Abstract
Deep learning (DL) and explainable artificial intelligence (XAI) have emerged as powerful machine-learning tools to identify complex predictive data patterns in a spatial or temporal domain. Here, we consider the application of DL and XAI to large omic datasets, in order to study biological aging at the molecular level. We develop an advanced multi-view graph-level representation learning (MGRL) framework that integrates prior biological network information, to build molecular aging clocks at cell-type resolution, which we subsequently interpret using XAI. We apply this framework to one of the largest single-cell transcriptomic datasets encompassing over a million immune cells from 981 donors, revealing a ribosomal gene subnetwork, whose expression correlates with age independently of cell-type. Application of the same DL-XAI framework to DNA methylation data of sorted monocytes reveals an epigenetically deregulated inflammatory response pathway whose activity increases with age. We show that the ribosomal module and inflammatory pathways would not have been discovered had we used more standard machine-learning methods. In summary, the computational deep learning framework presented here illustrates how deep learning when combined with explainable AI tools, can reveal novel biological insights into the complex process of aging.
Similar content being viewed by others
Introduction
Deep learning (DL) has emerged as a powerful machine learning framework in which to identify complex non-linear data patterns that are highly informative of outcomes, and which has been specially transformative in applications where these data patterns are of a sequential (i.e. spatial or temporal) nature1. Although this includes many applications in biology and medicine2,3,4,5,6, many outstanding challenges in the life-sciences domain remain that may also benefit from a DL based approach.
One of these outstanding challenges is the elucidation of molecular biological processes that are deregulated in aging and which could thus contribute causally to the aging process7. This has been a challenging endeavor for the following reasons. First, age-associated molecular changes (e.g. transcriptomic or DNA methylation (DNAm) changes) are characterized by small effect sizes8,9, hampering their reliable detection from noisy and often underpowered omic datasets9,10,11,12. Second, not until recently, most omic data has been generated in bulk-tissue, which is composed of many different cell-types, the implication being that measuring age-associated changes in such heterogeneous mixtures may mask the underlying changes in individual cell-types13,14. Fortunately, the single-cell omic data revolution has directly addressed this problem15,16, at least in the context of single-cell transcriptomics, although it should be noted that such data remains extremely noisy17,18. Whilst studies have begun to investigate aging in the context of single-cell RNA-Sequencing (scRNA-Seq) data8,19,20,21,22, the application of deep-learning methods to such data, or even to omic data from sorted (purified) cell populations, remains largely unexplored. Indeed, so far, DL methods have been applied to bulk-tissue DNA methylation (DNAm)23,24,25and RNA-Seq data26, building epigenetic and transcriptomic predictors of chronological age (termed “aging clocks”), respectively, but efforts to build such clocks from single-cell data have not yet applied a DL paradigm8,27, with the arguable exception of Yu et al.28, who applied a multi-layer-perceptron (MLP).
Here we address this gap, by developing and applying a novel DL approach to two distinct data-types, measured in purified immune cell populations (scRNA-Seq data and DNAm data of sorted cells), in order to build cell-type specific transcriptomic and epigenetic aging clocks. We focus on two of the largest available datasets, each profiling on the order of 1000 subjects29,30, which ensures adequate power to detect the aging effects at the molecular level, which, as we know, are characterized by small effect sizes. Because DL is specially powerful when the data is embedded in a spatial or temporal domain, we impart a spatial structure to both data-types, by integrating the scRNA-Seq and DNAm data with a prior-knowledge graph encoded by a protein-protein interaction (PPI) network31,32. This PPI network represents a crude, yet still highly informative, model of which genes (proteins) are more likely to interact33,34. In order to extract the complex patterns and biological gene modules driving the explanatory power of the DL aging clocks, we adapt advanced explainable artificial intelligence (XAI) tools23,35,36. Thus, our aim in applying a deep learning paradigm to predict chronological age from cell-type specific transcriptomic and DNA methylation data is two-fold. First, does a DL approach significantly outperform more traditional machine-learning methods (e.g. penalized multivariate regression37,38) in the task of predicting chronological age, a task which we note has been notoriously challenging in the context of both bulk and single-cell transcriptomic data. Second, can a combined deep-learning XAI method, when integrated with systems-level information (e.g. signaling pathways, protein-protein interactions), lead to important novel biological insights underlying the aging process?
As we shall see, a DL approach does not lead to substantial improvements in predicting chronological age, but in combination with XAI, does lead to critical novel biological insights, not obtainable using traditional penalized multivariate regression models.
Results
Building a DL-XAI clock to study aging at cell-type resolution
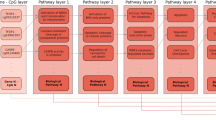
We collected one of the largest available scRNA-Seq datasets29, comprising over 1.2 million peripheral blood mononuclear cells (PBMCs) from 981 donors, displaying a wide age-range (20–100 years) (Fig. 1a, Methods). To reduce noise, we adopted the strategy of Buckley et al.8, forming meta-cells, each meta-cell being composed of 15 single cells (Fig. 1a, Methods). We posited that building an age predictor from this data using a deep learning explainable AI approach could help shed new insights into the molecular processes underlying aging, provided one embeds the features (i.e. genes) in a relevant spatial manifold. To give the genes a relevant spatial context, we consider the proteins they encode in the context of a highly curated PPI network (Fig. 1b, Methods). A key feature of the PPI network considered here is that every gene/protein maps to a specific signaling domain, including extracellular growth modulators (GM, e.g. cytokines), secreted factors (SF, e.g. ligands), membrane receptors (MR), intracellular receptor substrates (ICRS, e.g. kinases) and intracellular non-receptor substrates (ICNRS, e.g. nuclear transcription factors) (Methods, Fig. 1b). Thus, in the context of this network, proximity refers to gene pairs that belong to neighboring signaling domain layers. We posited that a DL-XAI approach, when applied to the mRNA expression meta-cell data overlayed onto this PPI network, would allow us to detect complex non-linear patterns that are predictive of age and thus highlight novel important gene modules that are deregulated in aging. Of note, the integration of the PPI network with molecular data (e.g. an mRNA expression data matrix) yields a topologically identical gene network for each sample, be it a single cell, meta-cell or cell population, but with the node (gene) attributes (e.g. expression levels) being meta-cell specific.
Deep-learning to build interpretable transcriptomic clocks from scRNA-Seq data. (a) Left panel: UMAP of the scRNA-Seq data of Yazar et al., encompassing over 1 million immune cell-types from 981 donors. CD4TCM = CD4 + central memory T-cells, nCD4T/CD8T = naïve CD4+/CD8 + T-cells, NK = natural killer, CD8TEM = CD8 + effector memory T-cells, CD14MC = CD14 + monocytes. Barplot displays the number of cells for each cell-type in units of 100,000. Middle panel: Construction of meta-cells from single-cells. Right panel: Example how the expression data matrix for CD4TCM meta-cells was split into training and test sets. Lower panels show the approximate meta-cell count distribution as a function of chronological age. Reason why training set is smaller is because we want a uniform chronological age distribution, to avoid biasing the clock to older age-groups. (b) For a given cell-type and training set composed of meta-cells, diagram depicts how this data is integrated with a protein-protein-interaction (PPI) network as prior biological information to yield meta-cell specific graphs where the node attributes are gene expression levels. This ensemble of graphs plus the chronological age information of a meta-cell defines the input to a multi-view graph-level representation learning (MGRL) framework that learns to predict chronological age. MGRL is a fusion between a graph convolutional neural network (DeeperGCN) and a multi-layer perceptron (MLP). PGExplainer is used to identify the PPI edges that contribute most to the predictions. The resulting age-predictor (the DL-XAI clock) is then applied to meta-cells from the test set to verify the predictions.
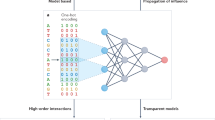
To build the chronological age predictor, we developed a novel multi-view graph-level representation learning (MGRL) algorithm that fuses a deep graph convolutional neural network (GCN) called DeeperGCN39 with a more traditional multi-layer perceptron (MLP) (Methods, Figs. 1b and 2a), the goal being to obtain an embedding of the whole graph that achieves an accurate prediction of a meta-cell’s chronological age. The rationale here is for DeeperGCN to capture local joint topological and gene expression information, with the MLP focusing solely on extracting gene expression information without regards to any underlying topology (Fig. 2a). We reasoned that this fusion of a state-of-the-art GCN with a MLP would enhance the model representation learning capability. Finally, in order to interpret the clock we adapt an advanced XAI algorithm (PGExplainer)36 that can extract the most predictive PPI edges and genes (Fig. 2b). Of note, in what follows we interchangeably refer to our DL-XAI clock also as MGRL.
Construction of the interpretable deep-learning age-predictor (DL-XAI clock). (a) Detailed diagram of the MGRL framework. A meta-cell’s input graph with a meta-cell’s expression values overlayed on this network defines the input to a graph convolutional neural network called DeeperGCN, to capture joint expression and topological features through two graph convolution layers that enable nodes to aggregate and update information from their neighbors. A mean pooling strategy is then applied to merge individual node representations into a unified graph-level feature vector as final topological information. Meanwhile, the MLP extracts node features from the expression data without consideration of network topology. The framework then fuses the joint topological-expression embeddings (topology view) with those derived from only expression (feature view) This fused embedding is finally fed into a fully-connected neural network to predict chronological age of the meta-cell. (b) Illustration of how PGExplainer is applied to our proposed MGRL method to extract the predictive subnetworks. The process begins by computing latent variables Ω from the edges of the original graph, which inform the edge distributions that underpin the explanatory framework. A random sampled graph is constructed by selecting the top-ranked edges based on Ω, and this graph is subsequently input into a trained MGRL model to generate the prediction (\(\:{y}_{s}\)). Finally, MLP parameters are optimized by minimizing the Mean Squared Error (MSE) between the original prediction (\(\:{y}_{o}\)) and the updated prediction (\(\:{y}_{s}\)).
