Abstract
This study investigates the relationship between Cyclin D1 (CCND1) gene and promoter methylation and liver fibrosis (LF)/liver cirrhosis (LC)induced by chronic hepatitis B (CHB). Peripheral blood mononuclear cells (PBMCs) are collected from patients diagnosed with chronic hepatitis B (CHB) and hepatitis B-related LF/LC, as well as from healthy individuals. The mRNA levels and promoter methylation of the CCND1 gene are measured. Single-cell analysis is performed to determine the cell types primarily expressing the CCND1 gene in LF/LC. The GSE84044 dataset is utilized to validate the experimental results. Single-gene GSEA and immune infiltration analyses are conducted to identify significant pathways and immune characteristics associated with the CCND1 gene. The mRNA level of CCND1 in PBMCs from patients with hepatitis B-related LF/LC is elevated compared to those with chronic hepatitis B (CHB) and healthy individuals, while the promoter methylation level of CCND1 is reduced. Single-cell analysis indicates high expression of CCND1 in M2 macrophages (M2) and T cells. The GSE84044 dataset confirms higher CCND1 mRNA levels in liver tissues from patients with CHB-related LF/LC compared to CHB patients. Single-gene GSEA analysis associates CCND1 expression with natural killer cell-mediated cytotoxicity, T cell receptor signaling, and B cell receptor signaling pathways. Increased expression of CCND1 enhances immune infiltration during the fibrosis/cirrhosis process of CHB. The CCND1 expression and promoter methylation may be involved in the process of LF/LC in CHB and may be related to the immune response in the course of the disease.
Similar content being viewed by others
Introduction
Hepatitis B virus (HBV), a non-cytotoxic hepatotropic virus, is frequently integrated into the genome of patients, which causes immune imbalance. A variety of immune cells, including T and B cells, monocyte-macrophages, etc., interact over the long term, which further exacerbates immune imbalance. This process leads to cirrhosis and eventually hepatocellular carcinoma (HCC), which poses a huge burden on global health1,2,3. Dynamic and prolonged, the process of LF/LC is characterized by the deposition of fibrous structures and vascular remodeling and transforms hepatic lobules into nodules. Historically, LF/LC was considered irreversible. However, recent research increasingly reveals the reversibility of CHB-related LF/LC under certain conditions. Despite these findings, effectively treating and assessing the degree of reversal in CHB-related LF/LC remains a great challenge2,4. The primary medications for treating chronic hepatitis B (CHB) are α-interferon and nucleos(t)ide analogs. Nevertheless, certain limitations exist in clinical application, like severe adverse reactions and the necessity for long-term medication. During CHB virus infection, the impaired functions of different immune cells are detected, which indicates that the immune response of hosts plays an important role in CHB progression. Therefore, manipulating the immune system is essential for treating patients with CHB5,6,7. Currently, various methods can be adopted to assess CHB-related LF/LC in clinical practice, including invasive and non-invasive techniques. Despite being accurate, invasive procedures are cumbersome to perform. Non-invasive methods encompass serum biomarkers and transient elastography (TE). Clinically, commonly used serum markers are albumin-to-globulin (AGR) and globulin-to-platelet ratios (GPR), aspartate aminotransferase-to-platelet ratio (APRI) and fibrosis-4 indexes (FIB-4). However, these non-invasive approaches have certain limitations. Convenient, rapid and accurate serum biomarkers are urgently needed to diagnose CHB-related LF/LC8,9,10,11. Cyclin D1 (CCND1) is a protein regulating the cell cycle by binding to cyclin-dependent kinase 4/6 (CDK4/6), facilitating the transition from the G1 to the S phase and promoting the phosphorylation of the retinoblastoma protein (Rb), which drives cells into the S phase. Studies indicate that the CCND1 gene is linked to liver and breast cancers, kidney fibrosis and other diseases. The phosphorylation of Rb releases Early 2 Factor (E2F) transcription factors, which activates the expression of genes involved in deoxyribonucleic acid (DNA) replication and thus propels cells into the DNA synthesis phase12,13,14. The overexpression of the CCND1 gene in various diseases results in reduced immune cell infiltration, the activation of immunosuppressive signaling pathways and the overexpression of immunosuppressive factors, which thereby exacerbates disease progression. Thus, understanding the mechanisms of CCND1 is crucial for the immunotherapy of CHB-related LF/LC15.
