Abstract
Alzheimer’s disease (AD) is a neurodegenerative disorder that begins with memory loss and can lead to death. In Puerto Rico, AD is the fourth leading cause of death, while in the United States, it ranks fifth. Research suggests the gut microbiome contributes to the pathophysiology of AD by promoting inflammation and amyloid buildup in the brain. This study examined the composition and diversity of the gut microbiota in Puerto Ricans with AD compared to controls and its relationship with cognitive impairment severity and APOE status. We recruited 100 participants (50 AD, 50 controls), who underwent clinical and cognitive assessments (MoCA/CDR). While overall microbial diversity and richness showed no significant differences, specific bacterial taxa exhibited subtle differential abundance. Faecalibacterium and Bacteroides were moderately significant and increased on controls compared to AD. Moreover, individuals carrying the APOE E4 allele exhibited lower abundances of butyrate-producing bacteria and an enrichment of potentially pro-inflammatory genera, including Eggerthella and Lachnoclostridium. These findings suggest that gut microbiota dysbiosis may contribute to cognitive decline and AD progression, highlighting the potential for microbiome-targeted therapeutic interventions in AD prevention and treatment.
Similar content being viewed by others
Introduction
Alzheimer’s disease (AD) is a neurodegenerative disease influenced by multiple factors that lead to the deterioration of various cognitive domains, particularly memory. It accounts for 60% to 80% of dementia cases, making it the leading cause of the condition currently affecting approximately 6.7 million Americans aged 65 and older. According to the Alzheimer’s Association Report, this number could grow to 13.8 million by 20601. Considering age as the most important risk factor for AD, the United States (U.S.) healthcare system is not equipped with the resources needed to attend to an increasing elderly population responding to a higher life expectancy and the Baby Boomers approaching the age of AD onset1. One of the fastest-growing populations in the U.S. is the Hispanic community, the largest ethnic minority group, comprising nearly 20% of the U.S. population, reaching 63.7 million in 2022 from 50.5 million in 2010. Indeed, the Hispanic community represents the highest expected increase in AD prevalence by 2060, with 423% when compared to 192% higher among Blacks and 63% higher among Whites2. Among Hispanics, Puerto Ricans constitute the second-largest population in the U.S., with a concerning trend in 2022 of approximately 23.5% of its population aged 65 and older3. In fact, in Puerto Rico, AD is the fourth leading cause of death3,4, whereas in the United States, it ranks fifth among individuals aged 65 and older in 20211.
Understanding the biological factors associated with AD is crucial, emphasizing physiological and other modifiable variables such as lifestyle, nutrition, and ancestry background. The most characteristic evidence in AD pathophysiology is the accumulation of misfolded proteins with the egregious effects of chronic oxidative and inflammatory damage, resulting in brain hypometabolism and neural death/atrophy5. Cerebral amyloid-β plaques (Aβ) accumulate as oligomers and hyperphosphorylated tau protein tangles, which choke neurons in the brain, inducing the cognitive deficits characteristic of AD6. The apolipoprotein E (APOE) gene is the most common genetic risk factor for AD, and it functions by binding lipids and transporting them through the lymphatic and circulatory systems. It influences the progression of associated pathological changes by forming neurofibrillary tangles and amyloid clearance processes. APOE is expressed in three isoforms (APOE E2, APOE E3, and APOE E4), each with varying abilities to regulate myelin levels and maintain neuronal membrane integrity7. The E2 allele is considered neuroprotective, E3 is neutral, and E4 significantly increases the risk of late-onset AD7. Individuals with one copy of the E4 allele face a two-to-four-fold increased risk, while those with two copies may have a 19-fold higher risk. The contribution of APOE E4 to cognitive decline may differ based on geographic background and ethnicity5,8,9. Previous studies have found that the association between APOE E4 and cognitive decline is strong and consistent in non-Hispanic White populations but appears weaker and more variable in Latinos. Such variability could relate to differences in genetic ancestry across Latino subgroups. When six different backgrounds were compared, the effect of APOE E4 on significant cognitive decline differed by background, with the strongest association seen in Cubans. Additionally, APOE E2 was found to be protective against MCI in Puerto Ricans, while higher Amerindian ancestry appeared to mitigate the risk associated with APOE E49.
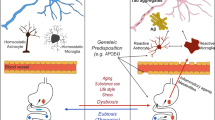
The gut microbiota (GM) intrinsically correlates with neurological diseases through the gut-brain axis (GBA). The microbiota consists of a symbiotic relationship among millions of beneficial microbes that have been shown to regulate neurochemical pathways10. The GBA is a two-way communication system mediated by the vagus nerve and enteric nervous system with various signaling mechanisms. Specific bacterial taxa, such as Bacteroides, may produce neurotransmitters, such as GABA (gamma-aminobutyric acid)10, glutamate, acetylcholine, norepinephrine, dopamine, and serotonin, and metabolites (e.g., brain-derived neurotrophic factor, short-chain fatty acids (SCFA), and cytokines) that may influence anxiety, cognition, and overall mood11,12,13. Research has indicated that GABA plays a role in regulating oxidative stress and pro-inflammatory agents (such as serum cholesterol, lipopolysaccharides, and cytokines), which are linked to amyloidosis and low-grade systemic inflammation—both recognized as pathological factors in AD13,14,15.
Furthermore, a recent discovery showed that modifying the gut microbiota can substantially reduce inflammation, tau pathology, and brain damage in a sex and APOE-dependent manner16. Microbial dysbiosis creates a pathogenic GBA, where variations in the abundance and diversity of specific species may contribute to neurological deficits through multiple mediator pathways. Additionally, microbiota-driven changes in inflammation have emerged as central mechanisms linking gut health and neurodegenerative diseases. Understanding changes in the microbiota with cognitive impairment and AD will reveal important insights into the potential modification of the microbiota through interventions, as restoration, a new hot topic of the human microbiome, has become a novel approach for mitigating and managing gut-related diseases17.
Numerous studies have reported altered gut microbiota profiles in AD patients compared to healthy individuals. For example, in a study with 25 AD and 25 controls, AD participants had decreased microbial richness and diversity and a distinct composition compared to controls18. They found that AD patients had a significantly reduced abundance of Firmicutes and Bifidobacterium, but increased Bacteroidetes and pro-inflammatory taxa such as Escherichia-Shigella, suggesting a microbial signature associated with systemic inflammation and neurodegeneration18. A more recent review identified taxonomic shifts in Blautia, Prevotella, and Alistipes related to changes in systemic inflammation and cognitive scores19. Moreover, research on frail elders, revealed an increased abundance of opportunistic pathogens such as Enterobacteriaceae and a decline in beneficial taxa, including Lactobacillus and Butyricicoccus, reinforcing the hypothesis that gut microbiota imbalance is linked to cognitive decline20. A review published in 2025 synthesized existing evidence on the GBA and therapeutic modulation of the microbiota, proposing that altering gut microbial composition may not only be a marker but a viable intervention pathway for AD treatment21.
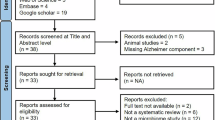
To our knowledge, this is the first study to examine the association between gut microbiota and cognitive impairment severity in both healthy individuals and AD patients within the Puerto Rican population. It provides novel evidence linking gut microbiota composition to pathogenic APOE isoforms and cognitive decline. Furthermore, it investigates the intricate relationship between AD and the gut microbiome while considering critical confounding factors. These findings are crucial for identifying potential targets related to AD risk assessment, prevention, early diagnosis, and microbiota-based therapeutic approaches.
Results
Study design and population description
One hundred participants, ages 55 to 94, were recruited from the University of Puerto Rico Medical Sciences Campus Internal Medicine and Geriatrics Outpatient Clinics. This cohort included AD patients (n = 50) and an unimpaired control group (n = 50) (Table 1). We compared the demographics and cognitive scales between the two groups. Most AD and control group participants were female, comprising 70% and 72% of the cohort, respectively. The most represented age group for AD patients was over 77 years (56%), while for controls, it was 66–76 years (54%). Only 14% of control subjects (7 individuals) were over 77 years, and 6% of AD subjects (3 individuals) were between 55 and 65 years old. Educational attainment above the 12th grade was higher in the control group (92%) compared to the AD group (58%) (p = 0.0002383). The median B12 level was 624 pg/mL for all participants, with AD patients having a median B12 level of 777 pg/mL and controls 454 pg/mL. Regarding body mass index (BMI), AD patients were primarily classified as overweight (42%), followed by normal weight (36%) and obese (22%). In contrast, the control group had a predominant BMI classification of obese (50%), followed by overweight (26%) and normal weight (24%). These inverted changes were significant (p = 0.01469).
