Abstract
An increasing number of studies indicate that long noncoding RNAs (lncRNAs) play important roles in tumour proliferation, migration and other vital processes and are expected to become novel biomarkers for early cancer screening. The expression of the lncRNA NBR2 (adjacent breast cancer suppressor BRCA1) has been found to decrease in several cancer types. However, it is still unknown whether the lncRNA NBR2 is involved in breast cancer and autophagy. According to the Kaplan–Meier plotter survival curve analysis, the survival rate of the group with high lncRNA-NBR2 expression was higher than that of the group with low lncRNA-NBR2 expression. The suppression of cancer cell proliferation, invasion and migration by the lncRNA NBR2 has been demonstrated, suggesting that this lncRNA is involved in the development and progression of cancer. Our subsequent study revealed that the lncRNA NBR2 inhibited autophagy in breast cancer cells, and that starvation conditions enhanced this inhibitory effect. Moreover, this lncRNA changed the proliferation ability of breast cancer cells by affecting protective autophagy. The aim of this study was to investigate the link between starvation and lncRNAs by evaluating changes in autophagy-related proteins, cell proliferation and other biological processes. Together, these studies provide strategies for the early screening of breast cancer and suggest that starvation therapy can be used as a new approach for the treatment of cancer.
Similar content being viewed by others
Introduction
Breast cancer refers to the cancerous transformation of normal mammary epithelial cells by a variety of tumour-causing factors and mainly manifests as a tissue mass in the breast, axillary lymph node enlargement and other symptoms. Cancer cells easily metastasize to multiple different organs, seriously threatening the life of patients1,2. Currently, surgical therapy, drug therapy, endocrine therapy and targeted therapy are the main therapeutic means for treating breast cancer. Due to the various types of breast cancer, drug resistance and postoperative recurrence, the mortality rate of breast cancer is high3,4. Therefore, identifying new combination treatments is urgently needed for increasing the cure rate of cancer.
RNA molecules with transcript lengths of more than 200 bases, known as long noncoding RNAs (lncRNAs), have been identified5,6. In recent years, lncRNAs have become one of the most popular topics in life science research. LncRNAs have a variety of biological functions, such as serving as molecular scaffolds in the nucleus, assisting in variable splicing, and regulating chromosome structure7,8,9. The regulation of tumour occurrence and development by lncRNAs, as well as their capacity to regulate gene expression, has been confirmed in previous studies10,11.
NBR2 (immediate breast cancer suppressor BRCA1) is produced under glucose starvation conditions12, and the expression of this gene is dysregulated in several types of cancer, such as osteosarcoma and lung cancer13. Studies have shown that the lncRNA NBR2 is downregulated in liver cancer tissues and liver cancer cell lines. In addition, the survival time and overall survival time of liver cancer patients with low lncRNA NBR2 expression are shorter. The expression of the lncRNA NBR2 can inhibit the proliferation, migration, and invasion of liver cancer cells, suggesting that it is a tumour suppressor in liver cancer14. However, the specific role of NBR2 in breast cancer remains unclear.
Recently, increasing evidence has revealed that protective autophagy is an important factor for the survival of cancer cells in adverse environments. The process of autophagy involves the encapsulation of ageing cells or misfolded proteins in vesicles, which are then joined with lysosomes to form autophagic lysosomes. These lysosomes then degrade their contents, leading to the removal of cells and proteins and enabling the reutilization of substances15,16. In recent years, “starvation” cancer therapy has garnered increasing interest among researchers. Many studies have reported that short-term starvation can increase autophagy in tumour cells17,18. Autophagy plays an important role in tumour development. Therefore, is there a direct link between NBR2 and autophagy in breast cancer cells, and does NBR2 influence the progression of breast cancer by regulating autophagy? Is the increase in autophagy in breast cancer cells under short-term starvation conditions caused by changes in the lncRNA NBR2? All of these factors need to be further studied.
Since its potential biological function in breast cancer has remained unexplored, further studies are needed to reveal the effect of interfering with NBR2 on tumours and its relationship with autophagy. A better understanding of the contributions of lncRNAs to protective autophagy will be beneficial for developing new approaches to achieve sustained remission in breast cancer patients.
Materials and methods
Cell lines and culture conditions
After resuspension, RPMI-1640 medium (Gibco, USA) supplemented with 10% foetal bovine serum (Clark, USA) was used to culture MDA-MB-231 and BT-549 cells. These cells were subcultured in an incubator at 37 °C with 5% CO2 every 2 to 3 days. Both cell lines were obtained from Procell Life Science and Technology (Wuhan, China).
Construction and chemicals
HBLV-ZsGreen-PURO-overexpressing NC (Lv-NC), HBLV-h-NBR2-Null-ZsGreen-PURO (Lv- NBR2), HBLV-ZsGreen-PURO NC (Lv-shNC) and HBLV-h-NBR2 shRNA1-ZsGreen-PURO (Lv-shNBR2) were purchased from Hanbio (Shanghai, China), and chloroquine was purchased from MedChem Express (Shanghai, China).
