Abstract
Coronary artery disease (CAD) commonly occurs and elevates the risk of cardiovascular events and mortality in chronic kidney disease (CKD) patients. The underlying pathogenesis of CKD-related CAD is believed to be closely linked to inflammatory responses. Here, we explored inflammation-related markers for early diagnosis and management of CAD in CKD patients. Through comprehensive bioinformatics analysis and machine learning techniques, glutamate cysteine ligase modifier subunit (GCLM), nuclear protein 1 (NUPR1), and prostaglandin E receptor 1 (PTGER1) were selected as hub biomarkers. Furthermore, GCLM and NUPR1 were demonstrated significantly upregulated in the two validation cohorts of CKD patients with or without hemodialysis, while the change in PTGER1 was not prominent. Additionally, GCLM and NUPR1 were identified as promising indicators to predict CAD in CKD patients. Our study deciphered the higher predictive genes for CAD associated with CKD that is related to inflammation, which provides novel insights into the diagnosis and therapeutic options.
Similar content being viewed by others
Introduction
Cardiovascular disease (CVD), which has a prevalence of up to 50% in advanced chronic kidney disease (CKD), is responsible for 40–50% of the deaths in this population1. As is well known, coronary artery disease (CAD), a CVD characterized by atherosclerotic lesions and blockage of vessels2, has a significantly higher incidence in patients with CKD than that in the general population3,4. Moreover, the clinical manifestations and main symptoms of CAD become less typical in the setting of renal dysfunction and the potential risks of adverse effects during interventions are also significantly higher, making the management of CAD particularly complex for patients with advanced CKD5. Thus, it is critical to achieve early diagnosis and management of CAD in CKD to reduce mortality and improve prognosis.
Mechanistically, it has been established that inflammation drives and orchestrates the early coronary artery disease6. Recently, anti-inflammatory treatments have been shown to have promising effects in regressing atherogenic plaques and altering the course of CAD7,8,9. In patients with CKD, as renal function deteriorates, uremia, oxidative stress, and reduced clearance of inflammatory factors often result in an increased inflammatory state, which may be one of the reasons for triggering and exacerbating the risk of CAD. Therefore, evaluating the increased inflammatory risk is expected to provide new perspectives for predicting and managing CAD in patients with CKD10.
In recent years, with the widespread application of bioinformatic technologies and machine learning in medicine, numerous novel genes that can serve as biomarkers and therapeutic targets have been discovered in various human diseases11,12. Although clinical research have shown that inflammation plays an important role in the pathophysiological process of CAD13, there is currently no study that has explored the inflammation-related genes associated with a high risk of CAD in patients with CKD through comprehensive bioinformatic methods combined with validation in real-world populations. In the present study, we analyzed databases GSE113079, GSE37171, and GSE70528 using multiple machine learning methods, screened for specific inflammation-related genes, and constructed an optimal diagnostic model for CAD in patients with CKD. Furthermore, we validated these genes in CKD patients at different stages, providing promising biomarkers and therapeutic targets for predicting the risk of CAD in CKD patients.
Results
Screening DEGs in CAD
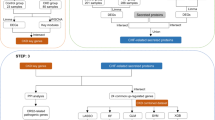
As shown in Fig. 1, the flowchart of bioinformatics analysis was performed. To detect differentially expressed genes (DEGs) related to CAD, the original dataset included 93 CAD and 48 healthy samples was collected from the Gene Expression Omnibus (GEO) repository. The box plot revealed the raw data before and after removing batch effect (Fig. 2A,B). The medial value of the box plot was at the same level, showing that the data preprocessing effect was effective. Differential analysis in CAD dataset revealed 2882 DEGs by p ≤ 0.05 and |log2 (fold change) |≥ 0.585, including 1544 upregulated genes and 1338 downregulated genes (Fig. 2C). Out of these, the top 20 upregulated genes and top 20 downregulated gens are presented in the form of heatmap (Fig. 2D).
The procession of CAD dataset and differentially expression analysis of CAD. (A) Box plot of normalization of data from 141 samples. (B) UMAP for GSE113079. (C) The volcano plot illustrating CAD DEGs in GSE113079. Upregulated gene were represented by red dots, while downloaded genes were represented by bule dots. (D) Heatmap of the top 20 upregulated and 20 downregulated DEGs in GSE113079. CAD, coronary artery disease; DEGs, differentially expressed genes.
