Abstract
Epithelial–mesenchymal transition (EMT) of retinal pigment epithelial (RPE) cells plays key roles in the pathogenesis of multiple vitreoretinal diseases, leading to profound and permanent vision loss. Circular RNAs (circRNAs) are widespread and functional endogenous RNAs that could regulate gene expression in eukaryotes. The functions of circRNAs in mediating EMT has been reported in several diseases. In the current study, we investigated the role of circRNA HIPK3 (circHIPK3) in EMT process of RPE cells (RPE-EMT). circHIPK3 is one abundant circRNA generated from the second exon of HIPK3 mRNA. We found that circHIPK3 expression was significantly increased in TGF-β1-induced RPE-EMT model. Silencing of circHIPK3 attenuated TGF-β1-induced RPE-EMT process, whereas forced expression of circHIPK3 could trigger EMT in RPE cells. Mechanistically, circHIPK3 regulates RPE-EMT process via sponging multiple microRNAs (miRNAs). This study provides novel insights into the mechanism of RPE-EMT. Targeting circHIPK3 might serve as a therapeutic strategy in RPE-EMT associated vitreoretinal diseases.
Similar content being viewed by others
Introduction
Retinal pigment epithelial (RPE) cells are crucial components of the outer blood-retinal barrier, maintaining the health and functional integrity of both photoreceptors and choroidal vasculature1. In pathological conditions, RPE cells could transform their epithelial characteristics into mesenchymal phenotype via epithelial-mesenchymal transition (EMT), resulting in resistance to apoptosis, increased cells migration ability, and production of extracellular matrix2. The process of EMT in RPE cells (RPE-EMT) contributes to the development of many vitreoretinal diseases, such as proliferative vitreoretinopathy, epiretinal membrane, subretinal fibrosis in age-related macular degeneration, and retinal fibrosis in proliferative diabetic retinopathy3. These diseases always lead to severe visual impairment and patients have to undergo surgery inevitably. Thus, revealing the molecular mechanisms of RPE-EMT is critical to the development of novel therapeutic modalities for these sight-threatening ocular diseases.
Circular RNAs (circRNAs) represent a class of single-stranded RNA with covalently closed-loop structure, which is highly conservative and stable in mammal genomes. In general, circRNAs exert regulatory functions through competing endogenous RNA (ceRNA) mechanism, binding to RNA-binding protein (RBP), or translation into peptides4,5. Numerous studies have shed light on the relationship between circRNAs and EMT-related diseases, especially in cancers. For example, circRIP2 influences bladder cancer progression through mediating EMT through miR-1305/TGF-β2/smad3 pathway6. circRNAs could also mediate the function of RPE cells. For example, circNR3C1 maintains RPE function by regulating miR-382-5p/PTEN/AKT/mTOR pathway7. However, whether circRNAs could regulate RPE-EMT has not been investigated.
Circular RNA HIPK3 (circHIPK3) is one abundant circRNA generated from the second exon of HIPK3 gene, which exerts ceRNA mechanism via sponging multiple miRNAs8. It has been reported that circHIPK3 could regulate EMT process in cancer cells9. The role of circHIPK3 has also been studied in several ocular cells, such as retinal vascular endothelial cells and lens epithelial cells10,11,12. In the current study, we explored the expression pattern, regulatory function, and regulatory mechanism of circHIPK3 in the EMT process of RPE cells. Our data suggest that circHIPK3 accelerates RPE-EMT by acting as multiple miRNA sponges.
Methods
Ethical approval and animal preparation
The study protocol was approved by the Animal Care and Use Committee of Yixing Eye Hospital and in accordance with Animal Research: Reporting In Vivo Experiments (ARRIVE) guidelines. All methods were carried out in accordance with relevant guidelines and regulations. Mice were purchased from Shanghai Laboratory Animal Center (Shanghai, China). Four-week-old male C57BL/6 mice were used and maintained in a 12-h light:12-h dark cycle with free access to food and water.
