Abstract
The decrease in sperm count and infertility is a global issue that remains unresolved. By screening environmental bacterial isolates, we have found that a novel lactic acid bacterium, Lactiplantibacillus plantarum SNI3, increased testis size, testosterone levels, sperm count, sexual activity and fertility in mice that have consumed the bacteria for four weeks. The abundance of L. plantarum in the colon microbiome was positively associated with sperm count. Fecal microbiota transplantation (FMT) from L. plantarum SNI3-dosed mice improved testicular functions in microbiome-attenuated recipient animals. To identify mediators that confer pro-reproductive effects on the host, untargeted in situ mass spectrometry metabolomics was performed on testis samples of L. plantarum SNI3-treated and control mice. Enrichment pathway analysis revealed several perturbed metabolic pathways in the testis of treated mice. Within the testis, a dipeptide, glutamyl-glutamate (GluGlu) was the most upregulated metabolite following L. plantarum SNI3 administration. To validate the pro-reproductive feature of GluGlu, systemic and local injections of the dipeptide have been performed. γ-GluGlu increased sperm count but had no effect on testosterone. These findings highlight the role of γ-GluGlu in mediating spermatogenetic effects of L. plantarum on the male mouse host and –following relevant human clinical trials- may provide future tools for treating certain forms of male infertility.
Similar content being viewed by others
Lactobacillus, mouse, testis, sperm, testosterone, in situ mass spectrometry imaging analysis.
Introduction
The gut microbiome, as a symbiotic ecosystem, synthesizes and releases a plethora of biologically active compounds (vitamins, amino acids and secondary metabolites) that are involved in microbe-microbe and microbe-host communication. Accumulating evidence suggests that the microbiome is involved in multiple diseases1,2 thus providing a putative platform for therapeutic interventions3. In addition to local actions, gut microorganisms and their metabolites might have direct and/or indirect effects on remote organs of the host, including liver4, pancreas5 heart6 and brain7,8.
Lactobacilli (Lb) are gram-positive, non-spore forming, aerotolerant/anaerobic bacteria, which are generally considered to confer beneficial metabolic adaptations in the host9,10, although they represent only a minor component of the human/mammalian gut microbiome11. Furthermore, significant anti-inflammatory and antioxidant effects of Lb have been identified12. Lactobacillus ssp. downregulate reactive oxygen species (ROS)-forming enzymes via nuclear factor erythroid 2 (NFE2)-related factor 2 (Nrf-2)-, nuclear factor kappa B (NF-κB)-and MAPK pathways13. Probiotic Lactobacilli immediately activate expression of anti-inflammatory cytokines such as interleukin (IL)-4, IL-10, IL11 and IL-13 and inhibit proinflammatoty pathways via Treg cells14. Furthermore, the anti-inflammatory effect of Lactobacilli might be related to production of short chain fatty acids, such as butyrate, which strengthens gut barrier function, lowers endotoxemia and subclinical inflammation15.
Certain Lactobacillus species promote reproductive health. For instance, in females, Lactobacilli in the vagina are important for preventing infection, clearing pathogenic microbes and modulating inflammation16. However, the reproductive impact of gut microbiome in general- and Lactobacilli in particular, on male host is much less understood. Recent recognition of the gut -testis axis highlights the impact of the gut microbiome on male reproductive organs17.
A well-balanced and diverse gut microbiome may have direct/indirect effects on male fertility18, by influencing factors like nutrient absorption, inflammation and hormonal regulation. However, the means through which certain probiotics affect the male host, remains to be fully elucidated.
Lactic acid bacteria are capable to produce numerous different metabolites (postbiotics) through which they affect host physiology. These include –but not limited to- organic acids, amino acids, dipeptides, short chain fatty acids, bacteriocins, conjugated linoleic acids, exopolysaccharides and vitamins19. Although Lactobacillus metabolites have been intensively studied20,21, Lactobacillus-induced metabolomic changes in the host remain largely unexplored.
In this report, first, we characterized the effects of a novel Lactobacillus strain referred to as Lactiplantibacillus plantarum SNI3 on gut microbiome composition, male reproduction and fertility. Fecal microbiota transplantation (FMT) was performed to confirm the role of Lactiplantibacillus plantarum SNI3 microbiome in mediating positive spermatogenic effect in the host. Then, using high resolution mass spectrometry imaging (MSI) analysis on testis samples we identified several metabolic pathways in the mouse testis, which are affected upon L. plantarum SNI3 administration. Finally, we validated a highly upregulated testicular dipeptide metabolite, γ-GluGlu in mediating L. plantarum SNI3 effect on spermatogenesis in the male mouse host.
Results
Isolation and characterization of lactiplantibacillus plantarum SNI3
In search for novel candidate probiotic Lactobacilli, we have screened a collection of more than 200 Lactobacillus strains isolated from farm- and zoo animals at Eszterházy Károly Catholic University (Eger, Hungary). In the frame of preliminary work, oligoxylan- and mannan-utilization capacity of these Lactobacillus strains was investigated and three strains, SK45, AT51 and SNI3, with the best prebiotic utilization capacity (Figure S1) and unique enzymatic profile (Figure S2), were selected for in vivo testing. Strain SK45 was isolated from the feces of Casuarius casuarius, AT51 from the feces of Aldabrachelys gigantea (Budapest Zoo) while SNI3 was isolated from feces of a breeding boar (Sus scrofa domestica, Sajóbábony, Hungary). Among the selected bacteria, we recognized a specific strain (SNI3), which was prominent in decreasing body fat content and increased testis weight in male mice (Figure S3). Based on molecular taxonomy (Table S1), this strain has been categorized as Lactiplantibacillus plantarum, strain SNI3 (Figure S4). The total sequence length of SNI3 genome was 3.057.331 bp and the GC content of 44.8%22.
In order to test a potential probiotic use of Lactiplantibacillus plantarum SNI3 acid- and bile tolerance data of the strain were obtained and shown in Figure S5.
L. Plantarum SNI3 affects body composition and reproductive physiology
First, we investigated the effect of L. plantarum SNI3 on body functions. Adult male mice were administered L. plantarum SNI3 suspension (5 × 107 CFU/mL, in autoclaved tap water) for 4 weeks. Control animals received autoclaved tap water to drink (Fig. 1). The mice tolerated the bacterial suspension, there were no changes in feces color or consistency and mice did not display any signs of gastrointestinal discomfort or sickness behavior. Body weight (BW), food consumption, fluid intake and were not different between L. plantarum SNI3-treated and control animals (Fig. 1A and Figure S6). However, lean body mass increased (t = 3.080, df = 8; p = 0.015) and fat mass decreased (t = 2.969, df = 8; p = 0.018) in treated animals compared to controls as measured by the EchoMRI™ body composition analyzer (Fig. 1B). The relative weights of the adrenal glands and thymus (indicative of chronic stress23) were not different (Figure S6). Splenomegaly, indicative of chronic immune stimulation was not detected in the treated group (Figure S6).
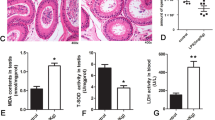
The most striking effect of L. plantarum SNI3 administration was the increased testis weight (t = 2.993, df = 35; p = 0.0050) (Fig. 1C). By contrast, the normalized epididymis weight was not different in control vs. treated animals (Fig. 1C). To reveal histomorphological changes underlying increased testis size, we compared the area of seminiferous tubules and the thickness of the seminiferous epithelium on hematoxylin-eosin stained sections of the testis from control and treated mice. Both measures were significantly higher (t = 3.221, df = 995; p = 0.0013; area) (t = 3.681, df = 456; p = 0.0003; thickness) following L. plantarum SNI3 administration, indicating increased spermatogenesis (Fig. 1D).
Next, we assessed the effect of L. plantarum SNI3 treatment on sperm count. The number of sperm collected from the cauda epididymis was significantly higher in treated animals (765.5 × 106 ± 0.60/mL) than in the controls (280.4 × 106 ± 0.26/mL) (t = 7.226 df = 35, p = 0.0001) (Fig. 1E). To detect any morphological alterations in sperms, smeared spermatozoa were stained with aniline-eosin. Immature acrosomes, crooked midpieces and hairpin tails were detected; however, the abundance of these defects was low and did not differ among experimental groups (3% in controls vs. 2,9% in treated animals). Triple fluorescence labeling (peanut lectin + phalloidin + DAPI) of sperm smears did not reveal overt morphological differences in sperm heads (acrosome) in control vs. L. plantarum SNI3 treated samples (Fig. 1E). Furthermore, computer-assisted sperm analysis (CASA) revealed a higher percentage of motile sperm in Lactobacillus treated mice (40.48 ± 2.73%) than in controls (26.59 ± 3.36%) (t = 3.238, df = 11; p = 0.0079) (Fig. 1E).
As expected from the higher sperm count and motility data of L. plantarum SNI3-treated mice, the fertilization ratio (the number of pregnant females following 5 days cohousing with experimental males) was higher in treated mice (37%) than in controls (16.6%). The reproductivity index (the total number of viable pups born per breeding pair cohoused for 5 days) was also higher in the case of L. plantarum SNI3-treated (2.5) than in the controls (1.5).
