Abstract
Ovarian Cancer (OC) is the most lethal gynecological malignancy, characterized by peritoneal metastasis, directly linked to most OC-related deaths. Here, by interrogating CRISPR-Cas9 loss-of-function genetic screen data, we identified a list of genes essential for metastatic OC, including several well-known oncogenes (PAX8, CCNE1, WWTR1, WT1, KAT6A, MECOM, and SOX17) and others whose roles in OC have not yet been explored. Protein-protein interaction analysis of the selected genes revealed the presence of a protein network participating in the epigenetic regulation of gene expression. For one of the network components, BRPF1, we found that its increased expression correlates with OC progression and a poor prognosis for OC patients. Functional assays demonstrated that BRPF1 inhibition significantly reduces cellular migration and invasion, supporting its role in metastatic progression. Pharmacological blockade of BRPF1 using small molecule inhibitors resulted in reduced proliferation of high-grade serous OC cells through mechanisms involving the activation of programmed cell death, cell cycle deregulation, and enhanced DNA damage. Furthermore, analysis of transcriptional changes induced by BRPF1 targeting showed that the growth inhibitory effects may be mediated by the deregulation of PPARα signaling. The obtained results indicate that BRPF1 represents a novel potential therapeutic target for metastatic OC treatment.
Similar content being viewed by others
Introduction
Ovarian cancer (OC) is the eighth most common cancer in women worldwide, ranking third among gynecological malignancies, after cervical and uterine cancers. Since the malignancy tends to manifest at a late stage and relapse occurs in more than 80% of patients, the 5-year survival rate is low, generally 30–40%1. Unsurprisingly, due to its high mortality rate, OC ranks as the fifth leading cause of cancer-related deaths globally2. The majority of OCs have epithelial origin and are classified into five types, according to their histopathological phenotype3. High-grade serous OC (HGSOC) is the most common and extremely aggressive epithelial OC subtype, primarily contributing to the poor prognosis of OC patients4. Due to the high metastatic potential of HGSOC, it is frequently diagnosed at late stages, when intraperitoneally disseminated tumors are already formed.
OC cells are prone to metastasize and known to possess a specific tropism for the adipose-rich omentum, which represents a favorable tumor microenvironment in the intraperitoneal cavity5. Metastasis formation is a poorly effective process because, during tumor evolution from primary to metastatic cancer, malignant cells encounter multiple obstacles, leading to the death of most of them. The surviving cells undergo a cascade of transformations required to adapt to the stress they are exposed to during cancer progression6. According to recent studies, a class of cancer-related genes called “cancer fitness genes” provide metastatic cancer cells the ability to survive under stress conditions during dissemination, promoting mechanisms enabling tumoral cells to overcome selective pressure6,7,8. It is important to notice that normal cells rarely experience such high stress levels during their lifetime, and are therefore less dependent on these genes under homeostatic conditions, which makes them a perfect therapeutic target since inhibition of their activity affects mainly cancer cells compared to normal ones8.
Genome-wide CRISPR screens have been widely applied to determine gene function in cancer and to identify fitness genes whose loss modulates the cancerous cell growth capacity9,10. Application of this method has already allowed to determine the involvement of several genes in the development of metastatic disease in different tumor types. In particular, it was used to establish the role of ADAMTSL3 and PTEN in inhibiting hepatocellular carcinoma proliferation and metastasis11, to reveal the involvement of GPS2 in suppressing the metastasis of uveal melanoma12, to discover that a novel tumor suppressor ZNF319 plays a critical role for breast cancer metastasis13, to detect genes promoting prostate cancer growth and metastasis14, and to find out genes regulating lung metastasis15. In OC, it allowed to identify PCMT1 as a critical driver of ovarian cancer metastasis16.
Here, using publicly available CRISPR-Cas9 screening data for OC cell lines, we identified a group of 75 genes characterized by higher essentiality in metastatic compared to primary OC, among which well-known oncogenes were present altogether with other ones whose role and targeting potential for OC treatment have been poorly addressed. Using protein interaction network analysis, we found a network of 14 proteins participating in the epigenetic regulation of gene expression, and since their activity may be blocked by small molecule inhibitors, these proteins could represent plausible therapeutic targets. Among the top-ranked genes was BRPF1, which encodes for Bromodomain and PHD Finger containing 1 protein, otherwise known as peregrine. Since small molecule inhibitors targeting its activity are available, this gene was chosen for further investigation. We found that BRPF1 expression correlates with clinicopathological features of OC tumors. Inhibition of the activity of BRPF1 negatively affects the proliferation, migration, invasion, and colony formation ability of OC cells. Furthermore, targeting BRPF1 with the small molecule inhibitor GSK6853 results in cell cycle deregulation, enhanced programmed cell death, and increased DNA damage. Finally, analysis of transcriptome changes induced by BRPF1 protein inhibition or gene silencing allowed us to determine that the PPARα signaling pathway may underlie the antiproliferative effect of BRPF1 targeting.
Methods
Identification of genes characterized by enhanced essentiality in metastatic ovarian cancer
The dependence scores of ovarian cancer cell lines were downloaded from the DepMap Portal (https://depmap.org/portal/) (version 22Q4)17; the scores were obtained from a series of loss-of-function genomic screenings in 54 ovarian cancer cell lines. A negative score represents an inhibitory effect of gene knockout on cell line survival, whereas a positive score value indicates a pro-proliferative effect of gene knockout. Genes characterized by elevated essentiality in metastatic OC cells were determined in the following way: for each gene present in the CRISPR-Cas9 screening dataset, mean scores for cells derived from Primary (SP) and Metastatic (SM) OC were calculated. Next, Δ(SM-SP) was calculated together with the statistical significance of the differences evaluated by applying the t-test. Genes with Δ(SM-SP) > 0 and p-value < 0.05 were considered more essential for the survival of metastatic cell lines than for primary tumor-derived cells. Genes with statistically significant differences in fitness scores and Δ(SM-SP) belonging to the Q4 were subjected to protein-protein interaction network analysis using the STRING database, v. 12.018. Only experimentally determined interactions and those from curated databases were taken into account.
Gene, protein expression and survival analysis in ovarian cancer
CSIOVDB, an ovarian cancer database of the Cancer Science Institute of Singapore (http://csibio.nus.edu.sg/CSIOVDB/CSIOVDB.html) containing microarray gene expression data for 3431 human OC tissues, together with clinicopathological characteristics and follow-up information, was used for gene expression analysis and its correlation with the pathological degree and clinical outcome of OC patients19. Values of BRPF1 protein expression in 15 OC cell lines, obtained by MS analysis20, were downloaded from https://www.ebi.ac.uk/gxa/experiments/E-PROT-28/Results.
Cell culture
Ovarian cancer cell line source and cultivation mode were previously described in Alexandrova et al.21.
Total protein and histone extraction
Total protein extraction was performed using the standard protocol, as previously described22. Histone isolation was accomplished using a histone extraction kit (ab113476, Abcam, Cambridge, UK) according to the manufacturer’s instructions.
Western blotting
SDS-PAGE and Western blot analysis were performed using standard protocols as previously described23. For proteins detection, the following primary antibodies were used: rabbit polyclonal BRPF1 antibody (PA5-27783, Thermo Fisher Scientific, Austin, TX, USA), rabbit monoclonal Bcl-2 antibody (Cat. 2870, Cell Signaling Technology, Denver, MS, USA), rabbit monoclonal cleaved Caspase-3 antibody (ab32042, Abcam), rabbit monoclonal Rad51 antibody (Cat. 8875, Cell Signaling Technology), rabbit monoclonal PARP antibody (Cat. 9532, Cell Signaling Technology), rabbit monoclonal p21 antibody (ab188224, Abcam), rabbit polyclonal phospho-histone H2A.X (Ser139) (γH2A.X) antibody (Cat. 2577, Abcam), rabbit polyclonal phospho-histone H2A.X (Tyr142) antibody (PA5-40153, Thermo Fischer Scientific), mouse monoclonal E-Cadherin antibody (Cat. 14472, Cell Signaling Technology), mouse monoclonal α-Tubulin antibody (T6074, Merck, Darmstadt, Germania), mouse monoclonal β-actin antibody (A1978, Marck) and rabbit monoclonal histone H3 antibody (Cat. 05-928, Merck).
Drug treatments
All cell lines were incubated with increasing concentrations of the BRPF1 inhibitors GSK5959 (S5916) or GSK6853 (S8265), both purchased from Selleckchem (Houston, TX, USA) or vehicle (DMSO, Merck, Darmstadt, Germany) as a control.