Primary cell culture and establishment of RPE-EMT model
Primary mouse RPE cells were isolated from male C57BL/6 mice at four-week-old. Mice were sacrificed by cervical dislocation under anesthesia induced by ketamine (80 mg/kg) and xylazine (10 mg/kg). The anterior sections and retinas were removed, and the eyecups were added with 0.25% trypsin for 10 min at 37 °C. Then the RPE sheets were peeled off gently. Single-cell suspension in the eyecup was transferred to DMEM/F12 culture medium (Gibco) supplemented with 10% fetal bovine serum (FBS) (Gibco) at 37 °C. RPE cells were passaged 1:2 using 0.25% trypsin every 3 days. Cells were seeded in 6-well plates for immunoblotting or PCR analysis, in 24-well plates for cellular migration assay, and in 96-well plates for immunofluorescence staining. After serum starvation for 24 h, RPE-EMT was induced by addition of TGF-β1 (10 ng/ml, Gibco) to the medium and incubation for 48 h.
Silencing of circHIPK3
Cells were transfected with small interfering RNAs (siRNAs, RiboBio) targeting circHIPK3 (100 nmol/L) using Lipofectamine RNAiMax (Life Technologies). The siRNA was synthesized targeting the backsplice sequence of circHIPK3.
Overexpression of circHIPK3
The entire sequence of circHIPK3 was synthesized and inserted into a lentivirus vector (RiboBio). Cells were transfected with lentivirus vector for 48 h according to the manufacturer’s protocols.
Quantitative reverse transcription PCR (qRT-PCR) analysis
TRIzol™ (Invitrogen) was employed to extract total RNA from RPE cells. qRT-PCR reactions were performed on StepOne Plus Real-Time PCR System (Applied Biosystems) according to the manufacturer's instructions. 18S ribosomal RNA (18S rRNA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), and U6 snRNA were used as housekeeping genes for relative quantification of circRNA, mRNA, and miRNA expression levels, respectively.
Western blot (WB) analysis
Protein was extracted from the treated cells with radioimmunoprecipitation (RIPA) lysis buffer on ice. Equal amounts of protein were resolved by 10% sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE), and then transferred to a polyvinylidene difluoride (PVDF) membrane. After blocking with 5% skim milk for 2 h at room temperature, the membrane blots were probed with primary antibodies overnight at 4 °C. The following day, membrane blots were rinsed in TBST, and then probed with horseradish peroxidase (HRP)-tagged secondary antibody for 1 h at room temperature. Finally, the protein was visualized with enhanced chemiluminescence detection reagent (Thermo Fisher Scientific) (Supplementary Information).
Immunofluorescence staining
Treated cells were fixed with 4% paraformaldehyde for 30 min, permeabilized with 0.2% Triton X-100/PBS for 1 h, and blocked with 5% BSA for 1 h at room temperature. Then cells were incubated with primary antibodies overnight at 4 °C. The following day, cells were washed and incubated with secondary antibodies for 2 h at room temperature. Cell nuclei were visualized with DAPI (Sigma-Aldrich).
Cell migration assay
Treated cells (1 × 105 cells/well) were seeded in a Transwell chamber of a 24-well plate, with 500 μl medium containing 10% FBS in the lower layer. After incubation for 24 h at 37 °C, cells in the upper layer were swabbed, and cells migrated across the membrane were fixed by methanol, stained with 0.5% crystal violet, and quantified using microscope.
Luciferase reporter assay
Entire circHIPK3 sequences (LUC-circHIPK3) or circHIPK3 sequences without putative miRNA binding sites (LUC-circHIPK3-MUT) were inserted into pGL3 luciferase reporter vector (Promega). RPE cells were seeded into 96-well plates (5 × 103 cells/well) and incubated for 24 h, then co-transfected with pGL3 luciferase reporter vector and miRNA mimics using Lipofectamine 2000 (Invitrogen). Forty-eight hours later, the luciferase activity was detected with the dual-luciferase reporter assay system (Promega).
Biotin-coupled miRNA capture
The 3ʹ end biotinylated miR-106b-5p or control miRNA (20 nM) was transfected into RPE cells for 24 h. The biotin-labeled complexes were isolated using streptavidin agarose beads (Invitrogen). The recruited transcripts were subjected to qRT-PCR analysis.