The DL-XAI clock compares favorably to competing machine learning methods
To ensure adequate power, we focused our investigation on PBMC subtypes with the highest cell number representation. For lymphoid cells, this included naïve CD4 + T-cells (nCD4T, n = 259,012), central memory CD4 + T-cells (CD4TM, n = 289,000) and natural-killer cells (NK, n = 164,933), whilst for myeloid cells we considered CD14 + monocytes (Mono, n = 36,130) (Fig. 1a). All of these cell-types play key roles in a wide range of age-related diseases and thus constitute a reasonable starting point for our analyses. The distribution of meta-cell number per donor and age indicated a larger number of cells and meta-cells in the older age groups (SI fig.S1). Thus, to ensure that our training process would not be skewed to donors of higher age, we built a training-test set partition in a way so as to ensure balanced numbers of meta-cells per age-group in the training set (Methods, SI fig.S1, Fig. 1a). Specifically, for naïve CD4T-cells, this resulted in a training set composed of 6,708 meta-cells from 360 donors, evenly distributed across age-groups (SI fig.S1, Fig. 1a). The test set, comprising 11,007 naïve CD4T meta-cells from 616 donors, is thus inevitably more skewed towards older age-groups (SI fig.S1, Fig. 1a), which we note allows rigorous assessment of the predictor in the age-group where biological age estimation would be of most interest. Hence, the DL-XAI clock was learned from the 6,708 naïve CD4T meta-cells making up the training set, and subsequently validated on the test-set (n = 11,007 meta-cells). As a benchmark, we considered two separate Elastic Net regression models, one restricted to the same genes on which the DL-XAI clock was trained on (i.e. after integration with the PPI network) and another that used the full set of available genes, a lasso regression model (GLMgraph) that takes the PPI network connectivity and topology into account, and finally also a Random Forest (RF) predictor (Methods). We followed the strategy of Buckley et al.8 to report prediction performance in the test set at two levels: (i) at the level of individual donors, by taking the median prediction over all meta-cells of a given donor, and (ii) over all meta-cells. Using the former, resulted in a mean absolute error (MAE) of 8.50 and R-value of 0.64, respectively (Fig. 3a). In contrast, the elastic net and Random Forest benchmarks restricted to the same genes obtained (MAE = 9.42, R = 0.67) and (MAE = 11.96, R = 0.53), respectively, which in the case of ElasticNet improved to MAE = 9.15 and R = 0.70 when trained on all genes (Fig. 3a, SI fig.S2a). Of note, for all clocks, performance measures dropped upon using the meta-cell level predictions instead of the donor-specific median values, indicating that there is substantial variability in the age-predictions made across all meta-cells of a given donor (Fig. 3b, SI fig.S2b). Importantly, regardless of whether we consider individual meta-cells or their donor-specific median values, the DL-XAI clock obtained improved MAEs, albeit at the expense of marginally lower R-values (Fig. 3c). It is noteworthy that the optimal performance of DL-XAI/MGRL was realized at an intermediate fusion parameter value (SI fig.S3). Very similar results were observed for the other immune cell-types (SI fig.S3-S6). Overall, these results indicate that our DL-XAI clock is comparable to the other methods in terms of chronological age prediction.
Predictive accuracy of DL-XAI clock and benchmarking. (a) Scatterplot of donors’ predicted age of naïve CD4 + T meta-cells (y-axis) against donors’ chronological age (x-axis). The predicted age of a given donor (taking the median prediction over all meta-cells of a given donor) was obtained by DL-XAI clock, the elastic net model with PPI genes, the elastic net model with all genes and the GLMgraph model, respectively. (b) Scatterplot of predicted age of naïve CD4T meta-cells (y-axis) against meta cells’ chronological age (x-axis). The predicted age was obtained by DL-XAI clock, the elastic net model with PPI genes, the elastic net model with all genes, and the GLMgraph model, respectively. PCC represents Pearson correlation coefficient, MAE represents mean absolute error, P-value was obtained from Pearson correlation analysis. (c) Barplots displaying the MAE and PCC of the 4 methods plus a Random Forest (RF) model, and for the analyses performed at the level of donors (left panels) and meta-cells (right panels).
DeeperGCN improves prediction compared to other GCN methods
Using the largest naïve CD4 + T-cell dataset, we also compared the predictive performance of the MGRL algorithm to different choices of the GCN. Specifically, we compared DeeperGCN to a classical GCN40, GraphSAGE41and GAT42(Methods). This revealed a marginally better performance for DeeperGCN (SI fig.S7a),consistent with the original study39. We also compared our MGRL framework to one that is purely based on DeeperGCN and which therefore does not fuse the DeeperGCN embedding with the MLP one (Fig. 2a). Without the MLP, performance dropped, thus highlighting the unique advantage of our multi-view fusion approach (SI fig.S7b).
The DL-XAI clock improves biological interpretability
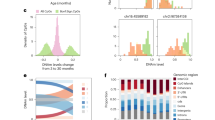
Next, we applied PGExplainer36, a general purpose explainable AI algorithm designed to robustly identify the features that drive the global explanatory power (Methods). For each meta-cell used in training, PGExplainer ranks the PPI edges according to their explanatory power. We ranked the PPI edges by the fraction of meta-cells in which they appeared among the top-100 selected edges (Fig. 4a). For naïve CD4T-cells, a total of 282 PPI edges were selected in at least 4.6% of meta-cells (Fig. 4a), representing a total of 199 unique genes. We verified that these 199 genes displayed significantly stronger associations with chronological age than the complement made up of all other genes (Fig. 4b). Among the 199 genes, we observed a strong enrichment for a small number of KEGG pathways43,44,45, including ribosome and splicesome (Fig. 4c). For the other 3 immune cell-types, thresholds were chosen to yield approximately 200 genes to ensure comparability of KEGG enrichment analyses (Fig. 4a). Broadly speaking, the enrichment of ribosome and splicesome was also seen in the other cell-types, whilst each cell-type also displayed enrichment of specific pathways (Fig. 4b-c). Importantly, performing the same KEGG enrichment analysis on a matched number of top-ranked genes from the Elastic Net, GLMgraph and RF predictors revealed significantly less enrichment (Fig. 4c-d). For instance, for all four immune cell-types, DL-XAI revealed strong enrichment of a ribosomal gene module, not found using an Elastic Net or graph lasso approach (Fig. 4c), whilst the RF predictor did retrieve it but at a lower level of statistical significance (SI fig.S2c-d). This highlights the advantage of our DL-XAI approach which can take full advantage of the underlying PPI network and complex interaction patterns to yield novel biological insight.
DL-XAI clock reveals enrichment of gene modules, not retrievable with penalized regression. (a) PPI edges sorted (x-axis) by the fraction of meta-cells in which a given edge is selected among the top-100 predictive edges of a meta-cell (y-axis), for each cell-type separately: naive CD4 + T cells, CD4 + central memory T cells (CD4TCM), natural killer (NK) cells, and monocytes (CD14MC). The horizontal red dashed line demarcates the threshold for final selection of edges, while the vertical grey dashed line indicates the final number of selected edges. The number of selected edges and unique genes is given in each panel. (b) Violin plot comparing the statistical significance levels (-log10P-values) of Spearman correlations between a gene’s expression level and age, for the genes selected by PGExplainer (PGE) versus the complement of other genes not selected by PGE (Other). The P-value given in the plot is derived from a one-tailed Wilcoxon-rank sum test, comparing the two distributions. (c) Balloon plot representing enrichment of KEGG pathways among the top genes extracted by DL-XAI clock, the elastic net models with PPI restricted and all genes, the GLMgraph and RF models, in each cell type. P-values have been adjusted using the Benjamini-Hochberg (BH) procedure. (d) The number of KEGG enrichment terms for the five models and for each cell type.
Ribosomal gene expression deregulation is an aging hallmark
A striking insight from our DL-XAI clock analyses is the enrichment of ribosomal genes in each of the 4 immune-cell types tested (Fig. 4c). As assessed over the 4 cell-types, a total of 78 ribosomal genes were captured by PGExplainer. We verified that these 78 ribosomal genes displayed stronger associations with age than other genes in the network (Fig. 5a). In each of the 4 immune cell types, ribosomal genes displayed preferential upregulation with age (Fig. 5b-c, SI fig. S8). We further verified that these 78 ribosomal genes displayed preferential deregulation with age in other immune cell-types, including naïve and memory CD8 + T-cells as well as naïve and memory B-cells, although in the case of memory B-cells the majority were downregulated with age, indicating that the direction of expression change may depend on the maturity status of the immune cell-type (SI fig. S9). Importantly, the ribosomal gene module also displayed preferential deregulation with age in brain cell subtypes from mouse8 (SI fig. S10), indicating that their deregulation with age is also seen in other cell-types and species. Thus, the expression levels of ribosomal genes are deregulated with age in a cell type and species independent manner, suggesting that this deregulation is a conserved and universal marker of aging.
