Abstract
We developed a practical framework to construct digital twins for predicting and optimizing triple-negative breast cancer (TNBC) response to neoadjuvant chemotherapy (NAC). This study employed 105 TNBC patients from the ARTEMIS trial (NCT02276443, registered on 10/21/2014) who received Adriamycin/Cytoxan (A/C)-Taxol (T). Digital twins were established by calibrating a biology-based mathematical model to patient-specific MRI data, which accurately predicted pathological complete response (pCR) with an AUC of 0.82. We then used each patient’s twin to theoretically optimize outcome by identifying their optimal A/C-T schedule from 128 options. The patient-specifically optimized treatment yielded a significant improvement in pCR rate of 20.95–24.76%. Retrospective validation was conducted by virtually treating the twins with AC-T schedules from historical trials and obtaining identical observations on outcomes: bi-weekly A/C-T outperforms tri-weekly A/C-T, and weekly/bi-weekly T outperforms tri-weekly T. This proof-of-principle study demonstrates that our digital twin framework provides a practical methodology to identify patient-specific TNBC treatment schedules.
Similar content being viewed by others
Introduction
Neoadjuvant therapy is considered the standard-of-care treatment of stage I-III, locally advanced triple-negative breast cancer (TNBC). Neoadjuvant therapy increases the success rate for breast conservation surgery by reducing tumor burden and provides the opportunity to treat micro-metastases in a naïve state, thereby improving the progression-free survival of patients1,2. TNBC patients who achieve a pathological complete response (pCR) in the neoadjuvant setting have a favorable recurrence-free survival; in contrast, patients who have residual disease after neoadjuvant chemotherapy (NAC) are at an increased risk of early recurrence and death3,4.
Current NAC regimens for TNBC are not without limitations. Less than half of TNBC patients treated with standard taxane/anthracycline-based NAC have a pCR or minimal residual disease at the time of surgical resection3. While emerging neoadjuvant therapies5,6,7 may improve treatment efficacy, tailoring treatments for individual patients remains a major barrier to improving the outcome of TNBC patients. For example, the immunotherapy pembrolizumab was recently approved for high-risk, early-stage TNBC in combination with chemotherapy8. The KEYNOTE-522 trial demonstrated that this new regimen achieved a 7.5% improvement in the pCR rate and an 8.0% improvement in the event-free survival rate in TNBC patients9,10. However, the combination is also associated with significant high-grade treatment-related toxicity8,9,10. Patients who do not respond well to pembrolizumab may suffer from (unnecessary) severe side effects, including increased likelihood of hospitalization, cardiac damage, leukemia, and even death11. Similarly, for the conventional therapies employed during NAC, the therapeutic regimen is far from optimal. NAC for TNBC generally combines two or three drugs (e.g., doxorubicin, paclitaxel, and docetaxel) administered in periodic cycles. For each chemotherapy, a few dose and schedule options are acceptable as determined by population-based studies of efficacy and toxicity. However, population-based approaches are intrinsically limited as they can only test a few dosing schedules; indeed, it is impossible for clinical trials to evaluate all possible drug doses, schedules, and combinations. Moreover, population-based approaches are, by definition, not designed to optimize interventions for an individual patient, and this can lead to sub-optimal outcomes12,13.
The need to optimize therapeutic schedules on a patient-specific basis in breast cancer has recently been well recognized across clinical and regulatory practice14,15. Within the last three years, multiple initiatives and workshops have been organized (by the FDA, AACR, SABCS, ASCO, etc.) to discuss how to design optimal dosing schedules in breast medical oncology16,17,18. Through these discussions, the lack of reliable mathematical models or biomarkers to guide individualized tailoring of therapy has been recognized as a fundamental barrier. Individualized dose optimization of systemic therapy is therefore a high-risk decision that must be supported with sufficient evidence. But the systematic comparison of multiple therapeutic regimens through conventional clinical trials is extraordinarily resource-intensive. Thus, there is an emerging need to develop individual-based approaches which can predict responses to various treatment schedules in a practical, rigorous, and resource-light fashion, so to guide the optimization.
As a potential solution to this challenge, biology-based mathematical models have recently been shown to accurately predict an individual’s spatiotemporal tumor response to therapy when calibrated with patient-specific imaging data19,20,21,22,23,24,25,26,27,28. The biology-based models can provide mechanistic interpretation of response during NAC via model parameters quantifying local tumor cell proliferation, cell mobility, parenchyma mechanics, therapy efficacy and pharmacokinetics22. Furthermore, once the model is calibrated with patient-specific data, it is capable of simulating a wide range of therapeutic regimens, thereby providing a unique opportunity to identify an optimal intervention in a patient-specific manner.
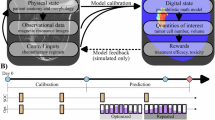
In this work, we developed practical pipelines to establish patient-specific digital twins29 based on a personalized biology-based model, and identified the therapeutic schedule(s) that can achieve the best TNBC response to NAC on a patient-specific basis (Fig. 1). The patient-specific digital twin was established by calibrating the biology-based model with an individual’s MRI data, and then used for predicting the patient’s response to various therapeutic schedules. Three optimization strategies were constructed and compared for their ability to improve outcome (i.e., pCR rate), as well as to provide guidance on the benefit of treatment escalation and de-escalation. We also performed a retrospective validation by comparing the treatment response predicted by the digital twin to the outcomes observed in previous clinical trials.
For individual TNBC patients (a), longitudinal MRIs (b) are collected pre- and early in the course of NAC, and then used to calibrate a biology-based mathematical model of tumor growth and treatment response, thereby establishing digital twins (c). The digital twins capture patient-specific tumor characteristics as well as response behaviors, so they can be used to systematically optimize NAC schedules (d). This digital twin-based optimization schema has the potential to improve pCR rates of TNBC (e, top), and to identify opportunities for therapy de-escalation (e, bottom) on a patient-specific basis.
This work is a proof-of-principle study with the overall goal of demonstrating the rationale and laying the technical foundation of our digital twin framework. In particular, we show how the digital twin can be applied in three use cases: 1) provide accurate predictions of the response of individual patients, 2) identify alternative therapeutic regimens hypothesized to outperform the standard-of-care regimen the patient actually received, and 3) recapitulate the results of three clinical trials that were central to establishing the current standard-of-care treatment regimens for TNBC. Meeting these goals will help motivate the design of future prospective trials and eventually clinical deployment of digital twins for the care of TNBC patients.
Results
Image-guided digital twins made accurate predictions of TNBC response to NAC
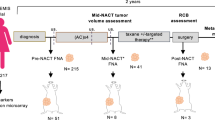
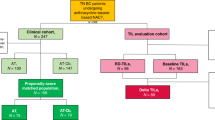
This study included treatment-naïve TNBC patients (n = 105) who participated in the ARTEMIS trial and received the standard two-component NAC: Adriamycin/Cytoxan (A/C) and Taxol (T). Longitudinal multi-parametric MRI characterizing morphology, blood flow, and cellularity were collected before, during, and after A/C therapy. Based on each patient’s MRI data, a digital twin was established by calibrating a biology-based model using previously described methods22,30 (Fig. 2a). As the calibrated digital twin captures tumor cell migration, proliferation and treatment efficacy with personalized parameters, it can be used to predict the spatiotemporal response of the patient’s tumor to the actual administered NAC (Fig. 2a).
The framework (a) consists of data preparation (i.e., image collection and processing pipeline), model personalization (i.e., calibration of the biology-based model using longitudinal MRI data), and response prediction. The prediction accuracy was evaluated by comparing the predicted final pathological status to the actual pathological status via ROC analysis (b). Applying both the actual schedule (navy curves in (c)) and the alternative schedule (red curves in (c)) of NAC to the digital twin allows for predicting the dynamics of tumor response (measured by the change of tumor volume over time; d) to each intervention. In this illustrative case, the actual therapeutic schedule leads to a predicted tumor volume after NAC (TVT) larger than the pCR/non-pCR differentiating threshold (TVT,J, determined from the optimal cutoff of ROC; see “Methods” section “Establishment of patient-specific digital twin to predict TNBC response to NAC” for details); thus, we predicted this patient as a non-pCR. In contrast, the alterative schedule leads to a predicted TVT less than the TVT,J; thus, we predicted the alternative schedule would lead to a pCR for this patient.