Statistical analysis
All results were statistically analyzed using GraphPad. T-test was applied between two independent groups and one-way ANOVA test was performed among various groups. P < 0.05 was thought as statistical significance. All data were presented as the mean ± s.d. of at least three independent experiments.
Results
Expression of circHIPK3 is increased in RPE-EMT model in vitro
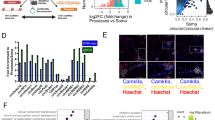
We treated RPE cells with TGF-β1 at a concentration of 10 ng/ml for 48 h to construct in vitro RPE-EMT model. After TGF-β1 treatment, the shape of RPE cells changed from epithelial pebbles to elongated mesenchymal spindles as shown by cell morphology images in bright field (Fig. 1A). Classical EMT related markers contain E-cadherin (E-Cad), zonula occludens-1 (ZO-1), vimentin (Vim), and N-cadherin (N-Cad)13. Downregulation of E-Cad and ZO-1, whereas upregulation of Vim and N-Cad after TGF-β1 treatment were revealed by qRT-PCR assay and immunofluorescence staining (Fig. 1B and C). Moreover, Transwell migration assay showed that, TGF-β1 treatment significantly enhanced cellular movement ability of RPE cells (Fig. 1D and E). These results indicate the successful construction of in vitro RPE-EMT model.
Expression of circHIPK3 is increased in RPE-EMT model in vitro. (A) Bright field images of RPE cells treated or untreated with TGF-β1. Scale bar, 100 μm. (B) Expression of EMT-related markers detected by qRT-PCR (normalized to GAPDH expression, n = 3). *P < 0.05 versus Ctrl group. (C) Expression of EMT-related markers detected by immunofluorescence staining (n = 3). Scale bar, 50 μm. (D) Transwell migration assays in RPE cells treated or untreated with TGF-β1 (n = 5). Scale bar, 50 μm. (E) Analysis of migrated cell number in the Transwell migration assays (n = 5). *P < 0.05 versus Ctrl group. (F) Expression pattern of circHIPK3 in RPE cells detected by RNA-FISH assay. Scale bar, 50 μm. (G) Expression of circHIPK3 treated with TGF-β1 for 0 h, 12 h, 24 h, and 48 h detected by qRT-PCR (normalized to 18S rRNA expression, n = 3). *P < 0.05 versus Ctrl group.
Next, we explored the expression pattern of circHIPK3 in RPE-EMT model. circHIPK3 was mainly expressed in the cytoplasm of RPE cells, as shown by fluorescence in situ hybridization analysis (Fig. 1F). qRT-PCR assay showed that, the expression level of circHIPK3 was significantly upregulated in RPE-EMT model (Fig. 1G). These results suggest that circHIPK3 might be a regulator during RPE-EMT process.
Silencing of circHIPK3 attenuates RPE-EMT process and inhibits cell migration
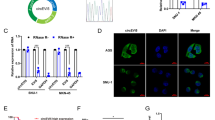
To explore the role of circHIPK3 in TGF-β1-induced RPE-EMT, we silenced circHIPK3 via small interfering RNAs (siRNAs) in RPE cells. Compared with cells transfected with scrambled siRNA (si-Scr), cells transfected with siRNA targeting circHIPK3 (si-circHIPK3) suppressed circHIPK3 expression by greater than 75% (Fig. 2A). qRT-PCR (Fig. 2B), immunoblot analysis (Fig. 2C and D), and immunofluorescence staining (Fig. 2E) showed that silencing of circHIPK3 significantly abrogated TGF-β1-induced downregulation of E-Cad and ZO-1, and abolished TGF-β1-induced upregulation of Vim and N-Cad. In addition, the number of cells migrating across the membrane was significantly lower in si-circHIPK3 groups compared to si-Scr groups (Fig. 2F and G). These results indicate that circHIPK3 could regulate RPE-EMT process and influence cellular migration ability of RPE cells.