The ribosomal gene module is associated with age independently of cell-type. (a) Violin plots display the -log10 transformed P-values with P-values obtained from a Spearman correlation test between gene expression and age, for two groups gene: Ribosome = the union of 78 ribosomal genes selected by PGExplainer across all cell-types, Other = all other genes. The P-value in the violin plot indicates whether the difference between the two groups is statistically significant, and is derived from a one-tailed Wilcoxon-rank sum test. (b) Heatmap of the correlation of 78 ribosomal genes with age in these four cell types. SCC denotes spearman’s rank correlation coefficient, p-values have been adjusted using the Benjamini-Hochberg (BH) procedure to yield FDR values. (c) Bar chart illustrates the fraction of 78 ribosomal genes that are either upregulated or downregulated across different cell types. The fraction is calculated by dividing the number of upregulated or downregulated ribosomal genes by the total of 78 ribosomal genes. The red bars represent upregulated genes, and the blue bars represent downregulated genes. The P-value is obtained from a one-tailed binomial distribution.
DL-XAI clock on DNA methylation data reveals enrichment of novel biological pathways
Next, we trained the same DL-XAI framework on the largest available DNAm dataset of sorted cells, encompassing 1202 monocyte samples (Reynolds et al.)30(Methods). Monocytes were also one of the cell-types analyzed earlier at the scRNA-Seq level, allowing us to assess if similar biological pathways are enriched at the transcriptomic and DNAm-levels. Because DNAm is measured at the level of individual CpGs, we applied a validated method to summarize DNAm at the level of gene-promoters, thus allowing integration with the PPI network used earlier (Methods). To test the DL-XAI clock, we applied it to an independent DNAm dataset of 139 monocyte samples (Methods)46. We found that the DL-XAI clock outperformed age-predictors built with Elastic Net or GLMgraph, but only when using the median absolute error as our evaluation metric, as the other models performed better in terms of Pearson Correlations (Fig. 6a). We verified that the 187 genes selected by PGExplainer were more correlated with chronological age than the complement of other genes (Fig. 6b-c). In line with the successful validation of the DL-XAI clock in the test set, these 187 genes also displayed a correlative association with age in the independent DNAm dataset (Fig. 6d), thus clearly demonstrating that these gene promoters do change their DNAm levels with age in monocytes. Next, we performed gene-set enrichment analysis of KEGG pathways among the 187 genes, which revealed significant enrichment for many pathways including notably cytokine cytokine-receptor interactions and JAK-STAT signaling (Fig. 6e, SI fig.S11). Of note, the corresponding Elastic Net and GLMgraph age predictors did not display enrichment for these same terms, nor of any other KEGG pathways, suggesting that the DL-XAI clock can significantly improve biological interpretability without compromising predictive accuracy (Fig. 6e). Cytokine and JAK-STAT signaling pathways play key roles in the aging immune system, controlling the balance of pro vs. anti-inflammatory responses47, and DNAm mediated deregulation in monocytes has been observed48. Consistent with this, we observed the expected anti-correlation between age-associated promoter DNAm levels and corresponding age-associated gene expression in the Reynolds et al. dataset (Fig. 6f), for which matched bulk RNA-Seq data of monocytes is available30. For instance, STAT4 displayed promoter hypomethylation and overexpression with increased age in monocytes, whilst our DL-XAI clock analysis implicates an epigenetically deregulated pathway involving IL4 and IL12 receptors, upstream of STAT4 (Fig. 6f-g).
Application of DL-XAI to DNAm in monocytes reveals enrichment of JAK-STAT signaling. (a) Scatterplot of predicted age (y-axis) against chronological age (x-axis) of independent Blueprint monocyte samples, obtained by interpretable MGRL model (DL-XAI), the elastic net models with PPI genes and all genes, and GLMgraph model, respectively. (b) Scatterplot illustrates the distribution of edge fractions for the edges derived from MGRL model trained on Reynolds et al. by using PGExplainer. (c) Violin plots comparing the significance levels of the top 187 genes to the complement of all other genes. The y-axis represents the -log10 transformed P-values, with P-values obtained from a t-test. The P-value indicating the difference between the two groups is obtained through one-tailed Wilcoxon-rank sum test. (d) Scatterplot of t-statistics for each gene derived from BluePrint DNAm data set against the corresponding t-statistic from Reynolds DNAm data set. t-statistics were obtained from linear regression of promoter DNAm against age. The gray line indicates the nominal significance level 0.05. The P-value is obtained from a one-tailed binomial distribution. (e) KEGG enrichment analysis displaying results for all 4 clock models, as shown. P-values have been adjusted using the Benjamini-Hochberg (BH) procedure. (f) Scatterplot of Spearman correlation coefficient(SCC) of the PGExplainer selected genes with age, as derived from Reynolds bulk RNA-seq data against the corresponding t-statistic from Reynolds DNAm data, with t-statistics obtained from a linear regression of promoter DNAm against age. The gray line indicates the nominal significance level 0.05. The P-value is obtained from a one-tailed Fisher exact test. (g) The top selected genes enriched in the JAK-STAT signaling pathway. The round nodes represent the genes, while the round rectangular node represents the KEGG term. The top selected genes are labeled with a bright green border. Magenta indicates top selected edges, light gray lines indicate remaining edges in the PPI network and dot line indicates KEGG enrichment. The t-statistic (t-stat) color the nodes and were obtained from a linear regression of promoter DNAm against age in the Reynolds DNAm data set.
Discussion
In this paper, we have explored the application of DL and XAI technology to build molecular predictors of chronological age in two distinct data-types (mRNA expression and DNAm) using omic data from approximately 1000 subjects, each one generated at cell-type resolution (single-cell for mRNA expression, sorted cells for DNAm), thus avoiding the confounding affect posed by cell-type heterogeneity49,50. In order to take advantage of a DL-paradigm, we considered gene expression and DNAm values embedded in a spatial manifold, as defined by a highly curated PPI network. We have seen that this approach marginally outperformed analogous age predictors developed within an Elastic Net penalized regression framework when using the median absolute error as the evaluation metric, but not when using Pearson correlations. This dependence on the evaluation metric could be related to subtle non-linearities, which the DL-based approach is better at capturing, rendering it less favorable when assessed using a Pearson (linear) correlation metric. On the other hand, a Random Forest age predictor performed significantly less well, even when compared to Elastic Net, which is probably related to the need to use larger ensemble of trees, which is computationally expensive. Of note, the marginal nature of the improvement of our DL-XAI clock in predicting chronological age is not entirely surprising: in the case of DNAm, current epigenetic clocks based on penalized regression already reach very high accuracies, for instance, Zhang’s clock can predict chronological age with a MAE of +/- 1–3 years51, hence the scope for improvement is narrow. In the case of scRNA-Seq data, the room for improvement in predicting chronological age is far greater. Although our DL-based clocks did not reach the R/PCC values recently reported by Buckley et al. for penalized linear regression based clocks in various brain cell subtypes8, our benchmarking against Elastic Net models suggests that the reason for these differences can be attributed to the different datasets and cell-types. In this regard, it is worth noting that we analyzed the largest available scRNA-Seq dataset, encompassing over 1 million single-cells from 981 donors, and that performance metrics obtained on much smaller scRNA-Seq datasets, such as the one considered in Buckley et al., could be subject to a larger sampling error and thus appear inflated. Overall, however, it would appear that the adoption of deep learning does not result in a substantially improved prediction accuracy for chronological age, consistent with recent studies23,24,25.
On the other hand, our DL-XAI/MGRL strategy has shed important novel biological insight into aging, highlighting the significance and added value of GCNs over the simpler penalized regression models or the lasso regression model defined over the same PPI network (i.e. GLMgraph). Indeed, the lack of interpretability of molecular clocks built with simple penalized regression models has been repeatedly emphasized52,53. The improvement over GLMgraph is particularly noteworthy as it highlights the importance of a deep-learning framework as both DL-XAI and GLMgraph use exactly the same underlying PPI network. In particular, using our DL-XAI clock we were able to identify a 78 ribosomal gene expression module that is consistently altered with age independently of cell-type and species, with most of these ribosomal genes displaying upregulation with age. This finding was not forthcoming from other scRNA-Seq aging studies19,21, although the upregulation of ribosomal genes has been observed in studies comparing young and old hematopoietic stem cells54. Moreover, whilst other studies have successfully identified specific biological processes (e.g. senescence, antigen and collagen processing, circadian rhythm, inflammation) that are altered in aging, often in a cell-type independent manner19,21, they did not identify any ribosomal gene module, highlighting the importance of using an advanced machine learning strategy. Previous single-cell studies that have explored age-associated transcriptomic changes have so far also been limited to profiling a relatively small number of subjects19,20, which points to another potential reason why our analysis has revealed novel biological insight.
Our MGRL-analysis on DNAm data in monocytes revealed epigenetic deregulation of cytokine and JAK-STAT signaling pathways, a result that was also not forthcoming using simpler penalized regression models or GLMgraph. In particular, the DL-XAI clock revealed a hierarchical pattern of age-associated deregulation involving the cytokine IL4, cytokine receptors IL12RB1/2 and STAT4, which could reflect an enhanced differentiation of monocytes towards M2 macrophages, and which is typically associated with anti-inflammatory responses and tissue repair55. It may thus indicate an age-related shift towards pro-repair mechanisms, potentially to counteract chronic inflammation or DNA damage accumulated over time56,57. Although the IL12 receptors were not individually altered, the deep-learning analysis implicates a role for them in mediating the deregulation of STAT4, which is entirely plausible since our GCNN is designed to capture complex interactive association patterns which are not evident at the single-node level. Consistent with this, IL12-signaling is known to promote the production of inflammatory cytokines57.
