Abstract
Insulin sensitivity (IS) is a key determinant of metabolic health and may share genetic factors with obesity-related traits. Previous large-scale genetic studies have identified variants associated with IS as well as obesity related traits like body mass index (BMI) and waist-to-hip ratio (WHR). Notably, many of these associations are shared across traits, indicating a potential genetic overlap. However, the genetic intersection between IS and obesity-related traits remains underexplored. To explore this gap, we investigated associations between six IS indices, including fasting and post-glucose load measures, and genetic variants linked to BMI and WHR to determine their influence on IS and related cardiometabolic traits. To achieve this, we calculated six IS indices using fasting and oral glucose tolerance test (OGTT) data from 5,007 non-diabetic individuals, grouping them into fasting, OGTT0,120, and OGTT0,30,120 categories. A total of 678 BMI-associated and 265 WHR-associated genetic variants were analysed using linear regression, adjusting for age and sex, with sex-specific analyses for WHR. Analyses were conducted with and without BMI adjustments and corrected for multiple testing (padj). Additionally, we explored the relationship between IS-linked variants and their associations with type 2 diabetes (T2D), coronary artery disease (CAD) and stroke. Among the 678 BMI-associated variants, 100 showed nominal associations (p < 0.05) with at least one IS index; and 20 remained significant after multiple testing correction (padj < 0.05) when not adjusting for BMI. After adjusting for BMI, 70 variants retained nominal associations, and six remained significant (padj < 0.05). In sex-specific analyses of the 265 WHR-associated variants, 12 variants were associated in females when adjusted for BMI, whereas no significant associations were observed in males. Furthermore, BMI- and WHR-associated variants linked to decreased IS, such as those in FTO and VPS13C loci, were also associated with increased T2D and stroke risk, whereas IS-increasing variants, including those in VPS13C and PPARG, were linked to lower T2D and stroke risk, with some, like THADA, showing opposing effects on CAD. This study offers insights into genetic variants that influence both IS and obesity-related traits, revealing BMI- and WHR-associated variants with both positive and negative effects on IS and their potential impact on cardiometabolic health.
Similar content being viewed by others
Introduction
The prevalence of obesity and type 2 diabetes (T2D) is rising, largely due to lifestyle changes and increased caloric intake1,2. While obesity and T2D co-occur, the relationship between them is complex; not all people with obesity develop T2D, and some with normal weight develop T2D3,4. Estimates suggest that between 15% and 45% of people with obesity are considered metabolically healthy despite excess body fat5. Adiposity is a critical factor linking obesity to T2D, primarily because obesity promotes insulin resistance (IR)6. However, evidence suggests that IR and hyperinsulinemia may also contribute to development of obesity7.
Decreased insulin sensitivity (IS), or IR, influences adipose tissue distribution and metabolic health, as seen in conditions like lipodystrophy. Given the role of IS in metabolic regulation, understanding its genetic determinants is crucial. Investigating the genetic basis of IS in relation to obesity metrics such as body mass index (BMI) and waist-to-hip ratio (WHR) may provide insights into the shared and distinct genetic factors as well as pathways influencing metabolic health. The relationship between IS and these obesity metrics remains inadequately characterized, underscoring the need to identify genetic variants associated with IS among those linked to BMI and WHR.
IS can be assessed using various methods, ranging from simple clinical indices to advanced techniques like the hyperinsulinemic-euglycemic clamp8. Clinical indices derived from fasting and oral glucose tolerance test (OGTT) measurements of blood glucose and insulin provide estimates of IS in both fasting and postprandial states. In this study, we selected six different IS indices from a previously reported set of 21 indices that demonstrated differential associations with T2D-associated genetic variants9. These six indices were chosen to capture the complex nature of IS influenced by genetics, which can vary depending on physiological conditions (fasting vs. postprandial) and the specific metabolic pathways involved. By calculating these six IS indices, each reflecting different aspects of glucose metabolism and insulin action, we aimed to capture diverse genetic associations with obesity metrices, as demonstrated in the previous study9.
Building on previous findings, we hypothesized that analyzing the associations between IS indices and obesity-related traits such as BMI and WHR could help uncover genetic factors that contribute to insulin resistance while also influencing obesity-related phenotypes in both beneficial and detrimental ways. Since BMI-increasing alleles are generally linked to insulin resistance through adiposity, we aimed to determine whether WHR-associated variants exhibit independent associations with IS indices, particularly those derived from an OGTT, after adjusting for BMI. Additionally, we investigated whether certain BMI-associated loci were unexpectedly linked to improved IS, potentially indicating a metabolically healthy phenotype.
