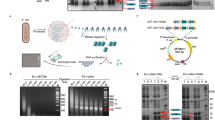
Abstract
As a fundamental unit for packaging genomic DNA into chromatin, the eukaryotic nucleosome core comprises a canonical octamer with two copies for each histone, H2A, H2B, H3, and H4, wrapped around with 147 base pairs of DNA. While H3 and H4 share structure-fold with archaeal histone-like proteins, the eukaryotic nucleosome core and the complete nucleosome (the core plus H1 histone) are unique to eukaryotes. To explore whether the eukaryotic nucleosome can assemble in prokaryotes and to reconstruct the possible route for its emergence in eukaryogenesis, we developed an in vivo system for assembly of nucleosomes in the model bacterium, Escherichia coli, and successfully reconstituted the core nucleosome, the complete nucleosome, and unexpectedly the non-canonical (H3-H4)4 octasome. The core and complete nucleosomes assembled in E. coli exhibited footprints typical of eukaryotic hosts after in situ micrococcal nuclease digestion. Additionally, they caused condensation of E. coli nucleoid. We also demonstrated the stable formation of non-canonical (H3-H4)2 tetrasome and (H3-H4)4 octasomes in vivo, which are suggested to be ‘fossil complex’ that marks the intermediate in the progressive development of eukaryotic nucleosome. The study presents a unique platform in a bacterium for in vivo assembly and studying the properties of non-canonical variants of nucleosome.

Similar content being viewed by others
Introduction
The nucleosome serves as a fundamental building block for packaging genome DNA of eukaryotes into chromatin. The core nucleosome comprises a canonical octamer of four histones - two copies for each of H2A, H2B, H3, and H4, and is tightly wrapped by 147 base pair (bp) of DNA segment in ~1.7 left-handed superhelical turns, forming a compact disc-like structure about 5.5 nm in height and 11 nm in diameter1,2. It is estimated that the structural configuration of the core nucleosome provides an initial ~7-fold linear compaction of genomic DNA3,4. Histone H1 is a primary component of the eukaryotic nucleosome, which functions in conjunction with the core nucleosome by binding to linker DNA at the entry-exit sites flanking the canonical octasome. Histone H1 is essential for folding the nucleosome strings into macromolecular structure known as chromatin fiber, the higher-order structure of the eukaryotic chromosome5,6,7. The nucleosomes and its higher-order structure play a pivotal role in eukaryotic cells in governing DNA accessibility and regulating essential cellular processes such as transcription, replication, and DNA repair8,9,10. Incorporation of different histone variants and covalent modifications of histones are the common mechanisms that modulate biophysical properties of histones, nucleosome assembling dynamics, and genome activities11,12,13.
In eukaryotes, the nucleosome complex’s structure is heterogeneous and dynamic. The canonical nucleosome is formed with the initial action of (H3-H4)2-tetramer being deposited onto a DNA template, creating a tetrasome platform for subsequent loading of H2A-H2B dimers, one-by-one individually to form the canonical octasome14,15. Genome activities often involve the dynamic assembly and disassembly of nucleosomes, and many intermediate stages, e.g. sub-nucleosomal structures including tetrasomes16,17,18, hexasomes19,20, di-nucleosomes21,22, etc, which are often transient and rely on the interactions of different combination of histone components. For example, transcription activities caused transient chromatin remodeling by partial unwrapping of genome DNA and release of a H2A-H2B dimer from core nucleosome, forming the ‘hexasome’ intermediates that were reported to accompany gene expression, DNA replication, and repair activities19,23.
The (H3-H4)-only octasome, an intriguing nucleosome structure, was first observed over four decades ago24,25. The formation of (H3-H4)4 octasome was reconstituted in vitro only at higher histones/DNA ratios. The (H3-H4)4 particles were directly observed with atomic force microscopy26. The high-resolution structure of (H3-H4)4 octasome was recently resolved using cryo–electron microscopy, suggesting a model of two stacked disks connected by a H4-H4’ four-helix bundle (FHB)27. In addition, evidence of crosslinking experiments that supported the possible existence of (H3-H4)4 octasomes in yeast cells. The existence of (H3-H4)4 octasomes within chromatin has the potential to modify both the structure and dynamics of chromatin, signifying a revolutionary change in how we understand the regulation of the epigenome.
