Abstract
Pregnancy poses unique hemodynamic and metabolic demands on the heart. Recent data suggests that a history of multiple pregnancies may be associated with a late-term increased risk of heart failure with preserved ejection fraction (HFpEF), however, the mechanistic basis is unknown. We postulated that the repetitive biological and hemodynamic impact of multiple pregnancies could cause persistent myocardial remodelling providing a substrate for subsequent HFpEF. Multiparous (MP) mice (C57Bl/6J) aged 24 months were compared to age-matched non-parous (NP) mice (n = 8) in a blinded analysis. We evaluated blood pressure, body composition, gross weight and histologic cardiac structure. Cardiac function was assessed by echocardiography together with transcriptomic analysis of the left ventricular myocardium. At 15 months post oestrous cycle disruption, MP mice continued to demonstrate a constellation of myocardial alterations including higher heart to tibial length ratio (P < 0.05), body weight (P < 0.01), and fat mass (P < 0.01) compared to NP mice. MP mice had increased isovolumetric relaxation time (P < 0.01), increased end-diastolic (P < 0.05) and systolic volumes (P < 0.005), and mildly reduced left ventricular ejection fractions (LVEF) (P < 0.005) compared to non-parous mice. This was accompanied by marked elevations in mRNA levels of cardiac markers; Myh6 (P < 0.01) and Nppa (P < 0.01). mRNA levels of Il18 (P < 0.05) and the basal extracellular matrix protein–fibronectin (P < 0.05) were also increased, with a significant increase in interstitial fibrosis (P < 0.05). Bulk RNA sequencing (RNA Seq) further revealed persistent differential expression of 128 genes with over/under-represented pathways involved in extracellular matrix (ECM) regulation and organic anion/ion transport, respectively. Multiparity alters cardiac gene profile and causes myocardial remodelling that persists into later life, providing a potential foundation for the HF phenotypes such as HFpEF. Further studies are required to investigate the nature of the interaction between pregnancy-associated remodelling and other factors known to be associated with HFpEF development.
Similar content being viewed by others
Heart failure (HF) with preserved ejection fraction (HFpEF) is now the dominant phenotype of HF, and it is projected that its prevalence will continue to rise substantially. Whilst some evidence-based treatments, such as SGLT2 inhibitors, have been recently identified1,2 and device-based therapies3 are being investigated, the impact of these approaches is limited. The pathogenesis of HFpEF is not well understood and, as such, HFpEF research is internationally acknowledged as a priority4. Strategies to prevent the development of HFpEF provide a major opportunity to reduce the HFpEF burden. HFpEF is linked epidemiologically to aging, female sex, hypertension, and obesity providing some insights into target populations5.
Women are over-represented among patients presenting with HFpEF. We previously showed that women have a more advanced cardiovascular HFpEF phenotype compared to men6,7, and hypothesised that several mechanisms could be responsible for the sex differences in HFpEF distribution. We proposed that obstetric history may be an important contributor to HFpEF severity, and we demonstrated that women with a history of 3 or more pregnancies had greater impairments in multiple physiologic parameters including diastolic reserve and pulmonary vascular resistance8.
Our studies demonstrated that elevation of the left ventricular filling pressure, most notably during exercise is a cardinal feature of HFpEF9. The cardiovascular mechanics that lead to this feature are complex, however, left ventricular diastolic dysfunction together with vascular stiffening are key components. At the tissue level, fibrosis and inflammation are critical contributors to HFpEF development, and studies in HF patients show that fibrosis is a key contributor to LV diastolic performance10. Consistent with this notion, anti-inflammatory interventions including drugs and diets have been shown to mitigate the extent of cardiac remodelling in experimental models11,12.
In the current study, we aimed to test the hypothesis that multiparity induces a persistent state of myocardial remodelling that could favour the development of HFpEF. To investigate this, we compared baseline myocardial structure and function in aged, post-menopausal mice who were either multiparous (ex-breeders with > 3 pregnancies) or non-parous (virgin mice) with no other experimental disease interventions. The use of multiparous mice in our study provides a novel model to tease out the differences in basal cardiac molecular and transcriptomics profiles of aged multiparous and virgin mice.
Results
Multiparous mice had marked differences in body weight and composition
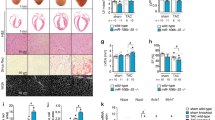
MP female mice had significantly increased body weight compared to NP mice (29.5 ± 0.5 vs. 26.5 ± 0.8 g, P < 0.005, Fig. 1A). This increase in body weight was coupled with a significant increase in fat mass (4.2 ± 0.4 vs. 2.2 ± 0.3 g, P < 0.005) and total water (19.7 ± 0.4 vs. 18.6 ± 0.4 mL, P < 0.05, Fig. 1B,C). There were no differences in lean mass and free water between MP and NP mice (Table 1). MP mice also had significantly greater heart (8.9 ± 0.3 vs. 7.9 ± 0.3 mg, P < 0.05) and lung (18.9 ± 0.8 vs. 13.1 ± 1.1, P < 0.001) weights when normalised to tibia length (Fig. 1D,E). Metabolic caging data showed that MP mice consumed more food (2.7 ± 0.2 vs. 1.4 ± 0.2 g/24 h, P < 0.001, Fig. 1F) and water (3.4 ± 0.3 vs. 2.2 ± 0.4 mL/24 h, P < 0.05, Fig. 1G) in 24 h than NP mice, which correlates with the increased body weight, fat mass and urine output (1.4 ± 1.2 vs. 0.7 ± 0.1 mL/24 h, P < 0.005, Fig. 1H). There were no differences in non-fasted blood glucose levels (Table 1).
Morphometric, metabolic, and physiological differences between age matched multiparous (MP) and nulliparous (NP) mice. MP mice had increased (A) body weight. EchoMRI analysis of body composition showed increased (B) fat mass and (C) total water (D). Quantitative analysis of heart weight-HW and (E) lung weight-LW normalised to tibia length-TL. Metabolic caging data showed increased (F) food, and (G) water intake and (H) urine output for MP mice within 24 h. MP, n = 8, NP, n = 8. *P < 0.05, **P < 0.005, ***P < 0.001, MP vs NP. Unpaired two tailed t-test, with Mann–Whitney test for post hoc analysis. Data presented as mean ± standard error of mean (SEM).
Multiparity elevates systolic blood pressure and alters cardiac function
Post-menopausal MP mice had significantly higher systolic blood pressure versus age-matched NP mice (119.8 ± 2.8 vs. 109.0 ± 3.7 mmHg P < 0.05, Fig. 2A). Multiparity increased both end-diastolic volume (EDV, 63.3 ± 6.3 vs. 45.2 ± 1.8 µL, P < 0.05) and end-systolic volume (ESV, 35.0 ± 3.5 vs. 21.8 ± 0.9 µL, P < 0.005) with a concurrently lower ejection fraction (44.7 ± 1.6% vs. 51.4 ± 1.5%, P < 0.005) and fractional shortening (FS, 24.3 ± 1.5% vs. 32.8 ± 2.3%, P < 0.05) (Fig. 2B–E). However, there were no changes in stroke volume (SV) or cardiac output (CO) (Supplementary Table S2). We detected evidence of diastolic dysfunction in MP mice as reflected by a prolongation of the isovolumetric relaxation time (IVRT, 21.0 ± 0.8 vs. 16.9 ± 0.8 ms, P < 0.005, Fig. 2F), whilst other measures including the mitral E and A wave velocities, E/A ratio, E/e’ ratio, GLS% or DT were not different in MP mice (Supplementary Table S2).