Silencing of circHIPK3 attenuates RPE-EMT process and inhibits cell migration. (A) Expression of circHIPK3 detected by qRT-PCR (normalized to 18S rRNA expression, n = 3). *P < 0.05. (B) Expression of EMT-related markers detected by qRT-PCR (normalized to GAPDH expression, n = 3). *P < 0.05. (C) Expression of EMT-related markers detected by WB (n = 3). The representative images were shown. (D) Analysis of band intensities in WB data (normalized to β-actin expression, n = 3). *P < 0.05. (E) Expression of EMT-related markers in RPE cells treated with TGF-β1 + si-Scr or TGF-β1 + si-circHIPK3 detected by immunofluorescence staining (n = 3). Scale bar, 50 μm. (F) Transwell migration assays in RPE cells treated with TGF-β1 + si-Scr or TGF-β1 + si-circHIPK3 (n = 5). Scale bar, 50 μm. (G) Analysis of migrated cell number in the Transwell migration assays (n = 5). *P < 0.05 versus Ctrl group.
Overexpression of circHIPK3 triggers RPE-EMT and accelerates cell migration
Next, we explored whether circHIPK3 acts as a trigger of RPE-EMT process. Overexpression of circHIPK3 was conducted using lentivirus vector (lv-circHIPK3). Compare to cells transfected with control lentivirus vector (lv-Scr), RPE cells transfected with lv-circHIPK3 showed eightfold increase of circHIPK3 expression (Fig. 3A). As expected, circHIPK3 overexpression significantly promoted the expression of Vim and N-Cad, and suppressed the expression of E-Cad and ZO-1 at the RNA (Fig. 3B) and protein levels (Fig. 3C). Furthermore, circHIPK3 overexpression significantly increased the number of cells migrating across the membrane (Fig. 3D and E). These results indicate that circHIPK3 acts as a trigger of EMT process in RPE cells.
Overexpression of circHIPK3 triggers RPE-EMT and accelerates cell migration. (A) Expression of circHIPK3 detected by qRT-PCR (normalized to 18S rRNA expression, n = 3). *P < 0.05. (B) Expression of EMT-related markers detected by qRT-PCR (normalized to GAPDH expression, n = 3). *P < 0.05 versus lv-Scr group. (C) Analysis of band intensities in WB data (normalized to β-actin expression, n = 3). *P < 0.05. (D) Transwell migration assays in RPE cells treated with lv-Scr or lv-circHIPK3 (n = 5). Scale bar, 50 μm. (E) Analysis of migrated cell number in the Transwell migration assays (n = 5). *P < 0.05 versus Ctrl group.
circHIPK3 regulates RPE-EMT process by sponging multiple microRNAs
Given that circHIPK3 has been reported to regulate pathological processes via ceRNA mechanism, we next constructed a ceRNA network of circHIPK3. Based on the EMT-related mRNAs published, we used the bioinformatics software ENCORI (The Encyclopedia of RNA Interactomes) and TargetScan to predict candidate miRNAs that might be involved in ceRNA network14,15,16. Ten miRNAs were predicted to be sponged by circHIPK3 and target EMT-related mRNAs simultaneously, including miR-124-3p, miR-302d-3p, miR-93-5p, miR-20a-5p, miR-20b-5p, miR-17-5p, miR-519d-3p, miR-106a-5p, miR-106b-5p, and miR-222-3p (Fig. 4A). Notably, three (miR-124-3p, miR-302d-3p, and miR-93-5p) of ten miRNAs have been reported to serve as inhibitors of RPE-EMT17,18. Other seven miRNAs have also been reported to be regulators of EMT in cancer cells19,20,21,22,23,24,25. To validate the miRNAs that bind to circHIPK3 in RPE cells, we conducted luciferase reporter assay. The entire sequence of circHIPK3 was inserted into pGL3 luciferase reporter vector, denoted LUC-circHIPK3. A significant decrease in luciferase activity was observed when cells were co-transfected with miR-124-3p, miR-302d-3p, miR-93-5p, miR-106a-5p, or miR-106b-5p mimics (Fig. 4B). The putative binding sites of these miRNAs associated with circHIPK3 were shown (Fig. 4C). Subsequently, LUC-circHIPK3 mutant without each miRNA binding sites was also constructed, denoted LUC-circHIPK3-MUT. Co-transfected with miR-124-3p, miR-302d-3p, miR-93-5p, miR-106a-5p, or miR-106b-5p mimics had no effect on the activity of LUC-circHIPK3-MUT (Fig. 4D). These results suggest that circHIPK3 might function as a sponge to these miRNAs.
