Abstract
Transcriptional and post-transcriptional regulation mediated by RNA binding proteins (RBPs) have essential influence on the progression of colorectal cancer (CRC), while the underlying mechanisms need to be clarified. In this study, we focused on RBP-RBM47 and overexpressed its expression in HCT116 cells to evaluate its influence on cellular phenotypes and performed transcriptome sequencing (RNA-seq) to identify the downstream targets. After RBM47 overexpression (RBM47-OE), we found the proliferation level was decreased while apoptosis level was increased. Meanwhile, the invasion and migration ability of HCT116 was also significantly inhibited by RBM47-OE. We identified 216 up and 97 down differentially expressed genes (DEGs) by RBM47-OE, and found they were significantly enriched in immune response, apoptosis, TNF signaling, and autophagy pathways. RBM47-OE can also regulate the splicing pattern of 2541 alternative splicing (AS) events. The regulated AS genes were enriched in cell cycle, DNA damage and repair, mRNA splicing, and cell division associated pathways. To validate the regulation on gene expression and AS, we selected several DEGs and AS events to perform RT-qPCR experiment. The results showed that the expression level of CASP3, CCN1, and ATF5, and the splicing pattern of CD44 and MDM2, were significantly regulated by RBM47-OE in HCT116 cells. In summary, our results demonstrated the anti-tumor function and the globally downstream targets of RBM47 in CRC cells. We extend the cellular and molecular cognition of RBM47 in tumor. The identified molecular targets can also be served as potential targets for CRC treatment in future.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) has emerged as one of the most malignant tumors that seriously affect the life quality and survival time of patients, ranking the third and second for morbidity and mortality of all cancer types, respectively1. At present, the treatment of colorectal cancer is mainly a comprehensive treatment consisting of surgical procedures and traditional radiotherapy and chemotherapy. However, it has many adverse reactions, long recovery time, and certain limitations. In addition, among newly diagnosed colorectal cancer patients, 20% of them have already metastasized2,3. Recently studies demonstrated the important roles of post-transcriptional (PT) regulation in the progression of cancers4. As the key regulators of PT regulation, RNA binding proteins (RBPs) coordinately regulate the biogenesis, progression, translation, and degradation of RNAs, thus participating in the development of multiple kinds of cancers5including CRC6,7. For example, ZMAT3, an RBP induced by p53, binds to the precursor RNA and represses oncogenic isoform of CD44 to inhibit growth of CRC cells8. Two RNA-binding motif proteins, RBM24 and RBM38, can bind to the 3’ untranslated region of PTEN to stabilize its expression, thus inhibiting tumorigenesis and progression of CRC, respectively9,10. Meanwhile, the functions and molecular mechanisms of most RBPs are largely unknown in CRC.
Recently, we focused on another RBP, RBM47, in CRC. One recent study has summarized the functions of RBM47 in vertebrate development and cancers by post-transcriptional regulation11. Meanwhile, RBM47 can also drive the cell fate decisions by regulating p53-p21 axis at transcriptional level12 indicating its multifaceted regulatory functions. In cancers, RBM47 has been recognized as a tumor suppressor in multiple tumor types, including breast cancer13 non-small cell lung carcinoma14 papillary thyroid carcinoma15 and hepatocellular carcinoma16. It is also reported that RBM47 can promote tumorigenesis of nasopharyngeal carcinoma through both transcriptional and post-transcriptional regulations by cooperating with hnRNPM17. In CRC, RBM47 knockdown has also been reported to promote cell migration, invasion and metastases formation in cancer cell lines18. RBM47 can also regulate intestinal injury and tumorigenesis of CRC by modifying proliferation, inflammation, and tumorigenic pathways19. In summary, these results demonstrate the important roles of RBM47 in inhibiting cancer progression, while the underlying mechanism are not well investigated.
In this study, to better understand how RBM47 regulates the biological processes of CRC cells, we enhanced the expression of RBM47 by transfecting the exogenous RBM47 plasmid in HCT116 cells and performing following experiments to decipher the function and regulatory mechanisms of RBM47. Cellular proliferation and apoptosis experiments were conducted to assess the influence of RBM47 on cell growth, and migration and invasion experiments showed the impact of RBM47 on cell metastasis. More importantly, we performed RNA-seq analysis to illustrate the underlying mechanism and downstream targets of RBM47 in HCT116 cells, including the differentially expressed genes and alternative splicing events. In summary, our study highlights the anti-cancer function of RBM47 in HCT116 cells, as well as the potential molecular targets, especially for the alternative splicing events and genes, regulated by RBM47. Our results extend the understanding of RBM47 function in cancer cells, and the identified molecular targets can be served as potential therapeutic targets for CRC in future.
Materials and methods
Cell culture and transfections
We selected the widely used human colorectal cancer HCT116 cell line as research object. The HCT116 cells (Procell, Wuhan, China) were validated by the short tandem repeat (STR) method, and cultured at 37℃ with 5% CO2 in McCoy’s 5 A with 10% fetal bovine serum (FBS), 100 µg/mL streptomycin, 100 U/mL penicillin. The RBM47 transcript NM_019027 was inserted into pIRES-hrGFP-1a plasmid, which was transfected into HCT116 cells by Lipofectamine 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocol. The transfected cells were cultured for 48 h for downstream experiments.