Of more theoretical interest, it is worth highlighting that our proposed MGRL framework, which combines DeeperGCN and MLP for chronological age prediction, and PGExplainer for interpretation, performed marginally better than more classical alternatives. In this regard, our interpretable MGRL method has two significant advantages compared to existing methods. First, the multi-view fusion strategy enables the model to fully utilize the joint topological-expression/DNAm information captured by DeeperGCN and the global expression/DNAm information extracted by MLP, which enhances the model’s representation learning capability. Indeed, removing the MLP part from the algorithm led to reduced performance, and optimal performance was observed for intermediate values of the fusion hyperparameter, indicating that the fusion approach is advantageous. Second, the interpretable MGLR employs PGExplainer to elucidate the factors affecting cell age prediction. PGExplainer is a perturbation-based interpretable method that parameterizes the interpretation process from a global perspective by a deep neural network, enabling multiple instances of interpretation and providing a comprehensive assessment of all genes and their expression values. In addition, PGExplainer has good generalization ability, which facilitates its application in an inductive environment. In contrast to the attention mechanism, PGExplainer is able to measure the significance of the same PPI in scRNA-seq datasets and avoids the linear, localized interpretation limitations imposed by gradients-based interpretable methods58,59.
Importantly, we have also shown that DeeperGCN is an improvement over more classical GCN alternatives40,41,42,60,61. There are two main reasons for this. First, DeeperGCN can more effectively utilize the joint topological and feature information of large input graphs, which is critical to improve the graph-level representation learning ability. Second, for large-scale graph-level representation learning, the limited number of layers of traditional GCNs is insufficient to expand its receptive field60,61, and increasing the number of layers of the network leads to over-smoothing and over-squeezing problems62, which further limit its representation capability. Thus, more advanced graph neural network models are needed to handle large-scale graph data. DeeperGCN uses a generalized differentiable message aggregation function, whilst also introducing a variant residual connection and message normalization.
Although the interpretable MGLR framework has shown good performance on scRNA-seq and DNAm datasets, one obvious limitation is the computational cost involved (SI fig.S12). For instance, the time taken to train a single epoch of the MGRL model on the 8511 node network and 10,000 meta-cells, using two NVIDIA A100 GPUs, is around a minute, which makes it much more computationally expensive than the Elastic Net models, when using 50 to 100 epochs. Nevertheless, our approach is computational feasible and leads to significantly improved biological insights compared to Elastic Net.
There are also two aspects that could further enhance the model’s learning and interpretability capabilities. First, an end-to-end interpretable GCN model could be developed that integrates prediction and interpretability in a single model. This would allow direct modeling of complex relationships in graph structures and generate globally consistent interpretations. This would not only help users understand the decision-making process of the model more deeply whilst enhancing the transparency and credibility of the model, but also simplify the complexity of the model and provide more intuitive explanations. Second, current large language models (LLMs)63 have achieved great success in the field of natural language processing. Applying LLMs in our context, could conceivably improve the identification of important factors and biological mechanisms affecting key aging processes, such as inflammation and immune-senescence.
Finally, we note that the deep-learning framework presented here could be applied to other single cell (or sorted) data-types, such as protein expression64or ATAC-Seq65. The only requirement is the need to summarize the measurements at the gene-level, which in the case of scATAC-Seq data, could be accomplished by computing gene chromatin accessibility scores.
In summary, this work points towards advanced graph convolutional neural networks and XAI, as a promising deep learning strategy for exploring complex biological phenomena such as aging. With ever larger omic datasets and ongoing improvements in mapping signaling pathway information, the application of these tools is likely to further unravel novel important features of biological aging.
Materials and methods
Data preprocessing procedures
scRNA-Seq dataset
The scRNA Seq dataset analyzed here is that of Yazar et al.29 who profiled a total of 1,248,980 peripheral blood mononuclear cells (PBMCs) from 981 donors. The dataset, provided as a raw Seurat Object, was downloaded from https://cellxgene.cziscience.com/collections/dde06e0f-ab3b-46be-96a2-a8082383c4a. This object contained the associated metadata encompassing cell type annotations to 29 PBMC subtypes, matched sample IDs, age, sex, and UMAP coordinates for all cells. Among the PBMC subtypes, we focused on lymphoid and myeloid cell-types that had highest cell numbers to ensure adequate power: these were naive CD4 + T-cells (nCD4T, n = 259,012), central memory CD4 + T-cells (CD4TM, n = 289,000), natural-killer cells (NK, n = 164,933) and CD14 + monocytes (Mono, n = 36,130).
DNA methylation (DNAm) datasets
We used an Illumina 450k DNAm dataset of 1202 sorted monocyte samples from Reynolds et al.30 for training and an independent Illumina 450k DNAm dataset of 139 monocyte samples from the BLUEPRINT consortium46 for testing. Analysis was restricted to the common set of 440,905 CpGs (after quality control), using processed normalized data as described previously66. Because the DNAm dataset is defined at the level of CpGs, in order to integrate with a PPI network (defined at the gene level), we summarized DNAm-values at the gene-level, by averaging the DNAm values of CpGs mapping to within 200 bp upstream of the gene’s transcription start-site (TSS). If no probes/CpGs map to this region, we use the average of 1st exon probes, and if these are also not present, we then average DNAm of CpGs 1500 bp upstream of the TSS. In case there are no such probes, the gene is not assigned a DNAm-value and is removed. This procedure of assigning DNAm-values to genes, has been validated by us previously67,68. At the end of this procedure, our DNAm data matrices were defined over 13,288 protein coding genes and 1202 (training) and 139 (testing) monocyte samples, respectively.
PPI network
As a scaffold for reported protein-protein interactions we used a highly curated integrated version from Pathway Commons31. Proteins in this network were assigned a main cellular localization, using cellular localization data from the Human Protein Reference Database (HPRD), as described by us previously69. We considered a total of 5 main cellular localizations or signaling domains: growth modulators (GM), secreted factors (SF), membrane receptors (MR), intracellular receptor substrates (ICRS) and intracellular non-receptor substrates (ICNRS). Specifically, we first defined an intra-cellular domain as all those GO-terms containing the following terms: Nucleus, Cytoplasm, Ribosome, Nucleolus, Mitochondri, Endoplasmic reticulum, Golgi, Lysosome, Cytosol, Cytoskeleton, Nuclear, Kinetochore, Chromosome, Endosome, Intracellular, Nucleoplasm, Perinuclear, Centrosome, Peroxisome, Microtubule, Microsome, Endosome, Centriole, Sarcoplasm, Secretory granule, Endocytic vesicle, Cytoskeleton, Peroxisomal membrane, Acrosome, Zymogen granule. For the membrane-receptor (MR) domain we used: Plasma membrane, Integral to membrane, Cell surface, Integral to plasma membrane, Cell projection, Basolateral membrane, Axoneme, Apical membrane and for the extra-cellular (EC) domain: Extracellular, Cell junction, Synapse, Dendrite, Secreted, Synaptic vesicle. The IC class was subdivided further into ICRS and ICNRS subclasses, according to whether the IC annotated protein interacts with a MR: if yes, it was assigned to ICRS, if not to ICNRS. Similarly, the EC class was subdivided further into GM and SF subclasses, according to whether the EC annotated protein interacts with a MR (SF) or not (GM). With this assignment, cytokines would map to GMs, ligands to SFs, kinases + phosphatases to ICRS and most transcription factors to ICNRS. The network was further filtered by removing edges for proteins within signaling domains or in non-neighboring signaling domains, e.g. a reported interaction between a GM and an IC protein was removed. In total this resulted in a sparse but connected network of 12,649 proteins, 464,091 edges (i.e. 0.2% of the total number of possible edges). Of the 12,649 proteins, 177 were unambiguously assigned to GM, 592 to SF, 2576 to MR, 4799 to ICRS and 2494 to ICNRS, with the rest being ambiguous and not assigned any domain.
Integration of PPI network with scRNA-Seq and DNAm datasets
scRNA-Seq data
We first removed genes expressed by less than 20 cells in these four cell types. Then, we mapped the remaining genes onto the PPI network. We further removed edges between genes (proteins) in the same signaling layer/domain. To ensure the connectivity of the entire network, the largest connected component in the mapped PPI network was selected as the integrated PPI network. As a result, the final PPI network has 8,511 nodes and 74,303 edges.
DNAm data
First, we summarized the Illumina 450k CpG-level DNAm monocyte data from Reynolds et al. at the gene-level, as described in the earlier subsection. We then identified the common genes between the DNAm data and the PPI network, removing any edges within the same signaling layer. We then extracted the largest connected component, resulting in an integrated PPI network containing 6,557 nodes and 37,866 edges.
Meta-cell construction
The use of meta-cells, defined as glomerates of single-cells (or pseudo single-cells) has been shown to help denoise scRNA-Seq data8. In building meta-cells, a key trade-off is the number of cells to use per meta-cell, since too many cells per meta-cell can mask biological variability and reduce power, whilst using too few cells would not help denoise the data. Given the extensive analysis performed by Buckley et al. in identifying the optimal trade-off regime8, we henceforth decided to use 15 cells per meta-cell. Since donors contribute widely different numbers of cells, the number of meta-cells per donor can vary. Specifically, the number of meta-cells per donor was determined by taking the integer ceiling of the ratio of number of cells divided by 15. For instance, for a donor contributing 100 cells, we built 7 (ceiling of 100/15 ~ 6.667) meta-cells. The cells making up each meta-cell were selected by randomly sampling without replacement 15 cells from all donor cells. This strategy optimized usage from as many cells as possible, whilst also minimizing redundancy from sampling the same cell multiple times. For each meta-cell, expression counts from all 15 cells were aggregated.
