Abstract
Several open-source oral-maxillofacial imaging datasets have been created but their characteristics, ethical clearance, and licensing for reuse remain unclear. This study aimed to systematically identify these datasets and investigate their characteristics, ethical approvals, and licensing requirements for reuse. Open-source oral-maxillofacial imaging datasets were identified through electronic databases and dataset platforms. 105 datasets with 437538 images and 100 intraoral videos from patients across twenty-one countries were included. The datasets comprise imaging modalities, including photographs, periapical, panoramic, and cephalometric radiographs, CBCT, MRI, surface scans, videos, and histopathological images. Nearly 80% of them provide annotations, but only 25.7% specified the annotators’ qualification. The majority (83.8%) did not disclose whether ethical approval was obtained, while 61.9% specified terms or licenses for dataset reuse. There is an urgent need to develop standardized guidelines for reusing image datasets and to establish AI-specific consents to fully inform patients about potential uses of their data in AI projects.
Similar content being viewed by others
Introduction
Dentistry has progressed swiftly towards digitalization in the last two decades, largely attributed to its significant dependency on advanced imaging techniques with computer-aided design and manufacturing. These technologies play a crucial role in various stages of dental practice, such as diagnosis, treatment planning, guided surgery, post-surgical evaluation, prosthodontic workflows including CAD/CAM applications, and follow-up assessment, and have even expanded to facilitate remote consultations through tele-dentistry1. The image data, generated during daily practices and easily accessible from dental clinic database systems, forms the backbone of most artificial intelligence (AI) models proposed in the field of dentistry2,3,4. Currently, many innovative dental AI models have been developed using images to automatically perform complex tasks, such as multimodal image registration3, segmentation of anatomical structures and pathologies in the oral and maxillofacial region5,6, detection of various dental diseases7, generation of 3D dental models8,9, and interpretation of dental radiographs10. Such AI tools have the potential to push the progression of digitalization in oral healthcare.
While certain dental AI models have demonstrated performance on par with or exceeding dental professionals with internal images, most of these models have not been externally validated due to a lack of external image data. This deficiency has been reported in previous studies highlighting a significant performance drop in dental AI models when tested with external images, likely a result of the absence of diverse image data used during model training11. The lack of large datasets, comprising images with varying conditions, greatly limits the development and validation of robust and widely applicable dental AI models. One potential solution to enhance the robustness and generalizability of AI models is to integrate images from multiple sources into the training and validation stages12.
In recent years, a growing number of publicly accessible datasets, such as TED36, Ctooth13, IO150K14, have been introduced. A previous study has identified 16 publicly available dental imaging datasets and summarized their characteristics to facilitate the use of dental imaging data in AI research15. More recently, an increasing number of AI studies have been published along with open-access oral-maxillofacial imaging datasets. However, the sources and characteristics of these recent public datasets for oral-maxillofacial imaging including annotation details, have not yet been systematically investigated. Without a thorough understanding of these datasets prior to their use in AI model training and testing, there is an increased risk of unintended biases, such as data leakage. This can occur when training and test sets contain duplicate images from various repackaged datasets, potentially leading to overly optimistic performance estimates as the model could learn from identical data in both phases. Moreover, the significance of understanding ethical considerations, specific terms, and licensing requirements for reusing these datasets is becoming more widely recognized16,17,18. Using these datasets without clear understanding of ethical and licensing information may incur substantial ethical and legal risks. The issue of whether AI models trained on datasets that prohibit commercial use can be licensed for commercial purposes remains controversial. Currently, the ethical clearance and specific terms regulating their reuse in AI projects are unclear. Therefore, the primary objective of this systematic review, reported in accordance with the PRISMA guideline19, was to provide a comprehensive summary of openly accessible datasets containing images from the oral-maxillofacial region, including details such as the year and purpose of dataset creation, creators, country and institution of origin, imaging modality, image type and format, patient and image count, imaging device manufacturer, image annotation details, annotators’ qualifications, and dataset access. The secondary objective was to investigate the ethical approvals, specific terms, and licenses for the reuse of these datasets.
Results
Image datasets included in this systematic review
The initial search conducted through PubMed and Google scholar yielded a total of 181 articles. After removing duplicates, 176 datasets remained. Following the screening of titles and abstracts, thirty-six studies were deemed eligible for full-text reading. Among these thirty-six studies, twelve were excluded due to the use of a blocking technique obscuring the oral cavity region (n = 5), issues with accessibility (n = 5), and unclear descriptions (n = 2). Consequently, twenty-four studies14,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42 providing information on the eligible datasets were included.
A total of 786 datasets were identified through Google Dataset Search, Kaggle, and Hugging Face. After removing duplicates, 614 datasets remained. Upon initial screening, 86 datasets were deemed eligible. However, seven of these datasets were subsequently excluded due to inaccessibility (n = 4), incorrect descriptions (n = 2), and degraded image quality (n = 1), resulting in 79 datasets included. Additionally, three datasets, which were recommended by experts in the field and met the inclusion criteria, were further included in this systematic review43,44,45,46,47.
A single duplicate was identified upon cross-checking between the literature and platform searches, resulting in a total of 105 datasets included in this systematic review. Figure 1 illustrates the flowchart of the study and dataset selection process. The two reviewers exhibited high inter reviewer agreement for the selection process with Cohen’s kappa values ranging from 0.83 to 0.92.
The flowchart illustrates the systematic process for selecting datasets relevant to the oral and maxillofacial region. Initially, records were identified from PubMed, Google scholar, Google Dataset Search, Kaggle, and Hugging Face, with duplicates removed. During screening, records were excluded for irrelevance, non-human subjects, or images outside the target region. The eligibility assessment further excluded datasets with obscured regions, unclear descriptions, or inaccessibility. Final inclusion involved expert recommendations and removal of duplicates, resulting in 105 records.