For Actinomycin D (ActD) treatment, after 48 h of transfection with RBM47 and control plasmids, the culture medium was removed and replaced with a medium containing 0.1 nM ActD (HY-17559, MCE). After continuing to culture for 48 h, the cells were collected.
Cell proliferation assay
The cell proliferation assay was performed using the cell counting kit-8 (CCK-8, CK04, Dojindo, Japan) with 0.5*105 cells as input according to the published procedure20. Subsequently, the optical density of the cells was measured with a microplate reader (ELX800, Biotek, USA) at an absorbance of 450 nm to calculate the cell proliferation rate.
Flow cytometric analysis of cell apoptosis
The Annexin V-FITC/PI cell apoptosis detection kit (40302ES60, Yeasen Biotechnology, China) was used for apoptosis experiment with 1.4*105 cells as input. The cells were mixed with 5 µl Annexin V-FITC incubated at room temperature in the dark for 5 min and 10 µl PI reagents incubated for 5 min as described above respectively. Then the samples were subjected to flow cytometry (FACSCanto, BD, USA) analysis to detect cell apoptosis levels.
Cell migration assay
In vitro migration assays were performed using transwell chambers (3422, Corning, USA). 3*105 HCT116 cells in 0.2 ml serum-free medium were added to the transwell chambers with 8 μm filter, then the chambers inserted in medium with 600ul 10% FBS (10091148, Gibco, China) served as a chemoattractant in the lower chamber, and incubated for 48 h at 37℃ and 5% CO2. Cells remaining on the upper membrane surface of the inserts were then removed with a cotton swab, and the total number of cells that migrated into the lower chamber were fixed by 4% paraformaldehyde (P0099, Beyotime, China) for 10 min, then stained with 0.1% crystal violet (C0121, Beyotime, China). The migration cells were observed and counted under inverted microscope (MF52-N, Mshot, China) at 200× magnification.
Cell invasion assay
In vitro invasion assays were performed using transwell chambers (3422, Corning, USA). The transwell chambers with 8 μm filter and precoated with a thin layer of Matrigel (356234, BD Biosciences, USA), diluted for 1:8 using serum-free medium, 100 µl diluted matrigel in chambers was incubated for 1 h at 37℃ and 5% CO2 and removed unsolidified supernatant. 3 × 105 HCT116 cells in 0.2 ml serum-free medium were added to the inserts, then the transwell chambers inserted in medium with 600 µl 10% FBS (10091148, Gibco, China) served as a chemoattractant in the lower chamber, and incubated for 48 h at 37℃ and 5% CO2. Cells remaining on the upper membrane surface of the inserts were then removed with a cotton swab, and the total number of cells that invaded into the lower chamber were fixed by 4% paraformaldehyde (P0099, Beyotime, China) for 10 min, then stained with 0.1% crystal violet (C0121, Beyotime, China). The invasion cells were observed and counted under inverted microscope (MF52-N, Mshot, China) at 200× magnification.
RNA extraction and sequencing
Total RNAs were extracted from HCT116 cells using TRIzol Reagent (Invitrogen, cat. NO 15596026)following the published method21. DNA digestion was carried out after RNA extraction by DNaseI. RNA quality was determined by examining A260/A280 with NanodropTM OneCspectrophotometer (Thermo Fisher Scientific Inc). RNA Integrity was confirmed by 1.5% agarose gel electrophoresis. Qualified RNAs were finally quantified by Qubit3.0 with QubitTM RNA Broad Range Assay kit (Life Technologies, Q10210).
We used 2 µg total RNAs for stranded RNA sequencing library preparation using KCTM Stranded mRNA Library Prep Kit for Illumina (Catalog NO. DR08402, Wuhan Seqhealth Co., Ltd. China) following the manufacturer’s instruction. PCR products corresponding to 200–500 bps were enriched, quantified and finally sequenced on Novaseq 6000 sequencer (Illumina) with PE150 model.
RNA-Seq raw data clean and alignment
Raw reads containing more than 2-N bases were first discarded. Then adaptors and low-quality bases were trimmed from raw sequencing reads using FASTX-Toolkit (Version 0.0.13). The short reads less than 16nt were also dropped. After that, clean reads were aligned to the human GRCh38 genome by HISAT222 allowing 4 mismatches. Uniquely mapped reads were used for gene reads number counting and fragments per kilobase of transcript per million fragments mapped (FPKM) calculation23. The statistical power of this experimental design, calculated in RNASeqPower is 1.
Differentially expressed genes (DEG) analysis
The R Bioconductor package DESeq224 was utilized to screen out the differentially expressed genes (DEGs). The thresholds P-value < 0.01 and fold change > 1.5 were set as the cut-off criteria for identifying DEGs.
Alternative splicing analysis
The alternative splicing events (ASEs) and regulated alternative splicing events (RASEs) between the samples were defined and quantified by using the ABLas pipeline as described previously25,26. In brief, ABLas detection of ten types of ASEs was based on the splice junction reads, including exon skipping (ES), alternative 5’ splice site(A5SS), alternative 3’splice site(A3SS), mutually exclusive exons (MXE), mutually exclusive 5’UTRs (5pMXE), mutually exclusive 3’UTRs (3pMXE), cassette exon, A3SS&ES, and A5SS&ES.
To assess RBP regulated ASE, Student’s t-test was performed to evaluate the significance of the ratio alteration of AS events. Those events which were significant at P-value cutoff corresponding to a false discovery rate cutoff of 5% were considered RBP regulated ASEs.