Based on the predicted tumor volume (TV) after the actual NAC schedule is administered, the image-guided digital twins achieved an area under the receiver operator characteristic (ROC) curve (AUC) (95% confidence interval) of 0.82 (0.73–0.88) for differentiating pCR (n = 64) from non-pCR cases (n = 41) (Fig. 2b). The optimal cutoff point of the ROC curve provided a sensitivity and specificity of 0.67 and 0.80, respectively, with a predicted pCR rate of 60.95% (51.43–70.48%) in the cohort. Compared to the actual pCR rate of 59.05%, the χ2 test detected no significant difference between the measured and digital twin-predicted pCR rates (P = 0.78).
Digital twin-based treatment optimization revealed individualized opportunities for outcome improvement and treatment de-escalation
In addition to predicting the response of a patient’s tumor to the actual therapeutic regimen, the digital twin was also used to systematically predict response to 128 alternative (but clinically feasible) NAC schedules (Table 1). The predicted responses to alternative schedules were then compared to the patient’s response to their actually received treatment to identify schedules that would hypothetically have outperformed the actual treatment (Fig. 2c, d).
Three optimization strategies were constructed to identify the optimal NAC regimen from all alternative NAC schedules: 1) “multi-step optimization” which sequentially optimizes the administration of each component of NAC (i.e., A/C and T, respectively) before its initiation (Fig. 3a), 2) “simultaneous optimization” which optimizes all NAC components at the same time before treatment initiation (Fig. 3b), and 3) “midway optimization” which begins with a standard schedule for the first NAC component, and optimizes subsequent components based on the outcome from the first component (Fig. 3c). We sought to determine whether each optimization strategy can significantly improve an individual patient’s post-NAC residual tumor volume (and therefore improve the possibility of achieving pCR), as well as provide guidance on de-escalation. For convenience, we refer to the predicted residual tumor volume at the end of the A/C and T components as TVA/C and TVT, respectively. (Note: since T is the last component of NAC, TVT is also the residual TV at the completion of all NAC).
In the multi-step optimization strategy (a), patient-specific response is predicted for all candidate A/C schedules (light blue area represents the range of tumor volume, TV, predicted from various A/C schedules), with the schedule yielding the minimized TVA/C identified as the optimal A/C schedule. Based on the minimized TVA/C, patient-specific response is then predicted for all candidate T schedules (light green area), and the one yielding the minimal TVT is identified as the optimal T schedule. Together, the optimal A/C and T schedules form the optimal NAC schedule (red area). In the simultaneous optimization strategy (b), patient-specific response is predicted for all candidate A/C and T schedules, with the one yielding the minimal TVT identified as the optimal NAC schedule (red area). In the midway optimization strategy (c), the A/C schedule is fixed to the actual schedule (black curve). Based on the TV achieved from the actual A/C schedule, patient-specific response is predicted to all candidate T schedules, with the one yielding the minimal TVT identified as the optimal T schedule.
Multi-step optimization identified opportunities for patient-specific treatment escalation or de-escalation
The multi-step optimization strategy illustrated that changing the A/C or T schedule can yield a significant difference in the predicted outcomes across the patients. Figure 4a shows an example (Patient 1), whereby the optimal A/C schedule that resulted in the minimal TVA/C had a duration of 56 days. Compared to the 63-day duration the patient actually received, the optimization suggested a potential benefit to delivering A/C treatment with a denser schedule (more aggressive, i.e., escalation). Conversely, Fig. 4b shows another example (Patient 2), whereby five different A/C schedules resulted in the same minimal TVA/C of 0 cm3, which had a duration within the 56–72 day range, thus all identified as optimal A/C schedules. Compared to the 59-day duration the patient actually received, the optimization revealed a potential benefit to deliver A/C with a less dense schedule (i.e., de-escalation). Interestingly, the optimized and actual duration of A/C were both 56 days for Patient 3 (Fig. 4c), suggesting that no alteration in the treatment schedule was needed for this patient. Of the 105 patients, the analysis identified 47 who would have had the opportunity for A/C escalation, 37 for A/C de-escalation, and 21 for whom no change in the A/C schedule was needed (Fig. 5a).
a–c and d–f show the results of first (i.e., A/C optimization) and second (i.e., T optimization) steps, respectively, of the multi-step optimization for each patient. In (a–c), the blue curves represent the outcomes from all candidate A/C schedules, with the black and red dots indicating the actual and the optimized outcomes, respectively. In (d–f), the light green curves/surfaces represent the outcomes of the candidate T schedules with 12 cycles, the navy curves/surfaces represent the outcomes of the candidate T schedules with 4 cycles, and the gray flat plane depicts the threshold of residual tumor volume (TVT,J) for differentiating pCR from non-pCR. g–i show the results of the simultaneous optimization in the same three patients, in which the light green surfaces represent the outcomes from all A/C and T candidates with 12 cycles, and the navy surfaces represent the outcomes from all A/C and T candidates with 4 cycles. j–l show the results of the midway optimization, in which the light green curves represent the outcomes from the candidate T schedules with 12 cycles, and the navy curves represent the outcomes from the candidate T schedules with 4 cycles. Patient 1 had a non-pCR with their actual treatment, and optimization of therapeutic schedule suggests this could be improved to pCR. Patient 2 had a pCR with their actual treatment, and optimization of schedule suggests an opportunity for de-escalation. Patient 3 had a non-pCR either with the actual or optimized therapeutic schedules.
a compares the duration of A/C that the individual patient actually received (horizontal axis) versus the duration optimized for the patient (vertical axis). Similarly, (b) compares the duration of Taxol that the individual patient actually received, and the duration optimized for the patient. Each gray dot represents one patient. Note that when multiple schedules lead to the optimal outcome for the patient (like Patient 2 in Fig. 4b), the “optimized duration” is shown as the longest one among the optimal schedules. The dots in the light blue areas indicate patients who had the opportunity for treatment de-escalation, whereas the dots in the light red areas indicate patients had the opportunity for treatment escalation. Dots falling on the diagonal indicate no change in the treatment schedules was suggested for the corresponding patients.
After identifying optimal TV outcomes from A/C, the optimal T schedules can then be found. As shown for Patient 1 in Fig. 4d, the optimal T schedule had a duration of 56 days, while the T schedule that the patient actually received had a duration of 84 days, suggesting potential benefits of escalation. A similar escalation benefit was observed for Patient 3 (Fig. 4f). Conversely, for Patient 2 (Fig. 4e), given the optimal A/C schedules, the optimal T schedules had a duration between 56–84 days compared to the 77-day duration the patient actually received, suggesting a potential benefit of de-escalation. Of the 105 patients, the analysis identified 82 patients who would have had the opportunity for T escalation, 10 for T de-escalation, and 13 for whom no change in the T schedule was needed (Fig. 5b).
Simultaneous optimization clearly identified patients who could benefit from altering the therapeutic schedules
The simultaneous optimization strategy also showed that changing the entire NAC schedule can yield a significant difference in predicted outcomes across the patients. For Patient 1 (Fig. 4g), the TVT was reduced from 6.90 cm3 to 0.03 cm3 by simultaneously optimizing the A/C and T schedules. In this case, the optimization suggested that delivering the treatment with a denser schedule changed the predicted outcome from non-pCR to pCR. For Patient 3 (Fig. 4i), the TVT was reduced from 4.31 cm3 to 3.22 cm3 by simultaneously optimizing the A/C and T schedules; however, this improvement did not change the predicted status of non-pCR after completion of NAC. Conversely, Patient 2 (Fig. 4h) achieved a pCR with the actual treatment schedule, and simultaneously optimizing the treatment schedule was able to find de-escalating options to maintain the pCR status after NAC.
Comparing the final outcomes from the multi-step (Fig. 4d-f) and simultaneous optimization strategies (Fig. 4g-i), we observed that both yielded the same optimal outcome (i.e., minimized TVT). However, when the optimal outcome can be achieved with multiple therapeutic schedules, the simultaneous optimization can provide more schedules that lead to this optimal outcome (Fig. 4h). We noted that the option(s) identified from the multi-step optimization strategy were always a subset of the option(s) identified from the simultaneous optimization strategy. Of the 41 patients who had a non-pCR outcome from their actual NAC, 26 (i.e., 63.41%) were predicted to achieve a pCR with treatment schedules identified by either the multi-step or simultaneous optimization strategies.