Reproductive functions are controlled by the hypothalamo-pituitary-gonad axis. To test whether L. plantarum SNI3 administration affects the synthesis of hypothalamic gonadotropin-releasing hormone (GnRH), pituitary follicle-stimulating hormone (FSH) and luteinizing-hormone (LH), we performed quantitative real-time PCR (qRT-PCR) measurements (Table S2) on hypothalamic and pituitary samples. Expression of GnRH, FSH- and LH- specific β subunits was not different between the control and treated mice (Fig. 1F) (GnRH: t = 0.057, df = 22; p = 0.954, FSHb: t = 0.445, df = 17; p = 0.661LHb: t = 0.304, df = 16; p = 0.764).
To further evaluate L. plantarum SNI3-dependent alterations in reproductive physiology, we measured serum testosterone (TESTO) levels. As shown in Fig. 1G, L. plantarum SNI3 treatment increased TESTO concentration (11.46 ± 3.40 ng/mL) compared to controls (1.46 ± 0.54 ng/mL; t = 2.751 df = 30; p = 0.014).
These results suggest that L. plantarum SNI3 acts on the periphery, rather than through the hypothalamo-pituitary axis to increase spermatogenesis and testicular hormone levels.
L. plantarum SNI3 affects body composition, increases testis weight, sperm count and testosterone level. Top left: Schematics of the experimental groups and design. (A) and (B). Body weight (BW) and body composition of mice in each group. Mean ± Standard Error of Mean (SEM). (C) Testis weight and epididymis weight of mice drinking autoclaved tap water (control) or L. plantarum SNI3 for 4 weeks. Mean ± SEM values. (D) Area of seminiferous tubuli and thickness of seminiferous epithelium in control and treated mice. Mean ± SEM values. Below: representative hematoxylin-eosin stained sections from the testes of control and L. plantarum SNI3-treated mice. (E) Sperm count in control and L. plantarum SNI3-treated mice as determined in Makler chamber; Percentage of motile sperms in control and treated groups assessed by CASA (computer-assisted sperm analysis) system. Fluorescent labeling of individual sperms obtained from control and L. plantarum SNI3-treated mice. Peanut lectin-FITC (green , acrosome) , phalloidin-Alexafluor595 (red , actin cytoskeleton) and DAPI (blue , DNA) labeling. (F) Expression of Gonadotropin-releasing hormone (GnRH) Follicle-stimulating hormone (FSH)- and Luteinizing hormone (LH)- β subunits in control and L. plantarum SNI3-treated mice. Mean ± SEM values. (G) Mean ± SEM of serum testosterone (TESTO) levels in control and L. plantarum SNI3-treated mice. On all graphs: *p < 0.05; ** p < 0.01; ****p < 0.0001 using unpaired , two-tailed Student t-test.
L. Plantarum SNI3 administration impacts male sexual behavior
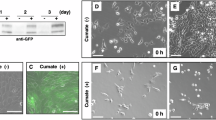
Because TESTO facilitates male sexual activity and performance, we next assessed the sexual behavior of L. plantarum SNI3-treated and control mice paired with estrus females. The usual sequence of the male mouse sexual behavior consists of (1) following the female, (2) anogenital sniffing, (3) mounting, (4) intromissions (5) ejaculation and (6) post-ejaculation grooming24. We found a significantly increased frequency and duration of nose-to-nose contacts, anogenital sniffing and mounting in L. plantarum SNI3-treated mice compared to control (Fig. 2A-C). There was no significant difference between the treated animals and controls in other sexual behavior elements.
L. plantarum SNI3 administration impacts sexual behavior. Top: Schematics of the experimental groups and design. (A) Color-coded ethograms of control and L. plantarum SNI3-treated mice. Quantification of sexual behavioral activity of control and L. plantarum SNI3-treated male mice. Six distinct behaviors that belong to sexual interaction sequence were analyzed. Each row represents one mouse. (B) and (C) Summary graphs showing frequency (B) and duration (C) of selected sexual behavior elements in control and SNI3-treated male mice. Mean ± SEM values. Unpaired t-test between control and SNI3- treated animals. *p < 0.05; **p < 0.01.
Gut microbiome is affected by L. Plantarum SNI3 administration
Next-generation sequencing (NGS) analysis of V3V4 16 S rRNA on proximal colon content samples revealed that L. plantarum SNI3 administration did not significantly affect the composition of the colon microbiome at the phylum level (Fig. 3A). The total number of OTUs was not significantly higher in L. plantarum SNI3-treated animals. Alpha diversity, based on observed OTUs, Shannon, Simpson and Chao indices was not significantly different in control vs. treated mice (Figure S7A). Although Firmicutes (F) increased and Bacteriodetes (B) decreased in treated animals (Fig. 3A), F/B ratios (1.72 ± 0.40 in controls vs. 2.42 ± 0.21 in treated animals) were not statistically different. The top 20 taxa at the genus level belong to Akkermansia, Lachnospiraceae NK4A136 group, Lactiplantibacillus, Prevotellaceae NK3B31 and UCG001 groups. In the microbiome of L. plantarum SNI3 (L. plantarum SNI3) treated animals the abundance of Lactiplantibacillus and Lachnospiraceae NK4A136 group increased and Akkermasia significantly decreased (Fig. 3A). At family level, abundance of Lactobacillaceae and Monoglobaceae was higher, while Akkermansiaceae and Eubacterium coprostanoligenes group was significantly lower in L. plantarum SNI3-treated mice compared to controls (Figure S7B). Lactiplantibacillus genus was not detected in control samples, while its abundance was 4.6% in SNI3-treated animals. Using L. plantarum SNI3- specific primers (Table S2) in PCR reaction on total DNA isolated from colon content samples, we found that 10 ng stool DNA contained 4.84 × 10−6 ng L. plantarum SNI3 specific DNA (R2-0.997, efficiency-90.6%) in the samples obtained from L. plantarum SNI3 treated mice. The calculated copy number was 463.3 (SD 317.7) in 1 ng stool DNA. Bacterial DNA isolated from control samples did not give any PCR product with L.plantarum SNI3- specific primers.
The abundance of L. plantarum and Lachnospiraceae NK4A136 group in colon content samples was positively correlated with sperm number (L. plantarumr = 0.5914, p = 0.0259; Lachnospiraceae NK4A136 r = 0.7735, p = 0.0012), while a negative correlation was seen between Akkermansia abundance in the colon microbiome and sperm number (r= -0.6609, p = 0.0101) (Fig. 3B).
Fecal microbiota transplantation from L. Plantarum SNI3 treated mice improves spermatogenesis in recipient animals
Next, we used fecal microbiota transfer (FMT) to investigate whether positive reproductive changes seen after L. plantarum SNI3 treatment is mediated by gut microbiome. Fecal microbiota, isolated from L. plantarum SNI3-treated donor mice was introduced to normal mice in which the microbiome was attenuated by antibiotic mixture. As shown in Fig. 3C, fecal microbiota isolated from L. plantarum SNI3 treated animals increased the sperm number [(F 2, 19) = 5.500, p = 0.0067] by 42% in antibiotic-treated recipient mice. FMT from L. plantarum SNI3 mice resulted in slightly but significantly increased testosterone levels [(F 2, 21) = 5.500, p = 0.0064]. By contrast, FMT from control animals did not affect either reproductive marker.
L. plantarum SNI3 affects colon microbiome. (A) Abundance of main phyla and top 20 taxa at genus level in the colon microbiome. Mean ± SEM values **** p < 0.0001 vs. control group. (B) Correlation between the abundance of different microbial populations in the colon and sperm count. Scatter plots showing the relationship between microbial population abundance and sperm number. Each point represents one individual sample (n = 14). (C) Impact of fecal material transplantation (FMT) from L.plantarum SNI3 treated mice on total bacterial DNA concentration in colon content, sperm count and serum testosterone level. No FMT: mice received antibiotic treatment and no FMT (n = 5), Control FMT: mice received antibiotic treatment and fecal microbiome from control, untreated animals (n = 6), SNI3 FMT: mice received antibiotic treatment and fecal microbiome from L. plantarum SNI3 treated mice (n = 13). All microbiome data from two independent cohorts, # FMT data were analyzed by one-way ANOVA followed by Dunnett’s multiple comparison test (n = 6–13 per group). Mean ± SEM values *p < 0.05, **p < 0.01.
L. Plantarum SNI3 administration affects the metabolome of testis
To explore metabolomic effects of L. plantarum SNI3 on male mice, in situ mass spectrometry imaging analysis (MSI) was performed on testis samples. Figure 4A shows testis sections (stained with hematoxylin-eosin) from control and L. plantarum SNI3-treated animals, which were included in the in situ mass spectrometry imaging. We performed hierarchical clustering analysis, component analysis by least squares discriminant analysis (sPLSDA) and ROC analysis for discriminant metabolic comparison of L. plantarum SNI3-treated and control groups.
Hierarchical clustering analysis illustrates a distinct separation between the control and L. plantarum SNI3-treated groups in the testes (Fig. 4B). Employing the sPLSDA algorithm, the data is depicted on a 2D score plot, with the x-axis representing component 1 and the y-axis representing component 2. This analysis reveals clear distinctions between the control and L. plantarum SNI3-treated groups (Fig. 4C).
