Abstract
Brain tumour segmentation is an important task in medical imaging, that requires accurate tumour localization for improved diagnostics and treatment planning. However, conventional segmentation models often struggle with boundary delineation and generalization across heterogeneous datasets. Furthermore, data privacy concerns limit centralized model training on large-scale, multi-institutional datasets. To address these drawbacks, we propose a Hybrid Dual Encoder–Decoder Segmentation Model in Federated Learning, that integrates EfficientNet with Swin Transformer as encoders and BASNet (Boundary-Aware Segmentation Network) decoder with MaskFormer as decoders. The proposed model aims to enhance segmentation accuracy and efficiency in terms of total training time. This model leverages hierarchical feature extraction, self-attention mechanisms, and boundary-aware segmentation for superior tumour delineation. The proposed model achieves a Dice Coefficient of 0.94, an Intersection over Union (IoU) of 0.87 and reduces total training time through faster convergence in fewer rounds. The proposed model exhibits strong boundary delineation performance, with a Hausdorff Distance (HD95) of 1.61, an Average Symmetric Surface Distance (ASSD) of 1.12, and a Boundary F1 Score (BF1) of 0.91, indicating precise segmentation contours. Evaluations on the Kaggle Mateuszbuda LGG-MRI segmentation dataset partitioned across multiple federated clients demonstrate consistent, high segmentation performance. These findings highlight that integrating transformers, lightweight CNNs, and advanced decoders within a federated setup supports enhanced segmentation accuracy while preserving medical data privacy.
Similar content being viewed by others
Introduction
Deep learning has significantly advanced medical imaging, especially in segmentation tasks that are crucial for disease diagnosis and treatment planning1. Convolutional Neural Networks (CNNs) such as U-Net have been extensively deployed for medical image segmentation, due to their capability to capture spatial hierarchies2,3. However, traditional CNN-based models exhibit several limitations when applied to brain tumour segmentation. The drawbacks are mainly due to the high variability in MRI scans arising from different scanning protocols, imaging hardware, and patient demographics4. These variations negatively impact model generalizability, leading to suboptimal performance across heterogeneous datasets. Brain tumour segmentation presents additional difficulties due to the heterogeneous type of tumour structures, which vary in shape, texture, and intensity. Conventional CNN architectures like U-Net and its variants rely on hierarchical feature extraction but often struggle with accurate tumour boundary delineation, leading to over- or under-segmentation. Hence, the limitations of CNN-based architectures hinder real-time clinical applications, necessitating the development of more effective and boundary aware segmentation models5.
A major drawback of centralized learning in medical image segmentation is data privacy concerns. Large-scale, high-quality datasets are essential for training robust segmentation models. However, sharing sensitive patient data across multiple institutions raises ethical and regulatory challenges. Federated Learning (FL) offers an alternative by enabling institutions to jointly train models without sharing raw data, while preserving patient privacy6. Despite its advantages, existing FL-based segmentation models face several constraints such as communication overhead, data heterogeneity, and inconsistency in local model updates across institutions7. Addressing these issues requires developing segmentation models that are efficient, privacy-preserving, and capable of handling diverse MRI datasets.
To mitigate these shortcomings, we propose a Federated Learning-based Dual Encoder-Decoder Segmentation Model that leverages the benefits of both convolutional and transformer-based architectures. This model integrates EfficientNet8 and Swin Transformer9, as dual encoders, and BASNet (Boundary-Aware Segmentation Network)10 decoder along with MaskFormer11 as dual decoders. EfficientNet offers lightweight yet powerful local feature extraction through compound scaling, while Swin Transformer introduces hierarchical self-attention for capturing global contextual information. The BASNet decoder incorporates a boundary-aware predict-and-refine mechanism, leveraging residual learning and edge-aware loss to progressively enhance tumour edge delineation. MaskFormer, on the other hand, reframes segmentation as a mask classification task, reducing noise and improving segmentation consistency. Together, these components improve tumour delineation, segmentation robustness, and efficiency in terms of total training time within federated learning environments.
The primary goals of this work are:
-
1.
Comparing traditional CNN-based segmentation models (U-Net12, UNet + + 13, ResUNet14) and transformer-based models with our proposed hybrid model integrating EfficientNet, Swin Transformer, BASNet decoder, and MaskFormer to quantify performance improvements.
-
2.
Optimizing federated learning (FL) training efficiency by integrating lightweight yet high-performing models to minimize training and communication costs.
-
3.
Evaluating privacy-preserving brain tumour segmentation in a federated setting, ensuring high segmentation accuracy while maintaining data security and regulatory compliance.
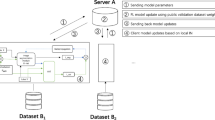
By addressing these challenges, our study advances privacy-preserving brain tumour segmentation, achieving enhanced segmentation accuracy, efficiency in terms of total training time, and generalizability across multi-institutional MRI datasets. Figure 1 illustrates the Federated Learning concept, where a central server coordinates learning from multiple hospital clients without accessing their raw MRI scans.
Background
Federated Learning (FL) has become a widely adopted strategy in medical imaging, allowing multi-institutional collaboration without compromising data privacy15. This decentralized learning paradigm enables local training on institutional data while sharing only updates of the model with a centralized server for aggregation. Studies have demonstrated that FL achieves competitive segmentation performance compared to centralized training while maintaining regulatory compliance and protecting patient confidentiality16. However, data heterogeneity remains a critical challenge in FL, where MRI scans from different hospitals may have variations in intensity, resolution, and scanner type. Personalized federated models and adaptive aggregation strategies have been proposed to mitigate these challenges, but further research is needed to improve model robustness in non-iid (non-independent and identically distributed) settings17.
CNN-based architectures have been increasingly used in medical image segmentation, with U-Net being one of the most popular and widely used model due to its skip-connection based encoder-decoder architecture18. UNet + + improves upon U-Net by incorporating dense, nested skip pathways, allowing the model to learn multi-scale features effectively19. ResUNet further refines this approach by introducing residual learning, mitigating vanishing gradient issues and enabling deeper networks to achieve superior segmentation accuracy20. However, CNN-based models have limitations in finding long-range dependencies, which are critical for accurately segmenting complex tumour structures21.
BASNet (Boundary-Aware Segmentation Network) was initially introduced for salient object detection but has exhibited robust performance in medical image segmentation, specifically for problems that need unambiguous boundary delineation22. Unlike traditional CNNs that rely on single-step feature extraction, BASNet utilizes a predict-and-refine mechanism that iteratively enhances segmentation masks by focusing on boundary refinement. This is especially useful in brain tumour segmentation, where accurate delineation between tumour and healthy tissue is crucial. Transformers have gained prominence in medical imaging due to their capability to interpret long-range dependencies using self-attention mechanisms. Vision Transformers (ViT) divide images into small patches and apply self-attention to learn contextual relationships between distant regions, making them highly effective for complex medical segmentation tasks23.
Swin Transformer, an extension of ViT, introduces hierarchical feature extraction through shifted window attention, significantly enhancing computational efficiency while preserving fine-grained spatial details. These transformer-based models have delivered exceptional performance in medical image analysis, surpassing traditional CNNs in tasks requiring contextual awareness24. EfficientNet has been proposed as a lightweight yet powerful CNN architecture that balances depth, width, and resolution scaling to achieve high accuracy with fewer parameters. In federated settings, EfficientNet reduces communication overhead by minimizing model update sizes while maintaining competitive segmentation performance25.