MP mice had elevated systolic blood pressure (SBP), chronic hypertension coupled with myocyte hypertrophy vs. NP mice. (A) MP mice had elevated SBP. Echocardiography analysis was suggestive of a chronic hypertension phenotype as indicated by (B) increased end diastolic volume- EDV, (C) end systolic volume-ESV, (D) reduced ejection fraction—EF%, (E) and fractional shortening-FS %, and (F) prolonged isovolumetric relaxation time- IVRT, in MP mice. (G) Myocyte size was increased with increased gene expression of (H) alpha myosin heavy chain- Myh6, and (I) atrial natriuretic peptide-Nppa. There were no significant changes in mRNA expression of (J) β myosin light chain- Myh7, and (K) brain natriuretic peptide- Nppb in MP mice. MP, n = 8, NP, n = 8. *P < 0.05, **P < 0.005, ns = P > 0.05, MP vs NP. Unpaired two tailed t-test, with Mann–Whitney test for post hoc analysis. Data presented as ± SEM.
Multiparity induces sustained cardiomyocyte hypertrophy
Cardiac myocyte size analysis through H&E staining showed MP mice had substantially greater cardiac myocyte size versus NP mice (574.1 ± 38.2 vs. 466.9 ± 31.3 µm2, P < 0.05, Fig. 2G). This was accompanied by increased mRNA expression of compensatory markers; alpha myosin heavy chain 6 (Myh6, P = 0.007) and atrial natriuretic peptide (Nppa, P = 0.001) (Fig. 2H–J), whilst left ventricular levels of mRNA for the known cardiac stress markers; brain natriuretic peptide (Nppb) and beta myosin heavy chain 7 (Myh7) were not different (Fig. 2J,K).
Multiparity has a synergistic effect on cardiac fibrosis, and gene expression of inflammatory markers
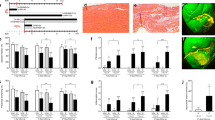
Multiparity was associated with histological evidence of persistent interstitial fibrosis (1.6 ± 0.1 vs. 1.2 ± 0.1%, P < 0.05, Fig. 3A, Supplementary Fig. 1A-B) and increased mRNA expression of the extracellular matrix protein, fibronectin, compared to NP mice (Fig. 3B). The slight increase in fibrosis is also observed in the cardiac sections of the MP mouse hearts, despite both NP and MP hearts showing presence of fibrosis (white arrows) (Supplementary Fig. 1A-B). In contrast it did not appear that perivascular fibrosis (Fig. 3C) was associated with MP mice. Collagen 1a1 (Col1a1, Fig. 3D), collagen 3a1 (Col3a1, Fig. 3E), alpha smooth muscle actin (Acta2, Fig. 3F), and transforming growth factor β1 (Tgfb1, Fig. 3G) gene expression levels were not altered. Similarly, regulators of extracellular matrix (ECM) turnover including tissue inhibitors of matrix metalloproteinase 1 and 2 (Timp1 & Timp2) and matrix metalloproteinase 9 and 2 (Mmp9, Mmp2) were not altered in MP mice (Supplementary Fig. 1C-H). Interestingly, MP mice had considerable elevations in interleukin 18 (Il18, P = 0.02, Fig. 3H) and interleukin 1β (Il1b, P = 0.05, Fig. 3I) mRNA expressions, whilst mRNA for other inflammatory mediators including NOD-like receptor family pyrin domain containing 3 (Nlrp3, Fig. 3J), tumour necrosis factor α (Tnfa, Fig. 3K), interleukin 6 (IL-6), and Il10 mRNA expressions (Fig. 3L,M) were not altered. The numbers of circulating mononuclear cells (Supplementary Table S3) were not different in MP mice.
MP mice had increased interstitial fibrosis & basal fibronectin mRNA, and IL-18 mRNA expression. (A) Blinded masson’s trichrome staining analysis of X2 sections on Aperio Image Scope showed increased % of interstitial fibrosis in the heart in MP compared to NP mice, (B) with no differences in perivascular staining as % of fibrosis to area of vessel wall. (C) RT-qPCR analysis showed increased basal fibronectin (Fn1), with no alteration in (D–G) Collagen (Col) 1a1, 3a1, alpha smooth muscle actin (acta2) & transforming growth factor β1 (Tgfb1) (mRNA expression. The inflammatory marker (H) Interleukin 18 (Il18) was increased, while there were no significant shift in (I) Il1b (J) Nod like receptor pryin domain containing 3 (Nlrp3), (K) tumor necrosis factor α (Tnfa), (L) Il6 & (M) Il10 mRNA expression in the mid-section of the heart. MP, n = 8, NP, n = 8. *P < 0.05, ns = P > 0.05 vs. NP group. Unpaired two tailed t-test, with Mann–Whitney test for post hoc analysis. Data are presented as mean ± SEM.
Cardiac transcriptomic profile changes in MP mice
RNA seq analysis to determine the differences in transcriptomic profiles of 24-month-old MP and NP mice revealed 128 differentially expressed genes (DEGs). Approximately 19,000 genes were identified from the RNA seq read set from C57Bl/6 J mouse heart mid-sections. Multi-dimensional scaling analysis (MDS) (Fig. 4A) elucidated that 42% of the variance between MP and NP mice could be attributed to the first two dimensions (Fig. 4B). Unsupervised hierarchical cluster analysis identified 2 distinct groups, with 88% identified as NP and 100% as MP clustering within their respective categories (Fig. 4C). Similarities in cardiac aging effects may have led to the NP mice clustering with the MP mice.
128 genes were differentially expressed in the MP mice compared to NP mice. (A,B) multi-dimensional (MDS) and scree plot showing 42% of the variance between MP (n = 8) and NP (n = 8) mice were due to dimension 1 and 2. (C) Heat map showing log CPM values of the top 100 differentially expressed genes (DEGS) ranked through hierarchical cluster profiling. Only 1 of the NP mice grouped into MP group. (D) Volcano plot analysis of DEGs showing 64 upregulated (red) and 64 down regulated genes (blue) in MP mice with P < 0.05, and false discovery rate (FDR) < 0.05, at logFC. (E) Significantly enriched gene ontology (GO) terms associated with biological processes (BP), cellular components (CC), and molecular function (MF) analysed through ShinyGO 0.8 ranked by FDR and fold change. (F) Chord Plot showing of CC GO terms and associated genes and their fold change. Graph generated through SR Plot.