circHIPK3 regulates RPE-EMT process by sponging multiple microRNAs. (A) Sankey diagram for the ceRNA network analysis of circHIPK3. (B) Luciferase activities of wide-type LUC-circHIPK3 after transfection with different miRNA mimics (n = 3). *P < 0.05 versus miR-NC group. (C) circRNA-miRNA binding sequences predicted by bioinformatics analysis. (D) Luciferase activities of mutant LUC-circHIPK3 after transfection with different miRNA mimics (n = 3). ns = not significant versus miR-NC group.
To confirm the role of circHIPK3-associated ceRNA network in RPE-EMT, we interfered the expression of circHIPK3 and detected the change of miRNA expression. qRT-PCR analysis showed that, silencing of circHIPK3 increased miRNA levels, whereas overexpression of circHIPK3 decreased miRNA levels (Fig. 5A). Given that the roles of miR-124-3p, miR-302d-3p, and miR-93-5p in RPE-EMT have been systematically investigated by other investigators previously, and miR-106b-5p had higher abundance than miR-106a-5p in normal RPE cells (Fig. 5B), thus in this study, we just explored the roles of miR-106b-5p in circHIPK3-associated ceRNA network17,18. Biotin-coupled miR-106b-5p mimic exhibited greater enrichment of circHIPK3 in captured fraction compared to the negative control (Fig. 5C). Transfection of miR-106b-5p mimic significantly attenuated TGF-β1-induced RPE-EMT process, which could partly mimic the role of circHIPK3 silencing (Fig. 5D–F). In addition, this phenotype could be reversed by ectopic expression of circHIPK3 (Fig. 5D–F). These data suggests that miR-106b-5p is involved in circHIPK3-associated ceRNA network in RPE-EMT. Taken together, these results indicate that circHIPK3 could serve as a sponge of multiple microRNAs to regulate RPE-EMT process.
circHIPK3 regulates RPE-EMT process partly through circHIPK3/miR-106b-5p axis. (A) qRT-PCR analysis of the expression of different miRNAs in RPE cells treated with si-circHIPK3 or lv-circHIPK3 (normalized to U6 snRNA expression, n = 3). *P < 0.05 versus Ctrl group. (B) qRT-PCR analysis of the expression of miR‐106a-5p and miR‐106b-5p in normal RPE cells (normalized to U6 snRNA expression, n = 3). *P < 0.05 versus miR‐106a-5p group. (C) qRT-PCR analysis of the expression of circHIPK3 in RPE cell lysates after biotin‐coupled miR‐106a-5p and miR‐106b-5p pull‐down assays (n = 3). *P < 0.05 versus miR‐106a-5p group. (D) Expression of EMT-related markers detected by qRT-PCR (normalized to GAPDH expression, n = 3). *P < 0.05. (E) Transwell migration assays in RPE cells treated with Scr mimic, miR-106b mimic, miR-106b mimic + lv-Scr, or miR-106b mimic + lv-circHIPK3 (n = 5). Scale bar, 50 μm. (F) Analysis of migrated cell number in the Transwell migration assays (n = 5). *P < 0.05 versus Ctrl group.