Age prediction based on multi-view graph-level representation learning.
-
a.
A. Notation and problem definition: In what follows we assume that X is the expression data matrix for a given cell-type (e.g. NK-cells) obtained after integration with the PPI network, with A representing the adjacency matrix of the PPI network. Similar considerations apply to the case where X is the DNAm data matrix. Thus, each cell is defined by a network whose topology is the same across all cells but where the node attributes (e.g. gene expression values) differ from cell to cell. Mathematically, for each cell we have a graph \(\:\rotatebox[origin=c]{25}{\calligra g}(\it{V},\:\it{X},\:\it{A})\)consisting of (i) a set of nodes \(\:\text{V}=\left({v}_{1},{v}_{2},\cdots\:{v}_{N}\right)\:\)where N is the number of nodes, (ii) a feature matrix X defined over these nodes and d node attributes, and (iii) an adjacency matrix \(\:A\). The feature matrix \(\:\text{X}\in\:{\mathbb{R}}^{N\times\:d}\), where each row \(\:{x}_{i}\) is a \(\:d\)-dimensional vector representing the attribute values of the node \(\:{v}_{\text{i}}\). In our applications, d=1, so the feature matrix is merely a feature vector containing the gene expression or DNAm values. The adjacency matrix \(\:\text{A}\in\:{\mathbb{R}}^{N\times\:N}\) is used to describe the connectivity between nodes in the graph. Specifically, if there is an edge connecting the node \(\:{v}_{i}\) to node \(\:{v}_{j}\), then \(\:{A}_{i,j}=1\), otherwise, \(\:{A}_{i,j}=0\). In the task of chronological age prediction, we utilize a protein-protein interaction (PPI) network as the adjacency matrix \(\:A\). The gene expression data matrix yields the feature vector of the nodes for each cell/graph. Given a dataset \(\:\text{D}=\left\{\left({\rotatebox[origin=c]{25}{\calligra g}}_{1},{y}_{1}\right),\left({\rotatebox[origin=c]{25}{\calligra g}}_{2},{y}_{2}\right),\cdots\:,\left({\rotatebox[origin=c]{25}{\calligra g}}_{n},{y}_{n}\right)\right\}\), where \(\:{\rotatebox[origin=c]{25}{\calligra g}}_{m}\) represents the graph for cell m and \(\:{\text{y}}_{m}\) is the corresponding chronological age value, the challenge is to learn a mapping function \(\:\text{f}:\rotatebox[origin=c]{25}{\calligra g}\to\:y\), which can accurately predict the chronological age of cells, using the PPI matrix and gene expression data matrix as input.
-
b.
B. The Overall Neural Network Architecture: The framework integrates two main views, topology and features, to achieve a comprehensive extraction and deep fusion of cellular network information. The topological view utilizes a deep graph convolutional network to deeply mine the topological structure information of the graph. Through a linear node encoding layer and two-layer DeeperGCN network architecture with residual connections, the model is able to pass and enhance the association features among nodes layer by layer, effectively capturing the complex interaction patterns and structural properties in cellular networks. The introduction of residual connections not only deepens the depth of the network, but also mitigates the common gradient vanishing problem of deep networks, ensuring the efficient transfer and utilization of information. Subsequently, through the average pooling technique, the model aggregates the fused node-level information into a graph-level embedding. The feature view is parallel to the topology view, and mainly utilizes a Multi-Layer Perceptron (MLP) to achieve high-level abstraction and representation learning of node features. Using a two-layer MLP architecture, the feature view is able to flexibly learn and extract biological features specific to each node, which are critical for understanding cell states and predicting their age. In order to achieve full fusion of information, the model efficiently fuses the information extracted from both topology and feature views. This process not only takes into account the topological information of the graph, but also the feature information of the nodes, thus constructing a richer and more comprehensive representation of the cellular network. The fused embedding vector comprehensively reflects the whole state and characteristics of the cellular network. Finally, the fused embedding is fed into a single fully connected layer, which is further processed and transformed to output the predicted value of cell age. The output dimension of the fully connected layer is set to 1, which directly corresponds to the predicted cell age, ensuring the intuition and accuracy of the prediction results. This design not only improves the prediction performance of the model, but also makes the results easy to interpret and apply.
-
c.
C. Topology View: The topological view is mainly used to extract the topological structure information of the graph using a deep graph convolutional neural network (GCNN). A GCNN is a neural network model specialized in processing graph-structured data in the field of deep learning. It uses the message-passing mechanism70 to realize the aggregation and update of information of nodes on the graph. This mechanism simulates the flow of information through the graph, i.e., each node receives information (messages) from its neighboring nodes through its connected edges and updates its state (feature representation) accordingly. This process can be carried out iteratively for many times, allowing messages to propagate throughout the graph, thus capturing information about the global structure of the graph. The message-passing graph-based neural network can be described as:
where ⊕ denotes a permutation invariant, differentiable function such as a maximum, average, or summation. \(\:{x}_{i}^{l}\) denotes the embedding of node \(\:{v}_{i}\) in layer \(\:l\), and \(\:{e}_{j,i}\:\)denotes the edge features between node \(\:{v}_{j}\) and node \(\:{v}_{i}\).
When dealing with large-scale graph data, traditional GCNs (e.g., GCN40, GraphSAGE41, and GAT42, etc.) tend to suffer from significant limitations, especially the overs-smoothing and over-squeezing problems, which constrain their ability to perform on complex graph structures60,61. These problems are particularly prominent on graph regression tasks, where the prediction goal needs to accurately capture the subtle dependencies between global and long-range nodes. However, the limited receptive field of GCNs is difficult to fully cover these long-range interactions, and simply expanding the receptive field by increasing the number of network layers often leads to over-smoothing and over-squeezing of information, which in turn weakens the expressive power of the model.
To break through this bottleneck, we adapt a Deep Graph Convolutional Network (DeeperGCN)71, an innovative framework designed to address the problem of limited expressiveness of graph neural networks on large-scale graph data. The key feature of DeeperGCN lies in its definition of a set of generalized differentiable information aggregation functions, which can not only effectively integrate the information of node neighborhoods, but also maintain the richness and variability of the information in the deep network, thus avoiding the over-smoothing problem that occurs in the deep stacking of traditional GCN. In addition, DeeperGCN elegantly incorporates variant residual connections with information normalization layers. The variant residual connection alleviates the problem of gradient vanishing in deep network training by directly transferring information across layers, ensuring the effective transfer and utilization of information. The information normalization layer further refines the feature representation and prevents extreme changes in the feature scale during propagation, i.e., the phenomenon of over-squeezing, thus maintaining the stability and accuracy of model learning. DeeperGCN significantly enhances the processing power on large-scale graph data through a well-designed information aggregation and updating mechanism. The key step of DeeperGCN can be represented as follows:
where \(\:{h}_{i}^{l}\) denotes the embedding of node \(\:{v}_{\text{i}}\:\)in the output of layer \(\:l\). \(\:{AGG}_{{SoftMax}_{\beta\:}}\left(\bullet\:\right)\) is an information aggregation function using a generalized SoftMax aggregation function that balances the averaging and maximization of node features. It flexibly controls the strength of the aggregation behavior by adjusting the inverse temperature parameter \(\:\beta\:\). \(\:\beta\:\) is a continuous variable that allows the model to adaptively adjust its aggregation features during training. Specifically, the \(\:{AGG}_{{SoftMax}_{\beta\:}}\left(\bullet\:\right)\) aggregation function is a generalization of the traditional SoftMax function, which is more effective in dealing with the complex relationships among graph nodes. \(\:{AGG}_{{SoftMax}_{\beta\:}}\left(\bullet\:\right)\) can be denoted as:
where \(\:{m}_{j,i}=\rho\:\left({x}_{i},{x}_{j},{x}_{{e}_{j,i}}\right)\), which is a learnable or differentiable function to calculate the correlation or importance weight between node \(\:{v}_{i}\) and its neighbor \(\:{v}_{j}\), taking into account the node’s features \(\:{x}_{i}\) ,\(\:{x}_{j}\), and the edge features \(\:{x}_{{e}_{j,i}}\).This design allows the \(\:{AGG}_{{SoftMax}_{\beta\:}}\left(\bullet\:\right)\) aggregation function to capture more subtle and rich inter-node dependencies. Once we have extracted the structural information of the graph using DeeperGCN, we need to summarize all the node information on the graph. By employing average pooling techniques, we effectively integrate the representations of individual nodes to generate a comprehensive and refined graph-level representation.
D. Feature View: Graph data not only contains rich structural information, but also carries a variety of feature information. Effective utilization of the feature information is crucial to enhance the graph representation72. In order to adaptively aggregate and refine the feature information, we adopt a MLP. The MLP, as a classical feed-forward artificial neural network model, has an architecture carefully constructed from input layers, several hidden layers, and output layers. The MLP is a classical feed-forward artificial neural network model whose architecture is carefully constructed from input layers, several hidden layers, and output layers. In the structure of the MLP, neurons in each layer are fully connected to all neurons in the next layer, and this dense connectivity pattern ensures comprehensive information transfer and fusion. At the same time, this connection mechanism is accompanied by the introduction of weights, which enables the network to learn and adapt to the complex characteristics of the data. By combining nonlinear activation functions (e.g., ReLU, Sigmoid, Tanh, etc.), the MLP exhibits a powerful nonlinear mapping capability that captures and approximates complex functional relationships. In the training phase, the backpropagation algorithm is cleverly applied to continuously optimize the weight parameters in the network based on the gradient descent method, aiming to minimize the error between the output prediction and the real labels, and thus to improve the performance and accuracy of the model. Due to the fully-connected nature of MLP, it demonstrates excellent results in handling tasks with fixed-size input vectors. Although MLP does not directly deal with the structural information of the graph, we can use the feature vectors of the nodes as inputs to MLP. Through the powerful nonlinear transformation capability of MLP, deeper feature representations are adaptively aggregated and extracted, which in turn enhances the understanding and analysis of the graph data. The feature information of the graph extracted by MLP can be denoted as:
where \(\:\delta\:\left(\bullet\:\right)\) is the activation function.