In recent years, numerous deep learning architectures have been studied for brain tumour segmentation. U-Net and its extensions (e.g., UNet + + , ResUNet) have remained popular due to their encoder-decoder structure with skip connections, enabling multi-scale feature learning and precise localization. However, their limited receptive fields constrain performance when segmenting irregularly shaped tumours or differentiating low-contrast boundaries. Advanced models such as DeepMedic26, 3D U-Net27, and V-Net28 have introduced volumetric segmentation and 3D convolutions, offering improved spatial coherence but often at a higher computational cost. More recently, attention mechanisms and transformer-based models like TransBTS29, UNETR30 and TFCNS31 have been proposed to deal with long-range dependencies in MRI volumes. These models leverage self-attention to model spatial context, showing promising results in tumour core and whole tumour segmentation tasks. Despite their effectiveness, these models are typically evaluated in centralized setups and rarely consider data privacy. Our work builds upon these advances by integrating both CNN and transformer components into a federated learning framework, addressing accuracy, boundary precision, and privacy simultaneously. Recent works have also proposed advancements in federated and semi-supervised learning for medical image analysis. For example 32, surveys FL across diagnostic contexts 33; proposes adaptive copy-paste supervision for tumour segmentation 34; reviews FL and ML methods for imaging tasks 35; introduces MSKI-Net for modality-specific glioma survival prediction and36 explores uncertainty-aware aggregation in histopathology segmentation.
Despite the success of FL and deep learning in medical segmentation, several challenges remain. There is limited research on BASNet in federated brain tumour segmentation. Existing studies mainly concentrate on CNN-based architectures, with minimal exploration of boundary-aware models in decentralized settings. There is a lack of comparative studies integrating Transformers and EfficientNet. Most studies compare FL-based segmentation using standard CNNs, but the effectiveness of hybrid Transformer-CNN architectures remains underexplored. While federated models preserve privacy, they often suffer from training inefficiency and performance degradation in heterogeneous data environments. Our study introduces lightweight and high-performance architectures to address these issues.
Methodology
This section describes the segmentation models evaluated, including both existing baselines and the proposed architecture, and outlines the federated learning framework used for training. The model architectures and FL setup are detailed. Three widely used CNN-based semantic segmentation models—U-Net, UNet + + , and ResUNet—were selected as baselines for comparison. These architectures have been extensively used in medical segmentation, but they exhibit limitations in boundary delineation, feature extraction efficiency, and computational scalability, especially in federated settings. U-Net has a symmetric encoder-decoder architecture with skip connections, where the encoder extracts multi-scale features through convolutional and pooling layers, and the decoder recreates spatial details using transposed convolutions. Skip connections aid in precise localization by directly passing low-level features to the decoder. The implementation includes a 5-level U-Net, where each downsampling operation halves the spatial resolution while doubling feature channels, and each upsampling operation reverses this process by concatenating encoder features to refine predictions.
UNet + + extends U-Net by introducing intermediate convolutional blocks between encoder and decoder, forming dense nested skip connections that enhance feature fusion. This improves segmentation accuracy in boundary regions and small structures but increases computational complexity. The architecture maintains the same depth as U-Net but incorporates additional convolutional layers and dense skip pathways. ResUNet incorporates residual learning into U-Net by adding residual (skip) connections in each convolutional block. These residual blocks help enhance gradient flow and stabilize training, reducing the vanishing gradient problem and improving optimization in deeper networks. Batch normalization is applied at each stage to further enhance convergence. All baseline models are trained within a federated environment using a uniform training pipeline. Federated Averaging (FedAvg) algorithm37 is used to aggregate model updates across multiple clients. In each FedAvg round, client models are locally trained on partitioned datasets, and updated model weights are transmitted to a centralized server. The server averages the weights to refine the global model, which is then resent to clients for the subsequent training iteration. This iterative process continues until convergence, ensuring improved model generalization without requiring raw data exchange.
Proposed dual encoder-decoder segmentation model
The proposed dual encoder-decoder segmentation model as shown in Fig. 2, is designed to leverage the benefits of both CNN-based local feature generation and Transformer-based global context modelling for high-accuracy brain tumour segmentation. The architecture follows a structured computational flow, beginning with input preprocessing, progressing through dual encoders (EfficientNet + Swin Transformer) for hybrid feature extraction, and concluding with dual decoders (BASNet decoder + MaskFormer) to refine segmentation quality and boundary precision. The pipeline begins with MRI brain scans, which undergo standardization, normalization (Min–Max scaling to [0,1]), and augmentation (random flips, rotations, and intensity normalization) to improve robustness. Images are resized to 256 × 256 pixels to ensure uniform input dimensions across the models. Both encoders (EfficientNet and Swin Transformer) process input images in parallel, enabling simultaneous extraction of low-level and global contextual features. Likewise, the decoder stage performs refinement in a parallel stream—BASNet for boundary enhancement and MaskFormer for semantic-level mask generation—before fusion at the output layer.
Encoder module: EfficientNet + swin transformer (hybrid feature extraction)
The EfficientNet encoder functions as a lightweight CNN-based feature extractor, efficiently capturing fine-grained spatial details using compound scaling (depth, width, resolution). This ensures computational efficiency while maintaining high-resolution local feature representation, making it well-suited for federated training environments. Complementing this, the Swin Transformer introduces self-attention-based global context extraction, addressing CNNs’ limited receptive field constraints. The shifted-window self-attention mechanism within Swin Transformer captures long-range dependencies, enhancing tumour shape recognition and segmentation of heterogeneous tumour textures. Together, these dual encoders ensure that both local feature hierarchies (EfficientNet) and global contextual relationships (Swin Transformer) are optimally leveraged for accurate segmentation.
EfficientNet employs compound scaling across depth d, width w, and resolution r. For a convolutional layer, the feature \({(F}_{cnn })\) extraction process can be expressed as given in Eq. (1).
where X is the input tensor, W is the weight matrix, b is the bias term, ∗ denotes convolution, and σ denotes the activation function (e.g., Swish). EfficientNet applies scaling as:
Subject to: \(\alpha \cdot {\beta }^{2}\cdot {\gamma }^{2}\approx 2\), where \(\alpha ,\beta ,\gamma >1\) are constants found through grid search.
The Swin Transformer operates on non-overlapping image patches using a shifted window-based self-attention mechanism. Let the input image be divided into a sequence of flattened patch embeddings, denoted as \(X\in {R}^{n\times d}\), where n is the number of patches, X is the input matrix to the transformer encoder and d is the feature dimension of each patch. The self-attention mechanism computes attention as shown in Eq. (2).
Here \(=X{W}_{Q}\), \(K=X{W}_{K}\), and \(V=X{W}_{V}\) are the query, key, and value matrices, respectively. The matrices \({W}_{Q}\), \({W}_{K}\), \({W}_{V}\) \(\in {R}^{d\times {d}_{k}}\) are learned projection weights, and \({d}_{k}\) is the dimensionality of the keys. This formulation allows the model to learn pairwise dependencies between different patches in the image. Swin Transformer introduces a computational optimization called shifted window self-attention, which partitions the input into smaller windows and shifts them between layers to enhance cross-window connections. This strategy reduces computational complexity from \(O({n}^{2})\) in standard global attention to \(O(n)\), significantly improving efficiency in high-resolution medical image segmentation while maintaining contextual modelling capabilities.