The construction of Weighted gene co-expression network analysis (WGCNA) and identification of key module genes
To detect the key genes in CAD, WGCNA was performed to analyze cohesive gene networks and their gene association patterns. We calculated the soft threshold power according to scale independence and mean connectivity, which was most suitable when set to 3 (Fig. 3A,B). As shown in Fig. 3C, 25 modules were selected while using the soft threshold power of 3. The blue module was found to have the most significant positive correlation with CAD according to the analysis of the relationship between these modules and CAD (Correlation = 0.85, P value = 5e-40) (Fig. 3D). The genes of this module were chosen as the main module. Comparing these genes to the DEGs, 995 overlapping genes were identified as crucial genes in CAD for subsequent analysis (Fig. 3E).
Identification of key module genes in CAD dataset by WGCNA and determination of key genes in CAD via overlapping DEGs and key module genes. (A, B) Identification of the best β value through scale independence and mean connectivity. (C) The dendrogram and module eigengenes. (D) Correlation between WGCNA modules and status of CAD. (E) The intersection of DEGs and key module gens through venn diagram. CAD, coronary artery disease; WGCNA, Weighted gene co-expression network analysis; DEGs, differentially expressed genes.
Functional enrichment analysis of crucial genes in CAD
To better understand the potential pathogenic mechanism in CAD, we conducted Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways enrichment analyses to explore the functions of crucial genes. According to the result presented in Fig. 4A–C, genetic biological processes are mainly including signal transduction, cell–cell signaling, cell–cell adhesion, leukocyte migration in inflammatory response. Moreover, the genetic cellular components were mainly localized to plasma membrane, extracellular space and cell face. The genetic molecular function mostly focused on G protein-coupled receptor activity, signaling receptor binding and cytokine activity. KEGG pathway analysis indicated that the crucial genes in CAD are mainly involved in cytokine-cytokine receptor interaction and leukocyte transendothelial migration (Fig. 4D). Following the GO and KEGG pathway enrichment analyses, it has been implicated that inflammatory response plays a fundamental role in CAD formation and development. Given the established role of inflammatory response, our research aims to unravel their significance in CAD process.
Functional enrichment analysis of key genes in CAD. (A) Bar plots of GO biological process enrichment analysis. (B) Bar plots of GO cell component enrichment analysis. (C) Bar plots of GO molecular function enrichment analysis. (D) The chord plot showing KEGG enrichment pathway results. CAD, coronary artery disease; GO, Gene Ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
Identification of IRGGs in CKD-related CAD
It is widely acknowledged that CAD was more prevalent in CKD than general populations, and the incidence of CAD increases with sustained decreases in kidney function17.To obtain the shared genes for CKD and CAD, we acquired the CKD PBMC dataset GSE37171 and GSE70528 from GEO dataset. We integrated the two datasets after removing the batch effect, and performed the principal component analysis of the integrated dataset. After batch effect removal, the differences between two datasets were significantly decreased, as demonstrated by the results (Fig. 5A,B). A total of 1721 genes were identified by adjusted p ≤ 0.05 and |log2 (fold change) |≥ 0.585 after pretreatment in integrated dataset, including 509 upregulated genes and 1212 downregulated genes (Fig. 5C). The top 20 significantly upregulated genes and downregulated genes were presented as heatmap (Fig. 5D). Using a relative score ≥ 5 as the screening threshold, we identified 4,238 IRGGs from GeneCards database. To shed more light on inflammatory response on their significance in CKD with CAD, a Venn diagram revealed 14 overlapping DEGs among CKD, CAD and IRRGs (Fig. 5E).
The integration of CKD datasets and identification of IRGGs in CKD related CAD. (A, B) PCA plots of two CKD datasets before and after batch-effect correction. (C) The volcano plot showing DEGs of integrated CKD dataset. Upregulated gene were represented by red dots, while downloaded genes were represented by bule dots. (D) The heatmap presenting the top 20 upregulated and 20 downregulated DEGs of integrated CKD dataset. (E) The intersection of key genes in CKD related CAD and IRGGs through venn diagram. CKD, chronic kidney disease; IRGGs, inflammatory response-related genes; CAD, coronary artery disease; DEGs, differentially expressed genes; PCA, principal component analysis.
Identification of hub genes by machine learning in CKD related CAD
To obtain the hub genes of CKD with CAD, two different machine learning algorithms were utilized to screen genes. The least absolute shrinkage and selection operator (LASSO) algorithm was employed to delve into potential inflammatory response genes in CAD (Fig. 6A,B). Additionally, we performed by random forest (RF) machine learning algorithm to rank the overlapping genes with the MeanDecreaseGini > 2 were extracted, as demonstrated in Fig. 6C. Identifying hub genes for CKD was carried out using the same methods (Fig. 6D–F). Based on the information contained in this, three hub genes nuclear protein 1 (NUPR1) , glutamate cysteine ligase modifier subunit (GCLM) and prostaglandin E receptor 1 (PTGER1) were identified that could serve as hub genes related inflammatory response for CKD and CAD (Fig. 6G).
