Abstract
Few effective treatments have been developed for intractable pancreatic exocrine disorders due to the lack of suitable disease models using human cells. Pancreatic acinar cells differentiated from human induced pluripotent stem cells (hiPSCs) have the potential to solve this issue. In this study, we aimed to elucidate the developmental mechanisms of pancreatic exocrine acinar lineages to establish a directed differentiation method for pancreatic acinar cells from hiPSCs. hiPSC-derived pancreatic endoderm cells were spontaneously differentiated into both pancreatic exocrine and endocrine tissues by implantation into the renal subcapsular space of NOD/SCID mice. Single-cell RNA-seq analysis of the retrieved grafts confirmed the differentiation of pancreatic acinar lineage cells and identified REG4 as a candidate marker for pancreatic acinar progenitor cells. Furthermore, differential gene expression analysis revealed upregulated pathways, including cAMP-related signals, involved in the differentiation of hiPSC-derived pancreatic acinar lineage cells in vivo, and we found that a cAMP activator, forskolin, facilitates the differentiation from hiPSC-derived pancreatic endoderm into pancreatic acinar progenitor cells in our in vitro differentiation culture. Therefore, this platform contributes to our understanding of the developmental mechanisms of pancreatic acinar lineage cells and the establishment of differentiation methods for acinar cells from hiPSCs.
Similar content being viewed by others
Introduction
The pancreas is mainly composed of acinar cells, duct cells, and islets of Langerhans and is involved with two principal exocrine and endocrine functions1. Acinar cells constitute most of the pancreas and are responsible for exocrine functions through the secretion of pancreatic juice containing digestive enzymes into the duodenum through the pancreatic duct2. Acinar cells synthesize various digestive enzymes, such as amylase, trypsin, and lipase, to digest carbohydrates, proteins, and fats, respectively2. Whereas the nucleus and endoplasmic reticulum (ER) for synthesizing digestive enzymes and proteins are localized on the basal side of acinar cells, zymogen granules for storage and secretion of digestive enzymes are on the apical side2. Several ion transporters are expressed in pancreatic duct cells, which serve the vital function of bicarbonate ion and water secretion3. Cells located at the junction of the acinus and duct are called centroacinar cells. Although these cells have the characteristics of duct cells, they may also function as pancreatic progenitor cells involved in regeneration4. On the other hand, the islets of Langerhans, which exert endocrine functions, occupy only about 1% of the total pancreas weight and consist of multiple endocrine cells, such as α, β, and δ cells that secrete different hormones5.
Diabetes is a typical endocrine disease, while common exocrine disorders include pancreatic cancers and pancreatitis, as well as congenital diseases, such as cystic fibrosis and hereditary pancreatitis. Detailed pathophysiological mechanisms of intractable exocrine pancreatic disorders remain scant, with few effective treatments having been developed, partly due to the lack of suitable disease models. While mice are mainly used as experimental animals, species differences often act as roadblocks to translation. Primary cultured cells derived from pancreatic cancer patients may behave differently once removed from the body6. In addition, isolated human pancreatic acinar cells undergo apoptosis rapidly during culturing, with some cells transdifferentiating spontaneously into mesenchymal and duct cells7. As isolated acinar cells in mice and rats also easily differentiate into duct-like cells8, difficulties associated with long-term acinar cell cultures limit their use6. Therefore, suitable disease models that reproduce the pathophysiological conditions of human exocrine pancreatic disorders are required.
Since pluripotent stem cells, such as embryonic stem cells (ESCs) and induced pluripotent stem cells (iPSCs), were established, substantial progress has been made in regenerative therapies, drug discovery, and disease modeling using them, including in the field of pancreatic diseases9,10,11,12. However, although there has been a gradual increase in reports on the generation of pancreatic acinar lineage cells from ESCs/iPSCs, the induction of acinar cells has yet to be fully established11,12,13,14,15,16,17,18,19,20. Efficient induction methods recapitulating the developmental process for differentiating human ESCs/iPSCs into pancreatic acinar lineage cells are necessary to elucidate the pathophysiology of exocrine pancreatic disorders and develop novel therapeutic agents.
After differentiating into embryonic germ layers via the anterior primitive streak, the definitive endoderm segments into the foregut, midgut, and hindgut in mice21. Subsequently, monolayer epithelial cells protrude from the posterior foregut expressing pancreatic and duodenal homeobox 1 (Pdx1) thicken and further differentiate to form pancreatic buds at embryonic day (E) 9.5, which express both pancreas-associated transcription factor 1a (Ptf1a) and NK6 homeobox 1 (Nkx6.1) and are crucial for the formation of pancreatic epithelia during early stages21,22,23,24,25,26. The primary transition in pancreatic development during E9.5-12.5 is an active and proliferative phase in which monolayer pancreatic buds become stratified squamous epithelium and further form multiple microlumens inside the epithelia21,25,26. At the end of the primary transition, the pancreatic epithelia begin dividing into the tip and trunk domains. Whereas multipotent progenitor cells (MPCs) are located at the tip domain, bipotent progenitor cells for endocrine and duct lineages comprise the trunk domain. The fate of each pancreatic lineage is decided during the secondary transition at E13.521,25,26. After E13.5, MPCs that dominate the tip domain and express marker genes Ptf1a and carboxypeptidase A1 (Cpa1), but not Nkx6.1, eventually differentiate into pancreatic acinar cells21,25,27. However, the differentiation mechanisms of acinar lineages in pancreatic development have not been fully elucidated, and the detailed expression pattern of pancreatic acinar markers, including those associated with digestive enzymes, remains unknown in both mice and humans.
In this study, we have created a differentiation system to form both exocrine and endocrine pancreatic lineages by combining in vitro and in vivo cultures of pancreatic endoderm cells derived from human iPSCs (hiPSCs). Single-cell RNA sequencing (scRNA-seq) of human pancreatic tissues revealed the differentiation mechanisms and temporal gene expression profiles of pancreatic acinar lineages during the differentiation process, which will contribute to the establishment of a selective induction method for pancreatic acinar lineage cells from hiPSCs.
Results
hiPSC-derived pancreatic endoderm cells differentiate into early embryonic pancreatic tissues in vivo
According to our previously reported method28, the hiPSC line 585A129 was differentiated into PDX1+NKX6.1+ pancreatic endoderm cells, which can differentiate into both pancreatic exocrine and endocrine lineage cells17,30 (Fig. 1A). While numerous studies have reported the generation of pancreatic endocrine lineages and β cells for diabetes treatment31,32,33,34,35,36,37,38,39, differentiation protocols for inducing hiPSC-derived pancreatic endoderm cells into pancreatic acinar lineage cells in vitro have not been fully established11,12,13,14,15,16,17,18,19,20. Thus, referring to previous studies reporting the implantation of pancreatic progenitor cells into immunodeficient mice that results in the differentiation of functional β cells in vivo28,31,34,38,39, we attempted to utilize the in vivo environment to facilitate the differentiation of hiPSC-derived pancreatic endoderm cells into pancreatic acinar lineages by implanting them into the renal subcapsular spaces of immunodeficient NOD.CB17-Prkdcscid/J (NOD/SCID) mice (Fig. 1A). We used aggregate cultures at Stage 4 to efficiently generate pancreatic endoderm as a previous study reported that the differentiation into pancreatic epithelia is facilitated by increasing cell–cell adhesion40. In addition, we assumed that cells in planar cultures may easily break apart and that cellular aggregates are easier to handle and would undergo less cell loss after being implanted into mouse renal subcapsules than cells grown in planar cultures. Immunostaining of pancreatic endoderm cell aggregates prepared for transplantation confirmed PDX1+NKX6.1+ cells in aggregates (Fig. 1B). Flow cytometry analysis also showed that PDX1+ and PDX1+NKX6.1+ cells accounted for approximately 90% and 35% of the aggregates, respectively (Fig. 1C).
In vivo differentiation of hiPSC-derived pancreatic endoderm cells mimics early pancreatic development. (A) A schematic representation of induction of hiPSC-derived pancreatic endoderm cells and implantation into immunodeficient mice. A, activin A; C, CHIR99021; Y, Y-27632; K, KGF; N, Noggin; KC, KAAD-cyclopamine; TT, TTNPB; E, EGF. (B) Immunostaining of hiPSC-derived cells on Stage 4 Day 6 (total day 17) before implantation for PDX1 (green), NKX6.1 (red), and nuclei (blue). (C) Induction efficiency of hiPSC-derived PDX1+NKX6.1+ pancreatic endoderm cells on Stage 4 Day 6 (total day 17) before implantation, as evaluated by flow cytometry. Data from three independent experiments are presented as mean ± SD (n = 3). (D) Macroscopic images of grafts under the left renal capsule of a NOD/SCID mouse sacrificed 30 days after implantation (upper panel) and following isolation from the host mouse kidney (lower panel). Blue arrows indicate grafts. (E) Immunostaining of grafts on the indicated days after implantation for CPA1 (green) and NKX6.1 (red; upper panels), Mucin 1 (green) and E-CADHERIN (red; middle panels), and PRSS1 (green) and nuclei (blue; lower panels). Note that background signals were found in the host mouse kidney (MK). Scale bars: 100 μm in (B) and (E) and 5 mm in (D).