General information on the datasets
The 105 datasets were created between 2018 and 2024, comprising a total of 437,538 images and 100 intraoral videos (Table 1; Fig. 2). The number of images per dataset ranged from 17 to 150,000 with 52 (49.5%) datasets containing over 1000 images. Only 13 (12.4%) datasets provided details about the imaging device manufacturer.
The bar chart illustrates the annual publication of datasets and images from 2018 to 2024. The left y-axis indicates the number of datasets, while the right y-axis indicates the number of images. Blue and red bars represent datasets and images, respectively. The chart reveals a notable upward trend, with significant increases in both datasets and images, particularly in 2023 and 2024, highlighting the growing interest and expansion in dataset and image publication during this period.
Regarding the imaging modality, 45 (43.2%) of the datasets contained panoramic radiographs, 24 (23.1%) photographs, 12 (11.5%) periapical radiographs, 8 (7.7%) histopathological images, 6 (5.8%) intra-oral/facial/model scans or images, 4 (3.9%) CBCT, with the remaining datasets including other modalities such as cephalometric radiographs, MRI, micro-CT, intraoral videos (Fig. 3). Notably, one dataset included both panoramic radiographs and CBCTs.
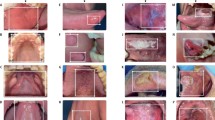
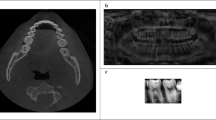
a Intraoral photograph of a patient missing two maxillary central incisors. b Periapical radiograph of the maxillary right posterior teeth and surrounding alveolar bone, displaying moderate horizontal bone loss and severe dental caries. c Panoramic radiograph providing a comprehensive view of the maxillary and mandibular teeth and jaw structure. d Lateral cephalometric radiograph illustrating a lateral perspective of the skull, teeth, and soft tissue profile. e Axial cone-beam computed tomography (CBCT) scan presenting detailed cross-sectional imaging. f Sagittal magnetic resonance imaging (MRI) scan presenting a sagittal view of craniofacial structures.
The image types across all datasets included 228,993 photographs, 125,975 panoramic radiographs, 29,390 histopathological images, 28,199 periapical radiographs, 12,860 intra-oral scans, 7990 head and face scans/images, 1097 model scans/images, 1031 micro-CT images, 709 CBCT scans, 392 MRI, 200 mid-sagittal CBCT, 702 cephalometric radiographs, and 100 intraoral videos (Table 2). All the access links to the datasets are provided in Supplementary Table S1.
Geographical contribution and institution of origin
Out of the 105 datasets, 66 (62.9%) did not report their origin. Of the remaining, 20 originated from Asia (10 from South Asia, 7 from East Asia, and 3 from Southeast Asia), seven from Europe, six from South America, four from North Africa and the Middle East, and two from North America. The geographical distribution of the datasets with known origin is demonstrated in Fig. 4. Only 38 (36.2%) of the datasets disclosed their institutional origin, with 24 originating from university research centres and 14 from local dental clinics (Table 1).
Purpose of dataset creation
The datasets included were created mainly for classification, segmentation, detection, and other specific tasks. For classification tasks, the datasets were designed to identify a wide range of oral conditions, including but not limited to oral cancer, oral mucosal lesions, gingivitis, calculus, ulcers, tooth discoloration, caries, missing teeth, as well as endodontic and periodontal diseases. For segmentation tasks, these datasets were used to develop AI models capable of delineating anatomical structures and pathologies, such as caries, teeth, maxilla, mandible, tongue, dental plaque, periapical lesions, mandibular canal, and oral epithelial dysplasia. Detection tasks involved the development of models to identify entities, such as dental implants, periapical lesions, alveolar bone loss, discoloured teeth, and carious lesions. The remaining datasets were created for specific tasks, such as anatomical landmark localization, volumetric mesh generation, cephalometric analysis, report generation, motion estimation, video stabilization, and the automated design of a complete denture metal base.
Annotations and annotators
Out of the 105 datasets, 83 (79.0%) included annotations, such as the delineation of teeth, caries, and dental restorations on periapical and panoramic radiographs, the delineation of teeth, tongue, mucosal lesions on photographs, the segmentation of teeth on CBCT, and the categorical label of the presence or absence of cancer cells on histopathological images (Table 3). However, only 27 (25.7%) datasets provided information about the qualification of the annotators. The annotators involved dental students, general dentists, and specialists such as endodontists, periodontists, orthodontists, radiologists, pathologists, ENT, craniofacial, and maxillofacial surgeons (Table 3).
Ethical approval, specific terms, and licenses of the included datasets
The majority of the included datasets (n = 88; 83.8%) did not indicate whether they had obtained ethical approval (Table 4). Out of the 105 datasets, 65 (61.9%) specified terms or licenses for their reuse (Table 4). The licenses attached to the datasets included CC BY 4.0 (n = 34; 52.3%), Apache 2.0 (n = 12; 18.5%), CC0 1.0 (n = 7; 10.8%), CC BY-NC 3.0/4.0 (n = 4; 6.2%), CC BY-SA 3.0/4.0 (n = 2; 3.1%), CC BY-NC-ND (n = 2; 3.1%), CC BY-NC-SA 4.0 (n = 1; 1.5%), and MIT (n = 1; 1.5%). One dataset specified dual licenses (CC0 1.0 and CC-BY), while another only provided the terms of reuse.