Identification of potential diagnostic genes for CKD related CAD via two machine learning methods. (A, B) The LASSO logistic regression algorithm was applied to determine the minimum and lambda values of diagnostic genes in CAD. (C) MeanDecreaseGini analysis was performed on 14 genes using the RF algorithm, and nine biomarkers with scores greater than 2 were selected in CAD. (D, E) The LASSO logistic regression algorithm was applied to determine the minimum and lambda values of diagnostic genes in CKD. (F) MeanDecreaseGini analysis was performed on 14 genes using the RF algorithm, and ten biomarkers with scores greater than 2 were selected in CKD. (G) Venn diagram showing the overlap in results between two algorithms. CKD, chronic kidney disease; CAD, coronary artery disease; LASSO, least absolute shrinkage and selection operator; RF, random forest.
Validation of hub genes
To evaluating the accuracy of the hub genes, receiver operating characteristic (ROC) curves were constructed utilizing both the training and validation sets. We split CAD dataset into a training set and a validation set at a 7:3 ratio. As we expected, three hub genes exhibited AUC values > 0.9 in training set (Fig. 7A,B). Consistently, the validation cohort presented AUC value > 0.9 (Fig. 7C,D). The result showed that the three hub genes have certain diagnostic value for CAD.
Diagnostic effectiveness of three hub genes in CAD dataset. (A, B) ROC curve of diagnostic biomarkers including NUPR1, GCLM and PTGER1 in training group. (C, D) ROC curve of diagnostic biomarkers including NUPR1, GCLM and PTGER1 in validation group. CAD, coronary artery disease; ROC receiver operating characteris tic.
Immune cell infiltration in CAD and CKD
Given that CKD with CAD is closely associated with inflammatory response, CIBERSORT was employed to analyze the proportion of immune infiltration cell. Figure 8A exhibited the distribution of 22 types immune cells in each sample. The result indicated that CAD exhibited higher T cells CD4 naive, T cells regulatory, T cells gamma delta, Monocytes, Macrophages M0, whereas lower T cells CD8, T cells CD4 memory resting, T cells CD4 memory activated, NK cells activated, Dendritic cells activated, Mast cells activated (Fig. 8B). We further explored the relationship between individual immune cells. As shown in Fig. 8C, T cells CD8 had a significant negative correlation with Monocytes (r = − 0.74), while Mast cells activated were positively related with NK cells activated (r = 0.47). To investigate the correlation between hub genes and immune cells. As presented in Fig. 8D, the hub genes were positively correlated with T cells regulatory, T cells gamma delta, T cells CD4 naive, Monocytes and Macrophages M0 in CAD. The findings suggested that three genes may be essential in the pathogenesis of CAD by affecting the infiltration of immune cells.
Immune cell infiltration analysis in CAD. (A) Stacked histogram showing the immune cell kinds and ratios in CAD. (B) Violin plot displaying the expression level of 22 types of immune cells between CAD and control group. (C) Heat map illustrating the correlation among 22 kinds of immune cells in CAD. The size of the colored squares represents the strength of the correlation, with red indicating positive and blue indicating negative correlations. Darker hues signify stronger associations. (D) Correlation map demonstrating the correlation between immune cells with GCLM, NUPR1 and PTGER1 in CAD. CAD, coronary artery disease. *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant.
External validation of hub genes
To further confirm the potential applicability and diagnostic efficacy of the three hub genes, we conducted validation experiments in CKD patients with or without CAD, consisting of 150 maintenance hemodialysis (MHD) patients and 42 non-MHD stage 3–5 CKD patients. As detailed in Tables 1 and 2, there were no statistically significant differences between the two groups in baseline characteristics except for age. In MHD patients, apart from PTGER1, both GCLM and NUPR1 were significantly elevated in CAD patients versus non-CAD patients (Fig. 9A–C). Notably, the ROC curves proposed that GCLM and NUPR1 harbored a promising performance to predict CAD. The AUCs of NUPR1 and GCLM were 0.744 and 0.823, respectively. The combined analysis of two genes further improved diagnostic accuracy for CAD, achieving an AUC of 0.860 (Fig. 9D,E). These findings were supported by analysis of the non-MHD stage 3–5 CKD population, which showed that the expression of GCLM and NUPR were increased in CAD group compared with the non-CAD group except for PTGER1 (Fig. 9F–H). ROC curves of NUPR1 (AUC = 0.736), GCLM (AUC = 0.734), and the combined two-gene (AUC = 0.831) demonstrated their promising diagnostic utility for CAD detection in CKD populations (Fig. 9I,J).
