Abstract
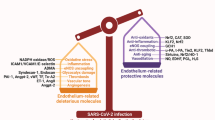
Vasculitis and vascular injury are induced in coronavirus disease 2019 (COVID-19) patients, suggesting an association between multi-organ failure and sequelae. Vascular endothelial dysfunction induced by severe acute respiratory syndrome-related coronavirus-2 (SARS-CoV-2) disrupts vascular barrier function, activates coagulation pathways, and induces extravasation of inflammatory cells. In addition to employing standard anti-inflammatory and antiviral agents, using treatments that stabilize and protect blood vessels following SARS-CoV-2 infection is a potentially effective strategy to relieve COVID-19 severity. Here, we focused on the vasoprotective effects of lysophosphatidic acid (LPA), a lipid mediator with diverse activities, and investigated whether it could be a novel therapeutic for COVID-19. Three-dimensional cultures of various human vascular endothelial cells that form luminal structures showed increased expression of LPA4 and entry receptors for SARS-CoV-2 infection, mimicking blood vessels in vivo. Validation using this culture system showed that LPA attenuated SARS-CoV-2 infection-induced vascular destruction and the activation of inflammatory signaling. In experiments with infected animals, LPA administration protected blood vessels and suppressed inflammation and vascular damage in lung tissue. The activation of LPA signaling in vascular endothelial cells may be an effective therapeutic approach for mitigating vascular injury in COVID-19.
Similar content being viewed by others
Introduction
In addition to respiratory symptoms such as pneumonia, severe coronavirus disease 2019 (COVID-19) is associated with tissue damage attributed to a cytokine storm triggered by the systemic release of pro-inflammatory cytokines. Characteristically, vasculitis and vasculopathy have been reported to be induced, suggesting an association with multi-organ failure in COVID-191,2,3. These symptoms are associated with the severity of COVID-19. In addition to these vascular disorders, vasculitis and vascular damage in the microvessels of systemic organs, such as the lungs and heart, may contribute to thrombus formation by activating coagulation4. The persistence of such microthrombi in systemic organs may cause various sequelae, including pulmonary fibrosis, neurological complications, and cardiovascular disorders, with medium- to long-term adverse effects5. Furthermore, the importance of protecting against vascular injury is evident from the fact that vascular dysfunction occurs under conditions of old age, obesity, hypertension, and diabetes mellitus, all of which are considered risk factors for COVID-196.
Severe-acute-respiratory syndrome-related coronavirus-2 (SARS-CoV-2) has been suggested to infect vascular endothelial cells via angiotensin-converting enzyme 2 (ACE2) and Nrp1, which are expressed on the surface of host cells7. Evidence of ACE2 expression and replicative infection by SARS-CoV-2 in human endothelial cells is lacking7,8. Nevertheless, multiple studies have reported that vascular endothelial dysfunction can occur during SARS-CoV-2 infection through direct and indirect mechanisms, including inflammatory cytokine effects9,10. Vascular endothelial cells form tubular structures in the lumen of blood vessels and exert their barrier function by controlling vascular permeability, maintaining appropriate cell-to-cell associations via adhesion molecules such as VE-cadherin. Vascular endothelial dysfunction during SARS-CoV-2 infection causes endothelial cell disruption, lysis, and death, leading to endothelial inflammation11. As this occurs, vascular barrier function is disrupted, resulting in tissue edema due to increased vascular permeability. The coagulation pathway is activated by the expression of P-selectin, von Willebrand factor, and fibrinogen12. Moreover, increased expression of leukocyte adhesion molecules in vascular endothelial cells leads to dysregulation of inflammatory cell infiltration, such as enhanced extravasation of neutrophils and other cells.
Lysophosphatidic acid (LPA) is a lipid mediator with various activities, such as cell proliferation, differentiation, and migration, and is involved in various biological phenomena, such as central nervous system development, blood vessel formation, and wound healing. LPA is biosynthesized by the hydrolysis of lysophosphatidine choline by autotaxin13. Mice lacking the autotaxin gene have abnormal blood vessel formation during the fetal stage, resulting in lethality around embryonic day 10.514,15. LPA signaling is activated by six subtypes of G protein-coupled transmembrane receptors: LPA1, LPA2, LPA3, LPA4, LPA5, and LPA6 and has an important role in angiogenesis16,17. We previously reported that the activation of LPA receptor 4 (LPA4) signaling in vascular endothelial cells induces the functional maturation of tumor vessels and improves drug delivery by enhancing VE-cadherin-mediated endothelial cell junctions18. Furthermore, using an orthotopic tumor transplantation model of brain tumors, it was clarified that the induction of vascular maturation by LPA improves the efficacy of immune checkpoint inhibitor therapy by improving immune cell infiltration into tumor tissue19. Few molecules are capable of vascular stabilization comparable to that exhibited by LPA, suggesting the potential of this unique molecule to be leveraged for treating conditions associated with vascular disorders. Especially, LPA is expected to be effective in the treatment of diseases characterized by pronounced vascular damage such as COVID-19. Hence, development of new LPA-based treatment modalities that differ from current antiviral and anti-inflammatory therapies is an intriguing area of research.
Here, we aimed to test the therapeutic efficacy of LPA on vascular injury and tissue damage seen in COVID-19. We analyzed pulmonary microvascular endothelial cells (HMVEC-Lung), brain microvascular endothelial cells (HMBECs), and renal glomerular endothelial cells (HRGECs) and conducted animal experiments using hamsters.
