Abstract
Isolation mechanisms between mosquito species of the Anopheles gambiae complex, which includes major malaria vectors, remain poorly understood. In some cases, pre-zygotic barriers have been shown to limit gene flow between species of the complex, leading to a low level of hybridisation in nature. Post-zygotic mechanisms manifest with F1 hybrid males fully sterile and F1 hybrid females with reduced fertility. Genetic approaches combined with DNA sequencing techniques have highlighted the involvement of genomic regions in hybrid incompatibility with a predominant role of the X chromosome. In addition, differences in the phenotype of F1 hybrid males have been identified depending on the directionality of the parental cross used to generate them. All these studies have focused on the interaction of nuclear DNA elements in hybrid individuals. Given the role that mitochondrial DNA plays in genetic incompatibilities within other organisms and its unique inheritance pattern, commonly maternal, we conducted a genetic study that relied on the introgression of mitochondrial DNA between Anopheles gambiae and Anopheles arabiensis. The findings indicate that the mitochondrial switch does not appear to restore the fertility of F1 hybrid males, suggesting that mitochondrial DNA may not play a role in hybrid incompatibilities in these Anopheles species.
Similar content being viewed by others
Introduction
Malaria continues to pose a substantial global health challenge, with an annual infection rate exceeding 200 million individuals and an estimated half a million fatalities reported each year. The African continent continues to experience the most significant impact of this burden1.
Major vectors of malaria in Africa belong to the Anopheles gambiae sensu lato (s.l.) (Diptera: Culicidae) species complex, which includes nine morphologically indistinguishable species of mosquitoes that significantly differ in their vectorial capacity2,3.
Prezygotic isolating mechanisms have been observed in some of these species, resulting in assortative mating. These factors, which are still poorly understood, include temporal, ecological, behavioural, and mechanical isolation4,5,6,7,8,9,10,11,12. In addition, post-zygotic isolation mechanisms, in the form of interspecific hybrid incompatibilities (HIs), have been identified and act to restrict gene flow: interspecific crosses between members of the complex produce sterile F1 hybrid males13,14,15,16,17. A notable exception to this pattern can be found in crosses between An. gambiae and An. coluzzii, the youngest species pair to diverge within the complex18. In accordance with Haldane’s rule, while F1 hybrid males express full sterility, homogametic F1 female hybrids between species of the complex are fertile, conditional on viability, providing still poorly understood opportunities for gene flow3,18,19,20,21,22,23. Nevertheless, both entomological and molecular methods suggest that hybridisation rates in African populations of Anopheles mosquitoes are relatively low9,24,25,26,27,28,29.
One of the most prominent models of speciation is based on the concept that mutations can occur and accumulate within the genomes of isolated populations. These mutations can confer an adaptive advantage or be neutral within these populations; however, if hybridisation between previously isolated populations occurs, the presence of these alleles in a hybrid genetic background could give rise to deleterious effects such as unviability, sterility or, more generally, fitness costs. The detrimental effects due to these genetic variants are known as Dobzhansky-Muller or Bateson-Dobzhansky-Muller incompatibilities (BDMIs), and examples of their significance in HIs and reproductive isolation are well documented30,31,32,33,34,35,36,37.
Several studies have attempted to elucidate the genetic mechanisms involved in HIs within the Anopheles gambiae complex. Introgression experiments have ruled out the role of the Y chromosome in HIs for An. gambiae and An. arabiensis hybrids15. On the other hand, in hybrids between these species, as well as between An. coluzzii and An. quadriannulatus, a prominent role of the X chromosome, has been highlighted38. In addition, autosomal regions have been identified as having an effect on hybrid fertility to various extents14,39,40.
A common feature of all genetic elements so far studied is that they are determined by nuclear DNA. However, several examples of reproductive barriers in eukaryotic organisms involve interactions between nuclear and mitochondrial genes41,42,43.
Mitochondria are membrane-bound cell organelles that, in eukaryotic cells, supply chemical energy for biological processes. These organelles carry a genome (mitogenome) that is inherited independently of the nucleus, and they are typically small and characterised by low levels of recombination. Interactions between mitochondrial and nuclear genes are crucial for proper cell function, and misregulation can cause severe pathological effects44. Importantly, mitochondrial genomes are uniparentally, usually maternally, inherited45. These unique features indicated that mitogenome-nuclear DNA should be considered as a candidate for the manifestation of interspecific BDMIs46. Furthermore, due to the maternal inheritance pattern, mutations in the mitochondrial genome are only subjected to natural selection in females. This means harmful mutations that affect males can persist in populations if beneficial or neutral in females. In males, selective pressure would favour the evolution of nuclear variants that restore mitonuclear interactions. This phenomenon, known as the Mother’s Curse47,48,49, could contribute to BDMIs.
Interestingly, Liang & Sharakhov (2019) and Liang et al., (2021) recently investigated F1 hybrid male sterility between An. merus and An. gambiae or An. coluzzii. In accordance with previous observations, the authors showed that while F1 hybrid males resulting from either reciprocal parental cross were fully sterile, an asymmetrical sex bias was observed in the offspring depending on the directionality of the parental cross50,51. In addition, an asymmetrical pattern also manifested in the cytological mechanisms underlying the sterility of F1 hybrid males (Darwin’s corollary)16,17. Given that autosomes are shared between male and female mosquitoes, the phenotype difference relative to the genetic crosses’ directionality could be attributed to the sex chromosomes. Alternatively, the mitogenome could be involved in these mechanisms.
Due to its high mutation rate (2 to 6 times faster than nuclear DNA in non-vertebrates), mitochondrial DNA has often been studied to investigate speciation and evolution52,53. In 2019, Hanemaaijer and colleagues identified 783 single nucleotide polymorphisms (SNPs) in the mitogenome of field-collected An. arabiensis, An. gambiae and An. coluzzii54. None of these SNPs was unique to either species, suggesting that there is no divergent selection in the mitogenome among these species of the Anopheles gambiae complex, possibly due to hybridisation events. These results suggest that the mitogenome may not play a role in HIs54.