Functional enrichment analysis
To sort out functional categories of DEGs, Gene Ontology (GO) terms and KEGG pathways were identified using KOBAS 2.0 server27. Hyper geometric test and Benjamini-Hochberg FDR controlling procedure were used to define the enrichment of each term.
Statistical analysis
For statistical analysis, error bars represent mean ± SEM. Unpaired and two-tail Student’s t-test was used to analyze the statistical difference between two groups.
Results
RBM47 acts as a tumor suppressor in CRC patients and HCT116 cell line
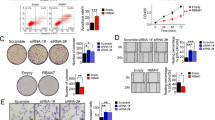
Previous study has demonstrated that RBM47 knockdown can promote cell migration, invasion and metastases formation of CRC cell lines18 while the underlying mechanism is largely unknown. By investigating the cancer genome atlas (TCGA) dataset of colon adenocarcinoma (COAD) patients, we found RBM47 was significantly downregulated in tumor samples compared with adjacent normal samples (Fig. 1A). At the same time, higher expression level of RBM47 in COAD patients was associated with better overall survival time compared with patients with lower RBM47 level (Fig. 1B). To further confirm the cellular functions of RBM47, we performed RBM47 overexpression (RBM47-OE) because of the low expression level of RBM47 in HCT116 cell line. Both RT-qPCR and western blot experiments confirmed the successful overexpression of RBM47 in HCT116 cells (Fig. 1C-D, Figure S1A-B). Then we explored the cellular functions of RBM47-OE by checking its influence on cell proliferation and apoptosis. The results demonstrated that RBM47-OE significantly inhibited the proliferation level of HCT116 cells at 48 and 72 h (Fig. 1E), while RBM47-OE significantly promoted the apoptosis level of HCT116 cells (Fig. 1F, Figure S1C). Then we performed cell invasion and migration experiments, and found that RBM47-OE significantly decreased the abilities of cell invasion (Fig. 1G) and migration (Fig. 1H) of HCT116 cells. These results were complementary to previous conclusion and further confirmed the tumor-suppressing function of RBM47 in CRC cell line.
RBM47 functions as a tumor suppressor in colorectal cancer. (A) Box plot showing the expression level of RBM47 in tumor and adjacent normal tissues of COAD patients from TCGA. (B) The survival curve of RBM47 in colorectal cancer in TCGA. (C) The histogram showed the RT-qPCR results of control and treatment samples. *** P-value < 0.001. (D) The result of western blot experiment showed that the RBM47 over expression was successful. (E) The proliferation results of HTC116 after RBM47 overexpression. * P-value < 0.05; ** P-value < 0.01. (F) The apoptosis results of HTC116 after RBM47 overexpression. *** P-value < 0.001. (G) The invasion results of HCT116 cells after RBM47 overexpression. ** P-value < 0.01. (H) The migration results of HCT116 cells after RBM47 overexpression. *** P-value < 0.001.
RBM47-OE globally modulated the transcriptome profile of HCT116 cells
To further explore how RBM47 modulates the cellular phenotypes of HCT116 cells, we performed global RNA-seq analysis and identified the transcriptome difference between RBM47-OE vs. NC samples. After calculating the FPKM values of expressed genes, we found that RBM47 expression was also significantly increased in OE samples (Fig. 2A), confirmed the detection by RT-qPCR. Principal component analysis (PCA) for all detected genes demonstrated the clear separation between RBM47-OE and NC samples at the first component (Fig. 2B), indicating that RBM47 can globally modulate the transcriptome profile of HCT116 cells. Thus, we predicted the differentially expressed genes (DEGs) using fold change > 1.5 and p-value < 0.01 as thresholds, and finally obtained 216 up and 97 down DEGs (Fig. 2C). The fact that more up DEGs than down DEGs indicates that RBM47-OE prefers to promote gene expression in HCT116 cells. By plotting the expression pattern of DEGs, we found they showed consistent expression pattern among the three replicates for each group (Fig. 2D).
RBM47-OE affect gene expression profiles of HCT116 cells. (A) Bar plot showing the expression pattern and statistical difference of DEGs for RBM47. Error bars represent mean ± SEM. *** P-value < 0.001. (B) PCA base on FPKM value of all detected genes RBM47 overexpression. The ellipse for each group is the confidence ellipse. (C) Volcano plot showing all differentially expressed genes (DEGs) between RBM47-OE and Ctrl samples. (D) Hierarchical clustering heat map showing expression levels of all DEGs.