Feature fusion layer
To integrate multi-scale features from both encoders, a feature fusion mechanism is employed. Let the output feature maps from EfficientNet and Swin Transformer be denoted as \({F}_{eff}\in {R}^{H\times w\times {c}_{1}}\) and \({F}_{swin}\in {R}^{H\times w\times {c}_{2}}\), where H and W are the spatial dimensions of the feature maps, and \({c}_{1}\) and \({c}_{2}\) represent the number of channels produced by each encoder. These feature maps are added along the channel axis to form a combined tensor: \({F}_{fused}= Concat({F}_{eff},{F}_{swin}),\) where \({F}_{fused}\in {R}^{H\times w\times {(c}_{1}+{c}_{2})}\). To harmonize channel dimensions and enhance representational learning, a 1 × 1 convolution followed by a non-linear activation is applied as shown in Eq. (3).
where W is a learnable 1 × 1 convolution kernel of shape (\({c}_{1}\)+\({c}_{2}\)) × \({c}_{f}\), b is a bias term, and σ denotes a non-linear activation function such as ReLU. The resulting fused output \(F_{{{\text{out}}}} \in R^{{H \times w \times c_{f} }}\) is passed to the decoder for final segmentation. This fused representation effectively combines high-resolution local textures from EfficientNet with global contextual information from the Swin Transformer, enabling robust and boundary-aware tumour segmentation, particularly in cases involving irregular shapes and heterogeneous intensity profiles.
Decoder module: BASNet + MaskFormer (boundary-aware segmentation)
The BASNet decoder is utilized for progressive boundary refinement, leveraging residual learning to sharpen tumour edges while suppressing false positives. This iterative feature refinement strategy.
ensures that the final segmentation map accurately delineates tumour boundaries, even in cases where tumour edges exhibit low contrast. In parallel, the MaskFormer decoder applies a transformer-based mask classification approach, where segmentation is treated as a mask-prediction problem instead of a pixel-wise classification task. This methodology significantly reduces noise, enhances segmentation robustness, and improves accuracy in regions with complex tumour textures and ambiguous boundaries.
BASNet performs boundary-aware segmentation using a residual refinement process. The iterative refinement is defined as given in Eq. (4).
where \({Y}_{t}\) is the predicted mask at time step t, and R is a residual function composed of convolutional operations guided by edge-aware supervision. The training loss combines binary cross-entropy, Dice loss, and edge loss as shown in Eq. (5), with tuneable weights \({\lambda }_{1}\), \({\lambda }_{2}\), \({\lambda }_{3}\).
MaskFormer treats segmentation as a mask classification problem, where each predicted mask is associated with a semantic class label. Given the decoder output vector \({z}_{i}\in {R}^{d}\), where \({z}_{i}\) denotes the embedding respective to the i-th predicted mask and d represents the dimensionality of the embedding, the class prediction is computed as given in Eq. (6).
In this formulation, X refers to the input image, \({M}_{i}\) is the i-th predicted segmentation mask, and \({f}_{cls}({z}_{i})\) is the class score vector obtained by passing \({z}_{i}\) through a classification head, typically a fully connected layer. The Softmax function \(Softmax\left(\bullet \right)\) transforms these class scores into normalized probabilities over all possible classes, enabling the model to assign each predicted mask a corresponding semantic category, such as tumour, non-tumour, or background. The segmentation mask is given by Eq. (7).
where \({f}_{cls}\) and \({f}_{mask}\) are the classification and mask prediction heads, respectively. The total loss for training the MaskFormer is defined as in Eq. (8).
where \({P}_{i}^{*}\) and \({M}_{i}^{*}\) are the ground truth labels and masks, Pi is the predicted class probability for instance i, Mi is the predicted segmentation mask and λ is a balancing hyperparameter.
To further enhance segmentation accuracy, particularly around object boundaries and semantically critical regions, an attention-based refinement loss (Latten) was integrated into the overall training objective. This component was designed to guide the attention mechanisms—particularly those in the Swin Transformer encoder and the MaskFormer decoder—toward more meaningful spatial regions. By encouraging alignment between attention maps and ground truth structures (such as edges or salient object interiors), the refinement loss helped suppress irrelevant background noise and sharpen boundary predictions. This led to noticeably cleaner and more accurate segmentation masks. In addition to improving Dice and IoU scores, the inclusion of this refinement term contributed to more stable training and faster convergence. Its role as a form of semantic regularization proved especially valuable in a hybrid setup involving both convolutional and transformer-based feature representations.
The overall loss function integrates Dice and BCE losses with boundary-aware and attention-guided terms to enhance both semantic accuracy and boundary sharpness as shown in Eq. (9).
Here \({L}_{Edge}\) stems from BASNet’s residual refinement (Eq. 5), while \({L}_{cls}\) and \({L}_{mask}\) are derived from MaskFormer’s mask classification strategy (Eq. 8). This formulation enables precise delineation of tumour margins while preserving contextual coherence. Similar trends have been observed in recent work that leverage enhanced boundary supervision and efficient architectures for early disease detection, such as the Force Map-Enhanced segmentation framework for cervical cancer proposed by38.
Output segmentation map generation
To effectively combine the strengths of both decoders in our brain tumour segmentation model, we used a learnable 1 × 1 convolution-based fusion strategy. Instead of simply averaging the outputs from BASNet and MaskFormer, this approach allows the model to intelligently decide how much importance should be given to each decoder at every pixel. BASNet contributes precise boundary details, while MaskFormer provides a broader understanding of tumour regions. By learning how to merge these two perspectives, the fusion layer helps produce more accurate and reliable segmentation masks. This is especially important for brain tumours, where capturing both the fine edges and the overall structure of the tumour is critical for clinical relevance. As shown in Fig. 2, the segmentation outputs from BASNet and MaskFormer are concatenated and fed through a 1 × 1 convolution after which ReLU activation is performed. This design balances architectural simplicity and computational efficiency, ensuring minimal overhead in the federated setting. Their outputs are fused to retain both spatial and contextual cues, improving segmentation fidelity.
The final segmentation output of the proposed model generates a high-resolution tumour mask that ensures precise tumour localization accuracy by leveraging the combined strengths of EfficientNet, Swin Transformer, BASNet, and MaskFormer. The boundary-aware refinement mechanism, driven by progressive residual learning in BASNet and mask classification-based segmentation in MaskFormer, effectively preserves tumour edges, reducing segmentation errors in complex regions. This refined output significantly minimizes false positives and false negatives, ensuring clinically reliable segmentation results suitable for diagnostic and treatment planning in medical imaging.
Federated learning framework
The proposed model is trained in a federated learning (FL) environment, enabling multi-institutional collaboration while preserving data privacy. Federated clients utilize MRI segmentation datasets partitioned across multiple institutions, simulating a real-world federated learning setting. Each client trains a locally hosted version of the dual Encoder-Decoder model on its respective dataset. After local training, model parameters are aggregated at a centralized server using the Federated Averaging (FedAvg) algorithm (Algorithm 1). Privacy-preserving techniques include Differential Privacy (DP), which adds noise to model updates before aggregation to avoid inference attacks, and Secure Aggregation (SA), which ensures encrypted parameter updates, preventing direct access to client-specific model weights.
The FL parameters such as the number of clients (M), number of rounds (R), number of local epochs (LE) and initial global weights (GW0) are initialized at the beginning of the training. In each round, all clients send their local weights (LW) to the server after training their local models with LE epochs. Each client (c) is assigned a relative weight (nc/n) proportional to its dataset size (nc) and considering total size of the dataset (n) of all clients. After each round, server does a weighted mean of local weights obtained from all clients to update global weights. The whole process is iterated for R rounds and the final federated model is obtained which is at par with the centralized model.