Construction of lentivirus packaged cell lines
MDA-MB-231 and BT-549 cells with suitable growth status were inoculated into 24-well plates. Lv-NC, Lv-NBR2, Lv-shNC and Lv-shNBR2 were added to the corresponding wells, and the cells were transfected overnight. On the second day of transfection, the medium of the cells was replaced with fresh complete medium, and the cells were cultured at 37 °C for another 48 h. The fluorescence intensity was preliminarily evaluated via fluorescence microscopy. The cells were seeded into six-well plates for culture according to cell growth, and stably transduced cell lines were screened with puromycin.
Quantitative real-time PCR (qRT-PCR)
An RNA extraction kit (Vazyme, Nanjing, China) was used to extract total RNA. A ChamQ Universal SYBR qPCR Master Mix qRT-PCR kit (Vazyme, Nanjing, China) was then used to reverse transcribe the RNA into cDNA, according to the manufacturer’s instructions, and this procedure was performed with QuantStudioDesign and Anailsis Software v1.4 (Thermo Company, China). The expression level of the lncRNA NBR2 relative to that of GAPDH was measured by PCR. The 2−ΔΔCt method was used to calculate the expression of target genes. Shanghai Sangon Company crafted the lncRNA NBR2 primer, the sequence of which is displayed in Table 1.
Western blot analysis
A protease inhibitor was added to the protein lysate to lyse the cells, thereby obtaining the total protein sample. Subsequently, a BCA kit was used to quantify the samples, followed by electrophoresis and membrane transfer. The membranes were incubated with primary antibodies against LC3B-II/I, P62, Beclin1 and β-actin (Proteintech, Wuhan, China) overnight at 4 °C, incubated with secondary antibodies for 2 h at room temperature, and visualized with an enhanced chemiluminescence (ECL) kit (Boster, USA); the bands were subsequently analysed with ImageJ software.
MTT cell proliferation assay
MDA-MB-231 and BT-549 cells transfected with the appropriate viral vectors were prepared as 6 × 103/mL cell suspensions, and 100 μL of each cell suspension was added to each 96-well plate. MTT reagent was added at 0 h, 24 h, 48 h and 72 h, and the culture was continued for 4 h. Then 200 μL of DMSO was added and the cells were incubated in the dark for 10 min. Detection was carried out using microplate reader at a wavelength of 490 nm.
Colony formation assay
MDA-MB-231 and BT-549 cells infected with virus were prepared as 1.5 × 103/mL cell suspensions and seeded into six-well plates for 15 days at 37 °C (the number of cells in each cell colony was approximately 100). The old medium was discarded, and the cells were washed twice with PBS. The cells were fixed with 1 mL of paraformaldehyde/well. Next, 1 mL of crystal violet was added to each well, and the number of cell colonies was counted.
Wound healing assay
The cells were seeded into a 6-well plate. When the cell density was close to 95%, a scratch was generated with a 10 μL pipette tip, and photographs were taken at 0 h and 24 h to record the degree of wound healing.
Cell apoptosis and cell cycle assays
The cells in each group were collected and washed with precooled PBS. Then, the supernatant was discarded, and 400 μL of binding solution was added to the cell precipitate to resuspend the cells. Then, 3 μL of Annexin V-APC and 3 μL of PI were added, and the mixture was incubated at 2–8 °C for 2–5 min. Flow cytometry was used to detect the percentage of apoptotic cells. The cells from each group were collected, washed with precooled PBS, and then rehydrated with precooled ethanol in a refrigerator at 4 °C overnight. On the second day, the cells were washed with precooled PBS before the supernatant was discarded. Flow cytometry was used to detect the cell cycle distribution after 400 μL of staining buffer, 25 μL of PI staining solution, and 10 μL of RNaseA were added to each tube of cell precipitate and incubated at 37 °C for 30 min.
Transwell experiment
The upper chamber was filled with 200 μL of cell suspension (3 × 104/mL) without FBS, diluted Matrigel was added prior to the invasion experiment, and 600 μL of fresh complete medium was added to the lower chamber. After 24 h of culture, the cells were fixed with paraformaldehyde for 30 min, and then stained with crystal violet for 20 min, and photographs were taken under a microscope.
Immunofluorescence staining
The confocal dishes were filled with cells in a healthy growth state, which were subsequently washed twice with PBS, fixed with methanol for 20 min and transferred to membranes for 20 min. After 3 washes with PBS, the cells were blocked for an hour, incubated with the primary antibody LC3B- II/I (Proteintech, Wuhan, China) overnight, and then washed 3 times with PBST. To prevent illumination, a secondary antibody was added, and the membrane was washed 3 times. Subsequently, the cells were stained with DAPI for two minutes, followed by a washing with PBST for 3 more minutes. Finally, the cells were observed and photographed under a laser scanning microscope. Tumour tissues were also stained with H&E.