Cognitive assessments using clinical dementia rating and montreal cognitive assessment
After analyzing the results for cognition assessment using Clinical Dementia Rating (CDR) and Montreal Cognitive Assessment (MoCA) scales, both were effective in distinguishing between groups (p = 2.2E-16 and p = 8.28E-09, respectively) (Table 1). Based on the CDR evaluations, 49 control subjects had no dementia (98%), while only one control subject suggested very mild dementia (2%). Among the AD participants, 32 subjects had moderate to severe dementia (64%), 14 had scores suggesting very mild dementia (28%), and 4 had a score of no dementia (8%). We noted that the AD subjects with a CDR score of no dementia (n = 4) were on dementia-related medications. Although being on medication could affect their performance and dementia severity, it is important to clarify that these patients were already classified as AD subjects with diagnoses based on clinical criteria22. In fact, only 8% of AD subjects were not on medication. A statistically significant difference was observed between AD patients and the control group across varying levels of CDR (p < 2.2E-16, Table 1), indicating a strong association between the grouping variable and CDR classifications, and suggesting that group distribution is influenced by disease severity.
Additionally, statistical analysis of MoCA scores revealed significant differences across cognitive impairment categories (p = 8.28E-09, Table 1). Among individuals with normal cognition (MoCA score 26–30), 6 participants (6.0% of the total sample) belonged to the control group, while no AD participants were in this category. In the mild cognitive impairment category (MoCA 18–25), 36 individuals (72.0%) were in the control group, while 13 participants (26.0%) were in the AD group, representing 49.0% of the total sample. In the moderate cognitive impairment category (MoCA 10–17), we identified eight control participants (16.0%) and 15 AD participants (30.0%), together comprising 23.0% of the total sample. The severe cognitive impairment category (MoCA ≤ 9) consisted exclusively of AD participants, with 22.0% of the total sample falling into this group. These results highlight a significant distributional difference in cognitive impairment severity between the groups, as 44 out of 50 control subjects (88%) exhibited some level of cognitive impairment based on MoCA performance, which was not reflected in CDR scale performance.
Apolipoprotein E (APOE) genotype-associated disease risks
We performed an APOE genotyping assay by real-time PCR to assess the combinatorial single nucleotide polymorphisms (SNPs) (Details of APOE genotyping can be found in the Methods section). We identified two homozygous states (E3/E3 and E4/E4), and two heterozygous (E2/E3 and E3/E4) genotypes, which we associated with the participant groups and used as variables for the microbiota analyses. The neuron-damaging APOE E4 variant was present in 33% of our participants, 38% of AD patients (19/50), and 28% of controls (14/50). The neuroprotective APOE E2 only had a 6% presence. The E3E3 genotype was the most prevalent in both groups, found in 54.0% of AD patients and 68.0% of controls. The E3E4 genotype, which is associated with an increased risk of AD, was present in 34.0% of AD patients compared to 26.0% of controls. The E2E3 genotype, often considered protective, was observed in 8.0% of AD patients and 4.0% of controls. Lastly, the E4E4 genotype, strongly linked to a higher risk of AD, was rare, appearing in only 4.0% of AD cases and 2.0% of controls. Although not statistically significant (p = 0.51), there was a slightly higher proportion of E3E4 and E4E4 genotypes among AD patients, reflecting known associations between APOE E4 and increased AD risk (Table 1).
Microbial composition analysis reveals subtle shifts in gut microbiota between AD patients and controls
After quality control of the gut microbiota analysis, 358,475 reads remained for 99 participants. One control participant was excluded from analyses due to potential contamination (AD = 50, control = 49). The comparison between AD participants and unimpaired controls revealed no statistically significant differences in the composition of species identified in either group (beta-diversity p > 0.05) (Fig. 1a). Similarly, no significant differences were observed in richness nor diversity between the two groups [alpha diversity (Observed (Obs) and Faith PD) p > 0.05] (Fig. 1b, c). Descriptive analysis of species composition and relative abundance at the phylum and genus levels exhibited slight variations within each group. At the phylum level, controls showed a higher abundance within the Bacteroidota phylum. In contrast, AD patients showed elevated relative abundance levels of Actinobacteriota, Verrucomicrobiota, and Euryarchaeota phyla (Fig. 1d). At the genus level, analysis of the top 10 taxa revealed that controls exhibited an abundance of Blautia, Agathobacter, and Faecalibacterium from the Firmicutes phylum, along with Bacteroides from the Bacteroidota phylum. In contrast, AD participants showed increased levels Clostridium spp. CAG 352, and Streptococcus from the Firmicutes phylum (Fig. 1e). For a more detailed overview of the taxa composition of individual participants at both the phylum and genus level, please refer to Supplementary Fig. S1. Furthermore, we conducted a Microbiome Multivariate Association Analysis with Linear Models 2 (MaAsLin2) controlling for BMI, age, and gender, and identified some bacterial shifts between groups. Although no bacterial biomarkers were suggested (p > 0.05), some of these taxa had significant raw-p-values (prior to correction). Controls had a higher abundance of taxa within Bacteroidota phylum (p = 0.046) (Fig. 2). At the genus level we identified Faecalibacterium (p = 0.015), including Bacteroides (p = 0.025) as well as Dialister (p = 0.0048). Moreover, AD participants had a slight increase in the Eggerthella genus (p = 0.034) and Holdemanella genus (p = 0.043) (Firmicutes phyla) (Fig. 2).
Community analyses comparing the gut microbiota of AD patients and healthy controls. (Panel a) Beta diversity analysis, 2D NMDS (PERMANOVA p = 0.034; ANOSIM p = 0.061). Alpha-diversity among species using Faith PD (Panel b: p = 0.839) and Evenness (Panel c: p = 0.511) indices. Taxonomic bar plots show the relative abundance of bacteria at the phyla (Panel d) and genus levels (top 10) (Panel e).
Reduced MoCA test scores are directly associated with low gut microbiota diversity
Participants exhibited distinct microbiota compositions based on their MoCA scores (beta-diversity, PERMANOVA p = 0.017); PERMANOVA was performed across the six subgroups stratified by diagnosis and MoCA severity (Fig. 3a). Notably, microbiota differences distinguished AD participants with moderate and severe cognitive impairment (ANOSIM p = 0.045) (Supplementary Table S1). Significant differences were also observed between AD participants with moderate cognitive impairment and controls with normal cognition (PERMANOVA p = 0.036; PERMDISP p = 0.013), as well as between AD participants with moderate cognitive impairment and controls with moderate cognitive impairment (PERMANOVA p = 0.040). Additionally, AD participants with severe cognitive impairment exhibited differences when compared to controls with normal cognition (PERMANOVA p = 0.022; PERMDISP p = 0.006), and controls with normal cognition also differed from those with moderate cognitive impairment (PERMANOVA p = 0.035). Significant differences in bacterial richness (Chao1 index) were detected among some groups. AD participants with mild cognitive impairment exhibited lower richness compared to controls with normal cognition (Chao1 p = 0.044), as did those with moderate cognitive impairment (Chao1 p = 0.010) (Fig. 3b, Supplementary Table S1). Further analysis of phylum composition revealed that controls with normal cognition had a higher abundance of Firmicutes than all other groups, while Verrucomicrobiota levels increased as cognitive function declined (Fig. 3c, h).