Cell proliferation and cell cycle analysis
Cell proliferation was evaluated at indicated time points by an MTT (3-(4,5-Dimethylthiazol-2-yr)-2,5-Diphenyltetrazolium Bromide)-based colorimetric assay (Invitrogen, Carlsbad, CA) as previously described21,22. Absorbance was measured by a Spark multimode microplate reader (Tecan, Männedorf, Switzerland) at 570 nm, subtracting background values read at 620 nm for each sample. Cell cycle analysis was performed on PEO4 and OVCAR-3 cells treated with 10µM and 20 µM GSK6853 for 12 days or DMSO as control. In brief, cells were harvested at indicated time point, fixed in cold 70% ethanol, treated with 50 µg/ml RNAse A (Thermo Fischer Scientiic) and stained with 15 µg/ml Propidium Iodide (Merck). Cell cycle data were acquired with the use of Sony MA900 Cell Sorter (Sony Biotechnology, San Jose, CA, USA) and subsequent analysis was performed using the FlowJo™ v10 Software (BD Biosciences, Franclin Lakes, NJ, USA).
Fluorimetric Caspase 3/7 assay
Activation of Caspase-3/7-mediated apoptosis was analysed using Amplite™ Fluorimetric Caspase 3/7 Assay (AAT Bioquest, Inc, Pleasanton, CA, USA), according to the protocol, provided by the manufacturer. Cells were plated in 96-well Clear Bottom Plates (CC3603, Corning, New York, NY, USA) and, after treatment with 10 µM BRPF1 inhibitor GSK6852 for 12 days, 100 µL of Caspase 3/7 working solution were added to each well. Plates were incubated at room temperature for 1 h, after that the fluorescence intensity was measured at Ex/Em = 490/525 nm (Cutoff = 515 nm) using Spark multimode microplate reader (Tecan). Fluorescent values were normalized to the number of viable cells measured by MTT assay conducted in parallel.
Transient small interfering RNA transfection
BRPF1 silencing was performed using BRPF1-specific siRNAs (IDs: s15421 and s15422, Ambion, Thermo Fisher Scientific), targeting different gene regions. Scrambled siRNA (ID: s813, Ambion, Thermo Fisher Scientific) was used as a negative control. Lipofectamine RNAiMAX transfection reagent (Invitrogen, Thermo Fisher Scientific) was used for siRNA delivery as previously described21. Each treatment was performed in 96-well plate wells in sextuplicate. After 96 h post-transfection, cells were harvested for total protein and RNA extraction.
Colony formation and wound healing assay
For analysis of the drug or BRPF1 knockdown effect on colony formation capability, 2000 non-treated or transfected a day before cells were seeded per well of a 6-well plate. Cells were grown for 14 days in the presence of 10 µM GSK6853 or DMSO or at standard culture conditions to evaluate the drug or BRPF1 knockdown effect on the clonogenic ability of OC cells, respectively. After 14 days, cells were fixed in a 4% paraformaldehyde solution and stained in 0.1% (w/v) crystal violet. The number of grown colonies was counted. In the case of wound healing assay, plates with cells grown for 12 days in the presence of 10 µM GSK6853 or DMSO or transfected one day before with BRPF1-targeting siRNA were scratched and photographed immediately after or 24 h post scratching. The gap width changes were measured using the Wound Healing Size Tool24 in ImageJ software.
Invasion assay
24-well inserts (Cat. 353097, Corning) were coated over-night at 37 °C, 5% CO2 using 100 µl of Matrigel (Cat. 356230, BD Biosciences) diluted to a concentration of 0.3 mg/ml in coating buffer (0.01 M Tris, pH 8.0, 0.7% NaCl). Cells were pre-incubated in serum-free medium for 24 h and then 105 cells were plated on Matrigel-coated and not coated inserts. Cells were allowed to invade for 24 h, after that, they were fixed using a 4% paraformaldehyde solution and stained in 0.1% (w/v) crystal violet. Non-invaded cells were swabbed from the apical membrane surface, and the percentage of invaded cells was calculated as a ratio of the mean number of cells invading through the Matrigel matrix-coated insert membrane to the mean number of cells migrating through the uncoated insert membrane.
Total RNA extraction
Total RNA was extracted from PEO4 and OVCAR-3 cell lines grown in the presence of 10 µM GSK6853 or vehicle (DMSO) for 3 days as previously described21,22. Total RNA from siRNA-transfected cells was extracted 96 hours post-transfection using the previously described method21. 10 µg of RNA were subjected to treatment with 2U of TURBO DNAse (AM2238, Invitrogen) in the presence of 1x reaction buffer for 30’ at 37 °C. After treatment, samples were purified using phenol: chloroform: isoamyl alcohol mix (25:24:1) and chloroform: isoamyl alcohol mix (24:1), followed by ethanol precipitation in the presence of sodium acetate. RNA purity, quantity, concentration, and integrity were assessed by NanoDrop™ 2000/2000c spectrophotometer (Thermo Fisher Scientific), Qubit RNA assay kit with a fluorimeter (Life Technologies, Carlsbad, CA) and Agilent 4200 Tapestation System (Agilent, Santa Clara, CA), respectively.
RNA sequencing and data analysis
Indexed libraries were prepared starting from 1 µg of DNAse-treated total RNA according to the Illumina Stranded Total RNA Prep Ligation with Ribo-Zero Plus kit (Illumina Inc., San Diego, CA, USA). Final libraries were sequenced at a concentration of 0.6 nM/lane on the Novaseq 6000 platform using S4 flowcell and v1.5 reagents (Illumina Inc.). The FASTQC tool (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) was used for quality control of the raw sequence files (.fastq files). Adapter sequences were removed using Trimmomatic version 0.3825. STAR v2.7.5a26 was used, with standard parameters, to align the filtered reads on human genome (version GRCh38.p12 - GenCode Release 37). Quantification of expressed genes was performed using featureCounts27 and DESeq228 to identify differentially expressed genes. Genes satisfying the following criteria were considered to be differentially expressed: |Fold Change (FC)| ≥ 1.2 and adjusted p-value ≤ 0.05.
Functional analyses
Functional analysis of differentially expressed transcripts has been performed using IPA (Ingenuity Pathway Analysis, QIAGEN, Hilden, Germania). GOplot29 was used for the Circos plot generation.
Statistical analyses
Comparisons between the two groups were performed by a Student’s t-test. Values of p ≤ 0.05 were considered statistically significant. Error bars represent the means ± SD of independent replicates.
Results
Identification of fitness genes representing candidate targets for metastatic ovarian cancer treatment
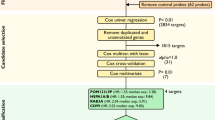
In order to determine genes characterized by elevated essentiality for survival of metastatic OC, we interrogated the data of genome-wide CRISPR-Cas9 loss-of-function genetic screens deposited in the dependency map (DepMap) portal17, the largest and latest database integrating three large-scale projects. The datasets analyzed comprises Chronos score values reflecting the knockdown effect of 17.453 genes in 54 OC cell lines derived from 25 primary and 29 metastatic tumors (Fig. 1a). Average fitness scores for primary (SP) and metastatic (SM) OC tumor-derived cells were calculated. 460 genes displayed elevated fitness properties in metastatic OC cells (SM < SP, p-value < 0.05). Among these, we selected 115 genes with the Δ(SM-SP) values belonging to the highest quartile (Q4) as the most promising therapeutic targets for metastatic OC treatment (the list of genes and fitness scores is available in Supplementary Table 1). Further analysis revealed that 40 among these represent common essential genes (Fig. 1a, Supplementary Table 1), namely genes ranking among the topmost depleted in at least 90% of cell lines tested in the large pan-cancer screening, indicating their ubiquitous essential activity in cancer cells. Interestingly, most of the first 20 genes characterized by maximum Δ(SM-SP) values, displayed in Fig. 1b, are known to be involved in transcriptional gene regulation, encoding for transcription factors or epigenetic regulators of gene expression. Among these are PAX8, CCNE1, WWTR1, WT1, IGF1R, KAT6A, MECOM, and SOX17, whose roles in OC progression and metastasis development have been widely demonstrated30,31,32,33,34,35,36 indicating the validity of the strategy applied here for target discovery. Moreover, MEN1 and KMT2A encoding for menin and MLL proteins, respectively, were among the genes found to be the most essential for metastatic OC. In our previous study, we examined the effect of inhibiting the interaction between these two proteins by small molecule inhibitors and showed that this caused a significant antiproliferative effect in OC cells, including metastasis-derived cell models21. To identify other functional protein complexes that may be targeted by small molecule inhibitors, we performed a protein-protein interaction network analysis using the STRING database and explored whether there is an interplay between 75 genes determined to be essential for metastatic OC. The analysis revealed a network of 14 proteins involved in epigenetic regulation of gene expression (listed in the order of decreasing Δ(SM-SP)): MEN1, KMT2A, BRPF1, KAT6A, MECOM, ASH1L, ARID1A, SAP130, APC, STT3A, MAPK14, H3-3B, CBX4, H2AJ (Fig. 1c). Since we have already addressed the effects of the first two genes, we focused our attention on the third one in the rank, BRPF1, which encodes a scaffold protein that forms a complex with the zinc finger protein MOZ, encoded by the KAT6A gene which is also present in the fitness genes list described here and displaying enhanced fitness properties in metastatic OC cells (Fig. 1b). The role of BRPF1 in OC development has been addressed in a study by Zhang et al., where the authors correlated high tumor BRPF1 expression with poor disease-free and overall survival and demonstrated that BRPF1 silencing exerts an antiproliferative effect on OC cell growth37. Considering that pharmacological blockade of BRPF1 protein activity has already been demonstrated as an effective strategy for the treatment of hepatocellular carcinoma38 and primary lower-grade gliomas39, we hypothesized that BRPF1 targeting by small molecule compounds may represent a novel therapeutic strategy for OC treatment.
