Abstract
Pine wilt disease (PWD) is a devastating disease caused by the pinewood nematode (Bursaphelenchus xylophilus). Its substantial ecological disruption harms global forestry and poses serious economic challenges. Although previous research has demonstrated that Bacillus subtilis JCK-1398 has the potential to induce systemic resistance in pine trees, the ecological mechanisms underlying its biocontrol efficacy remain underexplored. This study investigated how JCK-1398 treatment influences rhizosphere- and nematode-associated microbial communities to mitigate PWD. Metabarcoding analyses revealed that JCK-1398 treatment increased the abundance of beneficial microbial taxa (e.g., Nocardioides and Mesorhizobium) in the rhizosphere microbiome. Concurrently, nematode-associated microbial communities became dominated by Pantoea, a genus with known nematicidal properties. Isolation and characterization of Pantoea dispersa BC11 confirmed that it significantly limits nematode viability. These findings highlight the multifaceted defense that JCK-1398 offers, not only inducing systemic resistance, but also orchestrating beneficial microbiome dynamics. This study emphasizes the potential of manipulating a microbial holobiont for eco-friendly and sustainable disease management. The ability of JCK-1398 to recruit and enhance microbial allies offers a novel framework for developing biocontrol agents, with implications for managing PWD and other plant-pathogen systems.
Similar content being viewed by others
Introduction
Pine wilt disease (PWD), caused by the pinewood nematode (Bursaphelenchus xylophilus), is a devastating condition that rapidly colonizes and infests pine trees, leading to severe wilting and eventual tree death1. First identified as a significant threat in Japan during the early 20th century, PWD has since spread to Asia, Europe, and parts of North America, posing a critical challenge to global forestry2,3,4. The pinewood nematode is considered one of the most critical pests on international quarantine lists, highlighting its global significance. Ecologically, PWD disrupts forest ecosystems, indirectly accelerates deforestation, and compromises biodiversity. Economically, the disease imposes substantial burdens, including reduced timber yields and the high costs associated with tree removal, quarantine enforcement, and chemical treatments, which are often ineffective in halting its spread5. Growing environmental concerns over chemical control methods have underscored the urgent need for sustainable and effective management strategies, particularly given the far-reaching implications of the disease for forest health and economic6.
Microbial communities play a pivotal role in the progression and management of PWD. Although the pinewood nematode is the primary causal agent of the disease, accumulating evidence underscores the significant contributions of nematode-associated microbes to its pathogenicity. These microbes are known to influence disease severity by producing phytotoxins, enhancing nematode survival, and facilitating host colonization. Their role extends beyond virulence, as they can modulate plant defense responses and contribute to nematode adaptation within the pine environment7,8. Given their multifaceted contributions to PWD dynamics, studying the composition and functional interactions of these microbial communities is crucial for developing sustainable management strategies. Further details on the specific roles of nematode-associated microbes and their ecological implications are provided in the discussion section.
The rhizosphere microbiome, a dynamic community of microbes at the root-soil interface, plays important roles in shaping plant health and influencing the progression of PWD9,10. These microbial communities contribute to plant defense by inducing systemic resistance, promoting growth, and suppressing pathogens11. Notably, the physiological changes induced in pine trees during systemic resistance, such as those triggered by biocontrol agents, may also alter root exudate profiles12. As root exudates are critical determinants in recruiting and structuring rhizosphere microbial communities, these changes could drive shifts in the composition of the rhizosphere microbiome12,13. These findings highlight the intricate interactions between host physiology, microbial recruitment, and disease resistance, underscoring the critical ecological role of the rhizosphere in the PWD complex.
Biological control agents, particularly resistance-inducing microbes, offer a promising and sustainable approach to PWD management14,15. Unlike chemical treatments, which may lead to non-target effects, residual toxicity, and the disruption of ecological balance, biocontrol agents harness natural processes to suppress pathogens while preserving biodiversity. The biocontrol agents not only activate plant immune responses but also have the potential to restructure microbial communities in the rhizosphere, fostering conditions that favor disease suppression10. This study further explores their potential impact on nematode-associated microbiomes, presenting a novel aspect of biocontrol strategies. Understanding and leveraging these microbial interactions can provide novel insights into eco-friendly strategies for mitigating PWD while enhancing the resilience of host trees.
Previous studies have demonstrated the successful application of various microbial biocontrol agents in managing forest pathogens. For instance, Trichoderma species have been effectively utilized against Armillaria root rot, a devastating disease affecting numerous tree species16. In addition, Bacillus spp. have shown promise in promoting plant growth and suppressing a wide range of forest pests17. Furthermore, bacteria such as Aneurinibacillus migulanus have exhibited potent biocontrol activity through the production of antimicrobial compounds that effectively suppress various Phytophthora species—major threats to forest ecosystems18. These findings underscore the growing potential of microbial biocontrol agents in forestry, offering sustainable and environmentally friendly alternatives to conventional chemical treatments.
Building on previous findings by Mannaa et al.19, Bacillus subtilis JCK-1398 was demonstrated to induce systemic resistance in pine trees, leading to significant physiological changes. These include an increase in pinoresinol production, activation of defense-associated genes, and stimulation of key signaling pathways, which collectively enhance the tree defense mechanisms. This induced resistance was effective in limiting nematode colonization, as evidenced by a decrease in nematode migration and infestation levels. While these findings underscored the potential of JCK-1398 to reduce nematode infestation, its ecological impact on rhizosphere and nematode-associated microbial communities remains unexplored. The present study addresses this gap by exploring the broader ecological mechanisms underlying the biocontrol efficacy of B. subtilis JCK-1398. This investigation focuses on assessing the effects of JCK-1398 treatment on (1) the composition of the rhizosphere microbiome, (2) shifts within nematode-associated microbial communities in side pine tissues, and (3) the identification of microbial allies that may contribute to disease suppression. By integrating physiological, biochemical, and ecological perspectives, this research aims to provide a deeper understanding of the multifaceted role of JCK-1398 in managing PWD, with meaningful implications for the development of sustainable biocontrol strategies.