As a prevalent epigenetic modification, DNA methylation has been extensively documented to significantly influence multiple liver diseases like hepatitis C and non-alcoholic fatty liver disease16,17. Studies on the methylation of the CCND1 gene promoter, particularly about immunology, are crucial for elucidating the underlying mechanisms of CHB-related LF/LC, which ultimately contributes to novel anti-fibrotic therapeutic strategies.
This study aims to illuminate the critical role of CCND1 gene expression and promoter methylation in the immune response associated with CHB-related fibrosis/cirrhosis, which thereby provides theoretical support for the treatment of CHB-related LF/LC.
Experimental procedures
Sample collection
In the present study, 140 subjects were recruited from Qilu Hospital of Shandong University, including 53 CHB patients, 71 patients with CHB-related LF/LC and 16 healthy individuals. Those with CHB and CHB-related LF/LC were recruited according to standard clinical guidelines. The inclusion criteria for patients with chronic hepatitis B-related liver fibrosis/cirrhosis are as follows: 1. HBsAg positivity for ≥6 months; 2. Comprehensive assessment based on imaging, liver stiffness measurement, and laboratory indicators. The exclusion criteria for patients with chronic hepatitis B-related fibrosis/cirrhosis and chronic hepatitis B are as follows: (1) Patients with malignant tumors; (2) Hepatitis and fibrosis/cirrhosis caused by non-hepatitis B virus; (3)Patients with severe comorbidities involving other systems18,19. Clinical and laboratory examinations were collected from all participants. Table 1 summarizes the subjects’ basic clinical information.
Every participant signed an informed consent form, and this study gained the approval of the Ethics Committee of Qilu Hospital of Shandong University.
Data collection
The GSE84044 dataset, comprising liver biopsy samples from 43 CHB patients and 81 patients with CHB-related LF, was downloaded from the GEO database. First, the downloaded data underwent rigorous quality control. Based on the sample information, the data were categorized into two groups: patients with chronic hepatitis B and patients with chronic hepatitis B-related LF. Differential expression analysis was then performed using the limma package in R. A linear model was constructed to compare the expression levels of the CCND1 gene across thesee different groups. Additionally, single-cell data from five chronic hepatitis B-related liver fibrosis patients in the G2 phase were obtained from the GSE186343 dataset9,20.
DNA methylation and MethyLight analysis
Five ml of fasting blood was gathered from subjects, and peripheral blood mononuclear cells (PBMCs) were isolated from the blood through Ficoll-Plaque Plus gradient centrifugation. Genomic DNA was extracted using TRIzol reagent, and the extracted DNA was then modified by use of the EZ DNA Methylation-Gold Kit for bisulfite conversion. The total volume of the modified DNA was 20 µl. Finally, the samples underwent 15 minutes of processing at 95 °C as per the instructions of the reagent manufacturer, followed by 50 cycles of 95 and 60 °C for 15 seconds and 1 minute, respectively. MethyLight data were indicated by the percentage of methylated reference (PMR)21:
PMR = 100% × 2 exp− [Delta Ct (target gene in the sample−control gene in sample) −(Delta Ct 100% methylated target in reference sample−control gene in reference sample)]
Quantitative reverse transcription PCR
TRIzol reagent was used for the extraction of RNA from PBMCs. A complementary DNA (cDNA) synthesis kit was utilized to synthesize cDNA following the instructions of the manufacturer. Then, real-time polymerase chain reaction (RT-PCR) was performed, and the probes used are presented in Table 2.