Discussion
In normal eyes, the RPE is a highly polarized monolayer of pigmented cells, performing physiological processes necessary for the maintenance and support of photoreceptors and visual function26. During pathogenesis of many vitreoretinal diseases, the RPE cells can be reprogrammed into fibrotic cells through EMT process, resulting in the formation of contractile epiretinal or subretinal membrane27. circRNAs play important roles in multiple pathophysiological processes and human diseases via regulating gene expression at transcriptional and posttranscriptional levels. In this study, we investigated the expression pattern, regulatory functions, and underlying mechanism of circHIPK3 in the EMT process of RPE cells. The expression of circHIPK3 is significantly upregulated in RPE-EMT model. Intervention of circHIPK3 expression could influence the development of RPE-EMT process. Mechanismly, circHIPK3 regulates RPE-EMT via targeting multiple microRNAs.
Previous studies have demonstrated that circRNAs could modulate EMT process in cancers through ceRNA mechanism, in which they bind miRNAs to regulate EMT-related transcription factors (e.g., SNAIL, TWIST, and ZEB families) and signaling pathways (e.g., Wnt/β-catenin, TGF-β/SMAD, and Notch pathways)28. For example, SNAIL is one of EMT-related transcription factors, which is positively correlated with EMT progression14. circPRMT5 mediates SNAIL expression by sponging miR-30c, thereby facilitates EMT process in urothelial carcinoma of the bladder29. Similarly, TGF-β/SMAD pathway is a positive regulator of EMT14. In non-small-cell lung carcinoma, circPTK2 serves as a sponge for miR-429 and miR-200b-3p, promoting the expression of TIF1γ, which further represses the TGF-β/SMAD-related EMT progression30. Here, we found that circHIPK3 could regulate the progression of EMT in RPE cells. Considering the various miRNA binding sites within circRNA transcripts, sponging multiple miRNAs by a single circRNA is a general mechanism. Bioinformatics analysis and luciferase reporter assay confirm that circHIPK3 directly interacts with multiple miRNAs, and these miRNAs could further interact with EMT-related transcription factors and signaling pathways.
Based on the data of bioinformatics analysis, luciferase reporter assay, and qRT-PCR analysis, we demonstrated that circHIPK3 could sponge and inhibit at least five miRNAs during RPE-EMT process, including miR-124-3p, miR-302d-3p, miR-93-5p, miR-106a-5p, or miR-106b-5p. It has been reported that miRNA-124-3p controls RPE-EMT by targeting RHOG, while miR-302d-3p and miR-93-5p regulate RPE-EMT by suppressing TGF-β/SMAD pathway17,18. This suggests that circHIPK3 mediates RPE-EMT process might partly through circHIPK3/miRNA-124-3p/RHOG axis, circHIPK3/miR-302d-3p/TGF-β signaling axis, and circHIPK3/miR-93-5p/TGF-β signaling axis.
Given that the abundance of miR-106b-5p is much higher than that of miR-106a-5p in normal RPE cells, thus we wondered whether miR-106b-5p participates in circHIPK3-associated ceRNA network in RPE-EMT. miR-106b-5p is a highly conserved miRNA that regulating EMT progression in various types of cancers, such as breast cancer, non-small cell lung cancer, and gastric cancer31,32,33. A study analyzed the differentially expressed miRNAs during RPE-EMT process, which showed that miR-106b-5p is highly expressed in RPE cells, and is downregulated in RPE-EMT (17.30-fold in microarray data and 3.30-fold in qRT-PCR result)34. Here, we confirmed the interaction between circHIPK3 and miR-106b-5p via luciferase reporter assay and biotin-coupled miRNA capture. Transfection of miR-106b-5p mimic could partly imitate the phenotype of circHIPK3 silencing, which could be further rescued by ectopic expression of circHIPK3. This indicates that miR-106b-5p participates in circHIPK3-associated ceRNA network in RPE-EMT.
RPE-EMT is a troublesome pathological process in many vitreoretinal diseases. Here, we show that inhibition of circHIPK3 leads to repression of RPE-EMT process. The advantage of using circHIPK3 to inhibit RPE-EMT is that circHIPK3 sponges multiple EMT-related miRNAs and thus regulates the expression of EMT-associated mRNAs. Targeting circHIPK3 might serve as a promising therapeutic strategy for EMT-associated vitreoretinal diseases.