Patients and specimens
From May 2022 to November 2022 at the First Affiliated Hospital of Bengbu Medical College, we collected 30 sets of breast cancer tissue and paracancerous tissue samples from patients who had undergone surgery. The pathology department of the hospital verified that all of the patients with breast cancer had a pathological diagnosis. The tumour pathological data of the corresponding patients, including age, breast mass size, histopathological grade, TNM stage, distant lymph node metastasis status, molecular type, and Ki67 expression status, were collected. The inclusion criteria for patients were as follows: (1) pathological examination, such as a definite diagnosis of breast cancer; and (2) received an initial cancer diagnosis and had no history of radiotherapy, chemotherapy or targeted anticancer therapy. The exclusion criteria were as follows: (1) had another type of tumour; (2) had a previous history of breast cancer and antitumour therapy; (3) had a mental illness or serious heart, liver or other serious disease; and (4) had distant organ metastasis. This study strictly complied with the relevant medical ethical requirements of the school (All experiments involving animals were approved by the Ethics Committee of Bengbu Medical College).
Xenograft tumour model
All experiments involving animals were conducted in accordance with the guidelines of the Ethics Committee of Bengbu Medical College. Female BALB/ca-nu nude mice (3–4 weeks old) were purchased from Hefei Qingyuan Biotechnology. BT-549 cells (1 × 106) transfected with Lv-shNC or Lv-shNBR2 lentivirus were injected subcutaneously into mice and resuspended in 100 μL of PBS. The mice were euthanized, and the tumours were harvested within one month for subsequent analyses, including weight and volume analyses, immunohistochemistry, qPCR and western blotting.
Immunohistochemistry (IHC)
The tumours were fixed in 10% formalin solution, embedded in paraffin, and then cut into 5 μm-thick serial sections. After dewaxing in xylene and dehydrating in a gradient series of ethanol concentrations antigens were extracted with citrate buffer and blocked with 5% goat serum, followed by incubation with an anti-LC3B-II/I antibody overnight at 4 °C. Subsequently, the sections were incubated with peroxidase-conjugated goat anti-rabbit secondary antibody (Abcam, United States) for 2 h at room temperature and stained with a DAB kit (ZSGB-BIO, China).
Statistical methods
The experimental results are expressed as the mean ± SEM. Statistical analysis between experimental groups was performed using GraphPad Prism 8.0.2 software for tests or analysis of variance (ANOVA). P < 0.05 was considered to indicate statistical significance.
Results
LncRNA NBR2 expression
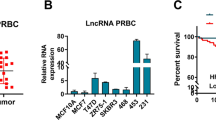
Gene expression data was downloaded from The TCGA database, and R language and other tools were subsequently used to construct a box plot. The results revealed that the expression level of the lncRNA NBR2 in the normal group was greater than that in the triple -negative breast cancer (TNBC) group and the triple-negative breast cancer (non-TNBC) group, and the difference was statistically significant (Kruskal–Wallis test, P < 0.001) (Fig. 1A). Kaplan Meier curve analysis of the effect of lncRNA NBR2 expression on the survival of patients with triple-negative breast cancer and patients with TNBC. The results of both groups showed that the five-year survival rate of the group with low lncRNA NBR2 expression was lower than that of the group with high lncRNA NBR2 expression. The P value of the survival rate in the group of patients with triple-negative breast cancer was 0.082 (P < 0.1). Our explanation was that the sample size of the group with high lncRNA NBR2 expression was lower than that of the group with low lncRNA NBR2 expression, and the survival time of the group with high lncRNA NBR2 expression was much greater than that of the group with low lncRNA NBR2 expression (Fig. 1B). Next, we used qRT-PCR to detect lncRNA NBR2 expression levels in cancer tissue and paracancerous tissue samples from 30 clinical breast cancer patients (Fig. 1C) and analysed the correlation between lncRNA NBR2 and clinically relevant indicators (Table 2). Then, we analysed the expression of the lncRNA NBR2 in nontumorigenic mammary epithelial cells and several breast cancer cell lines by qRT-PCR and found that the lncRNA NBR2 was generally underexpressed in breast cancer cell lines (Fig. 1D). We selected two breast cancer cell lines with different lncRNA NBR2 expression levels to study the biological function of lncRNA NBR2 in breast cancer cells, namely, MDA-MB-231 and BT-549 cells. A lentiviral vector with the lncRNA NBR2 sequence (Lv-NBR2) was used to upregulate the expression of the lncRNA NBR2 in MDA-MB-231 and BT-549 cells. Lentiviral vectors with shRNA targeting NBR2 (shNBR2) were used to downregulate the expression of the lncRNA NBR2 in MDA-MB-231 and BT-549 cells (Fig. 1E–F).
Expression of lncRNA NBR2. (A) The dataset was downloaded from the TCGA database, and a box plot was constructed with R to analyse the expression levels of the lncRNA NBR2 in the normal group, triple-negative breast cancer group (TNBC) and non-triple-negative breast cancer group (non-TNBC). (B) Kaplan‒Meier curve analysis of the relationship between the lncRNA NBR2 expression level and the survival of patients with triple-negative breast cancer and patients with non-triple-negative breast cancer. (C) LncRNA NBR2 expression levels in cancer tissue and paracancerous tissue samples from clinical breast cancer patients were analysed by qRT-PCR. (D) LncRNA NBR2 expression levels in breast cancer cells and MCF-10A cells were analysed by qRT-PCR. (E) The stable low expression of the lncRNA NBR2 in the BT-549 and MDA-MB-231 cell lines was verified by qRT-PCR. (F) The stable high expression of the lncRNA NBR2 in the BT-549 and MDA-MB-231 cell lines was verified by qRT-PCR. *P < 0.05, **P < 0.01, ***P < 0.001.