DEGs induced by RBM47-OE were enriched in cell cycle and apoptosis pathways
To investigate the functions of DEGs induced by RBM47-OE, we performed functional enrichment analysis using GO and KEGG databases. We found the up DEGs were enriched in cell projection organization, regulation of transcription by RNA polymerase II, cilium assembly, and endocytosis GO pathways (Fig. 3A), whereas the down DEGs were mainly enriched in defense response to virus, immune system process, apoptotic process, and negative regulation of apoptotic process pathways (Fig. 3B). For KEGG analysis, apoptosis-multiple species, TNF signaling pathway, and autophagy-animal pathways were emerged in the top pathways of up DEGs (Fig. 3C). Several cancer-associated pathways, including hepatocellular carcinoma and human papillomavirus infection were detected in down DEGs (Fig. 3D). Several apoptosis-associated genes, including CASP3, ACVR1C, LCN2, BIRC3, IL1A, and STK17B, were significantly upregulated by RBM47-OE, whereas several other apoptotic genes, including HSPA1B, IFI6, HMOX1, TMEM102, ATOX1, ATF5, CCN1, DNASE1, TERT, IFI27, BNIPL, and INPP5D, were downregulated by RBM47-OE (Fig. 3E). For cell cycle genes, INTS13, ABRAXAS2, BRCC3, POC5, THAP1, PIK3C3, STOX1, and BRCA2 were upregulated, whereas CDC20 was downregulated (Fig. 3F). To confirm the regulation of RBM47 on DEGs, we selected three DEGs and performed RT-qPCR experiment. CASP3 was upregulated, and CCN1 and ATF5 were downregulated after RBM47-OE by RT-qPCR (Fig. 3G). In addition, we also used Actinomycin D (actD) to inhibit the transcription process of HCT116 cells. Further RT-qPCR experiment demonstrated that RBM47-OE enhanced CASP3 expression, inhibited CNN1 expression, but not significantly changed the expression of ATF5 under ActD treatment (Fig. 3H). Meanwhile, we also tested the expression pattern of TERT and did not find significant difference between NC and RBM47-OE under ActD treatment (Fig. 3I). In summary, these results indicated that RBM47 can regulate gene expression at both transcriptional and post-transcriptional levels and were consistent with the discovery that RBM47-OE significantly changed the proliferation and apoptosis levels of HCT116 cells.
RBM47 broadly regulate expression of cell cycle and apoptosis associated genes in HTC116 cells. (A) The Scatter plot exhibiting the most enriched GO biological process results of the up-regulated DEGs. (B) The Scatter plot exhibiting the most enriched GO biological process results of the down-regulated DEGs. (C) The Scatter plot exhibiting the most enriched KEGG pathway results of the up-regulated DEGs. (D) The Scatter plot exhibiting the most enriched KEGG pathway results of the down-regulated DEGs. (E) Clustering heatmap showing the DEGs involved in apoptosis pathway. (F) Clustering heatmap showing the DEGs involved in cell cycle pathway. (G) Bar plot showing the expression pattern and statistical difference of DEGs for some important genes. (H) Bar plot showing the RT-qPCR results and statistical difference of DEGs in (G) by ActD treatment. (I) The same as (H) but for TERT gene. Error bars represent mean ± SEM. **** P-value < 0.0001, *** P-value < 0.001, ** P-value < 0.01, * P-value < 0.05.
RBM47-OE had profound impact on the alternative splicing pattern of HCT116 cells
RBM47 has been reported to regulate alternative splicing of cancer-related genes in nasopharyngeal carcinoma17. Thus, we dedicated to explore the AS profile alteration by RBM47-OE in HCT116 cells. We universally detected the known and novel AS events (ASEs) from the aligning result, and found that A3SS and A5SS were the two most detected ASEs (Fig. 4A). To further illustrate how RBM47-OE regulated AS profile, we predicted the regulated ASEs (RASEs) by RBM47, and finally detected 2541 RASEs. The results showed that A5SS and A3SS were also the top two RASE types if we divided cassette exon and exon skipping (ES) into two types (Fig. 4B). Meanwhile, we found more exons were included than excluded by RBM47-OE as we detected more excluded (down) ES and more included (up) cassette exon events (Fig. 4B), indicating that RBM47-OE had the ability to include exons during splicing progression. By plotting the ratios of these RASEs, we found these RASEs were consistently regulated in all the three replicates (Fig. 4C). Finally, we performed an overlap analysis for RASE genes (RASGs) and DEGs, and detected 30 overlapped genes (Fig. 4D). The much more RASG number indicates that RBM47 probably has more important roles in AS regulation that gene expression modulation in HCT116 cells.
RBM47-OE broadly change the alternative splicing patterns in HCT116 cells. (A) Bar plot showing the RBM47 all regulated alternative splicing event. (B) Bar plot showing the RBM47 regulated alternative splicing events (RASEs). (C) Hierarchical clustering heat map showing expression levels of RBM47 regulated alternative splicing event (RASE). (D) Venn diagram showing the overlapped genes between RASGs and DEGs. (E) Hierarchical clustering heat map showing expression levels of the overlapped 30 genes in (D).
RBM47 broadly regulates the AS profile of genes involved in cell cycle and DNA repair
AS RBM47-OE significantly modulated the AS pattern of 1716 RASGs (Fig. 4D), then we explored the enriched functions of these RASGs. We found these RASGs were dominantly enriched in cell cycle, cellular response to DNA damage stimulus, protein ubiquitination, mRNA processing, mRNA splicing via spliceosome, cell division, vesicle-mediated transport, chromatin organization, DNA repair, and base-excision repair GO BP pathways (Fig. 5A). KEGG enrichment analysis for these RASGs showed they were enriched in shigellosis, autophagy, and ubiquitin mediated proteolysis pathways (Figure S2A). Among the detected GO BP pathways, we found there were five pathways belonging to cell proliferation, thirteen pathways belonging to apoptosis, and sixteen pathways belonging to cell cycle (Fig. 5B-D, Figure S2B), although a lot of them were not significant. These results indicates that RASGs contained genes involved in cell proliferation, apoptosis, and cell cycle, which may have essential regulation on the cellular phenotypes modulated by RBM47. By clustering the RASE ratios of RASGs from proliferation, apoptosis, and cell cycle, we found these RASEs showed consistent dysregulation in three replicates by RBM47-OE (Fig. 5E). Meanwhile, we also found one RASE from CD44, an important factor in tumor metastasis28 was significantly dysregulated by RBM47-OE, and also confirmed by RT-qPCR experiment (Fig. 5F). MDM2, an E3 ubiquitin ligase protein that is a candidate target for tumor therapy29 also showed AS dysregulation with an A5SS event, and was confirmed by RT-qPCR (Figure S2C). Taken together, these results demonstrated that RBM47 can globally regulate the AS profile that are associated with cell cycle, DNA damage and repair, and mRNA splicing.