To address these questions, this study aimed to investigate the genetic relationship between IS and obesity-related traits by identifying genetic variants associated with IS among those previously linked to BMI and WHR. Using six IS indices derived from fasting and OGTT-based glucose and insulin measurements, we examined how BMI- and WHR-associated genetic variants influence IS across different metabolic states. Furthermore, we explored sex-specific effects of WHR-associated variants and assessed whether BMI adjustment alters these genetic associations. By integrating genetic data with IS indices, our goal was to elucidate the shared and distinct genetic architecture of obesity and IS, providing deeper insights into the mechanisms linking adiposity to metabolic health.
Results
Association between IS indices and BMI variants
Among the 678 BMI variants, 100 (15%) had a nominal association with at least one IS index (p < 0.05) in the analysis unadjusted for BMI (‘unadjusted’). In contrast, 70 (10%) of variants showed nominal associations in the analysis that adjusted for BMI (‘adjusted’) (Supplementary Table S3). We noted that 32 variants showed varying association across adjusted and unadjusted models (Fig. 1). In unadjusted models, 20 variants showed significant associations (padj < 0.05 based on a 5% false discovery rate) with at least one index from the fasting, OGTT0,120, or OGTT0,30,120 groups. While the number diminished to 6 when adjusted for BMI (Fig. 1).
Association between IS indices and BMI-related genetic variants. The left panel shows analyses adjusted for age, sex, and BMI, while the right panel shows analyses adjusted for age and sex only. All displayed associations are nominally significant (p < 0.05), highlighting variants linked to increased BMI. Variants are grouped using k-means clustering to demonstrate positive and negative effects on IS indices. The x-axis categorizes IS indices based on calculation time points (fasting, OGTT0,120, and OGTT0,30,120), and the y-axis lists genes, variants (rsIDs), and BMI-increasing alleles. Significance levels are indicated by asterisks: *padj < 0.05 and **padj < 0.01.
The identified variants broadly clustered into two groups: one where the BMI-increasing allele was associated with decreased insulin sensitivity (IS−) and another where the BMI-increasing allele was associated with increased insulin sensitivity (IS+). BMI adjustment influenced these associations, with fewer loci remaining significant compared to the unadjusted model. Specifically, the BMI-adjusted model identified four loci associated with IS + and two loci associated with IS−, while the unadjusted model identified eight loci for IS + and seven for IS−. Notably, rs1982441 (ELP3), rs9419958 (OBCF1), rs7692088 (CLNK), and rs1899951 (PPARG) remained significantly associated with IS + even after BMI adjustment (padj < 0.05). In contrast, rs16965062 (NOL4) and rs2228213 (HIVEP1) retained significantly associated with IS − following BMI adjustment (Figs. 1 and 2.
Venn diagrams illustrate the summary of the insulin sensitivity (IS)-associated genetic variants across different IS indices, comparing fasting indices (blue), OGTT0,120 (green), and OGTT0,30,120 (red). Variants are also labelled based on their association with increased (IS +) or decreased (IS -) insulin sensitivity. (A) BMI-associated variants in both sexes combined. (B) WHR-associated variants association in females. (C) WHR-associated variants association in males. Adjusted models (left column) accounting for BMI, while unadjusted models (right column) do not. The genetic variants displayed in each panel indicate loci associated with insulin sensitivity indices within each group. Missing circles indicate that no association was observed in the respective model.
We also observed that among the BMI variants, the majority were linked to OGTT0,30,120 indices, with fewer associations with fasting and OGTT0,120 indices when unadjusted. Upon adjustment, two of six variants were associated with fasting indices, one with OGTT0,120 and three with OGTT0,120 based indices (Figs. 1 and 2A).
Association between IS indices and WHR variants
In the combined analysis of both sexes, 41 variants showed nominal associations in the BMI-unadjusted analysis, and 52 variants showed nominal associations in the BMI-adjusted analysis (Supplementary Table S4). Only eight variants were significantly associated (padj < 0.05) after adjusting for BMI, and five variants were significantly associated without BMI adjustment (Supplementary Figure S1 and S2). Additionally, we conducted sex-specific analyses to determine if these associations differed between males and females. Among females, 26 (11%) of 246 variants exhibited nominal associations with at least one IS index (p < 0.05) when unadjusted, while 50 (20%) showed nominal associations when adjusted (Supplementary Table S5). In males, only 17 (6%) of the variants had nominal associations with at least one IS index (p < 0.05) when unadjusted, while 42 (16%) when adjusted (Supplementary Table S6).