Results
Confirmation of B. subtilis JCK-1398 protective role against pine wilt disease
The efficacy of B. subtilis JCK-1398 in controlling PWD in Pinus densiflora seedlings was assessed through a controlled seedling assay. Treatment with JCK-1398 resulted in a clear reduction in disease symptoms, as evident in Fig. 1. Seedlings inoculated with PWN but not treated with JCK-1398 exhibited typical PWD symptoms, including severe browning and wilting, indicating substantial disease progression. In contrast, seedlings treated with JCK-1398 maintained healthy foliage, suggesting effective disease suppression. Untreated control seedlings showed no disease symptoms (Fig. 1A). Quantitative assessment further validated these observations, with JCK-1398-treated seedlings displaying a significantly lower disease severity (P < 0.001) compared to the nematode-inoculated control group (Fig. 1B). This marked reduction in disease severity highlights the protective role of JCK-1398, demonstrating its potential as a biocontrol agent against PWD.
Suppression of pine wilt disease (PWD) severity in Pinus densiflora seedlings following treatment with Bacillus subtilis JCK-1398. (A) Representative images of pine seedlings displaying disease severity across different treatments: untreated control, pinewood nematode (Bursaphelenchus xylophilus)-inoculated, and nematode-inoculated seedlings treated with B. subtilis JCK-1398. The JCK-1398-treated seedlings exhibit notable disease suppression, maintaining healthy foliage, whereas nematode-inoculated seedlings without treatment show clear wilting and browning symptoms. (B) Quantitative assessment of PWD severity. The bar graph displays the mean disease severity scores for each treatment group. Error bars represent standard deviation (n = 3). The asterisk (P < 0.05) denote a statistically significant reduction in disease severity in JCK-1398-treated seedlings compared to the nematode-inoculated control group.
Impact of B. subtilis JCK-1398 treatment on the pine rhizosphere microbial composition
Metabarcoding of the P. densiflora rhizosphere yielded over 1,243,355 reads, averaging 103,613 reads per sample (Table S1). Rarefaction analysis confirmed adequate sequencing depth for assessing microbial diversity, plateauing at around 4000 reads (Fig. S1A). Alpha diversity did not vary significantly across treatments (Table S2). However, beta diversity analysis showed nuanced changes in microbial community structure due to treatment, as indicated by PCoA based on unweighted UniFrac distances (Fig. 2A).
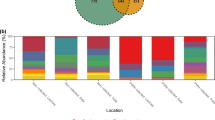
Effect of Bacillus subtilis JCK-1398 on the diversity and composition of the rhizosphere microbiota in pine seedlings. (A) Principal Coordinate Analysis (PCoA) of rhizosphere microbiota structure based on both unweighted and weighted UniFrac distance metrics, illustrating the beta diversity among treatments. Treatment groups: C (control seedlings), B (B. subtilis JCK-1398-treated seedlings), N (Bursaphelenchus xylophilus-inoculated seedlings), and BN (combined B. subtilis and nematode treatment). (B) Bar graph showing bacterial genera with significantly different relative abundances (P < 0.05) between nematode-inoculated samples and nematode-inoculated samples treated with B. subtilis JCK-1398, emphasizing the impact of B. subtilis JCK-1398 on microbial community composition.
In general, the bacterial community was dominated by Proteobacteria, Verrucomicrobia, Bacteroidetes, Acidobacteria and Actinobacteria in the four pine groups with over 84% relative abundance. JCK-1398 treatment notably impacted certain bacterial groups, such as Rhizobiales, and families, including Sinobacteraceae, that were significantly (P < 0.05) higher in B. subtilis JCK-1398-treated nematode-inoculated pine seedlings compared to untreated nematode-inoculated seedlings (Fig. S1B).
At the genus level, the heatmap constructed for the top 100 most abundant genera based on the Manhattan distance measurement and the average linkage hierarchical clustering shown in supplementary Fig. 2, confirmed that there were no drastic differences in the abundant microbial genera. The relative abundance of Asticcacaulis, Bordetella, Nocardioides, Mesorhizobium, Pseudoflavitalea, Solimonas, Methylosinus, Afifella, Methyloceanibacter, Solirubrobacter and Methyloligella was significantly (P < 0.05) higher in B. subtilis JCK-1398-treated nematode-inoculated pine seedlings compared to untreated nematode-inoculated seedlings whereas the relative abundance of Elstera, Racemicystis, Sphaerotilus, Oxalobacter, Amorphus, Aquaphilus, Terracidiphilus, Pelomonas, and Nitratireductor were significantly (P < 0.05) lower (Fig. 2B). These results indicate that while the overall microbial community composition remained relatively stable, JCK-1398 treatment induced significant shifts in specific microbial taxa, suggesting a potential influence of the biocontrol agent on rhizosphere microbial dynamics.
Impact of B. subtilis JCK-1398 treatment on the nematode-associated microbial composition inside pine tissues
Metabarcoding of nematodes isolated from JCK-1398 treated P. densiflora seedlings produced 1,242,068 reads, averaging 155,259 reads per sample, with quality metrics listed in Table S3. Rarefaction curves suggested sufficient sequencing depth to capture the core microbial diversity (Fig. S3). Alpha diversity metrics showed significantly greater microbial richness in nematodes from treated seedlings although the diversity was reduced (Fig. 3A). This suggests that Pantoea dominance in the treated group contributed to reduced diversity but was accompanied by an increase in less abundant taxa, reflecting a shift in community structure rather than a complete homogenization. Beta diversity assessments highlighted a marked difference in microbial structure, with PCoA based on UniFrac distances demonstrating distinct clustering (Fig. 3B).