Implementation
The implementation of the proposed model and other baseline models is carried out in PyTorch, ensuring compatibility with GPU-accelerated training. The federated training workflow follows a structured approach, beginning with data preprocessing and augmentation, where input MRI scans undergo normalization, resizing, and augmentation techniques such as rotation, flipping, and contrast adjustment to enhance generalization. In the federated training phase, each client trains its local model using EfficientNet and Swin Transformer as encoders, while BASNet decoder and MaskFormer function as decoders. The FedAvg algorithm aggregates model updates from all clients, iteratively refining the global segmentation model. Finally, model evaluation and testing are conducted on a test dataset, assessing segmentation performance using Dice Coefficient and IoU metrics. Visual outputs of predicted tumour masks are further analysed for qualitative assessment. Table 1 provides a list of hyperparameters used in federated training for baseline segmentation models (U-Net, UNet + + , ResUNet) versus the proposed dual encoder-decoder model. Although the baseline models were trained for 30 rounds, the proposed model achieved convergence significantly earlier—by approximately the 20th round—based on stabilization of validation metrics. Therefore, 20 rounds were used for final evaluation to reflect convergence efficiency and to avoid redundant training. Additional tests with 30 rounds for the proposed model showed negligible performance gain (< 0.2%), confirming the sufficiency of 20 rounds. Key hyperparameters such as learning rate (0.001), batch size (8), and local epochs (5) were selected based on preliminary tuning experiments conducted on a validation subset within the training data. Grid search was employed to evaluate convergence behaviour and stability under varying configurations, ensuring optimal trade-off between computational efficiency and segmentation accuracy. Sensitivity tests further demonstrated that the proposed model maintained high Dice performance (≥ 0.92) across a ± 25% range in learning rate, supporting robustness to moderate hyperparameter variations. These choices contribute to reproducibility and reliability in federated deployments.
Experimental setup
This section outlines the empirical procedures, including the dataset used and the preprocessing methods applied. The experiments are carried out using the publicly available Mateuszbuda LGG-MRI Segmentation Dataset39 in Kaggle, which includes 3,926 FLAIR-weighted contrast-enhanced MRI scans alongside their manually annotated segmentation masks delineating tumour regions. This dataset is specifically chosen due to its significant size, diversity of tumour shapes, textures, and sizes, as well as its practical relevance for clinical segmentation tasks. The augmented dataset is divided into training (80%) and testing (20%) subsets. The training and testing subsets are further partitioned across five clients, thereby simulating a realistic federated learning environment across multiple institutions. All MRI slices and masks are resized to 256 × 256 pixels to standardize input size while preserving anatomical details. Pixel intensities are scaled to [0,1] using min–max scaling to enhance training stability and prevent intensity-related biases. Training images undergo random horizontal/vertical flips, ± 10-degree rotations, and ± 10% scaling to improve model generalization. After preprocessing, each MRI image is treated as a single-channel 256 × 256 grayscale input, while the corresponding segmentation mask is a binary image of the same size. Models are trained using batch-based processing, with segmentation masks guiding the Dice loss function, optimizing the similarity between predicted and actual tumour regions. In addition to using pretrained EfficientNet and Swin Transformer modules, remaining convolutional layers were initialized with He Normal and linear layers with Xavier Uniform strategies. Experiments were conducted using NVIDIA A100 GPUs (40 GB VRAM), 256 GB RAM, and PyTorch 2.0. Each client trained independently in parallel, ensuring reproducibility of both training time and segmentation accuracy. While network latency was not emulated, communication cost was assessed through measured upload size per client and server aggregation time.
To reflect how federated learning would function in real-world medical environments, two types of data distribution across clients: IID (independent and identically distributed) and non-IID are implemented. In the IID setup, MRI slices and their corresponding tumour segmentation masks were randomly and evenly split among clients, ensuring that each client received a representative mix of tumour types and imaging characteristics. In contrast, the non-IID setting was specifically designed to mimic the challenges of real-world, multi-institutional deployment. A typical non-IID setting, quantity skew was implemented by assigning unequal amounts of data to each client—some clients had significantly fewer slices, while others had more—simulating the natural imbalance in data availability across hospitals. Additionally, feature distribution skew was incorporated to model the kind of visual variability caused by differences in scanners, acquisition settings, and local pre-processing protocols. To simulate this, the global dataset was first augmented using a variety of transformation techniques, including rotation, horizontal/vertical flips, contrast shifts, affine distortions, blurring etc. The augmented data was then split across clients such that each client received two specific types of augmentation. For example, one client received both blurred and horizontally flipped images, another received contrast-enhanced and rotated images, and so on. This dual-augmentation strategy better reflects real-world conditions, where multiple sources of variability may coexist within a single institution, resulting in heterogeneous but overlapping feature distributions across clients. This setup helps to investigate how well our proposed segmentation model could generalize and converge under diverse and challenging non-IID conditions that are representative of real-world federated healthcare applications.
Results and discussion
This section presents the evaluation metrics employed, and the comparative performance results of baseline models and the proposed dual encoder-decoder model. Results from both centralized and federated training environments are presented, along with comprehensive visual and statistical analysis highlighting performance improvements. Model performance is evaluated using standard segmentation metrics, focusing on overlap measures and pixel-wise accuracy. Dice Coefficient is the primary metric, defined as:
where P represents the predicted tumour region and G is the ground truth. A Dice score of 1 indicates accurate segmentation, while 0 means no overlap. Dice is susceptible to both false positives and false negatives, making it appropriate for medical segmentation where under-segmentation and over-segmentation are critical concerns. Intersection over Union (IoU), or the Jaccard Index, is given by:
IoU is stricter than Dice and provides an additional measure of segmentation accuracy, usually yielding lower values than Dice for the same prediction. Both Dice and IoU are reported as they are standard in tumour segmentation literature. Precision and Recall are computed to analyse the model’s ability to correctly identify tumour pixels. Precision, defined as:
Which indicates how many predicted tumour pixels are correct, while Recall, given by:
This measures how well the model detects tumour pixels without missing any. Higher recall ensures minimal false negatives, which is critical in medical imaging to avoid under-segmentation of tumours. F1-Score, computed as the harmonic mean of precision and recall, is also included as an additional verification, although it closely aligns with Dice in the segmentation context. F1-Score is given by:
Figure 3a,b illustrate the segmentation performance of the proposed model over 20 training rounds in IID setting. The blue line represents validation performance, demonstrating stable convergence and high segmentation accuracy in a federated learning setup. Training-time efficiency is assessed by tracking training time per round, total training time, and model update size. The server aggregation time per round and total communication overhead (sum of all model updates across all FL rounds) are recorded to evaluate feasibility in real-world federated networks. These factors are essential for determining the practicality of deploying large models in resource-constrained environments. Dice and IoU remain the primary metrics for comparing model performance on the test set, with higher Dice and IoU scores indicating superior segmentation quality.
The baseline models (U-Net, UNet + + , ResUNet) and the proposed dual encoder-decoder model are trained in a federated learning (FL) environment and evaluated on an independent test set comprising 20% of the dataset, which is never used in training. Table 2 summarizes the segmentation performance, highlighting Dice Coefficient, Intersection over Union (IoU), and training time per round.
Despite a higher per-round time (17 min), the proposed model required only 20 rounds to converge as observed through validation Dice and IoU stabilization in Fig. 3 and completed training in the shortest overall time (5.1 h), validating its training efficiency compared to baseline models. The proposed Dual Encoder-Decoder Model achieved the highest performance among all evaluated architectures, with a Dice Coefficient of 0.94 and IoU of 0.87, outperforming the best-performing baseline, UNet + + (Dice: 0.88, IoU: 0.79), by a margin of approximately 6–8 percentage points.