Results
Establishment of a 3D culture system for vascular endothelial cells
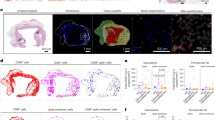
To analyze vascular injury, it is necessary to create 3D cultures of vascular endothelial cells that form structures resembling in vivo lumens. An in vitro vascular analysis system was established to investigate whether LPA protects against vascular damage caused by SARS-CoV-2 infection. Primary human intravascular cell lines were derived from the lung, heart, brain, kidney, liver, and peripheral tissues, where tissue damage occurs during COVID-19 (Table 1). Vascular endothelial cells were cultured in collagen gels at appropriate concentrations to form luminal vessels. The 3D luminal structure of blood vessels began to form 24 h after culture establishment and persisted for 72 h, as demonstrated by the CD31-positive luminal structures in Fig. 1A. In lumen-forming vascular endothelial cells, LPA4, a transmembrane receptor for LPA, was expressed in HMVEC-Lung, HMBEC, and HRGEC lines (Fig. 1B). In addition, the expression of ACE2 and TMPRSS2, entry receptors for SARS-CoV-2 infection, was significantly elevated when the endothelial cells were cultured in 3D to form luminal structures versus 2D planar cultures (Fig. 1C, D, E). Based on the criteria of high LPA4 and SARS-CoV-2 receptor expression (Table 2), we proceeded with SARS-CoV-2 infection experiments using three types of vascular endothelial cells: HMBECs, HMVEC-Lung, and HRGECs. However, during extended culture experiments, HMVEC-Lung tubes tended to collapse, making them unsuitable for long-term infection studies (Supplemental Fig. 1).
Expression of the genes for the LPA receptor, LPA4 and entry receptors for SARS-CoV-2 infection in various types of vascular endothelial cells. (A) Schematic diagram and image showing the formation of luminal structures in 3D cultures of vascular endothelial cells. The endothelial cells are stained with CD31 antibody, and the right image shows formed luminal structures of 3D-cultured HBMECs. Arrows indicate luminal structures. Scale bar, 100 μm. VEGF-A: vascular endothelial growth factor A. (B) Quantitative analysis of the mRNA expression of LPA4, a transmembrane receptor for LPA, in vascular endothelial cells cultured in planar culture (2D) or in three-dimensional culture in collagen gel (3D). The data are expressed as mean ± SD, n = 3. **P< 0.01 (C–E) Quantitative analysis of the mRNA expression of ACE2, TMPRSS2, and Nrp1, entry receptors for SARS-CoV-2 infection, in vascular endothelial cells cultured in planar culture (2D) or in three-dimensional culture in collagen gel (3D). The data are expressed as mean ± SD, n = 3. *P < 0.05, **P < 0.01.
Vasoprotective effects of LPA
The HMBEC and HRGEC lines were infected with the SARS-CoV-2 Wuhan strain at a multiplicity of infection of 0.1 after lumen construction by 3D culture on collagen gels. The infection caused disruption of the vascular structure, indicating that vascular damage had occurred. In contrast, the disruption of the vascular structure caused by SARS-CoV-2 infection was suppressed in the group treated with LPA, with only a slight decrease in tubular volume. In addition, the tubular volume of luminal vessels formed by HMBECs and HRGECs was significantly lower after SARS-CoV-2 infection relative to pre-infection volume (Fig. 2A-D). Similarly, infection with the Delta variant of SARS-CoV-2 caused the collapse of 3D HMBEC vessels, which was attenuated via LPA treatment (Fig. 2E, F). While HMVEC-Lung cells showed similar trends, their responses exhibited greater variability, producing less definitive results than those for HMBECs and HRGECs. Therefore, we focused our analysis on the latter two cell types. These results indicate that LPA treatment is effective in reducing vascular damage caused by SARS-CoV-2 infection by protecting blood vessels.
Analysis of vascular structure in a 3D human vascular culture system with SARS-CoV-2 infection and LPA treatment. (A, B) Representative image data of CD31-immunostained 3D-cultured HBMEC infected with SARS-CoV-2 Wuhan strain and the LPA-treated group. Pictures were taken from the bottom of the culture dish to 120 μm. The bar graph shows the measured volume of the 3D-cultured endothelial cell tube structure within a microscopic area. (C, D) 3D-cultured HRGEC infected with SARS-CoV-2 Wuhan strain and the LPA-treated group. (E, F) 3D-cultured HBMEC infected with Delta variant of SARS-CoV-2. The bar graph shows the measured volume of CD31 fluorescently labeled endothelial cell tube structures contained in the same amount of gel. All data are expressed as mean ± SD, n = 4 per group.**P < 0.01. ns, not significant. Scale bar, 100 μm.
Downregulation of pro-inflammatory genes by LPA
We analyzed gene expression to determine whether the activation of LPA signaling in vascular endothelial cells could suppress the inflammatory signaling pathways induced by SARS-CoV-2 infection. We conducted bulk RNA sequencing (RNA-seq) on HMBEC, HMVEC-Lung, and HRGEC, comparing SARS-CoV-2 infection with LPA treatment. The RNA-seq data on gene expression in HMVEC-Lung, comparing SARS-CoV-2 infection with the control, LPA treatment, are presented in Venn diagrams. The genes that were upregulated by more than 1.5 fold and downregulated by less than 1.5 fold (FDR < 0.05) are summarized in Fig. 3A. Of the 58 genes upregulated by infection, 6 were downregulated by LPA treatment; conversely, of the 48 genes downregulated by infection, 11 were upregulated by LPA treatment. Figure 3B shows a volcano plot of the specific genes. Transcript read counts for 15,156 genes in HMVEC-Lung were analyzed using k-means clustering of the 2,000 most variable genes (Fig. 3C). Of the 8 clusters, cluster 2 exhibited uniform transcriptional changes across 3 samples, with genes upregulated by SARS-CoV-2 infection and downregulated by LPA treatment. Based on the differentially expressed genes (DEGs), inflammation-related signals were examined using pathway analysis. The 116 genes in cluster 2 were mapped to Gene Ontology biological processes related to complement activation and immune reactions, and the associated KEGG pathways involved in infection and inflammatory signaling pathways were identified. TNF and IFN signals were activated by SARS-CoV-2 infection, but treatment with LPA effectively suppressed these signals (Fig. 3D). In addition, SARS-CoV-2 infection decreased the expression of genes related to cell adhesion, but their expression was restored by LPA treatment. These results were similar for HMBEC, HMVEC-Lung, and HRGEC. Overall, these gene expression analyses indicate that SARS-CoV-2 inoculation of 3D endothelial cell cultures results in upregulation of several key pro-inflammatory signaling pathways and downregulation of important cell adhesion genes, while LPA treatment downregulates these pro-inflammatory signaling pathways and restores the expression of cell adhesion genes.