Validation of the expression patterns of three hub genes in CKD patients and assessment of their diagnostic value for CAD in this population. (A–C) Expression levels of three hub genes from hemodialysis patients with or without CAD. (D, E) ROC curve of diagnostic performance including GCLM and NUPR1 in hemodialysis patients. (F, H) Expression levels of three hub genes from patients with CKD stage 3–5 with or without CAD. (I, J) The ROC curve of diagnostic performance including GCLM and NUPR1 in patients with CKD stage 3–5. CKD, chronic kidney disease; CAD, coronary artery disease. *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant.
Discussion
With increasing severity of renal dysfunction, the impact of CAD on patients with CKD gradually becomes heavier14,15. To reduce the burden, in our study, we for the first time screened the inflammation- related biomarkers to predict CAD in CKD patients by applying comprehensive bioinformatics analysis approaches. Totally, we identified three hub genes (GCLM, NUPR1 and PTGER1), and further validation work in two CKD cohorts confirmed that GCLM and NUPR1 have higher predictive value for CAD, providing novel insights into early diagnosis and management strategies.
Microinflammatory state disrupts immune tolerance and induces endothelial inflammation, promoting lipid deposition and platelet aggregation that culminate in atherosclerotic plaque formation and that impair coronary microvascular structure and microcirculatory function16. Consistent with previous studies, our GO and KEGG enrichment analysis of CAD-related crucial genes revealed significant enrichment in inflammatory pathways such as cytokine-cytokine receptor interaction and leukocyte transendothelial migration. Cytokines produced by activated inflammasomes drive inflammation, promote endothelial cell proliferation, and further facilitate the recruitment and activation of additional inflammatory cells17. This complex cascade underscores the critical role of cytokines in the pathophysiology of atherosclerosis. Extensive preclinical and clinical studies have revealed the infiltration of diverse immune cells within atherosclerotic plaques. In CAD patients, CD4 + T cells contribute to endothelial injury and macrophage activation through pro-inflammatory cytokine secretion. Under sustained local microenvironmental stimulation, these T cells differentiate into Th1, Th2, or Th17 subsets, thereby amplifying inflammatory responses18. Our study corroborates these findings by demonstrating significant differences in immune cell infiltration between CAD patients and healthy controls. Immune cell profiling in CAD patients revealed a predominance of monocytes, which serve as central drivers of atherosclerosis. Monocytes were significantly elevated in CAD patients and correlated with disease severity19. In independent models, reduction in circulating monocytes attenuates atherosclerotic lesion development20.
Accumulating evidence demonstrates that inflammation represents a fundamental pathological feature of CKD and a key pathogenic process in the progression to end-stage renal disease (ESRD)21. Indeed, this persistent microinflammatory state is a hallmark of CKD and contributes to adverse cardio renal outcomes22,23,24. Therefore, we further performed an intersection analysis among CAD-related genes, CKD-related genes, and inflammation-related genes, screening out 14 differentially expressed genes. Numerous studies have confirmed that RF and LASSO regression exhibit robust predictive performance in screening key genes11,25. Through in-depth analyses using these two machine learning algorithms, we ultimately identified three hub genes (NUPR1, GCLM, and PTGER1), which are of great significance for identifying inflammatory biomarkers of CAD and promising therapeutic intervention targets.
NUPR1 serves as a multifunctional stress-induced protein participating in various cellular stress responses26. NUPR1 promotes pathological inflammation in pancreatic ductal adenocarcinoma cells both in vivo and in vitro27. A recent study has found that NUPR1 is not only upregulated in the aorta of atherosclerotic mice but also significantly increased in peripheral blood mononuclear cells of CAD patients28. In line with the above findings, as a key regulator of inflammation and immunity, our results demonstrated that NUPR1 was upregulated in the peripheral blood of CKD patients with CAD. In vascular endothelial cells, the inflammatory inducer LPS significantly upregulates NUPR1 protein expression, and knockdown of NUPR1 inhibits LPS-induced autophagy in these cells29. This finding supports our conclusion that NUPR1 plays a crucial role as a regulatory molecule involved in CAD-associated inflammation. However, further investigation is needed to determine the exact molecular mechanism.