Of the DEGs, half were upregulated and half downregulated (Fig. 4D). Notably, genes such as centrosomal protein 128 (Cep128), thrombospondin type 1 domain containing 4 (Thsd4), and dipeptidyl peptidase 4 (Dpp4) were significantly upregulated (q < 0.01), whereas mitochondrial ribosomal protein L3 (Mrpl3), solute carrier family 22 member 6 (Slc22a6), transmembrane protein 174 (Tmem174), and serine peptidase inhibitor, clade A, member F (Serpina1f.) displayed significant downregulations (q < 0.02) in MP mice. c Slc22a6, Tmem174, Serpina1f. including kidney androgen-regulated protein (Kap) gene also had a greater fold change in the down-regulated genes. The neutrophil granule protein (Ngp), SH3 domain binding kinase family member 3 (Sbk3) and Iroquois homeobox 2 (Irx2) genes had the higher fold change in the upregulated genes. Cep128, Dpp4 and Thds4 upregulation were confirmed with real time PCR (Supplementary Fig. 1G-I).
Gene ontology (GO) term analysis through ShinyGO 0.7713 revealed a significant enrichment of biological processes and functions in the MP group. Noteworthy pathways included the neuregulin signalling pathways including epidermal growth factor 2–3, Erbb2-3, signalling, (Fig. 4E, and Supplementary Fig. 2A1). Enrichments were also observed in plasma membrane components like the basolateral plasma membrane (Supplementary Fig. 2A2) and in molecular functions including protein tyrosine kinase activity and Erbb3 class receptor binding (Supplementary Figs. 2A3, and 2C). Enriched biological processes in MP mice also encompassed those associated with cardiac contraction (e.g. actin polymerisation-dependent cell motility), kidney development (e.g. specification of the loop of Henley identity), and membrane transport (e.g. carboxylic acid transport;) (Fig. 4E). The extracellular region showed enrichment of genes (Fig. 4F, orange chord) that were also involved in fibronectin binding in the MP mice (Supplementary Fig. 2C, yellow chord), implicating key genes like myocilin (Myoc), insulin like growth factor binding protein 3 (Igfbp3), syndecan 4 (Sdc4), and fibulin 1 (Fbln1). Fbln1 (P < 0.001) upregulation in MP mice was confirmed by RT qPCR (Supplementary Fig. 1 J). Additionally, genes associated with membrane transport activities, particularly amino acid (e.g. L ornithine transmembrane transporter) and organic anion/ion transport (e.g. sodium independent organic anion transmembrane transporter activity), were downregulated (Supplementary Fig. 2C).
Multiparity led to differential expression of genes involved in fibrosis, contraction, and metabolic pathways
Biological pathways analysis of the 64 upregulated genes in MP mice through Bioplanet 201914 indicated the TGFβ regulation of the ECM pathway involving genes such as apolipoprotein E (Apoe), Dpp4, Fbln1, fibroblast growth factor 7 (Fgf7), protein kinase D1 (Prkd1), and Sdc4 were upregulated. Concurrently, the actin cytoskeleton organisation within adherent junctions was also enhanced in MP mice, evidenced by the upregulation of cell adhesion molecule 3 (Cadm3), and cadherin 11 (Cdh11) (Fig. 5A,B). In contrast, the pathways governing organic anion transport and actin cytoskeleton regulation were notably downregulated for MP mice and involved several SLC family genes (Slc22a6, Slc17a1, Slc22a8, and Slc22a7) and key regulatory genes such as myosin light chain 12A (Myl12a), ENAH action regulator (Enah), integrin subunit alpha 9 (Itga9) and epidermal growth factor (Egf) genes. Further pathway analysis revealed suppression of pathways in MP mice critical for mitochondrial metabolism, specifically the tricarboxylic acid (TCA) cycle and respiratory electron transport, implicating genes such as pyruvate dehydrogenase E1 subunit B (Phdb) and ubiquinone oxidoreductase subunit A10 (Ndufa10) genes (Fig. 5C). These findings suggest preliminary genetic signatures for cardiac fibrosis, hypertrophy, and metabolic insufficiency in the hearts of MP mice.
Cardiomyopathy related pathways were enriched by DE genes associated with ECM, cardiac contraction and membrane transport in MP mice. (A) Top 10 cellular signalling pathway analysis through Bio-Planet, 2019 for up regulated (red) and down regulated (blue) genes, P < 0.05, less stringent FDR of < 0.2. Applying stringent FDR of < 0.05 only produced top 2 pathways. (B,C) Sankey plot of genes involved in each of the top 10 upregulated and downregulated enriched Bio-Planet pathways within the 64 upregulated and 64 downregulated genes, respectively. (D) 5 of the top 10 disease perturbations in GEO UP cardiomyopathy related (red nodes). (E) Cluster map of top 40 genes associated with the top 10 disease perturbations, ranked by P value, FDR < 0.05. Enrichr was used for these enrichment analysis. Data can be found on https://maayanlab.cloud/Enrichr/enrich?dataset=cebccb1381901ed9014bc6aea9eabb9f.
To explore potential cardiac related disease perturbations within the 128 DEGs, further investigations were conducted using ENRCHR15,16 tool against the GEO UP dataset17,18. This analysis identified 5 cardiac-related disease perturbations (Fig. 5D). Dilated cardiomyopathy (No. 1 within red node, Fig. 5D) was identified as the most prominent and significant perturbation and involved upregulation of critical genes such as the transcription factor pleomorphic adenoma gene like-1 (Plagl1), Igfbp3, Fbln1, Slc7a1, C-type lectin domain family 3, member b (clec3b), proprotein convertase subtilisin/kexin type 6 (Pcsk6), Egrf pathway substrate 8 (Eps8), alongside down-regulation of the multifaceted STE20-like serine/threonine-protein kinase (Slk), the microtubule associated monooxygenase, calponin and LIM domain containing 2 (Mical2), tetraspanin 9 (Tspan9), and TNF receptor superfamily member 12a (Tnfrsf12a) (Fig. 5E, x axis column No. 1). Additionally, differential gene expression associated with ventricular hypertrophy, acute myocardial infarction, cardiac hypertrophy, and congestive heart disease were observed in the MP mice (No. 4, 6, 9 & 10 within red nodes, Fig. 5D). Notably, the downregulation of regulator of calcineurin 1 (Rcan1), and Tspan9 were associated with 4 of these cardiac related disease perturbations.
Discussion
Pregnancy poses unique hemodynamic and metabolic demands on the heart. During pregnancy expansion of the maternal circulating blood volume and cardiac output are associated with adaptive myocardial remodelling which peaks at around 28 weeks of gestation for women19,20. Observational clinical studies have suggested that multiparity is associated with hypertension, reduced vascular compliance and LV dysfunction21,22. A study conducted in multiparous rats demonstrated reduced vascular compliance and a hypercontractile response to phenylephrine23. In a recent study, we demonstrated that women with HFpEF and a history of 3 or more pregnancies had a more exaggerated hemodynamic phenotype7.