Gene expression analysis of 3D-cultured vascular endothelial cells following SARS-CoV-2 infection and LPA treatment. (A) Venn diagram of differentially expressed genes (DEGs) in vascular endothelial cells (HMVEC-Lung), comparing SARS-CoV-2 infection with control treatment (Cont) and LPA treatment with SARS-CoV-2 infection (FDR P < 0.05, fold change < 1.5). (B) Volcano plot showing upregulated (red) and downregulated (green) DEGs upon SARS-CoV-2 infection (left) and LPA treatment (right) in HMVEC-Lung. Purple spots indicate upregulated genes, and blue spots indicate downregulated genes when comparing SARS-CoV-2 with the control. (C) The k-means clustering analysis revealed 8 clusters in the 2,000 most variable genes in HMVEC-L. Pathway analysis of 116 genes in cluster 2 referred to GO biological processes and KEGG pathways. Transcriptional changes are heatmapped from green (strongly downregulated) to red (highly upregulated). Permission has been obtained from Kanehisa laboratories for using KEGG pathway database42. (D) Pathway analysis using KEGG database. A heatmap showing the activation of significantly enriched pathways, comparing SARS-CoV-2 infection with LPA treatment in HMVEC-Lung, HBMEC, and HRGEC. Pathways related to TNF signaling (highlighted in blue) and IL-17 signaling (highlighted in red) are shown. Enrichment values [-log(p-value)] are scaled from 0 to 1 (green to red). Permission has been obtained from Kanehisa laboratories for using KEGG pathway database42.
Therapeutic effects of LPA in an animal model
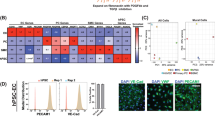
The therapeutic vasoprotective effects of LPA against COVID-19 were examined in an animal model. SARS-CoV-2 infection was confirmed by the development of characteristic respiratory symptoms and pulmonary pathology in infected animals, consistent with previously established infection models20,21. Daily doses of LPA (10 or 30 mg/kg body weight) were administered intraperitoneally to Syrian hamsters (n = 4 per group) that were intranasally infected with SARS-CoV-2 (1.0 × 106 PFU). Animals were housed under standard conditions (22 ± 2 °C, 12 h light/dark cycle, standard laboratory diet, and water ad libitum, housed in individually ventilated cages) and were euthanized by isoflurane overdose followed by cervical dislocation on day 5 after infection. (Fig. 4A). SARS-CoV-2 infection caused weight loss in hamsters, but LPA treatment did not inhibit this reduction in body weight (Fig. 4B). Lung tissue sections collected 5 days after infection were examined for inflammatory lesions by hematoxylin and eosin staining, and the tissue damage induced by SARS-CoV-2 infection was suppressed by LPA (Fig. 4C). Quantitative histopathological scoring revealed significantly reduced lung injury scores in LPA-treated groups compared to those in the SARS-CoV-2 infection group (P < 0.001), confirming the protective effect of LPA treatment. Furthermore, histological analysis of blood vessels in the lung tissue showed that vascular endothelial cells in the SARS-CoV-2-infected group were detached, as indicated by arrows in the high-magnification images (Fig. 4C), and the vascular structure was disorganized, whereas LPA administration improved vascular damage. These results suggest that LPA suppresses lung inflammation by inhibiting vascular injury caused by SARS-CoV-2 in an animal model.
Therapeutic efficacy of LPA in a SARS-CoV-2-infected hamster model. (A) Schematic diagram of the timeline of LPA administration and analysis in a model of SARS-CoV-2 infection using Syrian hamsters. The diagram shows experimental groups, including the mock infection control, SARS-CoV-2 infection, SARS-CoV-2 + LPA (10 mg/kg), and SARS-CoV-2 + LPA (30 mg/kg), with n = 4 hamsters per group. (B) Percent weight change from baseline in Syrian hamsters over time after SARS-CoV-2 infection with and without LPA administration. The data are expressed as mean ± SD, n = 4 hamsters per group. (C) Representative images of hematoxylin and eosin staining of lung tissue harvested 5 d after SARS-CoV-2 infection with or without LPA (30 mg/kg). Semi-quantitative lung injury scores were graded based on injury criteria described in the Materials and Methods. Data are expressed as mean ± SD, n = 20. ****P< 0.0001. ***P = 0.0002. Scale bar, 200 μm. (D) Representative images of vascular staining with CD31 (green) and TO-PRO-3 (blue) of lung tissue harvested 5 d after SARS-CoV-2 infection with or without LPA (30 mg/kg). Arrows in the SARS-CoV-2 panel indicate areas where endothelial cells have detached from the basement membrane, creating gaps in the vascular lining and resulting in a disorganized vascular structure compared to the continuous endothelial lining seen in control and LPA-treated tissues. Scale bar, 100 μm.
Expression of inflammation-related genes in infected tissues
Comprehensive gene expression analysis by RNA-seq was performed on the lung tissue 5 days after infection with SARS-CoV-2. The results showed that SARS-CoV-2 infection induced the expression of genes for pro-thrombotic regulators of hemostasis, such as von Willebrand factor, key pro-inflammatory cytokines implicated in the cytokine storm, and endothelial cell adhesion molecules that promote leukocyte migration, such as Vcam1 and Icam1. In contrast, LPA treatment was associated with decreased expression of these pro-thrombotic and pro-inflammatory genes. (Fig. 5A, B). Additionally, Gene Ontology analysis showed that the upregulation of pro-inflammatory genes in lung tissue was suppressed following LPA administration (Fig. 5C). Taken together, these results suggest that LPA may be an effective therapeutic for suppressing vascular injury associated with severe COVID-19 disease.