However, empirical studies that follow a traditional route of investigation that account for the complexity of biological factors have, to our knowledge, never been performed to investigate cytonuclear interactions in mosquitoes.
This study used a genetic cross scheme to introduce the mitogenome deriving from An. arabiensis into an An. gambiae genetic background and the mitogenome deriving from An. gambiae into an An. arabiensis genetic background. These genetic makeups allowed generations of F1 hybrids with a switch in their mitochondrial DNA composition and the possibility of exploring the role of mitochondria in HIs between these mosquito species.
Results
Cytology of reproductive organs in F1 hybrid males of an. Gambiae and An. Arabiensis
We employed two laboratory strains to investigate hybrid incompatibility in Anopheles mosquitoes: An. gambiae G3 (referred to as An. gambiae hereafter) is a hybrid strain of An. gambiae and An. coluzzii, and An. arabiensis Dongola (referred to as An. arabiensis hereafter).
We and others have previously shown that, in accordance with Haldane’s rule, F1 hybrid males generated between these strains displayed full sterility14,15,40,55.
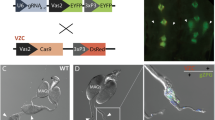
Interestingly, the severity of the morphological anomalies of the reproductive organs underlying this sterility varied according to the directionality of the parental cross. F1 hybrid males resulting from a cross of An. gambiae females to An. arabiensis males showed testes that were similar in shape and size to those of Wild Type (WT) males of either species but appeared to contain mostly large, round, undifferentiated cells. This phenotype was termed ‘marbled’ (Fig. 1A). When testes of this hybrid male cohort were squashed using a glass coverslip, some spermatozoa with partial tails were observed, though none were mature or motile (Figure S1).
Whole mount DAPI staining showing the cytological aspect of testes associated with reciprocal crosses between An. gambiae and An. arabiensis. Representative chromosomes are labelled with X (X chromosome), Y (Y chromosome) and A (autosome). Red indicates An. gambiae DNA, blue indicates An. arabiensis DNA. (A) WT An. arabiensis testis dissected from adult males show different stages of spermatogenesis. Large spermatocyte nuclei can be detected in the upper part of the testis. Mature, arrow-shaped spermatozoa are present in the sperm reservoir in the lower part of the organ. F1 hybrid males resulting from a cross of An. gambiae females to An. arabiensis males have ‘marbled’ testes where the characteristic spermatogenesis stages and mature spermatozoa are absent. (B) Testes dissected from WT An. gambiae adult males match the cytological phenotype described for WT An. arabiensis males. F1 hybrid males were generated from a cross of An. arabiensis females to An. gambiae males. At the pupal stage, cells at different stages of spermatogenesis can be seen, however, sperm cells show chromatin condensation defects and are larger in size compared to sperm in WT individuals. Following the transition from pupa to adult mosquito, testes undergo progressive degeneration, their size is significantly reduced (‘atrophic’) and the different stages of spermiogenesis are poorly defined.
Conversely, F1 hybrid males resulting from a cross of An. arabiensis females and An. gambiae males showed much smaller testes than WT males and were largely deprived of cellular components. This phenotype has been termed ‘atrophic’ (Fig. 1B). Notably, testes dissected from this cohort of hybrid males at the pupal stage seemed normal, indicating a progressive organ degeneration between the pupal stage and early adulthood (Fig. 1B). In pupae, DAPI staining revealed the presence of distinct developmental stages of spermatocytes and spermatozoa with chromatin condensation defects; none of these cells were motile (Figures S2, S3).
Generation of mosquito strains with interspecific mitochondrial and nuclear DNA
A scheme of introgressive hybridisation was employed to introduce An. arabiensis mitochondria into the genomic background of An. gambiae (Fig. 2A).
Schematics for the interspecific introgression of mitochondrial DNA. Representative chromosomes are labelled with X (X chromosome), Y (Y chromosome) and A (autosome). Red indicates An. gambiae DNA, blue indicates An. arabiensis DNA, grey indicates hybrid DNA and green denotes the X-linked GFP transgene. (A) Introgression of An. arabiensis mitochondria into an An. gambiae genetic background. WT An. arabiensis females were crossed to An. gambiae X-GFP males in cage. F1 hybrid females were backcrossed to WT An. gambiae males, and transgenic BC1 hybrid females were selected to continue the introgression by repeated backcrosses to WT An. gambiae males. The X-linked GFP transgene is no longer used for the selection of the individuals and is eventually lost. (B) Introgression of An. gambiae mitochondria into an An. arabiensis genetic background. X-GFP An. gambiae females were crossed to WT An. arabiensis males in cage. F1 hybrid females were backcrossed to WT An. arabiensis males and non-transgenic BC1 hybrid females were selected to continue the introgression by repeated backcrosses to WT An. arabiensis males.
WT An. arabiensis females were crossed en masse to transgenic An. gambiae males carrying a fluorescent marker on the X chromosome (X-GFP strain). Transgenic F1 hybrid females were then backcrossed to WT An. gambiae males en masse to generate backcross generation 1 (BC1) progeny. From this cohort, transgenic females, which had inherited one set of chromosomes from their WT An. gambiae fathers and one set of chromosomes that resulted from meiotic recombination in their F1 hybrid mothers, were selected.
Recombination rates between interspecific X chromosomes have been shown to be very low in anopheline mosquitoes14,38,39,40. As a result, the presence of the X-linked GFP marker was used to select BC1 females that had inherited the An. gambiae X chromosome from their F1 hybrid mothers, thus eliminating the An. arabiensis X chromosome from this introgression (Fig. 2A).
To further decrease the amount of nuclear An. arabiensis DNA in the introgressed strain, BC1 females were backcrossed to WT An. gambiae males, and this backcrossing scheme was employed for over 20 subsequent generations (more than one year). By selecting female progeny for each subsequent backcross, we ensured that the An. arabiensis mitochondrial DNA was retained and at generation BC26, this strain was expected to contain the An. arabiensis mitochondrial DNA within an An. gambiae genomic background. We refer to this strain as GamAm (Fig. 2A).