ROC curves allow one to see how the diagnostic performance varies along all possible threshold values. The area under the ROC curve (AUC) value is a robust measure for comparing the performance of different biomarker models25. Here, multivariate ROC curve exploration was performed to describe the trade-off between sensitivity and specificity with regard to biomarker performance. The processes of feature selection, model building and performance evaluation are performed multiple times by Monte-Carlo cross validation (MCCV) through balanced subsampling coupled with Random Forest (RF) algorithm. Figure 4D “ROC view” provides an overview comparing the ROC curves for all models created by MetaboAnalyst (Model 1–6 with 5, 10, 15, 25, 50 and 100 features respectively). Testis samples give high performance with AUC values > 0.965.
Hierarchical clustering analysis, sPLSDA analysis and ROC analysis for discriminant metabolic comparison. (A) Hematoxylin-eosin staining of testis samples from control and L. plantarum SNI3-treated mice. Each section represents an individual mouse. (B) Hierarchical clustering analysis demonstrating different metabolic profiles of L. plantarumSNI3-treated and control groups on testis samples. Colored cells correspond to intensity values of metabolites. Euclidean distance and Ward method were applied for cluster analysis. (C) Partial least squares discriminant analysis (sPLSDA) for discriminant metabolic comparison ofL. plantarumSNI3- treated and control groups. (D) An overview of ROC curves from different biomarker models using different numbers of features. ROC nalysis was performed to evaluate the discerning performances based on metabolic profiles. “ROC view” a provides an overview comparing the ROC curves for all models created by MetaboAnalyst (Model 1–6 with 5, 10, 15, 25, 50 and 100 features respectively). Testis samples give the performance with AUC values > 0.965.
Identification of L. Plantarum SNI3 affected metabolic pathways
A correlation network, illustrated in Fig. 5, has been built on the metabolite dataset obtained from untargeted in situ mass spectrometry imaging (MSI) to reveal functionally interconnected metabolites related to L. plantarum SNI3 treatment in testis. Several perturbed pathways from the enrichment pathway analysis are highlighted in the correlation network. Our results indicate that glucose, galactose, fructose, palmitoyl carnitine, phosphoenol pyruvate, succinate, fumarate and other metabolites, which can take part in central metabolic pathways, were down regulated in L. plantarum SNI3 treated animals, however, this process left the ATP level unchanged in the testis. Arachidonic acid metabolism, linoleic acid metabolism and glycerophospholipid metabolism were also found to be downregulated as response to L. plantarum SNI3 treatment. By contrast, fatty acids, like palmitic acid, palmitoleic acid, stearic acid and oleic acid were upregulated in L. plantarum SNI3 treated mice compared to controls. It is worth noting that palmitic glucuronide was one of the highly upregulated metabolites, whose function is unknown, but glucuronidated compounds generally are destined for secretion from the body. A couple of non-proteogenic amino acids (i.e. dimethylglycine, DMG and gamma aminobutyric acid, GABA) and peptides, such as glutamyl peptides (GluGlu and GluGln) and oxidized glutathione (GSSG) were also upregulated in the testis of L. plantarum SNI3 treated animals.
Metabolic correlation network of the testis in L. plantarum SNI3 treatment compared with control samples. Nodes represent metabolites , with size reflecting the log2 fold change between control and L. plantarum SNI3 treatment in testis samples. Node sizes increase with the log2 fold change , while the criteria for labelling nodes are a log2 fold change cutoff value greater than 0 for upregulation (green color) and less than 0 for downregulation (red color) , respectively. Edges represent shared KEGG pathways. Selected pathways are colored as follows: arachidonic acid metabolism (yellow), linoleic acid metabolism (pink), glycerophospholipid metabolism (blue), fatty acid metabolism (green), protein digestion and amino acid metabolism (orange). Metabolites in green and red squares highlight the most upregulated (Glutamyl glutamate) and the most downregulated metabolite (Phosphatidyl choline , PC(22:6(4Z , 7Z , 10Z , 13Z , 16Z , 19Z)/16:0 , respectively.
L. Plantarum SNI3 administration upregulates glutamyl-glutamate (GluGlu) in the testis
Volcano plots (Fig. 6A) and pathway enrichment analysis (Fig. 6B) were performed to further investigate the enriched metabolic pathways contributing to L plantarum SNI3 effect in the testis. The discriminative metabolites were selected using a volcano plot with fold change > 2 or < 0.5 and p-value < 0.05 and then subjected to metabolite annotation (HMDB database) and pathway analysis (KEEGG pathway). Figure 6B summarizes the discriminative pathways in the testis samples in response to L. plantarum SNI3 treatment. Pentose and glucuronate interconversions and dipeptides involved in amino acid degradation are upregulated pathways in L. plantarum SNI3-treated testis. In contrast, amino acid, carbohydrate and linoleic acid metabolism were downregulated upon L. plantarum SNI3 treatment.
Figure 6C shows MSI images of select down- (phosphatidylcholine) and upregulated (glutamyl-glutamate, GluGlu) metabolites in the testis. As shown on the image, MSI signal corresponding to GluGlu seems to be localized to the seminiferous tubules. Because GluGlu was the most upregulated metabolite in the testis of L. plantarum SNI3 treated mice, next, we identified the isomer form of the compound and then validated its’ physiological role in testicular function.
Identification of differential metabolites and pathways in control vs.L. plantarum SNI3-treated testes. (A) The volcano plot is a combination of fold change (FC) and t-tests. The x-axis is log (FC). Y-axis is log10 (p value) based on FDR adjusted p values. The discriminative metabolites were selected by a volcano plot with fold change > 2 or < 0.5 and p-value < 0.05. (B) Pathway enrichment analyses were performed by KEGG pathway analysis. Discriminative pathways are summarized here. (C) In situ mass spectrometry (MSI) images of select discriminative metabolites. On the left, MSI image of phosphatidylcholine , which is reduced upon L. plantarum SNI3 treatment; on the right, MSI image of glutamyl-glutamate, GluGlu, which is enhanced upon L. plantarum SNI3 treatment in the testis. Unpaired t-test was used for statistical analysis (*: p ≤ 0.05; ***: p ≤ 0.001).
To confirm the identity of GluGlu in the testes, LC-MS/MS analysis was performed using HILIC separation (Table S3). This method can separate the two isomers, α-GluGlu and γ-GluGlu, which were confirmed by chemical reference standards. Two peaks at m/z 275.088474 were detected, corresponding to the [M-H]- adduct of C10H16N2O7. Extracted ion chromatograms for GluGlu indicate that both α-GluGlu and γ-GluGlu were detected in testis tissue samples, but γ-GluGlu showed eight times higher intensity than α-GluGlu (Figure S7). Furthermore, in L. plantarum SNI3-treated testis, the ratio γ-GluGlu/ α-GluGlu was increased (Figure S9).
Since lactic acid bacteria are capable of synthesizing γ-glutamyl dipeptides26, we next checked whether L. plantarum SNI3 produces γ-GluGlu. HPLC analysis of the danzylated peptide revealed that the bacteria contained 188 pmol γ-GluGlu/mg of protein. γ-GluGlu was released into the culture medium which contained 555.92 nmol/mL after 24 h of culturing.
HPLC analysis also confirmed the enrichment of γ-GluGlu in the testes of mice treated with L. plantarum SNI3 for four weeks. The testicular γ-GluGlu content (1304 ± 107.7 pmol/mg protein in controls vs. 1794 ± 186.97 pmol/mg in SNI3-treated t = 2.27, df = 3.19; p = 0.051) showed a positive correlation with the abundance of Lactiplantibacilli in the colon microbiome. Furthermore, we observed a positive correlation between testicular γ-GluGlu content and sperm number (Figure S10).
γ-GluGlu administration affects sperm number in mice
Next, we investigated whether γ-GluGlu, the metabolite that has been significantly elevated in the testes of L. plantarum SNI3-treated mice, may recapitulate the effect of bacterial administration on testicular and hormonal changes. Oral administration of γ-GluGlu had no effect, whereas intraperitoneal injection of the dipeptide for 4 weeks resulted in a dose-dependent increase in sperm number (one-way ANOVA, F(5, 36) = 3.869; p = 0.006). Furthermore, direct intratesticular injection of γ-GluGlu in the dose range of 0,1–1 ng significantly increased the number of sperms isolated from the cauda epididymis compared to that of vehicle (PBS)-injected controls (one-way ANOVA, F(3, 11) = 6.973; p = 0.007) (Fig. 7). Systemic and local γ-GluGlu, however, did not significantly affect testosterone concentration (Figure S11). Following intraperitoneal injection of 10 and 50 mg/kg dose γ -GluGlu an increase of relative testis weight was detected.
Systemic and intratesticular γ-GluGlu increases spermatogenesis. C57Bl6 male mice received γ-GluGlu orally , intraperitoneally and locally (intratesticularly). Spermatozoa from the cauda epididymis were isolated and counted. Bar graphs showing the sperm number of mice injected with PBS or different doses of γ-GluGlu. Mean ± SEM values. One way ANOVA , *p < 0.05; **p < 0.01.
Discussion
Understanding the mechanisms through which microbiome in general-, and probiotic microorganisms in particular, affect host physiology and behavior, is essential to exploit trans-kingdom interactions.