RBM47 regulates alternative splicing of cell cycle, proliferation and apoptosis associated genes in HCT116 cells. (A) Scatter plot exhibiting the most enriched GO biological process results of the RASGs. (B) Scatter plot exhibiting all the proliferative pathways GO biological process results of the RASGs. (C) Scatter plot exhibiting all the apoptotic pathways GO biological process results of the RASGs. (D) Scatter plot exhibiting all the cycle pathways GO biological process results of the RASGs. (E) Hierarchical clustering heat map showing expression levels of all associated genes about (B), (C) and (D) in RBM47 regulated alternative splicing events (RASEs). (F) RBM47 regulates alternative splicing of CD44. Left panel: IGV-sashimi plot showing the regulated alternative splicing events and binding sites across mRNA. Reads distribution of RASE is plotted in the up panel and the transcripts of each gene are shown below. Right panel: The schematic diagrams depict the structures of ASEs. RNA-seq validation of RASEs was shown at the bottom of the right panel. Error bars represent mean ± SEM. *** P-value < 0.001, ** P-value < 0.01, * P-value < 0.05.
Discussion
The previous research suggests that RBM47 has an anti-proliferation and anti-metastasis effect in several cancer cells30,31,32. Our analysis of the TCGA database also confirmed that the expression of RBM47 was closely associated with the progression of CRC. Additionally, when we overexpressed RBM47 in the HCT116 cell line, we observed the inhibited cell proliferation and promoted apoptosis. These findings align with previous studies indicating that reducing RBM47 expression enhances CRC cell lines’ migration, invasion, and metastasis19. Moreover, we next performed RNA-seq analysis to decipher the underlying mechanisms and molecular targets about RBM47’s anti-cancer function in HCT116 cells. In summary, our results highlight the important roles in cellular phenotype and transcriptome modulation of RBM47, which extends our understanding about the regulatory mechanisms of RBM47 in CRC.
Through RNA-seq analysis, we found that RBM47-OE globally regulated the transcriptomic landscape of HCT116 cells. We identified 313 DEGs and 1716 RASEs. Functional enrichment analysis revealed that these DEGs and RASEs were primarily enriched in biological processes such as cell cycle, apoptosis, and DNA repair, which are closely related to the tumor-suppressive role of RBM47. RBM47 can act as an interferon-stimulating gene to enhance the antiviral effect of the host by stabilizing the mRNA of the target gene, revealing its role in antiviral immunity. The ISGylation of RBM47 can regulate immune activation, which is a well-established antiviral mechanism, and participate in DNA repair, autophagy, transcription, protein translation, and other cellular processes33,34. Interestingly, the DEGs of RBM47-OE are enriched in these related pathways, which implies RBM47 may regulate CRC cell proliferation by itself ISGylation, stabilizing mRNA, and alternative splicing of downstream immune response and tumor suppression-related genes.
We specifically focused on the effect of RBM47 on cell cycle and apoptosis-related genes. For instance, apoptosis-related gene such as CASP3, was significantly upregulated after RBM47-OE, while genes like CCN1 and ATF5 were downregulated. Cell cycle genes such as INTS13 and BRCA2 were also positively regulated by RBM47-OE. CCN1 is a cysteine-rich intercellular matrix protein, high expression of CCN1 is associated with the progression and poor prognosis of multiple cancers and is involved in tumor growth, angiogenesis, and cancer cell adhesion, migration, and invasion35,36,37,38. ATF5 modulates many genes’ expression and regulates the cell cycle, unfolded protein response (UPR), anti-apoptosis, immune, and other signaling pathways, playing a crucial role in the survival and proliferation of cancer cells. Its expression is negatively correlated with the survival rate of patients with various types of cancer, making it a popular target for cancer therapy39,40,41,42,43,44. CASP3 plays a key role in apoptosis and is a biomarker of apoptosis. Activation of the CASP3/GSDME signaling pathway can induce pyroptosis or apoptosis, which in turn affects the growth of tumors and the survival rate of cancer patients45,46,47. The expression of CASP3 is up-regulated in most cancers, but down-regulated in colon adenocarcinoma and rectum adenocarcinoma, indicating that CASP3 has different regulatory functions in various tumors48.