In parallel with this introgression experiment, the same strategy was used to generate a strain called AraGm, which carries An. gambiae mitochondrial DNA in an An. arabiensis genetic background (Fig. 2B).
Generation of F1 hybrids containing heterospecific X-chromosomes and mitochondrial DNA
This study aimed to investigate any possible effect of the presence of heterospecific X-chromosomes and mitochondrial DNA in the genomic context of F1 hybrid males. Therefore, we designed a scheme of genetic crosses utilising the introgressed strains GamAm and AraGm, as well as WT An. arabiensis and An. gambiae. By using this strategy, we produced hybrid males that inherited a set of chromosomes (autosomes/X chromosome) from one parental species in conjunction with a set of chromosomes (autosomes/Y chromosome) and mitochondrial DNA from the other species (Figs. 3A and 4A).
Genetic crosses performed to assess the effect of An. arabiensis mitochondria in F1 hybrid males containing the An. gambiae X chromosome. Representative chromosomes are labelled with X (X chromosome), Y (Y chromosome) and A (autosome). Red indicates An. gambiae DNA, blue indicates An. arabiensis DNA. (A) GamAm females were crossed in cage to WT An. arabiensis males to generate GamAm/Ara F1 hybrid males containing An. arabiensis mitochondria and An. gambiae X chromosomes. (B) GamAm males were crossed to WT An. gambiae females in cage to generate GamGm females which had the same genomic species-composition as the GamAm strain but contained An. gambiae mitochondria. These females were crossed to WT An. arabiensis males in cage to generate GamGm/Ara F1 hybrid males containing both the mitochondria and the X chromosome from An. gambiae. (C) As a control, F1 hybrid males were generated by crossing WT An. arabiensis males to WT An. gambiae females in cage. The Gam/Ara F1 hybrid males contained a set of autosomes from each species and mitochondria from An. gambiae.
Experimental crosses to assess the effect of An. gambiae mitochondria on testes phenotype and fertility in F1 hybrid males containing the An. arabiensis X chromosome. Representative chromosomes are labelled with X (X chromosome), Y (Y chromosome) and A (autosome). Red indicates An. gambiae DNA, blue indicates An. arabiensis DNA. (A) AraGm females were crossed in cage to WT An. gambiae males to generate AraGm/Gam F1 hybrid males containing An. gambiae mitochondria and the An. arabiensis X chromosome. (B) AraGm males were crossed to WT An. arabiensis females in cage to generate AraAm females which had the same genomic species composition as the AraGm strain but contained An. arabiensis mitochondria. These females were crossed to WT An. gambiae males in cage to generate AraAm/Gam F1 hybrid males containing both the mitochondria and the X chromosome from An. arabiensis. (C) As a control, F1 hybrid males were generated by crossing WT An. gambiae males to WT An. arabiensis females in cage. The Ara/Gam F1 hybrid males contained a set of autosomes from each species and mitochondria from An. arabiensis.
To ensure that any alteration of the sterility phenotype observed in these hybrid males was the result of the mitochondrial DNA switch rather than incomplete DNA introgression in the genome of GamAm and AraGm strains, we set up genetic crosses that aimed at generating hybrid males into which mitochondrial DNA had been reintroduced to match the species-of-origin of the X chromosomes (Figs. 3B and 4B).
In parallel, we generated F1 hybrid males from WT An. arabiensis and An. gambiae strains to serve as a control (Figs. 3C and 4C).
Hybrid males of the experimental and control groups were crossed en masse to WT An. gambiae females to assess their fertility. Moreover, testes were dissected and analysed to determine their phenotype.
Sterility in F1 hybrid males is not affected by the presence of heterospecific mitochondria and X chromosomes
Upon mating with hybrid males and blood feeding, WT An. gambiae female mosquitoes from the experimental and control cages were allowed to lay eggs. The number of eggs deposited and the hatching rate (number of larvae/number of eggs) were used to assess the fertility of the cohorts of F1 hybrid males.
A total of 4665 and 3738 eggs were recovered from the experimental cages of the GamAm and AraGm strains, respectively. No hatching of eggs was observed in either case, indicating the presence of full sterility in these males regardless of the mitochondrial genomes (Fig. 5A and D). In addition, testes in these hybrids displayed the phenotypes expected based on the species identity of the X chromosome. As such, hybrid males carrying the An. gambiae X chromosome displayed the marbled phenotype, while those carrying the An. arabiensis X chromosome were found to contain atrophic testes. Testes phenotypes, therefore, did not differ between the cohorts of introgressed-strain hybrids and the respective WT control hybrid males (Figs. 1 and 5B, C, E and F).
Result of fertility assays and analysis of testes phenotypes associated with F1 hybrids with different genetic background. Representative chromosomes are labelled with X (X chromosome), Y (Y chromosome) and A (autosome). Red indicates An. gambiae DNA, blue indicates An. arabiensis DNA. Testes were dissected at pupal stage for all the males analysed. (A, B) and C show the outcome of the genetic crosses described in Fig. 3. All cohorts of F1 hybrid males that were analysed showed full sterility (no eggs hatching) and the cytological phenotype of their testes was classified as ‘marbled’ with no evident differences to the control samples (C). D, E and F show the outcome of the crosses described in Fig. 4. AraGm/Gam and AraAm/Gam F1 hybrid males (D and E) showed full sterility (no eggs hatching) while from the cross of Ara/Gam F1 hybrid males (F) a single larva hatched, likely due to experimental error. The cytological phenotype of the testes in these male cohorts showed no evident differences relative to the control samples (F). Scale bar 100 μm.
Notably, a single larvae hatched from the cross of WT F1 hybrid males containing An. arabiensis X chromosomes (4161 eggs; HR: 0.00024) (Fig. 5F). The full sterility of F1 hybrid males with this genetic composition has been well-documented and established in countless experiments within our lab. We cannot rule out that this event was the result of contamination.