Our present research has uncovered a previously unknown Lactobacillus strain, Lactiplantibacillus plantarum SNI3, which has been found to have beneficial effects on male reproduction in a normal, healthy mouse model. Treatment with this strain resulted in increased testis weight, sperm number, sperm motility, testosterone levels and improvements in sexual behavior, ultimately leading to advanced male fertility. In this study, we also provide evidence that a specific dipeptide metabolite, γ-GluGlu mediated the increase in sperm number in male mice treated with L. plantarum SNI3.
The impact of Lactobacilli on female reproduction has been well established, however, the influence of probiotics on male fertility received less attention. A previous study found that a Lactobacillus reuteri strain was able to prevent gonadal ageing in obese male mice27. Additionally, other Lactobacillus strains, L. rhamnosus NCDC-610 and L. fermentum NCDC-400, have been shown to reduce chronic stress-induced reproductive deficits in C57Bl6 male mice28. A probiotic composition containing four Lactobacillus strains, two Bifidobacteria and Streptococcus thermophiles also increased sperm number in a clinical trial on human patients with idiopathic oligoasthenoteratozoospermia29. However, the mechanisms by which Lactobacilli promote testicular function and spermatogenesis remain unclear.
One possible explanation of the positive effect of L. plantarum SNI3 on sperm production would be direct colonization of Lactobacillus into testis. The notion that testes are sterile has been challenged by recent studies, which have uncovered a small community of microorganisms present in the human testes30 or testicular sperm samples31. Following rigorous decontamination steps, ten different genera were identified in human cultured testis samples31. However, we could not confirm colonization /or appearance of L. plantarum SNI3 in the testis after oral administration. By contrast, L. plantarum SNI3 was detected by PCR in the colon content of treated mice, indicating successful colonization in the gut. Thus, it is very likely that beneficial effects of L. plantarum SNI3 on male reproductive function are mediated through the gut-testis axis32,33.
The administration of L. plantarum SNI3 lasted for four weeks, encompassing almost a complete spermatogenesis cycle in mice (34,5 days)34. The abundance of L. plantarum was found to be positively correlated with sperm number. Fecal microbiome transplantation (FMT) from L. plantarum SNI3 donor mice improved the spermatogenesis in antibiotic-treated recipient animals. These findings support the relationship between gut microbiome, L. plantarum SNI3 and spermatogenesis.
It should be noted that L. plantarum SNI3 administration reshaped the gut microbiome. Abundance of Akkermansia decreased, whereas that of Lachnospiraceae NK4A136 group significantly increased after treatment. Lachnospiraceae NK4A136 group produces butyrate, a beneficial short chain fatty acid (SCFA)35, which improves gut barrier integrity, impacts the redox balance and have positive effect on sperm production36. Increased abundance of Lachnospiraceae NK4A136 group following L. plantarum SNI3 administration might be explained by crossfeeding between these bacteria37.
Although Akkermansia muciniphila has been associated with a healthier metabolic status and proven efficacy to improve obesity, diabetes and hepatic steatosis38,39, there are no reports on the relationship between A. muciniphila and reproductive function in healthy male subjects.
L. plantarum SNI3 administration increased the sperm motility as well. As the sperms acquire motile ability in the epididymis, further studies are required to reveal the potential impact of Lactobacilli on epididymal sperm maturation and antioxidative defense.
In search for specific metabolites that mediate L. plantarum SNI3 effect on male reproductive functions, present, non-targeted metabolomics has revealed that a total of 273 metabolites, which exhibit differential expression in the testes of mice treated with L. plantarum SNI3. Pentose glucuronate interconversion and dipeptide metabolism pathways were significantly upregulated, whereas the amino acid-, carbohydrate- and linoleic acid metabolism were the most downregulated pathways in the testes. It has been observed that a greater number metabolites were downregulated than upregulated.
One of our interesting observation on testicular metabolome was that several non-proteogenic amino acids and peptides (dimethylglycine, GABA, γ-GluGlu, γ-GluGln and GSSG) - all of them having possible regulatory function by binding to protein receptors - were upregulated.
The most upregulated metabolite in the testes of L. plantarum SNI3 treated animals was dipeptide glutamy-glutamate (GluGlu). GluGlu was present in α and γ form. The α form has peptide bond, while the γ form is synthesized by the transfer of glutamine to the carboxyl group at the gamma (γ) position of glutamic acid. Our LC-MS/MS analysis identified that the gamma form of GluGlu dominates the alpha form in L. plantarum SNI3-treated mouse testis. Previous studies have shown that some lactic acid bacteria are capable of producing various dipeptides, including γ-GluGlu26. In bacteria, two enzymes are responsible for γ-GluGlu synthesis: γ-glutamyl transferase (GGT) and γ-glutamyl-cysteine ligase (GCL)40. The impact of GGT on male reproductive functions is supported by the fact that GGT KO mice display reduced testis size, suppressed follicle- stimulating hormone (FSH) and are infertile41. Blast analysis of the L. plantarum SNI3 genome22 has revealed the presence of GCL protein (SNI3_14110) supporting the hypothesis of bacterial production of γ-GluGlu. HPLC analysis of L. plantarum SNI3 and its supernatant medium confirmed γ-GluGlu synthesis. With MSI imaging, GluGlu signal is seen in the seminiferous tubules of the testis however, the mechanisms how the dipeptide accumulates in the testicular tissue remains to be investigated.
γ-GluGlu may act locally within the testis, as local injection of this dipeptide reproduced the effect of orally administered L. plantarum SNI3 on sperm number. Oral administration of γ-GluGlu was not effective, probably because of the digestion of the dipeptide by enzymes of the gastrointestinal tract. Intraperitoneal application of the dipeptide resulted in a bell-shaped dose-response curve, with high dose (100 mg/kg) that was ineffective in stimulating sperm number.
Within the host testis, γ-GluGlu may enter into the γ-glutamyl cycle, which participates in amino acid transport across the cell membranes. γ-GluGlu through the glutamyl cycle facilitates the transport and availability of amino acids required for glutathione biosynthesis thus boosts antioxidant capacity and aid sperm production42. Indeed, protection of spermatozoa from oxidative damage is crucial for their viability and function29. In addition to the testis, antioxidant mechanisms are also important in the epididymis, where they support sperm maturation and motility.
The fact that intratesticular/intraperitoneal γ-GluGlu administration increased sperm number and testis weight, but not testosterone plasma concentration, suggests that the effect of L. plantarum SNI3 on male reproduction is complex and that hormonal changes are mediated by distinct mechanisms and may involve other metabolic pathways and metabolites. Neither hypothalamic GnRH nor pituitary FSH and LH mRNA levels changed after L. plantarum SNI3 administration, suggesting that peripheral rather than central regulatory mechanisms are involved in establishment of elevated TESTO concentration. For example, a combination of probiotic Lactobacilli and fructooligosaccharide upregulated the expression of steroidogenic acute regulatory protein (StAR), cytochrome P450 side-chain cleavage enzyme (P450scc) and 17β-hydroxysteroid dehydrogenase (17βHSD) in the testes, resulting in elevated testosterone levels in male mice28. In contrast, L. plantarum SNI3 administration did not affect host testicular expression of steroidogenic enzymes in our experiment (data not shown).
Additionally, certain intestinal bacteria can “recycle” inactive steroid glucuronates/sulphates into active hormones, which significantly contributes to steroid hormone action43,44,45. It should be noted however that Lactobacilli in general46 and L. plantarum SNI3 in particular, display low glucuronidase activity, therefore, other bacterial species of the host microbiome with high glucuronidase/sulphatase activity may be involved in elevation of host TESTO levels. Indeed, bacterial community isolated from the feces of L. plantarum SNI3-treated mice display strong β-glucuronidase activity (Figure S10). Further studies are necessary to reveal the precise peripheral molecular mechanisms that mediate the increased TESTO levels in L. plantarum SNI3-treated animals.
In conclusion, our present data highlight metabolic pathways through which the novel strain, L. plantarum SNI3 advances male host hormonal and reproductive functions and identifies γ-GluGlu as one of the metabolites, which may mediate these effects through the gut-testis axis. Additionally, our results strongly proved the concept of using “high throughput metabolome” data analysis of host animals in probiotic research, providing molecular background for health promoting effect of various bacterial strains.
Our results have a translational value, because in humans, one out of the six couples has difficulty conceiving. Half of these cases are caused by male infertility. Alarming declines in sperm count and quality have been reported in a comprehensive meta-regression analysis of men living in North America, Europe and Australia between 1973 and 201147. A recent (2022) examination of sperm counts revealed an accelerated pace of decline globally48. Although our recent findings were obtained on healthy male mice, present results, after successful clinical trials, can be exploited for future microbiome-based metabolite therapies of human male subfertility.
Limitation of the study
Our study was done on healthy male mice, even in these animals, L. plantarum SNI3 was able to increase sperm number. Clinical experiments are needed to show if our results are transferable to human with disrupted spermatogenesis.
Materials and methods
Lactiplantibacillus plantarum SNI3
L. plantarum SNI3, used in these experiments, was isolated from feces of a breeding boar in Sajóbábony, Hungary. The strain has been deposited in National Collection of Agricultural and Industrial Microorganisms, Institute of Food Science and Technology, Hungarian University of Agriculture and Life Sciences (NCAIM (P) B 001482).