Identification of candidate targets for metastatic ovarian cancer treatment. (A) Overview of CRISPR-Cas9 loss-of-function genetic screens analysis. The flow diagram depicts the criteria used for the selection of 75 genes with elevated essentiality in metastatic OC cells. (B) Histogram depicting the mean chronos score in primary (SP) and metastatic (SM) tumor-derived cell lines and the difference between the two (Δ(SM-SP)) for the top 20 genes characterized by maximum difference between the two values. (C) A network of 14 proteins involved in epigenetic regulation of gene expression determined by STRING protein-protein network interaction analysis.
Evaluation of BRPF1 expression and its clinical significance in OCs
To investigate BRPF1’s role in OC, we first extensively analyzed the clinical significance of BRPF1 expression in OC and evaluated whether there is a correlation between BRPF1 mRNA level and OC progression and prognosis. To this end, we utilized the CSIOVDB database containing transcriptomic microarray data for 3.431 human ovarian cancers. Results show that ovarian tumors are characterized by elevated BRPF1 expression, compared to normal ovarian surface epithelium (p-value = 1.37 × 10− 6) and stroma (p-value = 2.86 × 10− 2, Fig. 2a). Interestingly, high BRPF1 expression correlates with an advanced OC stage and grade (I vs. III, p-value = 8.02 × 10− 6; I vs. IV, p-value = 2.59 × 10− 4; G1 vs. G3, p-value = 2.06 × 10− 2; Fig. 2b, c). The comparison between different OC types revealed that clear cell and serous OC are characterized by the highest BRPF1 mRNA level (endometrioid vs. serous, p-value = 2.11 × 10− 2; mucinous vs. serous, p-value = 1.62 × 10− 2; serous vs. serous-low malignant potential, p-value = 1.62 × 10− 4; Fig. 2d). Finally, patients with tumors with high BRPF1 expression display worse overall survival (Fig. 2e), indicating a possible involvement of the BRPF1 protein in disease aggressiveness. Next, we analyzed BRPF1 protein expression in cell lines available in Coscia et al.20 and found out that metastatic tumor-derived OC cells are characterized by increased BRPF1 expression (Fig. 2f). Since BRPF1 displayed elevated expression in serous ovarian cancer, we next decided to adopt as an experimental model a panel of five cell lines isolated from HGSOC tumors, known to be the most frequent, aggressive and lethal form of OC, commonly associated with metastatic disease40. As a first step, we analyzed BRPF1 protein expression in the five OC cell lines representing models of primary non-metastatic (Caov-3) and metastatic (PEO14) OCs after a first recurrence (PEO1) and a second relapse due to the acquisition of chemotherapy-resistance (PEO4 and OVCAR-3) (Fig. 2g and Supplementary Fig. S2 ).
BRPF1 expression and prognostic significance in OC. Dot plots showing BRPF1 mRNA expression in (A) ovarian tumors, stroma, and normal ovarian surface epithelium; OC tumors of different stages (B) and grade (C) in the CSIOVDB database. (D) Dot plot showing BRPF1 expression in Clear Cell, Endometrioid, Mucinous, Mucinous-LMP, Serous, and Serous-LMP OC tumors. (* p < 0.05) (E) Kaplan-Meier curves showing overall survival of OC patients relative to BRPF1 mRNA expression (low expression values below the first quartile, high expression: values above the third quartile). (F) Histogram showing BRPF1 protein expression in 15 OC cell lines obtained by MS analysis by Coscia, F. et al.20. (G) BRPF1 protein expression in available OC cell lines determined by WB. β-Actin was used as loading control for immunoblot (Upper panel). Whole blots are shown in Supplementary Fig. S1, while two additional biological replicates are reported in Supplementary Fig. S2. Relative densities of the bands were normalized to ß-Actin from the same gel. Results are presented as the mean ± standard deviation of three biological replicates (Lower panel).
Collectively, these results suggest that BRPF1 is upregulated in ovarian cancer tissues, where this positively correlates with malignant progression, and that its high expression could represent a biomarker of poor prognosis for OC patients. The observed increase of BRPF1 expression in advanced-stage and higher-grade tumors suggests a potential involvement of this bromodomain protein in the molecular mechanisms of the metastasis and implies the possibility that targeting its activity may represent a valuable therapeutic strategy for OC treatment, which is worth investigating in detail.
BRPF1 pharmacological inhibition negatively affects OC cell proliferation, induces apoptosis, and reduces the metastatic potential of OC cells
We next tested the effect of the BRPF1-specific antagonist GSK6853, and its structural analog GSK5959, on OC cell proliferation. Both drugs target the BRPF1 bromodomain, preventing it from binding to acetylated histones41. A significant dose- and time-dependent antiproliferative effect of GSK6853 was observed in all experimental models, including the two metastatic and chemotherapy-refractory cell models, PEO4 and OVCAR-3 (Fig. 3a), as well as in the three other cell lines tested (Supplementary Fig. S3). This result was also confirmed for GSK5959 (Supplementary Fig. S4), a precursor of GSK6853 characterized by lower solubility and selectivity over other bromodomains41.
Impact of BRPF1 pharmacological blockade on metastatic potential, cell cycle, death, and DNA damage response of OC cells. (A) Relative cell viability was assessed in cells treated with indicated GSK6853 concentrations at displayed time points, determined by MTT assay in OVCAR-3 (left) and PEO4 (right) cells. Vehicle (DMSO) was used as control. Data represent the mean of six independent replicates ± SD (* p < 0.05). (B) Wound healing assay in OVCAR-3 and PEO4 after treatment with 10 µM GSK6853 or DMSO (CONTROL) as negative control for 12 days. Quantitative analysis of scratch width was performed at 0 and 24 h time points. Data are expressed as mean ± S.D. of five independent replicates (Supplementary Fig. S5). Stars indicate statistically significant differences (****p < 0.0001). (C) Colony formation assays were performed in OVCAR-3 (upper panel) and PEO4 (lower panel) cells after treatment with 10 µM GSK6853 or DMSO as control. Numbers displayed below images of cell culture plates indicate the number of colonies present in the corresponding well. The results reported on histograms represent an average of three independent measurements and are analyzed with respect to the DMSO (CONTROL). Stars indicate statistically significant differences (*p < 0.005, ****p < 0.0001). (D) Histograms displaying the results of cell invasion assay performed in OVCAR-3 (left panel) and PEO4 (right panel) grown in the presence of 10 µM GSK6853 or DMSO as control (CONTROL). The results reported on histograms represent the average of independent measurements and are analyzed with respect to the DMSO (CONTROL). Stars indicate statistically significant differences (*p < 0.05; ***p < 0.0001). (E) Histograms showing the results of cell cycle analysis performed after 12 days of GSK6853 (10 and 20 µM) treatment or DMSO as a control in OVCAR-3 (left panel) and PEO4 (right panel) cells. The reported results represent the average of three independent measurements. Stars indicate statistically significant differences (*p < 0.05; **p < 0.001). (F) Histograms displaying the results of the Caspase 3/7 activity assay following BRPF1 blockade with 10 µM GSK6853 in OVCAR-3 and PEO4 cells for 12 days. Data are presented as the mean ± S.D. of determinations from a representative experiment performed in six independent replicates after 12 days of treatment. All data are analyzed with respect to the DMSO (CONTROL). Asterisks indicate statistically significant differences (*p < 0.05, ****p < 0.0001). (G) Representative western blot showing cleaved Caspase 3, γH2A.X, pH2A.X (Tyr142) protein levels following 10 µM GSK6853 treatment for 12 days in OVCAR-3 and PEO4 OC cells. β-Actin and total Histone H3 were used as loading controls. (H) Representative western blot showing PARP, RAD51, Bcl-2, and p21 protein levels following 10 µM GSK6853 treatment for 12 days in OVCAR-3 and PEO4 OC cells. β-Actin was used as a loading control. The uncropped blots are presented in Supplementary Fig. S1.