E. Multi-view Fusion: In order to achieve the deep fusion of graph structure information and feature information and optimize the synergy of these two perspectives, we adopt a well-designed hyper-parameter adjustment mechanism. This mechanism can balance the graph structural information extracted by DeeperGCN with the graph feature information captured by MLP. By carefully tuning this hyperparameter, we are able to ensure that the two types of information are appropriately weighed in the model learning process, thus enhancing the overall representation capability. Specifically, this multi-view information fusion strategy can be formulated as follows:
where \(\:{f}_{i}\:\)denotes the embedding of node \(\:i\) after fusion. By tuning specific hyperparameters, we achieve precise control over the structural insights of DeeperGCN and the feature understanding of MLP, which in turn facilitates the fusion of the two types of information. This strategy not only enhances the model’s grasp of the intrinsic complexity of graph data, but also provides a richer and more precise view for graph analysis tasks.
F. Graph-level Prediction: The fused muti-view graph representations capture key features of cellular networks and provide us with an integrated view to understand the intrinsic properties of cells. After we obtain the fused information, we input these graph representations into an elaborate fully-connected network that is capable of further analyzing and learning the relationship between cellular features and age. With this structured method, we constructed a powerful predictive model that accurately predicts the age of cells, providing a new analytical tool for biological research and clinical applications.
Deep graph neural network-based interpretable prediction based on MGRL
In order to interpret biologically the age predictions obtained by the MGRL above, we adapt an XAI-tool called PGExplainer73. First, we decompose the original input graph into two subgraphs: the explanation graph \(\:{g}_{exp}\) and the residual graph \(\:{g}_{\text{r}\text{e}\text{s}}\). The explanation graph highlights the underlying structures that contribute significantly to the MGRL prediction results, while the residual graph contains the edge information that is not directly related to the prediction task. Next, we follow a procedure similar to the one used in GNNExplainer74, to maximize the mutual information between the prediction output of the MGRL with the explanation graph as input, and the explanation graph. Since directly optimizing the mutual information is computationally dense, we use an approximation strategy by assuming that the graph is a Gilbert random graph and selecting conditionally independent edges from it. We then use a neural network to predict the probability of an edge appearing in the interpretation graph and optimize the objective function through a reparameterization technique. To improve the global interpretability of the model, we further employ a multi-instance learning strategy. Specifically, we make predictions on the trained model and utilize a deep neural network to compute the latent variables of the edge distribution. By sampling the graphs from the latent variables and calculating the Mean Squared Error (MSE) loss between the predicted values of the sampled graph and the original predicted values, we can learn the optimal deep neural network parameters. Finally, the set of subgraph edges with potentially high contribution to the prediction task is selected based on the latent variables to obtain the important nodes and edges that affect the prediction results. Through these steps, we can effectively interpret the prediction results of the MGRL model and improve the trust and reliability of the model. The procedure is described mathematically below:
In the interpretation of the MGRL model predictions, we decompose the input graph \(\:g\) into two complementary subgraphs \(\:{g}_{S}\) and \(\:{g}_{\text{r}\text{e}\text{s}}\). \(\:{g}_{S}\) is the sampled subgraph and also the expected interpretation graph, which represents the underlying structures that contribute significantly to the MGRL prediction results. \(\:{g}_{\text{r}\text{e}\text{s}}\) contains the graphs of residual information, i.e., the structures that are not directly relevant to the current MGRL prediction task. To determine \(\:{g}_{exp}\), we use a method similar to GNNExplainer to maximize the mutual information between the predicted output of origin graph \(\:{\text{y}}_{\text{O}}\) and the predicted output of the sampled subgraph \(\:{\text{y}}_{S}\) when MGRL takes \(\:{g}_{S}\) as input. However, directly optimizing the mutual information is computationally dense, so we adopt an assumption-and-approximation strategy: For any nodes \(\:{v}_{i}\) and \(\:{v}_{j}\) in the graph, their node embeddings are \(\:{h}_{i}\) and \(\:{h}_{j}\), respectively. we utilize a deep neural network \(\:{f}_{\varphi\:}\left(\bullet\:\right)\:\)in which \(\:\varphi\:\) denotes its network parameters to predict the probability \(\:P({e}_{i,j}\in\:{g}_{S}|{h}_{i},{h}_{j})\)) of the edge\(\:\:{e}_{i,j}\) appearing in the interpreted graph \(\:{g}_{exp}\). Through the reparameterization technique, we introduce random variables to approximate the optimization objective and use MSE loss for optimization. Specifically, it first computes the latent edge selection variable \(\:{\Omega\:}\) through a deep neural network, which reflects which edges are more likely to form the explanatory subgraph \(\:{g}_{S}\). This process can be represented as:
where \(\:\mathcal{F}\varrho\:(\bullet\:)\) denotes the MGRL network and \(\:\varrho\:\) denotes its network parameters. Next, we sample subgraphs from \(\:{\Omega\:}\) and input them into a pre-trained MGRL model to obtain new predicted values and update the interpretation network parameters by using the difference with the original predicted values as a MSE loss:
where \(\:{\dot{\rotatebox[origin=c]{25}{\calligra g}}}_{S}^{k}\)denotes the \(\:k\)-th subgraph \(\:{\rotatebox[origin=c]{25}{\calligra g}}_{S}\) sampled from the edge probability distribution \(\:{\Omega\:}\), \(\:k\) denotes the subgraph index, and \(\:K\) is the total number of subgraphs. Then, in terms of global interpretability, through multi-instance learning, we iteratively learn the optimal explanatory network parameters from multiple instances (i.e., different input graphs). The Eq. (7) can be further represented as:
\(\:T\) denotes the set of multiple instances, \(\:{\rotatebox[origin=c]{25}{\calligra g}}_{O}^{t}\) denotes the original graph of the \(\:t\)-th instance, and \(\:{\dot{\rotatebox[origin=c]{25}{\calligra g}}}_{S}^{t,k}\) denotes the \(\:k\)-th sampled subgraph of the \(\:t\)-th instance. Finally, the deep neural network parameters are optimised through Eq. (8), and we select the set of subgraph edges with high potential contribution based on latent variables \(\:{\Omega\:}\). In this way, we can efficiently identify important nodes and edges that affect model predictions.
Construction and validation of the DL-XAI clock
We used the MGRL framework described above to train a corresponding predictor of chronological age (“DL-XAI-clock”). In the case of scRNA-Seq data, we illustrate the strategy for the naïve CD4 + T-cells. A similar approach was taken for all other immune cell-types. The 981 donors from the Yazar et al. data set29 were first divided into 6 age groups: (0,40), [40,50), [50,60), [60,70), [70,80), [80,100). The sample sizes of the six groups are 108, 76, 117, 261, 266, and 153 respectively. Five individuals contributing less than 15 naive CD4 T cells were excluded, resulting in sample sizes of 108, 76, 116, 261, 265, and 150 for each age-group, respectively. In order to ensure that the training is not biased towards older groups, we randomly selected 60 donors from each age group as the training set (360 donors in total), further requiring that the number of meta-cells of these donors to be between 5 and 40. The maximum of 40 was chosen to avoid certain donors from contributing too many meta-cells. The rest of donors, i.e. 616 in total, made up the test set. In the case of DNAm data, we used the 1202 sorted monocyte samples from Reynolds et al.30for training, and the 139 sorted monocyte samples from BLUEPRINT46 for testing.
We developed the interpretable MGRL framework utilizing PyTorch75and its PyTorch-geometric76 library on two NVIDIA A100 GPUs. In scRNA-Seq datasets, the embedding dimension of the node encoding layer in the topology view is 100 and our DeeperGCN implementation consists of two layers, each with a dimension of 100, to capture the complex relationships in the graph data. The MLP in our method comprises two hidden layers (with dimensions 1000 and 100) and a 100-dimensional output layer, to enhance the feature extraction capability. To prevent model overfitting, we set the dropout to 0.2 in the convolution and output in DeeperGCN. In order to achieve an optimal balance between the topology view information and the feature view information, we carefully tuned the hyperparameter \(\:\theta\:\) in Eq. (5) to 0.5, and this adjustment applies to all datasets except the CD14MC dataset, for which we set the value of \(\:\theta\:\) to 0.9. Furthermore, we optimized the training parameters by setting the batch size to 64 and the learning rate to 0.001. Finally, we configured the fully connected layer to output a dimension of 1, directly predicting age. Since the DNAm matrix is denser than the scRNA-Seq gene expression matrix and the PPI matrix of DNAm contains more complex relationships, in order to better extract the topology and feature information, our MGRL in DNAm datasets is set to 3-layer, containing three layers of DeeperGCN and three layers of MLP.
Construction and validation of the ElasticNet (PPI) & ElasticNet (all) clocks
We built the ElasticNet (PPI) model and ElasticNet (All) model for predicting chronological age using the R package glmnet (version 4.1.8)37, with the elastic net parameter \(\:\alpha\:\) set to 0.5. For both scRNA-seq data and DNAm data, the training set and test set used for both models are the same as those used in the DL-XAI model. However, the ElasticNet (PPI) model employs the same gene set as the DL-XAI model, while the ElasticNet (All) model utilizes all available genes. During the training of the models, the optimal penalty parameter (\(\:\lambda\:\)) was obtained by five-fold cross-validation.