L. plantarum SNI3 cultures were grown in sterile deMan, Rogosa and Sharpe (MRS) broth (VWR Chemicals) anaerobically at 37 oC for two days to reach bacterial density of 108 CFU/mL. Cultures (50mL) were then centrifuged, the pellet was washed 2x in autoclaved tap water and re-suspended in 100 mL autoclaved tap water.
Animal experiments
Experiments in this study were approved by the Animal Care and Use Committee of the Institute of Experimental Medicine, Hungarian Research Network (permit number: PEI/001/29 − 4/2013). All procedures were complied with the ARRIVE guidelines and performed in accordance with the guidelines of European Communities Council Directive (86/609 EEC), EU Directive (2010/63/EU) and the Hungarian Act of Animal Care and Experimentation (1998; XXVIII, Sect. 243/1998).
Adult (age 150–200 days) male C57BL/6J mice were used. Animals were bred at the Specific Pathogen Free (SPF) level in the animal facility of the Institute of Experimental Medicine, Budapest Hungary. Experiments were carried out at the Minimal Disease (MD) level of the same facility under standard controlled conditions with 12 h light/dark cycle (lights on from 6 a.m. to 6 p.m.) at 21–22 °C and 65% humidity. Animals received standard pelleted rodent chow (VRF1, special Diets Services (SDS), Witham, Essex, UK) containing 19.1 g% protein, 55.3 g% carbohydrate and 4.8 g% fat. Chow was provided ad libitum. Mice were randomly assigned to experimental groups. L. plantarum SNI3-treated mice (n = 20) received bacterial suspension as the sole drinking solution for four weeks. Based on the fluid consumption data (10 mL/day/animal), the average L. plantarum SNI3 consumption was ~ 5 × 108 CFU/mouse/day. Control animals (n = 17) drank autoclaved tap water. Fluid and chow consumption were measured daily, during the whole 4 weeks’ treatment. All drinking bottles were changed daily at the beginning of the dark (active) phase.
Body weight was measured weekly and at the end of the experiment. Whole body composition of the mice was measured before starting L. plantarum SNI3 treatment and at the end of the experiment using EchoMRI™ (Whole body composition analyzer; E26-233RM, Echo Medical Systems, Houston, TX, USA).
For testing the sexual behavior, female C57BL/6J mice were used.
Test of sexual behavior
A separate cohort of mice (n = 17) was used in this experiment. Estrus cycle of C57BL/6J female mice (age between 60 and 90 days) was followed by daily vaginal lavage and microscopic analysis of the smears for at least 2 consecutive cycles. Only females displaying normal 4 days’ cycles were included in the study. Both experimental males and females were separately habituated to a rectangular open field box (40 cm x 40 cm x 40 cm) for 30 min at two consecutive days prior to experiment. In the test, male mouse was placed first to the arena, left freely exploring for 5 min then estrus female was introduced. All tests were performed in the first hour of the dark phase of the circadian cycle. Sexual interactions were videotaped for 30 min with a Noldus Media recorder camera. Duration and frequency of behavioral elements were analyzed by Solomon coder.
To establish fertilization ratio and reproductivity index, randomly selected female mice were cohoused with experimental males for 5 consecutive days. The number of pregnant females was counted on day 15 and then the number of pups was also counted on the day of delivery.
Fecal microbiota transplantation
To establish if gut microbiome mediates pro-reproductive effects of L. plantarum SNI3, fecal microbiota transplantation (FMT) was done in a separate cohort of adult male mice. The FMT procedure was done as described by49. For donor stool preparation ~ 150 mg fecal pellet form control or L. plantarum SNI3-treated mice was collected into sterile Eppendorf tubes and soaked into 1 ml ice cold saline, homogenized via vortexing for 1 min and centrifuged (500x g, 5 min) to remove any particulate matter Then the bacterial suspension was washed twice in saline and the concentration was set to ~ 109 CFU/mL by measuring the OD of the solution at 600 nm.
Recipient mice were treated with an antibiotic mixture (ampicillin, 1 g/L; gentamycin, 0.5 g/L; vancomycin, 0.5 g/L; metronidazole, 1 g/L) through oral gavage (250 µl/animal) for three consecutive days. 24 h after the last antibiotic treatment mice were transplanted with donor bacterial samples via oral gavage (200 µl/animal, 5 × 108 CFU/animal. Recipient animals were given donor bacterial solution every day for 5 days, then once weekly until decapitation 36 days later. Recipient mice were divided into two groups: one group was transplanted with microbiota from control animals (Control FMT, n = 6), the other group received microbiota from L. plantarum SNI3 treated mice (SNI3 FMT, n = 13). An additional group of mice received antibiotic pretreatment, but did not receive any microbiome (No FMT, n = 5).
γ-GluGlu administration
Separate cohort of adult male C57BL/6 mice (n = 78) were used for testing the effects of γ-GluGlu. γ-GluGlu (Acros Organics) was either orally administered (n = 15) or intraperitoneally injected (n = 41) or directly given into the testis (n = 22). For oral administration, γ-GluGlu (1 mg/kg bw) was delivered via oral gavage (200 µL/ mouse), daily, for 4 weeks. Controls received 200 µL autoclaved phosphate buffered saline (PBS) via gavage. For intraperitoneal injection, γ-GluGlu was dissolved in PBS and used in 1 mg/kg – to 100 mg/kg dose range. 300 µL/mouse γ-GluGlu solution or PBS was injected daily. For intratesticular injections, animals were anesthetized with ketamine/xylazine cocktail (16.6 mg/mL ketamine and 0.6 mg/mL xylazine-hydrochloride in 0.9% NaCl, 10 ml/kg body weight i.p.). A midline incision was made through which testes were exposed and placed on moisturized sterile gauze and injected with γ-GluGlu, 100pg-10 ng/25µL in PBS) or with PBS vehicle (25µL/testis) using a Hamilton syringe. The abdominal muscles were sutured with absorbable yarn and the skin was closed with surgical clamps. Operated animals were checked daily for signs of behavioral discomfort (changes in home cage locomotor activity, grooming, writhe, appearance of fur50. Intratesticularly injected mice were sacrificed one week after injection, blood was collected for testosterone measurement and sperms were collected from the cauda epididymis and counted as described above.
Decapitation, autopsy and tissue collection
Animals were decapitated and trunk blood was collected, centrifuged and serum samples were stored at -20oC until hormone assay. Caudal part of the epididymis was dissected for sperm collection. Testes were collected, measured and frozen in liquid N2 for 30 min then kept at -70oC until MALDI-MSI analysis. Colon content (approx. 200 mg) from the proximal part was collected, frozen in dry ice then kept at -70oC until assay.
Thymus, spleen and adrenals were also dissected, measured and normalized organ weights were calculated as organ weight in mg/g body weight.
Brains and the whole pituitaries were also collected. Hypothalamus was dissected along the optic chiasm rostrally, the mammillary bodies caudally and the optic tract laterally. Tissues were collected on dry ice and stored at -70oC.
Serum hormone measurement
Serum testosterone was measured by ELISA (NovaTec, Immundiagnostica GmBH, Dietzenbach, Germany) according to the manufacturer’s instruction.
Sperm counting and staining
Groups of control and L. plantarum SNI3-treated mice were sacrificed for spermatozoa preparation. Scrotum was incised, left cauda of epididymis expelled and removed into 100 µl Card & Fertiup medium (CosmoBio Co, USA) minced to free spermatozoa at 37oC for 5 min. Solution was diluted 1:5 with saline and the number of spermatozoa was counted in Makler chamber and on smears made on defatted, uncoated slides51. Additional smears were stained with aniline-eosin (5% aniline and 0.5% eosin in 4% acetic acid). Slides were scanned with Pannoramic® MIDI II Slide Scanner. Images were analyzed with Caseviewer 2.3 software. Counting and quality analysis were performed by two independent investigators, who were blinded to treatments.
Sperm smears were briefly fixed with 4% paraformaldehyde, permeabilized with 0.1% TritonX 100 then incubated with AlexaFluor568-Phalloidin (1.200, ThermoFisher) and PNA (peanut agglutinin)–FITC (1:400, Sigma-Aldrich) and counterstained with DAPI-Fluoromount-G (1:1000, Southern Biotech). Smears were scanned with Pannoramic® MIDI II Slide Scanner. Images were analyzed with Caseviewer 2.3 software.
For computer assisted sperm analysis (CASA Microptic SCA), samples were prepared as above and transferred into pre-warmed counting chamber, where twenty images from each sample were obtained for evaluation of sperm count, motility and velocity.
Gene expression analysis
Frozen hypothalamic and pituitary tissue samples (n = 20) were homogenized in TRI reagent. Then, total mRNA was isolated from the homogenate using a Total mRNA Mini Kit (Geneaid) according to the manufacturer’s instructions. To eliminate genomic DNA contamination, DNase I (Fermentas) treatment was used. Sample quality control and the quantitative analysis were carried out by NanoDrop (Thermo Scientific). cDNA synthesis was performed with High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Real-Time PCR was carried out in ABI StepOnePlus instrument (Applied Biosystems) with Fast EvaGreen quantitative PCR master mix (Biotium) and gene-specific primers. Primers (Microsynth) were designed in our laboratory using Primer-BLAST software of the National Center for Biotechnology Information (NCBI). Forward and reverse primers used to quantify mRNA are listed in Table S2. Gene expression was analyzed by the 2-ΔΔCT method using the ABI StepOne Software v2.3 (Applied Biosystems). The amplicons were tested by melt curve analysis on ABI StepOnePlus instrument (Applied Biosystems). Relative changes in gene expression were normalized against GAPDH mRNA expression.