scRNA-seq analysis
This study utilized the dataset with the accession number GSE186343 from the GEO database, which comprises single-cell RNA sequencing (scRNA-seq) data from five patients at the G2 fibrosis stage: GSM5645899, GSM5645901, GSM5645903, GSM5645905, and GSM5645907. Data processing was primarily conducted using the Seurat package. The Harmony method was employed to correct for batch effects across the different patient samples. Subsequently, 2,000 highly variable genes were selected from the dataset, and principal component analysis (PCA) was performed on these genes for dimensionality reduction. The optimal number of principal components (PCs) was determined using an Elbow Plot. The R package singleR was then used to annotate cell types, assigning the most likely cell type to each cell by matching reference transcriptome data, thereby enabling cell classification and labeling.
Data acquisition and immune infiltration analysis
The immune infiltration of the dataset was analyzed using the CIBERSORT package to obtain the infiltration levels of 22 immune cell types in CHB-related LF/LC. After that, the relationship between CCND1 levels and the infiltration levels of different cell populations was determined by performing Spearman correlation analysis22.
GSEA analysis of CCND1
Based on the median expression level of CCND1, patients with CHB-related LF/LC in the dataset were classified into high and low CCND1 expression groups. Gene set enrichment analysis (GSEA) was performed on these two groups using the clusterProfiler package to compare the differences in enriched gene sets between high and low CCND1 expression groups23. Statistical significance was defined as a p-value < 0.05.
Statistical analysis
R (version 4.3.1) and GraphPad Prism (version 9.5.1) were used to perform all computations and statistical analyses. For data analysis, the non-parametric distribution of the data was assessed using Mann–Whitney U and Kruskal–Wallis tests. To evaluate the diagnostic performance of CCND1 promoter methylation, the receiver operating characteristic (ROC) curve was plotted in this study. Every two-sided p-value was less than 0.05.
Results
CCND1 mRNA expression and PMR values across different groups
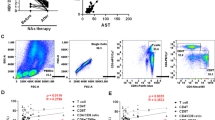
As illustrated in Fig. 1A, the cirrhosis group had a higher expression level of CCND1 mRNA than the CHB group and healthy controls (HCs), without a significant difference between CHB patients and HCs (p = 0.6210). As shown in Fig. 1B, the methylation level of the CCND1 promoter was measured in PBMCs from LF/LC, CHB and HCs. It was found that the cirrhosis group had a lower promoter methylation level than the CHB group and HCs, without significant differences between CHB patients and HCs (p = 0.6358). To validate the relationship between CCND1 methylation expression and mRNA expression, we used the Spearman rank correlation test and found that the PMR value of CCND1 was negatively correlated with mRNA expression (Fig. 1C). To further validate the differential expression of CCND1 between the chronic hepatitis B-related liver fibrosis/cirrhosis group and the chronic hepatitis B group, we used the public dataset GSE84044. In the validation cohort, CCND1 gene expression in CHB-related LF/LC patients was significantly higher than in CHB patients, as shown in Fig. 1D.
Expression of CCND1 in Different Groups. (A) mRNA levels of CCND1 in PBMCs from patients with HBV-related LF/LC, CHB patients, and HCs (***P < 0.001). (B) Methylation levels of the CCND1 promoter (***P < 0.001). (C) The relationship between CCND1 promoter and mRNA levels (Spearman’s r = −0.3556, P < 0.001) (D) The expression of CCND1 in different groups in GSE84044.
Single-cell analysis
To further evaluate the potential role of CCND1 in fibrosis pathology, this study analyzed single-cell RNA sequencing data from public databases. Initially, we corrected for batch effects in the single-cell dataset and performed manual annotation. A total of seven cell types were clustered (Fig. 2A), and the expression levels of the CCND1 gene in different cell types were analyzed using single-cell transcriptomic data. The results indicated higher expression in M2 macrophages and T cells (Fig. 2B).
