem. 171, 201–212 (2022).
Dong, X. M., Zhang, W. & Zhang, S. B. Selection and validation of reference genes for quantitative real-time PCR analysis of development and tissue-dependent flower color formation in Cymbidium lowianum. Int. J. Mol. Sci. 23, 738 (2022).
Yu, Y. et al. Selection of reference genes for qPCR analyses of gene expression in ramie leaves and roots across eleven abiotic/biotic treatments. Sci. Rep. 9, 1–13 (2019).
Sundari, B. K. R. & Dasgupta, M. G. Selection and validation of reference genes for real-time qRT-PCR normalization in different tissues of Eucalyptus tereticornis. Silvae Genet. 61, 280–286 (2012).
Boava, L. P. et al. Selection of endogenous genes for gene expression studies in Eucalyptus under biotic (Puccinia psidii) and abiotic (acibenzolar-S-methyl) stresses using RT-qPCR. BMC Res. Notes 3 (2010).
de Almeida, M. R. et al. Reference gene selection for quantitative reverse transcription-polymerase chain reaction normalization during in vitro adventitious rooting in Eucalyptus globulus Labill. BMC Mol. Biol. 11, 1–12 (2010).
Fernández, M., Villarroel, C., Balbontín, C. & Valenzuela, S. Validation of reference genes for real-time qRT-PCR normalization during cold acclimation in Eucalyptus globulus. Trees Struct. Funct. 24, 1109–1116 (2010).
Cassan-Wang, H. et al. Reference genes for high-throughput quantitative reverse transcription-PCR analysis of gene expression in organs and tissues of Eucalyptus grown in various environmental conditions. Plant Cell Physiol. 53, 2101–2116 (2012).
Martins, G. S., Freitas, N. C., Máximo, W. P. F. & Paiva, L. V. Gene expression in two contrasting hybrid clones of Eucalyptus camaldulensis × Eucalyptus urophylla grown under water deficit conditions. J. Plant Physiol. 229, 122–131 (2018).
Imai, T., Ubi, B. E., Saito, T. & Moriguchi, T. Evaluation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in Pyrus pyrifolia using different tissue samples and seasonal conditions. PLoS One 9 (2014).
Xiao, Z. et al. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Front. Plant Sci. 7 (2016).
Sundell, D. et al. The plant genome integrative explorer resource: PlantGenIE.org. New Phytol. 208, 1149–1156 (2015).
Hellemans, J., Mortier, G., De Paepe, A., Speleman, F. & Vandesompele, J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, 1–14 (2008).
Song, J., Cho, J., Park, J. & Hwang, J. H. Identification and validation of stable reference genes for quantitative real time PCR in different minipig tissues at developmental stages. BMC Genomics 23 (2022).
Kozera, B. & Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406 (2013).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622 (2009).
Mughal, B. B., Leemans, M., Spirhanzlova, P., Demeneix, B. & Fini, J. B. Reference gene identification and validation for quantitative real-time PCR studies in developing Xenopus laevis. Sci. Rep. 8 (2018).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25, 402–408 (2001).
Pfaffl, M. W. A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res. 29, e45–e45 (2001).
Lian, C. et al. Validation of suitable reference genes by various algorithms for gene expression analysis in Isodon rubescens under different abiotic stresses. Sci. Rep. 12, 1–13 (2022).
Yao, J. et al. Reference gene selection for qPCR analysis in Schima superba under abiotic stress. Genes (Basel) 13, 1887 (2022).
Ji, T. et al. Reference genes identification for qRT-PCR normalization of gene expression analysis in Cucumis sativus under Meloidogyne incognita infection and Pseudomonas treatment. Front. Plant Sci. 13, 4907 (2022).
Xu, H. et al. Screening of suitable reference genes for gene expression using quantitative real-time PCR in Gynura bicolor DC. ScienceAsia 48, 833 (2022).
Bai, S. et al. Selection and evaluation of reference genes for quantitative real-time PCR in tomato (Solanum lycopersicum L.) inoculated with Oidium neolycopersici. Agronomy 12, 3171 (2022).
Ling, H., Wu, Q., Guo, J., Xu, L. & Que, Y. Comprehensive selection of reference genes for gene expression normalization in sugarcane by real time quantitative RT-PCR. PLoS One 9, e97469 (2014).
Wang, G. et al. Identification and testing of reference genes for qRT-PCR analysis during pear fruit development. Biologia 2022(1), 1–15 (2022).
Wang, J. et al. Evaluation and selection of suitable qRT-PCR reference genes for light responses in tea plant (Camellia sinensis). Sci Hortic 289, 110488 (2021).
Berruien, N. N. A., Murray, J. F. & Smith, C. L. Pregnancy influences the selection of appropriate reference genes in mouse tissue: Determination of appropriate reference genes for quantitative reverse transcription PCR studies in tissues from the female mouse reproductive axis. Gene 801, 145855 (2021).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, 1–12 (2002).
Freitas, N. C. et al. Validation of reference genes for qPCR analysis of Coffea arabica L. somatic embryogenesis-related tissues. Plant Cell Tissue Organ Cult. 128, 663–678 (2017).
Abbas, A. et al. Selection and validation of reference genes for RT-qPCR analysis in Aegilops tauschii (Coss.) under different abiotic stresses. Int. J. Mol. Sci. 22, 11017 (2021).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. BMC Mol. Biol. 7, 33 (2006).