Microbiota community profiles comparing samples according to MoCA scores. (Panel a) Beta diversity analysis, 2D NMDS (PERMANOVA p = 0.017; ANOSIM p = 0.229; PERMDISP p = 0.136). (Panel b) Alpha-diversity among species using Chao1 (p = 0.263) index. Taxonomic bar plots show the relative abundance of bacteria at the phyla level (Panel c). MaAslin2 heatmap (Panel d), correcting for BMI, age range, and gender (p < 0.05). (Panel e) Beta diversity analysis of control participants, represented in a 2D NMDS (PERMANOVA p = 0.221; ANOSIM p = 0.662; PERMDISP p = 0.120). Alpha-diversity among species using Obs (Panel f: p = 0.935) and Faith PD (Panel g: p = 0.095) indices. (Panel h) Taxa bar plots show the relative abundance of bacteria at the phyla level.
Moreover, MaAsLin2 analysis, controlling for BMI, age, and gender, identified significant microbial differences among MoCA score groups (p < 0.05) (Fig. 3d). AD participants with moderate cognitive impairment exhibited an increased abundance of the Shuttleworthia genus, while this bacterium was reduced in the rest of the groups. Additionally, controls with moderate cognitive impairment had higher levels of the Clostridium innocuum species compared to controls with normal cognition as per MoCA scores.
The clinically defined control subjects (n = 49) were analyzed based on their MoCA results, as some exhibited cognitive impairment despite having intact CDR scores. To assess potential changes in the gut microbiota, we subdivided these participants into three groups: controls with normal cognition (n = 6), controls with mild cognitive impairment (n = 31), and controls with moderate cognitive impairment (n = 12). Beta diversity analysis revealed no significant differences in overall microbiota composition (ANOSIM p > 0.05) (Fig. 3e, Supplementary Table S2). However, a notable decrease in microbiota diversity, both in observed species and overall diversity (Faith PD index), was associated with cognitive decline (Fig. 3f–g). Pairwise comparisons showed significant differences between controls with normal cognition and those with moderate cognitive impairment (PERMANOVA p = 0.037; Obs p = 0.010; Faith PD p = 0.030), as well as between controls with normal cognition and those with mild cognitive impairment (Obs p = 0.031) (Supplementary Table S2).
Cognitive decline was further associated with an increase in Proteobacteria and a concomitant decrease in Firmicutes as we compare normal cognition with mild cognitive impairment within the control group (Fig. 3h). Besides, participants with moderate cognitive impairment exhibited increased abundances of the following genera: Clostridium innocuum, Ruminococcus gnavus, Eggerthella, Mauribaculaceae, and Mogibacterium, compared to those with normal cognition (Supplementary Fig. S2). For a comprehensive overview of gut microbiome characteristics in AD group according to their MoCA scores, refer to Supplementary Fig. S3.
Dementia severity according to CDR correlates with shifts in gut microbiota
To associate gut microbiota diversity with CDR, we analyzed samples from 94 participants (46 AD, 48 controls), excluding outliers with low sample numbers. This included one control participant with a CDR score of very mild dementia and four AD patients with a CDR score within the normal range. For analytical purposes, participants were grouped into CDR 0 (no dementia), CDR 0.5 (mild), and CDR 1–3 (moderate to severe dementia). Beta diversity analysis comparing controls with no dementia and AD with mild and moderate to severe dementia revealed significant differences in species composition (PERMANOVA p = 0.003). Specifically, there was a significant distinction between AD participants with mild dementia and those with moderate to severe dementia (PERMANOVA p = 0.016) (Fig. 4a, Supplementary Table S3). Further statistical differences were observed between AD participants with mild dementia and controls without dementia (PERMANOVA p = 0.020), as well as between AD participants with moderate to severe dementia and controls without dementia (PERMANOVA p = 0.011) (Supplementary Table S3).
Microbial analyses according to CDR scale. (Panel a) Beta diversity analysis, represented in a 2D NMDS (PERMANOVA p = 0.003; ANOSIM p = 0.084; PERMDISP p = 0.424). Alpha-diversity among species using Obs (Panel b: p = 0.333), Chao1 (Panel c: p = 0.391), and Faith PD (Panel d: p = 0.257) indices. (Panel e) Taxonomic bar plots show the relative abundance of bacteria at the phyla level (Panel e). (Panel f) MaAslin2 Heatmap, correcting for BMI, age range, and gender (p > 0.05).
Conversely, alpha diversity analyses revealed no significant differences (Obs p = 0.333; Chao1 p = 0.391; Faith PD p = 0.257) (Fig. 4b–d, Supplementary Table S3). Taxonomic bar plots illustrated the distribution of relative bacterial abundance at the phylum level comparing controls with AD patients with mild and moderate dementia (Fig. 4e). According to CDR scores, all participants had similar abundances of Firmicutes, with a slight decrease observed in AD participants with moderate to severe dementia (Fig. 4e). Additionally, controls showed a slightly higher abundance of Bacteroidota and Proteobacteria phyla. In contrast, moderate to severe AD participants exhibited higher levels of Verrucomicrobiota and Euryarchaeota phyla as compared with controls and mild AD. Those with mild AD showed an abundance of the Actinobacteriota phylum.
MaAslin2 analysis, adjusting for BMI, age, and gender, identified Eubacterium siraeum and UCG 005 genera (both are part of the Firmicutes phylum) as enriched in AD patients with mild dementia compared to controls (p < 0.05). Similarly, genera such as CAG 56, Eggerthella, Eubacterium brachy group, Fusobacterium, and Phascolarctobacterium were enriched in AD patients with moderate to severe dementia (p < 0.05) (Fig. 4f). Notably, AD patients with mild and moderate to severe dementia showed significant depletion of Bacteroides, Butyricicoccus, and Faecalibacterium, genera that represent SCFA-producing bacteria. When comparing AD patients across different CDR scores, significant differences in both alpha and beta diversity were observed (see Supplementary Fig. S4).
APOE genotypes influence gut microbiome composition
Overall, we found no significant differences in microbiota structure across APOE genotypes, (p > 0.05) (Fig. 5a–c). However, relative pairwise p values revealed a statistically significant difference between E2E3 and E3E3 genotypes (PERMANOVA p = 0.043) (Supplementary Table S4). Descriptive taxonomic bar plots at the phylum level showed a decreased abundance of Firmicutes phylum and an increased abundance of Verrucomicrobiota, Euryarchaeota, and Proteobacteria phyla when comparing participants with the E2 genotype, known for its protective effect, to those with E4 alleles, known for their increased risk for AD (Fig. 5d).
Microbial diversity analyses comparing the participants’ gut microbiota based on their APOE alleles. (Panel a) Beta diversity analysis, 2D NMDS (PERMANOVA p = 0.284; ANOSIM p = 0.051; PERMDISP p = 0.987). Alpha-diversity among species using Obs (Panel b: p = 0.658) and Faith PD (Panel c: p = 0.561) index. Taxonomic bar plots show the relative abundance of bacteria at the phyla (Panel d) and genus (Panel e) level. (Panel f) Variability in the Firmicute/Bacteroidota ratio.
At the genus level, descriptive taxonomic bar plots revealed that participants with the E2E3 genotype exhibited a higher abundance of Blautia and Streptococcus compared to those with the E4E4 genotype (Fig. 5e). When comparing the Firmicutes-to-Bacteroidota (F/B) ratio, a measure of the balance in gut bacteria, we found no significant differences. However, participants with the E4E4 genotype had a lower F/B ratio, indicating a lower abundance of Firmicutes (Fig. 5f). Pairwise Wilcoxon rank-sum tests revealed no significant differences between APOE genotypes (Supplementary Table S4).
Further analysis of microbiota composition at the genus level by APOE genotype revealed differences in specific taxa. Random Forest analysis showed, with high accuracy, that Clostridia UCG 014 was abundant in participants carrying the E3E3 and E3E4 alleles. Although with less accuracy, the model also identified a lower abundance of Roseburia and Dorea in subjects with the E4E4 genotype. Additionally, subjects with E2 alleles showed a decrease in Marvinbryantia and an increase in Solobacterium compared to those with E4 alleles (Fig. 6a).