GCLM, functioning as a modified subunit, along with the catalytic subunit GCLC, constitutes a crucial enzyme called glutamate cysteine ligase (GCL). GCL serves as the pivotal rate-limiting factor in the synthesis of glutathione (GSH), which plays a vital role as an antioxidant and scavenger of free radicals in vivo30. It is common knowledge that ROS directly induces and aggravates the inflammation responses in CAD31. The downregulation of GCLC/GCLM expression and subsequent decrease in total cellular GSH levels are strongly associated with microvascular dysfunction in atherosclerosis32. Conversely, administration of GSH precursors can stimulate intracellular synthesis, reduce ROS and LDL oxidation, increase nitric oxide (NO) production, and mitigate atherosclerosis and its complications33. These findings align with our results and suggest that GCLM may serve as a potential biomarker for premature coronary artery disease.
PTGER1, one of the receptor subtypes of prostaglandin E2 (PGE2), which is a G protein-coupled receptor-E type prostaglandin receptor34. Studies have found that PTGER1, expressed in the kidney and cardiovascular system, maintains their homeostasis35,36. Previous studies have shown upregulated expression of PTGER1 in peripheral blood mononuclear cells of patients with CAD and in the aortic tissues of ApoE−/− mice with atherosclerosis28. Contrary to their findings, PTGER1 showed no significant upregulation in our validation cohort. This discrepancy may be attributed to differences in the study populations. CKD is characterized by multidimensional imbalances in metabolism and inflammation, potentially leading to divergent PTGER1 expression and functions across different cohorts. In neuroinflammation and oncological diseases, PTGER1 promotes Th1 cell differentiation and exerts pro-inflammatory effects37,38. However, in doxorubicin-induced cardiac myocyte ferroptosis, activation of the PGE2/PTGER1 axis promotes antioxidant gene expression, protecting myocytes from ferroptosis and exerting anti-inflammatory effects in this context39. In type 1 diabetes, PTGER1 alleviates cytokine-induced β-cell dysfunction40. These findings suggested that PTGER1 may exhibit dual pro-inflammatory and anti-inflammatory roles depending on metabolic conditions and cellular microenvironments. Furthermore, medications commonly used to treat CAD may influence PTGER1 expression. A prior study revealed that atorvastatin can reduce PTGER1 expression41. Additionally, aspirin inhibits cyclooxygenase-2 (COX-2) to reduce PGE2 synthesis, thereby downregulating ligand-dependent expression of PTGER142. These pharmacological interventions may have masked the CAD-induced upregulation of PTGER1. Consequently, the biological effects of PTGER1 are likely subject to multifaceted regulation by metabolic status, cellular microenvironment, and pharmacological interventions. The pathophysiological role of PTGER1 in CAD still needs to be further explored.
This study is retrospective and based on public databases and clinical samples, which has several limitations. The lack of transcriptomic or multi-omics data from dual-disease cohorts may limit our ability to conclusively identify key genes driving CAD development in CKD patients. In the future, it is essential to increase the availability of sequencing datasets for CKD patients with concomitant cardiovascular disease to further investigate the genetic associations underlying the heart-kidney crosstalk. In addition, this study was conducted using a dataset comprising patients from three different countries. However, the validation of our results was limited to the Chinese population. Future studies should expand the validation cohort’s geographical coverage and ethnic diversity to enhance the generalizability of our findings.
Materials and methods
Data collection and process
The raw gene expression datasets of CAD and CKD, including GSE113079, GSE37171 and GSE70528 were obtained from the GEO database (https://www.ncbi.nlm.nih.gov/geo/). There exists a distinct batch effect in both diseases, we conducted the combat algorithm of “SVA” package (version 3.56.0) in R software (version 4.4.1) to remove batch effect. Meanwhile, IRRGs were downloaded from GeneCards database (https://www.genecards.org) by means of searching for the term “inflammation response.” The screening threshold was a relevance score of ≥ 5.
DEGs identification
The “Limma” package (version 3.62.2) was leveraged to identify DEGs with an adjusted p ≤ 0.05 and |log2 (fold change) |≥ 0.585 in CAD dataset and CKD integrated datasets (GSE37171 and GSE70528). Subsequently, the results of DEGs were visually presented in the form of volcano plots and heatmaps using online data analysis platform sangerbox (http://www.sangerbox.com/home.html).
WGCNA
WGCNA analysis enables the classification of genes into distinct gene modules based on their co-expression similarity in samples, and it can concurrently establish direct connections between these modules and clinical characteristics. To determine the correlation between gene modules and clinical features, outliers in the samples were initially identified through hierarchical clustering analysis using the “ggplot2” package (version 3.5.2), and an appropriate soft threshold was selected as the weight value. Once the modules were obtained, the correlation between gene modules and clinical phenotypes was evaluated using the Pearson correlation coefficient, and the module-trait relationship with the most significant correlation was screened out.