Construction and validation of the GLMgraph (PPI) clock
We used the R package glmgraph (version 1.0.3)77,78 to build a chronological age predictor with PPI network constraints. This penalized likelihood method has two penalty terms, one for sparsity \(\:{\lambda\:}_{1}\), and another for smoothness \(\:{\lambda\:}_{2}\), the latter being determined by the graph Laplacian L78. In more detail, if y encodes age and X the data matrix with rows labeling genes and columns the samples (meta-cells or sorted samples), then the penalized loss function is:
The smoothness penalty term \(\:{\beta\:}^{T}L\beta\:\) can be expressed as \(\:{\sum\:}_{i,\:j\le\:i}{A}_{ij}{\left({\beta\:}_{i}-{\beta\:}_{j}\right)}^{2}\) where \(\:{A}_{ij}\) is the adjacency matrix of the PPI network. For both the scRNA-seq data and DNAm data, the training set, test set and PPI network used for the model are the same as those used in the DL-XAI model. During the training of the model, the penalty parameters were optimized on grid with a five-fold cross-validation.
Construction and validation of the random forest model
To optimize the predictive performance of a Random Forest model79 for estimating chronological age, we utilized the randomForest package (version 4.7–1.2) in R. The model’s ‘ntree’ hyperparameter, which governs the number of trees in the Random Forest model, was calibrated using a five-fold cross-validation approach. We systematically incremented the ‘ntree’ value in steps of 50, starting from 50 until reaching the upper limit of 500 trees. Upon determining the optimal ‘ntree’ through this cross-validation procedure, we constructed the final Random Forest model using this value, ensuring that each tree had a minimum node size of five.
Gene Set Enrichment Analysis (GSEA)
The R package clusterProfiler (version 4.12.0)80 is used for KEGG enrichment analysis for top genes extracted from different models. For the top genes extracted by the ElasticNet (All) model, the background gene set consists of all genes expressed in the corresponding training set. For the top genes extracted by the other models, the background gene set comprises genes expressed in the PPI network of the respective training set.
Benchmarking against classical graph convolutional networks
We benchmarked the DeeperGCN method to more traditional graph neural net methods including the Graph Convolutional Network (GCN)40, GraphSAGE41, and the Graph Attention Network (GAT)42. The GCN applies convolution operations on graphs, allowing each node to aggregate features from its neighbors, thereby facilitating information propagation across the graph. GCN operates in the spectral domain, using a first-order approximation of the graph convolution filter for efficient computation of node representations. By stacking multiple layers of graph convolutions, GCN captures increasingly complex relationships between nodes, incorporating information from both direct neighbors and distant nodes. GraphSAGE addresses some of these limitations by being an inductive method. GraphSAGE is a more computationally efficient GCN. It uses a sampling-based approach: instead of considering all neighbors of a node, it samples a fixed-size neighborhood and aggregates their features, reducing both computational complexity and memory requirements, thus enabling the method to scale to larger graphs. GAT uses an attention mechanism to graph-based learning. Unlike GCN and GraphSAGE, which aggregate information uniformly from neighboring nodes, GAT allows each node to dynamically assign different levels of importance to its neighbors using a learnable attention mechanism. This enables the model to focus more on certain neighbors and disregard others based on their relevance, leading to more flexible and expressive node representations. GAT employs self-attention to compute attention scores between a node and its neighbors, which are then used to weight the contribution of each neighbor’s features in the aggregation process. Additionally, GAT utilizes multi-head attention, where multiple attention heads capture different aspects of node relationships. This attention mechanism allows GAT to handle graphs with nodes of varying degrees and adapt to different graph structures without the need for expensive preprocessing steps like computing the graph Laplacian. In summary, GCN, GraphSAGE, and GAT each offer distinct advantages: GCN provides an efficient spectral approach for semi-supervised learning on small-to-medium graphs, GraphSAGE scales well for large graphs through neighborhood sampling, and GAT introduces flexibility with its attention mechanism, allowing for adaptive aggregation of node information. However, when applying graph neural networks to complex graph structures, DeeperGCN offers compelling advantages over these classical methods, as described earlier.
Implementation of MGRL
The MGRL framework was implemented using PyTorch and its graph neural network extension library, PyTorch-Geometric. To compare the performance of GCN, GraphSAGE, and GAT against DeeperGCN, we replaced the DeeperGCN layers in our framework with the corresponding convolutional layers: GCNConv, SAGEConv, and GATConv. These convolutional layers are part of the PyTorch-Geometric library and have already been implemented within the framework. Apart from the different convolution types, the overall architecture, including the number of layers, hidden dimensions, and other components in MGRL, remained unchanged across all models.
Data availability
The main datasets used here are freely available from public repositories, the scRNA-Seq data from Yazar et al. is publicly available https://cellxgene.cziscience.com/collections/dde06e0f-ab3b-46be-96a2-a8082383c4a1. The bulk RNA-seq data and DNA methylation data from Reynolds et al. is publicly available from the NCBI GEO website under the accession number GSE56047(https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi? acc=GSE56047).
Code availability
The Python code used to generate DL-XAI frame work in the current study is available in the GitHub repository for this paper (https://github.com/Frank-qlu/agingXAI).
Change history
02 April 2025
A Correction to this paper has been published: https://doi.org/10.1038/s41598-025-95242-0
References
LeCun, Y., Bengio, Y. & Hinton, G. Deep learning. Nature 521, 436–444 (2015).
Webb, S. Deep learning for biology. Nature 554, 555–557 (2018).
Angermueller, C., Parnamaa, T., Parts, L. & Stegle, O. Deep learning for computational biology. Mol. Syst. Biol. 12, 878 (2016).
Maxmen, A. Deep learning sharpens views of cells and genes. Nature 553, 9–10 (2018).
Landhuis, E. Deep learning takes on tumours. Nature 580, 551–553 (2020).
Eraslan, G., Avsec, Z., Gagneur, J. & Theis, F. J. Deep learning: new computational modelling techniques for genomics. Nat. Rev. Genet. 20, 389–403 (2019).
Lopez-Otin, C., Blasco, M. A., Partridge, L., Serrano, M. & Kroemer, G. The hallmarks of aging. Cell 153, 1194–1217 (2013).
Buckley, M. T. et al. Cell-type-specific aging clocks to quantify aging and rejuvenation in neurogenic regions of the brain. Nat. Aging. 3, 121–137 (2023).
Teschendorff, A. E. On epigenetic stochasticity, entropy and cancer risk. Philos. Trans. R Soc. Lond. B Biol. Sci. 379, 20230054 (2024).
Palmer, D., Fabris, F., Doherty, A., Freitas, A. A. & de Magalhaes, J. P. Ageing transcriptome meta-analysis reveals similarities and differences between key mammalian tissues. Aging (Albany NY). 13, 3313–3341 (2021).
de Magalhaes, J. P., Costa, J. & Toussaint, O. HAGR: the human ageing genomic resources. Nucleic Acids Res. 33, D537–543 (2005).
de Magalhaes, J. P., Curado, J. & Church, G. M. Meta-analysis of age-related gene expression profiles identifies common signatures of aging. Bioinformatics 25, 875–881 (2009).
Jaffe, A. E. & Irizarry, R. A. Accounting for cellular heterogeneity is critical in epigenome-wide association studies. Genome Biol. 15, R31 (2014).
Liu, Y. et al. Epigenome-wide association data implicate DNA methylation as an intermediary of genetic risk in rheumatoid arthritis. Nat. Biotechnol. 31, 142–147 (2013).
Regev, A. et al. The Human Cell Atlas. Elife 6. (2017).
Rozenblatt-Rosen, O., Stubbington, M. J. T., Regev, A. & Teichmann, S. A. The human cell Atlas: from vision to reality. Nature 550, 451–453 (2017).
Brennecke, P. et al. Accounting for technical noise in single-cell RNA-seq experiments. Nat. Methods. 10, 1093–1095 (2013).
Stegle, O., Teichmann, S. A. & Marioni, J. C. Computational and analytical challenges in single-cell transcriptomics. Nat. Rev. Genet. 16, 133–145 (2015).
Tabula Muris, C. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Martinez-Jimenez, C. P. et al. Aging increases cell-to-cell transcriptional variability upon immune stimulation. Science 355, 1433–1436 (2017).
Maity, A. K., Hu, X., Zhu, T. & Teschendorff, A. E. Inference of age-associated transcription factor regulatory activity changes in single cells. Nat. Aging. 2, 548–561 (2022).
Maity, A. K. & Teschendorff, A. E. Cell-attribute aware community detection improves differential abundance testing from single-cell RNA-Seq data. Nat. Commun. 14, 3244 (2023).
Prosz, A. et al. Biologically informed deep learning for explainable epigenetic clocks. Sci. Rep. 14, 1306 (2024).
Galkin, F., Mamoshina, P., Kochetov, K., Sidorenko, D. & Zhavoronkov, A. DeepMAge: a methylation aging clock developed with deep learning. Aging Dis. 12, 1252–1262 (2021).
de Lima Camillo, L. P., Lapierre, L. R. & Singh, R. A pan-tissue DNA-methylation epigenetic clock based on deep learning. Npj Aging 8. (2022).
Holzscheck, N. et al. Modeling transcriptomic age using knowledge-primed artificial neural networks. NPJ Aging Mech. Dis. 7, 15 (2021).