Applicability concerns of the included datasets and the risk of bias in annotations
The evaluation of applicability concerns for the 105 datasets and the assessment of the risk of bias in annotations are presented in Table 4. Out of these datasets, only 12 (11.4%) were rated as having a “low” applicability concern due to their documentation of ethical approval and licensing. Conversely, 36 (34.3%) datasets were deemed to have a ‘high’ applicability concern due to the absence of reported ethical approval and licensing. Regarding the risk of bias in the ground truth annotations, out of the 83 annotated datasets, 59 (71.1%) were rated as “high” risk due to the lack of information about the annotators. Eighteen (21.7%) datasets were rated as “low” risk, attributed to the involvement of more than one annotator with explicit medical/dental qualifications. Furthermore, six (7.2%) datasets were rated as “moderate” risk, either because they were annotated by a single qualified annotator or by multiple annotators who lacked explicit qualifications.
Discussion
This study aimed to provide a comprehensive overview of the openly accessible oral-maxillofacial imaging datasets, their sources and characteristics of both the images and annotations. In addition, this study also investigated the ethical clearance, specific terms, and licenses concerning the reuse of these datasets. During full-text evaluation, three datasets21,31,48 required registered access. Access to the Tufts Dental Database21 and the dataset by Cipriano M was acquired by providing an email address, institutional affiliation, and the intended use of the data or by creating an account. However, no response was received from the owner of the dataset48 following multiple attempts to fulfil the access requirements. The datasets by Ramakrishnan et al.49, Chilamkurthy et al.50, and Iosifidis et al.51 could not be accessed as the specified download sites were not available both at the time of the initial search and at the time of manuscript submission. Access to the dataset by Ranjbar et al.52 can only be acquired by obtaining an affiliate appointment with the institution for collaborative projects. Moreover, three datasets identified on the Kaggle platform were not available and access to a dataset by Jian53 requires a subscription payment. Eventually, a total of 105 openly accessible datasets were identified from both electronic databases and dataset management platforms. The findings reveal a significant increase in the number of open-source datasets for oral-maxillofacial imaging since 2018.
Two previous review articles identified publicly available ophthalmological imaging datasets and skin cancer image datasets, both derived from searches on MEDLINE, Google, and Google Dataset Search54,55. Another study by Ni et al. identified publicly available datasets for health misinformation detection from searches on the Web of Science Core Collection and arXiv56. Uribe et al. identified sixteen publicly accessible dental imaging datasets, created from 2020 to 2023, containing intraoral photographs or radiographs, panoramic radiographs, cephalometric radiographs, CBCT, and intraoral 3D scans15. However, in contrast to their findings, this study identified a significantly higher number of datasets created between 2018 and 2024. This study identified 105 datasets containing not only dental images but also those from oral-maxillofacial regions, with a wider range of imaging modalities including intraoral and extraoral photographs, periapical radiographs, panoramic radiographs, cephalometric radiographs, histopathological images, CBCT, intraoral/facial/model scans or images, MRI, micro-CT, and intraoral videos. Moreover, this study included over fifty datasets each providing more than 1000 images while Uribe et al. reported only five datasets with over 1000 images.
Among all the datasets, panoramic radiography is the most prevalent imaging modality. The second most common imaging modality is photography, with 24 datasets consisting of images of the lips, oral cavity, teeth, buccal mucosa, and tongue. The largest dataset among those included comprised 150,000 photographic images, specifically created for tooth instance segmentation, annotated by orthodontists with the aid of a human-machine hybrid algorithm14. Compared to 2D images, datasets for 3D image volumes, including CBCT, MRI, intraoral, facial, and model scans, are limited and smaller probably due to the challenges associated with their acquisition, annotation, and storage. In public datasets, original 3D images are often converted into the NIfTI format to facilitate more straightforward analysis due to its superior compatibility with computational tasks.
In the literature, dental AI models were developed mainly for segmentation, detection, classification, and prediction tasks57. Segmentation tasks involve dividing an image into distinct sections based on variations in pixel intensity among different tissues. Detection tasks aim to localize objects within an image using class-labelled bounding boxes. Classification tasks assign a categorical label to an entire image, while prediction tasks estimate the likelihood of a certain event based on existing risk factors. Obtaining annotations for segmentation models is relatively straightforward as they can be completed through visual inspection of images58. On the contrary, obtaining annotations for the development of more clinically significant diagnostic models, such as models for detecting the onset of specific diseases or for diagnosing lesions that are indistinguishable from diagnostic images, are challenging. These annotations often rely on particular clinical, laboratory, or biopsy examinations11. Our findings reveal that the most common types of annotation from the included datasets are the mask (29%), bounding box (29%), and categorical label (20%). Notably, the annotations provided across these datasets for similar tasks differ significantly due to different labelling methods used. The diversity in annotation approaches can complicate the integration and use of annotations from datasets created for similar tasks. Moreover, the annotations for similar oral conditions differed across datasets and often lacked detailed descriptions. Thus, such annotations should be reused with caution to ensure their accuracy and precision.
While nearly 80% of the 105 datasets provided image annotations, only one-fourth of these datasets specified the annotators’ qualifications. Notably, even when qualifications were mentioned, detailed information regarding the annotators’ experience in dental specialties or annotation practices was rarely disclosed. The lack of this information increases the uncertainty of the annotation accuracy, affecting the reliability of open-access images and their corresponding ground truth annotations. Even though some annotations were carried out by specialists, the accuracy of these annotations might not be guaranteed or suitable for direct use in specific AI projects. Manual adjustments or re-annotations of these annotations may be necessary to meet the requirements for certain projects. This study included nine histopathological image datasets containing various cell types, including normal oral cavity epithelium, oral cancer cells, epithelial dysplasia and Leukoplakia cells. However, only four datasets explicitly stated that the annotations were performed by pathologists or specialists in pathology. Therefore, caution is advised when reusing these annotations, especially those with unknown origins or uncertainties for the development or validation of AI models.