Midway optimization identified different optimal outcomes than the multi-step and simultaneous optimizations
The midway optimization strategy again showed that changing the T schedule yielded a significant difference in predicted outcomes across the patients. As the initial conditions for the midway optimization strategy are different from the multi-step and simultaneous optimization strategies, the midway optimization can yield a different optimal outcome than the other two strategies. For example, the T schedule optimized with the midway strategy for Patient 1 (Fig. 4j) had a duration of 56 days, which was identical to both the multi-step and simultaneous optimized T schedules. However, the midway optimized outcome (i.e., TVT) was 0.30 cm3, which was an order of magnitude greater than the multi-step and simultaneous optimized outcome of 0.03 cm3. In contrast, a smaller TVA/C could not be achieved for Patients 2 and 3 by optimizing the A/C schedules (Fig. 4b, c); thus, the midway optimization provided the same optimal outcome (i.e., minimized TVT) as the other two strategies, though with fewer options to achieve this optimal outcome (Fig. 4k, l). Of the 41 patients who had a non-pCR from their actual NAC, 22 (i.e., 53.66%) were predicted to achieve a pCR with treatment schedules identified by the midway optimization strategy.
Digital twin-based treatment optimization significantly improved pCR rates in TNBC
The multi-step optimization resulted in a TVA/C with a median (interquartile range; IQR) of 0.32 (0.00–1.42) cm3 over the whole cohort, and a TVT of 0.26 (0.00–1.20) cm3 (Fig. 6a-b). In comparison, the TVA/C predicted with the actual treatment was 0.43 (0.00–1.66) cm3, and the TVT predicted with the actual treatment was 1.29 (0.01–2.72) cm3. Therefore, optimizing the A/C schedule caused a decrease in the TVA/C, though it was not significant (P = 0.28). However, optimizing the T schedule led to a significant decrease in the TVT (P < 0.01). Similarly, the simultaneous optimization resulted in a TVT of 0.26 (0.00–1.20) cm3 (Fig. 6c), which was a significant decrease (P < 0.01) from the TVT predicted with the actual treatment. The midway optimization resulted in a TVT of 0.45 (0.00–1.61) cm3 (Fig. 6d), which was also a significant decrease (P < 0.01) from the TVT predicted with the actual treatment.
a–d shows the improvement in outcomes for the whole cohort (n = 105) with the first step of multi-step optimization, second step of multi-step optimization, simultaneous optimization, and midway optimization, respectively. In each panel, the light blue and light red violin plots depict the distribution of predicted outcomes with the actual schedule and the optimized schedule, respectively. The boxplots show the median (red line) and IQR (blue box) of the corresponding outcomes. All three optimization strategies lead to a significant reduction of TVT when compared to the treatment schedule the patient actually received.
Both the multi-step and simultaneous optimization strategies predicted an improved pCR rate of 85.71%, representing a significant absolute improvement of 24.76% over that actually observed in the cohort (P < 0.001 by the χ2 test). Using the midway optimization strategy, the pCR rate was predicted to improve to 81.90%, representing a significant absolute improvement of 20.95% over that actually observed in the cohort (P < 0.001 by the χ2 test) (Table 2). No significant difference was observed between pCR rates predicted by all three optimization strategies (P > 0.4).
Validating digital twin-based predictions via historical clinical trial data
To further validate our optimization analysis, we conducted a retrospective study to determine if the digital twin-based methodology can recapitulate the findings observed in three landmark clinical trials that compared A/C and T administrative schedules.
INT C974131 determined that dose-dense A/C-T significantly improved clinical outcomes (determined by disease-free and overall survival; DFS and OS) compared to the conventional tri-weekly A/C-T. To reproduce this study with our digital twin system, we simulated the response of the individual patients in our cohort to the two regimens. Our simulation showed that the digital twin-based prediction yielded a pCR rate of 49.52% and 73.33% for the conventional and dose-dense regimens of A/C-T, respectively (Table 3). The χ2 test demonstrated a significant difference between these pCR rates (P < 0.001), consistent with the efficacy of the trial itself which found that the dose-dense regimen resulted in a significantly better outcome.
Sparano et al.32,33 (ECOG 1199) as well as more recent meta-analyses by Khan et al.34, established that weekly and bi-weekly Taxol provided similar outcomes (DFS and OS) superior to that provided by tri-weekly Taxol, especially for TNBC33. Using the digital twin system, we simulated individual patients’ responses to the weekly, bi-weekly, and tri-weekly T regimens in combination with a conventional regimen of A/C. Our digital-twin-based predictions yielded pCR rates of 49.52%, 55.24%, and 60.00%, respectively, for the tri-weekly, weekly, and bi-weekly T regimens (Table 3). The weekly and bi-weekly T regimens led to higher pCR rates than the tri-week regimen, as the trial observed. However, the χ2 test demonstrated no significant differences between the three regimens (P > 0.1).
Finally, SWOG S022 investigated four different A/C and T regimens with a 2 × 2 factorial design35. The trial observed that the four regimens resulted in similar disease-free survival over the population, while also suggesting a non-significant improvement of DFS and OS in the TNBC patients with bi-weekly A/C and T. Using the digital twin system, we simulated the response of the individual patients in our cohort to the four regimens investigated by SWOG S022. Our digital-twin-based predictions yielded pCR rates of 79.05%, 72.38%, 73.33%, and 69.52%, respectively, for the four regimens (Table 3). The χ2 test demonstrated no significant pair-wise differences (P > 0.1) between the arms, though Arm 1 had the highest pCR rate. Both results were consistent with the trial outcome.
Overall, our digital twin-based predictions on response to alternative treatment regimens had a high agreement with the observations in previous clinical trials.
Discussion
Existing methods of predicting response are insufficient for patient-tailored treatment. Numerous efforts have been devoted to correlating imaging36,37,38 and/or molecular biomarkers39,40 to the response of breast cancer to neoadjuvant therapy. More recently, machine learning techniques have been used to extract features from imaging or multi-modal data to build prognostic models for predicting response41,42. However, these population-based approaches rely on statistical inference from properties of large populations that can obscure conditions specific to the individual patient over time43. In contrast, biology-based mathematical models that explicitly account for tumor behavior with mechanism-related parameters (e.g., cell mobility, invasiveness, proliferation, and treatment-induced response) are interpretable and have the ability to be personally calibrated to an individual patient’s data, thus providing unique opportunities to guide personalized tailoring of treatment12.
To address the unmet needs for personalized NAC for TNBC, we established patient-specific digital twins by calibrating a biology-based mathematical model that explicitly accounts for cell mobility, invasiveness, proliferation, and treatment-induced response from the patient’s longitudinal MRI data. Once calibrated, the digital twin was able to predict the patient’s response to a range of therapeutic schedules, thereby providing the opportunity to identify optimal therapeutic regimens on a patient-specific basis. Through this process, we were able to identify scheduling strategies that we hypothesize would lead to absolute improvements in the pCR rate from the 60.95% observed in the 105-patient TNBC cohort to 81.90–85.71% predicted by the digital twin-based optimization. Importantly, it was observed that changing NAC regimens affected different patients differently. In particular, out of the 41 non-pCR patients in the study cohort, 26 patients were identified as being able to achieve a pCR through treatment optimization (e.g., Fig. 4g). Conversely, the effect of altering the NAC regimen was small and did not lead to a pCR outcome in the remaining 15 patients (e.g., Fig. 4i).
Three optimization strategies were developed to account for three different clinical scenarios. Multi-step optimization aims to optimize both components of NAC and allows for longitudinal updating of a treatment plan. Since this approach optimizes the NAC components sequentially, the schedule identified as optimal for each component also provides a practical way to systematically investigate treatment escalation or de-escalation for each NAC component. For the A/C component of our study, the multi-step optimization identified 47 of the 105 patients who had an opportunity to achieve a better outcome through escalation, while 37 patients had a possibility to preserve pCR with a de-escalation. However, multi-step optimization tends to favor early and aggressive options; In this regard, the simultaneous optimization has an advantage as it aims to make a one-time decision for the NAC regimen. Simultaneous optimization also allows for comprehensive interpretation on the combined effects of (and interactions between) different components of NAC. This is especially true when pCR can be achieved by multiple combinations of A/C and T schedules thereby providing clinicians and patients with scheduling flexibility.