Inhibitory effect of lncRNA NBR2 on breast cancer cells
We continued to explore the effects of the lncRNA NBR2 on the proliferation of breast cancer cells and used MTT, colony formation, cell cycle and apoptosis assays to detect changes in the biological behaviour of breast cancer cells after lncRNA NBR2 interference. We found that after downregulating the lncRNA NBR2, the formation of individual cell colonies increased (Fig. 2A), cell proliferation increased (Fig. 2B), and the percentage of cells in S phase increased (Fig. 2C), while the opposite results were obtained after upregulating the lncRNA NBR2. The flow cytometry results showed that the apoptotic effect of lncRNA NBR2 did not significantly change after upregulation or downregulation (Fig. 2D), which provided a new direction for our subsequent research.
Inhibition of the lncRNA NBR2 in breast cancer cells. (A) A colony formation assay was used to evaluate the proliferation of MDA-MB-231 and BT-549 cells after knockdown or overexpression of the lncRNA NBR2. (B) MTT proliferation experiments showed that overexpression of the lncRNA NBR2 limited the proliferation of BT-549 and MDA-MB-231 cells, while silencing lncRNA NBR2 promoted the proliferation of cells. (C) The cell cycle distribution of BT-549 and MDA-MB-231 cells was detected by flow cytometry. (D) The percentage of apoptotic cells was detected by Annexin V/PI double staining. The data are expressed as the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
The lncRNA NBR2 inhibits the invasion and migration of breast cancer cells
Wound healing and Transwell experiments revealed the influence of the lncRNA NBR2 on cancer cell invasion and migration. The results showed that downregulation of the lncRNA NBR2 promoted the invasion and migration of MDA-MB-231 and BT-549 cells (Fig. 3A,B), while upregulation of the lncRNA NBR2 had the opposite effect on the breast cancer cells.
Effects of the lncRNA NBR2 on the invasion and migration of breast cancer cells. (A) The healing ability of BT-549 and MDA-MB-231 breast cancer cells after knockdown or overexpression of the lncRNA NBR2 was evaluated by a scratch healing assay. (B) Transwell assays were used to detect the longitudinal migration and invasion ability of BT-549 and MDA-MB-231 breast cancer cells after knocking down or overexpressing the lncRNA NBR2. Each bar represents the mean ± SD of three independent experiments. *P < 0.05, **P < 0.01, ***P < 0.001.
The lncRNA NBR2 inhibits autophagy in breast cancer cells
Autophagy increased in the two breast cancer cell lines after medium starvation treatment with EBSS (Earle's balanced salt solution, with Ca2+ and Mg2+, autophagy induction reagent) for 0 h, 6 h, 9 h, or 12 h (Fig. 4A), and the lncRNA NBR2 expression decreased after short-term starvation for 6 h (Fig. 4B). Immunofluorescence assays revealed an increase in the number of autophagosomes in the two breast cancer cell lines after starvation for 6 h (Fig. 4C). Western blotting revealed increased autophagy in the downregulated lncRNA NBR2 group, and short-term starvation for 6 h promoted autophagy in the downregulated lncRNA NBR2 group (Fig. 4D). LncRNA NBR2 upregulation decreased autophagy in breast cancer cells, and starvation abrogated the inhibitory effect of upregulated lncRNA NBR2 on autophagy (Fig. 4E). Immunofluorescence experiments showed that under starvation conditions, autophagosome formation significantly increased after downregulation of the lncRNA NBR2, while the opposite effect was observed when the lncRNA NBR2 was upregulated (Fig. 4F,G). In conclusion, the lncRNA NBR2 inhibits autophagy in breast cancer cells, and starvation conditions can abrogate its effect on autophagy.
Changes in the expression of autophagy-related proteins and the lncRNA NBR2 and the number of autophagosomes in breast cancer cells treated with the lncRNA NBR2 under starvation conditions. (A) The relative levels of autophagy related proteins such as LC3 and P62 in BT-549 and MDA-MB-231 cells after 0 h, 6 h, 9 h and 12 h were detected by Western blotting. (B) Changes in lncRNA NBR2 expression were verified by qRT-PCR in BT-549 and MDA-MB-231 cells after 6 h of starvation. (C) Changes in the number of autophagosomes in BT-549 and MDA-MB-231 cells were detected by immunofluorescence experiments after 6 h of starvation. (D,E) Western blotting analysis was used to determine the changes in autophagy-related proteins such as LC3 and P62 in BT-549 and MDA-MB-231 cells after 6 h of starvation and downregulation/upregulation of the lncRNA NBR2. (F,G) Immunofluorescence assays were used to detect the number of autophagosomes in BT-549 and MDA-MB-231 cells after lncRNA NBR2 upregulation or downregulation after 6 h of starvation. *P < 0.05, **P < 0.01, ***P < 0.001.