Impact of Bacillus subtilis JCK-1398 on the diversity of microbiota associated with pine wood nematodes. (A) Alpha diversity metrics (ASV count, Chao1, Gini-Simpson, and Shannon index) of the nematode-associated microbiota, comparing treatments with and without B. subtilis JCK-1398. Error bars represent standard deviations (n = 4), and significant differences between treatments (P < 0.05) are indicated by different letters above the bars. (B) Principal Coordinate Analysis (PCoA) plots showing the beta diversity of nematode-associated microbiota based on unweighted and weighted UniFrac distances, highlighting differences in microbial community structure between treatments.
At the genus level, heatmap analysis further displayed clear distinctions in bacterial genera between groups (Fig. 4A). Microbial community composition analysis revealed that nematodes from JCK-1398 treated seedlings were overwhelmingly dominated by the genus Pantoea (97.44%). In contrast, untreated seedlings exhibited a more diverse microbial profile, with Erwinia and Kosakonia showing higher relative abundance in some samples; however, considerable variability was observed among replicates, indicating a heterogeneous microbial community within the untreated group (Fig. 4B).
Effect of Bacillus subtilis JCK-1398 on the composition of nematode-associated microbiota. (A) Heatmap showing the relative abundance of the top 25 bacterial genera in the microbiota associated with B. xylophilus nematodes. Manhattan distance was used for clustering, with Z-score standardization indicating the relative abundance of each genus across samples. (B) Stacked bar chart comparing the microbial composition at the genus level in nematodes isolated from untreated and B. subtilis JCK-1398-treated pine seedlings, illustrating the differences in microbial community structure following treatment.
The differential abundance analysis was performed to identify bacterial genera that differed significantly between treatment groups. The results revealed that nematodes from JCK-1398-treated seedlings exhibited a significantly higher relative abundance of Pantoea, Novosphingobium, Burkholderia, and Caballeronia, while untreated nematodes harbored significantly higher levels of Pseudomonas, Kosakonia, and Erwinia (Fig. 5). These findings suggest substantial alterations in the nematode microbiome following JCK-1398 treatment, despite the observed intra-sample variability in the untreated group.
Bar graphs showing the differential abundance analysis of nematode-associated bacterial genera between B. subtilis JCK-1398-treated and untreated groups. Bars represent the mean relative abundance (± standard error) of bacterial genera that were significantly different (P < 0.05) between the treatment groups. Red bars represent untreated samples, while blue bars represent JCK-1398-treated samples. *(P < 0.05), and ***(P < 0.001).
Isolation of microbes associated with nematodes recovered from pine tissues
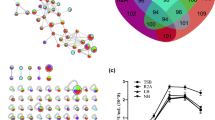
Since a significant shift in the microbiome of nematodes derived from pine tissues was observed after B. subtilis JCK-1398 treatment, we hypothesised that nematode-associated microbes might contribute to the observed reduction in nematode migration and, consequently, the mitigation of PWD symptoms. To explore this, we isolated bacteria from nematodes extracted from JCK-1398-treated and untreated seedlings, yielding 10 and 6 unique isolates, respectively. A 16 S rRNA gene-based maximum likelihood phylogenetic analysis incorporating type strain sequences that exhibited BLAST similarity to our isolates identified one strain, BC11, as closely related to Pantoea dispersa (Fig. 6A). Subsequent phylogenetic analyses, which included BC11 and other closely related Pantoea members, distinctly positioned BC11 within the Pantoea dispersa clade, corroborating its identification as Pantoea dispersa BC11 (Fig. 6B). The 16 S rRNA sequence of P. dispersa BC11 was deposited in GenBank under the accession number OR910539.
Identification and nematicidal activity of Pantoea dispersa isolated from pinewood nematode extracted from pine seedlings treated with Bacillus subtilis JCK-1398. (A) Phylogenetic analysis based on the 16 S rRNA sequence data of microbes isolated from nematodes in JCK-1398-treated seedlings (BC) and untreated seedlings (N) compared to the type of strain sequences. (B) Phylogenetic placement of the isolated microbial strain BC11 based on its 16 S rRNA sequence, showing its assignment to a separate cluster closely related to Pantoea dispersa. Bootstrap values (based on 1000 replicates) are shown at the nodes. GenBank accession numbers for each sequence are provided in parentheses, with reference sequences sourced from type strains. (C) Evaluation of nematicidal activity by the selected bioagent B. subtilis JCK-1398, the nematode isolated from P. dispersa strain BC11, using line and scatter graphs to represent nematode mortality rates following treatment with various concentrations of cell-free culture filtrates from P. dispersa BC11 compared to a control treatment with sterilised broth. Mortality was recorded two days post-treatment. An asterisk (*) above the error bar indicates a significant difference (P < 0.05).
Nematicidal activity assessment of Pantoea dispersa BC11
Following the isolation and taxonomic identification of strain BC11 as P. dispersa, we assessed its nematicidal activity. Cell-free culture filtrates demonstrated significant bioactivity against PWNs, with a notable effect observed at a concentration of 5%. This bioactivity increased markedly at 10% concentration and culminated in 100% nematode mortality at 20% concentration (Fig. 6C).