The nucleosome is a distinctive hallmark of eukaryotic organisms, which sets eukaryotes apart from bacteria that lack the intricate nucleosome complex and higher-order chromatin architecture in chromosome organization. Although archaea are known to have histone-like proteins that are widespread and share the folding of eukaryotic histones28,29,30, the higher-order structure of the eukaryotic nucleosome complex is unique to eukaryotic cells, which does not exist outside the eukaryotic domain. The archaea histone-like proteins have the same core fold as eukaryotic histones, but typically lack the N- and C-terminal tails, which are the principal substrates for post-translational modifications in eukaryotes28,29. In archaea, DNA binds to histone-like proteins to form homo-dimeric and homo-tetrameric structure that were revealed by recent crystal structure studies28,31. However, neither eukaryotic canonical octasomes nor eukaryotic type histone modifications have been found in archaea. Notably, Warnecke and colleagues recently expressed in Escherichia coli the archaeal histone-like protein, i.e. HMfA or HMfB, from Methanothermus fervidus, and successfully reconstituted their homo-oligomeric complexes on the E. coli chromosome in vivo32. Different from the eukaryotic nucleosomes, the archaeal histone complexes assembled in E. coli comprised only single-type histone molecules.
In this current study, we have undertaken a task to assemble the eukaryotic nucleosome complexes in vivo in the model bacterium, E. coli. This system for in vivo nucleosome assembly provided a platform to reconstitute the eukaryotic nucleosomes, and non-canonical variants in a ‘living’ prokaryote, and to facilitate studying the properties of canonical and non-canonical nucleosomes. This platform may also help shed light on the evolutionary origin and progressive development of the eukaryotic nucleosomes. We successfully assembled the eukaryotic nucleosome core and the complete nucleosome (the core plus H1 histone) in vivo in E. coli, which was confirmed by multi-lines of evidence, including micrococcal nuclease (MNase) digestion footprints, in vivo assembled core nucleosomes co-localizing with genome DNA in E. coli nucleoid, and abolished nucleosome formation upon subtracting one of the core histones. We further assembled the non-canonical nucleosome variants, i.e., the (H3-H4)2 tetrasomes and (H3-H4)4 octasomes, in E. coli, which had distinct MNase digestion profiles, were colocalization with genome DNA, and induced condensation of E. coli nucleoid. The creation of the non-canonical variants of eukaryotic nucleosome in a living bacterium adds evidence to support their in vitro reconstitution results, and presents a unique platform to study the properties of these non-canonical nucleosomes.
Results
In vivo assembled core nucleosomes co-localizing with genome DNA in E. coli nucleoid
We generated the construct pET29a-H2AH2BH3H4, which enabled poly-cistronic expression of xenopus core histones33, i.e., H2A, H2B, H3, and H4 (Supplementary Fig. 1a, and Supplementary Note 1). pET29a-H2AH2BH3H4 was transformed into E. coli BL21(DE3) Rosetta, generating the strain Ec-AB34. The expression of histones in Ec-AB34 was induced with IPTG (400 µM), and analyzed with SDS-PAGE. H2A, H2B, H3, and H4 were found primarily in the soluble fraction of the Ec-AB34 lysate (Supplementary Fig. 1b, c). While the histone H2A was among the highest expressed proteins compared to native proteins, H2B, H3 and H4’s levels were lower with significant amounts produced. To determine if the eukaryotic histones can assemble in vivo to form core nucleosomes in E. coli that has a cellular environment distinctly different from eukaryotes, we performed the in situ micrococcal nuclease digestion assay (herein named ecMNase) (Supplementary Fig. 1d), which was modified from the similar assay previously used to detect nucleosome formation in eukaryotic cells34,35. When expression of xenopus core histones was induced by IPTG (Supplementary Fig. 1e), ecMNase experiments showed it yielded a distinct DNA fragmentation pattern (Fig. 1a) resembling the fragmentation of nucleosome-protected DNA in eukaryotic cells by MNase digestion36,37,38. Note the protected DNA bands for mono-nucleosome (~147 bp) (Supplementary Figs. 2 and 3) and di-nucleosome (~300 bp) were absent for control strain Ec-pET29a.