Differential microbiome composition by APOE genotype. (Panel a) Random Forest analysis identified key bacterial genera with altered abundance across APOE genotypes. (Panel b) LEfSe analysis detected taxa enriched in specific genotypes. (Panels c–f) LEfSe box plots showing increased relative abundance of Corynebacterium, Gastranaerophilales, Lachnospiraceae NK4B4 group and Victivallis in participants with the E4E4 genotypes (p < 0.01). This was confirmed with a MaAsLin2 analysis after adjusting for group, gender, age, education, BMI, CDR, and MoCA (p < 0.001).
Linear discriminant analysis effect size (LEfSe) identified a predominance of the following genera: Lachnoclostridium and Tyzzerella in E4E4 carriers, Erysipelatoclostridium in E2E3, and Clostridia UCG 014 in E3E4 carriers (Fig. 6b). Furthermore, LEfSe box plots at the genus level revealed increased abundance of Corynebacterium (p = 0.01), Gastranaerophilales (p = 0.002), Lachnospiraceae NK4B4 group (p = 0.001), and Victivallis (p = 0.004) in participants with the E4E4 genotype (Fig. 6c–f). These findings were confirmed by MaAsLin2 analysis, which was adjusted for group, gender, age, education, BMI, CDR, and MoCA, showing a decreased log2 fold change values in E4E4 participants (log2FC: − 2.46, − 0.742, − 0.37, and − 0.643, respectively), with corresponding p < 0.001 (See table in Fig. 6).
Discussion
As alterations in the gut microbiome have been linked to changes in behavior and neurological diseases24, we investigated a Puerto Rican cohort with AD compared to cognitively unimpaired controls through detailed clinical and psychological evaluations, including cognitive impairment questionnaires (MoCA and CDR). While most studies report lower gut diversity in AD25, we found no statistically significant differences in microbial diversity between AD patients and controls. However, we identified slight compositional changes, notably higher levels of Proteobacteria phylum in controls with MoCA scores suggesting mild cognitive impairment and severe AD participants, consistent with previous reports26. This observation aligns with a study that found an enrichment of Proteobacteria and a reduction in beneficial taxa such as Lactobacillus in cognitively frail older adults, further implicating microbial shifts in neurodegeneration20. Controls with normal cognition exhibited particularly undetectable levels of Proteobacteria.
In contrast, controls exhibited a higher abundance of the Bacteroidota phylum, a major group, integral to maintaining various aspects of human health. Taxa within this phylum are known for their anti-inflammatory properties and roles in nutrient provision within the gut ecosystem. These taxa produce enzymes like glycosidases, hydrolases, and proteases that break down complex dietary molecules, promoting nutrition and gut health27.
Conversely, AD patients exhibited a higher relative abundance of taxa within Actinobacteria, Verrucomicrobiota, and Euryarchaeota phyla. While Actinobacteria can benefit immune regulation in certain contexts, they may act as opportunistic pathogens in immunocompromised individuals28. Verrucomicrobiota, a mucin-degrading bacterial group involved in glucose homeostasis, has been associated with promoting gut health and reducing obesity risk29,30. Euryarchaeota taxa, known as methanogens, are often related to constipation and irritable bowel syndrome (IBS)31 and play a critical role in fermentation processes and intestinal transit, highlighting their relevance to microbiome diversity32. The increased abundance of Euryarchaeota in this population could reflect genetic, environmental, or lifestyle factors unique to Puerto Ricans, emphasizing the need for population-specific studies to understand gut microbiota’s role in AD.
Although we did not detect clear bacterial biomarkers distinguishing AD from controls, some taxa exhibited fluctuations. Controls displayed higher levels of beneficial genera such as Blautia, Bacteroides, Agathobacter, and Faecalibacterium, which produce antibacterial compounds and metabolites like butyrate33,34. Butyrate plays a vital role in gut physiology due to its anti-inflammatory properties, intestinal barrier function enhancement, and mucosal immunity promotion35. Loss of Blautia and Faecalibacterium is often associated with compromised gut barrier integrity, increased systemic inflammation, and potentially accelerated AD progression36. In contrast, higher levels of Eggerthella, previously associated with severe gastrointestinal pathology, were detected in AD participants37.
Our study demonstrates that both the Clinical Dementia Rating (CDR) and the Montreal Cognitive Assessment (MoCA) were effective in distinguishing between Alzheimer’s disease (AD) patients and controls. However, a notable finding was that 44 control participants—classified as cognitively intact based on CDR (global score = 0)—scored in the mild or moderate impairment range on the MoCA. This discrepancy highlights important considerations in the interpretation of cognitive screening tools within diverse populations.
The CDR scale evaluates functional impairment, and is widely used in clinical diagnosis according to the 2011 NIA-AA criteria for dementia due to Alzheimer’s disease (McKhann et al., 2011). Our classification of AD cases and controls was based on this syndromic, functionally anchored framework, in which impairment in daily activities is required for a diagnosis of dementia. Control participants, by definition, showed no functional decline. In our study, MoCA served as a quantitative cognitive measure but not a standalone diagnostic tool. Importantly, MoCA scores are sensitive to sociodemographic variables, particularly education, cultural background, and language. In our Puerto Rican cohort, 25% of participants had 12 years of education or less, with Spanish as their primary language. Previous studies have reported reduced specificity of MoCA in similar populations, often identifying cognitive impairment where there is none [41]. This may explain the unexpectedly low MoCA scores observed in our control group and supports calls for culturally and linguistically adapted cognitive tools in underrepresented populations.
Interestingly, this MoCA variability had implications for our microbiome findings. The relationship between gut microbiota composition and cognition was evaluated using both MoCA and CDR scores. Our results align with previous research showing correlations between gut dysbiosis and MoCA-based cognitive decline [38,39], including studies of post-stroke cognitive impairment [40]. Notably, gut microbial profiles differed in control participants with lower MoCA scores, despite preserved functional status per CDR. These findings suggest that gut microbiota changes may be associated with early or subclinical cognitive decline and highlight the potential for microbiome profiling as a sensitive adjunct measure. Moreover, our data is consistent with reports of microbial dysbiosis in AD, including reduced microbial richness and altered abundances of Firmicutes, Proteobacteria, and Bacteroidetes [18], further supporting a biological link between cognition and the gut-brain axis.
A reduction in Firmicutes was associated with lower MoCA scores, suggesting a link between cognitive decline and microbiome composition. Low abundance of Firmicutes has been linked to impaired microbial reductase activity, and heightened DNA alterations and immune dysfunction42. The Bacteroidetes phylum, known for producing SCFAs, decreased with cognitive decline, while the Proteobacteria phylum increased, potentially contributing to AD pathogenesis through microglial phagocytic dysfunction and amyloid plaque accumulation26,43. An increase in Proteobacteria may indicate a potential association between oxidative stress and gut microbiota alterations, acting as early biomarkers for cognitive decline36. Cognitive decline in AD patients also includes a significant reduction in the Faecalibacterium genus with moderate cognitive impairment. Faecalibacterium, a butyrate-producer genus44, was significantly higher in controls with normal cognition.
Among controls, Clostridium innocuum genus (from the Firmicutes phylun) was notably abundant in those with moderate cognitive impairment. Clostridium innocuum is an opportunistic pathogen associated with various gastrointestinal conditions, including inflammatory bowel disease (IBD) and extraintestinal manifestations such as the “creeping fat” phenomenon observed in Crohn’s disease45. Population-specific factors (e.g., diet, lifestyle, environment) unique to Puerto Ricans may contribute to the observed microbiota differences in AD patients, as reported in other populations46. Further studies are needed to elucidate the underlying mechanisms responsible for these differences and to assess their potential impact on the progression and severity of AD in the Puerto Rican population.
Using the CDR scale, we found distinct microbial profiles associated with varying dementia severities, aligning with the former MoCA results and with other population findings47. Bacteroides were elevated in controls with no dementia in comparison to AD patients with mild and moderate to severe dementia. The Bacteroides genus, is known to reduce inflammation through propionate production and correlated with lower cognitive decline48. In contrast, the following genera: Eubacterium siraeum group and UCG 005 (from the Firmicutes phylum), were enriched in AD patients with mild dementia, whereas CAG 56 (Firmicutes), Eggerthella, and Fusobacterium genera were increased in those with moderate to severe dementia. These taxa have been commonly found in different gut pathologies, such as Fusobacterium nucleatum, which is clearly related to colorectal cancer, with direct identification in rectal biopsies49.