Downregulating the lncRNA NBR2-induced cytoprotective autophagy and promoted the proliferation of breast cancer cells in vitro
Autophagy plays dual roles in the emergence and growth of tumours by promoting the proliferation of tumour cells and impeding their growth. We conducted MTT, colony formation and flow cytometry experiments to explore the role of the lncRNA NBR2 in regulating autophagy. Autophagy was further regulated to observe the effect of the lncRNA NBR2 on the proliferation of breast cancer cells. The lncRNA NBR2 was knocked down in MDA-MB-231 and BT-549 cells, while chloroquine (CQ, a late autophagy inhibitor) was added to inhibit autophagy flow, and the proliferation of breast cancer cells was observed. We found that after chloroquine treatment, the proliferation of breast cancer cells decreased (Fig. 5A,C), the colony formation decreased (Fig. 5B,D), and the cell cycle was inhibited (Fig. 5E,F). These results imply that downregulating the lncRNA NBR2 can trigger autophagy in breast cancer cells, thereby stimulating cell proliferation and thereby protecting them. Blocking autophagy can weaken the proliferative effect of lncRNA NBR2 downregulation.
The lncRNA NBR2 was knocked down in BT-549 and MDA-MB-231 cells, and chloroquine was added to inhibit autophagic flow to observe the proliferation of breast cancer cells. (A,B) MTT and colony formation assays showed that the viability of BT-549 cells was significantly greater in the lncRNA-knockdown NBR2 group than in the control group, but this growth-promoting effect was weakened after CQ treatment. (C,D) MTT and colony formation assays were used to detect the proliferative ability of MDA-MB-231 cells after the addition of CQ. (E,F) Flow cytometry was used to detect changes in the cell cycle distribution of BT-549 and MDA-MB-231 cells after the addition of CQ. *P < 0.05, **P < 0.01, ***P < 0.001.
Tumour formation experiment in nude mice
In vitro-constructed BT-549 cells with low expression of the lncRNA NBR2 were implanted beneath the skin of nude mice, which were then split into two groups: the control group and the NBR2 low- expression group. The tumour size and weight of the latter group were greater than those of the former group (Fig. 6A,C). Moreover, protein and RNA were extracted from the tumour tissue, and PCR analysis revealed that lncRNA NBR2 expression was significantly downregulated in the low-expression group (Fig. 6B). Western blotting revealed increased expression of autophagy-related proteins in the group with low expression of the lncRNA NBR2 (Fig. 6D). Immunohistochemical analysis of tumour tissue sections revealed that downregulated lncRNA NBR2 expression promoted the malignant development of tumours and the expression of the autophagy-related index LC3 (Fig. 6E). In conclusion, the downregulation of lncRNA NBR2 expression can promote the development of tumours and autophagy in vivo.
Effects of the lncRNA NBR2 knockdown on tumorigenic ability and autophagy in vivo. (A) BT-549 cells with low lncRNA NBR2 expression and control cells were subcutaneously inoculated into nude mice. Representative images of isolated tumours showing significant growth. (B) RNA was extracted from tumour tissue, and the expression of the lncRNA NBR2 was analysed by qRT-PCR. (C) The volume and weight of subcutaneously transplanted tumours were measured in nude mice. (D) Protein was extracted from the tumour tissue, and autophagy-related protein expression was detected by western blotting. (E) Representative images of H&E staining and the expression of the autophagy-related index LC3 by immunohistochemistry in subcutaneous tumours. *P < 0.05, **P < 0.01, ***P < 0.001.
Discussion
The aetiology of breast cancer is relatively complex and involves factors such as genetics, hormones, reproduction, nutrition and other factors, which have a certain impact on its incidence. In recent years, many studies have shown that some RNA molecules are closely related to the proliferation, migration and invasion of tumour cells in vivo19,20,21, which makes lncRNAs attractive targets for identifying breast cancer intervention therapies. The TCGA BRCA gene expression data used in this study were downloaded from UCSC Xena (https://xenabrowser.net/). The data type was mRNA FPKM-UQ, and then tools such as R language were used to create a box plot. Next, we knocked down or overexpressed lncRNA NBR2 to detect changes in the proliferation, migration, invasion, and autophagy of breast cancer cells and in the formation of subcutaneous grafts in nude mice, to clarify the specific mechanism of the lncRNA NBR2 in the occurrence and development of breast cancer.
LncRNAs can have a great impact on the development of cancer, including the proliferation and metastasis of tumour cells22. Studies have shown that lncRNAs can regulate the expression of proteins related to epithelial mesenchymal transformation23 and that the expression of these proteins can affect the proliferation of tumour cells; therefore, lncRNAs may become key targets for future tumour therapy24,25. The expression of LINC01123 is upregulated in ovarian cancer cell lines, and silenced expression of LINC01123 inhibits the proliferation and metastasis of ovarian cancer cells and promotes apoptosis26. The overexpression of the lncRNA FOXD1-AS1 promotes the occurrence and self-renewal of pancreatic cancer tumours27. Knocking down TPT1-AS1 inhibits the proliferation and metastasis of liver cancer cells28, all of which suggest that lncRNAs play important roles in tumour development.