Verifying the species-of-origin of the mitochondrial DNA
The mitochondrial introgressions in this study relied on repeated backcrosses of hybrid female mosquitoes to An. gambiae WT males. We wanted to exclude the possibility that errors had occurred over the course of the introgression that may have contaminated the mitochondrial DNA content in the cohorts of hybrid males.
Mitochondrial DNA sequencing data, obtained via whole genome sequencing, was analysed from individual and pooled An. gambiae G3 and An. arabiensis Dongola strain mosquitoes from our laboratory colonies. A total of 5 single nucleotide polymorphisms (SNPs) were identified that were fixed within the respective strains and could serve as markers to identify the species-of-origin of the mitochondria (Table 1).
DNA was extracted from different cohorts of hybrid males (6 GamAm /Ara, 8 GamGm /Ara, 8 Gam/Ara, 8 AraGm/Gam, 10 AraAm/Gam and 10 Ara/Gam F1 hybrid males) and a region encompassing two of the SNP markers (at positions 7255 and 7654) was selected for PCR amplification and Sanger sequencing (Supplementary File 1). The investigation into the genotypes of the samples at the marker positions revealed a pattern for the mitochondrial DNA in line with the expectation, ruling out the possibility that contamination had taken place over the course of the introgression experiments.
Discussion
This study aimed to explore the role of mitochondrial DNA in hybrid incompatibility between An. gambiae and An. arabiensis.
It is important to note that the An. gambiae strain (G3) employed in these experiments is a hybrid of An. gambiae and Anopheles coluzzii. However, these two species are the most recently diverged in the Anopheles gambiae complex and the only exception to Haldane’s rule as they produce fertile hybrids of both sexes56,57. Therefore, it was not anticipated that this strain’s genetic background would compromise the study’s primary objective (assessing rescue of fertility in F1 hybrid males through a switch in the mitochondrial DNA).
In addition, our investigation focused on F1 hybrid males. In this genetic background, hybrid incompatibilities lead to full sterility. Any fertility restoration would have been evident through the production of offspring in crosses involving large numbers of males. On the other hand, assessing the impact on fertility in F1 hybrid females, which exhibit varying fertility levels, would have been challenging. Nevertheless, this aspect could be explored in future research.
The generation of F1 hybrid males between An. gambiae and An. arabiensis confirmed Haldane’s rule and full sterility19. However, cytological defects in their testes vary depending on the directionality of the parental cross. F1 hybrid males resulting from a cross of An. gambiae females to An. arabiensis males exhibit testes with a marbled phenotype. The reciprocal parental cross produces F1 hybrid males whose testes are atrophic16,17,55. While these findings have previously been described, we show that the atrophic testes phenotype of F1 hybrid males does not manifest until males reach adulthood. Notably, at the pupal stage, the testes of these hybrids clearly show the presence of meiotic chromosomes and sperm, which can develop despite DNA condensation defects. These observations suggest the involvement of genetic elements that operate late in development, potentially leading to the degeneration of the reproductive organs.
Regardless of the parental cross, F1 hybrid offspring inherit interspecific autosomes, one set from each parental species. This excludes interautosomal incompatibilities from being responsible for the difference in the testis’s phenotype. In addition, previous research has shown that the Y chromosome functions interchangeably between An. gambiae and An. Arabiensis, thus indicating that it does not harbour loci contributing to hybrid incompatibility15. F1 hybrid males produced from a cross of An. gambiae females to An. arabiensis males inherit the X chromosome from An. gambiae while F1 hybrid males produced from a cross of An. arabiensis females to An. gambiae males inherit the X chromosome from An. arabiensis. For this reason, the differences in the testes of these hybrid males have been attributed to X chromosome-linked genetic elements14,15. However, no specific factors involved in HIs have been identified so far.
In addition to the X chromosome, the mitochondria are also maternally inherited by F1 hybrid males and thus differ based on the directionality of the parental cross. Hanemaaijer et al. conducted a study where the complete mitochondrial sequences of 70 individual field-collected mosquito specimens were analysed in an attempt to identify species-specific genetic markers for An. arabiensis, An. coluzzii and An. gambiae. None of the identified SNPs were suitable as molecular markers to distinguish these species. The authors suggested that there is no divergent selection in the mitogenome, which episodic hybridizations between these species could explain54.
Recombination of mtDNA has been reported in several taxa, including plants, fungi, animals and humans58,59,60,61,62,63,64,65. For it to occur, more than one mitochondrial lineage must be present within a cell simultaneously. Due to the maternal inheritance of mtDNA, cells are generally homoplasmic59. However, paternal leakage of mtDNA from sperm has been demonstrated in several species66,67,68,69 and could occasionally cause heteroplasmy, providing the required conditions for recombination events to occur. Sperm contain mitochondria, and it is thought that up to 100 functional mitochondria (and their genomes) enter the oocyte cytoplasm at fertilisation in mammals70. In several mammalian species (including mice, cattle and humans), it has been shown that the mitochondria are tagged with ubiquitin, which enables recognition by oocyte proteolytic enzymes and commits sperm-derived mitochondria to destruction before or during the third proteolytic cleavage70. Rokas et al. theorised that survival of paternal mitochondria may be higher in the case of interspecific crosses than in within-species mating because molecular recognition systems may be less efficient in such genetic backgrounds71. Studies have demonstrated that there is a higher probability of paternal leakage in interspecific relative to intraspecific genetic backgrounds in mice72.
To our knowledge, these phenomena have never been investigated in Anopheles mosquitoes. In addition, although studies on the role of mtDNA in HIs have been performed using DNA sequencing data, no experimental studies on the matter have been conducted in these mosquito species.