Our analyses indicated shifts in the microbiota associated with APOE genotypes, with significant changes detected among the E2E3 and E3E3 groups. These findings align with previous research demonstrating that the APOE genotype is associated with specific gut microbiome profiles50, although some distinctions were noted. While we found no significant differences in overall microbiota diversity among aggregated APOE genotypes, specific variations in the relative abundance of certain bacterial taxa were observed between different APOE genotypes. Notably, participants carrying the E4 allele exhibited a reduction in the Firmicutes phylum and an increased abundance of Verrucomicrobiota, Euryarchaeota, and Proteobacteria phyla compared to those with at least one neuroprotective E2 allele. These alterations are consistent with previous studies suggesting that gut microbiota dysbiosis contributes to AD pathology and APOE-related variations18,50. Furthermore, E4E4 participants exhibited a lower F/B ratio compared to other genotypes, primarily due to a decreased abundance of Firmicutes, indicating potential gut dysbiosis.
Notably, our study observed a reduction in butyrate-producing genera, such as Roseburia, Dorea, and Marvinbryantia, in participants with the E4E4 phenotype, which supports prior findings linking butyrate-producing bacteria to APOE genotype variations and is consistent with reports of loss of SCFA-producing bacteria in E4 carriers. These findings propose a potential link between APOE-driven gut dysbiosis and neuroinflammation, metabolic dysfunction, and AD risk. The Blautia genus, highly enriched in association with the E2E3 genotype, has been previously correlated with anti-inflammatory properties and improved metabolic health33. Additionally, SCFA-producing Clostridium cluster IV (Clostridia UCG-014), which we found highly abundant in E2E3 and E3E4, decreased significantly in carriers of E3E3 and E4E4, which is often associated with gut dysbiosis51.
Lachnoclostridium and Tyzzerella genera were enriched in E4E4 individuals. Although we cannot predict direct functions, other studies have revealed these genera as predictors of amyloid and phosphorylated tau accumulation, suggesting a possible link to AD pathology52. Tyzzerella (from the Firmicutes phylum) has been linked to cardiovascular disease and gut dysbiosis, underscoring the systemic impact of microbial changes in APOE E4 carriers53. Corynebacterium genus, typically associated with skin and mucosal surfaces, has been detected in the gut microbiota and linked to immune modulation and potential pro-inflammatory effects54 and we found it mostly in association with E4E4. Gastranaerophilales and Lachnospiraceae NK4B4 group genera, both Firmicutes phylum members, have been implicated in metabolic regulation and SCFA production, although their precise roles in neurodegenerative conditions remain unclear. Victivallis, a genus within the Lentisphaerae phylum, has been associated with fiber metabolism and gut homeostasis, though its increase in E4E4 individuals may indicate a compensatory shift in microbial composition55. Overall, we find mostly a depletion of SCFA-producing bacteria in E4E4 individuals which aligns with previous reports of reduced butyrate synthesis in AD microbiomes, potentially exacerbating neuroinflammation. Meanwhile, the enrichment of pro-inflammatory and opportunistic taxa underscores the potential role of microbial dysbiosis in AD pathology. These findings highlight the complex relationship between APOE genotype variation, gut microbiome composition, and AD risk, supporting the growing body of evidence linking gut dysbiosis to neurodegenerative disorders.
This study has several limitations that should be acknowledged. Although dietary habits, environmental exposures, and the use of medications or supplements are recognized modulators of gut microbiota composition7, these variables were not incorporated into the present analyses. While information on these factors was collected via questionnaire and is intended for future investigation, the current models were adjusted only for age, sex, BMI, genotype, and cognitive performance (CDR and MoCA), which were the primary variables of interest. In addition, the potential effects of dementia-related medications on cognitive performance may have influenced clinical assessments, particularly among subjects with AD. These effects were not explicitly modeled, which may have attenuated differences in cognitive severity between groups. Some controls presented with lower-than-expected MoCA scores despite a CDR global score of 0, indicating intact daily functioning. This discrepancy can likely reflect the influence of demographic and psychosocial factors, including education, socioeconomic status, mood, and test-taking conditions—all of which may affect performance on cognitive screening tools and will be taken in care in future analysis. Moreover, the sample size and group distributions, particularly in subgroup analyses based on cognitive strata, may limit the generalizability of our findings. Biomarker confirmation of Alzheimer’s pathology (e.g., amyloid or tau) was not available, which constrains interpretation within a biological framework. Future studies from our team with a larger dataset, will benefit from the analysis of diet, medication use, sleep quality, and longitudinal sampling to better elucidate causal relationships between gut microbiota, cognition, and disease progression.
In conclusion, this study is the first to investigate the gut microbiome and AD association in a Puerto Rican cohort in relation to APOE status. While no significant differences in overall microbial diversity were observed, subtle compositional changes were detected. These findings emphasize the importance of population-specific research to understand the role of gut microbiota in AD and suggest that microbiota-targeted therapies could help mitigate neuroinflammation and slow disease progression in at-risk populations.
Materials and methods
Recruitment of participants and cognitive assessment
A total of 100 Puerto Rican participants were recruited from the Internal Medicine and Geriatrics outpatient clinics at the School of Medicine, Medical Sciences Campus, University of Puerto Rico. The sample included 50 participants with a clinical diagnosis of Alzheimer’s dementia and 50 cognitively unimpaired controls.
Participants with Alzheimer’s disease (AD) had been diagnosed by their treating physicians based on the 2011 National Institute on Aging and Alzheimer’s Association (NIA-AA) criteria for the clinical diagnosis of dementia due to Alzheimer’s disease (McKhann et al., 2011). No biomarker confirmation (e.g., amyloid PET, CSF or plasma tau/Aβ) was used; the diagnosis was based on clinical history, cognitive symptoms, and functional assessment in line with the NIA-AA syndromic framework.
Control participants were individuals without a clinical diagnosis of dementia or MCI and with no evidence of functional impairment, as indicated by a Clinical Dementia Rating (CDR) global score of 0. Although some controls had MoCA scores suggesting mild cognitive changes, they were included based on preserved daily functioning and the absence of clinical concern for cognitive decline. We acknowledge that variability in MoCA scores may be influenced by education, socioeconomic status, and mood, and this is addressed in the Discussion. Our study reinforces the importance of using multiple complementary assessments within an interdisciplinary team. While the CDR provides a robust clinical staging tool, MoCA contributes granularity to cognitive performance, though its interpretation must be nuanced within the context of cultural and educational variability.
Eligibility criteria
Inclusion criteria were Puerto Rican ethnicity, Spanish-speaking, age ≥ 55, ability to provide consent (or via Legally Authorized Representative), controlled hypertension and depression (if diagnosed), standard laboratory values, and availability of blood and stool samples. Exclusion criteria included: non-Puerto Rican ethnicity, bedridden status, uncontrolled stage 2 hypertension (≥ 140/90 mmHg), active psychotic depression, delirium, schizophrenia, neurologic disorders, history of gastrointestinal surgery, recent use of antibiotics or probiotics, and pregnancy. Informed consent and HIPAA authorization were obtained from all participants in accordance with the Declaration of Helsinki. The study protocol was approved by the Institutional Review Board of the Medical Sciences Campus (IRB #2,290,033,626).
Cognitive and functional assessment
All participants underwent a physical and neurological examination and were assigned unique study codes. Cognitive assessment was conducted using two validated tools. The Montreal Cognitive Assessment (MoCA) is a 30-point screening tool assessing multiple cognitive domains, including attention, memory, executive function, visuospatial skills, language, and orientation. A score < 26 indicated cognitive impairment, with interpretation adjusted for age and education where relevant. A score of 18–25 means mild cognitive impairment, 10–17 means moderate cognitive impairment and a scoreless or equal to 9 correlated with severe cognitive impairment58. The Clinical Dementia Rating (CDR) scale evaluates six cognitive and functional performance domains60. Subjects were categorized as follows: CDR 0: no cognitive impairment (controls), CDR 0.5: very mild dementia, and CDR 1–3: dementia with increasing severity. Both instruments are widely validated, used in clinical research on dementia, and administered by trained neuropsychologists who discuss doubtful cases with the geriatrics staff.