To further investigate the effect of BRPF1 inhibition on OC cells we evaluated the impact of GSK6853 treatment on the tumorigenic potential of PEO4 and OVCAR-3 cells. Our findings indicate that both cell lines exhibited reduced wound healing, colony formation, and invasion capabilities following BRPF1 targeting (Fig. 3b and d, Supplementary Fig. S5). We next assessed the effect of GSK6853 on cell cycle. In OVCAR-3 cells, GSK6853 treatment led to an accumulation of cells in G1 phase and a depletion of cells in the S phase (Fig. 3e, left histogram). In contrast, PEO4 cells displayed a distinct response, with a reduction of cell number in the G1 phase and an accumulation in S and pre-G1 phases (Fig. 3e, right histogram). To explore the role of BRPF1 inhibition in programmed cell death, we examined the activity of effector proteins involved in apoptosis and the expression of apoptosis-regulating proteins. Fluorimetric assay revealed an increase in substrate cleavage in both cell lines upon BRPF1 inhibition (Fig. 3f). Western blot analysis further confirmed an elevated expression of cleaved Caspase 3 (Fig. 3g) and a reduction in Bcl-2 levels (Fig. 3h), consistent with its known pro-survival role via inhibiting mitochondrial cytochrome c release42. Since we found that GSK6853 treatment induced apoptosis in OC cells, we next verified whether the phosphorylation status of histone variant H2A.X, a key regulator of the cellular response to DNA damage43,44 was affected. In particular, we analyzed phosphorylation at residues Ser139 (also known as γH2A.X) and Tyr 142, both of which are associated with DNA fragmentation during apoptosis45. We found that GSK6853 treatment leads to an increase of both γH2A.X and pH2A.X (Y142) levels in PEO4 cells, while in OVCAR-3 cells, only H2A.X (Y142) phosphorylation was elevated (Fig. 3g). These findings indicate the presence of DNA damage in both cell lines. We further explored the impact of BRPF1 inhibition on the expression of proteins involved in DNA damage repair via homologous recombination (RAD51) and base excision repair (PARP). Interestingly, the two cell lines displayed distinct responses, probably associated with different genetic backgrounds. In particular, GSK6853 induced PARP cleavage and an increase of RAD51 expression in the PEO4 cell line, whereas in OVCAR-3 cells, BRPF1 inhibition led to RAD51 depletion (Fig. 3h). Finally, analysis of p21 expression revealed its accumulation in both cell lines (Fig. 3h), partially explaining the changes in cell cycle progression described earlier (Fig. 3e).
BRPF1 inhibition alters the transcriptome in OC cells
Since BRPF1 is known to be an epigenetic factor involved in the regulation of gene expression, we next decided to explore transcriptome changes induced by its pharmacological blockade with GSK6853 in PEO4 and OVCAR-3 cells. As shown in Figs. 3 and 4a and b-day treatment with GSK6853 induced a profound effect on the transcriptome of both cell lines, resulting in 850 and 755 differentially expressed genes in the OVCAR-3 and PEO4, respectively (Supplementary Tables 2 and 3). Comparison of the GSK6853-induced transcription profile changes in the two cell lines revealed 38 and 66 concordantly up- and down-regulated genes respectively (Fig. 4c and d). IPA analysis performed on differentially expressed transcripts revealed that GSK6853 treatment induced concordant upregulation of L1CAM interactions and PPARα-regulated lipid metabolism, and down-regulation of signaling pathways involved in integrin cell surface interactions, collagen biosynthesis and modifying enzymes, antimicrobial peptides, NAD, Interferon Alpha, Beta and Gamma signaling, Post-translational protein phosphorylation, Insulin-Like Growth Factor (IGF)-regulated transport and IGF binding protein (IGFBP)-mediated uptake (Fig. 4e).
Impact of BRPF1 pharmacological blockade on the transcriptome of OC cells. (A, B) MA plots showing transcriptome changes, measured by RNA sequencing (RNA-seq) following BRPF1 pharmacological blockade with 10 µM GSK6853 in OVCAR-3 and PEO4 cells after 3-day treatment. RNA-seq was performed in biological triplicate. (C) Venn diagrams showing the number of up- (left) and down- (right) regulated transcripts in OVCAR-3 and PEO4 cells in result of 10 µM GSK6853 treatment. (D) Fold change (green and red) and expression (yellow and blue) heatmaps showing common concordantly deregulated transcripts in OVCAR-3 and PEO4 cells upon BRPF1 pharmacological inhibition with 10 µM GSK6853. (E) Lollipop chart showing IPA statistically significant pathways, concordantly deregulated upon BRPF1 pharmacological inhibition with GSK6853 in OVCAR-3 (left) and PEO4 (right) cell lines.
BRPF1 silencing reduces the metastatic potential of OC cells
In order to experimentally confirm our findings regarding the involvement of BRPF1 in OC cell metastasis, OC cells were transiently transfected with two siRNAs, targeting different regions of the BRPF1-encoding transcript, and a scrambled siRNA as a negative control (Fig. 5a, Supplementary Fig. S6). Changes in E-Cadherin expression, whose functional activity is known to control tumor cell metastasis at multiple steps46, were analyzed. We observed that E-cadherin levels increased upon BRPF1 knockdown in PEO14 and OVCAR-3 cells (Fig. 5a, Supplementary Fig. S6), indicating the acquisition of an epithelial-like phenotype. BRPF1 knockdown in PEO1 and PEO4 cells, instead, resulted in almost no detectable changes in E-Cadherin expression. This could be attributed to the very high BRPF1 protein levels in these models, coupled with the incomplete silencing achieved by siRNA transfection (Supplementary Fig. S6). We next analyzed the impact of BRPF1 knockdown on the clonogenic, migratory, and invasive potential of OVCAR-3 cells, which are characterized by the lowest BRPF1 expression among the metastatic tumor-derived cell lines tested. These cells represent a model of relapsed OC, in which siRNA-mediated BRPF1 knockdown significantly reduced target protein expression. Our results showed that BRPF1 depletion in these cells inhibited migration, invasiveness, and colony formation capabilities (Fig. 5b-d, Supplementary Fig. S7).
Effects of BRPF1 silencing on metastatic potential and transcriptome of OC cells. (A) Changes of BRPF1 and E-Cadherin protein expression in OVCAR-3 cells 96 h post transfection with two siRNAs targeting different regions of the BRPF1 mRNA. Scramble siRNA (C-) was used as the negative control. α-Tubulin was used as a loading control for immunoblot. Whole blots are presented in Supplementary Fig. S1. (B) Wound healing assay in OVCAR-3 cells performed 24 h post transfection with two siRNAs targeting different regions of the BRPF1 mRNA or scramble siRNA (CONTROL) as the negative control. Quantitative analysis of scratch width was performed at 0 and 24 h. Data are expressed as mean ± S.D. of five independent replicates (Supplementary Fig. S7). Stars indicate statistically significant differences (****p < 0.0001). (C) Colony formation assay performed in OVCAR-3 cells transfected with two siRNAs targeting different regions of the BRPF1 mRNA or scramble siRNA (CONTROL) as the negative control. Numbers displayed below images of cell culture plates indicate the number of colonies found in the corresponding well. The results reported on histograms represent the average of three independent measurements and are analyzed with respect to the scramble siRNA (CONTROL). Stars indicate statistically significant differences (**p < 0.001). (D) Histograms displaying the results of the cell invasion assay performed in OVCAR-3 cells 24 h post transfection with two siRNAs targeting different regions of the BRPF1 mRNA or scramble siRNA (CONTROL) as the negative control. The results reported on histograms represent the average of independent measurements and are analyzed with respect to the scramble siRNA (CONTROL). Stars indicate statistically significant differences (*p < 0.05; **p < 0.001). (E) MA plot showing transcriptome changes, measured by RNA-seq following BRPF1 silencing with siRNA #1 and #2. RNA-seq was performed in biological duplicates. (F) Circos plot showing up- and down-regulated transcripts following BRPF1 silencing in OVCAR-3 cells and belonging to the regulation of lipid metabolism by PPARα-mediated and NAD signaling pathways. (G) Histogram showing upstream regulators that could be responsible for the deregulation of PPARα signaling pathway upon BRPF1 pharmacological inhibition in OVRCAR-3 and PEO4 cells or silencing in OVCAR-3 cells. Dot plots showing PPARA mRNA expression in OC tumors of different stages (H) and grade (I) in the CSIOVDB database (* p < 0.05).
