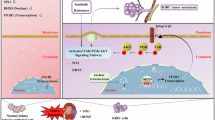
Abstract
Nasopharyngeal carcinoma (NPC) presents a significant medical challenge due to its high incidence rate and poor prognosis, which are attributed primarily to tumor metastasis and drug resistance. Sine oculis homeobox homolog 1 (SIX1) has been identified as a crucial target for cancer treatment. However, its role in NPC remains incompletely understood. This study investigated the mechanisms by which the degradation of the SIX1 protein, which is mediated by ubiquitin, affects the malignant characteristics of NPC throughout the cell cycle. Our findings reveal that reduced expression of the itchy E3 ubiquitin ligase E3 (ITCH) in NPC impedes the degradation of the SIX1 protein, leading to enhance oncogenic properties. Knockdown experiments which SIX1 was inhibited demonstrated a decrease in the proliferation, migration, and invasion of NPC cell lines, whereas overexpression of SIX1 yielded the opposite effects. Further experimental validation revealed that SIX1 promotes NPC progression via the cell division cycle 27 (CDC27)/cyclin B1 axis. These findings provide valuable insights into potential therapeutic targets and prognostic indicators for NPC treatment, emphasizing the ITCH/SIX1/CDC27/cyclin B1 axis as a promising target for novel therapies.
Similar content being viewed by others
Nasopharyngeal carcinoma (NPC) is a solid malignant tumor that arises in the nasopharyngeal cavity. Currently, chemoradiotherapy serves as the primary treatment for NPC1,2,3,4. However, it is associated with varying survival outcomes and significant side effects, including reduced white blood cell count, hair loss, and gastrointestinal reactions5,6. The poor prognosis of patients with NPC has underscored the urgent need for the development of novel therapeutic targets and alternative drugs.
Sine oculis homeobox homolog 1 (SIX1) is implicated in the growth of various organs7,8, inflammation9, glycolysis10, and tumor development11 by activating or repressing downstream transcription factors. Research has demonstrated a strong correlation between SIX1 and the malignant behavior of tumors, including Ewing sarcoma12, hepatocellular carcinoma13, cervical cancer14, and Wilms tumor15. Consequently, SIX1 is considered a significant factor in tumor development, leading to the development of targeted drugs aimed at inhibiting its activity. Understanding the mechanisms that regulate SIX1 is essential for the development of effective treatments for NPC.
The ubiquitination-proteasome degradation pathway is responsible for the degradation of substrate proteins by attaching ubiquitin to their lysine side chains and subsequently transferring them to lysosomes and proteasomes. This pathway is regulated by E3 ubiquitin ligases, which determine the specificity of the ubiquitination reaction16. The involvement of the ubiquitination-proteasome degradation pathway has been established in various diseases, including inflammation and tumors17. Itchy E3 ubiquitin ligaseE3(ITCH) is a HECT-type ubiquitin ligase, and its deletion both internal and external has been associated with the development of multiple diseases, such as chronic spontaneous urticaria18 and hepatocellular carcinoma19. However, no comprehensive study has directly linked ITCH to the occurrence of nasopharyngeal cancer.
Recent studies have indicated that the ubiquitination-mediated degradation of SIX1 is impaired in pancreatic cancer, which subsequently regulates tumor glycolysis and promotes the metastasis of pancreatic cancer cells20. Furthermore, MG132 administration has been shown to lead to the accumulation of the SIX1 protein. These findings suggest that SIX1 can be degraded through both proteasomal and lysosomal pathways. However, the relationship between SIX1 expression in NPC and the ubiquitination degradation pathway remains to be elucidated.
The cell cycle is a regulatory mechanism that governs the division of cells, and is divided into three phases: G0/G1 (DNA prophase)21, S phase (DNA replication)22, and G2/M (DNA separation)23. Cyclin-dependent kinases (CDKs) and cyclins are essential regulatory proteins that facilitate proper function of the cell cycle. Different cyclin/CDK complexes are responsible for driving various cell-cycle processes. Consequently, disrupting the normal function of the cell cycle in cancer cells is an effective strategy for impeding their growth. Recent studies have indicated that it plays a role in regulating the cell cycle and may promote disease progression under various conditions24,25,26. However, its specific role and the mechanism underlying its function in the cell cycle, particularly in NPC, remain unclear.
In this study, we aimed to investigate whether SIX1 acts as a substrate for ITCH-mediated protein degradation through ubiquitination. Our objective was to elucidate the specific mechanism by which SIX1 regulates the malignant behavior of NPC throughout the cell cycle. Our findings revealed a correlation between elevated levels of the SIX1 protein and decreased activity of the E3 ubiquitin ligase ITCH in NPC. Furthermore, we observed that high expression of SIX1 facilitated the malignant phenotype of NPC by modulating the cell cycle-related protein cell division cycle 27 (CDC27)/cyclin B1 axis. The ITCH/SIX1/CDC27-cyclin B1 axis may represent a potential target for the treatment and prognostic prediction of NPC.