Immune infiltration analysis of CCND1
To further determine the association of CCND1 gene expression with 21 immune cell types, the CIBERSORT algorithm was used for analyzing immune cell infiltration proportions, as shown in Fig. 3A. Figure 3B displays that CCND1 expression correlates with 11 immune cell types, including T cells gamma delta, macrophages M1 (M1), M2 and M0, natural killer (NK) cells resting, B cells memory, neutrophils, T cells regulatory (Tregs), B cells naive, plasma cells and T cells memory activated. M1, B cells naive, plasma cells, macrophages M0, T cells memory activated and T cells gamma delta are positively correlated with CCND1 expression, while M2, neutrophils, NK cells resting, B cells memory and Tregs are negatively correlated with it. This is consistent with the single-cell analysis results, which further indicate that the CCND1 gene may influence the activity of immune cells.
GSEA analysis of CCND1
To further explore the biological function and regulatory mechanisms of CCND1 in CHB-related LF/LC, we utilized the GSE84044 dataset, which contains gene expression data from CHB and CHB-related LF/LC patients, and performed single-gene enrichment analysis (GSEA). The analysis revealed that high expression of CCND1 is closely associated with multiple immune-related pathways, particularly NK cell-mediated cytotoxicity and T cell and B cell receptor signaling pathways. Specifically, as shown in Fig. 4A, B, and C, high expression of CCND1 is closely associated with enhanced NK cell activity and the upregulation of T cell and B cell receptor signaling pathways, suggesting that CCND1 may play a significant role in the occurrence and progression of liver fibrosis and cirrhosis by regulating these immune response pathways.
Diagnostic value of the CCND1 gene
To evaluate the diagnostic efficacy of CCND1 gene promoter methylation in CHB-related LF/LC, we plotted the receiver operating characteristic (ROC) curve and performed the corresponding analysis. As shown in Fig. 5, the methylation of the CCND1 promoter [95% confidence interval (CI): 0.872–0.963, area under the curve (AUC): 0.917] compared to FIB-4 [95% CI 0.760–0.901, AUC: 0.831] and APRI [95% CI 0.777–0.912, AUC: 0.844] demonstrated significantly higher diagnostic performance.
Discussion
Approximately 300 million people across the globe suffer from CHB, with around one million succumbing annually to end-stage liver disease, including decompensated LC, liver failure and HCC. The progression from CHB to cirrhosis typically takes several decades24. It is complex and widely believed to be primarily immune-mediated. HBV infection results in liver damage through the continuous infiltration of immune cells and the cytokines they secrete. This immune response damages liver lobules whose subsequent reconstruction induces cirrhosis7,25,26,27,28. It is important to develop effective therapeutic interventions against LF/LC through learning about the molecular mechanisms of immune dysfunction. Recent studies demonstrate that CCND1 plays a critical role in fibrotic diseases29,30. In LF, CCND1 is considered to influence the activation of hepatic stellate cells and stimulate the overproduction of the extracellular matrix. Previous animal experiments have shown that CCND1 is pivotal in renal fibrosis31,32. The role of CCND1 in CHB-related LF/LC remains to be explored.
Epigenetic dysregulation caused by methylation is recognized as a significant factor that contributes to liver diseases. Research has established that the methylation of the peroxisome proliferators-activated receptor-gamma (PPARγ) gene is crucial during the fibrosis process. Moreover, the dynamic characteristics of methylation make it a viable target for therapeutic intervention17,33,34,35. In our previous study, we have demonstrated that CCND1 methylation detection outperforms AFP levels, especially in AFP-negative HBV-HCC, and that the combination of CCND1 methylation and AFP can further improve diagnostic accuracy36.At present, no studies have indicated the relationship between the expression of the CCND1 gene and the status of promoter methylation in the PBMCs of CHB-related fibrosis/cirrhosis patients and immune response.