Evaluation of therapeutic efficacy of LPA by transcriptome analysis of lung tissue from hamsters challenged with SARS-CoV-2. (A) Dot plot showing DEGs in lung tissues from control vs. SARS-CoV-2-infected hamsters. RNA-seq of lung tissue harvested 5 d after SARS-CoV-2 infection reveals the upregulation of multiple genes (red dots), including several pro-inflammatory cytokines, while LPA administration results in the downregulation of a subset of pro-inflammatory genes (blue dots). (B) Heatmap showing gene expression patterns across experimental groups. Pro-inflammatory genes that were upregulated by SARS-CoV-2 infection and suppressed by LPA treatment are highlighted in red boxes. (C) Gene Ontology analysis visualization showing the activation of significantly enriched inflammatory pathways in lung tissue, comparing mock-infected controls, SARS-CoV-2-infected hamsters, and SARS-CoV-2-infected hamsters treated with LPA. Key inflammatory pathways affected by LPA treatment are highlighted. Permission has been obtained from Kanehisa laboratories for using KEGG pathway database42.
Discussion
In this study, we focused on the vascular stabilizing effects of LPA and examined whether it constitutes a novel therapeutic for COVID-19. We previously reported that LPA induces functional maturation of the tumor vasculature by inducing stabilization18,19. Tumor vessels are constantly exposed to an inflammatory environment that affects endothelial cells. Consequently, the junctions between the vascular endothelial cells are disrupted, leading to high leakage and reduced blood flow. The activation of LPA signaling enhances vascular endothelial cell adhesion by maintaining VE-cadherin on the plasma membrane and inhibiting vascular leakage. Analysis of gene-deficient mice showed that the induction of vascular stabilization by LPA is mediated by the LPA4 receptor. Although many molecules that activate vascular endothelial cells have been identified, molecules like LPA that stabilize blood vessels and protect them from inflammation are uncommon. The results of our gene expression and histopathological analyses suggest that LPA treatment preserves vascular integrity during SARS-CoV-2 infection, likely through the suppression of inflammatory signaling cascades. Administration of LPA did not prevent weight loss in our hamster infection model, possibly due to side effects caused by the action of LPA on cells other than vascular endothelial cells22. Therefore, an agonist that selectively acts on the LPA4 receptor is necessary to address LPA signaling as a therapeutic target for COVID-19. The vasoprotective effect of LPA on pulmonary inflammation in infected animal models suggests that LPA may be useful in suppressing the vascular injury observed in the tissues of COVID-19 patients and is expected to be a potential therapeutic agent.
COVID-19 is known to cause respiratory symptoms such as pneumonia and tissue damage due to systemic thrombus formation and cytokine storms. Vascular endothelial cell injury and inflammation have been implicated in the pathogenesis of these conditions1,3,23. Therefore, there is a need to develop therapeutic agents that inhibit not only viral proliferation but also vascular endothelial cell damage. LPA protects against SARS-CoV-2 infection-induced vascular damage, and its use in combination with antiviral drugs is expected to improve its therapeutic efficacy. In addition, many viruses that cause respiratory infections often have lingering effects, such as asthma, even after the virus has been cleared from the body, and there are concerns about the aftermath of COVID-1924,25. COVID-19 has been reported to cause significant vascular inflammation, leading to thrombus formation in the microvessels of the lungs, heart, and other systemic organs. It has been suggested that the persistence of such clots in systemic organs may have medium- to long-term adverse effects. Recent studies have further elucidated the mechanisms of vascular injury in COVID-19, confirming the central role of endothelial dysfunction in both acute infection and post-COVID syndrome26,27. These findings suggest that LPA may be particularly valuable for reducing the incidence of sequelae by inhibiting thrombus-related factors and suppressing inflammatory responses.
Gene expression and the properties of vascular endothelial cells differ between planar cultures for forming cell layers and 3D cultures for forming luminal structures28. To analyze the vascular injury caused by COVID-19 in vitro under conditions similar to those found in vivo, vascular endothelial cells that have formed a luminal structure are required. In this study, we constructed a 3D culture system capable of inducing luminal structure formation by culturing vascular endothelial cells in a collagen gel approximating physiological pressure (approximately 0.3 kPa). Under these culture conditions, both LPA4 receptors and entry receptors for SARS-CoV-2 infection, including ACE2, TMPRSS2, and Nrp1, were expressed by endothelial cells, indicating a phenotype similar to that of vascular endothelial cells in vivo. Multiple studies have reported the presence of SARS-CoV-2 viral particles in vascular endothelial cells in the lungs of patients with COVID-193,29,30, though debate remains about whether this represents productive infection or viral interaction without replication9,10. In addition, infection of cultured vascular endothelial cells with SARS-CoV-2 has been reported to disrupt vascular endothelial cell junctions31. Studies suggest that vascular damage may primarily result from inflammatory cytokine effects rather than direct viral infection6,32,33,34 highlighting the complexity of vascular pathology in COVID-19. The evaluation system employed in this study, using 3D cultures of vascular endothelial cells, facilitated the analysis of the luminal vessel structure. Moreover, it also allowed for a closer approximation of SARS-CoV-2 infection to that observed in vivo. The experiments using these 3D culture systems indicated that SARS-CoV-2 infection in endothelial cells results in the upregulation of several key pro-inflammatory signaling pathways, while LPA treatment effectively downregulates these pro-inflammatory pathways. In addition, our animal studies on infected Syrian hamsters showed that LPA protects and stabilizes blood vessels and suppresses the pulmonary inflammatory characteristics of COVID-19. Our pathway analysis identified significant transcriptional changes in inflammatory signaling pathways, though future studies should validate these findings at the protein level through western blot analyses of key signaling molecules. Based on the findings in this study, the activation of LPA signaling in vascular endothelial cells is expected to represent an effective therapeutic approach for mitigating vascular injury in COVID-19.
Additional therapeutic agents targeting blood vessels may also be effective against new mutant strains of SARS-CoV-2 and other emerging viruses. To prepare for future pandemics that may occur at any time, it is important to develop a vasoprotective drug that targets LPA as an immediate treatment until a vaccine is developed. Our previous work strongly implicated LPA4 as the primary receptor involved in the vasoprotective effect of LPA against SARS-CoV-218. However, LPA has been shown to exert various physiological effects through multiple receptors, and LPA administration may lead to effects other than vascular protection. Therefore, experiments using selective LPA4 agonists are needed to establish its role in protecting against SARS-CoV-2-induced vascular damage. Moreover, the relative contributions of different LPA receptors, including LPA4, to the vasoprotective effects we observed warrant further investigation. Translating these findings into clinical practice will require pharmacokinetic studies and safety trials with selective LPA4 agonists, followed by controlled clinical studies in patients with COVID-19 and early vascular dysfunction.