Results
SIX1 protein expression was increased in NPC tissues and correlated with poor prognosis
First, to identify differential genes in the NPC patients and normal patients, 14 normal nasopharyngeal tissue samples and 43 nasopharyngeal tumors samples were collected from the GSE64634 and GSE12452 datasets. Principal component analysis revealed that the mRNAs in the tumor and control groups could be classified into different clusters, indicating that the sequencing data was qualified for further analysis (Fig. 1A). A total of 1058 differentially expressed genes (DEGs) were identified: 377 upregulated and 681 downregulated genes (P. adj < 0.05 and |log2FC| ≥ 1; Fig. 1B). The log2FC of SIX1 mRNA was minus 1.2. A heatmap of the differentially expressed mRNAs is provided (Fig. 1C). The sequencing results of GSE13597 from other platforms were subsequently analyzed. Principal component analysis demonstrated heterogeneity in the data from 3 normal controls and 25 nasopharyngeal cancer patients (Fig. 1D). The expression of SIX1 mRNA (log2FC= − 1.67) also decreased (Fig. 1E). A significant correlation between SIX1 protein expression and overall survival was identified, indicating poor prognosis (P = 0.033) in NPC patients (Fig. 1F). Compared with those in normal nasopharyngeal epithelial cells (NP69), the expression levels of SIX1 mRNA were lower in NPC cell lines (6-10B vs. NP69, P = 0.0684; HK1 vs. NP69, P = 0.0002; HNE1 vs. NP69, P = 0.0029) (Fig. 1G). Interestingly, our results found that SIX1 protein expression was elevated in NPC cell lines compared with that in NP69 cells (Fig. 1H–I). Thus, SIX1 may play a significant role in the progression of NPC and its malignant behavior. We also hypothesize that the increased protein expression levels of SIX1 could be associated with post-translational modifications.
SIX1 protein expression was increased in NPC tissues and correlated with poor prognosis. (A) The GSE64634 and GSE12452 were analyzed for PCA. (B) Volcano diagram showed the DEGs between tumor(n = 43) and normal samples(n = 14). (C) Heat map showed clustering of differentially expressed genes in tumor and normal samples. (D) The GSE13597 were analyzed for PCA. (E) Volcano diagram showed the DEGs between tumor (n = 25) and normal samples (n = 3). (F) GEPIA2 analysis of the relationship of SIX1 expression on OS (The 208 patients with low SIX1 expression and 285 patients with high SIX1 expression). (G) The result of qPCR confirmed the SIX1 expression in NPC cell lines. (H,I) The SIX1 protein expression level in NP69 and NPC cell lines. The statistical analysis of band grey values was displayed on the side. The data are presented as the mean ± SD values. *P < 0.05; **P < 0.01; ***P<0.001.
Downregulation of E3 ubiquitination ligase ITCH maintained the stability of the SIX1 protein in NPC cells
To confirm our hypothesis that the increased expression level of SIX1 protein in NPC cell lines was associated with protein post-translational modification, we conducted an intervention study using the protein synthesis inhibitor cycloheximide (CHX) and the proteasome inhibitor MG132 in NPC cell lines. Our findings revealed that MG132 can inhibit the degradation of the SIX1 protein and exogenous transfection of the UB and SIX1 plasmids promoted the degradation of the SIX1 protein by ubiquitination (Fig. 2A–D). Subsequently, we utilized the human ubiquitin ligase-substrate interaction prediction and display system, UbiBrowse website (http://ubibrowser.bio-it.cn/ubibrowser_v3/), which displayed 12 proteins that may be involved in regulating SIX1 ubiquitin. These proteins were related to the ubiquitination pathway (Fig. 2E; Supplementary Table S3). Among these genes, ITCH exhibited the lowest expression level in NPC cell lines (Fig. 2F). Thus, we speculated that ITCH may play a role in regulating the degradation of SIX1. Molecular docking experiments were conducted to preliminarily confirm the interaction between ITCH and SIX1 (Fig. 3A). Immunofluorescence analysis demonstrated the colocalization of SIX1 and ITCH (Fig. 3B). Furthermore, as we gradually increased the expression of ITCH, we observed a time-dependent decrease in the protein expression of SIX1 (Fig. 3C, D). Additionally, a protein interaction between ITCH and SIX1 was observed (Fig. 3E, F). We constructed stable ITCH-overexpessing 6-10B cells. Immunofluorescence results revealed that after ITCH was overexpressed, its colocalization with SIX1 decreased significantly (Fig. 3G, H). Finally, after simultaneous exogenous transfection of ITCH and SIX1 plasmids, our experimental results revealed that the overexpression of ITCH promoted ubiquitination and degradation of the SIX1 protein (Fig. 3I, J). Thus, our study preliminarily confirmed that reduced ITCH expression may be the cause of SIX1 protein accumulation in NPC.
SIX1 could be modified by ubiquitination. (A,B) The western blot confirmed MG132 treatment could inhibit SIX1 protein degradation under the CHX intervention in HNE1 and 6-10B cells. (C,D) The western blot confirmed SIX1 protein undergo ubiquitination in 293T cells and 6-10B cells. (E) The picture showed the 12 E3 ubiquitination ligase genes predicted to be associated with SIX1 ubiquitination degradation by using UbiBrowser website. (F) The qPCR analysis of the expression of 12 E3 ubiquitination ligase genes in 6-10B cells versus NP69 cells. ns P>0.05; a*P < 0.05; b**P < 0.01; c***P<0.001.
ITCH was involved in ubiquitination degradation of SIX1 in NPC cell lines. (A) Molecular docking analysis of the interaction between ITCH and SIX1. (B) Immunofluorescence showed colocalization of ITCH (green) and SIX1 (Red) in 6-10B cells. (C,D) The western blot demonstrated the effect of gradient overexpression of ITCH on SIX1 protein expression in 6-10B cells. The statistical chart was displayed below it. (E,F) COIP assays showed the interaction between SIX1 and ITCH in 293T and 6-10B cells. G The western blot demonstrated the efficiency of ITCH overexpression in 6-10B cells. (H) Immunofluorescence showed the effect of ITCH overexpression on SIX1 colocalization in 6-10B cells. (I,J) COIP experiments confirmed that the E3 ubiquitination ligase ITCH promoted the ubiquitination level of SIX1. *P < 0.05.
SIX1 overexpression promoted the proliferation, invasion, and migration of NPC cell lines
To verify the effect of SIX1 on the malignant behavior of the tumors, we established stable NPC cell lines by overexpressing SIX1 (Fig. 4A). The results from the CCK8 and plate colony formation experiments revealed that the proliferation ability of NPC cell lines overexpressing SIX1 was significantly increased (Fig. 4B,C). Furthermore, the results of our invasion and migration assays showed that the overexpression of SIX1 substantially enhanced both the invasive and migratory capabilities of these cells (Fig. 4E–H). Consequently, our findings suggest that SIX1 overexpression can promote the proliferation, invasion, and migration of NPC cell lines.