The literature has shown that the lncRNA NBR2, which is located on human chromosome 17q21 and adjacent to the tumour suppressor gene BRCA1, is a product generated by a hypoglycaemic environment. Overexpression of the lncRNA NBR2 downregulated TGF-β1. However, overexpression of TGF-β1 does not affect the expression of the lncRNA NBR2, which can inhibit cell proliferation, differentiation and metastasis by inhibiting cell dryness in NSCLC29. Our results are consistent with the conclusions of these studies. In both the biogenic analysis and clinical breast cancer tissue sample detection, the expression of the lncRNA NBR2 in breast cancer tissues was downregulated, which is associated with poor prognosis. The expression of the lncRNA NBR2 is closely related to tumour size, histological grade, TNM stage, lymph node metastasis status and Ki67 expression. At the cellular level, lncRNA NBR2 expression was downregulated in a variety of breast cancer cell lines, inhibiting the proliferation, migration, invasion, cell cycle progression and other biological behaviours of breast cancer cells. Flow cytometry results showed that knocking down or overexpressing the lncRNA NBR2 did not change cell apoptosis. These results indicated that the lncRNA NBR2 did not change the biological behaviour of breast cancer cell proliferation by affecting cell apoptosis. To further verify this conclusion, we conducted tumour formation experiments in nude mice, and the results showed that the tumour volume and growth of nude mice in the NBR2 group increased compared with those in the NC group. The clinical sample collection in this study also has several limitations, such as a small sample size, but the above research results indicate that the lncRNA NBR2 plays an anticancer role in human breast cancer by inhibiting biological behaviours such as the growth, migration and invasion of breast cancer cells.
We know that autophagy in the current literature can be divided into two aspects: one is to protect tumour cells and promote the degradation of damaged substances in cells to achieve the reuse of substances and provide nutritional support for tumour cell proliferation; the other is to monitor the malignant changes of cells in the body and self-clear to damaged cells to protect the individual30,31. Autophagy and antiautophagic processes need to be balanced, and a change in a small molecule in the cell can alter this balance, so it is crucial to explore the factors that affect the balance of autophagy. The long noncoding RNA SNHG11 has been reported to be upregulated in gastric cancer, and its upregulation is associated with poor prognosis in patients. Functionally, SNHG11 increased the autophagy of gastric cancer cells and promoted their proliferation. Mechanistically, after the transcription of SNHG11, the expression of catenin β1 (CTNNB1) and autophagy -associated protein 12 (ATG12) was upregulated by miR-483-3p, and SNHG11 regulates autophagy in an ATG12-dependent manner32. From the above studies, we can see that lncRNAs can regulate the autophagy level of tumour cells, but what is the effect of lncRNA NBR2 on the autophagy of breast cancer cells? We detected the lncRNA NBR2 by western blot and immunofluorescence assays, and the results showed that the lncRNA NBR2 inhibited autophagy and the formation of autophagosomes in breast cancer cells. MTT, clonogenic and cell cycle assays were used for follow-up verification, and the results showed that cell proliferation was inhibited in the Lv-shNBR2 + CQ group compared with the Lv-shNBR2 group, and the cell cycle was blocked in the G1 phase. We can conclude that the lncRNA NBR2 inhibits cell proliferation, cell cycle progression and other biological behaviours by inhibiting protective autophagy in breast cancer cells. However, the specific mechanism by which lncRNA NBR2 affects protective autophagy in breast cancer cells still needs to be further explored (refer to the Western Blotting Results, Supplementary Information).
Glucose starvation induces the recruitment of the DNA damage receptor Mec1 protein to the mitochondrial surface, and the energy receptor Snf1 phosphorylates Mec1. Finally, phosphorylated Mec1 interferes with the induction of autophagy by affecting the formation of the ATG1 complex33. We further used western blotting, fluorescent quantitative PCR and immunofluorescence experiments to conduct studies under starvation conditions, and the results showed that lncRNA NBR2 expression was downregulated and autophagy in breast cancer cells was increased under starvation conditions. Compared with lncRNA NBR2 knockdown, lncRNA NBR2 knockdown combined with starvation significantly increased the expression of autophagy-related proteins and the formation of autophagosomes. This role in promoting autophagy may be realized by further downregulating lncRNA NBR2 expression under starvation treatment. In addition, starvation treatment alleviated the inhibitory effect of lncRNA NBR2 overexpression on cellular protective autophagy. These results indicate that starvation treatment and the lncRNA NBR2 have antagonistic effects on the expression of autophagy-related proteins to a certain extent, but the specific mechanism of action is still unclear. However, the results of this study provide a theoretical basis for the study of breast cancer.
Conclusion
In summary, we believe that the lncRNA NBR2 plays an antitumour role in breast cancer by inhibiting the proliferation, migration and invasion ability of breast cancer cells. The lncRNA NBR2 inhibits cell proliferation, cell cycle progression and other biological behaviours by reducing protective autophagy in breast cancer cells. Starvation can abrogate the inhibitory effect of the lncRNA NBR2 on protective autophagy in breast cancer cells.
Data availability
The data used in this study were public datasets, and the BRCA gene expression data were downloaded from UCSC Xena (https://xenabrowser.net) using The Cancer Genome Atlas (https://portal.GDC.cancer.gov). The corresponding data in the article are available from the corresponding author. All methods and public data were implemented according to the relevant guidelines and regulations.