In females, we identified 19 variants with varying association in both models. In the BMI-adjusted model, 13 variants remained significantly associated (padj < 0.05), all linked to OGTT-based IS indices, whereas no variants reached significance in the unadjusted model (Figs. 2B and 3). In males, 18 variants showed nominal associations; however, none remained significant in the adjusted model, while five variants retained significant associations (padj < 0.05) in the unadjusted model (Figs. 2C and 4). Similar to BMI-associated variants, WHR-associated variants exhibited associations with both increased IS and decreased IS, highlighting two distinct patterns of genetic influence on IS indices.
Association between IS indices and WHR-related genetic variants, analysed only in females. The left panel shows results adjusted for age and BMI, while the right panel shows results adjusted for age only. All displayed associations are significant (p < 0.05), highlighting variants linked to increased WHR. Variants are grouped using k-means clustering. The x-axis categorizes IS indices into three groups based on the time points used in their calculation, and the y-axis lists genes, variants (rsIDs), and WHR-increasing alleles. *padj < 0.05 and ** padj < 0.01.
Association between IS indices and WHR-related genetic variants, analysed only in males. The left panel shows results adjusted for age and BMI, while the right panel shows results adjusted for age only. All displayed associations are significant (p < 0.05), highlighting variants linked to increased WHR. Variants are grouped using k-means clustering. The x-axis categorizes IS indices into three groups, and the y-axis lists genes, variants, and WHR-increasing alleles. *padj < 0.05.
We further observed that variants with WHR-increasing alleles exhibited differing directions of association across IS indices groups. In females, variants such as rs6752964 (THADA locus), and rs10980797 (RP11-202G18.1) were positively associated with OGTT indices (padj < 0.05). Conversely, several variants showed strong negative association with OGTT indices, including rs3761706 (BCL2L11), rs3936510 (AC022431.2), rs72959041 (RSPO3), rs2277339 (HSD17B6, PRIM1), and rs113135335 (BCL2L11) (padj < 0.05). Additionally, rs2970332 (PRAS2), rs672356 (SHMT1), rs4964058 (ARNTL2), rs11204762 (PRUNE), rs28768122 (SETD8) and rs16976826 (RFX7) also demonstrated negative associations at padj < 0.05, albeit with slightly weaker effect sizes. In males, a similar pattern of both positive and negative associations was observed, though none reached statistical significance and effect sizes were generally weaker. Among WHR-associated variants, two loci were associated with IS + and ten loci with IS − in females when adjusted for BMI, whereas in males, three loci were linked to IS + and two to IS − in the unadjusted model.
Association of IS-associated obesity variants with T2D, CAD, and stroke
Genetic variants associated with BMI and WHR that showed significant associations with IS indices (padj < 0.05) were further analysed for their associations with T2D, CAD, and stroke using summary statistics from previously reported studies10,11,12. These variants were categorized into those linked to increasing or decreasing IS. An inconsistent pattern emerged, with variants showing associations with both increased and decreased risks of T2D, CAD, and stroke (Fig. 5). BMI-associated variants that reduce IS, such as those near FTO and VPS13C, were predominantly linked to an increased risk of T2D and stroke, while an IS-decreasing variant in GPR61 was associated with increased T2D and stroke risk but decreased CAD risk. Conversely, variants associated with increased IS exhibited diverse effects. A variant in VPS13C was linked to a lower risk of T2D and stroke with minimal impact on CAD, while a variant in PPARG was strongly associated with a lower risk of T2D but had no impact on CAD or stroke. However, variants in DPH6 and CLNK, despite being linked to increased IS, were still associated with a slightly higher risk of T2D and stroke. Meanwhile, variants in GNGT1, CDK5RAP3, and ANKS1A showed no significant associations with any of the three traits.
Association of BMI- and WHR-associated genetic variants significantly linked to insulin sensitivity (IS) and their association with type 2 diabetes (T2D) and coronary artery disease (CAD) and stroke. Forest plots display the odds ratios (OR) and confidence intervals (CI) for the association of BMI- and WHR-associated genetic variants with T2D (left panel) and CAD (middle) and stroke (right panel). Variants are categorized into four groups based on their effect on insulin sensitivity: BMI-linked variants associated with decreased IS, BMI-linked variants associated with increased IS, WHR-linked variants associated with decreased IS, and WHR-linked variants associated with increased IS. The color gradient reflects the -log10(p-value). Effect size are presented as odds ratio.