Both the multi-step and simultaneous optimizations begin prior to initiating NAC. Our current protocol requires at least one follow-up imaging session for model calibration; in practice, multiple imaging sessions are not usually performed during-treatment. To address this limitation, we developed the midway optimization strategy. Instead of beginning prior to initiating therapy, midway optimization allows the first component of NAC to be delivered according to the standard of care and then optimizes the remaining component(s) given the outcome of that first component. Midway optimization is more practical than the other two, though it may provide a less optimal outcome. Fortunately, in our study cohort, no significant difference in the improved pCR rates was observed between the three optimization strategies, which demonstrated that the midway optimization is an acceptable complementary alternative to the multi-step and simultaneous optimizations. One promising way to make the multi-step and simultaneous optimizations more clinically practical is to link the biology-based mathematical model with a deep learning network. This would allow for estimating model parameters to predict the spatio-temporal development of the tumor using only the pre-treatment imaging data; we have presented a preliminary study on this approach44.
The accuracy of the digital twin-based prediction was determined by comparing the predicted post-NAC response to the final pathological report, which yielded an AUC of 0.82 for differentiating pCR versus non-pCR, and a predicted pCR rate in the cohort that had no significant difference from the measured rate. Moreover, the reliability of the digital twin-based predictions of response to alternative therapeutic regimens was validated by virtually testing the effectiveness of various therapeutic regimens that had been investigated in previous clinical trials: INT C974131, ECOG 119932,33, and SWOG S022135. Based on the predicted pCR rates, we successfully observed that the dose-dense regimen of A/C-T leads to a significantly higher rate of pCR than the conventional regimen (INT C9741). We further observed that the weekly and bi-weekly Taxol schedules led to statistically identical pCR rates, and both were (not-significantly) higher than the pCR rate from the tri-weekly Taxol schedule (ECOG 1199). For the SWOG S0221 study, we observed that the four regimens based on combinations of weekly or bi-weekly A/C with weekly or bi-weekly Taxol led to statistically identical pCR rates, and that bi-weekly A/C + Taxol provided the highest pCR rate. All the observations from our digital-twin-based simulations matched the outcomes obtained by the actual trials.
We do, though, acknowledge that there are limitations in how our retrospective validation was executed. In particular, these three historical trials are not exactly the same as our study design; for example, they were designed for adjuvant therapy, used survival rates as the evaluation endpoints, and involved multiple breast cancer subtypes. Also, these trials did not collect longitudinal imaging data, so we used our own dataset to qualitatively reproduce the trial observations. A better cohort for the retrospective study would compare different neoadjuvant chemotherapy regimens for breast cancer, preferably early-stage TNBC, with longitudinal MRI collected before and during treatment for a range of treatment schedules, and with the data publicly available. Unfortunately, to the best of our knowledge, a cohort with these characteristics (that is publicly available) does not currently exist. However, the three historical clinical trials we used in our study were focused on comparing breast cancer chemotherapy regimens, and have directly led to the current clinical guidelines for the treatment of breast cancer in the neoadjuvant setting45,46, which are the directly related to our validation needs. Thus, we consider the agreement between our predictions and these trial observations an important retrospective validation of our digital twin methodology. Additionally, as the primary aim of this proof-of-principle study is to demonstrate the rationale and lay the technical foundations of our digital twin framework, the current retrospective results sufficiently serve this purpose.
The lack of trials comparing different neoadjuvant dosing schedules speaks directly to the need of our study: developing a practical methodology to support design of prospective trials that address the unmet need of patient-specific treatment tailoring. As essential next steps, practical—and low-risk—prospective studies to validate and eventually deploy digital twin frameworks can be designed on a cohort of early-stage TNBC patients who receive 1) longitudinal MRI, and 2) different NAC dosing schedules within the SOC options. We now describe two ways on how this next step could be achieved.
First, our digital twin can be included in “silent mode” for an observational trial in which the longitudinal MRI are collected to build the digital twins, but the model prediction and optimization results are not released to affect clinical decisions. Similar to the current study, the digital twins will be used to predict the individual patients’ response to the actual administered treatment and all possible SOC dosing schedules. Based on the prediction, the patients can be categorized into two groups:
-
1.
Escalation: if a patient is predicted to have a non-pCR response with the actual schedule, but predicted to have a pCR with an alternative SOC schedule.
-
2.
Non-escalation: If a patient is predicted to achieve a pCR with the actual schedule, or predicted to have a non-pCR with either the actual or alternative schedules.
Assuming the digital twins accurately predict the patients’ response to all NAC dosing schedules, patients in the Escalation group would have received less aggressive schedules than needed for achieving pCR, while patients in the Non-escalation group would have received the most aggressive schedules. Thus, we hypothesize that the actual pCR rate in the Escalation group will be significantly lower than that in Non-escalation group (P < 0.05 with the χ2 test). This presents a readily testable hypothesis within our digital twin framework.
In the current cohort of 105 TNBC patients, based on the multi-step and simultaneous optimizations, 26 patients were categorized into the Escalation group, while the remaining 79 patients were categorized into the Non-escalation group. The pCR rates were 30.77% and 68.35%, respectively, for the two groups (P < 0.01 with the χ2 test). This result supports the rationale of using digital twins to identify patients who will benefit from treatment escalation. Based on this internal evidence, our immediate next step is to perform prospective observational trials. Such a design has very low risk to the patients since the actual treatment decisions are made by the oncologists and not affected by the digital twins.
Second, after sufficient validation via observational studies, our digital twin can be further validated through a randomized adaptive trial. In the control arm, the treating oncologists would decide the dosing schedule without using digital twins. In the experimental arm, the oncologists will provide an initial plan for the patient’s NAC dosing schedule, and the digital twins will predict response to the initial plan as well as all other SOC dosing schedules. Patients in the experimental arm will be categorized into two groups based on the digital twin prediction:
-
1.
Escalation: if a patient is predicted to have a non-pCR response with the initial plan, but pCR with an alternative SOC schedule identified by the digital twin.
-
2.
Non-escalation: If a patient is predicted to have a pCR with the initial plan, or predicted to have non-pCR with either the initial plan or alternative schedules.
In the experimental arm, patients categorized into the Escalation group can be offered the digital-twin optimized dosing schedule, while patients categorized into the Non-escalation group will receive the initial plan. With patients randomized into the control and experimental arms, we hypothesize that the final pCR rate in the experimental arm will be significantly higher than the control arm (P < 0.05 with the χ2 test). This adaptive design allows for using novel statistical metrics like the “conditional average treatment effect” to analyze individualized treatment response47,48 while minimizing patient risk as no therapy reduction occurs based on the digital twin. Treatment de-escalation, though clinically important, is considered higher risk and would be investigated in later prospective studies.
Our digital twin-based treatment optimization schemes are practical, accurate, retrospectively validated, and built on key underlying characteristics of cancer; however, there are opportunities for improvement. The biology-based model used in our current digital twin pipeline only captures treatment efficacy, but no toxicity. Although the candidate NAC schedules we investigated are within a clinically practical range, and we empirically considered schedules with a shorter duration could lead to a higher chance of adverse toxicity under the same total dose, explicitly accounting for toxicity remains an open problem for model development. One way to address this limitation is to incorporate pharmacokinetic-pharmacodynamic modeling49,50. Additionally, the current mathematical model needs to be extended to account for the development of novel targeted- and immuno-therapies. Still, the chemo-backbone remains the most effective component of neoadjuvant therapy for early-stage TNBC including the latest update to that combines chemotherapy with Pembrolizumab8. Thus, the investigation of alternative, but clinically feasible, A/C + T schedules is of considerable clinical relevance.
Generalizability is an important consideration in evaluating clinical decision-making tools, and external validation is essential for reliable translation. We have shown the generalizability of our digital twin model’s ability to predict pCR on an external dataset from the multi-institutional trial, I-SPY251. In this case, our digital twins achieved an AUC of 0.78 for differentiating pCR from non-pCR in a cohort of 91 patients with various breast cancer subtypes52. Future validation through prospective trials (as described above) will also help to determine the generalizability of the proposed digital twin framework. Importantly, our digital twin is built on a biology-based, mechanistic model calibrated to individual patient imaging data, where the predictive process for each patient is independent of the results for the other patients. This is a fundamentally different from the “training-validating-testing” procedure in population-based approaches (like a statistical or machine learning model) and therefore provides the advantage of robust generalizability.