Despite the important findings of this study, there are certain limitations worth noting. First, while our 3D culture system provides valuable insights into vascular responses to SARS-CoV-2, it lacks the complex interactions between different cell types present in vivo, including pericytes and other supporting cells that contribute to vascular barrier function. Second, our animal model focused primarily on the pulmonary effects of SARS-CoV-2 infection, but COVID-19 is known to affect multiple organ systems. Future studies should explore the effects of LPA on vascular damage in other tissues affected by COVID-19. Finally, additional research is needed to understand the potential side effects of LPA treatment and to develop more targeted approaches, such as selective LPA4 receptor agonists, to maximize therapeutic benefits while minimizing adverse effects.
Methods
Viruses
Various SARS-CoV-2 strains, hCoV-19/Japan/QHN002/2021 (Alpha), hCoV-19/Japan/TY8-612/2021 (Beta), hCoV-19/Japan/TY7-503/2021 (Gamma), hCoV-19/Japan/TY11-927/2021 (Delta), and 2019-nCoV/Japan/TY38-873/2021 (Omicron), isolated at the National Institute of Infectious Diseases (Japan), were used in this study. We primarily used the Delta variant for our experiments because it was the predominant circulating strain during our study and has been associated with severe vascular complications35. SARS-CoV-2 was propagated in VeroE6/TMPRSS2 cells. The virus stock was generated from the supernatant of VeroE6/TMPRSS2 cells infected with SARS-CoV-2 at a multiplicity of infection of 0.1 and harvested 2 days after infection. The viral titer was determined using a plaque assay.
Plaque formation assay
Vero/TMPRSS2 (JCRB1818) was obtained from JCRB Cell Bank in National Institutes of Biomedical Innovation, Health and Nutrition. VeroE6/TMPRSS2 cells were seeded into 24-well plates (80,000 cells/well) at 37 °C in 5% CO2 overnight. The supernatants were serially diluted using the inoculated medium and incubated for 2 h. Next, the culture medium was removed, fresh medium containing 1% methylcellulose (1.5 mL) was added, and the cells were cultured for an additional 3 days. Finally, the cells were fixed with 4% paraformaldehyde in PBS (Nacalai Tesque, Inc. Kyoto, Japan), and the plaques were visualized using Giemsa’s azur-eosin-methylene blue solution (#109204, Merck Millipore, Darmstadt, Germany).
Reagents
The 1-oleoyl-LPA was purchased from Avanti Polar Lipids (Alabaster, AL, USA). LPA stock solution (10 mM) was prepared using 50% ethanol and stored at − 20 °C before use. LPA working solution was prepared by diluting the 50% stock solution in PBS at a ratio of 18:1. LPA (10 mg/mL) was used for study. For control experiments, 5% ethanol was used as the vehicle.
Cell culture
Human Coronary Artery Endothelial Cell (HCAEC), HMVEC-Lung, and Human Pulmonary Artery Endothelial Cell (HPAEC) lines were purchased from Lonza (Basel, Switzerland) and grown in EBM-2 supplemented with EGM-2 MV BulletKit. Human Cardiac Microvascular Endothelial Cell (HCMEC), Human Hepatic Sinusoidal Endothelial Cell (HHSEC), HRGEC, and HMBEC lines were purchased from ScienCell (Berlin, Germany) and grown in an endothelial cell medium containing FBS and endothelial cell growth supplement. The Human Saphenous Vein Endothelial Cell (HSaVEC) line was purchased from PromoCell (Heidelberg, Germany) and grown in an endothelial cell growth medium-2. Human vascular cells were maintained under conditions of 5% CO2 and 37 °C, below 80% confluency, and used in experiments at passage 7 or 8.
Endothelial cell 3D culture model
All eight human endothelial cell lines described above were cultured in collagen gels with 3D embedding and evaluated for their ability to form 3D luminal structures. Detailed characterization was performed on the three selected cell types (HMVEC-Lung, HMBEC, and HRGEC) based on receptor expression profiles. Cellmatrix Type I-A (Nitta Gelatin, Osaka, Japan) was diluted to 0.15% with dilute hydrochloric acid (Fujifilm Wako Pure Chemical Corporation, Osaka, Japan) at pH 3.0 and reconstituted according to the manufacturer’s protocol. The reconstituted collagen gel was mixed with cultured vascular endothelial cells at a concentration of 5 × 105 cells/150 µl. The collagen cell mixture solution was cultured at 37 °C in a CO2 incubator for 30 min in a glass-bottomed dish (Matsunami D141400, Osaka, Japan). After the gel hardened, 10 ng/ml of VEGF-A (PeproTech, Rocky Hill, NJ, USA) containing the culture medium was added. Following incubation in a humidified incubator at 37 °C and 5% CO2 for 2 days, CD31 staining was performed to identify luminal structures. Vascular endothelial cells cultured under 3D conditions were pretreated with 10 µM of LPA for 30 min, infected with SARS-CoV-2 (Wuhan strain and Delta strain) at a multiplicity of infection of 0.1, and incubated with 10 µM of LPA. Two days after infection, the 3D tubes were fixed with paraformaldehyde and stained with Alexa Fluor 488 anti-human CD31 antibody (BioLegend, San Diego, CA, USA). Images were obtained using a TCS SP8 confocal microscope (Leica Microsystems, Wetzlar, Germany). The 3D tube volume was analyzed using Volocity imaging software (Perkin Elmer, Waltham, MA, USA). The weakest 10% of Alexa Fluor 488 fluorescence signals were excluded from the analysis as gel autofluorescence signal noise. Furthermore, objects smaller than 500 µm3 were excluded from the volumetric analysis as endothelial cells that were not 3D tube structures.
Animals
Four-week-old male Syrian hamsters were purchased from Japan SLC (Shizuoka, Japan). The animals were housed in environmentally controlled rooms at the animal experimentation facility of Osaka University under standard conditions (22 ± 2 °C, 12 h light/dark cycle, standard laboratory diet, and water ad libitum). Animals were maintained in individually ventilated cages (ISOrat900 N, Tecniplast, Buguggiate, Italy) with appropriate bedding, enrichment, and air filtration. All experiments were performed according to the guidelines of the Osaka University Committee for Animal and Recombinant DNA Experiments (approval number 4062).