a DNA fragmentation profiles obtained from the xenopus histones-expressing strain (Ec-AB34), and the control strain (Ec-pET29a) using ecMNase assay. Arrows mark mono- and di- nucleosome bands, respectively. M, DNA standards. The complete gel pieces are provided in Supplementary Fig. 6. b Strain constructs containing different histone components. The symbols ‘+’ and ‘−’ denote the presence and absence of corresponding histone gene, whereas ‘+m’ indicates the histone gene is fused with a mCherry gene. c DNA fragmentation profiles of four strains: Ec-pET29a/21a, Ec-AB34/m*, Ec-A*B34, and Ec-AB*34 using ecMNase assay. Arrows mark mono- and di- nucleosome bands, respectively. M, DNA standards. The complete gel pieces are provided in Supplementary Fig. 6. d Confocal images of four strains Ec-pET29a/21a, Ec-AB34/m*, Ec-A*B34, and Ec-AB*34 using a Leica TCS SP8 STED 3× confocal laser scanning microscope. DAPI was used to stain E. coli DNA, and the ‘fire’ pseudocolor mode of ImageJ was applied to the DAPI images. BF, bright field. DAPI, mCherry and BF images are shown for the same field of cells. Scale bars = 2 μm.
To further characterize the assembled core nucleosomes in E. coli, we designed a set of experiments to visualize the nucleosome complexes using fluorescent protein-labeled histones. mCherry was either tethered to C-terminus of xenopus histones H2A (Ec-A*B34), H2B (Ec-AB*34), or independently expressed (Ec-AB34/m*) in E. coli cells (Fig. 1b). ecMNase assay showed that the fusion of mCherry to xenopus histones did not affect the assembly of nucleosome complexes, evidenced by the nucleosome-protected DNA fragments in both Ec-A*B34 and Ec-AB*34 (Fig. 1c). We next treated the different strains, Ec-pET29a/21a (negative control), Ec-AB34/m* (control with free-mCherry), Ec-A*B34 and Ec-AB*34, with IPTG induction and stained them with DAPI, before observing them using laser scanning confocal microscopy as previous described39. DAPI, a natural tracer of DNA, was commonly used to stain E. coli genome DNA and visualize E. coli nucleoid40. For the free-mCherry control strain, Ec-AB34/m*, the red fluorescence-protein was found to be broadly distributed within E. coli cells, while the blue fluorescence (DAPI stained-DNA) was concentrated to the restricted areas of nucleoid (Fig. 1d). In contrast, for strains Ec-A*B34 and Ec-AB*34, the red fluorescence (labeled core nucleosomes) and the blue fluorescence (stained genome DNA) were confined to the same restricted space of nucleoid, indicating co-localization of the core nucleosomes and chromosomal DNA in E. coli (Fig. 1d). The results provided corroborating evidence supporting the formation of nucleosome core complexes in E. coli. In addition, we noticed a comparably smaller nucleoid in strains Ec-AB34/m*, Ec-A*B34 and Ec-AB*34, than in Ec-pET29a/21a, which was further investigated next.