PCR measurement of lactiplantibacillus plantarum SNI3 from colon content samples
Total DNA was extracted from 220 mg of colon content using the QIAamp Fast DNA Stool Mini Kit. (Qiagen), following the manufacturer’s protocol. The QIAamp DNA Stool Mini Kit provides silica membrane-based purification of up to 30 µg genomic, bacterial, viral and parasite DNA from fresh or frozen human stool or other sample types with high concentrations of PCR inhibitors. The combined action of InhibitEX, a unique adsorption resin and an optimized buffer leads to removal of PCR inhibitors. The concentration and quality of the extracted genomic DNA were assessed using a NanoDrop spectrophotometer (Thermo Fisher). The sequence of L.plantarum SNI3- specific primers is listed in Table S2.
Statistical analysis of physiological/behavioral measurements
Data are expressed as mean ± SEM and analyzed by two-tailed, unpaired Student’s t-test (control vs. L. plantarum SNI3-treated) using GraphPad Prism software (version 9.4.0; San Diego, CA, USA). P-value < 0.05 was considered statistically significant. For FMT results and sperm counts after intraperitoneal and intratesticular γ-GluGlu injections, one-way ANOVA with Dunnett’s followed by multiple comparisons was performed.
Microbiome analysis
For microbiome analysis, DNA was extracted from the colon contents using NucleoMag DNA Microbiome kit (Machery-Nagel, Germany).
Library construction for Illumina MiSeq sequencing of the 16S rRNA amplicons. Standard library preparation was performed according to Illumina (San Diego, California, United States) 16S Metagenomic Sequencing Library Preparation protocol (15044223 Rev. B). The V3 and V4 hypervariable regions of bacterial 16S rRNA gene were sequenced with Illumina MiSeq benchtop sequencer generating amplicons of ~ 460 by using the universal primer set: 341F-5’ CCTACGGGNGGCWGCAG 3’and 785R-5’ GACTACHVGGGTATCTAATCC 3’ primers flanked by Illumina overhang adapter sequences (forward overhang: 5’ TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG 3’, reverse overhang: 5’ GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG 3’). After completion of the amplicon PCR with 2 x KAPA HiFi HotStart ReadyMix and dual indexing of the 96 with adaptor sequences (i7-N7xx-12 items, i5-S5xx-8 items) using the Illumina Nextera XT Index Kit (FC-131-1001/2). PCR cleanups and amplicon size selections were carried out with KAPA Pure Beads (KAPA Biosystems) based on the technical data sheet (KR1245 – v3.16) of the manufacturer resulting in final ~ 580–630 bp libraries. Every time, verifications were done with PCR Agilent D1000 screen tapes (5067–5582) and D1000 Reagents (5067–5583). The 16 S amplicon libraries for each sample were quantified with qPCR, normalised with respect to amplicon sizes and pooled into a single library in equal molar quantities. Finally, 5 µl of pooled 4 nM DNA library pool was prepared for sequencing on Illumina MiSeq platform. The library pool was denatured with 0.2 M NaOH and diluted to 8 pM final concentration. Sequencing was carried out with MiSeq Reagent Kit v3–618 cycle (MS-102-3003) following manufacturer’s protocols (Illumina, Inc., San Diego, CA, USA).
Sequencing read preparation for downstream analysis
Paired end reads were demultiplexed by the integrated software of the Illumina MiSeq sequencing machine. The FastQ files were imported into the Qiime 2 pipeline (https://qiime2.org/) according to the„ Atacama Soil microbiome” tutorial. Residual adapter sequences (CTGTCTCTTATACACATCT) were trimmed from the 3’ end of the reads with Cutadapt Software integrated into the Qiime 2 pipeline. Quality trimming was performed by using the DADA2 software52 and the denoising parameters were set as follows: forward read length was set to 299 bases; for the reverse reads the length was set to 249 bases. No trimming from the start of the reads was applied.
Operational taxonomic unit (OTU) generation
Multiple sequence alignment was performed with the Mafft software53 and reads were taxonomically classified using Naïve Bayesian classifier trained with the Greengenes reference database (ver13_8) by selecting mapping points according to the forward-reverse primer set that was used for amplifying the 16 S V3-V4 regions of the bacterial community (341 F, 806R). Aligned sequences were clustered into operational taxonomic units (OTU) with a threshold level of 99% sequence identity. Singletons were discarded in order to reduce the likelihood of sequence artefacts interfering with further downstream analysis. The phylogenetic tree was constructed with FastTree plugin54. Alpha diversity indices were calculated by R.
MALDI-MSI experiments
For MALDI-MSI analysis animals treated with L. plantarum SNI3 strain and showing obvious phenotypic changes were selected. Frozen testis samples (7 control and 7 L. plantarum SNI3-treated) were cut into 12 μm sections on a microtome (HM 355 S, Microm, ThermoScientific) and mounted onto indium-tin-oxide coated glass slides. The matrix solution consisted of 10 mg/ml 9-aminoacridine hydrochloride monohydrate (9-AA) (Sigma-Aldrich, Germany) in water/methanol 30:70 (v/v). SunCollect™ automatic sprayer (Sunchrom, Friedrichsdorf, Germany) was used for matrix application. The flow rates were 10, 20, 30 and 40 µL/min, respectively, for the first four layers. The other 4 layers were performed at 40 µL/min. The MALDI-MSI measurement was performed on a Bruker Solarix 7T FT-ICR-MS (Bruker Daltonik, Bremen, Germany) in negative ion mode using 100 laser shots per spot at a frequency of 1000 Hz. The MALDI-MSI data were acquired over a mass range of m/z 75-1000 with 50 μm lateral resolution. After MALDI-MSI measurements, acquired data underwent spectra processing in FlexImaging v. 5.0 (Bruker Daltonics, Bremen, Germany) and SCiLS Lab v. 2019 (Bruker Daltonics, Bremen, Germany). MALDI-MSI data were normalized to the root mean square of all data points.
Metabolite annotation and bioinformatics analysis
MATLAB® R2014b (v.7.10.0, Mathworks, Inc., Natick, MA) was used for pre-processing of MALDI spectra as described previously55,56. Mass spectra underwent resampling, smoothing and baseline subtraction to decrease the data dimensionality and to remove noise-level peaks and artefacts. Peak picking was performed using an adapted version of the LIMPIC algorithm57 with m/z 0.0005 minimum peak width. The signal-to-noise and intensity threshold was set to 2 and 0.01% respectively. Isotopes were automatically identified and excluded. Peaks in the mass range of m/z 75 to 1000 were resolved and annotated by accurate mass matching in Human Metabolome Database HMDB58, and METASPACE59. Pathway analysis was performed with the MetaboAnalyst 4.060 and Kyoto Encyclopedia of Genes and Genomes (KEGG) database61.
Metabolomic statistical analyses were performed using the MetaboAnalyst 4.0, which offers a variety of methods commonly used in metabolomic data analyses25. Component analysis by sparse Partial Least Squares Discriminant Analysis (sPLSDA) was used for discriminant analysis with data shown on a 2D score plot with the x-axis representing component 1 and the y-axis representing component 2. The volcano plot is a combination of fold change and t-tests. The x-axis is log (FC). Y-axis is log10 (p value) based on p values. The discriminative metabolites were selected by a volcano plot with fold change > 2 or < 0.5 and p-value < 0.05. Box plots were created with GraphPad PRISM v. 5.00 (GraphPad Software, Inc, La Jolla, USA). Statistical significance testing was performed using the unpaired t-test (two-tailed).
Metabolic interaction network
Metabolite correlation network was created with Cytoscape (v. 3.7.2)62. Nodes represent metabolites with a node size and color corresponding to the intensity log2 fold change between control and L. plantarum SNI3 treatment. Edges represent functional correlations. Metabolites with significant differences between control and L. plantarum SNI3 treatment (P ≤ 0.05) are shown. The network was visualized using the Compound Spring Embedder layout (Figure S3).
LC-MS/MS analysis
LC-MS/MS was used to confirm the identities of GluGlu. LC-MS/MS analysis was performed using a Sciex ExionLC AD system coupled to a Sciex ZenoTOF 7600 (Sciex, Darmstadt, Germany). Separation of metabolites was achieved on an Agilent InfinityLab Poroshell 120 HILIC-Z column (2.1 mm x 150 mm, 2.7 μm, PEEK-linked, Agilent Technologies, Waldbronn, Germany). Eluent A was 100% H2O / 10 mM ammonium acetate + 2.5 µM InfinityLab Deactivator Additive, pH 9 and eluent B was 15% H2O + 85% ACN / 10 mM ammonium acetate + 2.5 µM InfinityLab Deactivator Additive, pH 9. Separation was carried out at 50 °C with a flow rate of 0.25 mL/min. The exact gradient is summarized in Table S3 Detection was carried out in negative ionization mode. As chemical standards, α-GluGlu was obtained from Sigma-Aldrich and γ-GluGlu was obtained from Thermo Scientific. Both were dissolved in H2O at a concentration of 1 mg/mL and further diluted with 5% H2O / 95% ACN. Testis samples were homogenized in H2O/MeOH/CHCl3 (1/3/1, v/v/v) with 1 mL solvent per 50 mg tissue weight in a Precellys homogenizer (Bertin Technologies, Montigny-le-Bretonneux, France). After centrifugation the supernatant was evaporated and the residue dissolved in 50 µL 5% H2O / 95% ACN. Extracted ion chromatograms for GluGlu (Empirical Formula: C10H16N2O7, m/z 275.088474 ([M-H]-)) was constructed.