Animal infection experiments
Syrian hamsters were randomly assigned to experimental groups (n = 4 animals per group: mock infection control, SARS-CoV-2 infection, SARS-CoV-2 infection + LPA 10 mg/kg, and SARS-CoV-2 infection + LPA 30 mg/kg) following established protocols from our previous work20,21. Syrian hamsters were anesthetized with isoflurane and challenged with 1.0 × 106 PFU (in 60 µL) SARS-CoV-2 via intranasal routes. Infection was confirmed by the development of consistent respiratory symptoms and characteristic pulmonary pathology in SARS-CoV-2-inoculated animals, which were absent in mock-infected controls. To evaluate the therapeutic effects of LPA on vascular injury caused by SARS-CoV-2 infection, LPA (10 or 30 mg/kg, intraperitoneally) was administered daily from days 2 to 4 after infection. The selection of doses (10 and 30 mg/kg) was based on our previous studies with LPA in other mouse models18 and the treatment window (days 2–4) was chosen to reflect a clinically relevant scenario where treatment would begin after symptom onset. Five days post-infection, the hamsters were euthanized by isoflurane overdose followed by cervical dislocation, and the lungs were harvested for the experiments. All animal experiments with SARS-CoV-2 were performed at the Animal Biosafety Level 3 (ABSL3) facilities of the Research Institute for Microbial Diseases at Osaka University. Animal Experimentation, and the study protocol was approved by the Institutional Committee of Laboratory Animal Experimentation of the Research Institute for Microbial Diseases, Osaka University (approval number R02-08-0). All methods were performed in accordance with the relevant guidelines and regulations. All animal experiments were conducted in strict accordance with the guidelines for the care and use of laboratory animals established by the Ministry of Education, Culture, Sports, Science and Technology of Japan, and followed the ARRIVE guidelines for reporting animal research.
RNA isolation and expression analysis
Vascular endothelial cells cultured in planar or 3D cultures were collected, and RNA was isolated using the RNeasy Mini Kit (Qiagen, Hilden, Germany) according to the manufacturer’s protocol. Total RNA was extracted from the lung tissue using TRIzol reagent. RNA was transcribed into cDNA using the PrimeScript RT Reagent Kit (TaKaRa, Shiga, Japan) according to the manufacturer’s instructions. Quantitative real-time PCR was performed on a LightCycler 96 (Roche, Rotkreuz, Switzerland) using the TB Green™ Premix Ex Taq™ Kit (TaKaRa) and was run in triplicate. The results were normalized to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) levels using the comparative threshold cycle method. Specific primers used in this experiment were as follows: LPA4: 5’-CCTAGTCCTCAGTGGCGGTA-3’ (sense) and 5’-CTTCAAAGCAGGTGGTGGTT-3’ (anti-sense); GAPDH: 5’-GAAGGTGAAGGTCGGAGTC-3’ (sense) and 5’-GAAGATGGTGATGGGATTTC-3’ (anti-sense); ACE2: 5’-GGATTCCATGAAGCTGTTGG-3’ (sense) and 5’-TCGTGAGTGCTTGTTTGAGC-3’ (anti-sense); and TMPRSS2: 5’-TGGAGCCGGATACCAAGTAG-3’ (sense) and 5’-GTTGGGCAGACACACTGGTT-3’ (anti-sense). Three independent samples were collected from each group, and mRNA was extracted from endothelial cells using RNeasy Plus Micro Kits (Qiagen) according to the manufacturer’s protocol. RNA libraries were prepared using the TruSeq Sample Prep v2 kit and sequenced on a HiSeq 2500 (Illumina, San Diego, CA, USA) in the 75-base single-end mode. CASAVA 1.8.2 software (Illumina) was used for base calling. Sequenced reads were mapped to the mouse reference genome sequence (mm9) using TopHat v2.1.0. Fragments per kilobase of exons per million mapped fragments values were determined using Cuffnorm v.2.2.1.
Tissue staining
Lung tissue samples harvested from hamsters were fixed in 4% paraformaldehyde, and cryo-tissue sections were prepared. Hematoxylin and eosin staining was performed to evaluate immune cell infiltration and pathological changes in lung tissues, with 20 random high-power fields (400× total magnification) scored for each condition. Scores were assigned to each parameter, including neutrophil infiltration in the alveolar and interstitial spaces, hyaline membrane, proteinaceous debris filling the airspaces, and alveolar septal thickening36. Histopathological evaluation was performed by a pulmonologist who assessed the extent of inflammatory lesions and vascular damage in a blinded manner using a standardized scoring system. The blood vessels of the lung sections were stained with Lycopersicon esculentum Lectin, DyLight 488 conjugate (Vector Laboratories, Burlingame, CA, USA), and TO-PRO-3 stain (ThermoFisher, Waltham, MA, USA). Tissue preparation and staining were performed as previously described37.
DEG analysis
The RNA-seq data in Fig. 3 were analyzed using iDEP (ver2.01) (http://bioinformatics.sdstate.edu/idep/)38,39. DEGs were identified using DESeq40 with a maximum FDR p-value of < 0.05 and a fold change of at least 1.5 between groups. We performed k-means clustering40 on the 2,000 most variable genes using the Gene Ontology biological process41 and KEGG pathway databases42. We analyzed the RNA-seq data (Fig. 5) obtained under the three experimental conditions to identify genes with differential expression patterns. The conditions were as follows: (1) Samples collected 5 days after SARS-CoV-2 infection (SARS-CoV-2); (2) samples collected 5 days after SARS-CoV-2 infection with subsequent LPA stimulation on days 2, 3, and 4 post-infection (SARS-CoV-2 + LPA); and (3) samples collected 5 days after mock SARS-CoV-2 infection served as controls (mock). Four samples were analyzed for each condition. We then extracted genes exhibiting expression differences under the following conditions: (1) Genes with increased expression post-infection compared to non-infection (SARS-CoV-2) were selected if the log2 ratio of the average gene expression in SARS-CoV-2-infected samples to that in non-infected samples was ≥ log2(1.5), and the average expression in SARS-CoV-2-infected samples was at least 1.0. (2) Genes with decreased expression under LPA stimulation compared to SARS-CoV-2 infection (SARS-CoV-2 + LPA) were selected if the log2 ratio of the average gene expression in SARS-CoV-2 + LPA samples to that in SARS-CoV-2-infected samples was ≤ log2(0.75), and the average expression in SARS-CoV-2-infected samples was at least 1.0. Using these criteria, we identified specific gene sets that exhibited altered expression in response to SARS-CoV-2 infection and subsequent LPA stimulation.