Overexpression of SIX1 promoted proliferation, invasion and migration of NPC cell lines. (A) The western blot confirmed SIX1 overexpressed in NPC cell lines. The statistical analysis of band grey values was displayed on the side. (B) CCK8 assay showed the relative proliferation ability. (C,D) Representative images of the wells of plates in the colony formation assay. The statistical analysis was displayed on the side. (E,F) Transwell invasion assay in NPC cell lines. The statistical analysis was displayed on the side. (G,H) Transwell migrated assay in NPC cell lines. The statistical analysis was displayed on the side. *P < 0.05; **P < 0.01; ***P < 0.001. OENC-SIX1: Plasmid blank control group; OE-SIX1: overexpression SIX1 plasmid group.
.
SIX1 knockdown inhibited the proliferation, migration, and invasion of NPC cell lines
To investigate the significance of SIX1 in the development of NPC, we conducted a rescue assay utilizing NPC cell lines with silenced SIX1 (Fig. 5A). The results of the cck8 and plate cloning experiments demonstrated that the proliferative capacity of NPC cell lines with SIX1 knockdown was significantly impaired (Fig. 5B,C). Furthermore, our invasion and migration assays revealed that the loss of SIX1 resulted in a marked reduction in both invasion and migration abilities (Fig. 5E–H). The knockdown of SIX1 effectively inhibited the proliferation, invasion, and migration of NPC cell lines, underscoring the essential role of SIX1 in the malignant phenotype of NPC.
SIX1 knockdown alleviated proliferation, invasion and migration of NPC cell lines. (A) The western blot confirmed SIX1 knockdown in NPC cell lines. The statistical analysis of band grey values was displayed on the side. (B) CCK8 assay showed the relative proliferation ability. (C,D) Representative images of the wells of plates in the colony formation assay. The statistical analysis was displayed on the side. (E,F) Transwell invasion assay in NPC cell lines. The statistical analysis was displayed on the side. (G,H) Transwell migrated assay in NPC cell lines. The statistical analysis was displayed on the side. The data are presented as the mean ± SD values. *P < 0.05; ** P < 0.01; ***P < 0.001. shNC-SIX1: negative control; sh-SIX1:SIX1 knockdown.
SIX1 knockdown inhibited 6-10B tumorigenicity in vivo
To evaluate the tumorigenic effects of SIX1, we subcutaneously injected control 6-10B cells and 6-10B cells with SIX1 knockdown into nude mice. The results showed that the size, weight, and volume of the tumors were significantly reduced in the SIX1 knockdown group (Fig. 6A–E). Hematoxylin & eosin (HE) staining revealed a decrease in pathological nuclear pleomorphism and lighter nuclear staining in the tumor tissues of the SIX1 knockdown group (Fig. 6F). Additionally, the positive rate of Ki67, a marker associated with cell proliferation, was markedly lower in tumors from SIX1-knockdown mice than in those from control mice (Fig. 6G, H). These results demonstrated that inhibiting SIX1 expression effectively reduced tumor growth in vivo.
SIX1 knockdown inhibited 6-10B tumorigenicity in vivo. Four-week-old nude mice (six mice in each group) were injected subcutaneously with knockdown SIX 6-10B cells and control 6-10B cells (5 × 106 cells per mouse) and tumor size was measured starting on day 7. (A) Weight of nude mice, (B) photographs of 6-10B tumor formation in the xenografts of nude mice. (C–E) Picture of isolated tumors, tumor weight, and tumor volume. (F) The typical images -stained by H&E were presented, (G,H) Ki67 and SIX1 IHC staining, The statistical analysis was displayed on the side. The data are presented as the mean ± SD values. *P < 0.05; **P < 0.01; ***P < 0.001. shNC-SIX1: negative control; sh-SIX1:SIX1 knockdown.
SIX1 can upregulate CDC27 expression in NPC cell lines
To investigate the potential mechanisms of SIX1, we sequenced the transcriptomes of control cells and HNE1 cells that overexpressed SIX1 at BGI. The Sequencing analysis revealed 1067 DEGs (Fig. 7A). Kyoto Encyclopedia Genes and Genomes (KEGG) analysis indicated that the top 10 enriched pathways among these DEGs included endocytosis, ubiquitin-mediated proteolysis, the mTOR signaling pathway, the cell cycle, signaling pathways regulating the pluripotency of stem cells, protein processing in the endoplasmic reticulum, the MAPK signaling pathway, biosynthesis of amino acids, and axon regeneration (p < 0.05; Fig. 7B). We focused on the most statistically significant pathway, ubiquitin-mediated proteolysis (p = 0.0058), which involved a total of 17 genes including ANAPC5, KLHL13, TRIP12, TRIM32, UBE2M, CDC34, CDC23, SKP2, UBE4B, UBR5, BIRC6, UBC, HERC2, ANAPC1, CUL3, UBE2S, and CDC27 (Fig. 7C). Notably, CDC27 has been implicated not only in the ubiquitination pathway but also in the regulation of the cell cycle. Therefore, we further examined the relationship between CDC27 and SIX1. The qPCR results corroborated the sequencing findings (Fig. 7D–F), demonstrating that SIX1 promoted the malignant phenotype of NPC cell lines by upregulating CDC27 expression.