Another opportunity for future study is to develop robust methods of uncertainty quantification for establishing trust in the digital twin for clinical applications53. For example, due to the lack of imaging data acquired during the T regimen, we assigned model parameters describing the T effect based on parameters calibrated for A/C efficacy and decay (see the Methods section “Establishment of patient-specific digital twin to predict TNBC response to NAC” for more information). Even though the AUC achieved in this study indicated this assignment is reasonable to capture the patients’ response dynamics, it results in a source of uncertainty in predicted outcomes and such an uncertainty is not quantifiable with current approach. This limitation can be addressed by collecting additional MRI data early in the course of T. Furthermore, the digital twin implementation can be upgraded from the current deterministic parameter calibration to Bayesian calibration54. This would allow for directly providing quantification of uncertainty in personalized parameters and predicted outcomes, as well as assessment of sources of prediction bias. Quantifying the uncertainty of the predictions is central to providing both transparency and understanding when translating digital twins to guide clinical decision-making.
Beyond MRI-guided digital twins, future investigation may also involve integrating multi-modal data55 into the modeling56. Recently emerging efforts on multi-modal data fusion have shown strong potential on improving cancer diagnosis57, prognosis58,59,60,61, and response prediction62,63,64. However, these pioneering studies are based on population-based approaches (especially artificial intelligence techniques), which commonly face the question of interpretability and generalizability. Integrating multi-modal data through biology-based digital twins promises a more reliable approach in promoting precision medicine. Consideration of multi-modal data also provides the opportunity to address other clinically important questions which are not explicitly considered in our current model. Examples include the status of the axillary lymph nodes (by including ultrasound data) and metastasis (by including liquid biopsy data).
In conclusion, we established digital twin-based optimization schemes to identify NAC schedules that led to the optimal post-NAC outcome on a patient-specific basis. The prediction reliability was retrospectively validated and three optimization strategies providing similar optimization benefits ensured robust performance under different application scenarios. The development approaches provide a practical framework for the design of adaptive clinical trials and translating digital twin techniques for personalized optimization of NAC treatment plans in TNBC, which are also applicable to other types of cancer and therapies.
Methods
Patient population
Treatment-naïve stage I-III TNBC patients were enrolled in the prospective clinical trial, “ARTEMIS: A Robust TNBC Evaluation FraMework to Improve Survival” (ClinicalTrials.gov Identifier: NCT02276443, registered on October 21, 2014)65. The Institutional Review Board (IRB) of The University of Texas MD Anderson Cancer Center reviewed and approved the ARTEMIS study protocol (2014-0185). All patients were provided informed consent. All study procedures performed were in accordance with ethical standards of the IRB and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. All patients received four cycles of Adriamycin/Cytoxan (A/C) for the first component of NAC; each cycle was given once every two weeks (dose-dense) to three weeks. Patients who were chemo-insensitive (i.e., with disease progression, or < 70% reduction in tumor volume at the end of A/C) were offered the opportunity to enroll in a biomarker-guided clinical trial using targeted therapy. Patients who were chemo-sensitive (i.e., with ≥ 70% reduction in tumor volume at the end of A/C) were recommended to continue standard-of-care paclitaxel (Taxol; T) weekly for 12 cycles, or every 3 weeks for four cycles. (The exact T regimen for each patient was determined by their treating physician.) A total of 105 patients from the ARTEMIS trial were included in this study based on the following inclusion criteria: 1) signed informed consent between April 2018 to May 2021, 2) biopsy-confirmed stage I–III diseases, 3) completed NAC with T, 4) availability of the entire drug administration schedule, and 5) access to longitudinal multiparametric MRI scans without acquisition error or artifacts.
Imaging data and processing
All patients underwent longitudinal multiparametric MRI scans before treatment (MRI1), after two cycles of A/C (MRI2), and after four cycles of A/C (MRI3). The images collected at each scan included diffusion-weighted (DW-) MRI at two b-values (100, 800 s/mm2), and a median (range) of 42 (36-50) frames of dynamic contrast enhanced (DCE-) MRI with a temporal resolution of 11.1 (7.6–12.4) seconds. We have previously established a highly automated pipeline to process the longitudinal MRI data from each patient22,30 to yield tumor-tissue masks, pharmacokinetic maps from DCE-MRI, and tumor cellularity maps from DW-MRI, and image alignment across multiple visits. Details of image acquisition and processing can be found in prior work22.
Establishment of patient-specific digital twin to predict TNBC response to NAC
We have developed a biology-based mathematical model to represent the dynamics of tumor growth and response to NAC22. In particular, the model, which is governed by a reaction-diffusion partial differential equation, describes the spatiotemporal change in tumor cells, N(x,t), due to cell migration (1st term on the right-hand side of Eq. (1)), proliferation (2nd term), and treatment-induced death (3rd term):
with
where D(x,t) is tumor cell mobility determined by a diffusion coefficient, D0, with mechanical coupling via the von Mises stress, σ(x,t)66. k(x) is the cell proliferation rate at position x, and θ is the carrying capacity that defines that maximum number of cells that can occupy a voxel. αi and βi represent the efficacy (per unit dose) and decay rates of drug i, respectively, and τi,j indicates the time of the j-th administration of drug i at a total of Ji times. C(x) is the estimate of the spatial distribution of the drugs, which is derived from the DCE-MRI data22. A detailed list of the variables, parameters, and their definitions is given in Table 4.
For each patient, the geometry of the computational domain was determined by the segmented breast contour, tumor, fibroglandular, and adipose tissues. The tumor cellularity maps, {N(x, t = MRI1), N(x, t = MRI2), N(x, t = MRI3)} were derived from the apparent diffusion coefficient maps at each of the three imaging time points. These tumor cellularity maps, along with the therapeutic information, were used for model initialization and personalizing the model parameters in Eqs. (1) – (3). More specifically, k(x), D0, α1, α2, β1, and β2 are calibrated on a patient-specific basis. As no MRI data was available during the paclitaxel regimen, the efficacy of paclitaxel was assigned to a literature value67, α3 = 0.3 day-1; the decay rate of paclitaxel was assigned as the average of the calibrated A and C decay rates, β3 = (β1 + β2)/2. (The details of our numerical implementation has been previously described22).
Once the model was calibrated on a given patient’s specific data, it was referred as the digital twin as it can provide a prediction of how an individual’s tumor will respond to not only the actual NAC schedule, but also an array of alternative therapeutic regimens (Fig. 2a).
Given the actual treatment schedule, an individual patient’s digital twin predicted the residual tumor volume (TV) after completing NAC. We evaluated the accuracy of the digital twins by performing receiver operating characteristic analysis (ROC) on the predicted residual TV after NAC (TVT) to differentiate pCR and non-pCR for each patient in the cohort (Fig. 2b). From the ROC curve, the optimal cutoff point was determined by maximizing Youden’s J statistic68 (i.e., sensitivity + specificity – 1), from which the corresponding threshold for pCR versus non-pCR was determined and termed TVT,J = 1.90 cm3. That is, if we predicted a patient to have a TVT < TVT,J, then we were predicting that patient would have a pCR outcome after completing NAC. Given alternative treatment schedules (Fig. 2c), the individual patient’s digital twin predicted the tumor response dynamics as well as the TVT for each schedule. The predicted response to alternative therapeutic regimens was compared to the patient’s response to the treatment they actually received, thereby providing an opportunity to identify therapeutic regimens that could outperform the standard-of-care (Fig. 2d).
Selection of alternative therapeutic regimens of NAC
We evaluated the effect of altering the NAC regimens (i.e., the A/C-T schedules) on patient response. We note that the alternative therapeutic schedules were selected from other clinically feasible options. As the standard-of-care A/C therapy is generally administrated every 2-3 weeks for four cycles, we tested eight different therapeutic schedules of A/C with a duration between 56 and 84 days (Table 1). Similarly, the standard-of-care T therapy is generally administered weekly for 12 cycles, or every 2-3 weeks for four cycles. Thus, we tested 16 different therapeutic schedules of T consisting of eight options for 12 cycles with a duration between 56 and 84 days, and eight options for four cycles with a duration between 56 and 84 days (Table 1). In total, 128 clinically feasible schedules of NAC were investigated for each patient. To minimize the effect of altering the NAC regimens on toxicity, we kept the total dose constant for each schedule.