Unlabelled images can be effectively utilized in AI model training through self-supervised learning techniques, such as contrastive learning, mask image modelling59,60, and semi-supervised learning5,61. Self-supervised learning enables models to learn data distribution without manual labels by using pretext tasks that exploit the inherent structure of the data to generate labels. This method uses large amounts of unlabelled data to learn useful representations. Subsequently, a smaller set of labelled data is employed to fine-tune the model for specific tasks. This approach minimizes the dependence on extensive manual annotations and is beneficial for utilizing large unlabelled datasets efficiently.
The majority (83.8%) of the datasets did not disclose whether they had obtained ethical approval. This finding indicates a critical area in data usage and ethics that requires further attention. Some included studies have stated that their open datasets were derived from projects with ethical approval. However, this does not automatically grant permission for others to reuse the image data. Ethical approval confirms that the initial study is in compliance with ethical standards, but it does not extend to the subsequent use of the data by third parties62. Sharing patient data with either internal or external teams is often essential for AI project development and validation, which may not be explicitly covered in the original ethical approvals. The Europe General Data Protection Regulation legislation highlights the necessity of strict data processing regulations, which limit health data use unless explicit consent is given, ensuring that data processing aligns with protecting individuals’ vital interests63. However, details about patient consent are often missing in publicly accessible datasets. This situation raises serious ethical concerns about data sharing and patient consent, especially when developing AI applications in healthcare.
Public accessibility of datasets does not automatically grant unlimited usage rights, as licensing clearly defines the terms for data reuse. Dataset licenses allow creators to specify rights they reserve and those they waive. Without explicit licensing, even ethically approved datasets can still cause legal and ethical issues when reused. Common licenses include CC0-1.0 and various Creative Commons (CC) licenses, such as CC-BY, CC-BY-NC, CC-BY-SA, and CC-BY-ND64. The CC0-1.0 license permits creators to waive all their copyright and related rights in their works as much as legally possible. Other CC licenses provide options that retain copyright while allowing various levels of permission. For instance, CC-BY-NC allows non-commercial reuse, CC-BY permits modifications and commercial use with attribution, CC-BY-SA requires any adaptations to be shared under identical terms, and CC-BY-ND allows only unchanged and whole redistribution with proper credit. Of the datasets, 61.9% specified a license for their reuse, with over 50% licensed with CC BY, followed by Apache 2.0 (18.5%). In cases where a single dataset carries multiple licenses, such as one panoramic radiograph dataset with dual licenses (CC0 1.0 and CC-BY)65, the strictest of the licenses is applied.
While 61.9% of the datasets specified a license for reuse, some of them might have possibly mislabelled the license on dataset platforms. This contributes to the uncertainty regarding whether the openly accessible oral-maxillofacial imaging datasets were released with valid reuse terms or license, placing them in a legal grey area. Using copyrighted datasets for training AI models can potentially lead to legal issues62. The common practice of creating a training dataset by repackaging existing open-source datasets can be problematic. If a dataset is protected by NoDerivatives licenses, such as CC-BY-ND, it cannot be included in a dataset to train an AI model. In such case, the trained model could be considered a derivative of the training data, violating the exclusive rights of the copyright holders. Similarly, if an AI model is trained using a dataset protected by licenses permitting only non-commercial reuse, future commercialization of the trained model might be restricted62. These evolving legal issues regarding dataset reuse are gaining attention from academic organizations, industry labs, and research institutions. Therefore, reusing these datasets should be cautious due to potential legal issues. Schwabe et al. introduced the METRIC-framework, which provides a systematic approach for assessing training datasets, establishing reference datasets, and designing test datasets66. This framework proposes fifteen awareness dimensions across five data management clusters, including measurement process (device error, human-induced error, completeness, and source credibility), timeliness (timeliness), representativeness (variety, depth of data, target class balance), informativeness (understandability, redundancy, informative missingness, feature importance), consistency (rule-based consistency, logical consistency, and distribution consistency). These dimensions could contribute to the development of clear, standardized guidelines for the ethical reuse of publicly accessible medical and dental image datasets, while strictly complying with licensing requirements.
This systematic review has limitations. First, due to the large number of images from the included datasets, it is not practical to assess the quality of all images. Since image quality is often assessed for specific clinical indications, the quality of images from the included datasets should be evaluated by interested researchers based on their intended tasks. Second, some crucial factors such as metadata completeness, identification of data reuse issues, and data traceability were not included in the assessment of the risk-of-bias for the included datasets, which might not be able to fully account for all potential biases introduced into the datasets. Furthermore, this study excluded certain large, high-quality image datasets48 as the access could not be obtained due to a lack of response from the dataset owners, despite following the requirements for registered access. Moreover, the datasets released may be subject to continual updates without any official notification. Therefore, the changes in the number and annotations of images from the datasets should be confirmed with caution before reuse.
In conclusion, this study has systematically identified 105 public oral-maxillofacial imaging datasets and investigated their sources, characteristics, and ethical and licensing considerations. While the majority of the datasets included annotations, only some specified the annotators’ qualifications. Furthermore, more than half of the datasets specified the terms or licenses for reuse, but most did not disclose whether ethical approval was obtained. These findings highlight the need for careful consideration of ethical and legal implications when reusing these datasets and suggest the need to establish clear, standardized guidelines for reusing publicly accessible image datasets.