What’s more, we observed a broad effect of RBM47 on alternative splicing patterns, which may further regulate the functions of these genes. Cell cycle, DNA damage and repair, mRNA splicing, and cell division were the top enriched pathways for RASGs (Fig. 5A). It has been reported that RBM47 regulates the exon inclusion of TJP1 and inhibits expression of TJP1-α isoform, which in turn enhances the assembly of actin stress fibers and promotes cellular migration in A549 cells49. The identified RASGs in this study were tightly associated with the cellular functions of RBM47 in HCT116 cells. We propose that RBM47 can broadly alter the splicing pattern of these genes and modifies the expression level, amino acid sequences or structures of the encoded proteins, and finally changes their functions in tumor cells. RBM47 modulates the splicing of CD44, which is a marker of cancer stem cells. It is reported that CD44 has tumor-processing function and promotes EMT and metastasis of colon cancer cells28,50. RBM47 also altered the splicing of MDM2 (Figure S2C). Several studies have demonstrated that MDM2 is a new therapeutic option in colon cancer treatment51 as both the inhibitor and anti-sense oligonucleotide of MDM2 have anti-tumor activity by inhibiting growth and proliferation of colon cancer cells52,53. These results indicate that RBM47 inhibits growth and metastasis of HCT116 cells probably by regulating the splicing pattern of CD44 and MDM2 transcripts. Finally, the AS results were consistent with our RT-qPCR detection results, further confirming the anti-tumor function of RBM47 in CRC. Further studies are necessary to decipher the functions of the alternative spliced forms of CD44 or MDM2, which are not deeply investigated in this study.
Although our study provides insights into the potential mechanisms of RBM47’s role in CRC, it has some limitations. First, we conducted experiments using only one cell line, which may limit the generalizability of the results. In addition, we did not perform RBM47 knockdown experiments in cell and animal model to validate our findings. Furthermore, as an RNA-binding protein, it is worth exploring whether the regulatory effects of RBM47 may differ between left-sided and right-sided colorectal cancer due to the different primary sites54,55,56. Future studies should include multiple CRC cell lines and in vivo animal models to explore the function and molecular mechanism of RBM47.
Conclusions
In conclusion, our findings highlight the significance of RBM47 as a tumor suppressor gene in CRC. RBM47 plays a role in cell proliferation and apoptosis via regulating gene expression and alternative splicing, providing potential molecular targets and valuable information for the development of novel CRC treatment strategies.
Data availability
The raw RNA-seq data has been deposited in the NCBI GEO database with accession ID GSE282110. Editors and reviewers can contact the corresponding author Guangshi Liu for for the data availability request.
References
Bray, F. et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. https://doi.org/10.3322/caac.21834 (2024).
Biller, L. H. & Schrag, D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA 325, 669–685. https://doi.org/10.1001/jama.2021.0106 (2021).
Zhang, N. et al. Novel therapeutic strategies: targeting epithelial-mesenchymal transition in colorectal cancer. Lancet Oncol. 22, e358–e368. https://doi.org/10.1016/S1470-2045(21)00343-0 (2021).
Audic, Y. & Hartley, R. S. Post-transcriptional regulation in cancer. Biol. Cell. 96, 479–498. https://doi.org/10.1016/j.biolcel.2004.05.002 (2004).
Pereira, B., Billaud, M. & Almeida, R. RNA-Binding proteins in cancer: old players and new actors. Trends Cancer. 3, 506–528. https://doi.org/10.1016/j.trecan.2017.05.003 (2017).
Chatterji, P. & Rustgi, A. K. RNA binding proteins in intestinal epithelial biology and colorectal Cancer. Trends Mol. Med. 24, 490–506. https://doi.org/10.1016/j.molmed.2018.03.008 (2018).
Garcia-Cardenas, J. M. et al. Post-transcriptional regulation of colorectal cancer: A focus on RNA-Binding proteins. Front. Mol. Biosci. 6, 65. https://doi.org/10.3389/fmolb.2019.00065 (2019).
Muys, B. R. et al. The p53-induced RNA-binding protein ZMAT3 is a splicing regulator that inhibits the splicing of oncogenic CD44 variants in colorectal carcinoma. Genes Dev. 35, 102–116. https://doi.org/10.1101/gad.342634.120 (2021).
Xia, R. M., Liu, T., Li, W. G. & Xu, X. Q. RNA-binding protein RBM24 represses colorectal tumourigenesis by stabilising PTEN mRNA. Clin. Transl Med. 11, e383. https://doi.org/10.1002/ctm2.383 (2021).
Guan, B. et al. RNA-binding protein RBM38 inhibits colorectal cancer progression by partly and competitively binding to PTEN 3’UTR with miR-92a-3p. Environ. Toxicol. 36, 2436–2447. https://doi.org/10.1002/tox.23356 (2021).
Shivalingappa, P. K. M., Sharma, V., Shiras, A. & Bapat, S. A. RNA binding motif 47 (RBM47): emerging roles in vertebrate development, RNA editing and cancer. Mol. Cell. Biochem. 476, 4493–4505. https://doi.org/10.1007/s11010-021-04256-5 (2021).
Radine, C. et al. The RNA-binding protein RBM47 is a novel regulator of cell fate decisions by transcriptionally controlling the p53-p21-axis. Cell. Death Differ. 27, 1274–1285. https://doi.org/10.1038/s41418-019-0414-6 (2020).
Vanharanta, S. et al. Loss of the multifunctional RNA-binding protein RBM47 as a source of selectable metastatic traits in breast cancer. Elife https://doi.org/10.7554/eLife.02734 (2014).