Functional enrichment analysis
For a more detailed exploration of the biological function of the CAD pathogenic genes, GO and KEGG pathway enrichment43,44,45 were employed using DAVID database (https://davidbioinformatics.nih.gov), a biological information database for comprehensive gene and protein analysis. Moreover, the result of enrichment analysis was visually represented via bar and circus plot.
Machine learning
In order to identify potential candidate biomarkers and construct a diagnostic model for CKD with CAD, we applied two machine learning algorithms for screening hub genes, with LASSO and RF algorithm, respectively. The identification of candidate biomarkers was initially conducted through LASSO analysis, utilizing the “glmnet” package (version 4.1–8) in R software. Following this, the selection of genes was refined using the “randomforest” package (version 4.7–1.2), focusing on those with a MeanDecreaseGini > 2. Ultimately, the genes that overlapped between the two algorithms were identified as hub genes, which were employed to construct the diagnostic model for CKD-related CAD.
The evaluation of diagnostic efficacy of hub genes
To assess the diagnostic efficacy of hub genes in CAD, ROC and area under ROC curve (AUC) analysis were conducted using the “pROC” package (version 1.18.5) to estimate the predictive ability. In addition, the accuracy was evaluated through a calibration curve.
Immune infiltration analysis
To assess the level of immune cell infiltration in CKD and CAD gene expression data, the “CIBERSORT” package (version v1.03) was utilized. To compare the differences in the proportions of 22 different types of immune cells between disease and normal samples, the Wilcoxon test was employed, with a significance threshold set at p < 0.05. These differences were illustrated as stacked histograms, generated with the “ggplot2” package. Furthermore, the relationships among the 22 infiltrating immune cells were depicted using the “corrplot” software package (version 0.95). In the end, the Spearman rank correlation coefficient was applied to analyze the correlation between the levels of diagnostic biomarkers and the content of infiltrating immune cells, with statistical significance determined at p < 0.05.
Patients’ samples collection
Peripheral blood samples were collected from CKD patients who visited the Department of Nephrology at the Fourth Hospital of Hebei Medical University between January and December 2024. The inclusion criteria were an eGFR < 60 mL/ (min·1.73 m2) and age > 18 years. Exclusion criteria included acute inflammation, chronic infections (e.g., hepatitis B or C), current use of corticosteroids or immunosuppressants, and a history of malignancy. These samples served as the external training cohort. Informed written consent was obtained from all participants. Our study (No. 2020ky189) received approval from the Fourth Hospital of Hebei Medical University. CKD participants were categorized based on CAD status: the non-CAD group and the CAD group.
ELISA
The concentrations of GCLM, NUPR1, and PTGER1 in samples of participants were measured with commercially available kits (Elsbiotech, Shanghai, China). Initially, peripheral blood monocytes were treated with lysis buffer, followed by centrifugation at 12,000 × g for 20 min, according to the manufacturer’s instructions. The supernatant obtained after centrifugation was then used for detection. The absorbance of the samples was measured at a wavelength of 450 nm using a microplate reader, and these values were compared against a standard curve for quantification. The glutamate cysteine ligase modulating subunit (GCLM) concentrations were determined using the Human GCLM ELISA kit (EIA-3696-Hum; Elsbiotech, Shanghai, China), which has a detection range of 0.625–20 ng/mL. The nuclear protein1 (NUPR1) concentrations were determined using the Human NUPR1 ELISA kit (EIA-3701-Hum; Elsbiotech, Shanghai, China), which has a detection range of 3.12–100 ng/mL. The prostaglandin E receptor1 (PTGER1) concentrations were determined using the Human PTGER1 ELISA kit (EIA-2938-Hum; Elsbiotech, Shanghai, China), which has a detection range of 0.312–10 ng/mL. The intra-assay CV and inter-assay are less than 15%.
Statistical analysis
Statistical analyses were conducted using SPSS version 22.0 and GraphPad Prism 10 software. For categorical variables, either the chi-square test or Fisher’s exact test was applied, while continuous variables were analyzed using either Student’s t-test or the Mann–Whitney U test. Comparisons involving more than two groups were performed using one-way analysis of variance (ANOVA). Additionally, the Spearman method was employed to assess the correlation between two variables. Statistical significance was determined using two-sided p-values, with a threshold of less than 0.05 indicating significance. *p < 0.05; **p < 0.01; ***p < 0.001; ns not significant.
Data availability
The datasets used to support the current findings during the study are available from the corresponding authors on reasonable request.
References
Schuett, K. et al. The cardio-kidney patient: Epidemiology, clinical characteristics and therapy. Circ. Res. 132, 902–914 (2023).