Methods
This systematic review was conducted in accordance with the guidance of the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA)19. The PRISMA checklist used for this review is provided in Supplementary Table S2. The study protocol has been registered on the Open Science Framework (OSF) (Registration https://doi.org/10.17605/OSF.IO/SFN5C). The focused question guiding the search was, “Which open-source datasets related to images from the oral-maxillofacial region are available?”.
Search strategy and selection criteria
The search strategy consisted of two components, including the search of two electronic scientific literature databases (PubMed and Google scholar) and three widely-used dataset management platforms (Google Dataset Search, Kaggle, and Hugging face) to identify as many publicly accessible image datasets as possible. The search was conducted in September 2024. The literature search combined free-text terms of (“dentistry” OR “dental” OR “oral” OR “maxillofacial”) AND (“open source” OR “open access” OR “publicly available” OR “publicly accessible”) AND (“data” OR “dataset” OR “repository”) AND “images”. Vocabulary and syntax were adjusted accordingly for each database. The search terms used on dataset management platforms were “dentistry” OR “dental” OR “oral-maxillofacial” OR “dental image” OR “oral image”. The specific search strategies used for all databases are provided in Supplementary Table S3.
The electronic literature database search was conducted without any restrictions on the publication period. The criteria for inclusion were:
-
1.
Original and review articles published in English;
-
2.
Studies that report a dataset comprised of any type of image modalities generating images from the oral-maxillofacial region, including scans of dental models from patients; and
-
3.
Studies providing the access to the dataset.
Studies were excluded if one of the following exclusion criteria was met.
-
1.
Studies reporting a dataset consisting of images not from human subjects;
-
2.
Studies reporting a dataset consisting of images from cadavers or extracted teeth;
-
3.
Studies reporting a dataset consisting of images where the oral-maxillofacial region was obscured using a blocking technique;
-
4.
Studies reporting a dataset consisting of images that were included in the most recently updated dataset from the same source; or
-
5.
Studies where the full text is not available or accessible.
For the dataset management platforms search, all open-source datasets consisting of images from the oral-maxillofacial region were considered eligible. The exclusion criteria were:
-
1.
Datasets consisting of images not from human subjects;
-
2.
Dataset consisting of images from cadavers or extracted teeth;
-
3.
Datasets consisting of images where the oral-maxillofacial region was obscured using a blocking technique;
-
4.
Datasets consisting of images that were included in the most recently updated datasets from the same source;
-
5.
Datasets consisting of image files that were corrupted and could not be opened; or
-
6.
Datasets that require payment for access.
All records retrieved from the electronic literature database search were compiled using the reference manager software (EndnoteTM Version 21, Clarivate Analytics, New York, USA). The titles were automatically checked for duplicates. Two independent reviewers (J.H. and K.F.H.) screened the titles and abstracts of each record to select studies for further full-text evaluation. Reviewer K.F.H. is a professoriate faculty member in the subdivision of Oral-Maxillofacial Radiology with over ten years of experience in conducting diagnostic imaging studies. Reviewer J.H. is a PhD candidate at the same institution with more than five years of research experience in the development of artificial intelligence algorithms and is experienced in the collection and evaluation of AI-related public datasets. Additional manual searches on the reference lists of the included studies were conducted independently by two reviewers (J.H. and K.F.H.) to further identify potentially eligible studies that met the inclusion criteria. Subsequently, the two reviewers (J.H. and K.F.H.) independently assessed the full-texts of the included studies. The two reviewers compared the studies they identified as eligible, and then discussed their reasons for considering certain studies to be included based on the defined inclusion and exclusion criteria. Agreement was reached through discussion. In cases where agreement could not be achieved, a third experienced reviewer (Q.Y.H.A) was consulted to assist in reaching a consensus. Inter-reviewer agreement was evaluated by calculating Cohen’s kappa values. Eligible datasets identified from the dataset management platforms were organized using an Excel spreadsheet (Microsoft Corporation, Redmond, Washington). Any duplicates from the electronic literature database search and the dataset management platform search were eliminated.
Extraction of dataset characteristics and outcome of interest
Details regarding the year and purpose of dataset creation, creators, country and institution of origin, imaging modality, image type and format, the number of patients and images in the dataset, the manufacturer of the imaging device, image annotation details, the qualification of the annotators, and dataset access, were extracted by two reviewers (J.H. and K.F.H.) from the included studies and the metadata of the datasets. In addition, information pertaining to the acquisition of ethical approval for image collection as well as specific terms, conditions, and licensing requirements for reusing these datasets were collected. Any discrepancies detected in the extracted data were resolved through discussion. In the case of a discrepancy between the information provided in the included studies and the dataset repository, the information from the repository was used in this study. All data were systematically tabulated using a standardized template created in an Excel spreadsheet (Microsoft Corporation, Redmond, Washington).
Dataset accessibility
The accessibility of the datasets included in this study were divided into two categories as follows:
-
1.
Datasets that are readily accessible and can be directly downloaded without any requirement.
-
2.
Datasets that necessitate registered access, requiring submission of an email request or the creation of an account. Upon fulfilling these requirements, a download link for the dataset would be sent to the applicant’s email. The accessibility status of these datasets was re-confirmed at the time of manuscript submission.