We analysed the mtDNA derived from the laboratory mosquito colonies used in this study. Six SNPs were identified as different between and fixed within each strain. To investigate the potential role of mDNA in HIs, genetic crosses were used to introgress mitochondria from An. gambiae into An. arabiensis and vice versa. The species-specific markers identified were used to verify that introgression of interspecific mitochondria had been successful. These introgressed strains enabled us to generate F1 hybrid males in which the X chromosome and the mitochondria had originated from different species. Fertility assays and an investigation of testes phenotypes were performed to identify potential changes in the full sterility expected in these hybrids due to the presence of the interspecific mDNA and X chromosomes. No obvious fertility defect was detected in any of the experimental crosses, and the testes’ phenotype of F1hybrid males did not differ from that of their controls. F1 hybrid males with the X chromosome from An. gambiae and mitochondria from An. arabiensis showed testes with a marbled phenotype. Conversely, F1 hybrid males with the X chromosome from An. arabiensis and mitochondria from An. gambiae showed atrophic testes. These findings strongly suggest that mitochondrial DNA is unlikely to cause the observed sterility in F1 hybrid males between the species used in this study. Additionally, it does not appear responsible for the asymmetry in the testes’ phenotype.
The strain-specific SNPs used to monitor the genetic introgression aligned with the maternal inheritance of mitogenomes, confirming this pattern in these mosquito species and suggesting no mitochondrial recombination had occurred.
It is important to underline that these experiments were conducted in mosquito laboratory colonies, with a primary focus on the role of mtDNA in the context of post-zygotic His. For example, the impact of sexual selection was not explored. As a result, these findings may not entirely reflect what occurs in natural populations. It is possible that the presence of mitochondria within an interspecific genome may impose fitness costs in natural environments, which may not have been observed in the laboratory setting.
Finally, the recent report on a unique mtDNA lineage found in field-caught An. Coluzzii has brought attention to mtDNA variation within the Anopheles gambiae complex and the potential for complex coevolution between mitochondrial and nuclear DNA73. This presents an opportunity to expand studies similar to those outlined in this work to accommodate specific genetic backgrounds.
Materials and methods
Mosquitoes’ rearing
Mosquitoes were reared under standard conditions in an insectary at 28 °C and 80% relative humidity. Adult mosquitoes were housed in BugDorm-4M1515 insect-rearing cages (W17.5 x D17.5 x H17.5 cm), with standard cage crosses consisting of 50 virgin females and 50 virgin males sexed at the pupal stage. Larger crosses, consisting of more than 100 individuals, were instead housed in BugDorm-4M2222 insect rearing cages (W24.5 x D24.5 x H24.5 cm). For food, a 5% w/v glucose solution was added to a 25 ml glass bottle, and a Whatman grade 1 filter paper sheet was rolled up and partly submerged in the solution. The adult mosquitoes could land on the filter paper and feed on the glucose solution. Adult males and females were allowed to mate for approximately 6–7 days following emergence, at which point females were allowed to feed on bovine blood using a Hemotek membrane feeding system. Three days after the blood meal, egg bowls were added to the cage to allow females to lay eggs. Egg bowls consisted of 120 ml plastic cups to which rearing water (dH2O supplemented with 0.1% sodium chloride ) was added. In addition, the sides of each cup were covered with partially submerged filter paper in order to avoid eggs sticking to the plastic cup and drying out. Two days following oviposition, when the larvae had hatched, an average of 200 larvae were transferred to plastic trays (85(H) x 264(W) x 238(D)mm) containing rearing water and fish food pellets (NISHIKOI ™). Larvae were kept in trays until pupation. Pupae were collected and, following selection of a specific sex and/or screening for the presence of a transgenic marker, were placed into cups containing rearing water and allowed to emerge in cages.
Mosquito strains used for the experiments
The WT strains used in the experiments in this project, obtained from MR4, were the An. arabiensis Dongola strain (MRA-856) and the An. gambiae G3 strain (MRA-112). The G3 strain was isolated from the Gambia in 1975 and has since been maintained in a laboratory setting by the Malaria Research and Reference Reagent Resource Centre (MR4) (MRA-112). The strain is polymorphic for the 2La inversion. The An. arabiensis Dongola strain was collected in Sudan in 2004 and was also obtained from MR4 (MRA-856). As in all An. arabiensis strains, the 2La inversion is fixed in this strain. In addition, the strain is polymorphic for inversions 2Ra and 2Rb and has a fixed inversion on chromosome 3R. The X-insertion strain is a transgenic An. gambiae G3 strain expressing GFP under the control of the 3xP3 promoter from an insertion site on the X chromosome.
Hybrid fertility assessment (HFA)
To carry out a Cage HFA experiment, males or females of the introgressed strains were crossed to WT An. gambiae or An. arabiensis females in a standard cage cross to generate F1 progeny. F1 hybrid males were selected at the pupal stage (when the sexual dimorphism is apparent) and collected in cages until eclosion. Each group of F1 hybrid males was crossed to WT An. gambiae females in standard cage crosses. Following 6–7 days of mating, females were administered a blood meal and, two days later, allowed to oviposit in an egg bowl. The clutches were monitored for hatching for 4–6 days following oviposition.
DNA extraction and polymerase chain reaction (PCR)
According to the manufacturer protocol, genomic DNA was extracted using Qiagen DNeasy® Blood & Tissue Kit. DNA was isolated from pooled (6 individuals) F1 hybrids generated from each experimental condition (see Figs. 3 and 4). The region of interest containing the SNP was amplified from the mitogenome using the primers F: CGGTATTCTTATTCTTAAGCTTCCT and R: GCTCCTCCAACGTTAAATTTATTAG. The reaction was performed using Phusion® High-Fidelity PCR Master Mix following the manufacturer protocol. The PCR product was purified using the QIAquick® PCR Purification Kit, and the amplicons were sequenced using Eurofins.
Bioinformatic pipeline and tools
To identify species-specific mitochondrial SNP markers, whole genome sequencing (WGS) data of 10 pooled samples of 10 WT An. gambiae G3 adults was aligned to the AgamP4 reference genome, obtained from Vectorbase (Giraldo-Calderón et al., 2015), and variants were called. Fixed SNPs were identified and incorporated into the AgamP4 reference genome to create a G3 pseudoreference genome. WGS data from 2 individual males and a pooled sample of 38 WT An. arabiensis Dongola females was aligned to the G3 pseudoreference genome, variants were called and fixed SNPs for An. arabiensis were identified. The DNA sequencing data analysed are available in the NCBI Sequence Read Archive repository (Bioprojects PRJNA381033, PRJNA304755 and PRJNA1127581).