In kidneys collected by sacrificing NOD/SCID mice, grafts were confirmed as white protuberances under the renal capsule (Fig. 1D, blue arrows). Grafts were collected and histologically examined on days 0, 7, 14, 21, and 30 after implantation to evaluate the differentiation states (Figs. 1E, S1). Hematoxylin–Eosin (HE) stained sections at the central portions of grafts on days 0, 7, 14, 21, and 30 suggest that the cell density was higher on day 14 than on day 7. (Fig. S1A). By immunostaining, grafts exhibited disorganized structures on days 0 and 7 after implantation, with structural changes observed after 14 days and the cell polarity marker Mucin 1 more clearly expressed in the microlumen-like structures after 21 days, indicating the reproduction of the primary transition (Fig. 1E, S1B). After 30 days, branching structures were observed, in which tip-like cell populations expressing acinar markers, CPA1, PTF1A, and serine protease 1 (PRSS1), were separately developed from trunk-like cell populations expressing NKX6.1 and sex-determining region Y box 9 (SOX9) (Fig. 1E, S1B,C). In addition, cell populations expressing endocrine markers, such as INSULIN (INS) and GLUCAGON (GCG), were observed beside these branched structures (Fig. S1C). The findings on day 30 might mimic the process at the end of the primary transition to the secondary transition (Fig. 1E, S1B,C). These results indicate that grafts correspond to early embryonic pancreatic tissues and could be used to examine the differentiation process of pancreatic acinar lineages.
scRNA-seq analysis reveals differentiation dynamics during the transition from pancreatic endoderm cells to acinar lineage cells
Having observed the differentiation of hiPSC-derived pancreatic endoderm cells into pancreatic epithelia in vivo, we next examined the transition from pancreatic endoderm cells to acinar lineage cells. Structural changes were observed on day 14 after implantation, and the appearance of microlumen-like structures and the expression of PRSS1 was observed after 21 days (Fig. 1E). Based on these findings, we performed scRNA-seq at three timepoints: days 0 (before implantation), 11, and 25 after implantation. First, cellular aggregates formed from hiPSC-derived PDX1+NKX6.1+ pancreatic endoderm cells were used as day 0 samples. Then, the aggregates were implanted into renal subcapsules of NOD/SCID mice, and the grafts on days 11 and 25 after implantation were collected for analysis. After carefully isolating the grafts from renal parenchyma and capsule of host mice, contaminated mouse cells, including blood cells, were eliminated by flow cytometry sorting using mouse-specific CD45, H-2, and TER-119 antibodies and human-specific CD147 and CD298 antibodies41,42,43 (Fig. S2). The number of cells before and after sorting on days 0, 11, and 25 was 1.44 × 104, 12.5 × 104, and 20.2 × 104 and 0.25 × 104, 0.60 × 104, and 1.15 × 104, respectively. Therefore, the purification efficiency was 17.4%, 4.8%, and 5.7% for samples from days 0, 11, and 25, respectively (Fig. S2). Since we found that diverse tissues were organized over time (Fig. 1E) and assumed that more cells would be needed to capture the increased diversity at later time points, we collected 2500, 6000, and 11,500 cells on days 0, 11, and 25, respectively, totaling 20,000 human cells for scRNA-seq. From these 20,000 cells, 8222 cells were detected in micro-wells of the cartridge, of which 8189 cells were single-tagged. After quality control (QC; see Materials and Methods), we retained 6455 cells for subsequent analysis (Fig. S3).
To visualize cell populations from the randomly plotted cells by scRNA-seq analysis, we set the optimal dimension and resolution by referring to the elbow plot (dimension = 63, resolution = 0.8). As a result, a total of 6455 cells consisting of 1236, 2180, and 3039 cells on days 0, 11, and 25, respectively, were classified into fourteen clusters using the t-distributed Stochastic Neighbor Embedding (tSNE) method (Fig. S4A). As indicated by the heatmap that shows highly expressed genes representative of each cluster, pancreatic precursor cells (cluster 2), pancreatic exocrine cells (clusters 8 and 10), and pancreatic endocrine cells (clusters 0, 1, and 12) were roughly classified as pancreas-related cell populations (Fig. S4B, Supplementary Table 1). Although the majority of cells collected on day 0 were grouped into clusters 2 and 5 (Fig. S4A, right panel), cluster 5 was classified as a duodenal population due to the expression of enteroendocrine cell markers, NUROG3 and CCK44 and thus was excluded from subsequent analyses of pancreas-related cell populations (Fig. S4B, Supplementary Table 1).
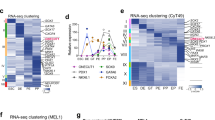
Next, to better understand the differentiation dynamics of pancreatic lineages, only the six clusters annotated as pancreatic cells (clusters 0, 1, 2, 8, 10, and 12) were reclustered. The optimal dimension and resolution were set by referring to the elbow plot (dimension = 23, resolution = 0.8), with a total of 3604 cells consisting of 820, 2020, and 764 cells on days 0, 11, and 25, respectively, classified into sixteen clusters using the tSNE method (Fig. 2A). We detected the expression of pancreatic endoderm markers, PDX1, NKX6.1, PTF1A, and SOX9, pancreatic endocrine cell markers, CHGA, INS, and GCG, and pancreatic acinar cell markers, PRSS1, SPINK1, CPA1, CPA2, CPB1, and CTRC, in different clusters (Fig. 2B). The expression levels of PDX1, NKX6.1, PTF1A, SOX9, CHGA, INS, PRSS1, and CPA1 in each cluster are also quantified as violin plots in Fig. S5A. Most INS- or GCG-expressing cells appeared on day 11 and were grouped into clusters 0 and 1 (Fig. 2A,B). We created feature plots to visualize the expression of INS and GCG in each single cell on days 0, 11, and 25 (Fig. S6). The plots show that whereas many cells expressed both INS and GCG on day 11, the number of cells expressing only INS or GCG was higher than those expressing both by day 25, indicating cells transitioned from poly- to mono-hormonal over time.
Transition dynamics of differentiation states from pancreatic endoderm cells to pancreatic acinar cells revealed by scRNA-seq. (A) Clustering of cells using the tSNE method (left panel) and distribution of cells on the indicated days (right panels). Only the six clusters annotated as pancreatic cells (clusters 0, 1, 2, 8, 10, and 12) in Fig. S4 (n = 96 for pancreatic endoderm aggregates, n = 8 for day 11 grafts, and n = 5 for day 25 grafts) were reclustered. (B) Distribution of cells expressing marker genes for pancreatic endoderm cells (PDX1, NKX6.1, PTF1A, and SOX9), pancreatic endocrine cells (CHGA, INS, and GCG), and pancreatic acinar cells (PRSS1, SPINK1, CPA1, CPA2, CPB1, and CTRC). (C) The direction and magnitude of differentiation stage transition among cell populations by RNA velocity. The lower boxed panel shows the velocity map by selecting clusters 5, 11, and 12. (D) Visualizing the differentiation stage transition from pancreatic endoderm to acinar lineage cells (upper panel) and changes in the expression of pancreatic acinar marker genes during the differentiation process (lower panel) by pseudotime analysis.
Differentiation dynamics were examined by RNA velocity analysis using scVelo, which predicts the fate of individual cells by focusing on the ratio of newly transcribed precursor-mRNAs (unspliced) to mature mRNAs (spliced) and indicates the direction and magnitude of changes of cellular state45,46 (Fig. 2C, upper panel). We also constructed the velocity map focused on clusters 5, 11, and 12 and confirmed that the streamlines of the velocity vector field are downward from cluster 12 to cluster 11 and rightward from 11 to 5, which indicates that transitions from cluster 12 to cluster 5 proceed through cluster 11 (Fig. 2C, lower panel). Although cluster 2 was categorized initially as a pancreatic precursor cell population before reclustering, the procedure reoriented these cells into new clusters: pancreatic endoderm cells (cluster 12: as determined by gene expression distribution and RNA velocity), acinar cells (cluster 5: higher expression of pancreatic acinar genes SPINK1, CPA1, CPA2, and CTRC than cluster 11), and acinar progenitor cells, which are cells at the intermediate stage between pancreatic endoderm and acinar cells (cluster 11: higher expression of SPINK1, PTF1A, CPA1, and CPA2 than cluster 12; Fig. 2A–C).
To characterize the differentiation dynamics of pancreatic acinar lineage cells, we performed pseudo-temporal ordering analysis by partition-based graph abstraction (PAGA)47 after narrowing down the clusters to 5, 11, and 12 (Figs. 2D, S5B). As a result, pseudotime analysis enabled the visualization of pancreatic acinar gene expression dynamics during the differentiation process from hiPSCs to pancreatic acinar lineage cells, showing the expression levels of CPA2, CTRC, and SPINK1 to peak relatively earlier than CPA1 and PRSS1 (Fig. 2D). We then constructed feature plots showing the expression of PRSS1, SPINK1, CTRC, CPA1, CPA2, and CPB1 along weeks post-conception and across cell types including early tip, tip, and acinar stages by reanalyzing a previously reported human fetal pancreas dataset48 (Fig. S7A–D). These plots revealed that while CPA2 and SPINK1 are expressed during relatively early developmental stages of acinar lineages, PRSS1, CTRC, and CPB1 are expressed in later stages (Fig. S7D), consistent with our results, thus indicating that our hiPSC-based acinar differentiation model reproduces aspects of acinar lineage development in human embryos. Therefore, we successfully established a system for analyzing human acinar lineage differentiation by facilitating in vivo differentiation of hiPSC-derived pancreatic endoderm cells generated in vitro.