Figure 4 further illustrates the distribution of Dice and IoU scores across 30 subjects. The proposed model achieved a median Dice score of 0.94 and IoU of 0.87, with an interquartile range (IQR) of approximately 0.02 for Dice and 0.025 for IoU, indicating high consistency across cases. In contrast, U-Net displayed a median Dice of 0.82 with a wider IQR of ~ 0.06, while ResUNet had a median Dice of 0.87 but with an IQR of ~ 0.045, suggesting higher performance variability. These distribution patterns confirm that the proposed architecture not only improves overall accuracy but also ensures stable and reliable segmentation. The results validate the contribution of transformer-based components (Swin Transformer and MaskFormer) for global context modelling, and EfficientNet for efficient and robust feature extraction, making the dual-stream design both accurate and consistent for brain tumour segmentation. Although the proposed dual encoder-decoder architecture integrates more components (EfficientNet, Swin Transformer, BASNet, and MaskFormer), it achieves relatively faster average training time per round (17 min) due to two key factors:
-
1.
EfficientNet Backbone: The use of EfficientNet as one of the encoders contributes significantly in reducing training time. EfficientNet is designed with compound scaling to maximize performance with fewer parameters, enabling faster forward and backward passes.
-
2.
Pretrained Modules and Parallelization: Both EfficientNet and Swin Transformer are initialized with pretrained weights, allowing the model to converge faster. Moreover, Swin Transformer’s shifted-window mechanism is optimized for computational efficiency, and its parallelization within the federated environment reduces per-client training overhead.
Together, these architectural choices provide a favourable trade-off between model complexity and execution efficiency, leading to a 24% reduction in total training time compared to ResUNet, despite the additional encoder-decoder layers. Table 3 presents an evaluation of the Precision, Recall, and F1-Score, metrics to assess clinical applicability of the segmentation models.
All evaluated models exhibited high recall, indicating accurate detection of tumour pixels. However, the proposed dual encoder-decoder model achieved an optimal balance of precision (0.93) and recall (0.95), significantly minimizing false positives and reducing the likelihood of over-segmentation. The baseline U-Net model, while showing good recall (0.89), presented lower precision (0.85), suggesting a tendency for false-positive predictions. The proposed architecture’s superior precision and recall indicate enhanced clinical reliability and usability.
Figure 5 illustrates a comparative assessment of precision, recall, and F1-score across four segmentation models. The proposed model achieved the highest performance, with a precision of 0.93, recall of 0.95, and F1-score of 0.94, indicating a strong balance between positive prediction accuracy and sensitivity. In contrast, UNet + + and ResUNet followed with F1-scores of 0.90 and 0.89, respectively, while U-Net showed the lowest performance with an F1-score of 0.87. The notable improvement of the proposed architecture—especially in recall—demonstrates its effectiveness in minimizing false negatives, which is crucial in medical imaging tasks such as tumour segmentation where missed regions can be clinically significant. Convergence analysis indicated that the proposed dual encoder-decoder model reached high validation Dice scores in fewer federated rounds than baseline models. EfficientNet’s pretrained weights provided a robust initialization, accelerating convergence and ensuring faster model training and superior generalization. Conversely, CNN-based baseline models, trained from scratch, required more federated rounds to achieve peak performance, underscoring the significant advantage of incorporating pretrained transformer and CNN architectures.
The training loss curves in Fig. 6 illustrate stable convergence in the federated learning environment across all models. Although all models achieved comparable final training loss values within the first 30 rounds, the proposed model maintained consistently higher validation Dice scores, signifying superior generalization performance. Importantly, federated training did not substantially diminish segmentation accuracy compared to centralized training. The proposed model achieved Dice score of approximately 0.94 in federated training, closely matching those obtained under centralized training conditions, confirming its robustness and effectiveness in decentralized learning setups.
To further strengthen the evaluation, additional metrics related to training efficiency and communication overhead are analysed. Table 4 and Fig. 7 present a comparative summary of the average client upload size, server aggregation time, and total training time for each model.
Despite its dual encoder-decoder structure, the proposed model demonstrated the shortest total training time of 5.1 h, compared to 5.8 h for U-Net, 6.0 h for UNet + + , and 6.6 h for ResUNet. This performance is attributed to the integration of EfficientNet and Swin Transformer, which offer high representational power with fewer parameters, as well as the use of pretrained weights that accelerate convergence. Although the proposed model incurs a higher average client upload size of 55 MB per round (versus 30–38 MB for baseline models), this marginal increase is offset by the reduced number of training rounds. These results validate the model’s practical applicability for deployment in communication-constrained federated settings. As shown in Fig. 7, the relationship between model complexity, communication cost, and training time reflects a favourable trade-off, with the proposed model achieving the best overall balance among all evaluated architectures.
Existing hybrid CNN–Transformer models, such as TransBTS, and UNETR, have demonstrated the benefits of combining convolutional feature extractors with transformer-based global context modelling for brain tumour segmentation. TransBTS employs a 3D CNN with a transformer bottleneck and reports similar performance. DeepMedic and V-Net have shown success in 3D medical segmentation, but they have high volumetric input constraints and memory requirements, and show limited adaptability in 2D federated learning setups. However, these models are computationally demanding and have not been evaluated in federated or privacy-preserving frameworks. In contrast, the proposed model integrates EfficientNet and Swin Transformer with BASNet and MaskFormer decoders, achieving a Dice coefficient of 0.94 and IoU of 0.87 in a federated learning environment. To further contextualize the results, Table 5 provides a comprehensive comparison of the proposed model with both baseline CNNs and existing hybrid models across centralized and federated training setups. The data demonstrate that the proposed model not only outperforms existing architectures in segmentation accuracy but also satisfies privacy, scalability, and efficiency requirements that are critical for real-world deployment in decentralized medical imaging systems.
Figures 8 and 9 visually compare the segmentation performance of the baseline models (U-Net, UNet + + , ResUNet) against the proposed dual encoder-decoder architecture, using representative MRI scans from the Mateuszbuda LGG-MRI Segmentation Dataset. The dataset encompasses 3,926 MRI scans with corresponding manual annotations. The proposed Dual Encoder-Decoder Segmentation Model, integrating EfficientNet and Swin Transformer as encoders with BASNet and MaskFormer as decoders, demonstrates superior segmentation accuracy, achieving enhanced boundary delineation and reduced false positive regions compared to baseline architectures. Visual assessments aligned closely with the quantitative metrics presented earlier in Tables 2 and 3, where the proposed model demonstrated superior precision (0.93), recall (0.95), Dice (0.94), and IoU (0.87). In particular, examples of segmentation failure from baseline models—such as boundary misclassification or incomplete tumour coverage—are visually apparent in Fig. 9. These failure patterns contrast with the proposed model’s more complete and anatomically consistent segmentation output. Notably, the segmentation masks generated by the proposed model exhibited significantly improved delineation of tumour boundaries with fewer false positives and reduced segmentation errors. These visual outcomes further validate the model’s enhanced capability for accurately localizing tumour boundaries compared to CNN-only architectures. This qualitative assessment reinforces the suitability and potential clinical effectiveness of the proposed federated learning-based segmentation approach.
(a) Brain tumour segmentation visualizations on the Mateuszbuda LGG-MRI dataset. Comparative results for identical brain slice across U-Net, UNet + + , ResUNet, proposed model. (b) Boundary-level visual comparison—Red contours denote predicted tumour boundaries; green contours show ground truth segmentation.