Evaluation of applicability concerns of the included datasets and the risk of bias in annotations
The applicability concerns of the included datasets and the risk of bias in annotations were assessed independently by two reviewers (J.H. and K.F.H.). Any discrepancies were resolved through discussion. A dataset was deemed to have a “low” applicability concern if it reported both ethical approval as well as the terms or licensing requirements for its reuse. If only either ethical approval or terms or licenses were reported, the concern was classified as “moderate”. If neither was reported, the concern was rated as “high”. The assessment of the risk of bias in annotations focused on the reliability of the reference standard (i.e., ground truth annotations), which is one of the four domains proposed by the Revised Tool for the Quality Assessment of Diagnostic Accuracy Studies (QUADAS-2), a tool widely used in diagnostic imaging studies67. According to the QUADAS-2, the risk of bias in the reference standard should be assessed by the signalling question “Is the reference standard likely to correctly classify the target condition?”. Based on the proposed signalling question, the risk of bias in the ground truth annotations was assessed by evaluating the reliability of the reference standard used for annotation. For datasets with annotations, the “low” risk-of-bias rating was assigned to datasets where ground truth annotations are confirmed by at least two annotators with explicit medical/dental qualifications, or those supported by clinically or pathologically confirmed results. Datasets with ground truth annotations determined by a single qualified annotator, and those involving at least two annotators identified as experts but without explicit qualifications, were given a “moderate” risk-of-bias rating. All remaining datasets were categorized as having a “high” risk of bias.
Data availability
Data sharing is not applicable to this article as no datasets were generated during the current study. All the access links to the datasets analysed in this study are provided in Supplementary Table S1.
Code availability
Not applicable.
References
Joda, T., Yeung, A., Hung, K., Zitzmann, N. & Bornstein, M. Disruptive innovation in dentistry: what it is and what could be next. J. Dent. Res. 100, 448–453 (2021).
Hung, K., Yeung, A. W. K., Tanaka, R. & Bornstein, M. M. Current applications, opportunities, and limitations of AI for 3D imaging in dental research and practice. Int. J. Environ. Res. Public Health 17, 4424 (2020).
Hung, K. F. et al. Current applications of deep learning and radiomics on CT and CBCT for maxillofacial diseases. Diagnostics 13, 110 (2022).
Hung, K., Montalvao, C., Tanaka, R., Kawai, T. & Bornstein, M. M. The use and performance of artificial intelligence applications in dental and maxillofacial radiology: a systematic review. DMFR 49, 20190107 (2020).
Hao, J. et al. A semi-supervised transformer-based deep learning framework for automated tooth segmentation and identification on panoramic radiographs. Diagnostics 14, 1948 (2024).
Hao, J. et al. T-Mamba: a unified framework with long-range dependency in dual-domain for 2D & 3D tooth segmentation. Preprint at https://arxiv.org/abs/2404.01065 (2024).
Razaghi, M., Komleh, H. E., Dehghani, F. & Shahidi, Z. Innovative diagnosis of dental diseases using YOLO V8 deep learning model. In IEEE 13th Iranian/3rd International Machine Vision and Image Processing Conference (MVIP), 1–5 (IEEE, 2024).
Xu, C. et al. TeethDreamer: 3D teeth reconstruction from five intra-oral photographs. In Medical Image Computing and Computer-Assisted Intervention — MICCAI 2024, 712–721 (Springer International Publishing, Cham, 2024).
Mei, L. et al. DTR-net: dual-space 3D tooth model reconstruction from panoramic X-Ray images. IEEE Trans. Med. Imaging 43, 517–528 (2023).
Gao, N. et al. Multi-level objective alignment transformer for fine-grained oral panoramic X-ray report generation. IEEE Trans. Multimed. 26, 7462–7474 (2024).
Hung, K. F., Yeung, A. W. K., Bornstein, M. M. & Schwendicke, F. Personalized dental medicine, artificial intelligence, and their relevance for dentomaxillofacial imaging. DMFR 52, 20220335 (2023).
Krois, J. et al. Generalizability of deep learning models for dental image analysis. Sci. Rep. 11, 6102 (2021).
Cui, W. et al. Ctooth: a fully annotated 3d dataset and benchmark for tooth volume segmentation on cone beam computed tomography images. In Intelligent Robotics and Applications, 191–200 (ICIRA, 2022).
Zou, B. et al. Teeth-SEG: an efficient instance segmentation framework for orthodontic treatment based on multi-scale aggregation and anthropic prior knowledge. In Proceedings of IEEE Conference on Computer Vision and Pattern Recognition, 11601–11610 (IEEE, 2024).
Uribe, S. E. et al. Publicly available dental image datasets for artificial intelligence. J. Dent. Res. 103, 1365–1374 (2024).
Ohmann, C. et al. Sharing and reuse of individual participant data from clinical trials: principles and recommendations. BMJ open 7, e018647 (2017).
Meystre, S. M. et al. Clinical data reuse or secondary use: current status and potential future progress. Yearb. Med. Inform. 26, 38–52 (2017).
Duke, C. S. & Porter, J. H. The ethics of data sharing and reuse in biology. BioScience 63, 483–489 (2013).
Moher, D., Liberati, A., Tetzlaff, J., Altman, D. G. & Group, P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. Int. J. Surg. 8, 336–341 (2010).
Abdi, A. H., Kasaei, S. & Mehdizadeh, M. Automatic segmentation of mandible in panoramic x-ray. J. Med. Imaging 2, 044003–044003 (2015).
Panetta, K., Rajendran, R., Ramesh, A., Rao, S. P. & Agaian, S. Tufts dental database: a multimodal panoramic x-ray dataset for benchmarking diagnostic systems. IEEE J. Biomed. Health Inform. 26, 1650–1659 (2021).
Zhang, Y. et al. Children’s dental panoramic radiographs dataset for caries segmentation and dental disease detection. Sci. Data 10, 380 (2023).