REG4 is a candidate marker for pancreatic acinar progenitor cells
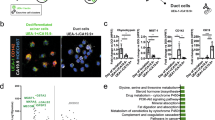
Next, we focused on pancreatic acinar progenitor cells to identify specific markers for this subpopulation for more efficient generation of pancreatic acinar cells from hiPSCs. To examine the expression of specific genes in acinar progenitor cells, we first extracted a group of differentially expressed genes (DEGs, |log2FC|> 1, P value< 0.01) common between acinar progenitor cells (cluster 11) and pancreatic endoderm cells (cluster 12), acinar cells (cluster 5), endocrine cells (cluster 2), or duct cells (cluster 3; Supplementary Table 2). Among the top 100 genes whose expression levels were significantly higher in acinar progenitor cells (cluster 11) than in other clusters evaluated using DotPlot, 34 genes were selected based on significantly lower expression levels in endocrine cells (cluster 2) and duct cells (cluster 3) than in acinar cells (cluster 5) (Figs. 3A, S5C, Supplementary Tables 2, 3). SPINK and CTRC were excluded from this list because their expression levels were lower than the top 100 genes. Nevertheless, their expression levels were higher in cluster 5 than in cluster 11 (Fig. 2A,B). Next, we compared gene expression between pancreatic endoderm cells (cluster 12) and acinar progenitor cells (cluster 11) and found 88 and 88 significantly up- and downregulated genes in acinar progenitor cells, respectively (Fig. 3B, left panel; Supplementary Table 3). Then, we compared gene expression between acinar progenitor cells (cluster 11) and acinar cells (cluster 5; Fig. 3B, middle panel; Supplementary Table 3). Most genes that continued to be upregulated from pancreatic endoderm cells to acinar cells via acinar progenitor cells were well-known pancreatic digestive enzyme-related genes (Supplementary Table 3). Among the genes whose expression levels were significantly higher in acinar progenitor cells (cluster 11) than in pancreatic endoderm cells (cluster 12; Fig. 3B, left panel), only four genes, REG4, BGN, COL1A2, and HAPLN1, were significantly upregulated in acinar progenitor cells than in acinar cells (cluster 5; Fig. 3B, middle and right panels). Since they were also among the 34 previously identified candidates (Fig. 3A, Supplementary Table 2), we continued to focus on these four genes. The expression distribution in clusters 2, 3, 5, 11, and 12 showed that COL1A2 and BGN were broadly expressed not only in acinar progenitor cells but also in pancreatic endoderm cells, duct cells, and endocrine lineage clusters (Fig. 3C). We then examined the expression of REG4 and HAPLN1 by reanalyzing previously reported expression profiles of human adult pancreas49,50,51,52,53,54 (Fig. S8) to find that while HAPLN1 was broadly expressed in multiple pancreatic cell types, REG4 expression was restricted to parts of acinar cells and gamma cells of islets in human adult pancreas (Fig. S8F, G). Therefore, we focused on REG4 as a candidate acinar progenitor cell marker. To clearly show the differential expression of REG4 and PRSS1 among pancreatic endoderm, acinar progenitor cells, and acinar cells, we compared DEGs (|log2FC|> 1, P value< 0.01, Supplementary Table 3), which showed that REG4 expression was substantially higher in acinar progenitor cells than in pancreatic endoderm cells and more than twice as high in acinar progenitor cells as in acinar cells (Fig. 3D). In contrast, the expression of PRSS1 was higher in acinar progenitor cells than in pancreatic endoderm cells but around twice as high in acinar cells than in acinar progenitor cells (Fig. 3D). These data suggest that REG4 is a potential marker for pancreatic acinar progenitor cells differentiated from hiPSCs.
Identification of novel marker candidates for pancreatic acinar progenitor cells. (A) Dot plots of DEGs (|log2FC|> 1, P value < 0.01) among clusters of pancreatic endoderm cells (cluster 12), acinar progenitor cells (cluster 11), acinar cells (cluster 5), endocrine cells (cluster 2), and duct cells (cluster 3). (B) Volcano plots of DEGs (|log2FC|> 1, P value < 0.01) comparing gene expression of pancreatic endoderm cells (cluster 12) versus acinar progenitor cells (cluster 11; left panel) and acinar progenitor cells (cluster 11) versus acinar cells (cluster 5; middle panel) and the Venn diagram (right panel). (C) Distribution of cells expressing the four candidate markers for acinar progenitor cells, REG4, BGN, COL1A2, and HAPLN1. (D) Expression pattern of REG4 and PRSS1 among the 3 clusters based on DEGs (|log2FC|> 1, P value < 0.01).
Then, we constructed feature plots showing the expression of REG4 by reanalyzing the previously reported human fetal pancreas dataset48 (Fig. S7A–C,E,F). The results showed that REG4 expression peaks at post-conception week (PCW)11 and is found in acinar cell type, thus supporting our conclusion that REG4 is a potential marker for pancreatic acinar progenitor cells during embryonic stages in hiPSC differentiation cultures.
In vitro treatment of hiPSC-derived pancreatic endoderm cells with forskolin facilitates the differentiation into pancreatic acinar progenitor cells
To identify factors and signaling pathways facilitating the differentiation of pancreatic acinar progenitor cells, we performed ingenuity pathway analysis (IPA) based on DEGs (|log2FC|> 1, P value< 0.01) obtained by comparing pancreatic endoderm cells (cluster 12) and acinar progenitor cells (cluster 11; Supplementary Table 4). We focused on the top eight candidate factors identified from this analysis: epidermal growth factor (EGF), insulin-like growth factor 1 (IGF1), interleukin-1 beta (IL-1B), hepatocyte growth factor (HGF), 8-(4-chlorophenylthio) adenosine 3′,5'-cyclic monophosphate sodium salt (pCPT-cAMP), forskolin, fibroblast growth factor 2 (FGF2), and lipopolysaccharide (LPS) (Fig. 4A, left panel). In addition, the retinoid X receptor (RXR) agonist bexarotene was added as a candidate factor based on activated pathways indicated by IPA (Fig. 4A, right panel). hiPSC-derived pancreatic endoderm cells on Stage 4 Day 6 (total day 17) were treated with the nine factors for 8 days (Fig. 4B). Three different concentrations of each factor were used to examine the expression of REG4 and PRSS1. We then narrowed down the factors that upregulated the expression of REG4 or PRSS1 by more than 1.5 times compared to the untreated control, which included EGF, IL-1B, HGF, pCPT-cAMP, forskolin, FGF2, and bexarotene (Fig. 4B, red arrows). Next, by adopting the concentration resulting in the highest induction efficiency for each factor, hiPSC-derived pancreatic endoderm cells were treated with the seven candidate factors (Fig. S9). In addition to REG4 and PRSS1, we examined the expression of other pancreatic acinar lineage markers, PTF1A and CPA1. As a result, the expression of these four markers trended higher with forskolin and pCPT-cAMP treatments than the control. Although we also tested combinatorial treatments with forskolin and each of the remaining six candidate factors (i.e., EGF, IL-1B, HGF, pCPT-cAMP, FGF2, and bexarotene), no combinations induced higher expression of REG4 or PRSS1 than treatment with forskolin alone (Fig. S10). Because both forskolin and pCPT-cAMP are involved in the cyclic AMP pathway55,56 and a combination of these two factors did not produce an additive effect compared to forskolin alone, we next examined the effects of forskolin treatment on hiPSC-derived pancreatic endoderm cells.
Induction from hiPSC-derived pancreatic endoderm to pancreatic acinar progenitor cells. (A) Candidate factors (left) and signaling pathways (right) involved in the differentiation of pancreatic acinar progenitor cells, as identified using pathway analysis by IPA. (B) Comparison of effects of 9 candidate differentiation-inducing factors on the expression of REG4 and PRSS1, as evaluated by qRT-PCR. The treatment started on Stage 4 Day 6 (total day 17) and continued for 8 days, ending on total day 25. The following three concentrations were examined for each factor: 10, 50, and 100 ng/mL EGF; 0.2, 1, and 2 ng/mL IL-1B; 10, 50, and 100 ng/mL HGF; 10, 50, and 100 µM pCPT-cAMP (pC); 2, 10, and 20 nM orskolin (FS); 10, 50, and 100 ng/mL FGF2; 0.02, 0.1, and 0.2 nM bexarotene (B); 5, 25, and 50 ng/mL IGF1; and 1, 5, and 10 µg/mL lipopolysaccharide (LPS). PE, pancreatic endoderm. Red arrows indicate factors that upregulated the expression of REG4 or PRSS1 more than 1.5 times compared to the untreated control (Control). Data from three independent experiments are presented as mean ± SD (n = 3). We normalized the expression values against the house keeping gene GAPDH and then against the untreated control. (C) Effects of treatment with 20 nM forskolin on the expression of acinar lineage markers, REG4, PTF1A, CPA1, PRSS1, SPINK1, and CTRC, as evaluated by qRT-PCR. The treatment started on Stage 4 Day 6 (total day 17) and continued for 8 days, ending on total day 25. Graft indicates one graft sample on day 30 after implantation, which differs from grafts used for scRNA-seq. The graft sample on day 30 after implantation was isolated from the host renal parenchyma and capsule, and cells were dispersed in a collagenase/dispase solution for 15 min at 37 °C, dissociated into single cells by gentle pipetting, and used for RNA isolation without sorting for human cells. Data from three independent experiments are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01 by two-tailed Student’s t-test. We normalized the expression values against GAPDH and then against the untreated control (Control). (D) Immunostaining of day 30 grafts from hiPSC-derived pancreatic endoderm cells with or without forskolin treatment for PRSS1 (green) and nuclei (blue; left and upper right panels), and CPA1 (green) and NKX6.1 (red; middle right panels) and quantification of the percentage (%) of PRSS1+ or CPA1+ cells from all cells within the engraftment region (lower right panel). Data from three independent experiments are presented as mean ± SD (n = 3). *p < 0.05, **p < 0.01 by two-tailed Student’s t-test. Scale bars: 300 μm in (D; left panels) and 100 μm in (D; right panels).