Abbreviations
- cDNA:
-
Complementary DNA
- DMSO:
-
Dimethyl sulfoxide
- ECL:
-
Electrochemical luminescence
- APC:
-
Allophycocyanin
- CQ:
-
Chloroquine
- MTT:
-
3-(4,5-Dimethyl-2-thiazolyl)-2–5-diphenyl-2H-terazolium bromide
- PBS:
-
Phosphate buffered saline
- PI:
-
Propidium iodide
- DAPI:
-
4′,6-Diamidino-2-phenylindole, dihydrochloride
- qRT-PCR:
-
Real-time quantitative polymerase chain reaction
- Rpm:
-
Rotation per minute
- RNA:
-
Ribonucleic acid
References
Wilkinson, L. & Gathani, T. Understanding breast cancer as a global health concern. Br. J. Radiol. 95, 20211033. https://doi.org/10.1259/bjr.20211033 (2022).
Barzaman, K. et al. Breast cancer: Biology, biomarkers, and treatments. Int. Immunopharmacol. 84, 106535. https://doi.org/10.1016/j.intimp.2020.106535 (2020).
Ren, C. et al. Ubiquitination of NF-κB p65 by FBXW2 suppresses breast cancer stemness, tumorigenesis, and paclitaxel resistance. Cell Death Differ. 29, 381–392. https://doi.org/10.1038/s41418-021-00862-4 (2022).
Xu, X., Zhang, M., Xu, F. & Jiang, S. Wnt signaling in breast cancer: Biological mechanisms, challenges and opportunities. Mol. Cancer 19, 165. https://doi.org/10.1186/s12943-020-01276-5 (2020).
Tan, Y. T. et al. LncRNA-mediated posttranslational modifications and reprogramming of energy metabolism in cancer. Cancer Commun. 41, 109–120. https://doi.org/10.1002/cac2.12108 (2021).
Xing, C., Sun, S.-G., Yue, Z.-Q. & Bai, F. Role of lncRNA LUCAT1 in cancer. Biomed. Pharmacother. 134, 111158. https://doi.org/10.1016/j.biopha.2020.111158 (2021).
Bridges, M. C., Daulagala, A. C. & Kourtidis, A. LNCcation: lncRNA localization and function. J. Cell Biol. 220, e202009045. https://doi.org/10.1083/jcb.202009045 (2021).
Ali, T. & Grote, P. Beyond the RNA-dependent function of LncRNA genes. ELife 9, e60583. https://doi.org/10.7554/eLife.60583 (2020).
Guo, C. et al. Pathophysiological functions of the lncRNA TUG1. Curr. Pharm. Des. 26, 688–700. https://doi.org/10.2174/1381612826666191227154009 (2020).
Abd El Fattah, Y. K., Abulsoud, A. I., Abdel Hamid, S. G. & Hamdy, N. M. Interactome battling of lncRNA CCDC144NL-AS1: Its role in the emergence and ferocity of cancer and beyond. Int. J. Biol. Macromol. 222, 1676–1687. https://doi.org/10.1016/j.ijbiomac.2022.09.209 (2022).
Zhang, L., Zhang, Z., Li, E. & Xu, P. c-Myc-regulated lncRNA-IGFBP4 suppresses autophagy in cervical cancer-originated HeLa cells. Dis. Mark. 2022, 7240646. https://doi.org/10.1155/2022/7240646 (2022).
Zhu, C. et al. lncRNA NBR2 attenuates angiotensin II-induced myocardial hypertrophy through repressing ER stress via activating LKB1/AMPK/Sirt1 pathway. Bioengineered 13, 13667–13679. https://doi.org/10.1080/21655979.2022.2062527 (2022).
Cai, W. et al. LncRNA NBR2 inhibits epithelial-mesenchymal transition by regulating Notch1 signaling in osteosarcoma cells. J. Cell. Biochem. 120, 2015–2027. https://doi.org/10.1002/jcb.27508 (2019).
Sheng, J.-Q. et al. LncRNA NBR2 inhibits tumorigenesis by regulating autophagy in hepatocellular carcinoma. Biomed. Pharmacother. 133, 111023. https://doi.org/10.1016/j.biopha.2020.111023 (2021).
Onorati, A. V., Dyczynski, M., Ojha, R. & Amaravadi, R. K. Targeting autophagy in cancer. Cancer 124, 3307–3318. https://doi.org/10.1038/nrc.2017.53 (2018).
Cao, W., Li, J., Yang, K. & Cao, D. An overview of autophagy: Mechanism, regulation and research progress. Bull. Cancer 108, 304–322. https://doi.org/10.1016/j.bulcan.2020.11.004 (2021).
Yang, B., Ding, L., Chen, Y. & Shi, J. Augmenting tumor-starvation therapy by cancer cell autophagy inhibition. Adv. Sci. 7, 1902847. https://doi.org/10.1002/advs.201902847 (2020).
Poillet-Perez, L. et al. Autophagy maintains tumour growth through circulating arginine. Nature 563, 569–573. https://doi.org/10.1038/s41586-018-0697-7 (2018).
Tang, Y. et al. Dissection of FOXO1-induced LYPLAL1-DT impeding triple-negative breast cancer progression via mediating hnRNPK/β-catenin complex. Research 6, 0289. https://doi.org/10.34133/research.0289 (2023).