Our study shows that the CCND1 gene promoter methylation level in PBMCs of HBV-related LF/LC patients is significantly lower than that in chronic hepatitis B (CHB) patients and healthy controls (HCs). In addition, as a hematological non-invasive biomarker for HBV-related LF/LC, CCND1 outperforms the clinically used FIB-4 and APRI scores. Through single-cell analysis and single-gene gene set enrichment analysis (GSEA), we found that CCND1 may play an important immunoregulatory role in various immune cell types. M2 macrophages are typically associated with immune suppression and tissue repair, whereas T cells play a central role in immune responses37,38. Meanwhile, the expression of CCND1 is closely associated with multiple immune-related signaling pathways, especially those involving NK cell-mediated cytotoxicity and T cell and B cell receptor signaling. These pathways are crucial in the immune response in liver diseases. We speculate that the high expression of CCND1 may play a significant role in the immune microenvironment of liver fibrosis and cirrhosis by enhancing the activity of these immune response pathways. Therefore, CCND1 has the potential to be a target for immunotherapy, offering new avenues for anti-fibrosis treatment. The study relies on data from public databases to suggest a potential association between the CCND1 gene and immune response but does not further investigate the underlying mechanisms. Additionally, the clinical samples are relatively few and originate from a single center.
In summary, it is evident that the CCND1 gene plays a significant role in the process of fibrosis/cirrhosis in CHB. The regulatory mechanism likely involves participation in the immune response, and the expression of the CCND1 gene may also be influenced by methylation. Furthermore, Hypomethylation of the CCND1 gene shows potential in the diagnosis of fibrosis/cirrhosis associated with chronic hepatitis B. Therefore, the expression and promoter methylation of the CCND1 gene can impact the immune response process in CHB-related LF/LC.
Data availability
The datasets analyzed and generated during the current study are available from the corresponding author on reasonable request.
References
Iannacone, M. & Guidotti, L. G. Immunobiology and pathogenesis of hepatitis B virus infection. Nat. Rev. Immunol. 22, 19–32. https://doi.org/10.1038/s41577-021-00549-4 (2022).
Rockey, D. C. Liver fibrosis reversion after suppression of hepatitis B virus. Clin. Liver Dis. 20, 667–679. https://doi.org/10.1016/j.cld.2016.06.003 (2016).
Chen, Y. & Tian, Z. HBV-induced immune imbalance in the development of HCC. Front. Immunol. 10, 2048. https://doi.org/10.3389/fimmu.2019.02048 (2019).
Bedossa, P. Reversibility of hepatitis B virus cirrhosis after therapy: Who and why?. Liver Int. 35(Suppl 1), 78–81. https://doi.org/10.1111/liv.12710 (2015).
Akbar, S. M. F., Yoshida, O. & Hiasa, Y. Immune therapies against chronic hepatitis B. J. Gastroenterol. 57, 517–528. https://doi.org/10.1007/s00535-022-01890-8 (2022).
Li, T. Y., Yang, Y., Zhou, G. & Tu, Z. K. Immune suppression in chronic hepatitis B infection associated liver disease: A review. World J. Gastroenterol. 25, 3527–3537. https://doi.org/10.3748/wjg.v25.i27.3527 (2019).
Cheng, L. S., Liu, Y. & Jiang, W. Restoring homeostasis of CD4⁺ T cells in hepatitis-B-virus-related liver fibrosis. World J. Gastroenterol. 21, 10721–10731. https://doi.org/10.3748/wjg.v21.i38.10721 (2015).
Chen, Y. P. et al. Stepwise application of fibrosis index based on four factors, red cell distribution width-platelet ratio, and aspartate aminotransferase-platelet ratio for compensated hepatitis B fibrosis detection. J. Gastroenterol. Hepatol. 33, 256–263. https://doi.org/10.1111/jgh.13811 (2018).
Bai, Y. M., Liang, S. & Zhou, B. Revealing immune infiltrate characteristics and potential immune-related genes in hepatic fibrosis: Based on bioinformatics, transcriptomics and q-PCR experiments. Front. Immunol. 14, 1133543. https://doi.org/10.3389/fimmu.2023.1133543 (2023).
Liao, M. J. et al. Novel index for the prediction of significant liver fibrosis and cirrhosis in chronic hepatitis B patients in China. World J. Gastroenterol. 28, 3503–3513. https://doi.org/10.3748/wjg.v28.i27.3503 (2022).