A total of 5 fixed SNP differences were identified between the mitochondrial genomes of An. arabiensis Dongola and An. gambiae G3 (Table 1). The two markers at positions 15,189 and 15,243 were located in a region of the mitogenome that was excluded from the analysis by Hanemaaijer et al. (2019), these markers were not selected for sequencing as primers design was hindered by the AT-rich nature of the region. Instead, primers were designed to amplify a region of DNA containing the SNP markers at positions 7255 and 7654 (Forward primer: CGGTATTCTTATTCTTAAGCTTCCT; Reverse primer: GCTCCTCCAACGTTAAATTTATTAG). DNA was extracted from 6 GamAm/Ara (hybrid males with An. gambiae X chromosome and mitochondria from An. arabiensis), 8 GamGm /Ara (hybrid males with An. gambiae X chromosome and mitochondria from An. gambiae), 8 Gam/Ara (control hybrid males with An. gambiae X chromosome and mitochondria from An. gambiae), 8 AraGm/Gam (hybrid males with An. arabiensis X chromosome and mitochondria from An. gambiae), 10 AraAm/Gam (hybrid males with An. arabiensis X chromosome and mitochondria from An. arabiensis),10 Ara/Gam (control hybrid males with An. arabiensis X chromosome and mitochondria from An. arabiensis) and used for Sanger sequencing and genotyping using Integrative Genomics Viewer (IGV). Sequencing results are available in Supplementary File 1.
The bioinformatic pipeline used to identify the species-specific SNPs in this study is described in Antonios Kriezis’s thesis available online (https://doi.org/10.25560/103207).
Testes dissection and whole-mount DAPI staining
Testes were dissected from pupal or adult stages from WT or F1 hybrid mosquitoes in a drop of fresh 1X PBS. For a quick analysis of the testis’s cytological phenotype (Fig. 5), testes were transferred in a clean drop of 1x PBS, and a coverslip was gently applied to the slide. Pictures of the testes were acquired using the cell imaging system EVOS (AMG) using phase contrast at 20x magnification. For Whole mount DAPI staining, following the dissection, testes were fixed in 3.7% Formaldehyde solution in 1X PBS for 10 min at Room Temperature (RT) and then washed 3 times in 0.1% Tween PBS 1X for 15 min. Testes were then mounted in a clean microscope slide in ProLong™ Gold Antifade Mountant with DAPI (ThermoFisher), and the coverslip was sealed using Cytobond (Scigene). Following at least 2 h of incubation at RT, testes were analysed using a Leica SP8 inverted confocal microscope and a 40x oil immersion objective was used.
Data availability
The datasets analysed during the current study are available in the NCBI Sequence Read Archive repository (Bioprojects PRJNA381033, PRJNA304755 and PRJNA1127581).
References
TEAM, W. World malaria report 2022. (2022).
Sinka, M. E. et al. A global map of dominant malaria vectors. Parasit. Vectors. 5, 69 (2012).
Barrón, M. G. et al. A new species in the major malaria vector complex sheds light on reticulated species evolution. Sci. Rep. 9, 1–13 (2019).
CHARLWOOD, J. D. & JONES, M. D. R. Mating behaviour in the mosquito, Anopheles gambiae s.1.Save. Physiol. Entomol. 4, 111–120 (1979).
Brogdon, W. G. Measurement of flight tone differentiates among members of the Anopheles gambiae species complex (Diptera: Culicidae). J. Med. Entomol. 35, 681–684 (1998).
WEKESA, J. W., BESANSKY, N. J. & BROGDON, W. G., HAWLEY, W. A. & Flight tone of field-collected populations of Anopheles gambiae and An. Arabiensis (Diptera: Culicidae). Physiol. Entomol. 23, 289–294 (1998).
Polerstock, A. R., Eigenbrode, S. D. & Klowden, M. J. Mating alters the Cuticular hydrocarbons of Female Anopheles gambiae Sensu Stricto and Aedes aegypti (Diptera: Culicidae). J. Med. Entomol. 39, 545–552 (2002).
Caputo, B. et al. Identification and composition of cuticular hydrocarbons of the major afrotropical malaria vector Anopheles gambiae s.s. (Diptera: Culicidae): analysis of sexual dimorphism and age-related changes. J. Mass. Spectrom. 40, 1595–1604 (2005).
Diabaté, A. et al. Mixed swarms of the molecular M and S forms of Anopheles gambiae (Diptera: Culicidae) in sympatric area from Burkina Faso. J. Med. Entomol. 43, 480–483 (2006).
Diabaté, A. et al. Spatial swarm segregation and reproductive isolation between the molecular forms of Anopheles gambiae. Proc. Biol. Sci. 276, 4215–4222 (2009).
Pennetier, C., Warren, B., Dabiré, K. R., Russell, I. J. & Gibson, G. Singing on the Wing’ as a mechanism for Species Recognition in the Malarial Mosquito Anopheles gambiae. Curr. Biol. 20, 131–136 (2010).
Dabire, K. R. et al. Assortative mating in mixed swarms of the mosquito Anopheles gambiae s.s. M and S molecular forms, in Burkina Faso, West Africa. Med. Vet. Entomol. 27, 298–312 (2013).
Davidson, G. Anopheles gambiae Complex. Nature 196, 907 (1962).
Slotman, M., Della Torre, A. & Powell, J. R. The genetics of inviability and male sterility in hybrids between Anopheles gambiae and An. Arabiensis. Genetics 167, 275 (2004).
Bernardini, F. et al. Cross-species Y chromosome function between malaria vectors of the Anopheles gambiae species complex. Genetics 207, 729–740 (2017).
Liang, J. & Sharakhov, I. V. Premeiotic and meiotic failures lead to hybrid male sterility in the Anopheles gambiae complex. Proc. R. Soc. B Biol. Sci. 286, (2019).
Liang, J., Hodge, J. M. & Sharakhov, I. V. Asymmetric phenotypes of sterile hybrid males from reciprocal crosses between species of the Anopheles gambiae Complex. Front. Ecol. Evol. 9, 375 (2021).