Rumberger, J. A. et al. Coronary artery calcium area by electron-beam computed tomography and coronary atherosclerotic plaque area: A histopathologic correlative study. Circulation 92, 2157–2162 (1995).
Lai, A. C. et al. A personalized approach to chronic kidney disease and cardiovascular disease. J. Am. Coll Cardiol. 77, 1470–1479 (2021).
Matsushita, K. et al. Epidemiology and risk of cardiovascular disease in populations with chronic kidney disease. Nat. Rev. Nephrol. 18, 696–707 (2022).
Sarnak, M. J. et al. Chronic kidney disease and coronary artery disease: JACC state-of-the-art review. J. Am. Coll. Cardiol. 74, 1823–1838 (2019).
Attiq, A. et al. Hegemony of inflammation in atherosclerosis and coronary artery disease. Eur. J. Pharmacol. 966, 176338 (2024).
Ridker, P. M. et al. Antiinflammatory therapy with canakinumab for atherosclerotic disease. N. Engl. J. Med. 377, 1119–1131 (2017).
Nidorf, S. M. et al. Colchicine in patients with chronic coronary disease. N. Engl. J. Med. 383, 1838–1847 (2020).
Ridker, P. M. et al. Inflammation and cholesterol as predictors of cardiovascular events among patients receiving statin therapy: A collaborative analysis of three randomised trials. Lancet 401, 1293–1301 (2023).
Jankowski, J. et al. Cardiovascular disease in chronic kidney disease: Pathophysiological insights and therapeutic options. Circulation 143, 1157–1172 (2021).
Liu, X. et al. Identification of 7 mitochondria-related genes as diagnostic biomarkers of MDD and their correlation with immune infiltration: New insights from bioinformatics analysis. J. Affect. Disord. 349, 86–100 (2024).
Zhang, Y. et al. Review of the applications of deep learning in bioinformatics. Curr. Bioinform. 15, 898–911 (2021).
Bäck, M. et al. Inflammation and its resolution in atherosclerosis: Mediators and therapeutic opportunities. Nat. Rev. Cardiol. 16, 389–406 (2019).
Briasoulis, A. et al. Chronic kidney disease as a coronary artery disease risk equivalent. Curr. Cardiol. Rep. 15, 340 (2013).
Burlacu, A. et al. Coronary artery disease in dialysis patients: Evidence synthesis, controversies and proposed management strategies. J. Nephrol. 34, 39–51 (2021).
Furman, D. et al. Chronic inflammation in the etiology of disease across the life span. Nat. Med. 25, 1822–1832 (2020).
Grebe, A. et al. NLRP3 inflammasome and the IL-1 pathway in atherosclerosis. Circ. Res. 122, 1722–1740 (2018).
Haybar, H. et al. T-bet transcription factor in cardiovascular disease: Attenuation or inflammation factor?. J. Cell Physiol. 234, 7915–7922 (2019).
Arnold, K. A. et al. Monocyte and macrophage subtypes as paired cell biomarkers for coronary artery disease. Exp. Physiol. 104, 1343–1352 (2019).
Stoneman, V. et al. Monocyte/macrophage suppression in CD11b diphtheria toxin receptor transgenic mice differentially affects atherogenesis and established plaques. Circ. Res. 100, 884–893 (2007).
Miguel, V. et al. Metabolism at the crossroads of inflammation and fibrosis in chronic kidney disease. Nat. Rev. Nephrol. 21, 39–56 (2025).
Gupta, J. et al. Association between albuminuria, kidney function, and inflammatory biomarker profile in CKD in CRIC. Clin. J. Am. Soc. Nephrol. 7, 1938–1946 (2012).
Eustace, J. A. et al. Prevalence of acidosis and inflammation and their association with low serum albumin in chronic kidney disease. Kidney Int. 65, 1031–1040 (2004).
Kittelson, K. S. et al. Cardiovascular-kidney-metabolic syndrome - An integrative review. Prog. Cardiovasc. Dis. 87, 26–36 (2024).
Zhu, E. et al. Screening of immune-related secretory proteins linking chronic kidney disease with calcific aortic valve disease based on comprehensive bioinformatics analysis and machine learning. J. Transl. Med. 21, 359 (2023).
Murphy, A. et al. Nuclear protein 1 imparts oncogenic potential and chemotherapeutic resistance in cancer. Cancer Lett. 494, 132–141 (2020).
Liu, J. et al. NUPR1 is a critical repressor of ferroptosis. Nat. Commun. 12, 647 (2021).
Liu, X. et al. Inflammatory gene signature identified by machine algorithms reveals novel biomarkers of coronary artery disease. J. Inflamm. Res. 18, 2033–2044 (2025).