Discussion
Building on the foundational insights from our previous study19, which established B. subtilis JCK-1398 as a potent inducer of systemic resistance in pine trees, the current study significantly advances our understanding of the ecological mechanisms underlying biocontrol efficacy of JCK-1398. The earlier work demonstrated that JCK-1398 treatment induced critical physiological changes within pine tissues, including increased pinoresinol production, the upregulation of defense-related genes and the activation of pathways such as MAPK signaling, which collectively strengthened pine defenses. This induced resistance effectively limited nematode colonization, evidenced by reduced nematode migration and infestation. Recognizing that disease progression or suppression is not only shaped by host resistance but also by the surrounding microbial communities, this study extends our investigation to include the rhizosphere and nematode-associated microbiomes as critical ecological factors influencing PWD dynamics.
Microbial communities in the plant root ecosystem play pivotal roles in disease suppression, nutrient availability, and growth promotion. The structure of the rhizosphere microbiome is largely dictated by root exudates, which modulate the soil physicochemical properties and determine the functional microbial community12. It is plausible that the induced resistance from JCK-1398 treatment could lead to alterations in rhizosphere microbiota composition through changes in root exudates11. Upon further investigation into the effects of B. subtilis JCK-1398 treatment on pine seedlings, we examined the rhizosphere microbiome. While no dramatic shift in microbial composition was observed, we noted a significant increase in the abundance of certain microbes in seedlings treated with JCK-1398.
Previous research by Mannaa et al.20 and Han et al.10 has indicated that pine seedlings treated for induced resistance with chemical or biotic elicitors may exhibit a modified rhizosphere microbiota. In this study, significant differences in relative abundance were observed in certain microbial groups between B. subtilis JCK-1398-treated seedlings and untreated controls, which may be indicative of an improved health status of the pine trees. Rhizobiales, including families such as Beijerinckiaceae and Rhodobiaceae, as well as Propionibacteriales from the Actinobacteria phylum including Nocardioidaceae and Solirubrobacteraceae, were notably more abundant in treated seedlings. These taxa are recognized for their beneficial roles in root-associated microbiota and in promoting root growth, largely through nitrogen fixation21. Members of the Actinobacteria, capable of producing a diverse array of bioactive natural products, exhibit plant growth-promoting activities and antibacterial properties against pathogens22. The Sinobacteraceae family was also found in higher relative abundance in treated seedlings, and these microbes are known to contribute to ammonia oxidation, potentially enhancing plant growth and acting as antagonists to plant pathogens such as Xanthomonadaceae23,24. The observed increase in beneficial bacterial genera such as Nocardioides, Mesorhizobium, Methylosinus, and Solirubrobacter in B. subtilis JCK-1398-treated seedlings further supports the hypothesis of a microbial-mediated enhancement of pine health. These genera, particularly from the Actinobacteria, are renowned for their production of plant growth-promoting substances and enzymes that degrade hydrocarbons25,26,27. Recent research has highlighted the role of Nocardioides spp. in alleviating stress and promoting plant growth through siderophore and phytohormone production28. The genera Mesorhizobium and Methylosinus, part of the Rhizobiales, are acknowledged for their plant growth promotion and nitrogen-fixing abilities, with additional attributes such as IAA production and nutrient solubilization29,30.
These findings resonate with previous studies that associate the resistance inducing acibenzolar-s-methyl treatment with an increased relative abundance of specific microbial genera capable of bolstering plant growth, including Nocardioides and Mesorhizobium20. Our results point to the potential of B. subtilis JCK-1398 treatment not only to induce resistance against PWD but also to possibly influence the recruitment of beneficial rhizosphere microbes that may contribute to an enhanced defense against the PWN. Strain JCK-1398 did not demonstrate direct nematicidal effects against PWN, indicating that its protective role is likely due to induced resistance within the host plant and shifts in microbial community composition, including nematode-associated microbes, rather than direct nematicidal action19.
The intricate interplay between PWD pathogenicity and the microbiota associated with the PWN has been increasingly recognized in recent literature. It has been well-documented that PWN-associated bacteria contribute to the disease virulence, with some capable of producing phytotoxins3. Certain nematode-associated bacteria, such as Burkholderia arboris and Pseudomonas fluorescens, produce phytotoxins, including pyochelin and phenylacetic acid, which exacerbate disease severity by damaging pine tissues31,32,33. Furthermore, axenic nematodes, or those devoid of their microbial partners, fail to cause typical disease symptoms when inoculated into pine trees, suggesting that these microbes are essential partners in the disease process31,34. Beyond their direct involvement in disease virulence, these associated microbes are believed to aid nematode survival within the hostile pine environment by neutralizing toxic compounds like α-pinene, a prevalent component of pine resin35. Specific nematode-associated bacteria, known for their tolerance to oxidative stress, are thought to reinforce nematode resistance against pine defense mechanisms36. Early signs of pine tissue alteration and physiological stress precede the proliferation of PWNs, suggesting a complex pathogenic process that encompasses both the nematodes and their bacterial symbionts37. This underlines the intricate and multifactorial nature of PWD, shaped by the interplay between the nematode and its associated microbial community.
Our approach, utilizing metabarcoding analysis for investigating the microbial communities from nematodes recovered from pine tissues, revealed a stark contrast in the microbial consortia of nematodes from JCK-1398-treated seedlings compared to those from untreated plants. The domination of Pantoea in nematodes isolated from treated samples, as opposed to the microbial diversity seen in untreated ones, indicates that JCK-1398 may foster a microbiome that is antagonistic to PWN. This is supported by previous findings where a strain of Pantoea was observed to completely inhibit PWN reproduction and reduce adult nematode size38, suggesting that the biocontrol effect of JCK-1398 might be mediated through microbial interactions that directly affect the nematode.
The potent nematicidal activity of P. dispersa BC11 isolated from nematodes associated with B. subtilis JCK-1398-treated seedlings highlighted a compelling biocontrol mechanism. Specifically, JCK-1398 treatment appears to induce a beneficial shift in the nematode-associated microbiome, resulting in diminished virulence and suppression of PWD symptoms. Such findings exemplify the holobiont concept, describing a cohesive unit of selection formed by a host organism and its associated microbial communities39,40.