Digital twin-based therapeutic schedule optimization strategies
We considered three optimization strategies (useful in three different clinical settings) to identify alternative therapeutic regimens, they consisted of 1) multi-step optimization, 2) simultaneous optimization, and 3) midway optimization. Multi-step optimization seeks to optimize the A/C component of the NAC regimen and then, from the optimal outcome of A/C, optimize the second T component. Simultaneous optimization attempts to optimize both A/C and T components at the same time. Midway optimization begins from the outcome of the actually administered A/C regimen and only optimizes the second regimen (i.e., T). Multi-step optimization corresponds to updating the treatment plan each time follow-up data becomes available, simultaneous optimization identifies a single pre-treatment plan, and midway optimization includes at least one image acquired after the initiation of therapy for digital twin calibration.
For the multi-step optimization (Fig. 3a), each patient’s digital twin was first used to predict the individual’s response to all eight candidate A/C schedules. The minimal TVA/C predicted by the digital twin from all investigated A/C schedules was considered the optimal outcome of A/C. The corresponding A/C schedule(s) resulting in this minimal TVA/C was(were) identified as the optimal A/C schedule(s). Note that if multiple schedules ended up with the same minimal TVA/C, multiple optimal A/C schedules are allowed (for example, the Patient 2 in Fig. 4b). Then, based on the optimal outcome of A/C, we used the patient’s digital twin to predict response to all 16 alternative T schedules. The minimum predicted TVT from all investigated T schedules was considered the optimal outcome of T; note that this procedure also produces the optimal outcome of the whole NAC procedure. The corresponding T schedule(s) resulting in this minimal TVT was (were) identified as the optimal T schedules. Moreover, the optimal pathological status was determined by evaluating if the predicted minimal TVT was below the differentiating threshold TVT,J (defined in Methods section “Establishment of patient-specific digital twin to predict TNBC response to NAC”), as illustrated in Fig. 2d.
For the simultaneous optimization (Fig. 3b), each patient’s digital twin was used to predict their response to all 128 combinations of alternative A/C and T schedules. The minimal TVT predicted from all combinations of A/C-T schedules was considered the optimal outcome of the whole NAC procedure. The corresponding A/C-T schedule(s) resulting in this minimal TVT was (were) identified as the optimal NAC schedule(s). Similarly, the optimal pathological status was determined by evaluating if the predicted minimal TVT was below TVT,J.
For the midway optimization (Fig. 3c), we begin with the predicted TVA/C from the actual A/C schedule and then used each patient’s digital twin to predict their response to all 16 candidate T schedules. Similar to the previous to optimization schemes, the minimal predicted TVT was considered the optimal outcome of the entire NAC procedure. The corresponding T schedule(s) resulting in this minimal TVT was(were) identified as the optimal NAC schedule(s). The optimal pathological status was determined by evaluating if the predicted minimal TVT was below TVT,J.
For each patient, we evaluated the effects of changing the therapeutic schedules (via each optimization strategy) on their outcome, and compared to the outcome from the therapeutic schedule the patient actually received. Additionally, we compared the optimal schedules and associated final outcomes (i.e., residual TV and pCR status after NAC) from each of the three optimization strategies to determine if they all produced statistically equivalent benefits.
Retrospective validation on digital twin-predicted response to alternative therapeutic regimens
We identified three seminal trials that compared A/C and T administrative schedules for our retrospective validation (Table 3). First, Citron et al.31 reported a randomized trial (INT C9741) investigating the effect of different A/C and T schedules on the response of axillary node-positive breast cancer response to adjuvant chemotherapy. The trial determined that the dose-dense regimen significantly improves clinical outcomes (defined by disease-free and overall survival; DFS and OS) compared to the conventional tri-weekly regimen. The conclusion that dose-dense regimens have superior performance was subsequently reported in multiple independent studies69. For retrospective validation, we used our patient-specific digital twin formalism to simulate individual patient response to the two regimens (Table 3) that were employed in the INT C9741 trial. We then derived the pCR rate for each regimen by evaluating if the predicted TVT was below TVT,J, and tested the following hypothesis, H1: the dose-dense regimen will lead to a significant higher rate of pCR than the conventional regimen in the cohort (n = 105).
The second trial (ECOG 1199) was carried out by Sparano et al.32,33 and investigated the efficacy of two different Taxol-based regimens in the adjuvant setting for breast cancer. The trial observed that weekly paclitaxel (Taxol; T) after the standard A/C improves DFS and OS in women32, and provided long-term benefits for TNBC33. A more recent meta-analysis reported the superiority of both the weekly and bi-weekly dose-dense T regimens over the conventional tri-weekly regimen, while also noting similar outcomes between the weekly and dose-dense T regimens34. For retrospective validation, we used our patient-specific digital twin formalism to simulate individual patient response to the three T regimens investigated in ECOG 1199 and the associated meta-analyses (Table 3). We then derived the pCR rate from each regimen by evaluating if the predicted TVT was below TVT,J, and tested the following hypothesis, H2: 1) the weekly and bi-weekly T regimens lead to significantly higher pCR rates than the tri-weekly T regimen, and 2) the weekly and bi-weekly T regimens have no significant difference in resulted pCR rates.
The third trial (SWOG S0221) we considered was performed by Budd et al.35, which sought to determine the optimal dose and schedule of A/C and T administration as the adjuvant therapy for early-stage breast cancer with a 2 × 2 factorial design. The trial observed that the four regimens resulted in statistically similar DFS, while in the subset of TNBC patients, a non-significant improvement of DFS and OS was suggested with bi-weekly dosing of both A/C and T. For retrospective validation, we used our patient-specific digital twin formalism to simulate individual patient responses to the four regimens in SWOG S0221 (Table 3). We then derived the pCR rate from each regimen by evaluating if the predicted TVT was below TVT,J, and tested the following hypothesis, H3: 1) No significant difference in pCR rates will be observed across Arm 1-4 regimens, and 2) Arm 1 regimen leads to the highest pCR rate in the cohort.
Statistical analysis
The confidence intervals of the AUC from the ROC analysis, and of pCR rates, were calculated with the Bootstrapping method with a resampling size of 1000. We statistically evaluated the effects that optimizing the therapeutic schedules had on the patient outcomes. For the multi-step optimization strategy, we first used the Wilcoxon test to compare the TVA/C predicted from the optimal schedule to the TVA/C predicted from the actual treatment. Second, we used the Wilcoxon test to compare the TVT predicted from the optimal schedule to the TVT predicted from the actual treatment. Third, we used the χ2 test to compare the predicted pCR rate from the optimal schedule to the predicted pCR rate from the actual treatment, as well as to the measured pCR rate. Similarly, for the simultaneous optimization and midway optimization strategies, we first used the Wilcoxon test to compare the predicted optimal TVT to the predicted TVT resulting from the actual treatment. Second, we used the χ2 test to compare the predicted pCR rate from the optimal schedule to both the measured and predicted pCR rate from the actual treatment. In all cases, statistical significance was defined as P-value < 0.05.
For the retrospective validation using the therapeutic regimens found in INT C9741, ECOG 1199 + , and SWOG S0221, we used the χ2 test to compare the pCR rates predicted from our digital twin formalism to the pCR rates found in the actual trials. Again, a P-value < 0.05 was considered significant.
Data availability
Raw data for this study were generated at The University of Texas MD Anderson Cancer Center. Raw data are not publicly available due to IRB restrictions of data containing information that could compromise research participant privacy and/or consent. The derived data that support the findings of this study are available from the corresponding author upon reasonable request.
Code availability
The underlying code for this study is not publicly available for proprietary reasons but may be made available to qualified researchers on reasonable request from the corresponding author.
References
Liedtke, C. et al. Response to neoadjuvant therapy and long-term survival in patients with triple-negative breast cancer. J. Clin. Oncol. 26, 1275–1281 (2008).