WHR-associated variants that decrease IS, such as those in AC022431, BCL2L11, SETD8, RSPO3, PRIM1, and SHMT1, were linked to an increased risk of T2D and CAD with no effect on stroke. Conversely, variants in RRAS2 and RFX7 were associated with a decreased risk of T2D (Fig. 5). WHR-associated variants that increase IS also displayed diverse effects. For instance, variants in JAZF1 and PIAS1 were strongly associated with an increased risk of T2D but had no observable effect on CAD or stroke. Meanwhile, variants in RP11 and PEPD were associated with a lower risk of T2D, without any significant effect on CAD or stroke. Most notably, a variant in THADA, which was associated with increased IS, was strongly linked to a decreased risk of T2D and stroke but to an increased risk of CAD.
Discussion
Previous large-scale GWAS studies have independently identified numerous genetic variants associated with the obesity-related traits such as BMI and WHR13,14 as well as IS indices15,16,17. In this study, we investigated the shared genetic factors between BMI/WHR and fasting and OGTT based IS indices and examined the impact of BMI adjustment on these associations. Our analysis showed that after adjusting for BMI, the number of BMI-associated variants significantly linked to IS indices, indicating that many genetic associations between BMI and IS are mediated through adiposity. The BMI and WHR variants were generally associated with both increased and decreased IS, with effects varying across fasting and OGTT-based IS indices. Additionally, WHR-associated variants exhibited sex-specific effects, with a stronger influence observed in females, particularly with postprandial IS indices following BMI adjustment.
Previous research has established that a high BMI polygenic risk score is a risk factor for T2D18, which is primarily mediated through onset of prediabetes and insulin resistance (IR)19. Consistent with this, our study reveals a pattern where genetic variants linked to obesity-related traits tend to be associated with reduced IS in models unadjusted for BMI. Many IS-associated variants, including those in FTO, BDNF and other loci, showed significant associations prior to BMI adjustment. However, after adjustment, most associations were attenuated, except for variants in PPARG and NOL4, which retained their significance. This is in line with prior studies where genetic variants in the FTO locus lost their association with IS index (Stumvoll index) after BMI adjustment20. These results support the hypothesis that genetic influences on IS, particularly postprandial IS, are largely mediated through BMI-dependent mechanisms, reinforcing the shared genetic predisposition between adiposity and glucose metabolism, as supported by studies showing a strong link between BMI and IR21.
Our results further highlight that fasting-based IS indices may have a stronger genetic component than postprandial IS indices. Associations that remained significant after BMI adjustment were primarily observed with fasting indices, suggesting that genetic factors may exert a more direct influence on fasting IS, whereas postprandial IS is more susceptible to environmental and BMI-mediated effects. This is consistent with previous study demonstrating that fasting IS has strong genetic component, while postprandial IS is more influenced by other non-genetic factors9. Notably, some variants became significantly associated with IS indices only after BMI adjustment, emphasizing the complexity of IS regulation. These findings suggest that different IS indices capture distinct physiological mechanisms, with fasting IS being more directly influenced by genetic predisposition, while postprandial IS is more intertwined with adiposity and fat distribution. Given the well-established link between higher BMI and increased IR due to greater adipose tissue accumulation, these results underscore the interplay between genetic susceptibility, body composition, and IS.
The presence of IS-improving variants among BMI-associated loci confirms the existence of “metabolically healthy obesity” genetic variants. Previous studies have shown that some genetic variants linked to increased adiposity confer favourable cardiometabolic profiles, implying a genetic basis for metabolically healthy obesity. These findings support the idea that certain individuals, due to their genetic makeup, can store fat in a way that minimizes ectopic lipid accumulation in vital organs, preserving metabolic health. This aligns with previous evidence that adiposity-promoting variants can also have protective effects on metabolism, highlighting the importance of understanding genetic heterogeneity within obesity-related traits22.
Interestingly, variants associated with both increased and decreased IS were linked to diverse metabolic consequences, challenging the assumption that all BMI- and WHR-associated variants generally contribute to metabolic dysfunction. For instance, PPARG, a key gene in lipid and glucose metabolism23,24, was positively associated with fasting-based IS and lower T2D risk, supporting its role in beneficial metabolic outcomes. Similarly, VPS13C variants exhibited protective effects on IS and were linked to decreased T2D and stroke risk. Conversely, variants in FTO, VPS13C, and GPR61 loci were associated with reduced IS and increased T2D risk, consistent with expected metabolic dysfunction. Interestingly, variants rs17238110 and rs12595158 within the same locus VPS13C, displayed opposite effects on IS as well as T2D, CAD and stroke, illustrating the complexity of genetic influences within the same genomic region.