In this scenario, the host pine tree, upon treatment with JCK-1398, appears to harness a microbial ally, P. dispersa, within the nematode microbiome. This strategic alteration of the nematode-associated microbiota underscores the dynamic interplay within the holobiont, where the induced resistance mechanisms in the host extend beyond physiological responses to actively reshape its associated microbial landscape. Thus, the JCK-1398 treatment not only strengthens the direct defenses of pine trees but also mobilizes its microbiome as an integral component of its resistance strategy, reinforcing the holobiont as a unified system in combating PWD. The biocontrol efficacy of B. subtilis JCK-1398 thus appears to be a symphony of interactions—ranging from induced systemic resistance within the host pine seedlings to the recruitment of beneficial rhizosphere microbes and modulation of nematode-associated microbiota (Fig. 7). However, given the limited sample size in the current study, further research is necessary to validate these findings on a larger scale and across diverse environmental conditions. Future studies should focus on conducting large-scale field trials to confirm the consistency of the microbial shifts observed and assess the long-term sustainability of the treatment. Additionally, exploring the molecular mechanisms underlying microbial recruitment and their interactions with the host plant will provide deeper insights into the holistic impact of JCK-1398 in PWD management.
Schematic representation of Bacillus subtilis JCK-1398’s microbiome-mediated defense strategy in mitigating pine wilt disease. This diagram illustrates the contrasting outcomes between untreated and B. subtilis JCK-1398-treated pine on disease progression and microbiome dynamics. On the left, untreated pine seedlings exhibit increased vulnerability to Bursaphelenchus xylophilus (pinewood nematode) infestation, which accelerates pine wilt disease symptoms. The nematode-associated microbiome is dominated by pathogenic bacterial genera such as Erwinia and Kosakonia, with a concurrent reduction in beneficial Pantoea. The rhizosphere microbiome reflects diminished levels of beneficial bacteria, including members of Rhizobiales. On the right, pine seedlings treated with B. subtilis JCK-1398 demonstrate enhanced resilience, with a shift in the nematode-associated microbiome towards nematicidal Pantoea dominance. In addition, the rhizosphere microbiome shows an increase in beneficial microbes, contributing to the plant’s overall health and defense system. This schematic highlights the broad-spectrum protective effects of B. subtilis JCK-1398, suggesting a multifaceted defense strategy that boosts the pine tree resistance to pine wilt disease by altering both nematode-associated and rhizosphere microbial communities.
In conclusion, this study built on the previously established capacity of B. subtilis JCK-1398 to induce systemic resistance, extending our understanding of its multifaceted role in combating PWD. By elucidating its impact on both the rhizosphere and nematode-associated microbiomes, we revealed how JCK-1398 fosters a dynamic defense strategy that integrates pine physiological resistance mechanisms and ecological reshaping of microbial communities. The promotion of beneficial microbes such as P. dispersa—a nematicidal agent within the nematode microbiome—emphasizes the intricate interplay between hosts and associated microbiota in disease suppression. Finally, the empirical evidence of B. subtilis JCK-1398 as an ecologically sustainable biocontrol agent provides a template for leveraging host-microbiome interactions in disease management strategies.
Materials and methods
Bacterial strain, nematode and pine seedlings
Bacillus subtilis JCK-1398 was previously isolated and identified as a potent inducer of systemic resistance in pine trees19. The strain was cultured overnight at 28 °C in Luria-Bertani (LB) broth, starting from a single colony grown on LB agar plates at 28 °C for 2 days. Bacterial cultures for seedling treatments were prepared in 1 mM MgSO4, and the concentration was adjusted to OD600 = 0.8.
A highly virulent strain of B. xylophilus, isolated from infected Pinus densiflora, was provided by the National Institute of Forest Research (Seoul, South Korea). Nematodes were maintained at 25 °C and fed Botrytis cinerea grown on potato dextrose agar plates.
Seedling assay and disease evaluation
The effect of B. subtilis JCK-1398 on PWD was evaluated using seedling assays, following a published protocol20. Three-year-old P. densiflora seedlings that were sourced from Daelim Sapling Farm (Okcheon, South Korea) grown in a greenhouse were sprayed with 5 mL of a bacterial suspension at OD600 = 0.8, twice at one-week intervals. To prepare nematode inoculum, B. xylophilus was extracted from B. cinerea cultures using a Baermann funnel after one week of growth at 25 °C. The concentration was adjusted to 20,000 nematodes/mL using sterile distilled water (SDW). One week after the final bacterial treatment, 100 µL of nematode suspension (2000 nematodes) was applied to each seedling. For inoculation, absorbent cotton soaked in nematode suspension was inserted into a small slit on the seedling stem, cut using a sterilized knife. The inoculation site was then sealed with Parafilm M (Heathrow Scientific, Vernon Hills, IL, USA). Three seedlings were used per treatment and control group was treated with sterilized 1 mM MgSO4 for the bacterial treatment and SDW for the nematode inoculation. Disease severity was evaluated at 30 days post-inoculation using a visual rating scale from 0 to 5, based on the extent of needle browning and wilting symptoms. A score of 0 represented the absence of symptoms, while a score of 1 corresponded to 1–19% needle browning. A score of 2 indicated 20–39% browning, and a score of 3 denoted 40–59% browning. Seedlings with 60–79% browning and terminal shoot bending were assigned a score of 4, whereas a score of 5 was given to seedlings exhibiting 80–100% needle browning and complete wilting.