During the first of two visits, the participant receives a stool specimen collection kit for self-collection. Adherence strategies include contacting patients over the phone by the study coordinator to ensure the biological specimen collection procedures are done correctly and confirm their appointment for the second visit. During the second visit, the patient arrived fasting to collect APOE E4 and standard blood tests, including fasting insulin and blood sugar levels. After a reasonable period for a snack (approximately 15 min), the participant underwent the memory scale evaluation by the neuropsychologist.
Genomic DNA extraction and 16S rRNA sequencing
Genomic DNA (gDNA) was extracted from fecal samples using the DNeasy PowerSoil Pro Kit (QIAGEN, Germantown, MD, USA) with specific manufacturer protocol modifications. (1) 200 mg of fecal matter was added to the PowerBead tubes. (2) After 10 min in the Vortex, the PowerBead Pro tubes were processed in a TissueLyser II at a frequency of 30.0 (1/s) for 3 min. (3) Lysate was placed in a spin filter membrane using a QIAvac Vacuum System (QIAGEN). And (4) elution was performed with 75 μL of warmed (55 °C) C6 solution. The 16S gene was amplified with universal 16S V4 primers 515F: (5’-GTGCCAGCMGCCGCGGTAA-3’) and 805R: (5’-GGACTACHVGGGTWTCTAAT-3’) (https://earthmicrobiome.org/protocols-and-standards/) and sequenced on an Illumina MiSeq platform. Demultiplexed 16S rRNA amplicons were deposited and quality-controlled in QIITA60 project ID PRJEB86129, followed by analysis with QIIME2-2023.7 and RStudio (https://www.r-project.org/). The resulting raw sequences were made available at the European Nucleotide Archive Project (ENA) under the access number ERP169503.
Quality control of sequencing reads and analyses of the microbial communities
Sequencing reads were demultiplexed and trimmed to 250 bp, followed by a deblur workflow (Deblur 2021.09)60. Silva taxonomy61,62 was used as a reference database with a minimum similarity threshold of 97%. The amplicon sequence variant (ASV) feature table was retrieved from QIITA; singletons, mitochondria, and chloroplasts were removed using QIIME2-2023.763. The rarefaction level for the analyses was set at 3700 reads per sample.
Samples were then analyzed for diversity and taxonomic composition using QIIME2-2023.7. Statistical analyses for beta diversity included PERMANOVA, ANOSIM, and PERMDISP tests within the QIIME2 pipeline, with pairwise community dissimilarities calculated using Bray–Curtis distances and visualized through non-metric multidimensional scaling (NMDS) using phyloseq64 and ggplot2 packages in RStudio (https://www.r-project.org/). Alpha diversity was evaluated using Obs, Chao1, Shannon, Evenness, and Faith PD (phylogenetic diversity) within the QIIME2 pipeline. Figures were generated using the phyloseq64, vegan65, and ggplot2 packages in RStudio (https://www.r-project.org/). Taxonomic bar plots at the phylum and genus levels and Pearson Correlation Coefficient were computed using Microbiome Analyst (http://www.microbiomeanalyst.ca/).
Statistical tests were performed using RStudio software packages, where high-performance computing servers are available to perform all bioinformatics analyses. Statistical p-value corrections for MaAsLin2 and LEfSe analyses were done using Benjamini–Hochberg correction to control for a false discovery rate.
To detect bacterial biomarkers, LEfSe algorithm was performed within Microbiome Analyst (http://www.microbiomeanalyst.ca/) with a p value cutoff of 0.0566. Linear discriminant analysis (LDA) identifies taxa characterizing their differences with metadata variables. This method emphasizes statistical significance, biological consistency, and effect relevance, allowing researchers to identify differentially abundant features that are also consistent with biologically meaningful categories (metadata), using non-parametric factorial Kruskal–Wallis (KW) sum-rank test, Wilcoxon rank-sum test, and LDA. Additionally, MaAsLin2 was used to identify enrichment in microbial taxa using a linear modeling approach in R67. MaAsLin2 was used to explore the relationships between microbial taxa and metadata variables including study group, MoCA, CDR, APOE genotypes while accounting for covariates such as age, gender and BMI. This helps ensure that observed associations are robust and not confounded by other cohort variables.
Random Forest classification analysis was performed using Microbiome Analyst (http://www.microbiomeanalyst.ca/). Taxonomic features were used as predictors to distinguish between APOE genotypes. The model used 500 trees and default parameters, and importance scores to identify top predictors. It is increasingly accepted that the F/B ratio could be used as a biomarker in microbiota analyses68,69,70. Boxplots depicting the F/B ratio were built using the vegan65 package in R Statistical significance was addressed with WRST.
APOE genotyping
Genomic DNA was first isolated from peripheral blood, specifically 200 μl EDTA anticoagulated venous blood collected from all study participants using a standardized extraction protocol with the QIAamp DNA Blood Mini Kit (Qiagen). The integrity and concentration of the DNA were assessed via spectrophotometry and agarose gel electrophoresis to ensure that the samples were of high quality and suitable for downstream applications.
The extracted DNA was then subjected to polymerase chain reaction (PCR) to amplify the regions of interest. Genotyping of SNPs was done using a real-time PCR (Applied Biosystems StepOne Real-Time PCR System, Thermofisher) using specific TaqMan genotyping assays, targeting the rs42958 and rs7412 polymorphisms, which define the APOE E2, E3, and E4 alleles71. Real-time PCR was made using 12.5 μL of 2 × TaqMan Genotyping Master Mix and 1.25 μL 2 × TaqMan genotyping assay mix (Thermofisher) using 25 μL reaction (48-well standard plate) and according to the manufacturer’s protocol. Amplified products subsequently confirmed via gel electrophoresis.
We targeted two key single nucleotide polymorphisms (SNPs) within the APOE gene, rs429358 and rs7412, which define the three major APOE alleles: E2 (rs429358, T; rs7412, T), E3 (rs429358, T; rs7412, C) and E4 (rs429358, C; rs7412, C)71. The real-time PCR process involved specific primer and probe pairs designed to distinguish between these SNPs based on fluorescence detection, allowing for automated genotype calling via endpoint genotyping software.
The APOE genotypes identified in the study included both homozygous and heterozygous combinations: E2/E2 (T/T, T/T), E2/E3 (T/T, T/C), E2/E4 (T/T, C/C), E3/E3 (T/C, T/C), E3/E4 (T/C, C/C), and E4/E4 (C/C, C/C). Among these, the E4 allele (rs429358 C and rs7412 C) was significantly associated with an increased risk of late-onset AD. The study found a distinct distribution of genotypes between cases and controls, with E3/E3 being the most common in controls, while E3/E4 and E4/E4 were more frequent in AD patients. This genotyping approach demonstrated high sensitivity and reliability, making it a suitable method for large-scale screening of APOE polymorphisms in AD research. Protocol followed was previously explained71.
General statistical analysis
Descriptive analysis for continuous variables (age, labs, memory results, microbial alpha diversity) includes means, standard deviations, medians, and range (min–max). Frequencies and percentages summarized categorical variables (gender, education, APOE E4). Chi-square and Fisher’s exact test compare categorical variables of demographics, lifestyle behaviors, and clinical characteristics between AD patients (overall and divided as mild, moderate and severe) and healthy controls. Categorical variables were compared with Fisher’s exact test and continuous variables with the Kruskal–Wallis test using library “table” in R. Also, as appropriate, the Student t-test and Mann–Whitney U test determined an association between continuous and count variables and overall AD (AD case vs. control). In addition, we explore the association between stages of AD and continuous variables such as alpha diversity using One-way ANOVA and the Kruskal–Wallis’s test, as appropriate. The normality of distributions and homoscedasticity of variables is evaluated through the Shapiro–Wilk and Bartlett’s tests, respectively. Since alpha diversity is a count variable, Poisson or negative binomial regression models (selected based on the outcome distribution and model fit) were used to assess the magnitude of association (β and 95% confidence intervals) between alpha diversity and disease status (i.e., healthy controls, mild AD, and moderate/severe AD) as the predictor. For the beta diversity (continuous variable) outcome, generalized linear regression models explored the associations with disease status; β and 95% confidence intervals were estimated. All regression models are adjusted for variables showing associations with the diversity and composition of the fecal microbiota. P-values less than 0.05 were considered statistically significant. Statistical tests were done using STATA v.16 (College Station, Texas 77,845 USA).