We also identified variants in AC022431.2 and BCL2L11 that were associated with decreased IS and an increased risk of T2D, CAD, and stroke, suggesting their potential role in glucose metabolism regulation. BCL2 has previously been linked to postprandial IS in BMI-adjusted models15. While the precise mechanism by which BCL2L11 influences IS remains unclear, the BCL2 family of proteins is known to regulate apoptosis by controlling mitochondrial permeability25. Similarly, AC022431.2, though not well-characterized, is highly expressed in adipose tissue, suggesting a possible role in metabolic function. Conversely, a variant in JAZF1, which was associated with increased IS, was linked to an elevated risk of T2D while showing no apparent effects on CAD or stroke. JAZF1 has previously been implicated in reduced insulin release and beta-cell dysfunction26. This paradoxical association of JAZF1 with both improved IS and higher T2D risk may be explained by a compensatory mechanism where reduced insulin release is offset by enhanced IS27, yet still contributes to increased T2D risk.
A variant in THADA was associated with increased IS but displayed a distinct risk profile for cardiometabolic diseases. THADA has been implicated in energy homeostasis, thermogenesis, pancreatic beta-cell function, and T2D28,29,30,31. Its dual role, enhancing IS while also increasing T2D risk, highlights its pleiotropic nature and diverse metabolic effects. The observed increase in IS could be explained by a compensatory mechanism, where improved insulin action offsets beta-cell dysfunction, potentially contributing to an increased risk of T2D. However, the inverse association with CAD remains unclear and warrants further investigation.
This study has some limitations. The sample size, while robust, may limit the statistical power of association analyses, and our findings have not been validated in diverse populations. Although the homogeneity of our sample minimizes ethnic confounders, it also restricts the generalizability of our results. Furthermore, adjusting for BMI in genetic analyses introduces complexities, as BMI itself is influenced by genetic variants and impacts IS, leading to potential collider bias32. This is particularly relevant for WHR-associated variants, given that they were identified in a BMI-adjusted framework in prior studies. While we acknowledge these potential biases, our results provide valuable insights into the genetic interplay between obesity and IS, warranting further validation and mechanistic exploration.
In summary, our findings demonstrate that genetic predisposition to higher BMI significantly impacts IS, with distinct effects on fasting and postprandial IS indices. Most BMI-linked variants were associated with IS in unadjusted models, but these associations largely diminished after BMI adjustment, suggesting their effects are mediated through adiposity. Conversely, WHR-associated variants, particularly in females, showed significant associations with OGTT-based indices after BMI adjustment, underscoring sex-specific genetic influences on IS. Notably, our results consolidate the notion that not all obesity-associated variants negatively impact metabolic health; some confer protective effects on IS, reinforcing the concept of metabolically healthy obesity. This study provides insights into the intricate genetic relationships between obesity and IS, offering a deeper understanding of the mechanisms driving metabolic health and disease. Understanding these relationships is crucial for the development of targeted interventions aimed at improving metabolic health through personalized genetic insights.
Methods
Study populations
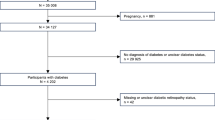
The Inter99 is a population cohort for primary cardiovascular disease prevention through lifestyle modifications (http://www.inter99.dk) originally comprising 6,784 individuals33,34. For this study we excluded individuals with known diabetes, siblings, and first-degree cousins, and those lacking complete phenotype data for IS calculations, with final sample size of 5,007 individuals (Table 1). The Inter99 study was approved by the Copenhagen Ethical Committee (reference numbers KA98155 and H-3-2012-155) and conducted in accordance with the ethical guidelines of the Declaration of Helsinki II. An informed consent was obtained from all participants of the study.
Clinical examinations
All participants in the study were thoroughly examined for demographic details, and measurements of height and weight were recorded to determine their BMI. Individuals without prior diagnosed T2D were subjected to a standardized oral glucose tolerance test (OGTT), following the World Health Organization’s guidelines35. Before undergoing the OGTT, fasting blood samples were collected from each participant after they had fasted overnight for 12 h. This was followed by the administration of a 75-gram glucose solution, with blood samples then being taken at intervals of 0, 30, and 120 min after consumption to assess plasma glucose and serum insulin levels. The clinical examination data was acquired from the baseline examination33 and a summary of the clinical parameters for the number of individuals used in this study is shown in Table 1.
Calculation of insulin sensitivity indices
We calculated six IS indices, using fasting and OGTT measures of plasma glucose (G0, G30 and G120) and serum insulin (I0, I30 and I120) using the formulas as described in Table 2. For HOMA-IR, we reversed its direction by multiplying with − 1 to represent IS. Therefore, we labeled it as “inv-HOMA-IR” to indicate its reflection of IS instead of IR.
Transformation of IS indices
To normalize IS indices, we used a rank-based inverse normal transformation, as described in our previous study9. This transformation converted raw IS indices to an approximately standard normal distribution, which also implicitly standardized them on a uniform scale. This approach enhances statistical robustness and facilitates effective comparison of IS indices.