We also generated a list of candidate differentiation-inducing factors by comparing DEGs (|log2FC|> 1, P value < 0.01) between pancreatic endoderm and endocrine cells and between pancreatic endoderm and duct cells (Supplementary Table 4). While forskolin was ranked as the 28th factor from the comparison between pancreatic endoderm and acinar progenitor cells, it ranked 75th and 55th in comparisons between pancreatic endoderm and endocrine cells and between pancreatic endoderm and duct cells, respectively. Although these results indicate that forskolin affects the differentiation of pancreatic endocrine and ductal cells as well as acinar progenitor cells, we confirmed by qRT-PCR analysis that forskolin treatment on hiPSC-derived pancreatic endoderm resulted in a decrease in the expression of a pancreatic endocrine marker, NKX6.1, with no substantial changes in the expression of another endocrine marker, NGN3, and ductal markers, KRT19, SOX9, and HNF1B (Fig. S9).
While in vitro forskolin treatment of hiPSC-derived pancreatic endoderm significantly upregulated REG4, PRSS1, and PTF1A, it did not substantially alter the expression of SPINK1, CPA1, and CTRC (Fig. 4C). By immunostaining, PRSS1 and CPA1 proteins were not detected in the cells after forskolin treatment. Then, to characterize forskolin-treated cells, we examined the ability of hiPSC-derived pancreatic endoderm cells, with or without forskolin treatment, to differentiate into acinar lineage cells by implanting them into immunodeficient mice in vivo. We conducted immunostaining for PRSS1 and CPA1 and quantified the number of PRSS1+ and CPA1+ cells in grafts (Fig. 4D). The proportion of PRSS1+ cells was significantly higher in the grafts from pancreatic endoderm cells with forskolin treatment (38.4 ± 5.6%; n = 3) than those without (6.2 ± 1.4%; n = 3; p < 0.01). The proportion of CPA1+ cells was also significantly higher in the grafts from pancreatic endoderm cells with forskolin treatment (8.7 ± 2.9%; n = 3) than those without (1.5 ± 0.4%; n = 3; p < 0.05), indicating that forskolin treatment induces hiPSC-derived pancreatic endoderm cells to differentiate into acinar lineages. These data indicate that while forskolin is a candidate inducer from hiPSC-derived pancreatic endoderm into pancreatic acinar lineage cells, further modification and optimization are required to generate pancreatic acinar cells from hiPSCs in vitro.
Discussion
Several studies have previously identified multiple cell populations by performing single-cell transcriptome analysis of embryonic mouse pancreas and hiPSC-derived pancreatic progenitor and endocrine cells57,58,59,60, thus indicating that in vitro differentiation of hiPSCs is a highly reproducible system for modeling human pancreatic development. The purpose of this study was to understand the mechanistic details of human pancreatic acinar cell differentiation to develop optimal hiPSC differentiation protocols for this cell type so we can ultimately achieve our goal of using hiPSC-based methods to model exocrine pancreatic disorders and design new therapies for their treatment. For this purpose, we first established a system that facilitates the in vivo differentiation of pancreatic endoderm cells induced from hiPSCs in vitro by implanting them into the renal subcapsular spaces of immunodeficient NOD/SCID mice. Although around 90% of hiPSC-derived cells used for implantation were PDX1+, around half of the engrafted cells were identified as non-pancreatic by scRNA-seq analysis (Figs. 1C, S4, Supplementary Table 1). Pdx1 is expressed not only in pancreatic cells but also in other lineages, such as duodenum, enteroendocrine cells, stomach, and bile duct, in mice61. Indeed, our scRNA-seq analysis revealed that in addition to pancreatic cells, grafts contained non-pancreatic lineage cells, such as intestinal cells (Fig. S4, Supplementary Table 1). Starting with a simple structural change 30 days after implantation, branching structures were formed, with tip-like cells expressing acinar markers, CPA1, PTF1A, and PRSS1, and trunk-like cells expressing NKX6.1 and SOX9. Although previous studies reporting on pancreatic differentiation showed the presence of both pancreatic acinar and endocrine lineage cell populations, few studies have managed to capture the process from the end of the primary transition to the secondary transition12,15,16,17,62. Therefore, the system established in the current study mimicking human pancreatic development in vivo enables the detailed mechanistic analysis of acinar lineage differentiation.
Although a previous study reported the expression of several pancreatic digestive enzyme-related genes by pancreatic acinar lineage cells during rat development63 and a recent study performed scRNA-seq on human fetal pancreas48, the expression patterns of pancreatic digestive enzyme-related genes in human embryos have not been examined in detail. In this study, we visualized for the first time the expression changes of pancreatic acinar genes during differentiation from hiPSC-derived pancreatic endoderm cells to acinar cells by pseudotime analysis, further validated by the human fetal pancreas dataset (Fig. S7C,D). Although the rat data were limited to the comparison of four time points (13 days post-coitus (dpc), 20 dpc, newborn, and adult), CPA1 was more highly expressed than CPA2 at 20 dpc63, which is inconsistent with our data (Fig. 2D). In contrast, consistent with our data, trypsin 1, corresponding to PRSS1, was expressed later than CPA163 (Fig. 2D). Thus, our hiPSC-based pancreatic differentiation culture system reproduces aspects of human embryonic acinar lineage development and helps order the developmental stages of pancreatic acinar lineage cells. However, future studies should examine temporal expression profiles during the differentiation process to pancreatic acinar cells in greater detail.
The induction of pancreatic acinar lineage cells from ESCs/iPSCs has not been fully established11,12,13,14,15,16,17,18,19,20. For the differentiation from ESCs/iPSCs to pancreatic acinar cells, including an intermediate acinar progenitor cell stage to induction methods may facilitate an increase in induction efficiency. Therefore, we aimed to identify novel markers for acinar progenitor cells and compared gene expression among pancreatic endoderm, acinar progenitor, and acinar cell populations. We selected four significantly upregulated genes for further analysis. We then focused on REG4, which was previously evaluated as a pancreatic cancer biomarker in humans and a regulator of pancreatic regeneration after pancreatitis in mice64,65. REG1-4 are members of the regenerating gene (REG) family and lectin-like proteins involved in cell proliferation and differentiation of the liver, stomach, intestines, and pancreas in humans66. Although a previous report examined the expression of REG genes in human fetal pancreas from gestational week (GW) 12 to 24 using in situ hybridization and found that mRNA levels of REG genes markedly increased from around GW16 only in acinar cells but not in islets67, their expression before GW12 and the cell types upregulating REG genes were not shown in the study. A more recent study reported REG4 as a lineage-specific target of acinar cell-specific transcription factors by a combined analysis of scRNA-seq and single-cell assay for transposase accessible chromatin sequencing (ATAC-seq) of PCW 4–11 human embryonic pancreas samples48. Moreover, other studies reported that REG4 is expressed in pancreatic acinar cells but not in duct or islet cells of the adult human pancreas68,69. In the current study, we showed that REG4 is a potential marker for acinar progenitor cells differentiated from hiPSCs by analyzing hiPSC-derived pancreatic cells rather than the human fetal pancreas. We also revealed that REG4 expression peaks at PCW11 and is found in acinar cell type by reanalyzing a reported human fetal pancreas dataset48 (Fig. S7C,E,F). In addition, the reanalysis also showed that REG4 is expressed in some acinar cells and gamma cells of islets in the human adult pancreas (Fig. S8C,F,G). Altogether, our data indicate that REG4 is a marker for pancreatic acinar progenitor cells during embryonic stages that may be used to induce the differentiation of pancreatic acinar lineage cells from human ESCs/iPSCs.
Although previous studies used multiple factors, such as activin A, FGF7, all-trans retinoic acid, FGF2, nicotinamide, forskolin, and dexamethasone, and more recent studies used FGF1, A83-01 (a TGFβ inhibitor), WNT1, Y-27632 (an ROCK inhibitor), SKL2001 (a β-catenin agonist), dexamethasone, FGF2, EGF, CHIR99021 (a WNT activator), FGF10, HPI-1 (a hedgehog inhibitor), XMU-MP-1 (a MST1/2 inhibitor), LDN193189 (a BMP inhibitor), CD3254 (an RXR selective agonist), and DBZ (a γ-secretase inhibitor) to generate pancreatic acinar cells from mouse or human ESCs/iPSCs11,12,13,14,15,16,17,18,19,20, inducing factors for acinar lineage cells have not been fully discerned. Therefore, we conducted IPA of DEGs obtained by comparing pancreatic endoderm and acinar progenitor cell populations and revealed candidate factors potentially involved in acinar progenitor cell differentiation. When hiPSC-derived pancreatic endoderm cells were treated with forskolin, REG4, PTF1A, and PRSS1 expression was significantly upregulated. These results are consistent with the previous report that treating pancreatic progenitor cells with three factors, including forskolin, can induce the differentiation into pancreatic acinar cells18 and further support forskolin as an inducing factor for pancreatic acinar lineage cells in hiPSC differentiation cultures. Forskolin is an inducer of intracellular cAMP production and activates pregnane X receptors (PXR) and farnesoid X receptors (FXR)70. Consistently, the PXR/RXR pathway was identified by IPA in this study (Fig. 4A, right panel). However, treating pancreatic endoderm cells induced from hiPSCs by our protocol with forskolin alone could not induce some acinar lineage markers, such as SPINK1 and CTRC (Fig. 4C), indicating that additional factors are required. When comparing the factors discovered in our analysis with previously reported factors, EGF and FGF2 are common and thus expected to be crucial. Although combined treatments with forskolin and either EGF or FGF2 only induced low expression of REG4 and PRSS1 in our study (Fig. S10), future examinations should test various combinations of these three factors, including different concentrations and treatment periods and timings to optimize the differentiation method for acinar lineage cells.