Impact of B. subtilis JCK-1398 treatment on the pine rhizosphere microbiome
To evaluate the effect of B. subtilis JCK-1398 foliar treatment on the rhizosphere microbiome, seedling assays were conducted as described above. Three seedlings were used per treatment and control group was treated with sterilized 1 mM MgSO4 for the bacterial treatment and SDW for the nematode inoculation. Sample collection involved taking composite rhizosphere samples from different sides and depths of the root zone and closely attached soils. A 10 g sample was dissolved in SDW, vortexed thoroughly, and centrifuged at 10,000 × g for 15 min. Approximately 250 mg of the collected pellet was used for metagenomic DNA extraction, using the PowerSoil® DNA Isolation Kit (MO BIO Laboratories, Carlsbad, CA, USA) according to the manufacturer’s protocol. The extracted DNA was evaluated for concentration and purity using a NanoDrop2000 spectrophotometer and further verified by agarose gel electrophoresis. Qualified samples were stored in Tris-EDTA buffer at − 20 °C until further use.
Metagenomic analysis of the pine rhizosphere microbiome focused on sequencing the V3 and V4 regions of the 16 S rRNA gene. PCR amplification and library preparation were conducted using the Herculase II fusion DNA polymerase Nextera XT Index Kit V2 (Illumina, USA) on an Illumina® MiSeq® platform at Macrogen (Seoul, South Korea).
Raw paired-end sequences obtained from the MiSeq platform were merged using FLASH software41. Adapters were removed, and and reads with a Phred quality score below 20 were trimmed to ensure high sequence accuracy. Sequences shorter than 200 bp after quality filtering were discarded to eliminate low-quality reads and artifacts. The processed sequences were analyzed with the DADA2 package42, which inferred amplicon sequence variants (ASVs) using high-resolution error correction. Sequences were clustered into ASVs with a 97% similarity threshold, using the UCLUST algorithm43. Bacterial taxonomic assignments were performed using the Greengenes database (version 13_8)44.
The analysis was managed through the Quantitative Insights Into Microbial Ecology version 2 (QIIME2) platform45,46, enabling calculations of alpha and beta diversity statistics, and taxonomic assignment of detected ASVs. All sequencing data have been deposited in the National Center for Biotechnology Information Sequence Read Archive under BioProject ID PRJNA641722.
Impact of B. subtilis JCK-1398 treatment on nematode-associated microbial composition
To examine the effect of B. subtilis JCK-1398 on the microbial community associated with nematodes from P. densiflora seedlings, a seedling assay was conducted as previously described. Two groups were evaluated: nematode-inoculated seedlings treated with B. subtilis JCK-1398 and untreated, nematode-inoculated control seedlings. Each treatment, including JCK-1398 and the control, was conducted with four biological replicates. Nematodes were extracted from the seedlings using a recently developed aseptic isolation method tailored for microbial studies47.
Nematodes isolated from pine tissues underwent mechanical disruption and were resuspended in 1 mM MgSO4 solution. For 16 S metabarcoding analysis of nematode-associated microbes, nematode pellets were washed, harvested, and subjected to DNA extraction using the PowerSoil® DNA Isolation Kit. The quality of DNA was assessed using a NanoDrop2000 spectrophotometer and agarose gel electrophoresis, and stored at − 20 °C in Tris-EDTA buffer. The 16 S rRNA gene metabarcoding sequencing and analysis were performed as described above for the rhizosphere microbiome. Sequence data were deposited in the NCBI Sequence Read Archive under BioProject ID PRJNA1032240.
Isolation and identification of nematode associated microbes
From the isolated nematodes that underwent mechanical disruption, a 200 µL aliquot of the diluted nematode suspension was plated on LB agar for single colony isolation, followed by incubation at 28 °C for 24–48 h. For further analysis, bacterial genomic DNA was extracted from overnight LB broth cultures using the Wizard Genomic DNA Purification Kit (Promega) in accordance with the manufacturer’s instructions. The 16 S rRNA gene region was amplified using universal primers: (fD1) 5ʹ-AGAGTTTGATCCTGGCTCAG-3ʹ and (rP2) 5ʹ-ACGGCTACCTTGTTACGACTT-3ʹ48. Amplified sequences were aligned with related species using the BLAST algorithm. Phylogenetic analysis and tree construction were conducted using the maximum likelihood method in MEGA 11 software, with bootstrap analysis (1000 replications) for validation49,50.
Nematicidal activity assessment of Pantoea dispersa BC11
Following the outcome of metabarcoding analysis, which revealed a predominance of certain microbes associated with nematodes from B. subtilis JCK-1398 treated pine seedlings, microbial strain BC11 was identified and selected for further study. This strain, belonging to the genus Pantoea and identified as Pantoea dispersa, was found to be a dominant microbe in the treated seedlings. The selected P. dispersa BC11 and the bioagent B. subtilis JCK-1398 were cultured in LB broth to obtain pure cultures. The cultures were centrifuged at 10,000 × g for 10 min to remove bacterial cells, and the supernatant was subsequently filtered through a 0.22 μm membrane filter to ensure sterility. To confirm the absence of viable bacterial cells, 200 µL of the filtered supernatant was plated onto LB agar and incubated at 28 °C for 24 h. The confirmed sterile cell-free culture filtrates were then prepared at concentrations of 0.3%, 0.6%, 1.2%, 2.5%, 5%, 10%, and 20% for nematicidal activity assessment.
The PWN, B. xylophilus were cultivated on PDA cultures of B. cinerea. The prepared filtrates of tested bacteria were applied to the nematodes, with sterilized LB broth serving as the control for dilution series. The mortality rate of the nematodes was recorded 48 h post-treatment. This assessment was repeated with each experiment consisting of three replicates.