BRPF1 silencing induces transcriptome deregulation in OC cells
To validate whether GSK6853-induced gene expression changes represent specific effects of BRPF1 inhibition, we next explored transcriptome changes induced by BRPF1 silencing in OVCAR-3 cells. The OC transcriptome was profoundly affected by BRPF1 knockdown (Fig. 5e), since the expression of 1326 transcripts (509 and 817 down- and up-regulated, respectively) was found to be modulated in this case (Supplementary Table 4). IPA analysis revealed the deregulation of multiple signaling pathways, including down-regulation of NAD signaling and up-regulation of lipid metabolism by PPARα, both observed also upon GSK6853 treatment of OVCAR-3 and PEO4 cell lines (Fig. 5f, Supplementary Fig. S8). Upstream regulators that could be responsible for the deregulation of identified signaling pathways were analyzed next, revealing that NUPR1 and PPARA: RXRA coactivators could potentially be in charge of the observed change of PPARα-mediated lipid metabolism signaling, in agreement with previously published results47,48 (Fig. 5g). Finally, we found that PPARA expression negatively correlates with an advanced tumor stage and grade of ovarian cancer (I vs. III, p-value = 3.55 × 10− 3; I vs. IV, p-value = 3.03 × 10− 4; II vs. IV, p-value = 4.31 × 10− 2; G1 vs. G3, p-value = 4.40 × 10− 2; G2 vs. G3, p-value = 3.70 × 10− 2; Fig. 5h and i), indicating the possible antiproliferative effect of its elevated expression in OC.
Discussion
OC is the most lethal gynecological malignancy that is frequently diagnosed at the advanced disease stage and characterized by peritoneal disseminated metastasis. Since most OC-related deaths are directly linked to diffused disease, a profound understanding of the cellular and molecular mechanisms underlying the development of ovarian cancer metastasis is indispensable for the development of novel therapeutic targets that will help contrast this life-threatening disease5,40.
In this study, we used publicly available genome-wide CRISPR-Cas9 loss-of-function genetic screens on the DepMap portal database to identify novel possible therapeutic targets for metastatic OC treatment. Comparing the fitness properties of 17.453 genes in metastatic and primary OC cells and selecting genes characterized by maximal differences of average fitness, we found 75 essential (fitness) genes as possible candidates. It should be noted that this approach allowed for the determination of genes involved in the adaptation of cancerous cells to stress conditions, such as oxygen and nutrition shortage, metabolic reprogramming, immunosurveillance, anoikis, and physical shearing stress observed during tumor progression7,8. At the same time, such genes are less essential for normal cells under physiological conditions since they do not have to cope with selective pressure during their lifetime8. For this reason, targeting determined genes may potentially be less toxic to normal cells, and theoretically allow for a wider therapeutic window of concentrations for treatment8.
Several of the top essential genes for metastatic OC selected here by this approach are well-known for their tumor- and metastatic-promoting roles in OC. Interestingly, the highest ranking gene, PAX8 (Fig. 1b), characterized by the highest essentiality for metastatic OC cells and displaying the maximum value of Δ(SM-SP), encodes for a transcription factor that belongs to the PAired-boX (PAX) gene family and is a well-known hallmark of OC. Indeed, strong PAX8 expression was found to be highly specific for ovarian serous tumors49, where it correlated with differentiation degree, FIGO stage and poor survival in OC patients50. Other genes found to be essential for metastatic OC and whose role has already been associated with OC development are: WT1 (Wilms Tumor 1), encoding for a zinc finger transcription factor and RNA-binding protein; transcriptional regulator MECOM (MDS1-EVI1 complex locus); cyclin E1-encoded by CCNE1 gene; and KAT6A, encoding for lysine acetyltransferase 6 A. Among these, WT1, similarly to PAX8, is known to be overexpressed in OC51 and to play an important role in OC development, being characterized by enhanced expression during OC progression to metastasis, and has been proposed as a potential therapeutic target for metastatic serous ovarian carcinoma32. CCNE1 was found to be frequently overexpressed in OC30, whereas SOX17, together with PAX8 and MECOM, were determined to be among the master transcription factors driving OC and regulator of the expression of WT1, MUC16 (CA125), and HE4, clinical biomarkers of the disease52. Finally, KAT6A (MOZ) was found to be upregulated in OC cells, and its high expression was associated with patient overall survival, and cisplatin sensitivity33.
Intriguingly, MECOM, SOX17, and WT1 activities were previously associated with those of PAX8. In particular, gynecological cancer patients with poor prognosis are characterized by a specific PAX8-MECOM gene signature34, whereas SOX17, in cooperation with PAX8, governs a tumorigenic transcriptional network in HGSOC35 and commonly regulates the expression of genes involved in blood vessel formation, therefore promoting OC development by driving angiogenesis36. MECOM, PAX8, SOX17, and WT1 were found to be lineage-enriched in HGSOC, and master regulators that cooperatively bind to DNA super-enhancer and, through regulation of target genes, govern tumor development53.
Of note, two additional candidate genes found here, MEN1 and KMT2A, encoding respectively for menin and MLL proteins, are known for their cooperative action in leukemia and several solid tumors. In a previous study, we demonstrated that inhibition of the activity of these two proteins by small molecule inhibitors exerts an antiproliferative effect on ovarian cancer cells, including metastatic-derived cell models21.
Altogether, these data indicate that the approach implemented here was successful in identifying strong candidate genes with a key role in OC metastasis development, as demonstrated by the presence of several genes already known for their role in OC dissemination. We therefore hypothesize that the remaining genes, whose role in OC has not been addressed yet, may represent a worth-exploring list of new therapeutic targets for metastatic OC.
In order to test whether genes determined by our approach act in collaboration with each other, we applied functional protein association network analysis to 75 members of our list. The analysis revealed a group of 14 proteins involved in epigenetic regulation of gene expression, among which BRPF1 ranked third after MEN1 and KMT2A. Its involvement in OC has already been proposed37, however the therapeutic effect of targeting BRPF1 in OC with small molecule inhibitors has not been explored yet. Therefore, we focused our study on the analysis of BRPF1 role as a prognostic factor and therapeutic target in OC.
BRPF1 encodes for a homonymous protein alternatively called peregrine, that represents one of 46 members of the bromodomain-containing proteins structurally clustered into eight distinct subfamilies54. BRPF1 represents a scaffolding subunit connecting MOZ or MORF proteins to ING5/6 and MEAF6 and contains multiple domains, including plant homeodomain-linked zinc finger (PHD), zinc knuckle (Zn), I and II, bromo domain and chromo/Tudor-related Pro-Tyr-Tyr-Pro (PWWP) domains55. BRPF1 bromodomain recognizes histone acetylation marks including H2AK5ac, H4K12ac, H4K8ac, H4K5ac, and H3K14ac, and is suggested to have the highest affinity towards H2AK5ac, followed by H4K12ac and H3K14ac. Mutations in BRPF1 are linked to developmental disorders, whereas MOZ-BRPF1 complex-mediated regulation of HOX genes and cell stemness plays an essential role in development55.