Fontaine, M. C. et al. Extensive introgression in a malaria vector species complex revealed by phylogenomics. Sci. (80-). 347, 1258524 (2015).
Cowell, F. 100 years of Haldane’s rule. J. Evol. Biol. https://doi.org/10.1111/jeb.14112 (2022).
Neafsey, D. E. et al. SNP genotyping defines complex gene-flow boundaries among African malaria vector mosquitoes. Science 330, 514–517 (2010).
Mallet, J., Besansky, N. & Hahn, M. W. How reticulated are species? BioEssays 38, 140–149 (2016).
Pombi, M. et al. Dissecting functional components of reproductive isolation among closely related sympatric species of the Anopheles gambiae complex. Evol. Appl. 10, 1102–1120 (2017).
Lee, Y. et al. Spatiotemporal dynamics of gene flow and hybrid fitness between the M and S forms of the malaria mosquito, Anopheles gambiae. Proc. Natl. Acad. Sci. 110, 19854–19859 (2013).
Temu, E. A., Hunt, R. H., Coetzee, M., Minjas, J. N. & Shiff, C. J. Detection of hybrids in natural populations of the Anopheles gambiae complex by the rDNA-based, PCR method. Ann. Trop. Med. Parasitol. 91, 963–965 (1997).
Toure, Y. T. et al. The distribution and inversion polymorphism of chromosomally recognized taxa of the Anopheles gambiae complex in Mali, West Africa. Parassitologia 40, 477–511 (1998).
Tripet, F. et al. DNA analysis of transferred sperm reveals significant levels of gene flow between molecular forms of Anopheles gambiae. Mol. Ecol. 10, 1725–1732 (2001).
Mawejje, H. D. et al. Insecticide resistance monitoring of field-collected anopheles gambiae s.l. populations from Jinja, eastern Uganda, identifies high levels of pyrethroid resistance. Med. Vet. Entomol. 27, 276–283 (2013).
Zouré, A. A. et al. Genetic analysis and population structure of the Anopheles gambiae complex from different ecological zones of Burkina Faso. Infect. Genet. Evol. J. Mol. Epidemiol. Evol. Genet. Infect. Dis. 81, 104261 (2020).
Lanzaro, G. C. & Lee, Y. Speciation in Anopheles gambiae — The Distribution of Genetic Polymorphism and Patterns of Reproductive Isolation Among Natural Populations. in (ed. Manguin, S.) Ch. 6IntechOpen, (2013). https://doi.org/10.5772/56232
Dobzhansky, T. Studies on hybrid sterility. II. Localization of sterility factors in Drosophila Pseudoobscura hybrids. Genetics 21, 113–135 (1936).
Coyne, J. A. & Charlesworth, B. Location of an X-linked factor causing sterility in male hybrids of Drosophila simulans and D. Mauritiana. Heredity (Edinb). 57 (Pt 2), 243–246 (1986).
Cabot, E. L., Davis, A. W., Johnson, N. A. & Wu, C. I. Genetics of reproductive isolation in the Drosophila simulans clade: complex epistasis underlying hybrid male sterility. Genetics 137, 175–189 (1994).
Coyne, J. A. & Orr, H. A. Speciation (Sinauer Associates, 2004).
Wu, C. I. & Ting, C. T. Genes and speciation. Nat. Rev. Genet. 5, 114–122 (2004).
Brideau, N. J. et al. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314, 1292–1295 (2006).
Sweigart, A. L., Fishman, L. & Willis, J. H. A simple genetic incompatibility causes Hybrid Male sterility in Mimulus. Genetics 172, 2465–2479 (2006).
Masly, J. P. & Presgraves, D. C. High-resolution genome-wide dissection of the two rules of speciation in Drosophila. PLOS Biol. 5, e243 (2007).
Deitz, K. C., Takken, W. & Slotman, M. A. The Genetic Architecture of Post-zygotic Reproductive isolation between Anopheles coluzzii and an. Quadriannulatus. Front. Genet. 11, p.925 (2020).
della Torre, A., Merzagora, L., Powell, J. R. & Coluzzi, M. Selective introgression of paracentric inversions between two sibling species of the Anopheles gambiae complex. Genetics 146, 239–244 (1997).
Slotman, M. A., Della Torre, A., Calzetta, M. & Powell, J. R. Differential introgression of chromsomal regions between Anopheles gambiae and An. Arabiensis. Am. J. Trop. Med. Hyg. 73, 326–335 (2005).
Burton, R. S., Pereira, R. J. & Barreto, F. S. Cytonuclear genomic interactions and hybrid breakdown. Annu. Rev. Ecol. Evol. Syst. 44, 281–302 (2013).
Gershoni, M., Templeton, A. R. & Mishmar, D. Mitochondrial bioenergetics as a major motive force of speciation. Bioessays 31, 642–650 (2009).
Ma, H. et al. Incompatibility between Nuclear and mitochondrial genomes contributes to an Interspecies Reproductive Barrier. Cell. Metab. 24, 283–294 (2016).
Cuperfain, A. B., Zhang, Z. L., Kennedy, J. L. & Gonçalves, V. F. The Complex Interaction of Mitochondrial Genetics and mitochondrial pathways in Psychiatric Disease. Mol. Neuropsychiatry. 4, 52–69 (2018).
Osellame, L. D., Blacker, T. S. & Duchen, M. R. Cellular and molecular mechanisms of mitochondrial function. Best Pract. Res. Clin. Endocrinol. Metab. 26, 711–723 (2012).
Sackton, T. B., Haney, R. A. & Rand, D. M. Cytonuclear coadaptation in Drosophila: disruption of cytochrome c oxidase activity in backcross genotypes. Evolution 57, 2315–2325 (2003).
Keaney, T. A., Wong, H. W. S., Dowling, D. K., Jones, T. M. & Holman, L. Mother’s curse and indirect genetic effects: do males matter to mitochondrial genome evolution? J. Evol. Biol. 33, 189–201 (2020).