Genotyping
The study participants were genotyped using the Illumina Human OmniExpress-24 (versions 1.0 A and 1.1 A) and analyzed using the Genotyping module of the GenomeStudio software (version 1.9.4, 2011.1; Illumina). To ensure data quality, we excluded any genotype samples that had a call rate below 95%, heterozygosity rates more than three interquartile ranges above the median ( > + 3(IQR = 3[IQ3-IQ1]), discrepancies between genetically inferred and reported sex, and duplicate samples with final sample size 5007 remaining in this study. Furthermore, we removed genetic variants with a call rate under 98%, heavily deviating from Hardy-Weinberg equilibrium (p < 10− 5), and monomorphic markers. To impute missing variants, we utilized the Michigan Imputation Server, using the Human Reference Consortium (HRC) panel on the GRCh37 genome build, 2016 release (HRC r1.1).
Variant selection
We selected 678 genetic variants associated with BMI and 265 variants associated with WHR from prior genome-wide association studies. These variants were selected based on a genome-wide significance threshold (p < 5 × 10− 8) and minor allele frequencies greater than 1% (Supplementary Tables 1–2) BMI-associated variants were sourced from Locke et al. (2018)13, which originally identified 941 independent variants through GCTA-COJO analysis. Filtering for MAF > 1% reduced this to 658 variants. The 678 BMI-associated variants composed of 660 non-coding, including: 26 in the 3’ untranslated regions, 4 in the 5’ untranslated regions, 440 within intronic regions, 3 nonsense-mediated decay transcripts, 14 within non-coding transcript exons, and 173 as non-coding transcript variants. There were 18 coding variants, encompassing 14 missense and 4 synonymous variants.
Similarly, WHR-associated variants were obtained from Pulit et al. (2019)14, which initially reported 463 independent variants, with the MAF > 1% filter reducing this to 265 variants. The 265 WHR-associated variants were composed of 23 coding, divided into 21 missense and 2 synonymous variants, and 242 non-coding variants, which included 10 in the 3’ untranslated regions, 2 in the 5’ untranslated regions, 176 in intronic regions, 4 associated with nonsense-mediated decay, 9 in non-coding transcript exons, and 41 as non-coding transcript variants (Supplementary Tables 1–2).
Genetic association analysis
Each BMI and WHR linked variant was tested for association with IS indices using two models: the first adjusting for age and sex, and the second adjusting for age, sex and BMI. To address potential population structure biases, 10 genetic principal components were included as covariates in both models, derived using PLINK v2.00a3.7LM. The RVTEST software (version: 20190205) was used for genetic association testing. P values < 0.05 were considered nominally statistically significant. We corrected the p value (padj) for multiple testing by applying a 5% false discovery rate using the Benjamini-Hochberg procedure, calculated for the number of variants tested in each test.
We tested the association between WHR-linked variants and IS indices following the same models as described above for BMI variants. We further conducted a sex-stratified analysis of WHR-linked variants with IS indices, assessing male and female groups separately using the same models as above, used for BMI variants analysis, but without sex as a covariate. The association tests between WHR variants and IS indices were adjusted for multiple testing, accounting for both the total number of variants in the combined-sex analysis and those tested separately in the sex-stratified analyses. The significance levels were reported accordingly after these corrections.
Variant clustering
Variants linked to BMI and WHR that were nominally associated with at least one IS index underwent hard clustering. This was achieved by applying the k-means clustering method using the standard Lloyd algorithm, which grouped the variants into clusters based on estimated association effects with respect to IS indices. Before applying K means clustering, we applied the elbow method to determine the optimal number of clusters for k-means clustering. The clustering was performed to delineate distinct clusters of variants associated with specific IS index groups. The clustered variants were visualized using the Complex Heatmap package (2.16.0) in R (version 4.3.2).
Association analysis of IS-associated obesity variants with T2D, CAD, and stroke
To assess whether IS-associated obesity variants are linked to T2D, coronary artery disease (CAD), and stroke, we conducted a lookup analysis using summary statistics from the largest available GWAS for these traits10,11,12. Genetic variants from BMI- and WHR-associated loci that remained significant for IS after a 5% FDR correction were examined for associations in European populations with T2D (242,283 cases and 1,569,730 controls), CAD (181,522 cases and 984,168 controls, and stroke (73,652 cases and 1,234,808 controls). Variants were categorized based on their obesity phenotype (BMI or WHR) and the direction of their IS beta estimates, distinguishing those associated with increased or decreased IS. To ensure consistency in effect direction, beta estimates were flipped for variants where the risk allele did not match the effect allele reported in the original GWAS. Odds ratios (ORs) were calculated as OR = eβ, and 95% confidence intervals (CIs) were computed as CI = e(β±1.96×SE).