Wu, S. et al. A novel axis of circKIF4A-miR-637-STAT3 promotes brain metastasis in triple-negative breast cancer. Cancer Lett. 581, 216508. https://doi.org/10.1016/j.canlet.2023.216508 (2024).
Wang, Z. et al. The circROBO1/KLF5/FUS feedback loop regulates the liver metastasis of breast cancer by inhibiting the selective autophagy of afadin. Mol. Cancer 21, 29. https://doi.org/10.1186/s12943-022-01498-9 (2022).
Wang, G. et al. A novel long noncoding RNA, LOC440173, promotes the progression of esophageal squamous cell carcinoma by modulating the miR-30d-5p/HDAC9 axis and the epithelial–mesenchymal transition. Mol. Carcinog. 59, 1392–1408. https://doi.org/10.1002/mc.23264 (2020).
Xu, B. et al. LncRNA SNHG3, a potential oncogene in human cancers. Cancer Cell Int. 20, 1–11. https://doi.org/10.1186/s12935-020-01608-x (2020).
Wang, Q., Li, K. & Li, X. Knockdown of LncRNA LINC00958 inhibits the proliferation and migration of NSCLC cells by miR-204-3p/KIF2A axis. Cell Transplant. 30, 09636897211025500. https://doi.org/10.1177/0963689721102550 (2021).
Yu, W. & Dai, Y. lncRNA LOXL1-AS1 promotes liver cancer cell proliferation and migration by regulating the miR-377-3p/NFIB axis. Oncol. Lett. 22, 1–11. https://doi.org/10.3892/ol.2021.12885 (2021).
Dong, B. et al. LncRNA LINC01123 promotes malignancy of ovarian cancer by targeting hsa-miR-516b-5p/VEGFA. Genes Genomics 46, 231–239. https://doi.org/10.1007/s13258-023-01440-3 (2024).
Ouyang, L. et al. LncRNA FOXD1-AS1 regulates pancreatic cancer stem cell properties and 5-FU resistance by regulating the miR-570–3p/SPP1 axis as a ceRNA. Cancer Cell Int. 24, 4. https://doi.org/10.1186/s12935-023-03181-5 (2024).
Li, H., Jin, J., Xian, J. & Wang, W. lncRNA TPT1-AS1 knockdown inhibits liver cancer cell proliferation, migration and invasion. Mol. Med. Rep. 24, 1–8. https://doi.org/10.3892/mmr.2021.12422 (2021).
Huang, J. et al. LncRNA NBR2 regulates cancer cell stemness and predicts survival in non-small cell cancer patients by downregulating TGF-β1. Curr. Pharm. Biotechnol. 24, 1059–1069. https://doi.org/10.2174/1389201023666220728143410 (2023).
Liu, J. et al. TMEM164 is a new determinant of autophagy-dependent ferroptosis. Autophagy 19, 945–956. https://doi.org/10.1080/15548627.2022.2111635 (2023).
Li, W. et al. Selective autophagy of intracellular organelles: Recent research advances. Theranostics 11, 222. https://doi.org/10.7150/thno.49860 (2021).
Wu, Q. et al. lncRNA SNHG11 promotes gastric cancer progression by activating the Wnt/β-catenin pathway and oncogenic autophagy. Mol. Ther. 29, 1258–1278. https://doi.org/10.1016/j.ymthe.2020.10.011 (2021).
Yao, W. et al. Mec1 regulates PAS recruitment of Atg13 via direct binding with Atg13 during glucose starvation-induced autophagy. Proc. Natl. Acad. Sci. USA 120, e2215126120. https://doi.org/10.1073/pnas.2215126120 (2023).
Funding
This work was supported by the Key Project of Natural Science Foundation of Anhui Provincial Department of Education (P. R. China) (2022AH051517), the Key Project of Natural Science Foundation of Anhui Provincial Department of Education (P. R. China) (2022AH051479) and the Graduate Research Innovation Plan of Bengbu Medical College (Byycx22041).
Author information
Authors and Affiliations
Contributions
J.Y., L.G., and Z.W. provided the idea and design of this article. J.Y., L.G., Z.W., and Y.X. wrote the main manuscript text and prepared Figs. 1, 2, 3, 4, 5, 6. L.Y. reviewed the revised paper. X.J. and Q.J. were responsible for clinical specimen collection. All of the authors reviewed the manuscript and agreed to publication.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
ARRIVE statement
All authors confirm that the research complies with the ARRIVE guidelines.
Ethics approval
All of the authors declare that all of the human experiments involved in this study were approved by the Ethics Committee of Bengbu Medical College, and informed consent was obtained from all of the subjects or their legal guardians.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, J., Gao, L., Wang, Z. et al. Effects of the lncRNA NBR2 on the proliferation and autophagy of breast cancer cells under starvation conditions. Sci Rep 14, 22624 (2024). https://doi.org/10.1038/s41598-024-72181-w
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-72181-w
Keywords
This article is cited by
-
Targeting the lncRNA RBM5-AS1/GCN5 axis under fasting conditions reprograms Glycolysis and induces apoptosis in ovarian cancer cells
Molecular Biology Reports (2025)