Conclusion
In conclusion, circHIPK3 acts as a modulator of RPE-EMT process through abolishing the functions of multiple miRNAs. This study provides novel insights into the mechanism of EMT process in RPE cells, and serves a promising therapy for EMT-associated vitreoretinal diseases.
Data availability
The data used and/or analyzed during the current study available from the corresponding author on reasonable request.
References
Lakkaraju, A. et al. The cell biology of the retinal pigment epithelium. Prog. Retin. Eye Res. 78, 100846 (2020).
Shu, D. Y., Butcher, E. & Saint-Geniez, M. EMT and EndMT: Emerging roles in age-related macular degeneration. Int. J. Mol. Sci. 21(12), 4271 (2020).
Zhou, M. et al. Role of epithelial–mesenchymal transition in retinal pigment epithelium dysfunction. Front. Cell Dev. Biol. 8, 501 (2020).
Chen, L. L. The biogenesis and emerging roles of circular RNAs. Nat. Rev. Mol. Cell Biol. 17(4), 205–211 (2016).
Thomson, D. W. & Dinger, M. E. Endogenous microRNA sponges: Evidence and controversy. Nat. Rev. Genet. 17(5), 272–283 (2016).
Su, Y. et al. circRIP2 accelerates bladder cancer progression via miR-1305/Tgf-beta2/smad3 pathway. Mol. Cancer 19(1), 23 (2020).
Chen, X. et al. Circular noncoding RNA NR3C1 acts as a miR-382-5p sponge to protect RPE functions via regulating PTEN/AKT/mTOR signaling pathway. Mol. Ther. 28(3), 929–945 (2020).
Zheng, Q. et al. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat. Commun. 7, 11215 (2016).
Qi, L. et al. circHIPK3 (hsa_circ_0000284) promotes proliferation, migration and invasion of breast cancer cells via miR-326. Onco Targets Ther. 14, 3671–3685 (2021).
Shan, K. et al. Circular noncoding RNA HIPK3 mediates retinal vascular dysfunction in diabetes mellitus. Circulation 136(17), 1629–1642 (2017).
Liu, X. et al. Circular RNA HIPK3 regulates human lens epithelial cells proliferation and apoptosis by targeting the miR-193a/CRYAA axis. Biochem. Biophys. Res. Commun. 503(4), 2277–2285 (2018).
Cui, G., Wang, L. & Huang, W. Circular RNA HIPK3 regulates human lens epithelial cell dysfunction by targeting the miR-221-3p/PI3K/AKT pathway in age-related cataract. Exp. Eye Res. 198, 108128 (2020).
Nieto, M. A. et al. Emt: 2016. Cell 166(1), 21–45 (2016).
Lamouille, S., Xu, J. & Derynck, R. Molecular mechanisms of epithelial-mesenchymal transition. Nat. Rev. Mol. Cell Biol. 15(3), 178–196 (2014).
Li, J. H. et al. starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 42(Database issue), D92-97 (2014).
Agarwal, V. et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife https://doi.org/10.7554/eLife.05005 (2015).
Jun, J. H. & Joo, C. K. MicroRNA-124 controls transforming growth factor beta1-induced epithelial-mesenchymal transition in the retinal pigment epithelium by targeting RHOG. Investig. Ophthalmol. Vis. Sci. 57(1), 12–22 (2016).
Fuchs, H. R. et al. The microRNAs miR-302d and miR-93 inhibit TGFB-mediated EMT and VEGFA secretion from ARPE-19 cells. Exp. Eye Res. 201, 108258 (2020).
Huang, Y. & Yang, N. MicroRNA-20a-5p inhibits epithelial to mesenchymal transition and invasion of endometrial cancer cells by targeting STAT3. Int. J. Clin. Exp. Pathol. 11(12), 5715–5724 (2018).
Qi, J. C. et al. miR-20b-5p, TGFBR2, and E2F1 form a regulatory loop to participate in epithelial to mesenchymal transition in prostate cancer. Front. Oncol. 9, 1535 (2019).