Statistical analysis
Seedling assay data were analyzed using the Wilcoxon Rank-Sum test to compare disease severity between treatment groups, as the data were measured on an ordinal scale. For the nematicidal activity assay, the Shapiro–Wilk test was used to assess data normality, followed by an independent two-sample t-test for normally distributed datasets. The significance level for all tests was set at P < 0.05. Metabarcoding data were processed using QIIME2 and analyzed with R software (version 3.1.3). Multivariate statistical analyses were conducted using PAST software (version 3.23)51. Beta diversity was assessed and visualized through PCoA using weighted and unweighted UniFrac distances. Heatmaps of bacterial genera relative abundance were generated using Manhattan distance and average linkage hierarchical clustering. Differential abundance analysis of nematode-associated microbiomes was performed using the Mann–Whitney U test to compare bacterial genera between treatment groups, with False Discovery Rate (FDR) correction applied to account for multiple comparisons. Genera with an FDR-adjusted P < 0.05 were considered significant.
Data availability
All raw sequences derived from this experiment were submitted to the Sequence Read Archive of NCBI and can be found under the BioProject ID PRJNA641722 (https://www.ncbi.nlm.nih.gov/bioproject/PRJNA641722/) and BioProject ID PRJNA1032240 (https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA1032240).
References
Tóth, Á. Bursaphelenchus xylophilus, the Pinewood nematode: its significance and a historical review. Acta Biol. Szeged. 55, 213–217 (2011).
Vicente, C., Espada, M., Vieira, P. & Mota, M. Pine wilt disease: A threat to European forestry. Eur. J. Plant. Pathol. 133, 89–99 (2012).
Mamiya, Y. History of pine wilt disease in Japan. J. Nematol. 20, 219 (1988).
Back, M. A., Bonifácio, L., Inácio, M. L., Mota, M. & Boa, E. Pine wilt disease: A global threat to forestry. Plant. Pathol. 73, 1026–1041 (2024).
Futai, K. Pine wood nematode, Bursaphelenchus Xylophilus. Annu. Rev. Phytopathol. 51, 61–83 (2013).
Jones, J. T. et al. Top 10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant. Pathol. 14, 946–961 (2013).
Xue, Q., Xiang, Y., Wu, X. Q. & Li, M. J. Bacterial communities and virulence associated with pine wood nematode Bursaphelenchus Xylophilus from different Pinus spp. Int. J. Mol. Sci. 20, 3342 (2019).
Proença, D. N., Grass, G. & Morais, P. V. Understanding pine wilt disease: Roles of the pine endophytic bacteria and of the bacteria carried by the disease-causing Pinewood nematode. Microbiol. Open. 6, e00415 (2017).
Wang, X., Yu, F., Ma, R., Zhang, L. & Liu, X. Effects of pine wilt disease on rhizosphere microbiota and fine root fungi. Forests 14, 1884 (2023).
Han, G. et al. Response of pine rhizosphere microbiota to foliar treatment with resistance-inducing bacteria against pine wilt disease. Microorganisms 9, 688 (2021).
Park, I., Seo, Y. S. & Mannaa, M. Recruitment of the rhizo-microbiome army: Assembly determinants and engineering of the rhizosphere Microbiome as a key to unlocking plant potential. Front. Microbiol. 14, 1163832 (2023).
Sharma, I., Kashyap, S. & Agarwala, N. Biotic stress-induced changes in root exudation confer plant stress tolerance by altering rhizospheric microbial community. Front. Plant. Sci. 14, 1132824 (2023).
Doornbos, R. F., van Loon, L. C. & Bakker, P. A. Impact of root exudates and plant defense signaling on bacterial communities in the rhizosphere: A review. Agron. Sustain. Dev. 32, 227–243 (2012).
Kim, N. et al. Induction of resistance against pine wilt disease caused by Bursaphelenchus Xylophilus using selected pine endophytic bacteria. Plant. Pathol. 68, 434–444 (2019).
Park, A. R. et al. A Diketopiperazine, cyclo-(L-Pro-L-Ile), derived from Bacillus Thuringiensis JCK-1233 controls pine wilt disease by elicitation of moderate hypersensitive reaction. Front. Plant. Sci. 11, 1023 (2020).
Chen, L. et al. Towards the biological control of devastating forest pathogens from the genus Armillaria. Forests 10, 1013 (2019).
Terhonen, E., Kovalchuk, A., Zarsav, A. & Asiegbu, F. O. Biocontrol potential of forest tree endophytes. In Endophytes of Forest Trees. Forestry Sciences 86, 297–318 (Pirttilä, A. & Frank, A., eds.)Springer, Cham, (2018).
Nowakowska, J. A., Belbahri, L. & Oszako, T. Biological control in forests protection. Forests 14, 446 (2023).
Mannaa, M. et al. Eco-friendly biocontrol of pine wilt disease: Enhancing tree defense with Bacillus subtilis JCK-1398 for sustainable forest management. Sci. Total Environ. 955, 177233 (2024).
Mannaa, M. et al. Influence of resistance-inducing chemical elicitors against pine wilt disease on the rhizosphere Microbiome. Microorganisms 8, 884 (2020).
Garrido-Oter, R. et al. Modular traits of the Rhizobiales root microbiota and their evolutionary relationship with symbiotic rhizobia. Cell. Host Microbe. 24, 155–167 (2018).
Qin, S., Xing, K., Jiang, J. H., Xu, L. H. & Li, W. J. Biodiversity, bioactive natural products, and biotechnological potential of plant-associated endophytic actinobacteria. Appl. Microbiol. Biotechnol. 89, 457–473 (2011).
Ho, A., Di Lonardo, D. P. & Bodelier, P. L. Revisiting life strategy concepts in environmental microbial ecology. FEMS Microbiol. Ecol. 93, fix006 (2017).