BRPF1 plays a cancer-promoting role in medulloblastoma, acute myeloid leukemia, hepatocellular carcinoma, and prostate cancer. In particular, in adult SHH medulloblastoma, overexpression of truncated BRPF1 and constitutively active Smoothened mutant was found to be involved in neuronal dedifferentiation and cancer development56, whereas in acute myeloid leukemia presenting MOZ-TIF2 fusion, BRPF1 interaction with the chimeric protein upregulates HOX expression, through MOZ-dependent histone acetylation, resulting in the promotion of tumor progression57. Another study determined that BRPF1 upregulation is associated with poor survival in liver cancer patients, where it regulates cell cycle progression, senescence, cancer stemness, and control of E2F2 and EZH2 oncogene expression through MOZ/MORF-mediated H3K14 acetylation on the promoter region of the two genes38. In prostate cancer, the USP35/BRPF1 axis was found to be involved in the control of SREBP2 expression, which in turn regulates mevalonate metabolism58, whereas expression of BRPF1 mRNA in urine was suggested as a potential biomarker of this cancer type59. In colon cancer, BRPF1 was found to be co-expressed with a coactivator of Wnt/b-catenin signaling, Pygopus 2 (Pygo2), and its targeting with GSK5959 induced suppression of cell growth60. Finally, in lower-grade gliomas, BRPF1 was found among potential epigenetic targets, and it was demonstrated that BRPF1 pharmacological blockade or silencing exerts an antiproliferative effect and reduces the colony formation ability of tumoral cells39.
In ovarian cancer, BRPF1 was found to be fucosylated in SKOV-3 and OVCAR-3 cells, and that ascites treatment of OC cells further stimulates protein fucosylation61. Another study explored the signature of bromodomain-containing proteins in OC and determined BRPF1 as a significant risk factor in OC patients’ OS and DFS37. Further investigation revealed that BRPF1 mRNA expression may represent a prognostic biomarker for OC patients. High BRPF1 expression is associated with a poor prognosis for OC patients37. Our results obtained using the CSIOVDB database demonstrate that OC tumors are characterized by increased BRPF1 mRNA expression and that high BRPF1 expression correlates with advanced OC stage and grade. Analysis of BRPF1 expression levels in different OC subtypes revealed that clear cell and serous OC are characterized by maximum BRPF1 expression levels, indicating the correlation between BRPF1 activity and disease aggressiveness. Finally, in line with the results of Zhang et al.37, we found that high BRPF1 mRNA expression in tumors correlates with worse overall survival in OC patients (Fig. 2e) and that its expression is elevated in cell lines derived from metastatic tumors compared to primary tumor-derived cells (Fig. 2f and g).
We next tested the effect of selective small molecule inhibitors of BRPF1 bromodomain41,62 on the phenotype of OC cells. In agreement with previously published results where the antiproliferative effect of BRPF1 knockdown was demonstrated37, we showed that BRPF1 pharmacological blockade exerts a dose-dependent growth inhibition effect in OC cells (Fig. 3a, Supplementary Fig. S3 and S4). It should be noted that the effect of BRPF1 inhibitor GSK5959 has already been tested in vivo, and no toxicity was observed in tumor-bearing nude mice38. Functional analysis of the effect of BRPF1 targeting revealed that inhibition of its activity or expression significantly reduces migratory, invasive, and colony formation potential of OC cells (Figs. 3b-d and 5b-d, Supplementary Fig. S5 and S7). Moreover, we observed that BRPF1 inhibition with GSK6853 leads to marked deregulation of the cell cycle, activation of programmed cell death, and increased DNA damage (Fig. 3e-h). Subsequently, analysis of transcriptional changes induced by the administration of BRPF1 inhibitor GSK6853 revealed the involvement of multiple metabolic and inflammation-related pathways (Fig. 4e). BRPF1 inhibition-induced transcriptome changes were further validated by RNA-seq after BRPF1 silencing by siRNAs, revealing that two signaling pathways, NAD signaling and regulation of lipid metabolism by PPARα, were concordantly deregulated indicating to possible involvement of these two signaling pathways in antiproliferative effect of BRPF1 inhibition (Fig. 5f). Additional analysis revealed that PPARA: RXRA activators or Nuclear Protein 1 (NUPR1), previously correlated to PPARα activity, were present among activated upstream regulators (Fig. 5g). In particular, retinoid X receptor α (RXRA) forms a heterodimer with agonist bound PPARα, binds to PPARA response elements (PPRE), and recruits co-activators to target gene promoters or enhancers48, whereas stress-activated NUPR1 induces PPARα signaling through Unfolded Protein Response (UPR)47.
PPARα belongs to the nuclear receptor superfamily and is known to be involved in the regulation of lipid metabolism, ketosis promotion, regulation of inflammation and endothelial nitric oxide synthase (eNOS), suppression of angiogenesis, and modulation of the immune system63. In cancer, PPARα activity plays a prevalently antitumoral role, however, tumor-promoting effects have also been reported. In ovarian cancer, both pro- and anti-proliferative effects of PPARα activity were observed. In particular, PPARα activation with agonists fenofibrate, clofibric acid, and clofibrate exerts an antiproliferative role64,65,66,67 that can be enhanced in the presence of ATM inhibitor, PPARγ ligand pioglitazone or clioquinol64,66,67. Apart from the growth inhibitory effect, PPARα activation was shown to reduce angiogenesis, and induce senescence and apoptosis64,65,66,68. On the other hand, upregulation of PPARα expression was found in OC effusions and solid metastases compared with primary carcinomas, and PPARα expression in effusions was associated with a less favorable response to chemotherapy and poor survival of OC patients56,69. Moreover, PPARα inhibition with GW6471 or knockdown abrogated the tumor-disseminating effect of mono-(2-ethylhexyl) phthalate70, whereas activation of PPARα with oleic acid stimulated OC growth by promoting glycolysis and cell cycle progression71.
In conclusion, the investigation of the role of BRPF1 in OC highlighted its potential as a prognostic biomarker for OC patients and a promising target for therapeutic interventions since its blockade induces modulation of genes involved in key cellular processes and significantly impairs the metastatic and growth potential of OC cells. The results presented here represent a solid starting point for further investigation on BRPF1 role in OC, aimed at confirming that the functional processes discovered here sustain the antiproliferative effect detected. Finally, further validations of BRPF1 inhibitors on patient-derived OC models and xenografts are required now to confirm the beneficial effect of the drugs in clinically relevant cancer models.
Data availability
Raw RNA sequencing data are deposited in the EBI ArrayExpress database (http://www.ebi.ac.uk/arrayexpress) with accession number E-MTAB-13663. Additional data generated and/or analysed during the current study are available from the corresponding authors upon reasonable request.
References
Reid, B. M., Permuth, J. B. & Sellers, T. A. Epidemiology of ovarian cancer: A review. Cancer Biology Med. 14, 9–32. https://doi.org/10.20892/j.issn.2095-3941.2016.0084 (2017).
Sung, H. et al. Global Cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. Cancer J. Clin. 71, 209–249. https://doi.org/10.3322/caac.21660 (2021).
Lim, D. & Oliva, E. Precursors and pathogenesis of ovarian carcinoma. Pathology 45, 229–242. https://doi.org/10.1097/PAT.0b013e32835f2264 (2013).
Bowtell, D. D. et al. Rethinking ovarian cancer II: Reducing mortality from high-grade serous ovarian cancer. Nat. Rev. Cancer. 15, 668–679. https://doi.org/10.1038/nrc4019 (2015).
Lengyel, E. Ovarian cancer development and metastasis. Am. J. Pathol. 177, 1053–1064. https://doi.org/10.2353/ajpath.2010.100105 (2010).
Shen, M. & Kang, Y. Stresses in the metastatic cascade: Molecular mechanisms and therapeutic opportunities. Genes Dev. 34, 1577–1598. https://doi.org/10.1101/gad.343251.120 (2020).
Esposito, M., Ganesan, S. & Kang, Y. Emerging strategies for treating metastasis. Nat. cancer. 2, 258–270. https://doi.org/10.1038/s43018-021-00181-0 (2021).
Shen, M. & Kang, Y. Cancer fitness genes: Emerging therapeutic targets for metastasis. Trends cancer. 9, 69–82. https://doi.org/10.1016/j.trecan.2022.08.007 (2023).
Katti, A., Diaz, B. J., Caragine, C. M., Sanjana, N. E. & Dow, L. E. CRISPR in cancer biology and therapy. Nat. Rev. Cancer. 22, 259–279. https://doi.org/10.1038/s41568-022-00441-w (2022).
Przybyla, L. & Gilbert, L. A. A new era in functional genomics screens. Nat. Rev. Genet. 23, 89–103. https://doi.org/10.1038/s41576-021-00409-w (2022).