A qualitative analysis was conducted on a difficult segmentation scenario selected from the Mateuszbuda LGG-MRI dataset, characterized by a tumour region comprising only 99 annotated pixels. This case represents a boundary-sensitive example where segmentation performance is likely to degrade due to limited spatial context and low contrast. As shown in Fig. 10, the MRI slice and corresponding ground truth highlight the small tumour region. Inclusion of such cases serves to evaluate model behaviour under edge conditions, which are clinically significant for early-stage lesion detection. The ability to maintain accurate delineation in this setting reflects the model’s sensitivity to fine-grained structural cues and supports its applicability to anatomically subtle segmentation tasks.
Analysis of boundary evaluation metrics
To rigorously examine the boundary delineation performance of the proposed hybrid segmentation model, three specialized metrics—Hausdorff Distance at the 95th percentile (HD95), Average Symmetric Surface Distance (ASSD), and Boundary F1-score (BF1)—were employed in addition to standard overlap-based metrics. HD95 measures the worst-case boundary deviation (excluding outliers) between the predicted and ground truth contours, capturing the extent of extreme errors. ASSD quantifies the average surface distance between corresponding contour points on the predicted and reference masks, offering insight into typical surface discrepancies. The BF1-score denotes the harmonic mean of boundary-level precision and recall, reflecting the spatial accuracy of contour prediction within a specified tolerance margin.
-
Hausdorff Distance (HD95): Measures the worst-case boundary deviation (excluding outliers) between predicted mask A and ground truth B.
$$HD95(A,B)=max\left\{\underset{a\in A}{\text{quantile}}95(\underset{b\in B}{\text{min}}\parallel a-b\parallel ),\underset{b\in B}{\text{quantile}}95\underset{a\in A}{(\text{min}}\parallel b-a\parallel )\right\}$$ -
Average Symmetric Surface Distance (ASSD): Quantifies the average surface deviation across all contour points on both prediction and ground truth.
$$ASSD(A,B)=\frac{1}{\mid A\mid +\mid B\mid }\left(\sum_{a\in A}\underset{b\in B}{\text{min}}\parallel a-b\parallel +\sum_{b\in B}\underset{a\in A}{\text{min}}\parallel b-a\parallel \right)$$ -
Boundary F1-score (BF1): Evaluates the harmonic mean of precision and recall for pixels near the boundary within a tolerance margin.
$$BF1=\frac{2\cdot {Precision}_{boundary}*{Recall}_{boundary} }{{Precision}_{boundary}+{Recall}_{boundary}}$$
Table 6 presents the results of the boundary evaluation metrics on the Mateuszbuda LGG-MRI dataset. The proposed model shows superior boundary delineation performance compared to baseline architectures. It achieves the lowest HD95 (1.61) and ASSD (1.12) values, indicating highly accurate and tight boundary alignment with the ground truth. Additionally, it attains the highest BF1-score (0.91), reflecting excellent boundary precision and recall. In contrast, traditional models like UNet, UNet + + , and ResUNet show higher boundary deviations, with HD95 values of 6.74, 5.07, and 3.97, and BF1-scores of 0.81, 0.84, and 0.83 respectively. This underscores the effectiveness of the proposed model in precise boundary segmentation. The high boundary evaluation metrics achieved by our model—such as low Hausdorff Distance (HD95), low Average Symmetric Surface Distance (ASSD), and high Boundary F1-score (BF1)—highlight its strong clinical relevance and interpretability. Accurate boundary delineation is critical in neuro-oncology, especially for surgical resection and radiotherapy, where precise tumour margins guide treatment. These metrics indicate that the model captures fine anatomical details and closely adheres to expert-drawn contours, reducing the risk of under- or over-segmentation. The high BF1 score further supports its reliability in real-world settings by showing consistency along tumour edges, thereby enhancing clinician trust and enabling safer integration into diagnostic workflows.
Supplementary dataset evaluation
The proposed model and baseline models are additionally evaluated on a supplementary dataset from the Kaggle repository to demonstrate the generalizability and robustness of segmentation performance across different data distributions. The Nikhilroxttomar dataset43 has 3064 pairs of brain MRI images and their corresponding binary masks indicating tumour. This dataset consists of brain MRI slices containing various tumour shapes and sizes. It serves as a strong benchmark for model validation under moderately complex segmentation conditions. Quantitative results for this dataset are shown in Table 7. The proposed model outperformed baseline architectures—including U-Net (Dice: 0.82, IoU: 0.75), UNet + + (0.84, 0.78), and ResUNet (0.85, 0.79)—by achieving a significantly higher Dice score of 0.93 and IoU of 0.85. This substantial improvement highlights the model’s superior capability in accurately segmenting tumour regions with enhanced boundary precision and spatial consistency.
Handling data and computational heterogeneity
To assess model robustness under realistic deployment scenarios, we conducted experiments in a non-IID federated setting incorporating both quantity skew and feature skew. These reflect typical challenges encountered in multi-institutional medical imaging, such as uneven data volume and scanner-specific variations in image appearance. In the presence of quantity skew, where clients received differing amounts of data but with similar feature distributions, baseline models showed moderate performance degradation as shown in Table 8. Our proposed model showed greater stability, achieving 0.92 (Dice) and 0.85 (IoU), indicating resilience to sample imbalance during training. The effect of feature skew, however, was more severe. Here, each client received data augmented with a distinct visual transformation (e.g., blur, noise, contrast shift), leading to domain-level feature divergence. This significantly reduced the performance of baseline models. In contrast, our proposed model maintained relatively better performance with a Dice Score of 0.89 and IoU of 0.81, showing only a marginal decline from its IID performance. This robustness is primarily attributed to the model’s architectural design, which combines a hybrid feature extraction backbone with a dedicated boundary delineation module. The hybrid backbone facilitates the model to extract both global context and localized semantic features, while the boundary-aware component enhances spatial precision in segmenting fine tumour structures. Together, these innovations allow the model to maintain performance even under significant domain shifts and inter-client variability. In non-IID setting involving both quantity and feature skew, convergence to a target Dice score required approximately 40–70% more communication rounds compared to the IID setting, aligning with established observations in federated learning literature. In non-IID setting, the data is non-uniformly partitioned among clients, resulting in each client possessing dataset of varying size. In such cases, clients with smaller local datasets exhibited slightly higher validation loss fluctuations. However, the Dice scores across clients remained within ± 0.015 of the global mean, confirming consistent performance and strong generalizability of the proposed model across heterogeneous client data distributions. Overall, the results demonstrate that our proposed model effectively mitigates the impact of data heterogeneity in federated brain tumour segmentation, offering both improved accuracy and greater reliability in non-IID environments.
To simulate computational heterogeneity, each client randomly selected a local epoch count between 1 and 5 in each communication round. Compared to the homogeneous setting where all clients trained for 5 epochs in every round, this variation led to slower convergence and minor fluctuations in performance. The final segmentation accuracy showed a slight decline, with Dice scores reducing from 0.94 (homogeneous) to approximately 0.91–0.93 in the heterogeneous case. This highlights the sensitivity of federated optimization to uneven computational loads across clients.