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on a reasonable request and according to Danish data protection regulations. The Inter99 data sets may be obtained by a third party by contacting Allan Linneberg at allan.linneberg@regionh.dk and/or Torben Hansen at torben.hansen@sund.ku.dk.
References
Blüher, M. Obesity: global epidemiology and pathogenesis. Nat. Rev. Endocrinol. 15, 288–298 (2019).
Loos, R. J. F. & Yeo, G. S. H. The genetics of obesity: from discovery to biology. Nat. Rev. Genet. 23, 120–133 (2022).
Vecchié, A. et al. Obesity phenotypes and their paradoxical association with cardiovascular diseases. Eur. J. Intern. Med. 48, 6–17 (2018).
Blüher, M. Metabolically healthy obesity. Endocr. Rev. 41, bnaa004 (2020).
Loos, R. J. F. & Kilpeläinen, T. O. Genes that make you fat, but keep you healthy. J. Intern. Med. 284, 450–463 (2018).
Ghaben, A. L. & Scherer, P. E. Adipogenesis and metabolic health. Nat. Rev. Mol. Cell. Biol. 20, 242–258 (2019).
James, D. E., Stöckli, J. & Birnbaum, M. J. The aetiology and molecular landscape of insulin resistance. Nat. Rev. Mol. Cell. Biol. 22, 751–771 (2021).
Gastaldelli, A. Measuring and estimating insulin resistance in clinical and research settings. Obesity 30, 1549–1563 (2022).
Suleman, S. et al. Genetic underpinnings of fasting and oral glucose-stimulated based insulin sensitivity indices. J. Clin. Endocrinol. Metab. dgae275 https://doi.org/10.1210/clinem/dgae275 (2024).
Suzuki, K. et al. Genetic drivers of heterogeneity in type 2 diabetes pathophysiology. Nature 1–11. https://doi.org/10.1038/s41586-024-07019-6 (2024).
Aragam, K. G. et al. Discovery and systematic characterization of risk variants and genes for coronary artery disease in over a million participants. Nat. Genet. 54, 1803–1815 (2022).
Mishra, A. et al. Stroke genetics informs drug discovery and risk prediction across ancestries. Nature 611, 115–123 (2022).
Yengo, L. et al. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 27, 3641–3649 (2018).
Pulit, S. L. et al. Meta-analysis of genome-wide association studies for body fat distribution in 694 649 individuals of European ancestry. Hum. Mol. Genet. 28, 166–174 (2019).
Walford, G. A. et al. Genome-wide association study of the modified Stumvoll insulin sensitivity index identifies BCL2 and FAM19A2 as novel insulin sensitivity loci. Diabetes 65, 3200–3211 (2016).
Chen, J. et al. The trans-ancestral genomic architecture of glycemic traits. Nat. Genet. 53, 840–860 (2021).
Williamson, A. et al. Genome-wide association study and functional characterization identifies candidate genes for insulin-stimulated glucose uptake. Nat. Genet. 55, 973–983 (2023).
Khera, A. V. et al. Polygenic prediction of weight and obesity trajectories from birth to adulthood. Cell 177, 587–596e9 (2019).
Miao, Z. et al. The causal effect of obesity on prediabetes and insulin resistance reveals the important role of adipose tissue in insulin resistance. PLOS Genet. 16, e1009018 (2020).
Do, R. et al. Genetic variants of FTO influence adiposity, insulin sensitivity, leptin levels, and resting metabolic rate in the Quebec family study. Diabetes 57, 1147–1150 (2008).
Kolb, H. Obese visceral fat tissue inflammation: from protective to detrimental? BMC Med. 20, 494 (2022).
Huang, L. O., Loos, R. J. F. & Kilpeläinen, T. O. Evidence of genetic predisposition for metabolically healthy obesity and metabolically obese normal weight. Physiol. Genomics. 50, 169–178 (2018).
Kim, H. & Ahn, Y. Role of peroxisome proliferator-activated receptor-γ in the glucose-sensing apparatus of liver and β-cells. Diabetes 53, S60–S65 (2004).
Francque, S. et al. Nonalcoholic steatohepatitis: the role of peroxisome proliferator-activated receptors. Nat. Rev. Gastroenterol. Hepatol. 18, 24–39 (2021).
Brenner, D. & Mak, T. W. Mitochondrial cell death effectors. Curr. Opin. Cell. Biol. 21, 871–877 (2009).