In the current study, we aimed to further establish whether multiparity establishes a persistent phenotype that may provide a substrate for the development of HFpEF later in life when exposed to other known risk factors such as hypertension. We compared the cardiac structure, function, and transcriptomic profile of multiparous mice at 24 months of age (approximately 69 years old in human years- post-menopausal) vs non-parous age-matched mice. Multiparity in the context of this study is defined as being a retired/ex-breeder, which are known to have > 5 litters before retirement at 10–11 months of age. We specifically investigated the effect of parity per se in the context of aging alone, rather than adding further experimental interventions for potential HFpEF triggers, such as hypertension or obesity, to allow for a focused analysis of parity’s contribution to the pathology. Mice were examined at 24 months of age, representing a substantial period (approximately 15 months) post-oestrous cycle disruption which typically occurs at 10 months in mice.
Our findings for the first time provide evidence for multiparity priming the heart for HF in older females. The distinct cardiac transcriptomic profile and enriched pathway signatures observed in the aged multiparous mice were accompanied by significant phenotypic changes that align with the pathophysiology of HF. Echocardiographic investigation demonstrated elements of diastolic dysfunction as exemplified by prolonged isovolumic relaxation time, in the absence of any ongoing stimulus. Left ventricular volumes were increased in the MP mice, potentially of relevance as potential contributors to the pathophysiology of HF with mildly reduced EF (HFmrEF) noting that 25% of HFmrEF patients do transition to HFpEF in females with less severe HF and comorbidities24. Transcriptomic analysis identified the downregulation of the myosin light chain regulatory gene, Myl12a, which has been associated with prolonged QRS complex duration in genome-wide association studies25. Clinical observations support our findings and have shown shifts in EF, and diastolic function in multiparity and twin pregnancies22,26,27. This contrasts with singleton pregnancies where EF and contractility rarely change28. The reduced EF in our multiparous mice is reflected in older women (66 years and over) who have had ≥ 3 pregnancies22, therefore highlighting the novelty of the genomic differences we have shown here. At a histological level, MP hearts were characterised by cardiomyocyte hypertrophy accompanied by an increase in the compensatory cardiac gene marker- Nppa, encoding atrial natriuretic peptide (ANP). This increase was not observed in our RNA seq data, perhaps associated with technical differences in qPCR and RNAseq rather than biological. However, RNA seq did reveal the downregulation of genes involved in actin cytoskeleton regulation (Itga9, Enah, and Egf). Both Itga9 and Nppa have been linked to changes in blood pressure29,30, while deletion of Enah and downregulation of Egf receptor result in cardiac hypertrophy and contractile dysfunction in mice31,32. Plasma ANP levels are known to increase by 40% during pregnancy and up to 148% one week postpartum. This is driven by increased signalling through components of the renin–angiotensin–aldosterone system (RAAS), aimed at regulating BP, peripheral vascular resistance, and hydro-electrolyte balance33,34. Accumulating research indicates that an overactive RAAS in aging is a key contributor to heart disease in the elderly35,36. Aging-related cellular senescence also contributes to myocyte hypertrophy37 which could be exacerbated by multiparity given the significant increase in the atrial natriuretic peptide gene, Nppa. Multiparity could also establish an energy-deficient environment indicated by downregulation of Pdhb, Ndufa10, and the monocarboxylic (pyruvate, lactate etc.) and the carboxylic acid transmembrane transporter genes; Slc22a19, Slco1a6, Slc22a6 and Slc22a838,39. The organic anion transporters 1 (OAT1) and 3 (OAT3) are encoded by the genes Slc22a6 and Slc22a8. Inhibition of OAT1/3 has been shown to reduce fatty acid oxidation in a chronic heart failure model40. However, the parallel reductions in Pdhb encoding pyruvate dehydrogenase E1 subunit Beta, which through its multienzyme complex catalyses pyruvate into acetyl-CoA linking glycolysis and the TCA cycle41, and Nduaf10- an accessory subunit of the mitochondrial respiratory chain complex 1, responsible for its assembly38, imply fluctuations in rate and relative oxidation of fatty acids and glucose in the MP mice hearts. It is worth mentioning here the upregulated Prkd1 and Erbb3 genes are associated with gestational obesity and have been indicated as important female-specific risk factors for HF42. Moreover, it is possible that subtle diet changes could have led to these metabolic changes in the heart given the increased weight of the MP mice. Interestingly, there was a small but significant increase in the Myh6 gene, encoding alpha myosin heavy chain (αMHC), in the MP mice hearts at 15 months post oestrous cycle disruption. Since these mice do not have experimental interventions, the only distinctive factor is multiple pregnancies, therefore this increase may be a lingering effect of increased cardiac contractility during repeated pregnancies43, indicating a compensatory response. Contractility has been observed to increase with raised Myh6 levels44.
In addition to myocardial hypertrophy and altered cardiac energetics, we also detected genetic and phenotypic evidence of interstitial fibrosis in MP mouse hearts. MP mice hearts had significantly increased heart weights. LV wall thickness and mass do increase by 28–52% above pre-pregnancy levels in humans as the hemodynamic load increases indicating the presence of cardiac hypertrophy and remodelling19,28. This eventually leads to a temporal increase in mRNA levels of fibrotic markers and increased tissue and perivascular fibrosis45,46. Perivascular fibrosis is often noted in animal models where hypertensive stimuli such as L-NAME is employed47, therefore it was not observed in the current study which has no added hypertensive stimuli. As the myocardial performance returns to pre-gravid structural function after birth, the myocardium experiences reverse remodelling, which includes a delicate balancing of Mmp and Timp Transcriptomics of MP mice hearts indicate a state of incomplete reverse remodelling with fibrotic gene signatures in the ECM still differentially expressed at 15 months post oestrous cycle disruption. These signatures included the genes encoding the ECM proliferating proteins such as Ngp, Sdc4, Fbln1, neuregulin 1 (Nrg1), and modular calcium binding protein 1 (Smoc1). Increased Nrg1 mRNA expression in the cardiac microvasculature is triggered by neurohormones such as Ang II and mechanical pressure, leading to NRG1 forward signaling through the Nrg1/ErbB pathways affecting cardiomyocyte growth and cardiovascular processes48,49. Additionally, through their protein products, Fbln1 induces ECM deposition, while Sdc4 and Smoc1 enhance myocardial stiffness in Ang II-induced and pressure-overloaded heart dysfunction50,51,52.
The matrix metalloproteases Thsd4/ A disintegrin and metalloproteinase with thrombospondin motifs 6 (Adamst6) and ADAMST like 3 (Adamstl3) were also upregulated and both function in regulating the cardiac ECM53,54. However, there were no significant differences in known fibrotic markers including collagens, Acta2, Mmps, and Timps genes, in both RNA Seq and qPCR data. A similar Mmp landscape has been noted 7 days post-partum in rats55, indicating how quickly they revert to pre-pregnancy levels, thus affirming our findings showing no changes at 15 months post oestrous cycle disruption. Several studies have reported concerted switching of Mmps and Timps before, during, and later in pregnancy45,46,55,56. The Mmps act as collagenases and play an important role in the degradation of the ECM57. In normal aging, Mmp9 levels are elevated and are involved in the modulation of cytokines, chemokine, hormones, growth factors, and angiogenic factors58. Interestingly we observed a significant increase in Il18 mRNA, which may be a lingering evidence of pregnancy-induced compensatory myocyte hypertrophy in the MP mice59,60, which is consistent with the increase in stress induced mRNA expression of ANP.