Ålund, M. et al. Tracking hybrid viability across life stages in a natural avian contact zone. Evol. (N Y). 78, 267–283 (2024).
Munasinghe, M., Haller, B. C. & Clark, A. G. Migration restores hybrid incompatibility driven by mitochondrial–nuclear sexual conflict. Proc. R. Soc. B Biol. Sci. 289, 20212561 (2022).
DAVIDSON, G. Anopheles gambiae, a complex of species. Bull. World Health Organ. 31, 625 (1964).
Davidson, G. & The five mating-types in the anopheles gambiae complex. Riv Malariol 43, 167-183 (1964).
Lynch, M., Koskella, B. & Schaack, S. Mutation pressure and the evolution of organelle genomic architecture. Science 311, 1727–1730 (2006).
Allio, R., Donega, S., Galtier, N. & Nabholz, B. Large variation in the ratio of mitochondrial to Nuclear Mutation Rate across animals: implications for genetic diversity and the Use of mitochondrial DNA as a molecular marker. Mol. Biol. Evol. 34, 2762–2772 (2017).
Hanemaaijer, M. J. et al. Mitochondrial genomes of Anopheles arabiensis, an. Gambiae and An. Coluzzii show no clear species division. F1000Research 7, 1–14 (2019).
Bernardini, F., Kriezis, A., Galizi, R., Nolan, T. & Crisanti, A. Introgression of a synthetic sex ratio distortion system from Anopheles gambiae into Anopheles arabiensis. Sci. Rep. 2019. 91 9, 1–8 (2019).
Aboagye-Antwi, F. et al. Experimental swap of Anopheles gambiae’s assortative mating preferences demonstrates key role of X-Chromosome Divergence Island in Incipient Sympatric Speciation. PLOS Genet. 11, e1005141 (2015).
Diabaté, A., Dabire, R. K., Millogo, N. & Lehmann, T. Evaluating the effect of postmating isolation between Molecular forms of Anopheles gambiae (Diptera: Culicidae). J. Med. Entomol. 44, 60–64 (2007).
Lunt, D. H. & Hyman, B. C. Animal mitochondrial DNA recombination. Nature 387, 247 (1997).
Birky, C. W. The inheritance of genes in Mitochondria and chloroplasts: laws, mechanisms, and models. Annu. Rev. Genet. 35, 125–148 (2001).
Ladoukakis, E. D. & Zouros, E. Recombination in animal mitochondrial DNA: evidence from published sequences. Mol. Biol. Evol. 18, 2127–2131 (2001).
Hoarau, G., Holla, S., Lescasse, R., Stam, W. T. & Olsen, J. L. Heteroplasmy and evidence for recombination in the mitochondrial Control Region of the Flatfish Platichthys flesus. Mol. Biol. Evol. 19, 2261–2264 (2002).
Kraytsberg, Y. et al. Recombination of human mitochondrial DNA. Science 304, 981 (2004).
Gantenbein, B., Fet, V., Gantenbein-Ritter, I. A. & Balloux, F. Evidence for recombination in scorpion mitochondrial DNA (Scorpiones: Buthidae). Proceedings. Biol. Sci. 272, 697–704 (2005).
Piganeau, G., Gardner, M. & Eyre-Walker, A. A. Broad survey of recombination in Animal Mitochondria. Mol. Biol. Evol. 21, 2319–2325 (2004).
Ciborowski, K. L. et al. Rare and fleeting: an example of interspecific recombination in animal mitochondrial DNA. Biol. Lett. 3, 554–557 (2007).
Gyllensten, U., Wharton, D., Josefsson, A. & Wilson, A. C. Paternal inheritance of mitochondrial DNA in mice. Nature 352, 255–257 (1991).
Magoulas, A. & Zouros, E. Restriction-site heteroplasmy in Anchovy (Engraulis encrasicolus) indicates incidental biparental inheritance of mitochondrial DNA. Mol. Biol. Evol. 10, 319 (1993).
Schwartz, M. & Vissing, J. Paternal inheritance of mitochondrial DNA. N Engl. J. Med. 347, 576–580 (2002).
Kvist, L., Martens, J., Nazarenko, A. A. & Orell, M. Paternal leakage of mitochondrial DNA in the great tit (Parus major). Mol. Biol. Evol. 20, 243–247 (2003).
Sutovsky, P. et al. Ubiquitinated sperm mitochondria, selective proteolysis, and the regulation of mitochondrial inheritance in mammalian embryos. Biol. Reprod. 63, 582–590 (2000).
Rokas, A., Ladoukakis, E. & Zouros, E. Animal mitochondrial DNA recombination revisited. Trends Ecol. Evol. 18, 411–417 (2003).
Shitara, H., Hayashi, J. I., Takahama, S., Kaneda, H. & Yonekawa, H. Maternal inheritance of mouse mtDNA in interspecific hybrids: segregation of the leaked paternal mtDNA followed by the prevention of subsequent paternal leakage. Genetics 148, 851–857 (1998).
Amaya Romero, J. E. et al. Mitochondrial variation in Anopheles gambiae and Anopheles coluzzii: Phylogeographic Legacy and Mitonuclear associations with Metabolic Resistance to pathogens and insecticides. Genome Biol. Evol. 16(9), p.evae172 (2024).
Acknowledgements
We thank the whole Crisanti lab for experimental assistance and Jhon Connolly and members of the Target Malaria teams for useful suggestions. This work was supported by a grant from the Bill & Melinda Gates Foundation and Open Philanthropy. We thank the Facility for Imaging by Light Microscopy (FILM) at Imperial College London for the microscopy analysis.
Author information
Authors and Affiliations
Contributions
F.B. and A.K. designed the research, A.K., G.M. and M.V. performed the research, F.B., A.K. and M.V. analysed the data. A.K., G.M. and M.V. wrote the paper with the input from A.C.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Kriezis, A., Vitale, M., Morselli, G. et al. Unravelling the role of mitochondrial DNA in hybrid incompatibility within species of the Anopheles gambiae complex. Sci Rep 14, 29467 (2024). https://doi.org/10.1038/s41598-024-80887-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-024-80887-0