Data availability
Sequence data that support the findings of this study have been deposited in the European Nucleotide Archive, Project: PRJEB86129ENA (https://www.ebi.ac.uk/ena/browser/view/PRJEB86129). Metadata can also be found in the QIITA study (Study ID 14,626) (https://qiita.ucsd.edu/study/description/14,626).
References
2023 Alzheimer’s disease facts and figures. Alzheimer’s and Dementia 19, (2023).
Rajan, K. B. et al. Population estimate of people with clinical Alzheimer’s disease and mild cognitive impairment in the United States (2020–2060). Alzheimer’s Dementia 17, 1966–1975 (2021).
Friedman, D. B. et al. Increasing community awareness about Alzheimer’s disease in Puerto Rico through coffee shop education and social media. J. Commun. Health 41, 1006–1012 (2016).
Figueroa, R., Steenland, K., MacNeil, J. R., Levey, A. I. & Vega, I. E. Geographical differences in the occurrence of Alzheimer’s disease mortality: United States versus Puerto Rico. Am. J. Alzheimers Dis. Other Demen. 23, 462–469 (2008).
Querfurth, H. W. & LaFerla, F. M. Alzheimer’s disease. N. Engl. J. Med. 362, 329–344 (2010).
Condello, C., Yuan, P. & Grutzendler, J. Microglia-mediated neuroprotection, TREM2, and Alzheimer’s disease: Evidence from optical imaging. Biol. Psychiatry https://doi.org/10.1016/j.biopsych.2017.10.007 (2018).
Yamazaki, Y., Zhao, N., Caulfield, T. R., Liu, C. C. & Bu, G. Apolipoprotein E and Alzheimer disease: Pathobiology and targeting strategies. Nat. Rev. Neurol. https://doi.org/10.1038/s41582-019-0228-7 (2019).
Raulin, A. C. et al. ApoE in Alzheimer’s disease: Pathophysiology and therapeutic strategies. Mol. Neurodegen. https://doi.org/10.1186/s13024-022-00574-4 (2022).
Granot-Hershkovitz, E. et al. APOE alleles’ association with cognitive function differs across Hispanic/Latino groups and genetic ancestry in the study of Latinos-investigation of neurocognitive aging (HCHS/SOL). Alzheimer’s Dementia 17, 466–474 (2021).
Chen, Y., Xu, J. & Chen, Y. Regulation of neurotransmitters by the gut microbiota and effects on cognition in neurological disorders. Nutrients https://doi.org/10.3390/nu13062099 (2021).
Nicholson, J. K. et al. Host-gut microbiota metabolic interactions. Science https://doi.org/10.1126/science.1223813 (2012).
Shin, J. H. et al. Consumption of 85% cocoa dark chocolate improves mood in association with gut microbial changes in healthy adults: A randomized controlled trial. J. Nutr. Biochem. 99, 108854 (2022).
Luca, M., Mauro, M. D., Mauro, M. D. & Luca, A. Gut microbiota in Alzheimer’s disease, depression, and type 2 diabetes mellitus: The role of oxidative stress. Oxid. Med. Cell. Longev. https://doi.org/10.1155/2019/4730539 (2019).
Zhao, Y. & Lukiw, W. J. Microbiome-generated amyloid and potential impact on amyloidogenesis in Alzheimer’s disease (AD). J Nat Sci 1, (2015).
Cattaneo, A. et al. Association of brain amyloidosis with pro-inflammatory gut bacterial taxa and peripheral inflammation markers in cognitively impaired elderly. Neurobiol. Aging 49, 60–68 (2017).
Seo, D. O. et al. ApoE isoform- and microbiota-dependent progression of neurodegeneration in a mouse model of tauopathy. Science (1979) 379, eadd1236 (2023).
Acevedo-Román, A., Pagán-Zayas, N., Velázquez-Rivera, L. I., Torres-Ventura, A. C. & Godoy-Vitorino, F. Insights into gut dysbiosis: Inflammatory diseases, obesity, and restoration approaches. Int. J. Mol. Sci. https://doi.org/10.3390/ijms25179715 (2024).
Vogt, N. M. et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 7, 13537 (2017).
Chandra, S., Sisodia, S. S. & Vassar, R. J. The gut microbiome in Alzheimer’s disease: What we know and what remains to be explored. Mol. Neurodegener. https://doi.org/10.1186/s13024-023-00595-7 (2023).
Haran, J. P. & McCormick, B. A. Aging, frailty, and the microbiome—how dysbiosis influences human aging and disease. Gastroenterology 160, 507–523 (2021).
Zhou, X. P. et al. The complex relationship between gut microbiota and Alzheimer’s disease: A systematic review. Ageing Res. Rev. https://doi.org/10.1016/j.arr.2024.102637 (2025).
McKhann, G. M. et al. The diagnosis of dementia due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimer’s Dementia 7, 263–269 (2011).
Dickerson, F. et al. The microbiome and mental health across the lifespan. in Current Topics in Behavioral Neurosciences vol. 61 (2023).
Bairamian, D. et al. Microbiota in neuroinflammation and synaptic dysfunction: a focus on Alzheimer’s disease. Mol. Neurodegener. https://doi.org/10.1186/s13024-022-00522-2 (2022).
Hung, C. C., Chang, C. C., Huang, C. W., Nouchi, R. & Cheng, C. H. Gut microbiota in patients with Alzheimer’s disease spectrum: A systematic review and meta-analysis. Aging 14, 477 (2022).
Seo, D. O. & Holtzman, D. M. Current understanding of the Alzheimer’s disease-associated microbiome and therapeutic strategies. Exp. Mol. Med. https://doi.org/10.1038/s12276-023-01146-2 (2024).
Zafar, H. & Saier, M. H. Gut Bacteroides species in health and disease. Gut Microbes https://doi.org/10.1080/19490976.2020.1848158 (2021).
Pickard, J. M., Zeng, M. Y., Caruso, R. & Núñez, G. Gut microbiota: Role in pathogen colonization, immune responses, and inflammatory disease. Immunol. Rev. https://doi.org/10.1111/imr.12567 (2017).
Gryaznova, M. et al. Dynamics of changes in the gut microbiota of healthy mice fed with lactic acid bacteria and bifidobacteria. Microorganisms 10, 1020 (2022).
Ulker, I. & Yildiran, H. The effects of bariatric surgery on gut microbiota in patients with obesity: A review of the literature. Biosci. Microbiota Food Health https://doi.org/10.12938/bmfh.18-018 (2019).
Kim, G. et al. Methanobrevibacter smithii is the predominant methanogen in patients with constipation-predominant IBS and methane on breath. Dig. Dis. Sci. 57, 3213–3218 (2012).
Sofer, T. et al. A polygenic risk score for Alzheimer’s disease constructed using APOE-region variants has stronger association than APOE alleles with mild cognitive impairment in Hispanic/Latino adults in the U.sS. Alzheimers Res. Ther. 15, 146 (2023).
Liu, X. et al. Blautia—a new functional genus with potential probiotic properties?. Gut Microbes https://doi.org/10.1080/19490976.2021.1875796 (2021).
Hua, X. et al. The gut microbiota and associated metabolites are altered in sleep disorder of children with autism spectrum disorders. Front Psychiatry 11, 855 (2020).
Liu, H. et al. Butyrate: A double-edged sword for health?. Adv. Nutr. https://doi.org/10.1093/advances/nmx009 (2018).
Schattke, K. Conciliating the dimensionality debate surrounding SDT and redefining motivation? A comment on Meyer et al.’s article ‘motivational mindsets versus reasons for action’. Zeitschrift fur Sportpsychologie https://doi.org/10.1026/1612-5010/a000380 (2022).