HPLC analysis
γ-Glutamyl-glutamate (γ-GluGlu) concentration in bacteria, culture media and tissue extracts were measured by high performance liquid chromatography (HPLC). Samples were homogenized by ultra-sonication in ice-cold 0.01 M per chloric acid solution, centrifuged at 3510 g for 10 min at 4oC and the pellet was saved for protein measurement according to63. Perchloric anion from the supernatant was precipitated by 4 M dipotassium phosphate and removed by centrifugation. Samples were stored at -20oC until analysis. To measure γ-GluGlu concentration, the dansyl derivative formation method was used as described by64. HPLC separation was performed by Shimadzu LC-20 AD Analytical System, using UV (Agilent 1100 VW set at 253 nm) and Fluorescence (Gilson model 121 fluorimeters, at 340 nm excitation and 450 nm emission wavelengths) detection. Solid phase extraction (SPE) was carried out on a HALO Phenyl-Hexyl (75 × 2.1 mm I.D., 5 μm) column and for separation, it was coupled to an ACE Ultra Core Super C-18 (150 × 2.1 mm I.D., 5 μm) analytical column. The flow rate of mobile phases (A: 10 mM ammonium formate, 16.8% acetonitrile, methanol 4.8%; B: 10 mM ammonium formate, 70% acetonitrile, methanol 20%, 3 pH) was 400 µl/minute in a linear gradient mode. Concentrations were calculated by a two-point calibration curve internal standard method: (Ai * f * B)/(C * Di * E) (Ai: Area of Glu-Glu component; B: Sample volume; C: Injection volume; Di: Response factor of 1 pmol Glu-Glu standard; E: Protein content of sample; f: recovery factor of Internal Standard (IS area in calibration/IS area in actual). Data are expressed as mean ± standard error of mean at pmol/mg protein concentration. The enrichment and stripping flow rate of the buffer (10 mM ammonium formate, 1.9% acetonitrile, 1.1% methanol) was 300 µl/minute for 8 min, with total runtime of 55 min. The efficiency of the determination was calculated based on the area under the curve of the Nor-valine internal standard, which included the derivatization, SPE and separation factors; the value was R = 88.4 ± 9.1% (n = 25).
Data availability
Metabolomic data have been deposited to https://zenodo.org/records/10473905. Video files of the behavior are found at https://figshare.com/s/9f17be7dbbeade9098d8. Further information, accession codes and requests for resources and reagents should be directed to and will be fulfilled by the lead c, ontact, Krisztina J. Kovács (kovacs@koki.hu). All other data reported in this paper will be shared by the lead contact upon request.
References
Vich Vila A, Imhann F, Collij V, Jankipersadsing SA, Gurry T, Mujagic Z, et al. Gut microbiota composition and functional changes in inflammatory bowel disease and irritable bowel syndrome. Sci Transl Med. 2018;10.
Ke S, Hartmann J, Ressler KJ, Liu YY, Koenen KC. The emerging role of the gut microbiome in posttraumatic stress disorder. Brain Behav Immun. 2023;114:360–70.
Sun J, Chen F, Wu G. Potential effects of gut microbiota on host cancers: focus on immunity, DNA damage, cellular pathways, and anticancer therapy. ISME J. 2023;17:1535–51.
Saeedi BJ, Liu KH, Owens JA, Hunter-Chang S, Camacho MC, Eboka RU, et al. Gut-Resident Lactobacilli activate hepatic Nrf2 and protect against oxidative Liver Injury. Cell Metab. 2020;31:956–68 e5.
Perry RJ, Peng L, Barry NA, Cline GW, Zhang D, Cardone RL, et al. Acetate mediates a microbiome-brain-beta-cell axis to promote metabolic syndrome. Nature. 2016;534:213–7.
Ahmad AF, Dwivedi G, O’Gara F, Caparros-Martin J, Ward NC. The gut microbiome and cardiovascular disease: current knowledge and clinical potential. Am J Physiol Heart Circ Physiol. 2019;317:H923-H38.
Wang Y, Kasper LH. The role of microbiome in central nervous system disorders. Brain Behav Immun. 2014;38:1–12.
Borrego-Ruiz A, Borrego JJ. An updated overview on the relationship between human gut microbiome dysbiosis and psychiatric and psychological disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2023;128:110861.
Clarke G, Cryan JF, Dinan TG, Quigley EM. Review article: probiotics for the treatment of irritable bowel syndrome–focus on lactic acid bacteria. Aliment Pharmacol Ther. 2012;35:403–13.
Khalesi S, Bellissimo N, Vandelanotte C, Williams S, Stanley D, Irwin C. A review of probiotic supplementation in healthy adults: helpful or hype? Eur J Clin Nutr. 2019;73:24–37.
Walter J. Ecological role of lactobacilli in the gastrointestinal tract: implications for fundamental and biomedical research. Appl Environ Microbiol. 2008;74:4985–96.
Ayyanna R, Ankaiah D, Arul V. Anti-inflammatory and antioxidant properties of Probiotic Bacterium Lactobacillus mucosae AN1 and Lactobacillus fermentum SNR1 in Wistar albino rats. Front Microbiol. 2018;9:3063.
Kong Y, Olejar KJ, On SLW, Chelikani V. The potential of Lactobacillus spp. for modulating oxidative stress in the gastrointestinal tract. Antioxidants (Basel). 2020;9.
Virk MS, Virk MA, He Y, Tufail T, Gul M, Qayum A, et al. The anti-inflammatory and curative exponent of Probiotics: a Comprehensive and authentic ingredient for the sustained functioning of Major Human organs. Nutrients. 2024;16.
Kumar A, Alrefai WA, Borthakur A, Dudeja PK. Lactobacillus acidophilus counteracts enteropathogenic E. Coli-induced inhibition of butyrate uptake in intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2015;309:G602-7.
DiGiulio DB, Callahan BJ, McMurdie PJ, Costello EK, Lyell DJ, Robaczewska A, et al. Temporal and spatial variation of the human microbiota during pregnancy. Proc Natl Acad Sci U S A. 2015;112:11060–5.
Zhang P, Feng Y, Li L, Ge W, Yu S, Hao Y, et al. Improvement in sperm quality and spermatogenesis following faecal microbiota transplantation from alginate oligosaccharide dosed mice. Gut. 2021;70:222–5.
Magill RG, MacDonald SM. Male infertility and the human microbiome. Front Reprod Health. 2023;5:1166201.
Tang H, Huang W, Yao YF. The metabolites of lactic acid bacteria: classification, biosynthesis and modulation of gut microbiota. Microb Cell. 2023;10:49–62.
Li Z, Li Y, Xiao C, Yan Z, Pan R, Gao Y, et al. Genomic and metabolic features of the Lactobacillus sakei JD10 revealed potential probiotic traits. Microbiol Res. 2022;256:126954.
Teame T, Wang A, Xie M, Zhang Z, Yang Y, Ding Q, et al. Paraprobiotics and postbiotics of Probiotic Lactobacilli, their positive effects on the Host and action mechanisms: a review. Front Nutr. 2020;7:570344.
Otto MO, M.; Kovács K.J.; Ferenczi Sz.; Bata-Vidács I.; Kosztik, J.; Nagy, I.; Kukolya, J. Complete genome sequence of Lactiplantibacillus plantarum SNI3, a potential reprobiotic strain. International Scientific Conference, Probiotics, Prebiotics, Gut Microbiota and Health. Bratislava2023.
Scribner KA, Walker CD, Cascio CS, Dallman MF. Chronic streptozotocin diabetes in rats facilitates the acute stress response without altering pituitary or adrenal responsiveness to secretagogues. Endocrinology. 1991;129:99–108.
Liu ZW, Jiang N, Tao X, Wang XP, Liu XM, Xiao SY. Assessment of sexual behavior of male mice. J Vis Exp. 2020.
Xia J, Broadhurst DI, Wilson M, Wishart DS. Translational biomarker discovery in clinical metabolomics: an introductory tutorial. Metabolomics. 2013;9:280–99.
Zhao CJ, Ganzle MG. Synthesis of taste-active gamma-glutamyl dipeptides during Sourdough fermentation by Lactobacillus reuteri. J Agric Food Chem. 2016;64:7561–8.
Poutahidis T, Springer A, Levkovich T, Qi P, Varian BJ, Lakritz JR, et al. Probiotic microbes sustain youthful serum testosterone levels and testicular size in aging mice. PLoS One. 2014;9:e84877.
Akram M, Ali SA, Kaul G. Probiotic and prebiotic supplementation ameliorates chronic restraint stress-induced male reproductive dysfunction. Food Funct. 2023.