Despotovic, J., Dragicevic, S. & Nikolic, A. Effects of chemotherapy for metastatic colorectal cancer on the TGF-beta signaling and related miRNAs hsa-miR-17-5p, hsa-miR-21-5p and hsa-miR-93-5p. Cell Biochem. Biophys. 79(4), 757–767 (2021).
Jin, W. et al. Long noncoding RNA regulator of reprogramming regulates cell growth, metastasis, and cisplatin resistance in gastric cancer via miR-519d-3p/HMGA2 axis. Cancer Biother. Radiopharm. 38, 122–131 (2020).
Zheng, Y. J. et al. Long noncoding RNA SMAD5-AS1 acts as a microRNA-106a-5p sponge to promote epithelial mesenchymal transition in nasopharyngeal carcinoma. FASEB J. 33(11), 12915–12928 (2019).
Liu, S. et al. miR106b5p targeting SIX1 inhibits TGFbeta1induced pulmonary fibrosis and epithelialmesenchymal transition in asthma through regulation of E2F1. Int. J. Mol. Med. https://doi.org/10.3892/ijmm.2021.4857 (2021).
Fan, L. et al. Non-canonical signaling pathway of SNAI2 induces EMT in ovarian cancer cells by suppressing miR-222-3p transcription and upregulating PDCD10. Theranostics 10(13), 5895–5913 (2020).
Baba, K., Goyal, V. & Tosini, G. Circadian regulation of retinal pigment epithelium function. Int. J. Mol. Sci. 23(5), 2699 (2022).
Yang, S. et al. Long noncoding RNA ERLR mediates epithelial-mesenchymal transition of retinal pigment epithelial cells and promotes experimental proliferative vitreoretinopathy. Cell Death Differ. 28(8), 2351–2366 (2021).
Shang, B. Q. et al. Functional roles of circular RNAs during epithelial-to-mesenchymal transition. Mol. Cancer 18(1), 138 (2019).
Chen, X. et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR-30c to induce epithelial-mesenchymal transition. Clin. Cancer Res. 24(24), 6319–6330 (2018).
Wang, L. et al. Circular RNA hsa_circ_0008305 (circPTK2) inhibits TGF-beta-induced epithelial-mesenchymal transition and metastasis by controlling TIF1gamma in non-small cell lung cancer. Mol. Cancer 17(1), 140 (2018).
Smith, A. L. et al. The miR-106b-25 cluster targets Smad7, activates TGF-beta signaling, and induces EMT and tumor initiating cell characteristics downstream of Six1 in human breast cancer. Oncogene 31(50), 5162–5171 (2012).
Savita, U. & Karunagaran, D. MicroRNA-106b-25 cluster targets beta-TRCP2, increases the expression of Snail and enhances cell migration and invasion in H1299 (non small cell lung cancer) cells. Biochem. Biophys. Res. Commun. 434(4), 841–847 (2013).
Kim, Y. K. et al. Functional links between clustered microRNAs: Suppression of cell-cycle inhibitors by microRNA clusters in gastric cancer. Nucleic Acids Res. 37(5), 1672–1681 (2009).
Chen, X. et al. Differentially expressed microRNAs in TGFbeta2-induced epithelial–mesenchymal transition in retinal pigment epithelium cells. Int. J. Mol. Med. 33(5), 1195–1200 (2014).
Funding
This work was supported by General Program of Wuxi Municipal Health Commission (No. M202224 to Qian Shi), Science and technology project of Wuxi Health (No. 2020S20247 to Qian Shi), Foundation of Yixing Innovation and technology Project (No. 2020SF12 Lijun Shi), and Foundation of Yixing Innovation and technology Project (No. 2020SF27 to Qian Shi).
Author information
Authors and Affiliations
Contributions
QS designed the study. YF and FY drafted the manuscript. YF, FY, and JZ conducted all the experiments. LS, TX, and YL helped to collate the data. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Feng, Y., Yang, F., Zheng, J. et al. Circular RNA HIPK3 mediates epithelial–mesenchymal transition of retinal pigment epithelial cells by sponging multiple microRNAs. Sci Rep 14, 28872 (2024). https://doi.org/10.1038/s41598-024-71119-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71119-6