Mallet, V. et al. The accuracy of the FIB-4 index for the diagnosis of mild fibrosis in chronic hepatitis B. Aliment Pharmacol. Ther. 29, 409–415. https://doi.org/10.1111/j.1365-2036.2008.03895.x (2009).
Osaki, Y. et al. Blocking cell cycle progression through CDK4/6 protects against chronic kidney disease. JCI Insight https://doi.org/10.1172/jci.insight.158754 (2022).
Shi, M. J. et al. A network pharmacology approach to investigating the mechanism of Tanshinone IIA for the treatment of liver fibrosis. J. Ethnopharmacol. 253, 112689. https://doi.org/10.1016/j.jep.2020.112689 (2020).
Katoh, M. Multi-layered prevention and treatment of chronic inflammation, organ fibrosis and cancer associated with canonical WNT/β-catenin signaling activation (Review). Int. J. Mol. Med. 42, 713–725. https://doi.org/10.3892/ijmm.2018.3689 (2018).
Chen, Y. et al. CCND1 amplification contributes to immunosuppression and is associated with a poor prognosis to immune checkpoint inhibitors in solid tumors. Front. Immunol. 11, 1620. https://doi.org/10.3389/fimmu.2020.01620 (2020).
Luo, B. et al. Cell-free DNA methylation markers for differential diagnosis of hepatocellular carcinoma. BMC Med. 20, 8. https://doi.org/10.1186/s12916-021-02201-3 (2022).
Zeybel, M. et al. DNA methylation profiling identifies novel markers of progression in hepatitis B-related chronic liver disease. Clin. Epigenet. 8, 48. https://doi.org/10.1186/s13148-016-0218-1 (2016).
Xu, X. Y. et al. Chinese guidelines on the management of liver cirrhosis (abbreviated version). World J. Gastroenterol. 26, 7088–7103. https://doi.org/10.3748/wjg.v26.i45.7088 (2020).
Terrault, N. A. et al. Update on prevention, diagnosis, and treatment of chronic hepatitis B: AASLD 2018 hepatitis B guidance. Clin. Liver Dis. (Hoboken) 12, 33–34. https://doi.org/10.1002/cld.728 (2018).
Shao, L. et al. Biopsy-based single-cell transcriptomics reveals MAIT cells as potential targets for controlling fibrosis-related liver inflammation due to chronic hepatitis-B infection. Clin. Transl. Med. 12, e1073. https://doi.org/10.1002/ctm2.1073 (2022).
Gao, S. et al. Aberrant GSTP1 promoter methylation predicts short-term prognosis in acute-on-chronic hepatitis B liver failure. Aliment Pharmacol. Ther. 42, 319–329. https://doi.org/10.1111/apt.13271 (2015).
Chen, D. et al. KLF4 loss in hepatocellular carcinoma: Improving prognostic prediction and correlating immune infiltrates. Front. Genet. 14, 1106952. https://doi.org/10.3389/fgene.2023.1106952 (2023).
Ren, H. et al. Apolipoprotein C1 (APOC1) promotes tumor progression via MAPK signaling pathways in colorectal cancer. Cancer Manag. Res. 11, 4917–4930. https://doi.org/10.2147/cmar.S192529 (2019).
Lin, J., Wu, J. F., Zhang, Q., Zhang, H. W. & Cao, G. W. Virus-related liver cirrhosis: Molecular basis and therapeutic options. World J. Gastroenterol. 20, 6457–6469. https://doi.org/10.3748/wjg.v20.i21.6457 (2014).
Lee, J. et al. Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc. Natl. Acad. Sci. USA 103, 1828–1833. https://doi.org/10.1073/pnas.0510801103 (2006).
Diebold, S. S., Kaisho, T., Hemmi, H., Akira, S. & Reis e Sousa, C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science 303, 1529–1531. https://doi.org/10.1126/science.1093616 (2004).