Helli B, Kavianpour M, Ghaedi E, Dadfar M, Haghighian HK. Probiotic effects on sperm parameters, oxidative stress index, inflammatory factors and sex hormones in infertile men. Hum Fertil (Camb). 2022;25:499–507.
Alfano M, Ferrarese R, Locatelli I, Ventimiglia E, Ippolito S, Gallina P, et al. Testicular microbiome in azoospermic men-first evidence of the impact of an altered microenvironment. Hum Reprod. 2018;33:1212–7.
Molina NM, Plaza-Diaz J, Vilchez-Vargas R, Sola-Leyva A, Vargas E, Mendoza-Tesarik R, et al. Assessing the testicular sperm microbiome: a low-biomass site with abundant contamination. Reprod Biomed Online. 2021;43:523–31.
Hao Y, Feng Y, Yan X, Chen L, Zhong R, Tang X, et al. Gut microbiota-testis axis: FMT improves systemic and testicular micro-environment to increase semen quality in type 1 diabetes. Mol Med. 2022;28:45.
Leelani NB, P., Parekh, N., Vij, S.C., Lundy,S.D. The emerging role of the gut testis axis in male reproductive health and infertility. Fertil Steril Rev. 2023;4:131–41.
Oakberg EF. Duration of spermatogenesis in the mouse and timing of stages of the cycle of the seminiferous epithelium. Am J Anat. 1956;99:507–16.
Vital M, Howe AC, Tiedje JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. 2014;5:e00889.
Lin Y, Wang K, Che L, Fang Z, Xu S, Feng B, et al. The improvement of Semen Quality by Dietary Fiber Intake is positively related with gut microbiota and SCFA in a Boar Model. Front Microbiol. 2022;13:863315.
Culp EJ, Goodman AL. Cross-feeding in the gut microbiome: Ecology and mechanisms. Cell Host Microbe. 2023;31:485–99.
Dao MC, Everard A, Aron-Wisnewsky J, Sokolovska N, Prifti E, Verger EO, et al. Akkermansia muciniphila and improved metabolic health during a dietary intervention in obesity: relationship with gut microbiome richness and ecology. Gut. 2016;65:426–36.
Cirstea M, Radisavljevic N, Finlay BB. Good Bug, Bad Bug: Breaking through Microbial Stereotypes. Cell Host Microbe. 2018;23:10 – 3.
Kobayashi S, Ikeda Y, Shigeno Y, Konno H, Fujii J. Gamma-glutamylcysteine synthetase and gamma-glutamyl transferase as differential enzymatic sources of gamma-glutamylpeptides in mice. Amino Acids. 2020;52:555–66.
Kumar TR, Wiseman AL, Kala G, Kala SV, Matzuk MM, Lieberman MW. Reproductive defects in gamma-glutamyl transpeptidase-deficient mice. Endocrinology. 2000;141:4270–7.
Castellon EA. Glutathione and gamma-glutamyl cycle enzymes in rat testis during sexual maturation. Arch Androl. 1994;33:179–85.
Basit A, Amory JK, Mettu VS, Li CY, Heyward S, Jariwala PB, et al. Relevance of human aldoketoreductases and microbial beta-glucuronidases in Testosterone Disposition. Drug Metab Dispos. 2023;51:427–35.
Winter J, Bokkenheuser VD. Bacterial metabolism of natural and synthetic sex hormones undergoing enterohepatic circulation. J Steroid Biochem. 1987;27:1145–9.
Collden H, Landin A, Wallenius V, Elebring E, Fandriks L, Nilsson ME, et al. The gut microbiota is a major regulator of androgen metabolism in intestinal contents. Am J Physiol Endocrinol Metab. 2019;317:E1182-E92.
Hawksworth G, Drasar BS, Hill MJ. Intestinal bacteria and the hydrolysis of glycosidic bonds. J Med Microbiol. 1971;4:451–9.
Levine H, Jorgensen N, Martino-Andrade A, Mendiola J, Weksler-Derri D, Mindlis I, et al. Temporal trends in sperm count: a systematic review and meta-regression analysis. Hum Reprod Update. 2017;23:646–59.
Levine H, Jorgensen N, Martino-Andrade A, Mendiola J, Weksler-Derri D, Jolles M, et al. Temporal trends in sperm count: a systematic review and meta-regression analysis of samples collected globally in the 20th and 21st centuries. Hum Reprod Update. 2022.
Barcena C, Valdes-Mas R, Mayoral P, Garabaya C, Durand S, Rodriguez F, et al. Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat Med. 2019;25:1234–42.
Turner PV, Pang DS, Lofgren JL. A review of Pain Assessment methods in Laboratory rodents. Comp Med. 2019;69:451–67.
Somfai T, Bodo S, Nagy S, Gocza E, Ivancsics J, Kovacs A. Simultaneous evaluation of viability and acrosome integrity of mouse spermatozoa using light microscopy. Biotech Histochem. 2002;77:117–20.
Callahan BJ, McMurdie PJ, Rosen MJ, Han AW, Johnson AJ, Holmes SP. DADA2: high-resolution sample inference from Illumina amplicon data. Nat Methods. 2016;13:581–3.
Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013;30:772–80.
Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490.
Sun N, Ly A, Meding S, Witting M, Hauck SM, Ueffing M, et al. High-resolution metabolite imaging of light and dark treated retina using MALDI-FTICR mass spectrometry. Proteomics. 2014;14:913–23.
Ly A, Buck A, Balluff B, Sun N, Gorzolka K, Feuchtinger A, et al. High-mass-resolution MALDI mass spectrometry imaging of metabolites from formalin-fixed paraffin-embedded tissue. Nat Protoc. 2016;11:1428–43.
Mantini D, Petrucci F, Pieragostino D, Del Boccio P, Di Nicola M, Di Ilio C, et al. LIMPIC: a computational method for the separation of protein MALDI-TOF-MS signals from noise. BMC Bioinformatics. 2007;8:101.
Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vazquez-Fresno R, et al. HMDB 4.0: the human metabolome database for 2018. Nucleic Acids Res. 2018;46:D608-D17.
Palmer A, Phapale P, Chernyavsky I, Lavigne R, Fay D, Tarasov A, et al. FDR-controlled metabolite annotation for high-resolution imaging mass spectrometry. Nat Methods. 2017;14:57–60.
Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, et al. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res. 2018;46:W486-W94.
Kanehisa M, Sato Y, Furumichi M, Morishima K, Tanabe M. New approach for understanding genome variations in KEGG. Nucleic Acids Res. 2019;47:D590-D5.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–504.
Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951;193:265–75.
Baranyi MSB. Optimization and validation of online column-switching assisted HPLC-spectrometric method for quantificationof dansylated endocannabinoids andneurotransmitters in animal models ofbrain diseases open Jounal of Analytical and Bioanalytical Chemistry. 2019:p83-93.
Acknowledgements
Acknowledgement: The study was supported by grants from the Hungarian National Research, Development and Innovation Office PD 143386 and National Brain Research Program 2017-1.2.1-NKP-2017-00002. Project number RRF-2.3.1-21-2022-00011 titled National Laboratory of Translational Neuroscience has been implemented with the support provided by the Recovery and Residence Facility of the European Union within the framework of Programme Széchényi Plan Plus for KJK, Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) SFB 824 C04 and SFB/Transregio 205 S01 for AW. The authors thank to Dr. Paul E. Sawchenko for his valuable advises on the manuscript.
Author information
Authors and Affiliations
Contributions
Balázs Juhász: conceptualization, data curation, formal analysis, investigation, visualization, writing-review and editing; Krisztina Horváth: data curation, formal analysis, investigation, writing-review and editing; Dániel Kuti: data curation, formal analysis, investigation, writing-review and editing; Ildikó Bata-Vidács: investigation, data curation; Jian Shen: investigation, data curation; Annette Feuchtinger: investigation, data curation; Chaoyang Zhang: investigation, data curation; István Nagy: conceptualization, data curation, writing-review and editing; József Kukolya: conceptualization, data curation, writing-review and editing; Michael Witting: data curation, formal analysis, writing-review and editing; Mária Baranyi: data curation, formal analysis, writing-review and editing; Szilamér Ferenczi: conceptualization, investigation, data curation; Axel Walch: conceptualization, resources, writing-review and editing; Na Sun*: conceptualization, data curation, formal analysis, investigation, visualization, writing-original draft, writing-review and editing, *Equally contributing senior author; Krisztina J. Kovács: conceptualization, data curation, formal analysis, investigation, visualization, writing-original draft, writing-review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. The Lactiplantibacillus plantarum SNI3 and its use on male subjects has been filed for patent application on June 4, 2021. in Hungary. PCT procedure and national phases of PCT are under progress.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Juhász, B., Horváth, K., Kuti, D. et al. Dipeptide metabolite, glutamyl-glutamate mediates microbe-host interaction to boost spermatogenesis. Sci Rep 14, 21864 (2024). https://doi.org/10.1038/s41598-024-73216-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-73216-y
This article is cited by
-
Integrated profiling of semen microbiota and metabolome reveals dysregulation in primary idiopathic male infertility
Journal of Translational Medicine (2025)
-
The Promising Biological Role of Postbiotics in Treating Human Infertility
Probiotics and Antimicrobial Proteins (2025)