Despite the lack of change in mRNA levels of Col1a1, Col3a1, and Acta2, we did observe a marked elevation of Fn1 mRNA indicating an increase in basal collagen levels. Fn1 polymerisation has been shown to be critical in ECM composition, organisation, and stability, and is linked to increased fibrosis in an HF model61,62. Fn1 encodes the large basal glycoprotein-fibronectin, which interacts with lysly oxidase to encourage collagen cross linking63. This is supported by the RNA seq analysis indicating enrichment of fibronectin binding function with upregulation of Fbln1, Sdc4, Igfbp3 and Nrg1 genes. The activation of Nrg1/Erbb signalling process plays a modulatory role in physiological hemodynamic overload such as pregnancy48, and it’s activation months after the last pregnancy maybe linked to epigenetic changes. This signalling pathway is also activated as a compensatory mechanism in response to a failing heart64, linked especially to maintaining systolic function and has recently being targeted as therapy for HF65,66. Therefore, a reduced Collagen deposition in the aged mice is also gradual and below (1–4% of LV area) the levels seen in pathological states such as a myocardial infarction67,68, although the current observations were in the absence of other pro-fibrotic, active stimuli. However, increased cross-linking and reduced degradation due to aging69 can result in increase in interstitial fibrosis. The histological features observed in the MP mice in our study are similar to those observed in autopsies of HFpEF hearts which had more interstitial fibrosis than controls with increased hypertrophy70. The average age at HFpEF for these patients was 75 years old. Weight gain and increase in fat mass in the MP mice could also have influenced these findings71.
Taken together, the findings of this study provide a transcriptomic, functional, and structural basis for clinical observations of an association between multiparity as a potential risk factor for HFpEF. Future studies are required to further establish the nature and extent of a potential interaction between multiparity and other conventional risk factors for HFpEF. From the clinical perspective, our observations may also provide support for closer longitudinal follow-up of multiparous women, particularly in the setting of exposure to other ongoing cardiovascular risk factors.
Limitations
In the present study, we do not have detailed data regarding the effects of litter size, and number of pregnancies, which could have impacted the results and could account for variations in the clusters for RNA seq data. Therefore, these could have impacted the interpretation of the data provided in this study. Additionally, due to the explorative nature of this study, there is no baseline (ie, at last pregnancy) echocardiography data to compare the echocardiography results at 24 months of age. Additional confirmatory cardiac function assessment such as pressure volume catheterisation and left atria volume index were not performed in this study. We also recommend our findings be interpreted keeping in mind the differences between mice and humans in terms of the number of offspring in a single pregnancy and the gestational periods.
Materials and methods
Experimental design for animal studies
All animal experiments were performed in accordance with the National Health and Medical Research Institute Animal Welfare Committee guidelines and the study protocols were approved by the Alfred Medical Research and Education Precinct (AMREP) ethics committee (E/1829/2018/B & E/8252/2022/B). The authors complied with the ARRIVE guidelines. All animals were obtained at 22–23 months of age from Animal Resource Centre, Perth, WA and housed on a 12 h light/dark cycle with food (Irradiated Rat & Mouse Diet-SF00-100, Specialty Feeds, WA) and water provided ad libitum. The MP and NP mice were maintained on SF00-100 normal chow diet consisting of 4.2% fat, 19.6% protein and 59.3% carbohydrates during their aging period and study period. Both MP (n = 8) and NP (n = 8) mice were aged to 24 months of age and were considered post-oestrous cycle disruption by 10 months given that female mice cease reproductive cycles at 16 months72. MP mice were selected from the ex-breeding stock whilst NP mice were selected from non-breeding stock. Multiple pregnancies in this study is defined as having > 3 pregnancies determined from first pregnancy at 6–8 weeks, and the last pregnancy before retirement at approximately 40 weeks of age. There were no other experimental interventions conducted on these mice except for the differences in their obstetric history and procedures to measure their morphometric and cardiac differences. Both groups under-went echoMRI testing for body composition analysis, and food and water consumption were assessed through metabolic caging. Both NP and MP mice received normal rodent chow diet. Body weights of all groups were recorded prior to euthanasia. In addition, systolic blood pressure was measured via non-invasive tail cuff plethysmography. Mice were sacrificed by euthanasia using carbon dioxide (CO2) exposure, plasma and tissues were taken and stored at − 80 °C until use.
EchoMRI body composition analysis
EchoMRI was performed on the EchoMRI 4 in 1 body composition analyser (EchoMRI™, TX, USA), following a previously published protocol73. Fat mass, lean mass, free water, and total water were determined during the scan which lasted approximately 1 min.
Metabolic caging
24 h metabolic caging was performed using single mouse metabolic cages (Techniplast, MI, Italy). Mice were housed on a 12 h light/dark cycle with food and water provided ad libitum. Body weight, food weight, and water weight and urine volume were recorded over the 24 h period of metabolic caging for quantitative analysis of differences in energy and fluid homeostasis.
Tail cuff plethysmography for systolic blood pressure measurement
Briefly, mouse was placed in the restrainer, the tail threaded through the tail cuff, and stabilised with the sensory assembly unit on the MC4000 non-invasive multi-channel blood pressure analysis system (Hatteras Instruments, NC, USA). The platform temperature was acclimatised and monitored at 37 °C prior to restraining the mice. 5 preliminary readings were taken before the 10 actual reading cycles by inflating the tail cuff. Pressure readings were taken every minute for 10 min.
Echocardiography analysis
To assess cardiac function, echocardiography was performed on mice under anaesthesia (Ketamine, Xylazine, Atropine (KXA), 80/8/0.96 mg/kg, i.p.). Echocardiography was performed using a Vevo 2100 (Fujifilm Visualsonics Inc., ON, Canada) echocardiography system with a 40 MHz MS55OD transducer, with mice imaged on an ECG-, temperature-, and respiration-recording platform. Parasternal long axis imaging of the left ventricle was obtained by 2D B-mode and analysed using the VevoLab VevoStrain analysis package to measure left ventricular volumes, ejection fraction (EF) and global longitudinal strain (GLS %). Trans-mitral flow velocity was assessed using Doppler where the early phase (E wave) and late phase (A wave) were examined to determine E/A ratio and deceleration time (DT) was measured from the peak E wave to it’s projected baseline. Similarly, tissue Doppler was performed to assess early diastolic mitral annular velocity (e’), and the ratio E/e’. All traces were analysed in a blinded fashion using the VevoLab analysis software-VisualSonics.