Gene enrichment analysis
To explore the functional characteristics of the genes altered by SARS-CoV-2 infection, we performed gene enrichment analysis on two groups: those with increased expression following SARS-CoV-2 infection and those exhibiting decreased expression upon LPA stimulation. Fisher’s exact test was used to identify functionally enriched gene groups using Gene Ontology sets from MsigDB (c5.all.v7.2. symbols.gmt). This approach delineates the functional pathways and biological processes involved in these gene sets.
Statistical analysis
All data are presented as mean ± SD. No data were excluded from the analyses. For the in vivo studies, age-matched and weight-matched hamsters were randomized into experimental or control groups. Statistical analysis was performed using the statcel4 software package (OMS, Saitama, Japan) with analysis of variance for all data, followed by the Tukey–Kramer multiple comparisons test. Two-sided Student’s t-tests were used to compare two groups. P < 0.05 was considered to indicate statistical significance.
Data availability
The datasets generated and analyzed in this study are available from the corresponding author upon request. The RNA-seq data used in this study are available from GEO under the accession number PRJDB18309 (https://ddbj.nig.ac.jp/search/entry/bioproject/PRJDB18309).
References
Ackermann, M. et al. Pulmonary vascular endothelialitis, thrombosis, and angiogenesis in Covid-19. N Engl. J. Med. 383, 120–128. https://doi.org/10.1056/NEJMoa2015432 (2020).
Becker, R. C. COVID-19-associated vasculitis and vasculopathy. J. Thromb. Thrombolysis. 50, 499–511. https://doi.org/10.1007/s11239-020-02230-4 (2020).
Varga, Z. et al. Endothelial cell infection and endotheliitis in COVID-19. Lancet 395, 1417–1418. https://doi.org/10.1016/S0140-6736(20)30937-5 (2020).
Jose, R. J. & Manuel, A. COVID-19 cytokine storm: the interplay between inflammation and coagulation. Lancet Respir Med. 8, e46–e47. https://doi.org/10.1016/S2213-2600(20)30216-2 (2020).
Levy, J. H. et al. COVID-19: thrombosis, thromboinflammation, and anticoagulation considerations. Int. J. Lab. Hematol. 43 (Suppl 1), 29–35. https://doi.org/10.1111/ijlh.13500 (2021).
Teuwen, L. A., Geldhof, V., Pasut, A. & Carmeliet, P. COVID-19: the vasculature unleashed. Nat. Rev. Immunol. 20, 389–391. https://doi.org/10.1038/s41577-020-0343-0 (2020).
McCracken, I. R. et al. Lack of evidence of Angiotensin-Converting enzyme 2 expression and replicative infection by SARS-CoV-2 in human endothelial cells. Circulation 143, 865–868. https://doi.org/10.1161/CIRCULATIONAHA.120.052824 (2021).
Cantuti-Castelvetri, L. et al. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science 370, 856–860. https://doi.org/10.1126/science.abd2985 (2020).
Lei, Y. et al. SARS-CoV-2 Spike Protein Impairs Endothelial Function via Downregulation of ACE2. bioRxiv, (2020). https://doi.org/10.1101/2020.12.04.409144
Evans, P. C. et al. Endothelial dysfunction in COVID-19: a position paper of the ESC working group for atherosclerosis and vascular biology, and the ESC Council of basic cardiovascular science. Cardiovasc. Res. 116, 2177–2184. https://doi.org/10.1093/cvr/cvaa230 (2020).
Giannotta, M., Trani, M. & Dejana, E. VE-cadherin and endothelial adherens junctions: active guardians of vascular integrity. Dev. Cell. 26, 441–454. https://doi.org/10.1016/j.devcel.2013.08.020 (2013).
Pober, J. S. & Sessa, W. C. Evolving functions of endothelial cells in inflammation. Nat. Rev. Immunol. 7, 803–815. https://doi.org/10.1038/nri2171 (2007).
Moolenaar, W. H., van Meeteren, L. A. & Giepmans, B. N. The Ins and outs of lysophosphatidic acid signaling. Bioessays 26, 870–881. https://doi.org/10.1002/bies.20081 (2004).
Tanaka, M. et al. Autotaxin stabilizes blood vessels and is required for embryonic vasculature by producing lysophosphatidic acid. J. Biol. Chem. 281, 25822–25830. https://doi.org/10.1074/jbc.M605142200 (2006).
van Meeteren, L. A. et al. Autotaxin, a secreted lysophospholipase D, is essential for blood vessel formation during development. Mol. Cell. Biol. 26, 5015–5022. https://doi.org/10.1128/MCB.02419-05 (2006).
Noguchi, K., Herr, D., Mutoh, T. & Chun, J. Lysophosphatidic acid (LPA) and its receptors. Curr. Opin. Pharmacol. 9, 15–23. https://doi.org/10.1016/j.coph.2008.11.010 (2009).
Yasuda, D. et al. Lysophosphatidic acid-induced YAP/TAZ activation promotes developmental angiogenesis by repressing Notch ligand Dll4. J. Clin. Invest. 129, 4332–4349. https://doi.org/10.1172/JCI121955 (2019).
Takara, K. et al. Lysophosphatidic acid receptor 4 activation augments drug delivery in tumors by tightening endothelial Cell-Cell contact. Cell. Rep. 20, 2072–2086. https://doi.org/10.1016/j.celrep.2017.07.080 (2017).