SIX1 can upregulate CDC27 expression in NPC cell lines. (A) Volcano plot showing differential genes in control and overexpressed SIX1 in NPC cell lines. (B) Top 10 KEGG pathway enrichment results for differential genes in the module. (C) Functional enrichment analysis of differential genes involved in the ubiquitin-mediated proteolysis in modules. (D–F) The result of qPCR confirmed the regulatory relationship between SIX1 and CDC27. The data are presented as the mean ± SD values. *P < 0.05; **P < 0.01; ***P < 0.001. OENC-SIX1: Plasmid blank control group; OE-SIX1: overexpression SIX1 plasmid group.
SIX1 promoted the malignant behavior of NPC cell lines by regulating the CDC27/cyclin B1 axis
To validate sequencing results indicating that the SIX1/CDC27 pathway is involved in the cell cycle, we conducted flow cytometry assays on SIX1 knockdown NPC cells and control cells. The results of these assays demonstrated that the knockdown of SIX1 inhibited progression through the G2/M phase (P6-10B = 0.0001; PHK1 < 0.0001, PHNE1 = 0.0046), but had no significant effect on the G0/G1or S phase (Fig. 8A–F). In this study, we identified cell cycle proteins associated with the G2/M phase and subsequently verified them using Western blotting. The results revealed that SIX1 knockdown led to a decrease in the expression levels of CDC27 and cyclin B1 (Fig. 9A–C). Co-immunoprecipitation (COIP) experiments confirmed the interaction between SIX1 and CDC27 (Fig. 9D). To further elucidate the relationship between SIX1 and CDC27, we overexpressed CDC27 in SIX1 knockdown NPC cell lines and assessed its impact on cellular phenotype (Fig. 10A). CCK-8 and plate colony formation assays demonstrated that the overexpression of CDC27 partly counteracted the inhibitory effects of SIX1 knockdown on the proliferation of NPC cell lines (Fig. 10B, C). Additionally, subsequent invasion and migration assays indicated that CDC27 overexpression alleviated the inhibitory effects of SIX1 knockdown on the invasion and migration capacities of NPC cell lines (Fig. 10D, E). Our findings also revealed that CDC27 overexpression rescued the cell cycle arrest induced by SIX1 knockdown at the G2/M phase (Fig. 10F). The molecular mechanism by which SIX1 regulated the malignant phenotype of NPC cell lines is schematically illustrated in Fig. 11.
Effects of SIX1 knockdown on the cell cycle of NPC cell lines. (A,B) Representative diagram was presented showing the percentage of cells in each phase of the cell cycle in control 6-10B cells and SIX1 Knockdown 6-10B cells. The statistical analysis was displayed on the side. (C,D) Representative diagram was presented showing the percentage of cells in each phase of the cell cycle in control HK1 cells and SIX1 knockdown HK1 cells. The statistical analysis was displayed on the side. (E,F) Representative diagram was presented showing percentage of cells in each phase of the cell cycle in control HNE1 cells and SIX1 Knockdown HNE1 cells. The statistical analysis was displayed on the side.
.
SIX1 Knockdown decreased the expression of CDC27/cyclinB1 protein axis in NPC cell lines. (A) The western Blot results presented the relationship between SIX1 and CDC27/cyclinB1 axis in 6-10B cells. The statistical analysis was displayed on the side. (B) The western Blot results presented the relationship between SIX1 and CDC27/cyclinB1 axis in HK1 cells. The statistical analysis was displayed on the side. (C) The western Blot results presented the relationship between SIX1 and CDC27/cyclinB1 axis in HK1 cells. The statistical analysis was displayed on the side. (D) COIP assays showed an interaction between SIX1 and CDC27. **P < 0.01; ***P<0.001. shNC-SIX1: negative control; sh-SIX1:SIX1 knockdown.
SIX1 Knockdown inhibited malignant behavior of NPC cell lines by downregulating CDC27 expression. (A) Typical Western blot experiment demonstrated the efficiency of SIX1 knockdown and CDC27 overexpression in NPC cell lines. (B) CCK8 assays revealed the impact of SIX1 knockdown and CDC27 overexpression on the proliferation of NPC cell lines. (C) Plate cloning experiments illustrated the effects of SIX1 knockdown and CDC27 overexpression on the clonal proliferation capacity of NPC cell lines. (D) Transwell invasion assay demonstrated the effects of knocking down SIX1 and overexpressing CDC27 on the invasion ability of NPC cell lines. (E) Transwell migrated assay demonstrated the effects of knocking down SIX1 and overexpressing CDC27 on the migration ability of NPC cell lines. The statistical analysis was displayed on the side. (F) Representative flow chart showing the effects of SIX1 knockdown and CDC27 overexpression on the cell cycle in NPC cell lines. The statistical analysis was displayed on the side. The data are presented as the mean ± SD values. *P<0.05, **P<0.01, ***P<0.001. sh-SIX1:SIX1 knockdown, OE-CDC27: overexpression CDC27 plasmid group.
Discussion
Previous studies have identified various mechanisms that regulate SIX1 expression in tumor cells, such as transcription factor regulation, epigenetics, signaling pathways, and post-translational modifications27,28,29,30. With its HD and SD domains, SIX1 can interact with key genes in cancer-promoting pathways31. Upregulation of SIX1 expression has been linked to the development of different tumors, including endometrial cancer32, esophageal squamous cell carcinoma33, and prostate cancer34. Elevated intracellular SIX1 levels can trigger extracellular signals that increase oncogenic potential, tumor progression, and invasiveness. However, the role of SIX1 in NPC remains unclear. Our study reveals the involvement of the ITCH/SIX1/CDC27-cyclinB1 signaling axis in regulating the malignant behavior of NPC, addressing a research gap and offering a potential treatment approach for this type of cancer.
Compared with other reports in tumor, our study found that the expression level of SIX1 mRNA was decreased in NPC cell lines. However, expression at the protein level is increased, which indicates that some regulatory mechanism in NPC tumors promotes the stable expression of the SIX1 protein. Several previous studies have shown that SIX1 has post-translational modifications such as phosphorylation35, methylation28, glycosylation and ubiquitination13,36 in different tumors. Therefore, we speculate that modifications that mediate the stable expression of SIX1 in NPC may exist.