Quantitative real-time PCR
To assess changes in gene expression, total RNA was isolated from left ventricular tissue using RNeasy Mini Kit (Qiagen, NRW, Germany) following manufacturer instructions. RNA samples were reverse-transcribed into cDNA using High-Capacity cDNA Reverse Transcription Kit on the Veriti Dx 96-well Thermal Cycler (Applied Biosystems, MA, USA). Reverse transcribed cDNA from each sample were plated on a 384-well plate and run for PCR amplification with specific primer pairs for genes of interest (Sigma-Aldrich, MA, USA; Supplementary Table S1). Real-time PCR was performed using Fast SYBR Green Master on the QuantStudio 7 Flex Real-Time PCR System (Applied Biosystems, MA, USA) to determine mRNA levels of the target genes. Gene expression was calculated using comparative CT (ΔΔCT) method normalised to the housekeeper gene, Glyceraldehyde 3-phosphate dehydrogenase (GAPDH).
RNA sequencing and bioinformatics analysis
To complement the targeted qPCR studies, RNA seq was also performed on mid sections of the left ventricular tissue. Total RNA was extracted, and cDNA was generated using Illumina Stranded Total RNA Prep kit with Ribo-Zero Plus depletion. Sequencing was completed on a NovaSeq X with 150 paired-end reads and sequenced to a depth of a minimum of 50M reads per sample.
Data quality control and preprocessing
Upon sequencing, raw reads were demultiplexed by bcl2fastq version 2.20 (RRID:SCR_015058) and fastq files resulted were subjected to preprocessing by TrimGalore suite v0.6.10 (RRID:SCR_011847), involving Illumina adapter and priming site removal using Cutadapt version 4.8 (DOI:10@@@.14806/ej.17.1.200), followed by quality control assessments with FastQC v0.12.1 (http://www.bioinformatics.babraham.ac.uk/projects/fastqc). Before further demultiplexing the splits of fatsq files were concatenated back into 23 samples for further data processing, and Multi-QC version 1.2174 was performed to validate the sequencing-quality and -consistency for each split.
Alignment and quantification
The filtered reads were mapped to the mouse reference genome (GRCm38.p6.vM25) using STAR aligner version 2.7.11b75. BAM files were processed and indexed using Samtools version 1.19.276 and Bamtools version 2.5.277 to provide mapping statistics. HTSeq v0.11.178 was employed for constructing counting matrices, and an aggregated quality control summary was compiled with Multi-QC.
Differential expression analysis
Count matrices were processed using base-R version 4.2.2 (https://www.R-project.org) for metadata integration, gene annotation, filtering of lowly expressed genes. After Trimmed Mean of M-values (TMM) normalization by EdgeR version 3.40.279, procedures including mean–variance dependence elimination, empirical Bayes moderation, and quality-weighted VOOM transformation were implemented using limma package version 3.54.280. Glimma version: 2.8.081 was applied for unsupervised sample clustering and other data visualization for the differential gene expression analyses.
Downstream analyses
Differentially expressed genes (DEGs) were identified using predefined criteria of adjusted P-values (< 0.05). Enrichment and Gene Set Enrichment Analysis (GSEA) of DEGs in context of Gene Ontology (GO)82 or KEGG pathway annotations were performed using clusterProfiler version 4.10.1 83,84, Enrichr (https://maayanlab.cloud/Enrichr/#libraries) 15,16,85 for Bioplanet 2019 14 and Disease Perturbations in GEO UP 17,18 and Pathview version 3.18 86, ShinyGO 0.77 (http://bioinformatics.sdstate.edu/go/) 13 and visualised using SR Plot87. All enrichment analysis files are supplied in Supplementary Excel Files S4.
Histological assessment of collagen staining and myocyte size
To assess collagen deposition, 4 mm thick whole heart tissue sections (excluding atria) were subjected to Masson’s Trichrome staining. This was performed by the Monash Histology Platform, Clayton, VIC, Australia. Scanned bright field images from Aperio Slide Scanner using the Olympus FSX100 bright field microscope at X 20 magnification. Analysis was conducted using Aperio Image Scope software version 12.4.6 (Leica Biosystems, HE, Germany). 2 cardiac tissue cross sections per heart were annotated and blood vessels and cavities were excluded before analysing for positive pixel intensity of aniline blue stained collagen fibres. Similarly, to assess cardiomyocyte width, cardiac tissue sections were processed and stained with Hematoxylin & Eosin (H&E). The size of 20 cardiomyocytes/section randomly selected within the left ventricle of 2 cardiac cross sections were analysed by two blinded scientists from slide scanned images.
Data and statistical analysis
All data analysis were conducted in a blinded manner, with analyst blinded to group allocation for echocardiography, PCR, histology, and morphometrics data analysis. Sample size estimates were based on our prior studies investigating the effects of angiotensin II as a stimulus for cardiac remodelling11. Statistical significance was determined using a 2-tailed Student’s t-test, where normality was tested using Shapiro–Wilk test. Mann–Whitney test was used for post hoc analysis. A P-value of less than 0.05 was deemed statistically significant, exclusive of RNA seq data which were deemed statistically significant at an adjusted P-value of < 0.05 after correcting for multiple testing. All data are expressed as mean ± standard error of the mean (SEM), unless otherwise noted, and statistical analysis was carried out using GraphPad Prism 10.0 software for all other data analysis, while R (version 4.2.2) was used for RNA seq data analysis.
Data availability
All data supporting the findings of this study are available upon request. RNA sequencing dataset generated during the current study are available in the Gene Expression Omnibus (GEO) repository and can be accessed here https://urldefense.com/v3/https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE289, with the token- ircdaaauhnwzpyz. Contact Dr. Bing Wang to access this data if needed.
Abbreviations
- DEGs:
-
Differentially expressed genes
- HF:
-
Heart failure
- HFpEF:
-
Heart failure with preserved ejection fraction
- MP:
-
Multi-parous/multiple pregnancies
- NP:
-
No pregnancy (virgin)
- RNA Seq:
-
RNA sequencing
References
Anker, S. D. et al. Empagliflozin in heart failure with a preserved ejection fraction. N. Engl. J. Med. 385(16), 1451–1461 (2021).
Solomon, S. D. et al. Dapagliflozin in heart failure with mildly reduced or preserved ejection fraction. N. Engl. J. Med. 387(12), 1089–1098 (2022).
Shah, S. J. et al. Atrial shunt device for heart failure with preserved and mildly reduced ejection fraction (REDUCE LAP-HF II): A randomised, multicentre, blinded, sham-controlled trial. Lancet 399(10330), 1130–1140 (2022).
Shah, S. J. et al. Research priorities for heart failure with preserved ejection fraction: National heart, lung, and blood institute working group summary. Circulation 141(12), 1001–1026 (2020).
Owan, T. E. et al. Trends in prevalence and outcome of heart failure with preserved ejection fraction. N. Engl. J. Med. 355(3), 251–259 (2006).
Beale, A. L. et al. Sex differences in cardiovascular pathophysiology: Why women are overrepresented in heart failure with preserved ejection fraction. Circulation 138(2), 198–205 (2018).
Beale, A. L. et al. Sex differences in heart failure with preserved ejection fraction pathophysiology: A detailed invasive hemodynamic and echocardiographic analysis. JACC Heart Fail. 7(3), 239–249 (2019).