Gardiner, B. J. et al. Clinical and microbiological characteristics of eggerthella lenta bacteremia. J. Clin. Microbiol. 53, 626–635 (2015).
Meyer, K. et al. Association of the gut microbiota with cognitive function in midlife. JAMA Netw. Open https://doi.org/10.1001/jamanetworkopen.2021.43941 (2022).
Qu, L. et al. Gut microbiome signatures are predictive of cognitive impairment in hypertension patients—a cohort study. Front Microbiol. 13, 841614 (2022).
Huang, Y., Shen, Z. & He, W. Identification of gut microbiome signatures in patients with post-stroke cognitive impairment and affective disorder. Front Aging Neurosci. 13, 706765 (2021).
Zhou, Y. et al. Use of the MoCA in detecting early Alzheimer’s disease in a spanish-speaking population with varied levels of education. Dement Geriatr. Cogn. Dis. Extra 5, 85–95 (2015).
Jandhyala, S. M. et al. Role of the normal gut microbiota. World J. Gastroenterol. 21, 8787 (2015).
Wasén, C. et al. Bacteroidota inhibit microglia clearance of amyloid-beta and promote plaque deposition in Alzheimer’s disease mouse models. Nat. Commun. 15, 3872 (2024).
Rey, F. E. et al. Dissecting the in vivo metabolic potential of two human gut acetogens. J. Biol. Chem. 285, 22082–22090 (2010).
Bhattacharjee, D., Flores, C., Woelfel-Monsivais, C. & Seekatz, A. M. Diversity and prevalence of clostridium innocuum in the human gut microbiota. mSphere 8, e00569-22 (2023).
Jemimah, S., Chabib, C. M. M., Hadjileontiadis, L. & AlShehhi, A. Gut microbiome dysbiosis in Alzheimer’s disease and mild cognitive impairment: A systematic review and meta-analysis. PLoS ONE 18, e0285346 (2023).
Yıldırım, S. et al. Stratification of the gut microbiota composition landscape across the Alzheimer’s disease continuum in a Turkish cohort. Systems 7, e00004-22 (2022).
Haran, J. P. et al. Alzheimer’s disease microbiome is associated with dysregulation of the anti-inflammatory P-glycoprotein pathway. MBio 10, 10–1128 (2019).
Kim, M. et al. Fusobacterium nucleatum in biopsied tissues from colorectal cancer patients and alcohol consumption in Korea. Sci. Rep. 10, 19915 (2020).
Tran, T. T. T. et al. APOE genotype influences the gut microbiome structure and function in humans and mice: Relevance for Alzheimer’s disease pathophysiology. FASEB J. 33, 8221 (2019).
Lopetuso, L. R., Scaldaferri, F., Petito, V. & Gasbarrini, A. Commensal Clostridia: Leading players in the maintenance of gut homeostasis. Gut Pathogens https://doi.org/10.1186/1757-4749-5-23 (2013).
Verhaar, B. J. H. et al. Gut microbiota composition is related to AD pathology. Front Immunol. 12, 794519 (2022).
Kelly, T. N. et al. Gut microbiome associates with lifetime cardiovascular disease risk profile among bogalusa heart study participants. Circ. Res. 119, 956–964 (2016).
Zhu, G., Zhao, J., Zhang, H., Wang, G. & Chen, W. Gut microbiota and its metabolites: Bridge of dietary nutrients and Alzheimer’s disease. Adv. Nutr. https://doi.org/10.1016/j.advnut.2023.04.005 (2023).
Dissanayaka, D. M. S., Jayasena, V., Rainey-Smith, S. R., Martins, R. N. & Fernando, W. M. A. D. B. The role of diet and gut microbiota in Alzheimer’s disease. Nutrients https://doi.org/10.3390/nu16030412 (2024).
Nova, E., Gómez-Martinez, S. & González-Soltero, R. The influence of dietary factors on the gut microbiota. Microorganisms 10, 1368 (2022).
Rodríguez-Santiago, M. A., Wojna, V., Miranda-Valentín, E., Arnold, S. & Sepúlveda-Rivera, V. Diagnosing Alzheimer’s disease: Which dementia screening test to use in elderly Puerto Ricans with mild cognitive impairment and early Alzheimer’s disease?. Alzheimer’s Dementia Diagn. Assess. Dis. Monitor. 16, e12554 (2024).
Nasreddine, Z. S. et al. The Montreal cognitive assessment, MoCA: A brief screening tool for mild cognitive impairment. J. Am. Geriatr. Soc. 53, 695–699 (2005).
Morris, J. C. The clinical dementia rating (cdr): Current version and scoring rules. Neurology 43, 2412–2412 (1993).
Gonzalez, A. et al. Qiita: Rapid, web-enabled microbiome meta-analysis. Nat. Methods 15, 796–798 (2018).
Quast, C. et al. The SILVA ribosomal RNA gene database project: Improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596 (2013).
Pruesse, E. et al. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196 (2007).
Bolyen, E. et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. https://doi.org/10.1038/s41587-019-0209-9 (2019).
McMurdie, P. J. & Holmes, S. Phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217 (2013).
Oksanen, J. et al. Vegan: Community ecology package. R package version 2.5–7. Commun. Ecol. Package 10, (2020).
Segata, N. et al. Metagenomic biomarker discovery and explanation. Genome Biol. 12, R60 (2011).
Mallick, H. et al. Multivariable association discovery in population-scale meta-omics studies. PLoS Comput. Biol. 17, e1009442 (2021).
Bahar-Tokman, H. et al. Firmicutes/bacteroidetes ratio in the gut microbiota and IL-1β, IL-6, IL-8, TLR2, TLR4, TLR5 gene expressions in type 2 diabetes. Clin. Lab. 68 (2022).
Petakh, P., Oksenych, V. & Kamyshnyi, A. The F/B ratio as a biomarker for inflammation in COVID-19 and T2D: Impact of metformin. Biomed. Pharmacother. 163, 114892 (2023).
An, J., Kwon, H. & Kim, Y. J. The firmicutes/bacteroidetes ratio as a risk factor of breast cancer. J. Clin. Med. 12, 2216 (2023).
Yi, L., Wu, T., Luo, W., Zhou, W. & Wu, J. A non-invasive, rapid method to genotype late-onset Alzheimer’s disease-related apolipoprotein E gene polymorphisms. Neural Regen. Res. 9, 69–75 (2014).
Acknowledgements
We would like to acknowledge the support of Dr. Michel Santiago-Berrios, Mr. Fabián J. Pérez-Luzunaris and Ms. Jalexca Piñero-Cifuentes
Funding
Research reported in this publication was supported by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Number U54GM133807 (CRRF), P20GM103475 (PR-INBRE), P20GM156713-01 (COBRE PR-CMS)and the UPRMSC Hispanics-In-Research Capability (HiREC) Endowment (S21MD001830) by the NIH National Institute of Minority Health and Health Disparities. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
Conceptualization, V.S.-R. and F.G.-V.; patient recruitment and clinical coordination, V.S.-R., H.M.-G., C.H.-R., J.R.-A., A.R.-E., C.S.-Ll., G.P.-M., V.C.-B. and E.B.-R.; psychological examination, A.C.S.-M.; data curation, G.O.-H., H.M.-G. and C.H.-R.; formal analysis, V.S.-R., G.O.-H. and F.G.-V.; funding acquisition, V.S.-R. and F.G.-V.; DNA extraction, G.O.-H.; methodology, V.S.-R. and F.G.-V.; project administration, V.S.-R., F.G.-V.; resources, V.S.-R. and F.G.-V.; software, G.O.-H. and F.G.-V.; supervision, V.S.-R. and F.G.-V.; visualization, G.O.-H.; writing – original draft, V.S.-R., G.O.-H., H.M.-G., C.H.-R., A.R.-E., G.P.-M., E.B.-R. and F.G.-V.; writing – review & editing: all authors. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Sepúlveda-Rivera, V., Olivieri-Henry, G., Morales-González, H. et al. Gut microbiota distinguishes aging hispanics with Alzheimer’s disease: associations with cognitive impairment and severity. Sci Rep 15, 28505 (2025). https://doi.org/10.1038/s41598-025-13262-2
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-13262-2