Grarup, N. et al. Association testing of novel type 2 diabetes risk alleles in the JAZF1, CDC123/CAMK1D, TSPAN8, THADA, ADAMTS9, and NOTCH2 loci with insulin release, insulin sensitivity, and obesity in a population-based sample of 4,516 glucose-tolerant middle-aged Danes. Diabetes 57, 2534–2540 (2008).
Ahrén, B. & Thorsson, O. Increased insulin sensitivity is associated with reduced insulin and glucagon secretion and increased insulin clearance in man. J. Clin. Endocrinol. Metab. 88, 1264–1270 (2003).
Moraru, A. et al. THADA regulates the organismal balance between energy storage and heat production. Dev. Cell. 41, 72–81e6 (2017).
Zhang, Y. et al. THADA Inhibition in mice protects against type 2 diabetes mellitus by improving pancreatic β-cell function and preserving β-cell mass. Nat. Commun. 14, 1020 (2023).
Simonis-Bik, A. M. et al. Gene variants in the novel type 2 diabetes loci CDC123/CAMK1D, THADA, ADAMTS9, BCL11A, and MTNR1B affect different aspects of pancreatic beta-cell function. Diabetes 59, 293–301 (2010).
Zeggini, E. et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat. Genet. 40, 638–645 (2008).
Sun, Y. V. et al. Identification and correction for collider bias in a genome-wide association study of diabetes-related heart failure. Am. J. Hum. Genet. 111, 1481–1493 (2024).
Jørgensen, T. et al. A randomized non-pharmacological intervention study for prevention of ischaemic heart disease: baseline results Inter99 (1). Eur. J. Cardiovasc. Prev. Rehabil. 10, 377–386 (2003).
Kolberg, J. A. et al. Development of a type 2 diabetes risk model from a panel of serum biomarkers from the Inter99 cohort. Diabetes Care. 32, 1207–1212 (2009).
Alberti, K. G. M. M. & Zimmet, P. Z. Definition diagnosis and classification of diabetes mellitus and its complications part 1: diagnosis and classification of diabetes mellitus provisional report of a WHO consultation original articles. https://doi.org/10.1002/(SICI)1096-9136(199807)15:7 (1997).
Katz, A. et al. Quantitative insulin sensitivity check index: A simple, accurate method for assessing insulin sensitivity in humans. J. Clin. Endocrinol. Metab. 85, 2402–2410 (2000).
Matthews, D. R. et al. Homeostasis model assessment: insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28, 412–419 (1985).
Stumvoll, M., Van Haeften, T., Fritsche, A. & Gerich, J. Oral glucose tolerance test indexes for insulin sensitivity and secretion based on various availabilities of sampling times [11]. Diabetes Care. 24, 796–797 (2001).
Gutt, M. et al. Validation of the insulin sensitivity index (ISI0,120): comparison with other measures. Diabetes Res. Clin. Pract. 47, 177–184 (2000).
Matsuda, M. & DeFronzo, R. A. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 22, 1462–1470 (1999).
Hansen, T. et al. The BIGTT test: a novel test for simultaneous measurement of pancreatic beta-cell function, insulin sensitivity, and glucose tolerance. Diabetes Care. 30, 257–262 (2007).
Acknowledgements
The authors thank C. F Rundsten, M. Schubert, and J. Bork-Jensen for their technical assistance in the project. We also thank L. Ryborg and K. N. Kaadtmann for their administrative and research support.
Funding
The study was supported by a grant from Independent Research Fund Denmark 1030-00280B to NG. Further, S.S, L.A, T.H, and N.G were also supported by an unrestricted donation to the Novo Nordisk Foundation Center for Basic Metabolic Research (https://cbmr.ku.dk/) at the University of Copenhagen by the Novo Nordisk Foundation (Grant No. NNF18CC0034900 and NNF23SA0084103).
Author information
Authors and Affiliations
Contributions
S.S, N.G and T.H proposed and designed the overall study. S.S conducted the statistical analysis, reported, and interpreted the findings and wrote the manuscript. A.L is responsible for the curation and documentation of data from the Inter99 study and provides advice on the dataset. L.A provided statistical guidelines for study models and reviewed the manuscript thoroughly. N.G and T.H are the guarantors of this work and have full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis. All authors reviewed and approved the final version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
N.G is currently employed at Novo Nordisk A/S. T.H owns stocks in Novo Nordisk A/S. Remaining authors declared no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Suleman, S., Ängquist, L., Linneberg, A. et al. Exploring the genetic intersection between obesity-associated genetic variants and insulin sensitivity indices. Sci Rep 15, 15761 (2025). https://doi.org/10.1038/s41598-025-98507-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-025-98507-w