Trapp, A., Kerepesi, C. & Gladyshev, V. N. Profiling epigenetic age in single cells. Nat. Aging. 1, 1189–1201 (2021).
Yu, D. et al. CellBiAge: improved single-cell age classification using data binarization. Cell. Rep. 42, 113500 (2023).
Yazar, S. et al. Single-cell eQTL mapping identifies cell type-specific genetic control of autoimmune disease. Science 376, eabf3041 (2022).
Reynolds, L. M. et al. Age-related variations in the methylome associated with gene expression in human monocytes and T cells. Nat. Commun. 5, 5366 (2014).
Rodchenkov, I. et al. Pathway Commons 2019 Update: integration, analysis and exploration of pathway data. Nucleic Acids Res. 48, D489–D497 (2020).
Cerami, E. G. et al. Pathway Commons, a web resource for biological pathway data. Nucleic Acids Res. 39, D685–690 (2011).
Teschendorff, A. E. & Feinberg, A. P. Statistical mechanics meets single-cell biology. Nat. Rev. Genet. 22, 459–476 (2021).
Flint, J. & Ideker, T. The great hairball gambit. PLoS Genet. 15, e1008519 (2019).
Kalyakulina, A., Yusipov, I., Moskalev, A., Franceschi, C. & Ivanchenko, M. eXplainable Artificial Intelligence (XAI) in aging clock models. Ageing Res. Rev. 93, 102144 (2023).
Luo, D. et al. Parameterized Explainer for Graph Neural Network. In 34th Conference on Neural Information Processing Systems (NeurIPS 2020); Vancouver, Canada. arXiv; (2020).
Friedman, J., Hastie, T. & Tibshirani, R. Regularization paths for generalized Linear models via Coordinate Descent. J. Stat. Softw. 33, 1–22 (2010).
Horvath, S. DNA methylation age of human tissues and cell types. Genome Biol. 14, R115 (2013).
Li, G., Xiong, C., Qian, G., Thabet, A. & Ghanem, B. DeeperGCN: training deeper GCNs with generalized aggregation functions. IEEE Trans. Pattern Anal. Mach. Intell. 45, 13024–13034 (2023).
Kipf, T. N. & Welling, M. Semi-supervised classification with graph convolutional networks. arXiv Preprint arXiv :160902907 (2016).
Hamilton, W., Ying, Z. & Leskovec, J. Inductive representation learning on large graphs. Adv. Neural. Inf. Process. Syst. 30. (2017).
Veličković, P. et al. Graph attention networks. arXiv Preprint arXiv :171010903 (2017).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951 (2019).
Kanehisa, M., Sato, Y. & Kawashima, M. KEGG mapping tools for uncovering hidden features in biological data. Protein Sci. 31, 47–53 (2022).
Kanehisa, M., Sato, Y., Kawashima, M., Furumichi, M. & Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 44, D457–462 (2016).
Chen, L. et al. Genetic drivers of epigenetic and transcriptional variation in Human Immune cells. Cell 167, 1398–1414e1324 (2016).
Fulop, T. et al. Cytokine receptor signalling and aging. Mech. Ageing Dev. 127, 526–537 (2006).
Shchukina, I. et al. Enhanced epigenetic profiling of classical human monocytes reveals a specific signature of healthy aging in the DNA methylome. Nat. Aging. 1, 124–141 (2021).
Teschendorff, A. E. & Zheng, S. C. Cell-type deconvolution in epigenome-wide association studies: a review and recommendations. Epigenomics 9, 757–768 (2017).
Qi, L. & Teschendorff, A. E. Cell-type heterogeneity: why we should adjust for it in epigenome and biomarker studies. Clin. Epigenetics. 14, 31 (2022).
Zhang, Q. et al. Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 11, 54 (2019).
Bell, C. G. et al. DNA methylation aging clocks: challenges and recommendations. Genome Biol. 20, 249 (2019).
Mei, X., Blanchard, J., Luellen, C., Conboy, M. J. & Conboy, I. M. Fail-tests of DNA methylation clocks, and development of a noise barometer for measuring epigenetic pressure of aging and disease. Aging (Albany NY). 15, 8552–8575 (2023).
Sun, D. et al. Epigenomic profiling of young and aged HSCs reveals concerted changes during aging that reinforce self-renewal. Cell. Stem Cell. 14, 673–688 (2014).
Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 3, 23–35 (2003).
Goronzy, J. J. & Weyand, C. M. Successful and maladaptive T cell aging. Immunity 46, 364–378 (2017).
Franceschi, C. & Campisi, J. Chronic inflammation (inflammaging) and its potential contribution to age-associated diseases. J. Gerontol. Biol. Sci. Med. Sci. 69 (Suppl 1), S4–9 (2014).
Yuan, H., Yu, H., Gui, S. & Ji, S. Explainability in graph neural networks: a taxonomic survey. IEEE Trans. Pattern Anal. Mach. Intell. 45, 5782–5799 (2023).
Hao, M. et al. Large-scale foundation model on single-cell transcriptomics. Nat. Methods. 21, 1481–1491 (2024).
Rampášek, L. et al. Recipe for a general, powerful, scalable graph transformer. Adv. Neural. Inf. Process. Syst. 35, 14501–14515 (2022).
Chen, M., Wei, Z., Huang, Z., Ding, B. & Li, Y. Simple and deep graph convolutional networks. In International conference on machine learning. PMLR; : 1725–1735. (2020).
Oono, K., Suzuki, T., GRAPH NEURAL NETWORKS & EXPONENTIALLY LOSE EXPRESSIVE POWER FOR NODE CLASSIFICATION. In 8th International Conference on Learning Representations, ICLR 2020, April 30, 2020; Addis Ababa, Ethiopia. International Conference on Learning Representations, ICLR; (2020).
Zhao, W. X. et al. A Survey of Large Language Models. ArXiv abs/2303.18223. (2023).
Schoof, E. M. et al. Quantitative single-cell proteomics as a tool to characterize cellular hierarchies. Nat. Commun. 12, 3341 (2021).
Cao, J. et al. Joint profiling of chromatin accessibility and gene expression in thousands of single cells. Science 361, 1380–1385 (2018).
Teschendorff, A. E., Jing, H., Paul, D. S., Virta, J. & Nordhausen, K. Tensorial blind source separation for improved analysis of multi-omic data. Genome Biol. 19, 76 (2018).
Jiao, Y., Widschwendter, M. & Teschendorff, A. E. A systems-level integrative framework for genome-wide DNA methylation and gene expression data identifies differential gene expression modules under epigenetic control. Bioinformatics 30, 2360–2366 (2014).
Teschendorff, A. E., Zhu, T., Breeze, C. E. & Beck, S. EPISCORE: cell type deconvolution of bulk tissue DNA methylomes from single-cell RNA-Seq data. Genome Biol. 21, 221 (2020).
Gao, Y. & Teschendorff, A. E. Epigenetic and genetic deregulation in cancer target distinct signaling pathway domains. Nucleic Acids Res. 45, 583–596 (2017).
Gilmer, J., Schoenholz, S. S., Riley, P. F., Vinyals, O. & Dahl, G. E. Neural message passing for quantum chemistry. In International conference on machine learning. PMLR; : 1263–1272. (2017).
Li, G., Xiong, C., Thabet, A. & Ghanem, B. Deepergcn: all you need to train deeper gcns. arXiv Preprint arXiv :200607739 (2020).
Li, Z-P. et al. Hierarchical Multi-view Top-k pooling with deep Q-networks. IEEE Trans. Artif. Intell. (2023).
Luo, D. et al. Parameterized explainer for graph neural network. Adv. Neural. Inf. Process. Syst. 33, 19620–19631 (2020).
Ying, Z., Bourgeois, D., You, J., Zitnik, M. & Leskovec, J. Gnnexplainer: Generating explanations for graph neural networks. Adv. Neural. Inf. Process. Syst. 32. (2019).
Paszke, A. et al. Automatic differentiation in PyTorch. In.; (2017).
Fey, M. & Lenssen, J. E. Fast Graph Representation Learning with PyTorch Geometric. (2019).
Chen, L., Liu, H., Kocher, J. P., Li, H. & Chen, J. Glmgraph: an R package for variable selection and predictive modeling of structured genomic data. Bioinformatics 31, 3991–3993 (2015).
Li, C. & Li, H. Network-constrained regularization and variable selection for analysis of genomic data. Bioinformatics 24, 1175–1182 (2008).
Breiman, L. Random Forests (Springer, 2001).
Wu, T. et al. clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innov. (Camb). 2, 100141 (2021).
Funding
This work was supported by STI 2030—Major Projects, under Grant 2021ZD0200403, National Science Foundation of China (NSFC) grants, grant numbers 32170652, 32370699, 62333018, 62372255, and Key Research and Development Program of Ningbo City under Grant Nos.2023Z219, 2023Z226, 2024Z112.
Author information
Authors and Affiliations
Contributions
AET conceived the study. ZPL and DSH designed the mult-view graph-level representation learning framework. Statistical analyses were performed by ZPL and DZ. Final manuscript was written by ZPL, AET, DSH and DZ.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable here since we have only analyzed existing publicly available data.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The original online version of this Article was revised: The original version of this Article contained errors. Modifications have been made to in the Materials and methods section and the Funding section. Full information regarding the corrections made can be found in the correction for this Article.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, ZP., Du, Z., Huang, DS. et al. Interpretable deep learning of single-cell and epigenetic data reveals novel molecular insights in aging. Sci Rep 15, 5048 (2025). https://doi.org/10.1038/s41598-025-89646-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-89646-1