The ubiquitination-proteasome pathway is a common method of protein degradation in cells, impacting cellular biological processes37. To validate this hypothesis, CHX and MG132 were used as protein synthesis and proteasome inhibitors, respectively, confirming the ubiquitination of SIX1. The UbiBrowser 2.0 system was subsequently employed to identify potential targets associated with the ubiquitination and degradation of SIX1. The analysis revealed that the HECT family E3 ubiquitin ligase ITCH played a crucial role in mediating this process. Experimental evidence demonstrated that increased ITCH expression triggered SIX1 ubiquitination and degradation, suggesting low level expression of ITCH was beneficial to maintaining the stability of SIX1 protein in NPC. Therefore, our study attributed the ability of SIX1 protein to stably exert its biological role in NPC to the restriction of its proteasome degradation pathway.
Further studies on the role of SIX1 in NPC have demonstrated that its overexpression can enhance malignant behaviors such as proliferation, invasion, and migration. Conversely, silencing SIX1 not only reversed these effects but also inhibited tumorigenicity in nude mice. These findings underscore the tumor-promoting role of SIX1 in NPC and align with previous studies regarding the pathological functions of SIX1 in various tumors12,38,39.
To further investigate the precise mechanism by which SIX1 facilitates the development of NPC, we sequenced cells stably overexpressing SIX1 and control cells. The results of the sequencing analysis revealed significant enrichment of the KEGG pathway associated with the cell cycle. Previous studies have shown that abnormalities in the cell cycle are a manifestation of cancer progression23,40. To explore the regulatory mechanism of the SIX1-associated cell cycle in conjunction with sequencing results, cell cycle-related genes were detected in NPC cell lines overexpressing SIX1. The results indicated that overexpression of SIX1 mRNA significantly enhanced the expression of cell cycle-related protein cell division cycle 27 (CDC27) mRNA in cells. Subsequently, knockdown of SIX1 suppressed the expression of CDC27, indicating that CDC27 may serve as a critical the link between SIX1 and the cell cycle. Next, we further explored the relationship between SIX1 and CDC27. Our study demonstrated that CDC27 overexpression can reverse the inhibitory effects of SIX1 knockdown on the proliferation, invasion, and migration of NPC cell lines. Thus, these findings confirmed the regulatory axis between SIX1 and CDC27 and its impact on the malignant behavior of NPC cell lines.
Interestingly, our flow cytometry analysis revealed that the regulation of the cell cycle by the SIX1-CDC27 signaling axis predominantly impacts the G2/M phase in NPC cell lines. Additionally, Western blot experiments provided preliminary evidence that this regulation may be mediated through alterations in the expression of cyclin B1. However, one study demonstrated that knocking down SIX1 in breast cancer inhibited tumor proliferation by reducing the expression of cyclinA1 protein41. Another study indicated that SIX1 enhanced tumor prognosis by regulating the expression of cyclinD1 in pancreatic cancer42. We speculate that the observed differences may be attributed to variations in the signaling molecules of SIX1 within the microenvironments of different tumor types, which in turn influences the selection of cell cycle proteins by SIX1.
In summary, we propose a novel signaling axis ITCH/SIX1/CDC27/cyclinB1 that regulates the development of NPC. The underlying mechanism involves a decrease in ITCH expression in NPC, which inhibits the ubiquitination of SIX1. This allows for the stable expression of SIX1 in NPC, further promoting cell cycle progression through the regulation of the CDC27/cyclinB1 signaling axis, thereby enhancing the malignant phenotype of the tumor. Our study indicates that the ITCH/SIX1/CDC27/cyclinB1 pathway may play a crucial role in tumor proliferation in nasopharyngeal carcinoma, offering new insights for clinical targeted therapies.
Materials and methods
Bioinformatics analysis
We integrated GEO series (GSE) datasets from the same GEO platform (GPL). The datasets GSE64634 and GSE12452 from GPL570, contained 14 cases of normal nasopharyngeal epithelial tissues and 43 cases of nasopharyngeal tumor tissues. The original count matrix rows were analyzed via the DESeq2 package of R software. |log2(fold change) | >1 and P. adj < 0.05 were used as the significance criteria. Functional clustering was conducted using online analysis software (Microbiology Letter; http://www.bioinformatics.com.cn/). The dataset GSE13597 from GPL96, contained 3 cases normal nasopharyngeal tissue samples and 25 nasopharyngeal tumor tissues samples. |log2(fold change) | >1 and P < 0.05 were used as the significance criteria. Poor prognostic outcome analysis was performed via GEPIA 2 (http://gepia2.cancer-pku.cn/#index).
Cell culture
NP69(C1280, WHELAB, Shanghai, China), 6-10B (C1651, WHELAB, Shanghai, China), HK1(WARNER, Wuhan, Hubei, China), and HNE1(C1794, WHELAB, Shanghai, China) cell lines were obtained from Shanghai Meiwan Biotechnology Co. The 6-10B, and HNE1 cells were grown in RPMI-1640 (Servicebio), and the HK1 cells were grown in high-glucose DMEM (Servicebio) supplemented with FBS at a final concentration of 10%. The cells were cultured in an incubator under standard conditions. Cell passaging and seeding were conducted when the cell coverage reached 70–85%.
Western blot
Western blot assays were performed as previously described. Proteins were detected using the following antibodies: anti-SIX1 antibodies (dilution, 1:1000; cat.16960 S; Cell Signaling Technology, Danvers, MA, USA), anti-CDC27 antibodies (dilution 1:1000; cat.sc9-972; Santa Cruz Biotechnology, Dallas, Texas, USA), anti-cyclin B1 (dilution 1:1000; 4138-T; Cell Signaling Technology, Danvers, MA, USA), anti-β-Tubulin(dilution 1:20000, ANT326, Antgene, Wuhan, Hubei, China) and anti-ITCH (dilution 1:3000; ER1901-94; Huabio; Wuhan, Hubei, China).