Shen, D. J., Jiang, Y. H., Li, J. Q., Xu, L. W. & Tao, K. Y. The RNA-binding protein RBM47 inhibits non-small cell lung carcinoma metastasis through modulation of AXIN1 mRNA stability and Wnt/beta-catentin signaling. Surg. Oncol. 34, 31–39. https://doi.org/10.1016/j.suronc.2020.02.011 (2020).
Qin, Y. et al. RBM47/SNHG5/FOXO3 axis activates autophagy and inhibits cell proliferation in papillary thyroid carcinoma. Cell. Death Dis. 13, 270. https://doi.org/10.1038/s41419-022-04728-6 (2022).
Guo, T. et al. RBM47 inhibits hepatocellular carcinoma progression by targeting UPF1 as a DNA/RNA regulator. Cell. Death Discov. 8, 320. https://doi.org/10.1038/s41420-022-01112-3 (2022).
Xu, X. C. et al. RNA-binding motif protein RBM47 promotes tumorigenesis in nasopharyngeal carcinoma through multiple pathways. J. Genet. Genomics. 48, 595–605. https://doi.org/10.1016/j.jgg.2021.05.006 (2021).
Rokavec, M., Kaller, M., Horst, D. & Hermeking, H. Pan-cancer EMT-signature identifies RBM47 down-regulation during colorectal cancer progression. Sci. Rep. 7, 4687. https://doi.org/10.1038/s41598-017-04234-2 (2017).
Soleymanjahi, S. et al. RBM47 regulates intestinal injury and tumorigenesis by modifying proliferation, oxidative response, and inflammatory pathways. JCI Insight https://doi.org/10.1172/jci.insight.161118 (2023).
Saiding, A. et al. PCMT1 knockdown attenuates malignant properties by globally regulating transcriptome profiles in triple-negative breast cancer cells. PeerJ 11, e16006. https://doi.org/10.7717/peerj.16006 (2023).
Chomczynski, P. & Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 162, 156–159. https://doi.org/10.1006/abio.1987.9999 (1987).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods. 12, 357–360 (2015).
Trapnell, C. et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 1–21 (2014).
Jin, L. et al. Transcriptome analysis reveals the complexity of alternative splicing regulation in the fungus verticillium dahliae. BMC Genom. 18, 1–14 (2017).
Xia, H. et al. CELF1 preferentially binds to exon-intron boundary and regulates alternative splicing in hela cells. Biochim. Et Biophys. Acta (BBA)-Gene Regul. Mech. 1860, 911–921 (2017).
Xie, C. et al. KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res. 39, W316–W322 (2011).
Su, Y. J., Lai, H. M., Chang, Y. W., Chen, G. Y. & Lee, J. L. Direct reprogramming of stem cell properties in colon cancer cells by CD44. EMBO J. 30, 3186–3199. https://doi.org/10.1038/emboj.2011.211 (2011).
Vassilev, L. T. MDM2 inhibitors for cancer therapy. Trends Mol. Med. 13, 23–31. https://doi.org/10.1016/j.molmed.2006.11.002 (2007).
Li, Z. et al. Deubiquitinating enzyme OTUD4 stabilizes RBM47 to induce ATF3 transcription: a novel mechanism underlying the restrained malignant properties of ccrcc cells. Apoptosis https://doi.org/10.1007/s10495-024-01953-6 (2024).
Su, Q. et al. RBM47 restrains renal cell carcinoma progression and chemoresistance through interacting with lncrna HOXB-AS1. Cell. Death Discov. 9, 329. https://doi.org/10.1038/s41420-023-01623-7 (2023).
Wang, C. et al. Downregulation of RNA binding protein 47 predicts low survival in patients and promotes the development of renal cell malignancies through RNA stability modification. Mol. Biomed. https://doi.org/10.1186/s43556-023-00148-w (2023).
Wang, K. et al. RNA-binding protein RBM47 stabilizes IFNAR1 mRNA to potentiate host antiviral activity. EMBO Rep. 22, e52205. https://doi.org/10.15252/embr.202052205 (2021).
Ding, S. et al. Dynamic RBM47 isgylation confers broad immunoprotection against lung injury and tumorigenesis via TSC22D3 downregulation. Cell. Death Discov. 9, 430. https://doi.org/10.1038/s41420-023-01736-z (2023).
Espinoza, I. et al. Depletion of CCN1/CYR61 reduces triple-negative/basal-like breast cancer aggressiveness. Am. J. Cancer Res. 12, 839–851 (2022).
Yan, J., Yang, B., Lin, S., Xing, R. & Lu, Y. Downregulation of miR-142-5p promotes tumor metastasis through directly regulating CYR61 expression in gastric cancer. Gastric Cancer. 22, 302–313. https://doi.org/10.1007/s10120-018-0872-4 (2019).
Tanaka, F. et al. Snail promotes Cyr61 secretion to prime collective cell migration and form invasive tumor nests in squamous cell carcinoma. Cancer Lett. 329, 243–252. https://doi.org/10.1016/j.canlet.2012.11.023 (2013).
Maity, G. et al. The MAZ transcription factor is a downstream target of the oncoprotein Cyr61/CCN1 and promotes pancreatic cancer cell invasion via CRAF-ERK signaling. J. Biol. Chem. 293, 4334–4349. https://doi.org/10.1074/jbc.RA117.000333 (2018).
Angelastro, J. M. Targeting ATF5 in Cancer. Trends Cancer. 3, 471–474. https://doi.org/10.1016/j.trecan.2017.05.004 (2017).