Xie, F., Wang, J. & Zhang, B. RefFinder: A web-based tool for comprehensively analyzing and identifying reference genes. Funct. Integr. Genomics 23, 1–5 (2023).
Xie, F., Xiao, P., Chen, D., Xu, L. & Zhang, B. miRDeepFinder: A miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84 (2012).
Coelho, T. C. et al. Reference gene selection for quantitative PCR in liver, skeletal muscle, and jejunum of Bos indicus cattle. Revista Brasileira de Zootecnia 51 (2022).
Xie, C. Di et al. Validation of the reference genes for the gene expression studies in different cell lines of pig. Biomed. Res. Int. 2021 (2021).
Ni, M. et al. Selection and validation of reference genes for the normalization of quantitative real-time PCR in different muscle tissues of rabbits. BMC Zool. 7 (2022).
Daúde, M. M. et al. Selection and validation of reference genes for RT-qPCR gene expression studies in Candida viswanathii cultivated under different grown conditions. J. Microbiol. Methods 211, 106777 (2023).
Li, J. Y. et al. Screening of reference genes in real-time PCR for Radopholus similis. PeerJ 7, e6253 (2019).
Pant, N., Rush, C., Warner, J. & Eisen, D. P. Effect of savirin or ticagrelor treatment on the expression of commonly used reference genes in Staphylococcus aureus. Microorganisms 11 (2023).
Volland, M., Blasco, J. & Hampel, M. Validation of reference genes for RT-qPCR in marine bivalve ecotoxicology: Systematic review and case study using copper treated primary Ruditapes philippinarum hemocytes. Aquatic Toxicol. 185, 86–94 (2017).
Zheng, H., Zhao, H., Zhang, X., Liang, Z. & He, Q. Systematic identification and validation of suitable reference genes for the normalization of gene expression in Prunella vulgaris under different organs and spike development stages. Genes (Basel) 13, 1947 (2022).
Huang, P. Y., Catinot, J. & Zimmerli, L. Ethylene response factors in Arabidopsis immunity. J. Exp. Bot. 67, 1231–1241 (2016).
Chen, L. et al. Expansion and stress responses of AP2/EREBP superfamily in Brachypodium distachyon. Sci. Rep. 6, 1–14 (2016).
Liu, C. & Zhang, T. Expansion and stress responses of the AP2/EREBP superfamily in cotton. BMC Genomics 18, 1–16 (2017).
Nakano, T., Suzuki, K., Fujimura, T. & Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 140, 411–432 (2006).
Licausi, F., Ohme-Takagi, M. & Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 199, 639–649 (2013).
Xu, Y., Zhan, C. & Huang, B. Heat shock proteins in association with heat tolerance in grasses. Int. J. Proteomics 2011, 1–11 (2011).
Pinsupa, S. et al. Transcriptome analysis reveals genes involved in responses of Eucalyptus to gall wasp infestation. Horticulturae 9 (2023).
Siviero, A. [UNESP]. Avaliação de métodos de inoculação de Phytophthora parasitica e mapeamento de QTLs de resistência em híbridos de Citrus sunki vs. Poncirus trifoliata a gomose. https://repositorio.unesp.br/server/api/core/bitstreams/0b640b16-0da3-47f4-b3e0-693e353fd172/content (2001).
Chang, S., Puryear, J. & Cairney, J. A simple and efficient method for isolating RNA from pine trees. Plant Mol. Biol. Rep. 11, 113–116 (1993).
Gonçalves, R. C. et al. Evaluation of extraction methods for obtaining high-quality RNA from sweet potato. Genet. Mol. Res. 20 (2021).
Paysan-Lafosse, T. et al. InterPro in 2022. Nucleic Acids Res. 51, D418–D427 (2023).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Acknowledgements
We thank the “Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq)” (Process number 405279/2023-0), the “Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES) - PROCAD Amazônia" (Process number 306011/2022-0), the "Fundação Amazônia de Amparo a Estudos e Pesquisas (FAPESPA)" (Process number 066/2023 - PPG/BIONORTE), the “Rede de Biodiversidade e Biotecnologia da Amazônia Legal (Bionorte)”, the "Fundação de Amparo à Pesquisa do Estado do Tocantins (FAPT)”, the “Programa de Pós-Graduação em Agroenergia Digital” and the “Universidade Federal do Tocantins (PROPESQ-UFT)” for the financial support.
Author information
Authors and Affiliations
Contributions
Conceived and designed the experiments R.A.S., H.G.B., M.I.S. & S.A.S.; Experiment, data collection & Software analysis, M.M.D., J.N.R. & N.M.P.L.; R.A.S., H.G.B.; M.I.S.; Supervision, H.G.B. & R.A.S.; Writing-original draft, M.M.D.; Made critical revisions of the content of the paper, H.G.B., R.A.S., S.A.S., A.A.L. & M.I.S. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Daude, M.M., Ságio, S.A., Rodrigues, J.N. et al. Reference genes for Eucalyptus spp. under Beauveria bassiana inoculation and subsequently infestation by the galling wasp Leptocybe invasa. Sci Rep 14, 2556 (2024). https://doi.org/10.1038/s41598-024-52948-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-52948-x