PCR assay
TRIzol reagent was used to extract total RNA, and cDNA synthesis was performed according to the protocol of the transcriptase kit (Vazyme Biotech, Nanjing, China). Quantitative reverse transcription polymerase chain reaction (qRT-PCR) was conducted on a CFX96 Connect instrument (Bio-Rad, Hercules, CA) and a SYBR Green PCR kit (Servicebio, Hubei, China). The mRNA expression levels were calculated by the 2−ΔΔCt method. The primer sequences are provided in Supplementary Table S1.
Molecular docking
The predicted structures of ITCH and SIX1 were generated using AlphaFold. Subsequently, AutoDockTools-1.5.7 was employed to manually optimize the two protein structures through processes such as dehydration and hydrogenation. The docking server ZDock was subsequently utilized for protein-protein docking. The resulting protein-protein complex underwent further manual optimization, which included water removal and hydrogenation via AutoDockTools-1.5.7. Finally, PyMOL was employed to predict protein interactions and create diagrams of the protein-protein interaction. In PyMOL, we selected the highest ZDock score of 1634.178 for visualization. ITCH is represented as a purple model, while SIX1 is depicted as a cyan model with their binding sites illustrated as stick structures in the corresponding colors.
Immunofluorescence
The cells were fixed following membrane rupture and subsequently blocked with 5% BSA for one hour. In accordance with the antibody instructions, SIX1 (dilution 1:200, cat.16960s, Cell Signaling Technology, Danvers, MA, USA) and ITCH (dilution 1:200, cat. 67757-1-Ig, Proteintech, Hubei, China) antibodies were prepared. After an overnight incubation with the antibodies at 4 °C, the cells were washed with PBS, incubated for 1 h with a fluorescent secondary antibody at room temperature, and were washed again with PBS. The cell nucleus was stained with DAPI, and after a final wash with PBS, the slides were mounted and photographed using a Zeiss 980 confocal camera(Carl Zeiss AG, Jena).
Transient transfection
The plasmid information was presented in Supplementary Table S2. Plasmid transfection was conducted according to the instructions of PEI 40 K Transfection Reagent (Servicebio, G1802, Wuhan, Hubei, China). After 48 h of transfection, Western Blot was conducted to verify the transfection efficiency.
Stable transfection
The plasmid information was presented in Supplementary Table S2. Plasmid transfection was carried out following the instructions of PEI 40 K Transfection Reagent (Servicebio, G1802, Wuhan, Hubei, China). After 48 h of lentiviral transduction, 1 µg/ml puromycin was added for 24–48 h of screening. Western blotting was used to verify whether the cells successfully overexpressed or silenced SIX1.
Co-immunoprecipitation and immunoblot assay
The cells were inoculated into 6 cm dishes and transfected with plasmids. The ratio of plasmid to PEI was 1:3. After 6 h of transfection, the medium was replaced with a fresh medium, and the cells were cultured for 48 h. The cells were collected for COIP. Proteins were extracted according to the steps of the kit used (Protein A/G Magnetic Beads; cat. HY-K0202; MedChemExpress) and verified by Western blotting.
Cell counting kit-8
NPC cell lines which SIX1 was knocked down or overexpressed and control cells were seeded into 96-well plates at 1000 cells per well. The medium was replaced with a fresh medium containing 10% cell CCK-8 reagent (Biosharp, cat.BS350A, Hefei, China), and the cells were incubated at 37 °C for 1 h. Then, cell viability was assessed at 450 nm.
Colony formation
NPC cell lines in which SIX1was knocked down or overexpressed and control cells were seeded at 500 cells per well into six-well plates. The cell colonies were fixed and stained with 0.3% crystal violet in ethanol for 15 min until the colonies were visible to the naked eye. Finally, the whole well was photographed.
Transwell invasion assay
Cell invasion was measured using Matrigel-precoated Transwell inserts (cat. 353097; Corning; Becton Dickinson and Company, New Country West, USA). The chambers were pretreated with Matrigel (cat.354234; Becton, Dickinson and Company, New Country West, USA). The cells were seeded in the Transwell inserts at a concentration of 1 × 106 cells per well and incubated for 48 h. Each assay was performed in triplicate.
Transwell migration assay
Cell migration was assessed via Transwell inserts. NPC cell lines with knocked down or overexpressed SIX1 and control cells were seeded into Transwell inserts at a concentration of 5 × 105 cells per well and incubated overnight. Each assay was performed in triplicate.
Animal study
Three- to four-week-old male nude mice were bred at the animal center of Zhongnan Hospital of Wuhan University. The procedures were approved by the institutional animal care and use committees. A total of 6-10B cells at a concentration of 5 × 106 were subcutaneously inoculated into each mouse, including SIX1-knockdown and control 6-10B cells. After 7 days, the tumor volume was recorded every 2 days and calculated using the formula a × a × b × 0.5 (a represents the prolate axis and b represents the brachy axis). All mice were euthanized and weighed after CO2 inhalation and cervical dislocation. The tumors were extracted from the subcutaneous tissues and weighed. The tumor tissues were fixed with 4% paraformaldehyde, embedded in paraffin, and sliced for HE and immunohistochemistry staining.
Hematoxylin & eosin staining
Hematoxylin & eosin staining was performed using an HE staining kit (G1076, Servicebio).