Zhou, X., Li, R., Jing, R., Zuo, B. & Zheng, Q. Genome-wide CRISPR knockout screens identify ADAMTSL3 and PTEN genes as suppressors of HCC proliferation and metastasis, respectively. J. Cancer Res. Clin. Oncol. 146, 1509–1521. https://doi.org/10.1007/s00432-020-03207-9 (2020).
Shi, S. et al. Genome-wide CRISPR knockout screening identified G protein pathway suppressor 2 as a novel tumor suppressor for uveal melanoma metastasis. J. Cancer Res. Clin. Oncol. 149, 3349–3360. https://doi.org/10.1007/s00432-022-04160-5 (2023).
Wang, L. et al. Genome-wide CRISPR/Cas9 knockout screening uncovers ZNF319 as a novel tumor suppressor critical for breast cancer metastasis. Biochem. Biophys. Res. Commun. 589, 107–115. https://doi.org/10.1016/j.bbrc.2021.12.023 (2022).
Wang, W. et al. Genome-Wide CRISPR-Cas9 screening and identification of potential genes promoting prostate Cancer growth and metastasis. Curr. Cancer Drug Targets. 23, 71–86. https://doi.org/10.2174/1568009622666220615154137 (2022).
Chen, S. et al. Genome-wide CRISPR screen in a mouse model of tumor growth and metastasis. Cell 160, 1246–1260. https://doi.org/10.1016/j.cell.2015.02.038 (2015).
Zhang, J. et al. Genome-wide CRISPR/Cas9 library screen identifies PCMT1 as a critical driver of ovarian cancer metastasis. J. Exp. Clin. Cancer Res. 41 https://doi.org/10.1186/s13046-022-02242-3 (2022).
Pacini, C. et al. Integrated cross-study datasets of genetic dependencies in cancer. Nat. Commun. 12, 1661. https://doi.org/10.1038/s41467-021-21898-7 (2021).
Szklarczyk, D. et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 43, D447–452. https://doi.org/10.1093/nar/gku1003 (2015).
Tan, T. Z. et al. CSIOVDB: A microarray gene expression database of epithelial ovarian cancer subtype. Oncotarget 6, 43843–43852. https://doi.org/10.18632/oncotarget.5983 (2015).
Coscia, F. et al. Integrative proteomic profiling of ovarian cancer cell lines reveals precursor cell associated proteins and functional status. Nat. Commun. 7, 12645. https://doi.org/10.1038/ncomms12645 (2016).
Alexandrova, E. et al. Combinatorial targeting of Menin and the histone methyltransferase DOT1L as a novel therapeutic strategy for treatment of chemotherapy-resistant ovarian cancer. Cancer Cell. Int. 22, 336. https://doi.org/10.1186/s12935-022-02740-6 (2022).
Alexandrova, E. et al. Interaction proteomics identifies erbeta association with chromatin repressive complexes to inhibit cholesterol biosynthesis and exert an oncosuppressive role in Triple-negative breast Cancer. Mol. Cell. Proteom. 19, 245–260. https://doi.org/10.1074/mcp.RA119.001817 (2020).
Stellato, C. et al. Identification of cytoplasmic proteins interacting with unliganded Estrogen receptor alpha and beta in human breast cancer cells. Proteomics 15, 1801–1807. https://doi.org/10.1002/pmic.201400404 (2015).
Suarez-Arnedo, A. et al. An image J plugin for the high throughput image analysis of in vitro scratch wound healing assays. PLoS One. 15, e0232565. https://doi.org/10.1371/journal.pone.0232565 (2020).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for illumina sequence data. Bioinformatics 30, 2114–2120. https://doi.org/10.1093/bioinformatics/btu170 (2014).
Dobin, A. et al. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. https://doi.org/10.1093/bioinformatics/bts635 (2013).
Liao, Y., Smyth, G. K. & Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930. https://doi.org/10.1093/bioinformatics/btt656 (2014).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. https://doi.org/10.1186/s13059-014-0550-8 (2014).
Walter, W., Sanchez-Cabo, F. & Ricote, M. GOplot: An R package for visually combining expression data with functional analysis. Bioinformatics 31, 2912–2914. https://doi.org/10.1093/bioinformatics/btv300 (2015).
Kanska, J., Zakhour, M., Taylor-Harding, B., Karlan, B. Y. & Wiedemeyer, W. R. Cyclin E as a potential therapeutic target in high grade serous ovarian cancer. Gynecol. Oncol. 143, 152–158. https://doi.org/10.1016/j.ygyno.2016.07.111 (2016).
Jeong, G. O. et al. TAZ mediates lysophosphatidic acid-induced migration and proliferation of epithelial ovarian cancer cells. Cell. Physiol. Biochem. 32, 253–263. https://doi.org/10.1159/000354434 (2013).
Barbolina, M. V., Adley, B. P., Shea, L. D. & Stack, M. S. Wilms tumor gene protein 1 is associated with ovarian cancer metastasis and modulates cell invasion. Cancer 112, 1632–1641. https://doi.org/10.1002/cncr.23341 (2008).
Liu, W. et al. KAT6A, a novel regulator of beta-catenin, promotes tumorigenicity and chemoresistance in ovarian cancer by acetylating COP1. Theranostics 11, 6278–6292. https://doi.org/10.7150/thno.57455 (2021).
Bleu, M. et al. PAX8 and MECOM are interaction partners driving ovarian cancer. Nat. Commun. 12, 2442. https://doi.org/10.1038/s41467-021-22708-w (2021).
Lin, L. et al. SOX17 and PAX8 constitute an actionable lineage-survival transcriptional complex in ovarian cancer. Oncogene 41, 1767–1779. https://doi.org/10.1038/s41388-022-02210-3 (2022).
Chaves-Moreira, D. et al. The transcription factor PAX8 promotes angiogenesis in ovarian cancer through interaction with SOX17. Sci. Signal. 15, eabm2496. https://doi.org/10.1126/scisignal.abm2496 (2022).
Zhang, J. et al. Identification of bromodomain-containing proteins prognostic value and expression significance based on a genomic landscape analysis of ovarian serous cystadenocarcinoma. Front. Oncol. 12, 1021558. https://doi.org/10.3389/fonc.2022.1021558 (2022).
Cheng, C. L. et al. Bromodomain-containing protein BRPF1 is a therapeutic target for liver cancer. Commun. Biol. 4, 888. https://doi.org/10.1038/s42003-021-02405-6 (2021).
Xia, M. et al. Transcriptional networks identify BRPF1 as a potential drug target based on inflammatory signature in primary Lower-Grade gliomas. Front. Oncol. 11, 766656. https://doi.org/10.3389/fonc.2021.766656 (2021).
Masoodi, T. et al. Genetic heterogeneity and evolutionary history of high-grade ovarian carcinoma and matched distant metastases. Br. J. Cancer. 122, 1219–1230. https://doi.org/10.1038/s41416-020-0763-4 (2020).
Bamborough, P. et al. GSK6853, a chemical probe for Inhibition of the BRPF1 bromodomain. ACS Med. Chem. Lett. 7, 552–557. https://doi.org/10.1021/acsmedchemlett.6b00092 (2016).
Murphy, K. M., Ranganathan, V., Farnsworth, M. L., Kavallaris, M. & Lock, R. B. Bcl-2 inhibits Bax translocation from cytosol to mitochondria during drug-induced apoptosis of human tumor cells. Cell. Death Differ. 7, 102–111. https://doi.org/10.1038/sj.cdd.4400597 (2000).
Rogakou, E. P., Pilch, D. R., Orr, A. H., Ivanova, V. S. & Bonner, W. M. DNA double-stranded breaks induce histone H2AX phosphorylation on Serine 139. J. Biol. Chem. 273, 5858–5868. https://doi.org/10.1074/jbc.273.10.5858 (1998).
Lu, C. et al. Cell apoptosis: Requirement of H2AX in DNA ladder formation, but not for the activation of caspase-3. Mol. Cell. 23, 121–132. https://doi.org/10.1016/j.molcel.2006.05.023 (2006).
Rogakou, E. P., Nieves-Neira, W., Boon, C., Pommier, Y. & Bonner, W. M. Initiation of DNA fragmentation during apoptosis induces phosphorylation of H2AX histone at Serine 139. J. Biol. Chem. 275, 9390–9395. https://doi.org/10.1074/jbc.275.13.9390 (2000).