Eino, D. et al. LPA4-Mediated vascular network formation increases the efficacy of Anti-PD-1 therapy against brain tumors. Cancer Res. 78, 6607–6620. https://doi.org/10.1158/0008-5472.CAN-18-0498 (2018).
Higuchi, Y. et al. Engineered ACE2 receptor therapy overcomes mutational escape of SARS-CoV-2. Nat. Commun. 12, 3802. https://doi.org/10.1038/s41467-021-24013-y (2021).
Ikemura, N. et al. An engineered ACE2 decoy neutralizes the SARS-CoV-2 Omicron variant and confers protection against infection in vivo. Sci. Transl Med. 14, eabn7737. https://doi.org/10.1126/scitranslmed.abn7737 (2022).
Yanagida, K. & Shimizu, T. Lysophosphatidic acid, a simple phospholipid with myriad functions. Pharmacol. Ther. 246, 108421. https://doi.org/10.1016/j.pharmthera.2023.108421 (2023).
Kang, S. et al. IL-6 trans-signaling induces plasminogen activator inhibitor-1 from vascular endothelial cells in cytokine release syndrome. Proc. Natl. Acad. Sci. U S A. 117, 22351–22356. https://doi.org/10.1073/pnas.2010229117 (2020).
Gonzalez-Parra, G. & Dobrovolny, H. M. The rate of viral transfer between upper and lower respiratory tracts determines RSV illness duration. J. Math. Biol. 79, 467–483. https://doi.org/10.1007/s00285-019-01364-1 (2019).
Bellanti, J. A. The long COVID syndrome: A conundrum for the allergist/immunologist. Allergy Asthma Proc. 43, 368–374. https://doi.org/10.2500/aap.2022.43.220059 (2022).
Qiao, H. et al. SARS-CoV-2 induces blood-brain barrier and choroid plexus barrier impairments and vascular inflammation in mice. J. Med. Virol. 96, e29671. https://doi.org/10.1002/jmv.29671 (2024).
Thomas, D. et al. CCL2-mediated endothelial injury drives cardiac dysfunction in long COVID. Nat. Cardiovasc. Res. 3, 1249–1265. https://doi.org/10.1038/s44161-024-00543-8 (2024).
Murikipudi, S., Methe, H. & Edelman, E. R. The effect of substrate modulus on the growth and function of matrix-embedded endothelial cells. Biomaterials 34, 677–684. https://doi.org/10.1016/j.biomaterials.2012.09.079 (2013).
Deng, H. et al. Endothelial dysfunction and SARS-CoV-2 infection: association and therapeutic strategies. Pathogens 10 https://doi.org/10.3390/pathogens10050582 (2021).
Colmenero, I. et al. SARS-CoV-2 endothelial infection causes COVID-19 chilblains: histopathological, immunohistochemical and ultrastructural study of seven paediatric cases. Br. J. Dermatol. 183, 729–737. https://doi.org/10.1111/bjd.19327 (2020).
Thacker, V. V. et al. Rapid endotheliitis and vascular damage characterize SARS-CoV-2 infection in a human lung-on-chip model. EMBO Rep. 22, e52744. https://doi.org/10.15252/embr.202152744 (2021).
Jin, Y. et al. Endothelial activation and dysfunction in COVID-19: from basic mechanisms to potential therapeutic approaches. Signal. Transduct. Target. Ther. 5, 293. https://doi.org/10.1038/s41392-020-00454-7 (2020).
Iwasaki, M. et al. Inflammation triggered by SARS-CoV-2 and ACE2 augment drives multiple organ failure of severe COVID-19: molecular mechanisms and implications. Inflammation 44, 13–34. https://doi.org/10.1007/s10753-020-01337-3 (2021).
Pelle, M. C. et al. Endothelial dysfunction in COVID-19: potential mechanisms and possible therapeutic options. Life (Basel). 12 https://doi.org/10.3390/life12101605 (2022).
Manzur-Pineda, K. et al. COVID-19-related thrombotic complications experience before and during delta wave. J Vasc Surg 76, 1374–1382 e1371, (2022). https://doi.org/10.1016/j.jvs.2022.04.053
Matute-Bello, G. et al. An official American thoracic society workshop report: features and measurements of experimental acute lung injury in animals. Am. J. Respir Cell. Mol. Biol. 44, 725–738. https://doi.org/10.1165/rcmb.2009-0210ST (2011).
Kidoya, H. et al. APJ regulates parallel alignment of arteries and veins in the skin. Dev. Cell. 33, 247–259. https://doi.org/10.1016/j.devcel.2015.02.024 (2015).
Ge, S. X., Son, E. W. & Yao, R. iDEP: an integrated web application for differential expression and pathway analysis of RNA-Seq data. BMC Bioinform. 19, 534. https://doi.org/10.1186/s12859-018-2486-6 (2018).
Ge, X. iDEP web application for RNA-Seq data analysis. Methods Mol. Biol. 2284, 417–443. https://doi.org/10.1007/978-1-0716-1307-8_22 (2021).
Love, M. I., Huber, W. & Anders, S. Moderated Estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Ashburner, M. et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 25, 25–29. https://doi.org/10.1038/75556 (2000).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Acknowledgements
We thank Dr. D. Motooka and D. Okuzaki (RIMD, Osaka University, Japan) for performing RNA-seq analysis and Mr. R. Sasaki (Microeyes, Japan) for his help with microscopic analysis. We are grateful to Ms. M. Kuwabara, Ms. T. Matsuyma, Ms. Y. Mori, and Ms. N. Aikawa for their technical assistance.
Author information
Authors and Affiliations
Contributions
F.M., T.Su., T.O., and H.K. designed and performed most of the experiments. T.Su., Y.H., K.T., L.L., A.S., W.J., Y.N., T.O., and N.T. supervised assays and performed some experiments. N.H., and T.Sh. analyzed and interpreted the data. H.K., N.H., and F.M. analyzed the results and wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Muramatsu, F., Hosoe, N., Suzuki, T. et al. Vasoprotective effects of lysophosphatidic acid inhibit vascular injury caused by SARS-CoV-2 infection. Sci Rep 15, 24622 (2025). https://doi.org/10.1038/s41598-025-06569-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-06569-7