Immunohistochemistry
The tumor tissues were fixed and dehydrated using gradient concentrations of ethanol. Then, IHC staining was conducted according to the instructions provided in the Maixin IHC kit, Fuzhou, China). Anti-Ki-67 antibodies (dilution, 1:500; cat.GB111141; 100; Servicebio, Hubei, China), anti-SIX1 antibodies (dilution, 1:100; cat.16960 S; Cell Signaling Technology, Danvers, MA, US), anti-CDC27 antibodies (dilution 1:500; cat.sc-9972; Santa Cruz Biotechnology, Dallas, Texas, US), and anti-cyclin B1 (dilution, 1:100; cat.4-138T; Cell Signaling Technology, Danvers, MA, US) were incubated overnight at 4 °C. Finally, ImageJ v1.8.0 (National Institutes of Health, Bethesda, MA) was used for analysis.
RNA‑seq
Total RNA was extracted from SIX1-overexpressing and control cells for transcriptome sequencing at BGI (Shenzhen, China; three independent replicates per group). Relative analysis was carried out using the Tom system (https://biosys.bgi.com).
Cell cycle assay
NPC cell lines with knocked down or overexpressed SIX1 and control cells from the six-well plates were collected and stained according to the instructions of the kits used (cat. CCS012; MULTI SCIENCES, Hangzhou, China). After processing, the samples were transferred to flow tubes for machine testing (Cytoflex, BECKMAN).
Data analysis
The Data are presented as means and standard deviations from at least three repeats. To determine the statistical probabilities, we used unpaired Student’s t-tests or one-way ANOVA. Statistical analysis was performed with GraphPad Prism 9.0. Differences between groups were considered statistically significant when p < 0.05.
Data availability
Sequence data that support the findings of this study have been deposited in the Genome Sequence Archive with the accession numbers HRA006795.A direct link to the deposited data: https://ngdc.cncb.ac.cn/gsa-human/s/u4ltOu54.
Abbreviations
- CDC27:
-
Cell division cycle 27
- CDKs:
-
Cyclin-dependent kinases
- CHX:
-
Cycloheximide
- GPL:
-
GEO platforms
- GSE:
-
GEO series
- ITCH:
-
Itchy E3 ubiquitin ligaseE3
- NPC:
-
Nasopharyngeal carcinoma
- SIX1:
-
SIX homeobox 1
References
You, R. et al. Hyperfractionation compared with standard fractionation in intensity-modulated radiotherapy for patients with locally advanced recurrent nasopharyngeal carcinoma: a multicentre, randomised, open-label, phase 3 trial. Lancet. 401, 917–927 (2023).
Chua, M. L. K., Lee, V. H. F. & Lee, A. W. M. Hyperfractionation for reirradiation of recurrent nasopharyngeal carcinoma. Lancet. 401, 878–879 (2023).
Mao, Y. P. et al. Medial retropharyngeal nodal region sparing radiotherapy versus standard radiotherapy in patients with nasopharyngeal carcinoma: open label, non-inferiority, multicentre, randomised, phase 3 trial. BMJ. 380, e072133 (2023).
Lv, J. et al. The tumor immune microenvironment of nasopharyngeal carcinoma after gemcitabine plus cisplatin treatment. Nat. Med. 29, 1424–1436 (2023).
Kang, Y. et al. Advances in targeted therapy mainly based on signal pathways for nasopharyngeal carcinoma. Signal. Transduct. Target. Ther. 5, 245 (2020).
Thornton, J. Oncology at CERN: accelerating radiotherapy. Lancet. 401, 92–93 (2023).
Perillo, M., Swartz, S. Z., Pieplow, C. & Wessel, G. M. Molecular mechanisms of tubulogenesis revealed in the sea star hydro-vascular organ. Nat. Commun. 14, 2402 (2023).
Fujimoto, Y. et al. Homeoproteins Six1 and Six4 regulate male sex determination and mouse gonadal development. Dev. Cell. 26, 416–430 (2013).
Liu, Z. et al. A NIK-SIX signalling axis controls inflammation by targeted silencing of non-canonical NF-κB. Nature. 568, 249–253 (2019).
Li, L. et al. Transcriptional regulation of the Warburg effect in cancer by SIX1. Cancer Cell. 33, 368–385e367 (2018).
Liu, W. et al. Homeoprotein SIX1 compromises antitumor immunity through TGF-β-mediated regulation of collagens. Cell. Mol. Immunol. 18, 2660–2672 (2021).
Hughes, C. J. et al. SIX1 and EWS/FLI1 co-regulate an anti-metastatic gene network in Ewing Sarcoma. Nat. Commun. 14, 4357 (2023).
Liu, Y. et al. SNS-023 sensitizes hepatocellular carcinoma to sorafenib by inducing degradation of cancer drivers SIX1 and RPS16. Acta Pharmacol. Sin. 44, 853–864 (2023).
Sui, H. Y. et al. LRRC75A-AS1 delivered by M2 macrophage exosomes promotes cervical cancer progression via enhancing SIX1 expression. Cancer Sci. 114, 2634–2649 (2023).
Stevenson, M. J. et al. Altered binding affinity of SIX1-Q177R correlates with enhanced WNT5A and WNT pathway effector expression in Wilms tumor. Dis. Model. Mech. 16, dmm050208 (2023).
Li, T. et al. Nucleosome assembly protein 1 like 1 (NAP1L1) promotes cardiac fibrosis by inhibiting YAP1 ubiquitination and degradation. MedComm. 4, e348 (2023).
Cockram, P. E. et al. Ubiquitination in the regulation of inflammatory cell death and cancer. Cell. Death Differ. 28, 591–605 (2021).
Trier, A. M. et al. IL-33 potentiates histaminergic itch. J. Allergy Clin. Immunol. 153, 852–859e853 (2023).
Patel, T. H. et al. FDA approval summary: tremelimumab in combination with durvalumab for the treatment of patients with unresectable hepatocellular carcinoma. Clin. Cancer Res. 30, 269–273 (2023).
Zhou, B. et al. TRIM16 promotes aerobic glycolysis and pancreatic cancer metastasis by modulating the NIK-SIX1 axis in a ligase-independent manner. Am. J. Cancer Res. 12, 5205–5225 (2022).