Impact of differential privacy and secure aggregation
In the federated training process, privacy was reinforced through the combined use of differential privacy and secure aggregation. The global model was trained locally by each client on its private dataset. Before updates were transmitted to the central server, differential privacy was applied by clipping gradients and adding calibrated Gaussian noise, ensuring that individual client data could not be inferred from the shared updates. Following this, secure aggregation was employed by masking each noisy update, allowing only the aggregated result of all clients’ contributions to be accessed by the server. At no point were individual updates exposed or examined. Through this dual-layered privacy mechanism, sensitive client data was effectively protected, making the approach well-suited for applications involving confidential information, such as medical image analysis. Incorporating differential privacy resulted in a moderate reduction in segmentation accuracy (Dice score dropped by ~ 4% compared to baseline), likely due to the noise injected into gradients. When combined with secure aggregation, no further degradation in accuracy was observed, indicating that SA does not interfere with model convergence quality, as it only protects communication-level confidentiality. Differential privacy introduced noticeable training instability in the early rounds due to noise, requiring approximately 30% more FL rounds to reach convergence compared to the baseline. Secure aggregation, being computational but not algorithmic in nature, did not significantly affect the number of rounds to convergence. Applying DP added negligible overhead on client-side computation (Gaussian noise addition is a lightweight operation). However, secure aggregation significantly increased computation overhead due to key exchange and encryption steps. Specifically, the training time per round increased by ~ 1.7 × in the DP + SA setting compared to the baseline FedAvg.
Ablation study
In our proposed image segmentation framework, we adopted a dual-encoder architecture that brings together EfficientNet and the Swin Transformer to balance efficiency and representational power—both critical in federated learning settings. EfficientNet was chosen for its lightweight design and ability to deliver strong performance with fewer parameters, making it ideal for client devices with limited computational resources. Complementing this, the Swin Transformer contributes by modelling both local and global features using a hierarchical attention mechanism that is particularly effective in segmentation tasks. To further enhance learning efficiency and generalization, both encoders are initialized with pre-trained weights from ImageNet. This use of transfer learning proved especially valuable in federated scenarios, where data heterogeneity and limited local data are common challenges. To understand the impact of these pre-trained components, an ablation study is carried out. The proposed model with both encoders pre-trained achieved a Dice score of 0.94 and an IoU of 0.87. When EfficientNet was trained from scratch, the Dice score dropped to 0.91; removing Swin pre-training resulted in a further decline to 0.90. Training both encoders from scratch led to the lowest performance, with a Dice score of 0.88 and IoU of 0.82. These results highlight the critical role of pre-trained weights, contributing nearly a 7% improvement in segmentation accuracy and noticeably faster convergence. Overall, this combination of EfficientNet and Swin Transformer backed by transfer learning proved to be highly effective in federated image segmentation.
Training and communication efficiency
Despite incorporating more complex model components, the proposed architecture remained efficient in terms of total training time in federated setups. Our proposed model which has a larger parameter count (~ 25 M), incorporates a dual-encoder structure that benefits from pretrained initialization, enabling rapid convergence with fewer communication rounds—ultimately reducing total training time. Although the parameter count exceeded simpler baseline models (U-Net: 7.8 M; UNet + + : 9.0 M; ResUNet: 8.5 M), the federated training efficiency remained practical. Model parameter updates averaged around 55 MB per client per round compared to approximately 30 MB per round for simpler U-Net architectures, comfortably within typical hospital IT infrastructure limits. Federated per-round training time averaged approximately 17–18 min for the proposed model per federated client, marginally longer than simpler CNN models (around 12–14 min). Nevertheless, the complete federated training process still completed in under a few hours, demonstrating training efficiency and practicality for real-world clinical deployments. EfficientNet’s lightweight convolutional backbone significantly contributed to reducing computational load, whereas transformer-based global context modelling ensured fewer rounds for model convergence compared to baseline CNNs. In real-world deployments of federated learning, communication challenges such as latency and packet loss can have a notable impact on model performance and training efficiency. When clients are distributed across different locations or operate in low-bandwidth environments, high latency can slow down the synchronization of model updates. This often results in delayed or stale updates, which may reduce the overall effectiveness of global aggregation. In some cases, slower clients may even be dropped from participation, introducing potential bias into the model. Packet loss adds another layer of complexity—missing or corrupted transmissions can lead to incomplete updates or force repeated communication attempts, further increasing training time and communication overhead. While our current implementation assumes stable and reliable connections, we recognize that such assumptions may not always hold true in practice. As a part of future work, we aim to incorporate network latency simulation, further addressing potential communication disruptions like packet loss to improve robustness in real-world federated settings.
Scalability and adaptability
Scalability assessments involving increased number of federated clients confirmed that federated learning remains robust and scalable for realistic multi-institutional deployment. However, increased data heterogeneity across sites naturally demanded additional training rounds to achieve convergence and optimal accuracy. Increasing number of clients to 7 and 10 leads to a marginal difference in performance due to reduced data per client and increased update variance. Under an IID setup, the Dice score value remains within 0.93–0.935 for 7 clients and 0.91–0.93 for 10 clients, with corresponding IoU values between 0.855–0.865 and 0.83–0.855 respectively. In the non-IID scenario, as the number of clients increases to 7 and 10 under the same heterogeneity conditions, the segmentation performance moderately drops due to increased data fragmentation. Dice score value ranges between 0.865–0.88 for 7 clients and 0.84–0.865 for 10 clients, with corresponding IoU values in the range of 0.78–0.80 and 0.75–0.78, respectively. Future implementations can explore Personalized Federated Learning or site-specific model fine-tuning techniques. These approaches would explicitly mitigate domain shifts and data distribution differences inherent in distributed datasets, thereby improving local model performance and adaptability.
Effectiveness of federated learning in medical segmentation
This study confirms that Federated Learning (FL) is a highly effective and practical framework for training advanced segmentation models in distributed healthcare environments while ensuring patient data privacy. Despite decentralized data distribution, the FL approach demonstrated negligible accuracy loss compared to centralized model training. The results indicate that federated training does not inherently compromise performance, provided models are carefully selected and optimized. Moreover, federated learning, when combined with differential privacy and secure aggregation, enables robust, privacy-preserving training across institutions with no significant performance degradation, making it well-suited for secure and collaborative medical AI development.
Conclusion and future work
This research introduced a dual encoder-decoder segmentation model combining EfficientNet and Swin Transformer as encoders with BASNet decoder and MaskFormer as decoders, demonstrating exceptional accuracy in brain tumour segmentation tasks. The proposed hybrid dual encoder-decoder model achieved Dice and IoU scores of 0.94 and 0.87, respectively, along with superior boundary evaluation metrics (HD95 = 1.61, ASSD = 1.12), demonstrating its robustness, precision, and potential clinical applicability. The hybrid transformer-CNN architecture enabled effective extraction of both local and global spatial features, particularly improving boundary delineation through the boundary-aware BASNet decoder, and mask-level classification refinement with MaskFormer. Furthermore, federated learning combined with differential privacy and secure aggregation demonstrated robust, privacy-preserving training capabilities without significant accuracy degradation, underscoring its suitability for secure, multi-institutional collaboration in medical AI. Future directions will explore advanced hyperparameter optimization to further enhance model accuracy and computational efficiency. Additionally, the research aims to integrate multi-modal MRI data (T1, T2, FLAIR) to enhance segmentation reliability in diverse clinical contexts. Efforts will also focus on real-time inference optimization to facilitate practical deployment within hospital networks, and exploring personalized federated learning to effectively address data heterogeneity among institutions, thus progressing towards clinically reliable, privacy-preserving AI solutions in medical imaging. Future extensions may also incorporate network latency simulation and dynamic client partitioning to better reflect real-world federated environments and communication constraints.
Data availability
The datasets generated and/or analysed during the current study are available in the Kaggle repository, https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation. https://www.kaggle.com/datasets/nikhilroxtomar/brain-tumor-segmentation
References
Bakator, M. & Radosav, D. Deep learning and medical diagnosis: A review of literature. Multimodal Technol. Interact. (2018).
Yadav, S. S. & Jadhav, S. M. Deep convolutional neural network based medical image classification for disease diagnosis. J. Big Data 113 (2019).