Na, T. Y., Schecterson, L., Mendonsa, A. M. & Gumbiner, B. M. The functional activity of E-cadherin controls tumor cell metastasis at multiple steps. Proc. Natl. Acad. Sci. U S A. 117, 5931–5937. https://doi.org/10.1073/pnas.1918167117 (2020).
Teresa Borrello, M. et al. NUPR1 protects liver from lipotoxic injury by improving the Endoplasmic reticulum stress response. FASEB J. 35, e21395. https://doi.org/10.1096/fj.202002413RR (2021).
Berger, J. & Moller, D. E. The mechanisms of action of PPARs. Annu. Rev. Med. 53, 409–435. https://doi.org/10.1146/annurev.med.53.082901.104018 (2002).
Laury, A. R. et al. A comprehensive analysis of PAX8 expression in human epithelial tumors. Am. J. Surg. Pathol. 35, 816–826. https://doi.org/10.1097/PAS.0b013e318216c112 (2011).
Chai, H. J. et al. PAX8 is a potential marker for the diagnosis of primary epithelial ovarian cancer. Oncol. Lett. 14, 5871–5875. https://doi.org/10.3892/ol.2017.6949 (2017).
Taube, E. T. et al. Wilms tumor protein 1 (WT1)-- not only a diagnostic but also a prognostic marker in high-grade serous ovarian carcinoma. Gynecol. Oncol. 140, 494–502. https://doi.org/10.1016/j.ygyno.2015.12.018 (2016).
Reddy, J. et al. Predicting master transcription factors from pan-cancer expression data. Sci. Adv. 7, eabf6123. https://doi.org/10.1126/sciadv.abf6123 (2021).
Nameki, R. A. et al. Rewiring of master transcription factor cistromes during high-grade serous ovarian cancer development. BioRxiv https://doi.org/10.1101/2023.04.11.536378 (2023).
Barbieri, I., Cannizzaro, E. & Dawson, M. A. Bromodomains as therapeutic targets in cancer. Brief. Funct. Genom. 12, 219–230. https://doi.org/10.1093/bfgp/elt007 (2013).
Viita, T. & Cote, J. The MOZ-BRPF1 acetyltransferase complex in epigenetic crosstalk linked to gene regulation, development, and human diseases. Front. Cell. Dev. Biol. 10, 1115903. https://doi.org/10.3389/fcell.2022.1115903 (2022).
Aiello, G. et al. Truncated BRPF1 cooperates with smoothened to promote adult Shh medulloblastoma. Cell Rep. 29, 4036–4052 https://doi.org/10.1016/j.celrep.2019.11.046 (2019).
Shima, H. et al. Bromodomain-PHD finger protein 1 is critical for leukemogenesis associated with MOZ-TIF2 fusion. Int. J. Hematol. 99, 21–31. https://doi.org/10.1007/s12185-013-1466-x (2014).
Lin, G., Huang, T., Zhang, X. & Wang, G. Deubiquitinase USP35 stabilizes BRPF1 to activate mevalonate (MVA) metabolism during prostate tumorigenesis. Cell. Death Discov.. 8, 453. https://doi.org/10.1038/s41420-022-01231-x (2022).
Sole, C. et al. The urinary transcriptome as a source of biomarkers for prostate Cancer. Cancers (Basel). 12 https://doi.org/10.3390/cancers12020513 (2020).
Ling, J., Tang, Z., Yang, W., Li, Y. & Dong, X. Pygo2 activates BRPF1 via Pygo2-H3K4me2/3 interaction to maintain malignant progression in colon cancer. Exp. Cell. Res. 431, 113696. https://doi.org/10.1016/j.yexcr.2023.113696 (2023).
Alberto-Aguilar, D. R. et al. Ascites from ovarian cancer induces novel fucosylated proteins. Cancer Microenviron. 12, 181–195. https://doi.org/10.1007/s12307-019-00227-z (2019).
Demont, E. H. et al. 1,3-dimethyl benzimidazolones are potent, selective inhibitors of the BRPF1 bromodomain. ACS Med. Chem. Lett. 5, 1190–1195. https://doi.org/10.1021/ml5002932 (2014).
Tan, Y. et al. PPAR-alpha modulators as current and potential cancer treatments. Front. Oncol. 11, 599995. https://doi.org/10.3389/fonc.2021.599995 (2021).
Chen, C. W., Buj, R., Dahl, E. S., Leon, K. E. & Aird, K. M. ATM Inhibition synergizes with Fenofibrate in high grade serous ovarian cancer cells. Heliyon 6, e05097. https://doi.org/10.1016/j.heliyon.2020.e05097 (2020).
Yokoyama, Y. et al. Clofibric acid, a peroxisome proliferator-activated receptor alpha ligand, inhibits growth of human ovarian cancer. Mol. Cancer Ther. 6, 1379–1386. https://doi.org/10.1158/1535-7163.MCT-06-0722 (2007).
Shigeto, T., Yokoyama, Y., Xin, B. & Mizunuma, H. Peroxisome proliferator-activated receptor alpha and gamma ligands inhibit the growth of human ovarian cancer. Oncol. Rep. 18, 833–840 (2007).
Tuller, E. R. et al. PPARalpha signaling mediates the synergistic cytotoxicity of clioquinol and docosahexaenoic acid in human cancer cells. Biochem. Pharmacol. 77, 1480–1486. https://doi.org/10.1016/j.bcp.2009.02.002 (2009).
Zhou, J. et al. Activation of peroxisome proliferator-activated receptor alpha (PPARalpha) suppresses hypoxia-inducible factor-1alpha (HIF-1alpha) signaling in cancer cells. J. Biol. Chem. 287, 35161–35169. https://doi.org/10.1074/jbc.M112.367367 (2012).
Davidson, B., Hadar, R., Stavnes, H. T., Trope, C. G. & Reich, R. Expression of the peroxisome proliferator-activated receptors-alpha, -beta, and -gamma in ovarian carcinoma effusions is associated with poor chemoresponse and shorter survival. Hum. Pathol. 40, 705–713. https://doi.org/10.1016/j.humpath.2008.09.019 (2009).
Leng, J. et al. Low-dose mono(2-ethylhexyl) phthalate promotes ovarian cancer development through PPARalpha-dependent PI3K/Akt/NF-kappaB pathway. Sci. Total Environ. 790, 147990. https://doi.org/10.1016/j.scitotenv.2021.147990 (2021).
Kado, T. et al. Oleic acid stimulates cell proliferation and BRD4-L-MYC-dependent glucose transporter transcription through PPARalpha activation in ovarian cancer cells. Biochem. Biophys. Res. Commun. 657, 24–34. https://doi.org/10.1016/j.bbrc.2023.03.051 (2023).
Acknowledgements
E.A., F.Ri. and F.Ru. are residents of the Postgraduate School in Clinical Pathology and Clinical Biochemistry of the University of Salerno. D.D.R is PhD student of the Research Doctorate “Molecular and Translational Oncology and Innovative Medical-Surgical Technologies”, of the University of Catanzaro “Magna Graecia”. E. A. was supported by Fondazione Umberto Veronesi.
Funding
This study was supported by AIRC Foundation for Cancer Research (AIRC: grant IG-23068), University of Salerno (FARB), Ministry of University and Research (PNRR-MUR NextGenerationEU PRIN 2022 cod. 2022Y79PT4), Regione Campania (Progetto GENOMAeSALUTE, POR Campania FESR2014/2020; CUP: B41C17000080007 and Progettto Rare-Plat-Net ‘La Campania lotta contro il cancro’, CUP: B63D18000380007).
Author information
Authors and Affiliations
Contributions
Conceptualization: E.A., A.W. and F.Ri.; investigation: E.A., M.S., D.D.R., R.P, F.Ru. and R.N.B.; writing and original draft preparation: E.A. and F.Ri.; software and formal analysis: G.G., G.N. and R.N.B.; writing and editing: E.A., F.Ri., R.T. and A.W.; funding acquisition: F.Ri., and A.W. All the authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Alexandrova, E., Smal, M., Di Rosa, D. et al. BRPF1 inhibition reduces migration and invasion of metastatic ovarian cancer cells, representing a potential therapeutic target. Sci Rep 15, 7602 (2025). https://doi.org/10.1038/s41598-025-92438-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-92438-2
Keywords
This article is cited by
-
BRPF1 in cancer epigenetics: a key regulator of histone acetylation and a promising therapeutic target
Cell Death Discovery (2025)