Liu, S. V., Melstrom, L., Yao, K., Russell, C. A. & Sener, S. F. Neoadjuvant therapy for breast cancer. J. Surg. Oncol. 101, 283–291 (2010).
Wu, K., Yang, Q., Liu, Y., Wu, A. & Yang, Z. Meta-analysis on the association between pathologic complete response and triple-negative breast cancer after neoadjuvant chemotherapy. World J. Surg. Oncol. 12, 95 (2014).
Spring, L. M. et al. Pathologic complete response after neoadjuvant chemotherapy and impact on breast cancer recurrence and survival: a comprehensive meta-analysis. Clin. Cancer Res. 26, 2838–2848 (2020).
Nabholtz, J. M. et al. Multicentric neoadjuvant pilot phase II study of cetuximab combined with docetaxel in operable triple negative breast cancer: phase II Study of Cetuximab Combined with Docetaxel for TNBC. Int. J. Cancer 138, 2274–2280 (2016).
Silver, D. P. et al. Efficacy of neoadjuvant cisplatin in triple-negative breast cancer. J. Clin. Oncol. 28, 1145–1153 (2010).
Schmid, P. et al. Pembrolizumab plus chemotherapy as neoadjuvant treatment of high-risk, early-stage triple-negative breast cancer: results from the phase 1b open-label, multicohort KEYNOTE-173 study. Ann. Oncol. 31, 569–581 (2020).
Shah, M. et al. FDA approval summary: pembrolizumab for neoadjuvant and adjuvant treatment of patients with high-risk early-stage triple-negative breast cancer. Clin. Cancer Res. 28, 5249–5253 (2022).
Schmid, P. et al. Event-free survival with pembrolizumab in early triple-negative breast cancer. N. Engl. J. Med. 386, 556–567 (2022).
Schmid, P. et al. Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020).
McAndrew, N. & DeMichele, A. Neoadjuvant chemotherapy considerations in triple-negative breast cancer. J. Target. Ther. Cancer 7, 52–69 (2018).
Yankeelov, T. E. et al. Designing clinical trials for patients who are not average. iScience 27, 108589 (2024).
Hormuth, D. A. et al. Math, magnets, and medicine: enabling personalized oncology. Expert Rev. Precis. Med. Drug Dev. 6, 79–81 (2021).
Loeser, A. et al. The right dose: results of a patient advocate–led survey of individuals with metastatic breast cancer regarding treatment-related side effects and views about dosage assessment to optimize quality of life. JCO Oncol. Pract. 20, 972–983 (2024).
FDA. Optimizing the Dosage of Human Prescription Drugs and Biological Products for the Treatment of Oncologic Diseases: Guidance for Industry. https://www.fda.gov/media/164555/download (2024).
San Antonio Breast Cancer Symposium. Clinical Workshop to review dosage, regimen optimization in breast medical oncology. (2024).
FDA-ASCO. Second Annual FDA ASCO WORKSHOP on Getting the Dosage Right. (2023).
AACR. FDA-AACR Public Workshop on Optimizing Dosages for Oncology Drug Products: Quantitative Approaches to Select Dosages for Clinical Trials. (2024).
Baldock, A. L. et al. From patient-specific mathematical neuro-oncology to precision medicine. Front. Oncol. 3, 62 (2013).
Brady-Nicholls, R. et al. Prostate-specific antigen dynamics predict individual responses to intermittent androgen deprivation. Nat. Commun. 11, 1750 (2020).
Weis, J. A. et al. A mechanically coupled reaction–diffusion model for predicting the response of breast tumors to neoadjuvant chemotherapy. Phys. Med. Biol. 58, 5851–5866 (2013).
Wu, C. et al. MRI-based digital models forecast patient-specific treatment responses to neoadjuvant chemotherapy in triple-negative breast cancer. Cancer Res. 82, 3394–3404 (2022).
Jarrett, A. M. et al. Evaluating patient-specific neoadjuvant regimens for breast cancer via a mathematical model constrained by quantitative magnetic resonance imaging data. Neoplasia 22, 820–830 (2020).
Hormuth, D. A., Al Feghali, K. A., Elliott, A. M., Yankeelov, T. E. & Chung, C. Image-based personalization of computational models for predicting response of high-grade glioma to chemoradiation. Sci. Rep. 11, 8520 (2021).
Woodall, R. T. et al. Patient specific, imaging-informed modeling of rhenium-186 nanoliposome delivery via convection-enhanced delivery in glioblastoma multiforme. Biomed. Phys. Eng. Express 7, 045012 (2021).
Whitmire, P., et al. Sex-specific impact of patterns of imageable tumor growth on survival of primary glioblastoma patients. BMC Cancer 20, 447 (2020).
Scheufele, K., Subramanian, S. & Biros, G. Fully automatic calibration of tumor-growth models using a single mpMRI scan. IEEE Trans. Med. Imaging 40, 193–204 (2021).
Le, M. et al. MRI based Bayesian personalization of a tumor growth model. IEEE Trans. Med. Imaging 35, 2329–2339 (2016).
Wu, C. et al. Integrating mechanism-based modeling with biomedical imaging to build practical digital twins for clinical oncology. Biophys. Rev. 3, 021304 (2022).
Jarrett, A. M. et al. Quantitative magnetic resonance imaging and tumor forecasting of breast cancer patients in the community setting. Nat. Protoc. 16, 5309–5338 (2021).
Citron, M. L. et al. Randomized trial of dose-dense versus conventionally scheduled and sequential versus concurrent combination chemotherapy as postoperative adjuvant treatment of node-positive primary breast cancer: first report of Intergroup Trial C9741/Cancer and Leukemia Group B Trial 9741. J. Clin. Oncol. J. Am. Soc. Clin. Oncol. 21, 1431–1439 (2003).
Sparano, J. A. et al. Weekly paclitaxel in the adjuvant treatment of breast cancer. N. Engl. J. Med. 358, 1663–1671 (2008).
Sparano, J. A. et al. Long-term follow-up of the E1199 phase III trial evaluating the role of taxane and schedule in operable breast cancer. J. Clin. Oncol. 33, 2353–2360 (2015).
Khan, U., DeRosa, A., Xu, J. & Patel, T. A. Comparing efficacy and toxicity profile of weekly paclitaxel to dose-dense paclitaxel (every two weeks) in treatment of breast cancer: A meta-analysis and systematic review. J. Clin. Oncol. 38, e12634 (2020).
Budd, G. T. et al. SWOG S0221: a phase III trial comparing chemotherapy schedules in high-risk early-stage breast cancer. J. Clin. Oncol. 33, 58–64 (2015).
Marinovich, M. L. et al. Early prediction of pathologic response to neoadjuvant therapy in breast cancer: Systematic review of the accuracy of MRI. Breast 21, 669–677 (2012).
Khairalseed, M. et al. Monitoring early breast cancer response to neoadjuvant therapy using h-scan ultrasound imaging: preliminary preclinical results. J. Ultrasound Med. 38, 1259–1268 (2019).
Wang, Y., Zhang, C., Liu, J. & Huang, G. Is 18F-FDG PET accurate to predict neoadjuvant therapy response in breast cancer? A meta-analysis. Breast Cancer Res. Treat. 131, 357–369 (2012).
Weigelt, B., Baehner, F. L. & Reis-Filho, J. S. The contribution of gene expression profiling to breast cancer classification, prognostication and prediction: a retrospective of the last decade. J. Pathol. 220, 263–280 (2010).
Dressman, H. K. et al. Gene expression profiles of multiple breast cancer phenotypes and response to neoadjuvant chemotherapy. Clin. Cancer Res. 12, 819–826 (2006).
Sammut, S. -J. et al. Multi-omic machine learning predictor of breast cancer therapy response. Nature 601, 623–629 (2022).
Tahmassebi, A. et al. Impact of machine learning with multiparametric magnetic resonance imaging of the breast for early prediction of response to neoadjuvant chemotherapy and survival outcomes in breast cancer patients. Investig. Radiol. 54, 110–117 (2019).
Coveney, P. V., Dougherty, E. R. & Highfield, R. R. Big data need big theory too. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 374, 20160153 (2016).
Stowers, C. E. et al. Combining biology-based and MRI data-driven modeling to predict response to neoadjuvant chemotherapy in patients with triple-negative breast cancer. Radiol. Artificial Intell. 7, e240124 (2024).