Beale, A. L. et al. The effect of parity on exercise physiology in women with heart failure with preserved ejection fraction. ESC Heart Fail. 7(1), 213–222 (2020).
Maeder, M. T. et al. Hemodynamic basis of exercise limitation in patients with heart failure and normal ejection fraction. J. Am. Coll. Cardiol. 56(11), 855–863 (2010).
Lewandowski, D., et al., Relation of Left ventricular diastolic function to global fibrosis burden: Implications for heart failure risk stratification. JACC Cardiovasc. Imaging 16(6), 783–796 (2023).
Kaye, D. M. et al. Deficiency of prebiotic fiber and insufficient signaling through gut metabolite-sensing receptors leads to cardiovascular disease. Circulation 141(17), 1393–1403 (2020).
Chu, P. Y. et al. CXCR4 antagonism attenuates the cardiorenal consequences of mineralocorticoid excess. Circ. Heart Fail. 4(5), 651–658 (2011).
Ge, S. X., Jung, D. & Yao, R. ShinyGO: A graphical gene-set enrichment tool for animals and plants. Bioinformatics 36(8), 2628–2629 (2019).
Huang, R., et al. The NCATS BioPlanet—An integrated platform for exploring the universe of cellular signaling pathways for toxicology, systems biology, and chemical genomics. Front. Pharmacol. 10 (2019).
Xie, Z. et al. Gene set knowledge discovery with enrichr. Curr. Protoc. 1(3), e90 (2021).
Kuleshov, M. V. et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 44(W1), W90–W97 (2016).
Barrett, T., et al. NCBI GEO: Archive for functional genomics data sets—Update. Nucleic Acids Res. 41(Database issue), D991–5 (2013).
Dudley, J. T. et al. Disease signatures are robust across tissues and experiments. Mol. Syst. Biol. 5, 307 (2009).
Sanghavi, M. & Rutherford, J. D. Cardiovascular physiology of pregnancy. Circulation 130(12), 1003–1008 (2014).
Klein, H. H. & Pich, S. Cardiovascular changes during pregnancy. Herz 28(3), 173–174 (2003).
Ogunmoroti, O., et al. Multiparity is associated with poorer cardiovascular health among women from the multi-ethnic study of atherosclerosis. Am. J. Obstet. Gynecol., 221(6), 631 e1–631 e16 (2019).
Kim, H. J. et al. Effects of multiparity on left ventricular diastolic dysfunction in women: cross-sectional study of the KoRean wOmen’S chest pain rEgistry (KoROSE). BMJ Open 8(12), e026968 (2018).
Dhawan, V., Brookes, Z. L. & Kaufman, S. Long-term effects of repeated pregnancies (multiparity) on blood pressure regulation. Cardiovasc. Res. 64(1), 179–186 (2004).
Savarese, G. et al. Prevalence and prognostic implications of longitudinal ejection fraction change in heart failure. JACC Heart Fail. 7(4), 306–317 (2019).
Evans, D. S. et al. Fine-mapping, novel loci identification, and SNP association transferability in a genome-wide association study of QRS duration in African Americans. Hum. Mol. Genet. 25(19), 4350–4368 (2016).
Ghi, T. et al. The effect of chorionicity on maternal cardiac adaptation to uncomplicated twin pregnancy: A prospective longitudinal study. Fetal Diagn Ther 45(6), 394–402 (2019).
Orabona, R., et al. Maternal left ventricular function in uncomplicated twin pregnancies: a speckle-tracking imaging longitudinal study. J. Clin. Med. 11(18) (2022).
Ducas, R. A. et al. Cardiovascular magnetic resonance in pregnancy: insights from the cardiac hemodynamic imaging and remodeling in pregnancy (CHIRP) study. J. Cardiovasc. Magn. Reson. 16(1), 1 (2014).
Sun, G., et al. A bioinformatics perspective on the links between tetraspanin-enriched microdomains and cardiovascular pathophysiology. Front. Cardiovasc. Med. 8 (2021).
Castro, L. C., Hobel, C. J. & Gornbein, J. Plasma levels of atrial natriuretic peptide in normal and hypertensive pregnancies: a meta-analysis. Am. J. Obstet. Gynecol. 171(6), 1642–1651 (1994).
Aguilar, F. et al. Mammalian enabled (Mena) is a critical regulator of cardiac function. Am. J. Physiol. Heart Circ. Physiol. 300(5), H1841–H1852 (2011).
Guo, S. et al. Epidermal growth factor receptor-dependent maintenance of cardiac contractility. Cardiovasc. Res. 118(5), 1276–1288 (2022).
Leal, C. R. V. et al. Renin-angiotensin system in normal pregnancy and in preeclampsia: A comprehensive review. Pregnancy Hypertens. 28, 15–20 (2022).
Yu, L. et al. Angiotensin-(1–5), an active mediator of renin-angiotensin system, stimulates ANP secretion via Mas receptor. Peptides 86, 33–41 (2016).
Conti, S., Cassis, P. & Benigni, A. Aging and the renin-angiotensin system. Hypertension 60(4), 878–883 (2012).
Ferrario, C. M. ACE2: More of Ang-(1–7) or less Ang II?. Curr. Opin. Nephrol. Hypertens. 20(1), 1–6 (2011).
Tang, X., Li, P. H. & Chen, H. Z. Cardiomyocyte senescence and cellular communications within myocardial microenvironments. Front. Endocrinol. (Lausanne) 11, 280 (2020).
Guo, P. et al. CAV3 alleviates diabetic cardiomyopathy via inhibiting NDUFA10-mediated mitochondrial dysfunction. J. Transl. Med. 22(1), 390 (2024).
Tu, C. et al. Tachycardia-induced metabolic rewiring as a driver of contractile dysfunction. Nat. Biomed. Eng. 8(4), 479–494 (2024).
Lai, Q. et al. Inhibition of OAT1/3 and CMPF uptake attenuates myocardial ischemia-induced chronic heart failure via decreasing fatty acid oxidation and the therapeutic effects of ruscogenin. Transl. Res. 261, 1–15 (2023).
Li, A. et al. Molecular characterization and transcriptional regulation analysis of the bovine PDHB gene. PLoS ONE 11(7), e0157445 (2016).
van der Kemp, J. et al. Women-specific risk factors for heart failure: A genetic approach. Maturitas 109, 104–111 (2018).
Melchiorre, K. et al. Maternal cardiovascular function in normal pregnancy. Hypertension 67(4), 754–762 (2016).
Gacita, A. M. et al. Genetic variation in enhancers modifies cardiomyopathy gene expression and progression. Circulation 143(13), 1302–1316 (2021).
Parrott, M. E. et al. Maternal cardiac messenger RNA expression of extracellular matrix proteins in mice during pregnancy and the postpartum period. Exp. Biol. Med. (Maywood) 243(15–16), 1220–1232 (2018).
Umar, S., et al. Cardiac structural and hemodynamic changes associated with physiological heart hypertrophy of pregnancy are reversed postpartum. J. Appl. Physiol. 113(8), 1253–9 (1985).