Lastly, we note several limitations of our work. First, we used the combination of PDX1 and NKX6.1 as a maker for multipotent pancreatic endoderm cells for this study. However, during our investigation, it was shown that Glycoprotein 2 (GP2) can act as a specific cell surface marker for human pancreatic endoderm cells and that GP2-enriched PDX1+NKX6.1+ pancreatic endoderm cells have multipotency20,71, thus suggesting further improvements to our in vivo differentiation protocol described here. Second, because we did not conduct lineage tracing experiments for REG4, we could not fully establish the gene as a marker for pancreatic acinar progenitor cells. Third, the exact function of REG4 in pancreatic development remains unclear, although this study showed that it is upregulated in acinar progenitor cells differentiated from hiPSCs. Fourth, for the identification of signaling cues, a strong focus was set in the current study on autocrine signaling to identify upregulated factors in pancreatic endoderm, acinar progenitors, and acinar cells. Paracrine signaling from neighboring cells, such as trunk cells, later endocrine precursor cells, and duct cells, or non-parenchymal cells, including stromal cells, is likely also involved in guiding acinar specification. Future studies with ligand-receptor analysis might also help elucidate specification mechanisms. Fifth, since forskolin treatment alone did not result in sufficient induction of pancreatic acinar cells from hiPSCs in vitro, further studies should optimize the differentiation method for acinar lineage cells by testing various culture conditions, including the combinations of forskolin with other factors used in previous reports11,12,13,14,15,16,17,18,19,20. Forskolin acts by upregulating the second messenger cAMP, which has broad, context-dependent effects on cells55. Further investigation should be performed to investigate the effects of forskolin on the cell cycle, cell activation, and other cellular processes during acinar differentiation. Finally, while a total of only 440 cells was used for the main analysis of clusters 5, 11, and 12 in our scRNA-seq analysis, an analysis using a larger number of cells should improve the accuracy.
In conclusion, by constructing a differentiation system for pancreatic acinar lineages from hiPSCs, we identified REG4 as a candidate marker for acinar progenitor cells and forskolin to facilitate differentiation into acinar lineage cells in hiPSC differentiation cultures. These findings contribute to the detailed mechanistic elucidation of pancreatic acinar cell development and the establishment of induction methods to generate pancreatic acinar cells from hiPSCs.
Materials and methods
Ethics statement
All experiments using hiPSCs were approved by the Ethics Committee of the Department of Medicine and Graduate School of Medicine, Kyoto University, and performed in accordance with institutional guidelines. All donors from whom hiPSCs were generated provided informed consent. All animal experiments were approved by the CiRA Animal Experiment Committee and performed in accordance with institutional guidelines, including the ARRIVE guidelines (https://arriveguidelines.org).
hiPSC maintenance
The hiPSC line 585A129 was cultured in feeder-free conditions using Essential 8 medium (Thermo Fisher Scientific) supplemented with Essential 8 medium supplement. hiPSCs at 80% confluency were dissociated into a single-cell suspension by incubation with 0.5 mM EDTA/PBS (Thermo Fisher Scientific) for 8–13 min at 37 °C and gentle pipetting and passaged every 3–4 days. Cells were routinely checked for mycoplasma contamination using Mycoprobe Detection Kit (R&D Systems).
In vitro hiPSC differentiation
hiPSCs were sequentially differentiated into four stages of pancreatic development: definitive endoderm (Stage 1), primitive gut tube (Stage 2), posterior foregut (Stage 3), and pancreatic endoderm (Stage 4), as described previously28. At Stage 1, 80% confluent hiPSCs were dissociated into single cells by incubation with 0.5 mM EDTA/PBS for 8–13 min at 37 °C and gentle pipetting and resuspended in RPMI 1640 medium (Nacalai Tesque) supplemented with 2% (vol/vol) growth factor-reduced B27 (GFR-B27; Thermo Fisher Scientific), 50 U/mL penicillin/streptomycin (P/S; Thermo Fisher Scientific), 100 ng/mL activin A (R&D Systems), 3 μM CHIR99021 (Axon Medchem), and 10 μM Y-27632 (Wako). Then, cells were seeded into 6-well plates coated with GFR Matrigel (Becton Dickinson) according to the manufacturer’s recommendations at a density of 1 × 106 cells/well and cultured for one day. For the next two days, cells were cultured in RPMI 1640 medium supplemented with 2% GFR-B27, 50 U/mL P/S, 100 ng/mL activin A, and 1 μM CHIR99021. For the next day, cells were cultured in RPMI 1640 medium supplemented with 2% GFR-B27, 50 U/mL P/S, and 100 ng/mL activin A. At Stage 2, cells were cultured in Improved MEM Zinc Option (iMEM) medium (Thermo Fisher Scientific) supplemented with 1% GFR-B27, 100 U/mL P/S, and 50 ng/mL keratinocyte growth factor (KGF; R&D Systems) for four days. At Stage 3, cells were cultured in iMEM medium supplemented with 1% GFR-B27, 100 U/mL P/S, 50 ng/mL KGF, 100 ng/mL NOGGIN (Peprotech), 0.5 μM 3-Keto-N-aminoethyl-N′-aminocaproyldihydrocinnamoyl cyclopamine (KAAD-CYC; Toronto Research Chemicals) and 0.5 nM 4-[(E)-2-(5,6,7,8-Tetrahydro-5,5,8,8-tetramethyl-2-naphthalenyl)-1-propenyl]benzoic acid (TTNPB; Santa Cruz Biotechnology) for three days. At Stage 4, cells were dissociated into single cells by incubation with 0.25% trypsin–EDTA (Invitrogen) for 15 min at 37 °C and gentle pipetting and resuspended in iMEM medium supplemented with 1% GFR-B27, 100 U/mL P/S, 100 ng/mL KGF, 100 ng/mL NOGGIN, 50 ng/mL EGF (R&D Systems), and 10 μM Y-27632. For aggregation cultures, cells were seeded into low-binding 96-well plates (Greiner Bio) at a density of 2 × 104 cells/well and cultured for an additional six days.
For the differentiation into acinar lineage cells, cellular aggregates of hiPSC-derived pancreatic endoderm on Stage 4 Day 6 formed in low-binding 96-well plates were cultured in iMEM medium supplemented with 1% GFR-B27, 100 U/mL P/S, and candidate factors for eight days with a medium change every 4 days. A total of 24 and 48 cellular aggregates formed in the 96-well plates were used to test each factor in the experiments of Figs. 4B, S9, and S10 and those of Fig. 4C, respectively. Negative controls were samples without any candidate factors. The following factors and concentrations were examined: 10, 50, and 100 ng/mL EGF; 0.2, 1, and 2 ng/mL IL-1B (Wako); 10, 50, and 100 ng/mL HGF (Peprotech); 10, 50, and 100 µM pCPT-cAMP (Abcam); 2, 10, and 20 nM forskolin (Wako); 10, 50, and 100 ng/mL FGF2 (R&D Systems); 0.02, 0.1, and 0.2 nM bexarotene (Tocris Bioscience); 5, 25, and 50 ng/mL IGF1 (Peprotech); and 1, 5, and 10 µg/mL LPS (Santa Cruz Biotechnology). For experiments in Figs. S9 and S10, 20 nM forskolin; 100 µM pCPT-cAMP; 0.02 nM bexarotene; 100 ng/mL EGF; 10 ng/mL FGF2; 100 ng/mL HGF; and 2 ng/mL IL-1B were used.
Flow cytometry
We dissociated Stage 4 Day 6 hiPSC-derived pancreatic endoderm cell aggregates into single cells by incubation with 0.25% trypsin–EDTA for 10 min at 37 °C and gentle pipetting, fixed cells with BD Cytofix/Cytoperm Kit (Becton Dickinson) and blocked cells with 2% donkey serum (Merck Millipore) in permeabilization solution for 15 min at room temperature (RT). Then, cells were incubated with primary antibodies diluted with 2% donkey serum in permeabilization solution overnight at 4℃. After washing with permeabilization solution, cells were incubated with secondary antibodies diluted with 2% donkey serum in permeabilization solution for 1 h at RT.
hiPSC-derived pancreatic endoderm cell aggregates (day 0 sample) were dissociated into single cells by incubation with 0.25% trypsin–EDTA for 10 min at 37 °C and gentle pipetting to prepare for scRNA-seq. Graft samples on days 11 and 25 after implantation were isolated from the host renal parenchyma and capsule and chopped using a razor. Then, cells were dispersed in a collagenase/dispase solution consisting of 3.5 mg collagenase type II (GIBCO) and 2 mg dispase II (Wako) dissolved in 1 mL collagenase buffer for 15 min at 37 °C and dissociated into single cells by gentle pipetting. Staining with TotalSeq™-A0951 PE-Streptavidin (BioLegend), TotalSeq™-A0952 PE-Streptavidin (BioLegend), and TotalSeq™-A0953 PE-Streptavidin (BioLegend) was used to identify cells on days 0, 11, and 25, respectively. TotalSeq™-A0951 PE-Streptavidin, TotalSeq™-A0952 PE-Streptavidin, and TotalSeq™-A0953 PE-Streptavidin were diluted to 0.2 µg/mL (approximately 500-fold dilution) in cold Cell Staining Buffer (BioLegend), centrifuged at 14000×g for 10 min at 4 °C, and stored on ice in the dark. Single-cell suspensions were collected by centrifugation at 300×g for 5 min at 4 °C, with supernatants removed and cells resuspended in 200 μL of PBS. Cells were immediately resuspended by pipetting and left on ice for 10 min after adding 1 μL of EZ-Link™ Sulfo-NHS-LC-Biotin (Thermo Fisher Scientific) dissolved in DMSO (100 mg/mL). After incubation, 900 μL of 3% FBS/PBS was added. Cells were resuspended in 1 mL of 3% FBS/PBS after collection by centrifugation and mixed by pipetting after adding 1 mL of Cell Staining Buffer. Cells were collected by centrifugation and mixed by pipetting with 50 μL of the supernatant from the TotalSeq-PE-Streptavidin reaction solution prepared earlier, then incubated on ice for 20 min in the dark. Cells were washed thrice with 1 mL of Cell Staining Buffer after biotin-streptavidin binding. FITC-conjugated mouse-specific CD45, H-2, and TER-119 antibodies that stain mouse white blood cells, a wide range of mouse MHC class I+ cells, and mouse red blood cells, respectively42 and APC-conjugated human-specific CD147 and CD298 antibodies that both stain a wide range of human cells including white blood cells42,43 were added to 2% FBS/PBS to make 500 μL of antibody solution (1:40). Cells were resuspended in the antibody solution at 1 × 106 cells per 50 μL and incubated for 20 min at 4 °C. After adding 1 mL of 2% FBS/PBS to the cell suspension, cells were collected by centrifugation at 500×g for 3 min at 4 °C and resuspended in 500 µL of 2% FBS/PBS with 0.5 μL of 4′,6-diamidino-2-phenylindole (DAPI, Thermo Fisher Scientific, 1:1,000). Lastly, 2% FBS/PBS was added to make the final cell suspension at 1 × 104 cells per 1 mL.