Meng, N. et al. A butyrolactone derivative suppressed lipopolysaccharide-induced autophagic injury through inhibiting the autoregulatory loop of p8 and p53 in vascular endothelial cells. Int. J. Biochem. Cell Biol. 44, 311–319 (2012).
Lu, S. C. et al. Glutathione synthesis. Biochim. Biophys. Acta 1830, 3143–3153 (2013).
Schmitz, G. et al. Role of redox regulation and lipid rafts in macrophages during Ox-LDL–mediated foam cell formation. Antioxid. Redox Signal. 9, 1499–1518 (2007).
Jain, S. K. et al. Hydrogen sulfide upregulates glutamate-cysteine ligase catalytic subunit, glutamate-cysteine ligase modifier subunit, and glutathione and inhibits interleukin-1b secretion in monocytes exposed to high glucose levels. Metab Syndr. Relat. Disord. 12, 299–302 (2014).
Rom, O. et al. Induction of glutathione biosynthesis by glycine-based treatment mitigates atherosclerosis. Redox Biol. 52, 102313 (2022).
Tober, K. L. et al. Possible cross-regulation of the E prostanoid receptors. Mol. Carcinog. 46, 711–715 (2007).
Amadio, P. et al. Production of prostaglandin E2 induced by cigarette smoke modulates tissue factor expression and activity in endothelial cells. FASEB J. 29, 4001–4010 (2015).
Breyer, M. D. et al. Prostaglandin receptors: their role in regulating renal function. Curr. Opin. Nephrol. hy. 9(1), 23–29 (2000).
Tsai, Y.-T. et al. Reprogramming of arachidonate metabolism confers temozolomide resistance to glioblastoma through enhancing mitochondrial activity in fatty acid oxidation. J Biomed. Sci. 29(1), 21 (2022).
Zhao, X. et al. Toxic role of prostaglandin E2 receptor EP1 after intracerebral hemorrhage in mice. Brain Behav. Immun. 46, 293–310 (2015).
Wang, B. et al. EP1 activation inhibits doxorubicin-cardiomyocyte ferroptosis via Nrf2. Redox Biol. 65, 102825 (2023).
Martin Vázquez, E. et al. NR5A2/LRH-1 regulates the PTGS2-PGE2-PTGER1 pathway contributing to pancreatic islet survival and function. iScience 25(5), 104345 (2022).
Gómez-Hernández, A. et al. Atorvastatin reduces the expression of prostaglandin E2 receptors in human carotid atherosclerotic plaques and monocytic cells: Potential implications for plaque stabilization. J. Cardiovasc. Pharm. 47(1), 60–69 (2006).
Hua, H. et al. Complex roles of the old drug aspirin in cancer chemoprevention and therapy. Med. Res. Rev. 39(1), 114–145 (2019).
Kanehisa, M. & Goto, S. Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000).
Kanehisa, M. et al. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51(D1), D587–D592 (2023).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28(11), 1947–1951 (2019).
Acknowledgements
We thank all subjects who participated in this study.
Funding
This work was supported by the project of Natural Science Foundation of Hebei Province (H2023206385).
Author information
Authors and Affiliations
Contributions
Binhong Yang, Xinyue Yang and Haoqi Sun conceived and designed the research, performed the bioinformatics analysis and analyed the data. Binhong Yang, Xinyue Yang, Haoqi Sun, Meijuan Cheng and Jingjing jin performed the experiments. Binhong Yang, Meijuan Cheng and Jingjing Jin wrote the manuscript. Yunhui Wu, Qi An and Kaixing Yan provided the methodology of sample collection. Shenglei Zhang and Yaling Bai supervised the study and revised the manuscript. Jinsheng Xu received the funding and managed the project.
Corresponding author
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethics approval
The study was conducted in accordance with Declaration of Helsinki principles, and was approved by the Ethics Committee of the Fourth Hospital of Hebei Medical University (No. 2022ky033) with obtaining written informed consent from all the participants.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, B., Yang, X., Sun, H. et al. Identification and validation of inflammatory response genes linking chronic kidney disease with coronary artery disease based on bioinformatics and machine learning. Sci Rep 15, 19184 (2025). https://doi.org/10.1038/s41598-025-03622-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-03622-3
Keywords
This article is cited by
-
Enhanced early chronic kidney disease prediction using hybrid waterwheel plant algorithm for deep neural network optimization
Scientific Reports (2025)
-
From kidney injury to cardiac dysfunction: the central role of oxidative stress in diabetes and CKD
Basic Research in Cardiology (2025)