Sanhueza-Olivares, F., et al. Vascular remodelling in a mouse model of heart failure with preserved ejection fraction. J. Physiol. (2025).
Lemmens, K. et al. Role of neuregulin-1/ErbB2 signaling in endothelium-cardiomyocyte cross-talk. J. Biol. Chem. 281(28), 19469–19477 (2006).
Wang, Y. et al. Neuregulin-1, a potential therapeutic target for cardiac repair. Front. Pharmacol. 13, 945206 (2022).
Ge, Q. et al. Fibulin1C peptide induces cell attachment and extracellular matrix deposition in lung fibroblasts. Sci. Rep. 5(1), 9496 (2015).
Herum, K. M. et al. Syndecan-4 is a key determinant of collagen cross-linking and passive myocardial stiffness in the pressure-overloaded heart. Cardiovasc. Res. 106(2), 217–226 (2015).
Li, Y. et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521(7553), 520–524 (2015).
Mead, T. J. ADAMTS6: Emerging roles in cardiovascular, musculoskeletal and cancer biology. Front. Mol. Biosci. 9, 1023511 (2022).
Rypdal, K., et al. Deletion of the extracellular matrix glycoprotein ADAMTSL3 increases pro-fibrotic signalling and exacerbates heart failure in mice. Eur. Heart J. 43(Supplement_2) (2022).
Virgen-Ortiz, A., et al. Matrix metalloproteinases system and types of fibrosis in rat heart during late pregnancy and postpartum. Medicina (Kaunas) 55(5) (2019).
Chung, E., Heimiller, J. & Leinwand, L. A. Distinct cardiac transcriptional profiles defining pregnancy and exercise. PLoS ONE 7(7), e42297 (2012).
Visse, R. & Nagase, H. Matrix metalloproteinases and tissue inhibitors of metalloproteinases: structure, function, and biochemistry. Circ. Res. 92(8), 827–839 (2003).
Meschiari, C. A. et al. The impact of aging on cardiac extracellular matrix. Geroscience 39(1), 7–18 (2017).
Abbate, A. et al. Interleukin-1 and the inflammasome as therapeutic targets in cardiovascular disease. Circ. Res. 126(9), 1260–1280 (2020).
O’Brien, L. C. et al. Interleukin-18 as a therapeutic target in acute myocardial infarction and heart failure. Mol. Med. 20(1), 221–229 (2014).
Sottile, J. & Hocking, D. C. Fibronectin polymerization regulates the composition and stability of extracellular matrix fibrils and cell-matrix adhesions. Mol. Biol. Cell 13(10), 3546–3559 (2002).
Valiente-Alandi, I. et al. Inhibiting fibronectin attenuates fibrosis and improves cardiac function in a model of heart failure. Circulation 138(12), 1236–1252 (2018).
Fogelgren, B. et al. Cellular fibronectin binds to lysyl oxidase with high affinity and is critical for its proteolytic activation. J. Biol. Chem. 280(26), 24690–24697 (2005).
Keulenaer, G. W. D. et al., Mechanisms of the multitasking endothelial protein NRG-1 as a compensatory factor during chronic heart failure. Circ. Heart Fail. 12(10), e006288 (2019).
Mikami, Y. et al. Compensatory role of neuregulin-1 in diabetic cardiomyopathy. J. Pharmacol. Sci. 153(3), 130–141 (2023).
Cools, J. M. T. et al. Small-molecule-induced ERBB4 activation to treat heart failure. Nat Commun 16(1), 576 (2025).
Chiao, Y. A. et al. Matrix metalloproteinase-9 deletion attenuates myocardial fibrosis and diastolic dysfunction in ageing mice. Cardiovasc. Res. 96(3), 444–455 (2012).
Lin, J. et al. Age-related cardiac muscle sarcopenia: Combining experimental and mathematical modeling to identify mechanisms. Exp. Gerontol. 43(4), 296–306 (2008).
Hecker, L., et al. Reversal of persistent fibrosis in aging by targeting Nox4-Nrf2 redox imbalance. Sci. Transl. Med., 6(231), 231ra47 (2014).
Mohammed, S. F. et al. Coronary microvascular rarefaction and myocardial fibrosis in heart failure with preserved ejection fraction. Circulation 131(6), 550–559 (2015).
Che, C., Dudick, K. & Shoemaker, R. Cardiac hypertrophy with obesity is augmented after pregnancy in C57BL/6 mice. Biol. Sex Differ. 10(1), 59 (2019).
Nelson, J. F. et al. A longitudinal study of estrous cyclicity in aging C57BL/6J Mice: I. Cycle frequency, length and vaginal cytology1. Biol. Reprod. 27(2), 327–339 (1982).
Lancaster, G. I. & Henstridge, D. C. Body composition and metabolic caging analysis in high fat fed mice. J. Vis. Exp. (135) 2018.
Ewels, P. et al. MultiQC: Summarize analysis results for multiple tools and samples in a single report. Bioinformatics 32(19), 3047–3048 (2016).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29(1), 15–21 (2013).
Danecek, P., et al. Twelve years of SAMtools and BCFtools. Gigascience 10(2) (2021).
Barnett, D. W. et al. BamTools: A C++ API and toolkit for analyzing and managing BAM files. Bioinformatics 27(12), 1691–1692 (2011).
Anders, S., Pyl, P. T. & Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31(2), 166–169 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1), 139–140 (2010).
Ritchie, M. E. et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43(7), e47 (2015).
Su, S. et al. Glimma: Interactive graphics for gene expression analysis. Bioinformatics 33(13), 2050–2052 (2017).
Harris, M. A., et al. The Gene Ontology (GO) database and informatics resource. Nucleic Acids Res. 32(Database issue), D258–61 (2004).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28(1), 27–30 (2000).
Yu, G. et al. clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 16(5), 284–287 (2012).
Chen, E. Y. et al. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinf. 14, 128 (2013).
Luo, W. et al. Pathview Web: User friendly pathway visualization and data integration. Nucleic Acids Res. 45(W1), W501–W508 (2017).
Tang, D. et al. SRplot: A free online platform for data visualization and graphing. PLoS ONE 18(11), e0294236 (2023).
Funding
DK is the recipient of an NHMRC Investigator Grant ID 2008017.
Author information
Authors and Affiliations
Contributions
Conceptualization – R. M, D. M. K; Methodology – R. M; Investigation and assisted with experiments – R. M, S. B, D.G. D, H. K, K. G, R. N, H.R. L, S. S, M. N, L. S; B. H. W; Resources – B. H. W, D. M. K; Statistical analysis for RNA sequencing Data- W. H, F. T. Manuscript Original Draft – R. M; Manuscript Review & Editing – All authors.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Magaye, R.R., Wang, B.H., Liu, H.R. et al. Multiparity induces persistent myocardial structural, functional and transcriptomic remodelling in mice. Sci Rep 15, 24254 (2025). https://doi.org/10.1038/s41598-025-08248-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-08248-z