Han, H. S. et al. Early-stage triple-negative breast cancer journey: beginning, end, and everything in between. Am. Soc. Clin. Oncol. Educ. Book e390464 10.1200/EDBK_390464 (2023).
Asaoka, M., Gandhi, S., Ishikawa, T. & Takabe, K. Neoadjuvant chemotherapy for breast cancer: past, present, and future. Breast Cancer Basic Clin. Res. 14, 117822342098037 (2020).
Imai, K. & Li, M. L. Experimental evaluation of individualized treatment rules. J. Am. Stat. Assoc. 118, 242–256 (2023).
Künzel, S. R., Sekhon, J. S., Bickel, P. J. & Yu, B. Metalearners for estimating heterogeneous treatment effects using machine learning. Proc. Natl Acad. Sci. 116, 4156–4165 (2019).
Wu, C. et al. Towards patient-specific optimization of neoadjuvant treatment protocols for breast cancer based on image-guided fluid dynamics. IEEE Trans. Biomed. Eng. 69, 3334–3344 (2022).
Helmlinger, G. et al. Quantitative systems pharmacology: an exemplar model-building workflow with applications in cardiovascular, metabolic, and oncology drug development. CPT Pharmacomet. Syst. Pharmacol. 8, 380–395 (2019).
Barker, A. et al. I-SPY 2: an adaptive breast cancer trial design in the setting of neoadjuvant chemotherapy. Clin. Pharmacol. Ther. 86, 97–100 (2009).
Patel RJS, Wu C, Stowers CE, Mohamed RM, Ma J, Rauch GM, Yankeelov TE. Predicting the response of locally advanced breast cancer to neoadjuvant therapy using MRI-based mathematical modeling of the I-SPY 2 dataset. in San Antonio Breast Cancer Symposium 2024 (2024).
Committee on Foundational Research Gaps and Future Directions for Digital Twins et al. Foundational Research Gaps and Future Directions for Digital Twins. 26894 (National Academies Press, 2024). https://doi.org/10.17226/26894.
Chaudhuri, A. et al. Predictive digital twin for optimizing patient-specific radiotherapy regimens under uncertainty in high-grade gliomas. Front. Artif. Intell. 6, 1222612 (2023).
Shaik, T., Tao, X., Li, L., Xie, H. & Velásquez, J. D. A survey of multimodal information fusion for smart healthcare: Mapping the journey from data to wisdom. Inf. Fusion 102, 102040 (2024).
Kazerouni, A. S. et al. Integrating quantitative assays with biologically based mathematical modeling for predictive oncology. iScience 23, 101807 (2020).
Cui, C. et al. Deep multimodal fusion of image and non-image data in disease diagnosis and prognosis: a review. Prog. Biomed. Eng. 5, 022001 (2023).
Shao, W. et al. Multi-task multi-modal learning for joint diagnosis and prognosis of human cancers. Med. Image Anal. 65, 101795 (2020).
Madabhushi, A., Agner, S., Basavanhally, A., Doyle, S. & Lee, G. Computer-aided prognosis: Predicting patient and disease outcome via quantitative fusion of multi-scale, multi-modal data. Comput. Med. Imaging Graph. 35, 506–514 (2011).
Yin, X. -X., Hadjiloucas, S., Zhang, Y. & Tian, Z. MRI radiogenomics for intelligent diagnosis of breast tumors and accurate prediction of neoadjuvant chemotherapy responses-a review. Comput. Methods Prog. Biomed. 214, 106510 (2022).
Braman, N. et al. Deep Orthogonal Fusion: Multimodal Prognostic Biomarker Discovery Integrating Radiology, Pathology, Genomic, and Clinical Data. in Medical Image Computing and Computer Assisted Intervention—MICCAI 2021 (eds. De Bruijne, M. et al.) vol. 12905 667–677 (Springer International Publishing, Cham, 2021).
Rathore, S., Chaddad, A., Iftikhar, M. A., Bilello, M. & Abdulkadir, A. Combining MRI and histologic imaging features for predicting overall survival in patients with glioma. Radiol. Imaging Cancer 3, e200108 (2021).
Shao, L. et al. Multiparametric MRI and whole slide image-based pretreatment prediction of pathological response to neoadjuvant chemoradiotherapy in rectal cancer: a multicenter radiopathomic study. Ann. Surg. Oncol. 27, 4296–4306 (2020).
Niecikowski, A. et al. A multi-modal deep learning-based decision support system for individualized radiotherapy of non-small cell lung cancer. Int. J. Radiat. Oncol. 114, e100–e101 (2022).
Yam, C. et al. Immune phenotype and response to neoadjuvant therapy in triple-negative breast cancer. Clin. Cancer Res. 27, 5365–5375 (2021).
Hormuth, D. A., Eldridge, S. L., Weis, J. A., Miga, M. I. & Yankeelov, T. E. Mechanically Coupled Reaction-Diffusion Model to Predict Glioma Growth: Methodological Details. in Cancer Systems Biology (ed. Von Stechow, L.) vol. 1711 225–241 (Springer New York, 2018).
Lorenzo, G. et al. Abstract PS13-44: Identifying relevant parameters that characterize the early response to NAT in breast cancer patients using a novel personalized mechanistic model integrating in vitro and in vivo imaging data. Cancer Res. 81, PS13-44–PS13-44 (2021).
Ruopp, M. D., Perkins, N. J., Whitcomb, B. W. & Schisterman, E. F. Youden index and optimal cut-point estimated from observations affected by a lower limit of detection. Biom. J. 50, 419–430 (2008).
Bonilla, L. et al. Dose-dense chemotherapy in nonmetastatic breast cancer: a systematic review and meta-analysis of randomized controlled trials. JNCI J. Natl. Cancer Inst. 102, 1845–1854 (2010).
Acknowledgements
We thank the National Institutes of Health for funding through NCI U01CA142565 (T.E.Y and G.S.K.), U01CA174706 (T.E.Y.), U24CA226110 (T.E.Y.). We thank the Cancer Prevention and Research Institute of Texas for support through CPRIT RR160005 (T.E.Y.). T.E.Y. is a CPRIT Scholar in Cancer Research. This material is based upon work supported by the National Science Foundation Graduate Research Fellowship under Grant No. DE2137420 (C.E.S.). We thank the Oncological Data and Computational Sciences collaboration between the Oden Institute for Computational Engineering and Sciences at The University of Texas at Austin, the MD Anderson Cancer Center, and the Texas Advanced Computing Center for providing seed funding on this project (C.W. and E.A.B.F.L). We thank the generous philanthropic contributions of the University of Texas MD Anderson Cancer Center Moon Shots Program™.
Author information
Authors and Affiliations
Contributions
C.W.: Conceptualization, resources, software, formal analysis, investigation, visualization, methodology, writing–original draft, writing–review and editing. E.A.B.F.L.: Conceptualization, resources, software, funding acquisition, investigation, methodology, writing–review and editing. C.E.S.: Conceptualization, resources, software, investigation, methodology, writing–review and editing. Z.X.: Resources, data curation, formal analysis, investigation, methodology, writing–review and editing. C.Y.: Data curation, investigation, writing–review and editing. J.B.S.: Data curation, investigation, writing–review and editing. J.M.: Resources, investigation, methodology, writing–review and editing. G.M.R.: Resources, funding acquisition, investigation, methodology, writing–review and editing. T.E.Y.: Conceptualization, supervision, funding acquisition, investigation, methodology, writing–original draft, writing–review and editing.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wu, C., Lima, E.A.B.F., Stowers, C.E. et al. MRI-based digital twins to improve treatment response of breast cancer by optimizing neoadjuvant chemotherapy regimens. npj Digit. Med. 8, 195 (2025). https://doi.org/10.1038/s41746-025-01579-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-01579-1
This article is cited by
-
AI-powered in silico twins: redefining precision medicine through simulation, personalization, and predictive healthcare
Saudi Pharmaceutical Journal (2026)
-
Predicting the response of triple negative breast cancer to neoadjuvant systemic therapy via biology-based modeling and habitat analysis
Scientific Reports (2025)
-
Digital twins for the personal touch
Nature Medicine (2025)
-
Computational Modeling and Digital Twin Technologies in Medical Device Development
Biomedical Materials & Devices (2025)