To sort cells for scRNA-seq by flow cytometry (Fig. S2), human cells positive for either human CD147 or CD298 but not mouse CD45, H-2, or TER-119 were first sorted from a mixture of human and mouse cells based on fluorescence intensity using a combination of FITC and APC. Blood cells were also sorted and removed using mouse-specific CD45, H-2, and TER-119 antibodies. For isolation, gating was set such that mouse cells had a positive fraction of less than 1% based on a negative control experiment using only NOD/SCID mouse kidney cells. Second, DAPI- live cells were sorted from human cells, with damaged cells and doublets removed using a combination of FSC and SSC gating. Third, only Streptavidin–Biotin-tagged cells positive for TotalSeq™-A0951, A0952, or A0953 were isolated based on PE fluorescence intensity. For isolation, gating was set such that non-fluorescent cells had a positive fraction of less than 10% based on a negative control experiment using unstained hiPSC-derived pancreatic endoderm cells.
Flow cytometry was performed using a FACSAriaII or LSRFortessa (BD Biosciences). Antibodies used in this study are listed in Supplementary Table 5.
Animal experiments
We used 6- to 14-week-old male NOD.CB17-Prkdcscid/J (NOD/SCID) mice (Charles River Laboratories Japan) for implantation experiments. Mice were anesthetized with inhalable isoflurane and maintained at 37 °C in the right lateral position, with the left kidney pulled out of the abdominal cavity through a flank incision. After a small incision in the left renal capsule, a part of the renal capsule at the implant site was peeled off gently with a thin glass rod made in-house. We implanted hiPSC-derived Stage 4 Day 6 pancreatic endoderm cell aggregates under the left renal capsules using a slim micropipette. A total of 192 aggregates containing a total of 3.84 × 106 cells were implanted per mouse. We harvested 192 aggregates directly from the culture dishes, removed the culture medium by suction, and implanted the aggregates into the renal subcapsular spaces without any buffer, solution, or Matrigel. Grafts were harvested at 7–30 days after implantation. For collecting tissues after animal sacrifice, euthanasia was performed by cervical dislocation after the mice had lost consciousness rapidly due to an overdose inhalation of isoflurane in a small cage filled with the volatile anesthetic.
Immunostaining
Differentiated cells and grafts were fixed with 4% paraformaldehyde (PFA) (Nacalai Tesque)/PBS for 20 min and from 3 h to overnight at 4 °C, respectively. Then, grafts were sequentially dehydrated with 10% sucrose/PBS solution for over 1 h, 20% sucrose/PBS solution for over 2 h, and 30% sucrose/PBS solution from over 3 h to overnight at RT, frozen at − 80 °C, and sectioned at a thickness of 10 µm by Cryostat (CM1520; Leica). Fixed cells and frozen sections of grafts were blocked with 5% donkey serum (Millipore)/0.4% Triton X-100 (Nacalai Tesque)/PBS (PBT) for 30 min at RT and incubated with primary antibodies from 1 h to overnight at RT. Then, samples were washed with PBT once and incubated with secondary antibodies for 1–2 h at RT. Finally, samples were washed with PBT three times and observed using a fluorescence microscope (BZ-9000; Keyence). Immunostaining images of graft sections were quantified using ImageJ macros (National Institutes of Health). Antibodies used in this study are listed in Supplementary Table 5.
Single-cell RNA sequencing (scRNA-seq)
Mice were sacrificed to obtain the left kidneys with grafts on days 11 and 25 after implantation. We manually isolated grafts from the renal subcapsular space and used eight and five grafts on days 11 and 25, respectively, for the analysis. The procedures of cell preparation and human cell isolation by flow cytometry are described above. scRNA-seq was performed by ImmunoGeneTeqs, Inc. (Chiba, Japan) using protocols described previously72. Cells with RNA count per cell (nCount_RNA ≧ 20,000, the number of genes per cell (nFeature_RNA) ≦ 2000 or ≧ 15,000, mitochondrial gene ratio (percent.mt) ≧ 10%, and Human_Mouse_nCount_ratio.log2 ≦0.6 were removed as quality control (QC) for scRNA-seq (Fig.S3). After QC, 6,455 cells were normalized and analyzed by Seurat package version 4.73 in R version 4. We set the optimal dimension and resolution by referring to the elbow plot. We performed principal component analysis (PCA) and determined the k-nearest neighbor graph using the first 63 PCs. We found clusters of cells using the FindClusters function (resolution = 0.8) in Seurat. Differentially expressed genes (DEGs) for each cluster were identified using the FindAllMarkers function with thresholds of |log2FC|≥ 0.25 and a minimum percentage of cells expressing the gene ≥ 25% and applying the Wilcoxon test. Genes with an adjusted p-value < 0.01 were selected. The top 10 genes for each cluster, ranked by expression, were extracted, and a heatmap was generated using the pheatmap () function. We used the FeaturePlot function in Seurat to visualize the expression of known marker genes for each cell on tSNE plots. To compare the expression of known markers in each dataset or cluster, we used the VlnPlot function in Seurat. Each cluster was annotated based on CellMarker2024 data using Enrichr.
RNA velocity and pseudo-temporal ordering analysis were performed by scvelo 0.2.4 and scanpy 1.9.1 packages in Python 3.9. The dot plot was created using scanpy 1.9.1 packages in Python 3.9. IPA analysis (Qiagen) was conducted based on 176 DEGs (|log2FC|> 1, P value < 0.01) obtained by comparing pancreatic endoderm (cluster 12) and acinar progenitor cells (cluster 11).
For scRNA-seq of a previously published human adult pancreatic dataset (Fig. S8), we obtained a curated, quality controlled and cell type-annotated scRNA-seq dataset (panc8) using an R package SeuratData, originally derived from five datasets (GSE81076, GSE85241, GSE86469, E-MTAB-5061, and GSE84133)50,51,52,53,54. Murine samples (GSM2230761 and GSM2230762) in GSE84133 were not included. Among human samples, 13 samples were from deceased organ donors with and without type 2 diabetes (T2D), 11 samples were from deceased organ donors without T2D, and 7 samples were from deceased organ donors with T2D (Fig. S8A). For batch correction, to deal with the heterogeneity of diabetic conditions and the composition of cell types across datasets, we utilized the reciprocal PCA (RPCA) method, which allows the non-overlapping of cell types across datasets73. We added QC metrics in Fig. S8B. Although the number of genes per cell varies depending on each single-cell technology platform (Fig. S8A), we could integrate the data and visualize with UMAP each cell type cluster according to the original annotation using Seurat’s SCTransform and RPCA method (dims = 1:30, k.anchor = 20) (Fig. S8C–E), suggesting successful batch correction among five datasets with heterogeneous single-cell technology platforms and diabetic conditions. To visualize the expression of known marker genes for each cell on UMAP plots or to compare the expression of known markers across clusters, we used batch-uncorrected, normalized (SCTransform) gene expression data.
For scRNA-seq analysis of previously published human fetal pancreatic datasets48, we obtained a processed and cell type-annotated scRNA-seq dataset including 17,135 pancreatic cells from PCW4 to 11. We normalized the data with Seurat’s SCTransform function to regress out cell cycle scores (calculated by the CellCycleScoring function) together with sequencing depth and batch as described previously48. We performed PCA and constructed the k-nearest neighbor graph using the first 50 PCs. We found clusters of cells using the FindClusters function (resolution = 1.0) in Seurat. UMAP was depicted based on the dimensionalities of the previously reported analysis48. Monocle374 was used to calculate pseudotime. Multipotent pancreas progenitor, early tip, tip, and acinar cell types were selected to depict pseudotemporal patterns of exocrine-related gene expression using geom_smooth function of ggplot2 (generalized additive model).
Real-time quantitative RT-PCR (qRT-PCR)
Total RNA was extracted using the RNeasy Mini Kit (Qiagen) according to the manufacturer’s instructions, and the concentration of total RNA was measured using Nanodrop 8000 (Thermo Fisher Scientific). cDNA was obtained by reverse transcription using ReverTra Ace qPCR Master Mix (Toyobo), dNTP Mix (Qiagen), and oligo dT primer (Fasmac). Quantitative PCR was performed using SYBR Premix Ex Taq II (Takara) and a StepOnePlus Real-Time PCR System (Applied Biosystems). Denaturation was performed at 95 °C for 10 s and annealing by 40 cycles at 95 °C for 5 s and at 60 °C for 30 s and dissociation at 95 °C for 15 s, 60 °C for 1 min and 95 °C for 15 s. Expression of target genes was normalized to housekeeping genes ACTB or GAPDH. Primer sequences used in this study are listed in Supplementary Table 6.