Hamamci, I. E. et al. Dentex: An abnormal tooth detection with dental enumeration and diagnosis benchmark for panoramic x-rays. Preprint at https://arxiv.org/abs/2305.19112 (2023).
Zhou, W. et al. A dual-labeled dataset and fusion model for automatic teeth segmentation, numbering, and state assessment on panoramic radiographs. BMC Oral. Health 24, 1201 (2024).
Ke, W. et al. Biological gender estimation from panoramic dental x-ray images based on multiple feature fusion model. Sens. Imaging 21, 1–11 (2020).
Murga & Stefan. RGB-D tongue state classification dataset. Borealis V1 https://doi.org/10.5683/SP2/5T2RD9 (2019).
Rashid, J. et al. Mouth and oral disease classification using InceptionResNetV2 method. Multim. Tools Appl. 83, 33903–33921 (2024).
Kusakunniran, W. et al. Deep Upscale U-Net for automatic tongue segmentation. MBEC 62, 1751–1762 (2024).
Liao, C. W. et al. Self-assembled micro-computed tomography for dental education. PLoS One 13, e0209698 (2018).
Gholamalizadeh, T. et al. Open-Full-Jaw: an open-access dataset and pipeline for finite element models of human jaw. Comput. Methods Prog. Biomed. 224, 107009 (2022).
Cipriano, M. et al. Deep segmentation of the mandibular canal: a new 3D annotated dataset of CBCT volumes. IEEE Access 10, 11500–11510 (2022).
Wang, Y. et al. STS MICCAI 2023 challenge: grand challenge on 2D and 3D semi-supervised tooth segmentation. Preprint at https://arxiv.org/abs/2407.13246 (2024).
Andlauer, R. et al. 3D-guided face manipulation of 2D images for the prediction of post-operative outcome after cranio-maxillofacial surgery. IEEE Trans. Image Process. 30, 7349–7363 (2021).
Ben-Hamadou, A. et al. Teeth3DS+: an extended benchmark for intraoral 3D scans analysis. Preprint at https://arxiv.org/abs/2210.06094v1 (2022).
Abdel-Alim, T. et al. Quantifying dysmorphologies of the neurocranium using artificial neural networks. J. Anat. 245, 903–913 (2024).
Ribeiro-de-Assis, M. C. F. et al. NDB-UFES: an oral cancer and leukoplakia dataset composed of histopathological images and patient data. Data Brief. 48, 109128 (2023).
Rahman, T. Y., Mahanta, L. B., Das, A. K. & Sarma, J. D. Automated oral squamous cell carcinoma identification using shape, texture and color features of whole image strips. Tissue Cell 63, 101322 (2020).
Silva, A. B. et al. Oralepitheliumdb: A dataset for oral epithelial dysplasia image segmentation and classification. J. Imaging Inf. Med. 37, 1691–1710 (2024).
Hsieh, H.-C. et al. Deep learning-based automatic image classification of oral cancer cells acquiring chemoresistance in vitro. PloS One 19, e0310304 (2024).
Rönnau, M. M. et al. Automatic segmentation and classification of Papanicolaou-stained cells and dataset for oral cancer detection. Comput. Biol. Med. 180, 108967 (2024).
Ruthven, M., Peplinski, A. M., Adams, D. M., King, A. P. & Miquel, M. E. Real-time speech MRI datasets with corresponding articulator ground-truth segmentations. Sci. Data 10, 860 (2023).
Zeng, M., Yan, Z., Liu, S., Zhou, Y. & Qiu, L. Cascaded convolutional networks for automatic cephalometric landmark detection. Med. Image Anal. 68, 101904 (2021).
Adnan, N. & Umer, F. Orthopantomogram teeth segmentation and numbering dataset. Data Brief. 57, 111152 (2024).
Román, J. C. M. et al. Panoramic dental radiography image enhancement using multiscale mathematical morphology. Sensors 21, 3110 (2021).
Wang, C.-W. et al. A benchmark for comparison of dental radiography analysis algorithms. Med. Image Anal. 31, 63–76 (2016).
Wang, C.-W. et al. Evaluation and comparison of anatomical landmark detection methods for cephalometric x-ray images: a grand challenge. IEEE Trans. Med. Imaging 34, 1890–1900 (2015).
Lindner, C. et al. Fully automatic system for accurate localisation and analysis of cephalometric landmarks in lateral cephalograms. Sci. Rep. 6, 33581 (2016).
Shi, J. et al. Semantic decomposition network with contrastive and structural constraints for dental plaque segmentation. IEEE Trans. Med. Imaging 42, 935–946 (2022).
Ramakrishnan, D. et al. A large open access dataset of brain metastasis 3D segmentations on MRI with clinical and imaging information. Sci. Data 11, 254 (2024).
Chilamkurthy, S. et al. Deep learning algorithms for detection of critical findings in head CT scans: a retrospective study. Lancet 392, 2388–2396 (2018).
Iosifidis, A., Marami, E., Tefas, A., Pitas, I. & Lyroudia, K. The MOBISERV-AIIA eating and drinking multi-view database for vision-based assisted living. J. Inf. Hiding Multim. Signal Process. 6, 254–273 (2015).
Ranjbar, S. et al. Weakly supervised skull stripping of magnetic resonance imaging of brain tumor patients. Front. Neuroimaging 1, 832512 (2022).
Jian, G. Aoralscan3 tooth segmentation dataset. IEEE Dataport. https://doi.org/10.21227/w9mp-5w63 (2023).
Wen, D. et al. Characteristics of publicly available skin cancer image datasets: a systematic review. Lancet Digit. Health 4, e64–e74 (2022).