Minaee, S. et al. Image segmentation using deep learning: A survey. IEEE Trans. Pattern Anal. Mach. Intell. 44(7), 3523–3542 (2022).
Jiang, B., et. al. Deep learning for brain tumour segmentation in multimodal MRI images: A review of methods and advances, Image Vis. Comput. 156 (2025).
Bandyk, M. G., et al. MRI and CT bladder segmentation from classical to deep learning based approaches: Current limitations and lessons. Comput. Biol. Med. 134 (2021).
Yang, Q., Liu, Y., Chen, T. & Tong, Y. Federated machine learning: Concept and applications. ACM Trans. Intell. Syst. Technol. (TIST), 1–19 (2019).
Ahamed, M. F. et al. A review on brain tumour segmentation based on deep learning methods with federated learning techniques. Comput. Med. Imaging Graph. 110 (2023).
Tan, M. et al. EfficientNet: Rethinking model scaling for convolutional neural networks. arxiv.org/abs/1905.11946 (2019).
Liu, et al. Swin transformer: Hierarchical vision transformer using shifted windows. Proceedings of the IEEE/CVF International Conference on Computer Vision (ICCV) (2021).
Qin, X., et al., Boundary-aware segmentation network for mobile and web applications. arxiv.org/abs/2101.04704 (2021).
Cheng, B., Schwing, A. G. & Kirillov, A. Per-pixel classification is not all you need for semantic segmentation, arxiv.org/abs/2107.06278 (2021).
Ronneberger, O. et al. U-Net: Convolutional networks for biomedical image segmentation. arXiv: 1505.04597 (2015).
Zhou, Z., et al. UNet++: A nested U-Net architecture for medical image segmentation. arxiv.org/abs/1807.10165, 2018.
Diakogiannis, F. I., et al. ResUNet-a: A deep learning framework for semantic segmentation of remotely sensed data. arxiv.org/abs/1904.00592 (2019).
Guan, H., Yap, P.-T., Bozoki, A. & Liu, M. Federated learning for medical image analysis: A survey. Pattern Recognit. 151 (2024).
Sandhu, S. S., Gorji, H. T., Tavakolian, P., Tavakolian, K. & Akhbardeh, A. Medical imaging applications of federated learning. Diagnostics (2023).
Yang, L., He, J., Fu, Y. & Luo, Z. Federated learning for medical imaging segmentation via dynamic aggregation on non-IID data silos. Electronics (2023).
Azad, R. et al. Medical image segmentation review: The success of U-Net. IEEE Trans. Pattern Anal. Mach. Intell. 46, 10076–10095 (2024).
Micallef, N., Seychell, D. & Bajada, C. J. Exploring the U-Net++ model for automatic brain tumour segmentation. IEEE Access 9, 125523–125539 (2021).
Rahman, H., Bukht, T. F. N., Imran, A., Tariq, J., Tu, S. & Alzahrani, A. A deep learning approach for liver and tumour segmentation in CT images Using ResUNet. Bioengineering (2022).
Yao, W., Bai, J., Liao, W. et al. From CNN to transformer: A review of medical image segmentation models. J. Digit. Imaging. Inform. Med. (2024).
Siva, R., et al. Polyp tumour segmentation using basnet. Grenze Int. J. Eng. Technol. (GIJET, (2024).
Xiao, H., Li, L., Liu, Q., Zhu, X. & Zhang, Q. Transformers in medical image segmentation: A review. Biomed. Signal Process. Control (2023).
Wei, C., Ren, S., Guo, K., Hu, H. & Liang, J. High-resolution swin transformer for automatic medical image segmentation. Sensors (2023).
Lin, S. & Lin, C. Brain tumour segmentation using U-Net in conjunction with EfficientNet. PeerJ Comput. Sci. (2024).
Kamnitsas, K. et al. DeepMedic for brain tumour segmentation, Lecture Notes in Computer Science, vol 10154, Springer (2016).
Pranjal Agrawal, Nitish Katal, Nishtha Hooda, Segmentation and classification of brain tumour using 3D-UNet deep neural networks, Int. J. Cogn. Comput. Eng. (2022).
Milletari, F., et al. V-Net: Fully convolutional neural networks for volumetric medical image segmentation. In: Fourth International Conference on 3D Vision (2016).
Wang, W. et al. TransBTS: Multimodal brain tumour segmentation using transformer (Springer, 2021).
Hatamizadeh, A., et. al. UNETR: Transformers for 3D medical image segmentation. IEEE/CVF Winter Conference on Applications of Computer Vision (WACV) (2022).
Li, Z., Li, D., Xu, C., Wang, W., Hong, Q., Li, Q. & Tian, J. Tfcns: A cnn-transformer hybrid network for medical image segmentation. In International conference on artificial neural networks (pp. 781–792). Cham: Springer Nature (2022).
Guan, H., Yap, P. T., Bozoki, A. & Liu, M. Federated learning for medical image analysis: A survey. Pattern Recognit. 110424 (2024).
Jin, Q., Cui, H., Wang, J., Sun, C., He, Y., Xuan, P. & Su, R. Iterative pseudo-labeling based adaptive copy-paste supervision for semi-supervised tumour segmentation. Knowl. Based Syst. 113785 (2025).
Hernandez-Cruz, N., Saha, P., Sarker, M. M. K. & Noble, J. A. Review of federated learning and machine learning-based methods for medical image analysis. Big Data Cogn. Comput. 8(9), 99 (2024).
Su, R., Xiao, J., Cui, H., Xuan, P., Feng, X., Wei, L. & Jin, Q. (2024). MSKI-Net: Towards modality-specific knowledge interaction for glioma survival prediction. In 2024 IEEE International Conference on Bioinformatics and Biomedicine (BIBM) (pp. 2438–2445). IEEE.
Jin, Q., Cui, H., Sun, C., Song, Y., Zheng, J., Cao, L., & Su, R. Inter-and intra-uncertainty-based feature aggregation model for semi-supervised histopathology image segmentation. Expert Syst. Appl. 238, 122093 (2024).
McMahan, B. et al. Communication-efficient learning of deep networks from decentralized data 1273–1282 (Artificial intelligence and statistics, 2017).
Umirzakova, S., Muksimova, S., Baltayev, J. & Cho, Y. I. Force map-enhanced segmentation of a lightweight model for the early detection of cervical cancer. Diagnostics 15(5) (2025).
https://www.kaggle.com/datasets/mateuszbuda/lgg-mri-segmentation.
Elbachir, Y. M., et al. Federated learning for multi-institutional on 3D brain tumour segmentation. International Conference on Pattern Analysis and Intelligent Systems (PAIS), 2024.
Yi, L., et al. SU-Net: An efficient encoder-decoder model of federated learning for brain tumour segmentation. Lecture Notes in Computer Science, vol 12396. Springer (2020).
Vaibhav, S. et al. Multiencoder-based federated intelligent deep learning model for brain tumour segmentation. Int. J. Imag. Syst. Technol. (2023).
https://www.kaggle.com/datasets/nikhilroxtomar/brain-tumour-segmentation
Funding
This work did not receive any specific grant from any funding agency.
Author information
Authors and Affiliations
Contributions
K. Narmadha contributed to the conceptualization, methodology, and writing of the manuscript, while P. Varalakshmi provided supervision and conducted the review.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Narmadha, K., Varalakshmi, P. Enhanced brain tumour segmentation using a hybrid dual encoder–decoder model in federated learning. Sci Rep 15, 34416 (2025). https://doi.org/10.1038/s41598-025-17432-0
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-17432-0