Statistics
All quantitative data are presented as mean ± SD. Data were analyzed for statistical significance using Microsoft Excel for Windows version 2312. The two-tailed Student’s t-test was used to compare the means of two comparison groups. p < 0.05 was considered statistically significant for all analyses. * and ** indicate p < 0.05 and p < 0.01 in the figures, respectively.
Data availability
The analysis datasets used in this study are available from the corresponding author (K.O.) upon reasonable request. The NCBI GEO accession number for single-cell RNA sequencing data reported in this paper is GSE 264026.
References
Leung, P. S. Overview of the pancreas. Adv. Exp. Med. Biol. 690, 3–12. https://doi.org/10.1007/978-90-481-9060-7_1 (2010).
Pandol, S. J. The Exocrine Pancreas (Morgan & Claypool Life Sciences, 2010).
Steward, M. C., Ishiguro, H. & Case, R. M. Mechanisms of bicarbonate secretion in the pancreatic duct. Annu. Rev. Physiol. 67, 377–409. https://doi.org/10.1146/annurev.physiol.67.031103.153247 (2005).
Beer, R. L., Parsons, M. J. & Rovira, M. Centroacinar cells: At the center of pancreas regeneration. Dev. Biol. 413, 8–15. https://doi.org/10.1016/j.ydbio.2016.02.027 (2016).
In’t Veld, P. & Marichal, M. Microscopic anatomy of the human islet of Langerhans. Adv. Exp. Med. Biol. 654, 1–19. https://doi.org/10.1007/978-90-481-3271-3_1 (2010).
Ulrich, A. B., Schmied, B. M., Standop, J., Schneider, M. B. & Pour, P. M. Pancreatic cell lines: A review. Pancreas 24, 111–120. https://doi.org/10.1097/00006676-200203000-00001 (2002).
Fanjul, M. et al. Evidence for epithelial-mesenchymal transition in adult human pancreatic exocrine cells. J. Histochem. Cytochem. 58, 807–823. https://doi.org/10.1369/jhc.2010.955807 (2010).
Rooman, I., Heremans, Y., Heimberg, H. & Bouwens, L. Modulation of rat pancreatic acinoductal transdifferentiation and expression of PDX-1 in vitro. Diabetologia 43, 907–914. https://doi.org/10.1007/s001250051468 (2000).
Maehr, R. et al. Generation of pluripotent stem cells from patients with type 1 diabetes. Proc. Natl. Acad. Sci. U. S. A. 106, 15768–15773. https://doi.org/10.1073/pnas.0906894106 (2009).
Teo, A. K. et al. Derivation of human induced pluripotent stem cells from patients with maturity onset diabetes of the young. J. Biol. Chem. 288, 5353–5356. https://doi.org/10.1074/jbc.C112.428979 (2013).
Tulpule, A. et al. Pluripotent stem cell models of Shwachman-Diamond syndrome reveal a common mechanism for pancreatic and hematopoietic dysfunction. Cell Stem Cell 12, 727–736. https://doi.org/10.1016/j.stem.2013.04.002 (2013).
Huang, L. et al. Ductal pancreatic cancer modeling and drug screening using human pluripotent stem cell- and patient-derived tumor organoids. Nat. Med. 21, 1364–1371. https://doi.org/10.1038/nm.3973 (2015).
Skoudy, A. et al. Transforming growth factor (TGF)beta, fibroblast growth factor (FGF) and retinoid signalling pathways promote pancreatic exocrine gene expression in mouse embryonic stem cells. Biochem. J. 379, 749–756. https://doi.org/10.1042/BJ20031784 (2004).
Nakanishi, M., Hamazaki, T. S., Komazaki, S., Okochi, H. & Asashima, M. Pancreatic tissue formation from murine embryonic stem cells in vitro. Differentiation 75, 1–11. https://doi.org/10.1111/j.1432-0436.2006.00109.x (2007).
Shirasawa, S. et al. Pancreatic exocrine enzyme-producing cell differentiation via embryoid bodies from human embryonic stem cells. Biochem. Biophys. Res. Commun. 410, 608–613. https://doi.org/10.1016/j.bbrc.2011.06.036 (2011).
Takizawa-Shirasawa, S. et al. FGF7 and cell density are required for final differentiation of pancreatic amylase-positive cells from human ES cells. Cell Tissue Res. 354, 751–759. https://doi.org/10.1007/s00441-013-1695-6 (2013).
Hohwieler, M. et al. Human pluripotent stem cell-derived acinar/ductal organoids generate human pancreas upon orthotopic transplantation and allow disease modelling. Gut 66, 473–486. https://doi.org/10.1136/gutjnl-2016-312423 (2017).
Ito, K. et al. Delivery of pancreatic digestive enzymes into the gastrointestinal tract by pancreatic exocrine tissue transplant. Sci. Rep. 9, 5922. https://doi.org/10.1038/s41598-019-42362-z (2019).
Huang, L. et al. Commitment and oncogene-induced plasticity of human stem cell-derived pancreatic acinar and ductal organoids. Cell Stem Cell 28, 1090–1104. https://doi.org/10.1016/j.stem.2021.03.022 (2021).
Merz, S. et al. Single-cell profiling of GP2-enriched pancreatic progenitors to simultaneously create acinar, ductal, and endocrine organoids. Theranostics 13, 1949–1973. https://doi.org/10.7150/thno.78323 (2023).
Pan, F. C. & Wright, C. Pancreas organogenesis: from bud to plexus to gland. Dev Dyn 240, 530–565. https://doi.org/10.1002/dvdy.22584 (2011).
Hald, J. et al. Generation and characterization of Ptf1a antiserum and localization of Ptf1a in relation to Nkx6.1 and Pdx1 during the earliest stages of mouse pancreas development. J Histochem Cytochem 56, 587–595. https://doi.org/10.1369/jhc.2008.950675 (2008).
Kawaguchi, Y. et al. The role of the transcriptional regulator Ptf1a in converting intestinal to pancreatic progenitors. Nat. Genet. 32, 128–134. https://doi.org/10.1038/ng959 (2002).
Jensen, J. Gene regulatory factors in pancreatic development. Dev. Dyn. 229, 176–200. https://doi.org/10.1002/dvdy.10460 (2004).
Villasenor, A., Chong, D. C., Henkemeyer, M. & Cleaver, O. Epithelial dynamics of pancreatic branching morphogenesis. Development 137, 4295–4305. https://doi.org/10.1242/dev.052993 (2010).
Marty-Santos, L. & Cleaver, O. Progenitor epithelium: Sorting out pancreatic lineages. J. Histochem. Cytochem. 63, 559–574. https://doi.org/10.1369/0022155415586441 (2015).
Zhou, Q. et al. A multipotent progenitor domain guides pancreatic organogenesis. Dev. Cell 13, 103–114. https://doi.org/10.1016/j.devcel.2007.06.001 (2007).
Toyoda, T. et al. Rho-associated kinases and non-muscle Myosin IIs Inhibit the differentiation of human iPSCs to pancreatic endoderm. Stem Cell Rep. 9, 419–428. https://doi.org/10.1016/j.stemcr.2017.07.005 (2017).
Okita, K. et al. An efficient nonviral method to generate integration-free human-induced pluripotent stem cells from cord blood and peripheral blood cells. Stem Cells 31, 458–466. https://doi.org/10.1002/stem.1293 (2013).
Hogrebe, N. J., Maxwell, K. G., Augsornworawat, P. & Millman, J. R. Generation of insulin-producing pancreatic beta cells from multiple human stem cell lines. Nat. Protoc. 16, 4109–4143. https://doi.org/10.1038/s41596-021-00560-y (2021).
Rezania, A. et al. Maturation of human embryonic stem cell-derived pancreatic progenitors into functional islets capable of treating pre-existing diabetes in mice. Diabetes 61, 2016–2029. https://doi.org/10.2337/db11-1711 (2012).
Assady, S. et al. Insulin production by human embryonic stem cells. Diabetes 50, 1691–1697. https://doi.org/10.2337/diabetes.50.8.1691 (2001).
D’Amour, K. A. et al. Production of pancreatic hormone-expressing endocrine cells from human embryonic stem cells. Nat. Biotechnol. 24, 1392–1401. https://doi.org/10.1038/nbt1259 (2006).
Kroon, E. et al. Pancreatic endoderm derived from human embryonic stem cells generates glucose-responsive insulin-secreting cells in vivo. Nat. Biotechnol. 26, 443–452. https://doi.org/10.1038/nbt1393 (2008).
Zhang, D. et al. Highly efficient differentiation of human ES cells and iPS cells into mature pancreatic insulin-producing cells. Cell Res. 19, 429–438. https://doi.org/10.1038/cr.2009.28 (2009).
Schulz, T. C. et al. A scalable system for production of functional pancreatic progenitors from human embryonic stem cells. PLoS One 7, e37004. https://doi.org/10.1371/journal.pone.0037004 (2012).
Bruin, J. E. et al. Maturation and function of human embryonic stem cell-derived pancreatic progenitors in macroencapsulation devices following transplant into mice. Diabetologia 56, 1987–1998. https://doi.org/10.1007/s00125-013-2955-4 (2013).
Pagliuca, F. W. et al. Generation of functional human pancreatic beta cells in vitro. Cell 159, 428–439. https://doi.org/10.1016/j.cell.2014.09.040 (2014).
Rezania, A. et al. Reversal of diabetes with insulin-producing cells derived in vitro from human pluripotent stem cells. Nat. Biotechnol. 32, 1121–1133. https://doi.org/10.1038/nbt.3033 (2014).
Marty-Santos, L. & Cleaver, O. Pdx1 regulates pancreas tubulogenesis and E-cadherin expression. Development 143, 1056. https://doi.org/10.1242/dev.135806 (2016).