Khan, S. M. et al. A global review of publicly available datasets for ophthalmological imaging: barriers to access, usability, and generalisability. Lancet Digit. Health 3, e51–e66 (2021).
Ni, Z., Bousquet, C., Vaillant, P. & Jaulent, M.-C. Rapid review on publicly available datasets for health misinformation detection. Healthc. Transform. Inf. Artif. Intell., 305, 123–126 (2023).
Hung, K. F., Ai, Q. Y. H., Leung, Y. Y. & Yeung, A. W. K. Potential and impact of artificial intelligence algorithms in dento-maxillofacial radiology. Clin. Oral. Investig. 26, 5535–5555 (2022).
Hung, K. F. et al. Automatic detection and segmentation of morphological changes of the maxillary sinus mucosa on cone-beam computed tomography images using a three-dimensional convolutional neural network. Clin. Oral. Investig. 26, 3987–3998 (2022).
Xie, Z. et al. Simmim: a simple framework for masked image modelling. In Proceedings of IEEE Conference on Computer Vision and Pattern Recognition, 9653–9663 (IEEE, 2022).
Chen, Z. et al. Masked image modeling advances 3d medical image analysis. In Proceedings of IEEE Winter Conference on Applications of Computer Vision, 1970–1980 (IEEE, 2023).
Jing, Y. et al. USCT: uncertainty-regularized symmetric consistency learning for semi-supervised teeth segmentation in CBCT. Biomed. Signal Process. Control. 91, 106032 (2024).
Longpre, S. et al. A large-scale audit of dataset licensing and attribution in AI. Nat. Mach. Intell. 6, 975–987 (2024).
Dziedzic, A., Issa, J., Chaurasia, A. & Tanasiewicz, M. Artificial intelligence and health-related data: the patient’s best interest and data ownership dilemma. Proc. Inst. Mech. Eng. H. 238, 1023–1028 (2024).
Kim, M. The Creative Commons and copyright protection in the digital era: Uses of Creative Commons licenses. JCMC 13, 187–209 (2007).
Rahman, R. B. et al. Dental OPG XRAY dataset. Mendeley Data, V4, https://doi.org/10.17632/c4hhrkxytw.4 (2024).
Schwabe, D., Becker, K., Seyferth, M., Klaß, A. & Schaeffter, T. The METRIC-framework for assessing data quality for trustworthy AI in medicine: a systematic review. NPJ Digit. Med. 7, 203 (2024).
Whiting, P. F. et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann. Intern. Med. 155, 529–536 (2011).
Abdi, A. & Kasaei, S. Panoramic dental X-rays with segmented mandibles. Mendeley Data V2 https://doi.org/10.17632/hxt48yk462.2 (2020).
Brahmi, W. & Jdey, I. Automatic tooth instance segmentation and identification from panoramic X-Ray images using deep CNN. Multimed. Tools Appl. 83, 55565–55585 (2024).
Farzi, I., Kazemi, A. & Hosseini, M. Panoramic radiography Images with diagnosis of need for Apicoectomy surgery as label. Mendeley Data, V1, https://doi.org/10.17632/9d8mcyp284.1 (2023).
Budagam, D. et al. Instance segmentation and teeth classification in panoramic X-rays. Preprint at https://arxiv.org/abs/2406.03747 (2024).
Waqas, M., Hasan, S., Khurshid, Z. & Kazmi, S. OPG dataset for Kennedy classification of partially edentulous arches. Mendeley Data, V1, https://doi.org/10.17632/ccw5mvg69r.1 (2024).
Sengupta, N., Sarode, S. C., Sarode, G. S. & Ghone, U. Scarcity of publicly available oral cancer image datasets for machine learning research. Oral. Oncol. 126, 105737 (2022).
Chandrashekar, H. S.,Geetha Kiran, A., Murali, S., Dinesh, M. S. & Nanditha, B. R. Oral images dataset. Mendeley Data, V2, https://doi.org/10.17632/mhjyrn35p4.2 (2021).
Hoang B. D. A Dental intraoral image dataset of gingivitis for image captioning. Mendeley Data, V1, https://doi.org/10.17632/3253gj88rr.1 (2024).
Nisrean, T. A dataset of dental periapical X-ray. Mendeley Data, V1, https://doi.org/10.17632/8ys8jssm9k.1 (2023).
Viet, D. Panoramic radiographs with periapical lesions Dataset. Mendeley Data, V3, https://doi.org/10.17632/kx52tk2ddj.3 (2024).
Li, C. et al. Efficient complete denture metal base design via a dental feature-driven segmentation network. Comput. Biol. Med. 175, 108550 (2024).
Acknowledgements
This study was supported by the Seed Fund for Basic Research for New Staff at the University of Hong Kong (103036002).
Author information
Authors and Affiliations
Contributions
Conceptualisation: K.F.H., Q.Y.H.A., M.M.B. and J.K.H.T.; Methodology: K.F.H., Q.Y.H.A., A.W.K.Y, M.M.B. and J.K.H.T.; Data curation and investigation: J.H., K.F.H., and Q.Y.H.A.; Writing—original draft preparation, J.H. and K.F.H.; writing—review and editing, A.N., A.W.K.Y, R.T., Q.Y.H.A., W.Y.H.L., Y.Y.L., Z.S., A.A., M.M.B., C.M. and J.K.H.T. All authors have read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hao, J., Nalley, A., Yeung, A.W.K. et al. Characteristics, licensing, and ethical considerations of openly accessible oral-maxillofacial imaging datasets: a systematic review. npj Digit. Med. 8, 412 (2025). https://doi.org/10.1038/s41746-025-01818-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41746-025-01818-5