Liu, Y. & Ludewig, U. Nitrogen-dependent bacterial community shifts in root, rhizome, and rhizosphere of nutrient-efficient Miscanthus × giganteus from long-term field trials. GCB Bioenergy. 11, 1334–1347 (2019).
Prauser, H. Nocardioides, a new genus of the order actinomycetales. Int. J. Syst. Evol. Microbiol. 26, 58–65 (1976).
Ikunaga, Y. et al. Nocardioides Sp. strain WSN05-2, isolated from a wheat field, degrades Deoxynivalenol, producing the novel intermediate 3-epi-deoxynivalenol. Appl. Microbiol. Biotechnol. 89, 419–427 (2011).
Franke-Whittle, I. H., Manici, L. M., Insam, H. & Stres, B. Rhizosphere bacteria and fungi associated with plant growth in soils of three replanted Apple orchards. Plant. Soil. 395, 317–333 (2015).
Meena, K. K. et al. Mitigation of salinity stress in wheat seedlings due to the application of phytohormone-rich culture filtrate extract of Methylotrophic actinobacterium Nocardioides Sp. NIMMe6. Front. Microbiol. 11, 412 (2020).
Laranjo, M., Alexandre, A. & Oliveira, S. Legume growth-promoting rhizobia: An overview on the Mesorhizobium genus. Microbiol. Res. 169, 2–17 (2014).
Menéndez, E. et al. Plant growth promotion abilities of phylogenetically diverse Mesorhizobium strains: Effect in the root colonization and development of tomato seedlings. Microorganisms 8, 412 (2020).
Kawazu, K., Zhang, H., Yamashita, H. & Kanzaki, H. Relationship between the pathogenicity of the pine wood nematode, Bursaphelenchus xylophilus, and phenylacetic acid production. Biosci. Biotechnol. Biochem. 60, 1413–1415 (1996).
Le Dang, Q. et al. Pyochelin isolated from Burkholderia arboris KRICT1 carried by pine wood nematodes exhibits phytotoxicity in pine callus. Nematology 13, 521–528 (2011).
Guo, Q., Guo, D., Zhao, B., Xu, J. & Li, R. Two Cyclic dipeptides from Pseudomonas fluorescens GcM5-1A carried by the pine wood nematode and their toxicities to Japanese black pine suspension cells and seedlings in vitro. J. Nematol. 39, 243–247 (2007).
Han, Z. M., Hong, Y. D. & Zhao, B. G. A study on pathogenicity of bacteria carried by pine wood nematodes. J. Phytopathol. 151, 683–689 (2003).
Cheng, X. Y. et al. Metagenomic analysis of the Pinewood nematode Microbiome reveals a symbiotic relationship critical for xenobiotics degradation. Sci. Rep. 3, 1869 (2013).
Vicente, C. S., Ikuyo, Y., Mota, M. & Hasegawa, K. Pinewood nematode-associated bacteria contribute to oxidative stress resistance of Bursaphelenchus Xylophilus. BMC Microbiol. 13, 1–8 (2013).
Oku, H., Shiraishi, T., Ouchi, S., Kurozumi, S. & Ohta, H. Pine wilt toxin, the metabolite of a bacterium associated with a nematode. Naturwissenschaften 67, 198–199 (1980).
Zhao, B., Liu, Y. & Lin, F. Mutual influences in growth and reproduction between pine wood nematode Bursaphelenchus Xylophilus and bacteria it carries. Front. Forestry China. 1, 324–327 (2006).
Bordenstein, S. R. & Theis, K. R. Host biology in light of the microbiome: Ten principles of holobionts and hologenomes. PLoS Biol. 13, e1002226 (2015).
Mannaa, M. & Seo, Y. S. Plants under the attack of allies: Moving towards the plant pathobiome paradigm. Plants 10, 125 (2021).
Magoč, T. & Salzberg, S. L. FLASH: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Callahan, B. J. et al. DADA2: High-resolution sample inference from illumina amplicon data. Nat. Methods. 13, 581–583 (2016).
Edgar, R. C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461 (2010).
McDonald, D. et al. An improved greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618 (2012).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–336 (2010).
Bolyen, E. et al. Reproducible, interactive, scalable, and extensible Microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857 (2019).
Mannaa, M. & Seo, Y. S. Improved and simplified method for aseptic isolation of nematodes and nematode-endosymbiotic bacteria from pine seedlings. MethodsX 11, 102421 (2023).
Weisburg, W. G., Barns, S. M., Pelletier, D. A. & Lane, D. J. 16S ribosomal DNA amplification for phylogenetic study. J. Bacteriol. 173, 697–703 (1991).
Felsenstein, J. Evolutionary trees from DNA sequences: A maximum likelihood approach. J. Mol. Evol. 17, 368–376 (1981).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3037 (2021).
Hammer, Ø., Harper, D. A. T. & Ryan, P. D. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electron. 4, 9 (2001).
Acknowledgements
This research was supported by Learning and Academic research institution for Master’s PhD students, and Postdocs (LAMP) Program of the National Research Foundation of Korea (NRF) grant funded by the Ministry of Education (No. RS-2023-00301938).
Author information
Authors and Affiliations
Contributions
M.M.: Data collection, protocol development, data analysis, manuscript writing; A.R.P.: Data collection, manuscript editing; J.C.K.: Data management, manuscript editing; Y.S.S.: Project development, data analysis, manuscript writing, manuscript editing. All authors read and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Mannaa, M., Park, A.R., Kim, JC. et al. Microbial allies recruited by Bacillus subtilis JCK-1398 to defend pine trees against pinewood nematode. Sci Rep 15, 9670 (2025). https://doi.org/10.1038/s41598-025-94434-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-94434-y