Deng, P. & Haynes, C. M. Mitochondrial dysfunction in cancer: potential roles of ATF5 and the mitochondrial UPR. Semin Cancer Biol. 47, 43–49. https://doi.org/10.1016/j.semcancer.2017.05.002 (2017).
He, F., Xiao, H., Cai, Y. & Zhang, N. ATF5 and HIF1α cooperatively activate HIF1 signaling pathway in esophageal cancer. Cell. Commun. Signal. 19, 53. https://doi.org/10.1186/s12964-021-00734-x (2021).
Chen, M. et al. Emerging roles of activating transcription factor (ATF) family members in tumourigenesis and immunity: implications in cancer immunotherapy. Genes Dis. 9, 981–999. https://doi.org/10.1016/j.gendis.2021.04.008 (2022).
Zhou, J. et al. Activating transcription factor 5 (ATF5) promotes tumorigenic capability and activates the Wnt/b-catenin pathway in bladder cancer. Cancer Cell. Int. 21, 660. https://doi.org/10.1186/s12935-021-02315-x (2021).
Cao, M. et al. ATF5 promotes malignant T cell survival through the PI3K/AKT/mTOR pathway in cutaneous T cell lymphoma. Front. Immunol. 14, 1282996. https://doi.org/10.3389/fimmu.2023.1282996 (2023).
Bhat, A. A. et al. The pyroptotic role of Caspase-3/GSDME signalling pathway among various cancer: A review. Int. J. Biol. Macromol. 242, 124832. https://doi.org/10.1016/j.ijbiomac.2023.124832 (2023).
Hu, Y. et al. The multifaceted roles of GSDME-mediated pyroptosis in cancer: therapeutic strategies and persisting obstacles. Cell. Death Dis. 14, 836. https://doi.org/10.1038/s41419-023-06382-y (2023).
Pu, X. et al. Caspase-3 and caspase-8 expression in breast cancer: caspase-3 is associated with survival. Apoptosis 22, 357–368. https://doi.org/10.1007/s10495-016-1323-5 (2017).
Zhou, Z., Xu, S., Jiang, L., Tan, Z. & Wang, J. A. Systematic Pan-Cancer analysis of CASP3 as a potential target for immunotherapy. Front. Mol. Biosci. 9, 776808. https://doi.org/10.3389/fmolb.2022.776808 (2022).
Kim, Y. E. et al. RBM47-regulated alternative splicing of TJP1 promotes actin stress fiber assembly during epithelial-to-mesenchymal transition. Oncogene 38, 6521–6536. https://doi.org/10.1038/s41388-019-0892-5 (2019).
Cho, S. H. et al. CD44 enhances the epithelial-mesenchymal transition in association with colon cancer invasion. Int. J. Oncol. 41, 211–218. https://doi.org/10.3892/ijo.2012.1453 (2012).
Hav, M. et al. MDM2 gene amplification and protein expressions in colon carcinoma: is targeting MDM2 a new therapeutic option? Virchows Arch. 458, 197–203. https://doi.org/10.1007/s00428-010-1012-7 (2011).
Rigatti, M. J., Verma, R., Belinsky, G. S., Rosenberg, D. W. & Giardina, C. Pharmacological inhibition of Mdm2 triggers growth arrest and promotes DNA breakage in mouse colon tumors and human colon cancer cells. Mol. Carcinog. 51, 363–378 (2012).
Tortora, G. et al. A novel MDM2 anti-sense oligonucleotide has anti‐tumor activity and potentiates cytotoxic drugs acting by different mechanisms in human colon cancer. Int. J. Cancer. 88, 804–809 (2000).
Hong, Z. et al. Proteomic profiling and tumor microenvironment characterization reveal molecular and immunological hallmarks of Left-Sided and Right-Sided Colon cancer tumorigenesis. J. Proteome Res. 22, 2973–2984. https://doi.org/10.1021/acs.jproteome.3c00302 (2023).
Zhang, L. et al. Immune landscape of colorectal Cancer tumor microenvironment from different primary tumor location. Front. Immunol. 9, 1578. https://doi.org/10.3389/fimmu.2018.01578 (2018).
Guo, W. et al. Resolving the difference between left-sided and right-sided colorectal cancer by single-cell sequencing. JCI Insight https://doi.org/10.1172/jci.insight.152616 (2022).
Acknowledgements
We are grateful for the help and discussion about the manuscript preparation from the colleagues of Wuhan Nissi Biotechnology.
Funding
This study was supported by the “Tianshan Talents” Training Program for High level Talents in Medicine and Health (NO. TSYC202301A031).
Author information
Authors and Affiliations
Contributions
Guangshi Liu: Writing-original draft, Supervision, Formal analysis, Data curation, Conceptualization. Tao Li: Writing-review & editing, Supervision, Conceptualization, Visualization, Resources, Methodology, Funding acquisition. Peng Li: Visualization, Resources, Methodology, Formal analysis. Suyan Wei: Writing-original draft, Validation, Resources, Methodology, Investigation. Kamuran Abulizi: Writing – review & editing, Supervision, Resources, Data curation.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, G., Li, T., Li, P. et al. RBM47 functions as an anti-oncogene by regulating expression and alternative splicing of cell proliferation and apoptosis associated genes in colorectal cancer cells. Sci Rep 15, 26685 (2025). https://doi.org/10.1038/s41598-025-05151-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-05151-5