Chang, J., Block, T. M. & Guo, J. T. The innate immune response to hepatitis B virus infection: Implications for pathogenesis and therapy. Antiviral Res. 96, 405–413. https://doi.org/10.1016/j.antiviral.2012.10.001 (2012).
Zheng, P., Dou, Y. & Wang, Q. Immune response and treatment targets of chronic hepatitis B virus infection: Innate and adaptive immunity. Front. Cell Infect. Microbiol. 13, 1206720. https://doi.org/10.3389/fcimb.2023.1206720 (2023).
Alu’datt, M. H. et al. Royal jelly mediates fibrotic signaling, collagen cross-linking and cell proliferation in cardiac fibroblasts. Biomed. Pharmacother. 164, 114922. https://doi.org/10.1016/j.biopha.2023.114922 (2023).
Zhang, P. et al. Identification of shared molecular mechanisms and diagnostic biomarkers between heart failure and idiopathic pulmonary fibrosis. Heliyon 10, e30086. https://doi.org/10.1016/j.heliyon.2024.e30086 (2024).
Tsuchiya, Y. et al. Fibroblast growth factor 18 stimulates the proliferation of hepatic stellate cells, thereby inducing liver fibrosis. Nat. Commun. 14, 6304. https://doi.org/10.1038/s41467-023-42058-z (2023).
Mullany, L. K. et al. Distinct proliferative and transcriptional effects of the D-type cyclins in vivo. Cell Cycle 7, 2215–2224. https://doi.org/10.4161/cc.7.14.6274 (2008).
Murphy, S. K. et al. Relationship between methylome and transcriptome in patients with nonalcoholic fatty liver disease. Gastroenterology 145, 1076–1087. https://doi.org/10.1053/j.gastro.2013.07.047 (2013).
Zeybel, M. et al. Differential DNA methylation of genes involved in fibrosis progression in non-alcoholic fatty liver disease and alcoholic liver disease. Clin. Epigenet. 7, 25. https://doi.org/10.1186/s13148-015-0056-6 (2015).
Zhao, Q. et al. DNA methylation patterns of peroxisome proliferator-activated receptor gamma gene associated with liver fibrosis and inflammation in chronic hepatitis B. J. Viral Hepat. 20, 430–437. https://doi.org/10.1111/jvh.12048 (2013).
Liu, H. H. et al. Hypomethylation of the cyclin D1 promoter in hepatitis B virus-associated hepatocellular carcinoma. Medicine (Baltimore) 99, e20326. https://doi.org/10.1097/md.0000000000020326 (2020).
Tacke, F. & Zimmermann, H. W. Macrophage heterogeneity in liver injury and fibrosis. J. Hepatol. 60, 1090–1096. https://doi.org/10.1016/j.jhep.2013.12.025 (2014).
Koda, Y., Nakamoto, N. & Kanai, T. Regulation of progression and resolution of liver fibrosis by immune cells. Semin. Liver Dis. 42, 475–488. https://doi.org/10.1055/a-1957-6384 (2022).
Acknowledgements
We thank the GEO database for providing their platforms and contributors for their meaningful datasets to the general public.
Funding
This work was supported by the National Key Research and Development Program of China (2021YFC2301801) and the National Natural Science Foundation of China (82272313).
Author information
Authors and Affiliations
Contributions
N.C. designed the study, performed the research, analyzed the data, and drafted the paper. P.-Y.L. performed the research and revised the paper. Y.-N.T., P.L. revised the paper. J.W., Y.-C.F. supervised this study. L.-Y.H., K.W. supervised, designed the study, and revised the paper. All authors reviewed and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interest
The authors declare no competing interests.
Ethics approval
The study was approved by the Ethics Committee of Qilu Hospital of Shandong University, according to the guidelines of the 1975 Declaration of Helsinki. All participants provided written informed consent.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, N., Luo, P., Tang, Y. et al. Accelerators of chronic hepatitis B fibrosis cirrhosis CCND1 gene expression and promoter hypomethylation. Sci Rep 15, 10630 (2025). https://doi.org/10.1038/s41598-025-93778-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41598-025-93778-9