Jiang, S. et al. A polysaccharide from native Curcuma kwangsiensis and its mechanism of reversing MDSC-induced suppressive function. Carbohydr. Polym. 297, 120020 (2022).
Solvie, D. et al. MYC multimers shield stalled replication forks from RNA polymerase. Nature. 612, 148–155 (2022).
Matthews, H. K., Bertoli, C. & de Bruin, R. A. M. Cell cycle control in cancer. Nat. Rev. Mol. Cell. Biol. 23, 74–88 (2022).
Dong, B., Yi, M., Luo, S., Li, A. & Wu, K. RDGN-based predictive model for the prognosis of breast cancer. Exp. Hematol. Oncol. 9, 13 (2020).
Wu, K. et al. EYA1 phosphatase function is essential to drive breast cancer cell proliferation through cyclin D1. Cancer Res. 73, 4488–4499 (2013).
He, A. et al. Polycomb repressive complex 2 regulates normal development of the mouse heart. Circ. Res. 110, 406–415 (2012).
Yang, X. et al. miR-489-3p/SIX1 axis regulates melanoma proliferation and glycolytic potential. Mol. Ther. Oncolytics. 16, 30–40 (2020).
Wei, D. et al. Transcriptional regulation by CpG sites methylation in the core promoter region of the bovine SIX1 gene: roles of histone H4 and E2F2. Int. J. Mol. Sci. 19, 213 (2018).
Feng, D. et al. CARM1 drives triple-negative breast cancer progression by coordinating with HIF1A. Protein Cell. 13, pwae010 (2024).
Ropa, J. et al. SETDB1 mediated histone H3 lysine 9 methylation suppresses MLL-fusion target expression and leukemic transformation. Haematologica. 105, 2273–2285 (2020).
Rafiq, A., Aashaq, S., Jan, I. & Beigh, M. A. SIX1 transcription factor: a review of cellular functions and regulatory dynamics. Int. J. Biol. Macromol. 193, 1151–1164 (2021).
Zhang, Z. et al. Novel LncRNA LINC02936 suppresses ferroptosis and promotes tumor progression by interacting with SIX1/CP axis in endometrial cancer. Int. J. Biol. Sci. 20, 1356–1374 (2024).
Zhang, P. et al. BCLAF1 drives esophageal squamous cell carcinoma progression through regulation of YTHDF2-dependent SIX1 mRNA degradation. Cancer Lett. 591, 216874 (2024).
Liao, Y. et al. A new role of GRP75-USP1-SIX1 protein complex in driving prostate cancer progression and castration resistance. Oncogene. 40, 4291–4306 (2021).
Rafiq, A. et al. GSK3β phosphorylates Six1 transcription factor and regulates its APC/C(Cdh1) mediated proteosomal degradation. Cell. Signal. 115, 111030 (2024).
Chu, Y. et al. O-GlcNAcylation of SIX1 enhances its stability and promotes hepatocellular carcinoma proliferation. Theranostics. 10, 9830–9842 (2020).
Yang, M. et al. A converged ubiquitin-proteasome pathway for the degradation of TOC and TOM tail-anchored receptors. J. Integr. Plant. Biol. 66, 1007–1023 (2024).
Liu, D. et al. Correction: SIX1 promotes Tumor Lymphangiogenesis by coordinating TGFβ signals that increase expression of VEGF-C. Cancer Res. 79, 1715 (2019).
Hsu, J. Y. et al. SIX1 reprograms myogenic transcription factors to maintain the rhabdomyosarcoma undifferentiated state. Cell. Rep. 38, 110323 (2022).
Jessen, M. et al. Inhibition of the YAP-MMB interaction and targeting NEK2 as potential therapeutic strategies for YAP-driven cancers. Oncogene. 43, 578–593 (2024).
Coletta, R. D. et al. The Six1 homeoprotein stimulates tumorigenesis by reactivation of cyclin A1. Proc. Natl. Acad. Sci. U S A. 101, 6478–6483 (2004).
Li, Z. et al. Six1 promotes proliferation of pancreatic cancer cells via upregulation of cyclin D1 expression. PLoS One. 8, e59203 (2013).
Funding
This work was supported by grants from the National Natural Science Foundation of China (Grant number 8207040451); Zhongnan Hospital of Wuhan University, Excellent Doctor Fund Project (Grant number ZNYB2022004); Zhongnan Hospital of Wuhan University, Technology and Innovation Seed Fund (Grant number CXPY2020046), and Zhongnan Hospital of Wuhan University Excellent Doctor (Postdoctoral) Fund (ZNYB2022004).
Author information
Authors and Affiliations
Contributions
Z.L., and W.C. designed the project and performed experiments, analyzed data, and wrote the paper; B.H., and Y.S. carried out molecular biology and bioinformatic analyses and in vitro and vivo experiments; Y.Hu., and S.H. analyzed data; J.L., and X.C. supervised the progress of the study. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
The study was approved by the Experimental Animal Ethics Committee of Zhongnan Hospital of Wuhan University (NO.02523039 A), and we confirm that all experiments were performed in accordance with relevant named guidelines and regulations complied with the ARRIVE guidelines.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lin, Z., Cai, W., Sun, Y. et al. Implications of ITCH-mediated ubiquitination of SIX1 on CDC27-cyclinB1 signaling in nasopharyngeal carcinoma. Sci Rep 14, 24140 (2024). https://doi.org/10.1038/s41598-024-73239-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-73239-5
This article is cited by
-
Identification and validation of CDC20 and ITCH as ubiquitination related biomarker in idiopathic pulmonary fibrosis
Hereditas (2025)
-
Necroptosis in cancer: insight from epigenetic, post-transcriptional and post-translational modifications
Journal of Hematology & Oncology (2025)