Weisel, N. M. et al. Surface phenotypes of naive and memory B cells in mouse and human tissues. Nat Immunol 23, 135–145. https://doi.org/10.1038/s41590-021-01078-x (2022).
Sperber, H. S. et al. mu-Lat: A mouse model to evaluate human immunodeficiency virus eradication strategies. FASEB J 34, 14615–14630. https://doi.org/10.1096/fj.202001612RR (2020).
Hamelin, B. et al. Single-cell analysis reveals inter- and intratumour heterogeneity in metastatic breast cancer. J Mammary Gland Biol Neoplasia 28, 26. https://doi.org/10.1007/s10911-023-09551-z (2023).
Zeve, D. et al. Robust differentiation of human enteroendocrine cells from intestinal stem cells. Nat Commun 13, 261. https://doi.org/10.1038/s41467-021-27901-5 (2022).
Bergen, V., Lange, M., Peidli, S., Wolf, F. A. & Theis, F. J. Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol. 38, 1408–1414. https://doi.org/10.1038/s41587-020-0591-3 (2020).
La Manno, G. et al. RNA velocity of single cells. Nature 560, 494–498. https://doi.org/10.1038/s41586-018-0414-6 (2018).
Wolf, F. A. et al. PAGA: Graph abstraction reconciles clustering with trajectory inference through a topology preserving map of single cells. Genome Biol. 20, 59. https://doi.org/10.1186/s13059-019-1663-x (2019).
Ma, Z. et al. Deciphering early human pancreas development at the single-cell level. Nat. Commun. 14, 5354. https://doi.org/10.1038/s41467-023-40893-8 (2023).
Stuart, T. et al. Comprehensive integration of single-cell data. Cell 177, 1888–1902. https://doi.org/10.1016/j.cell.2019.05.031 (2019).
Baron, M. et al. A single-cell transcriptomic map of the human and mouse pancreas reveals inter- and intra-cell population structure. Cell Syst. 3, 346–360. https://doi.org/10.1016/j.cels.2016.08.011 (2016).
Lawlor, N. et al. Single-cell transcriptomes identify human islet cell signatures and reveal cell-type-specific expression changes in type 2 diabetes. Genome Res. 27, 208–222. https://doi.org/10.1101/gr.212720.116 (2017).
Grun, D. et al. De novo prediction of stem cell identity using single-cell transcriptome data. Cell Stem Cell 19, 266–277. https://doi.org/10.1016/j.stem.2016.05.010 (2016).
Muraro, M. J. et al. A single-cell transcriptome atlas of the human pancreas. Cell Syst. 3, 385–394. https://doi.org/10.1016/j.cels.2016.09.002 (2016).
Segerstolpe, A. et al. Single-cell transcriptome profiling of human pancreatic islets in health and type 2 diabetes. Cell Metab. 24, 593–607. https://doi.org/10.1016/j.cmet.2016.08.020 (2016).
Yan, K., Gao, L. N., Cui, Y. L., Zhang, Y. & Zhou, X. The cyclic AMP signaling pathway: Exploring targets for successful drug discovery (Review). Mol. Med. Rep. 13, 3715–3723. https://doi.org/10.3892/mmr.2016.5005 (2016).
Christensen, A. E. et al. cAMP analog mapping of Epac1 and cAMP kinase. Discriminating analogs demonstrate that Epac and cAMP kinase act synergistically to promote PC-12 cell neurite extension. J. Biol. Chem. 278, 35394–35402. https://doi.org/10.1074/jbc.M302179200 (2003).
Weng, C. et al. Single-cell lineage analysis reveals extensive multimodal transcriptional control during directed beta-cell differentiation. Nat. Metab. 2, 1443–1458. https://doi.org/10.1038/s42255-020-00314-2 (2020).
Krentz, N. A. J. et al. Single-cell transcriptome profiling of mouse and hESC-derived pancreatic progenitors. Stem Cell Rep. 11(1551), 1564. https://doi.org/10.1016/j.stemcr.2018.11.008 (2018).
Sharon, N. et al. Wnt signaling separates the progenitor and endocrine compartments during pancreas development. Cell Rep. 27, 2281–2291. https://doi.org/10.1016/j.celrep.2019.04.083 (2019).
Veres, A. et al. Charting cellular identity during human in vitro beta-cell differentiation. Nature 569, 368–373. https://doi.org/10.1038/s41586-019-1168-5 (2019).
Chen, C., Fang, R., Davis, C., Maravelias, C. & Sibley, E. Pdx1 inactivation restricted to the intestinal epithelium in mice alters duodenal gene expression in enterocytes and enteroendocrine cells. Am. J. Physiol. Gastrointest Liver Physiol. 297, G1126-1137. https://doi.org/10.1152/ajpgi.90586.2008 (2009).
Castaing, M., Duvillie, B., Quemeneur, E., Basmaciogullari, A. & Scharfmann, R. Ex vivo analysis of acinar and endocrine cell development in the human embryonic pancreas. Dev. Dyn. 234, 339–345. https://doi.org/10.1002/dvdy.20547 (2005).
Han, J. H., Rall, L. & Rutter, W. J. Selective expression of rat pancreatic genes during embryonic development. Proc. Natl. Acad. Sci. U. S. A. 83, 110–114. https://doi.org/10.1073/pnas.83.1.110 (1986).
Takayama, R. et al. Serum tumor antigen REG4 as a diagnostic biomarker in pancreatic ductal adenocarcinoma. J. Gastroenterol. 45, 52–59. https://doi.org/10.1007/s00535-009-0114-y (2010).
Dai, J. et al. Reg4 regulates pancreatic regeneration following pancreatitis via modulating the Notch signaling. J. Cell. Physiol. 236, 7565–7577. https://doi.org/10.1002/jcp.30397 (2021).
Zhang, Y. W., Ding, L. S. & Lai, M. D. Reg gene family and human diseases. World J. Gastroenterol. 9, 2635–2641. https://doi.org/10.3748/wjg.v9.i12.2635 (2003).
Mally, M. I., Otonkoski, T., Lopez, A. D. & Hayek, A. Developmental gene expression in the human fetal pancreas. Pediatr. Res. 36, 537–544. https://doi.org/10.1203/00006450-199410000-00022 (1994).
Takehara, A. et al. Novel tumor marker REG4 detected in serum of patients with resectable pancreatic cancer and feasibility for antibody therapy targeting REG4. Cancer Sci. 97, 1191–1197. https://doi.org/10.1111/j.1349-7006.2006.00297.x (2006).
Zheng, H. C., Xue, H. & Zhang, C. Y. REG4 promotes the proliferation and anti-apoptosis of cancer. Front. Cell Dev. Biol. 10, 1012193. https://doi.org/10.3389/fcell.2022.1012193 (2022).
Howard, W. R., Pospisil, J. A., Njolito, E. & Noonan, D. J. Catabolites of cholesterol synthesis pathways and forskolin as activators of the farnesoid X-activated nuclear receptor. Toxicol. Appl. Pharmacol. 163, 195–202. https://doi.org/10.1006/taap.1999.8869 (2000).
Aghazadeh, Y. et al. GP2-enriched pancreatic progenitors give rise to functional beta cells in vivo and eliminate the risk of teratoma formation. Stem Cell Rep. 17, 964–978. https://doi.org/10.1016/j.stemcr.2022.03.004 (2022).
Shichino, S. et al. TAS-Seq is a robust and sensitive amplification method for bead-based scRNA-seq. Commun. Biol. 5, 602. https://doi.org/10.1038/s42003-022-03536-0 (2022).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 e3529, https://doi.org/10.1016/j.cell.2021.04.048 (2021).
Cao, J. et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature 566, 496–502. https://doi.org/10.1038/s41586-019-0969-x (2019).
Acknowledgements
The authors would like to thank Ms. Hiromi Tanaka, CiRA, Kyoto University, for technical support of the culture and animal experiments, Dr. Kanae Mitsunaga, CiRA, Kyoto University, for the flow cytometry analysis, ImmunoGeneTeqs, Inc., Chiba, Japan, for the scRNA-seq analysis, and Dr. Kelvin Hui, for critically reading the manuscript. This study was partially supported by the Japan Agency for Medical Research and Development (AMED; JP23bm1123002, JP24bm1123002, JP23bm1323001, JP24bm1323001) to K. Osafune, and the iPS Cell Research Fund.
Author information
Authors and Affiliations
Contributions
A.M., A.K., T.T., and K.O. designed this study. A.M. performed all the experiments. A.M. wrote the original draft. A.M. and K.O. edited this manuscript. A.M., A.K., R.I., Y.H., H.T., S.-I.M., J.Y., N.U., and T.T. contributed to planning and performing each experiment. J.Y. performed scRNA-seq. Y.H., H.T., and W.F. provided consultation on scRNA-seq data analysis. R.I. and Y.H. provided consultation on the statistical analysis methods. N.U., T.T., H.S., and K.O. supervised all aspects of this project. All authors participated in the preparation of the manuscript, discussed the results, and commented on the manuscript.
Corresponding authors
Ethics declarations
Competing interests
K.O. is a founder and member of the scientific advisory boards of iPS Portal, Inc., and a founder and chief scientific advisor of RegeNephro Co., Ltd. T.T. is a scientific adviser of Orizuru Therapeutics, Inc. A.K. and H.T. are an employee of RegeNephro Co., Ltd. There are no competing interests to declare by the other authors.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mima, A., Kimura, A., Ito, R. et al. Mechanistic elucidation of human pancreatic acinar development using single-cell transcriptome analysis on a human iPSC differentiation model. Sci Rep 15, 4668 (2025). https://doi.org/10.1038/s41598-025-88690-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-88690-1