In vivo assembled core nucleosomes causing condensation of E. coli nucleoid
The formation of nucleosomes condensed DNA into a smaller volume, helping package chromosomal DNA into chromatin in eukaryotic cells3. Intrigued by the observation of smaller nucleoid in nucleosome-forming E. coli, we looked more closely into the changes to E. coli nucleoid induced by formation of nucleosomes. Under the confocal microscope, the DAPI-stained nucleoids in the nucleosome-forming strain, Ec-AB34, appeared shrunk than those in the control strain, Ec-pET29a, which indicated a more condensed nucleoid in the nucleosome-forming strain, Ec-AB34 (Fig. 2a).
a Confocal images of the nucleosome-forming strain (Ec-AB34) and the control strain (Ec-pET29a) using a Leica TCS SP8 STED 3× confocal laser scanning microscope. DAPI was used to stain E. coli DNA, and the ‘fire’ pseudocolor mode of ImageJ was applied to the DAPI images. BF, bright field. DAPI and BF images are shown for the same field of cells. Scale bars = 3 μm. b Thin-section transmission electron photomicrographs of the nucleosome-forming strain (Ec-AB34) and the control strain (Ec-pET29a) using a H7650 electron microscope. Scale bars = 500 nm.
We then used transmission electron microscopy (TEM) to examine the thin sections of E. coli cells from strains, Ec-AB34, and Ec-pET29a. For the control strain Ec-pET29a, the nucleoid exhibited a dispersed and lobular structure, scattered with granules of uneven size and density inside the nucleoid space. For the nucleosome-forming strain, Ec-AB34, the nucleoid appeared organized in more compacted structure with string-like substance intertwined together and tightly packed into a more restricted nucleoid space (Fig. 2b). It was known that nucleosomes themselves generate negative supercoiled DNA41, thus driving toward higher DNA superhelical density in the nucleosome-forming E. coli. The condensation of the E. coli nucleoids corroborated the formation of nucleosomes in E. coli, and also to certain degree, may reflect how genome DNA is constrained in the eukaryotic chromatin.
Subtracting each individual histone abolishing formation of nucleosome in E. coli
To investigate the dependency of nucleosome formation on different histone species, we designed a series of in vivo experiments by subtracting one histone species each time from the nucleosome constructs (Fig. 3a). mCherry was fused to the C-terminus of H3 for constructs Ec-B3*4, Ec-A3*4 and Ec-AB3*, and of H4 for Ec-AB4* in the case H3 was removed. The results showed the formation of nucleosomes was abolished, losing the characteristic 147-bp canonical octasome-protected DNA fragments, when one single core histone was subtracted each time (Fig. 3b). These results served as an important control, indicating all four histone proteins are essential for in vivo formation of the nucleosomes in E. coli. More interestingly, we observed some vague bands at a size around 70 bp in Ec-B3*4 and Ec-A3*4, but not in Ec-AB4* or Ec-AB3* cells (Fig. 3b), which were further explored.
a Strain constructs containing different histone components. The symbols ‘+’ and ‘−’ denote the presence and absence of corresponding histone gene, whereas ‘+m’ indicates the histone gene is fused with a mCherry gene. b DNA fragmentation profiles of six strains Ec-AB*34, Ec-pET29a/21a, Ec-B3*4, Ec-A3*4, Ec-AB4*, and Ec-AB3* using ecMNase assay. The green arrow marks the (H3-H4)2 tetrasome, while the red arrow marks mononucleosome particle. M, DNA standards. The complete gel pieces are provided in Supplementary Fig. 6. c Confocal images of five strains Ec-AB*34, Ec-pET29a/21a, Ec-B3*4, Ec-A3*4, Ec-AB4*, and Ec-AB3* using a Leica TCS SP8 STED 3× confocal laser scanning microscope. DAPI was used to stain E. coli DNA, and the ‘fire’ pseudocolor mode of ImageJ was applied to the DAPI images. BF bright field. DAPI mCherry and BF images are shown for the same field of cells. Scale bars = 2 μm.
The E. coli cells with different histone combinations, and Ec-AB*34 cells (positive control) were stained with DAPI and analyzed using Laser scanning confocal microscopy. We observed, however, two groups with distinct distribution patterns of blue fluorescence that represents DAPI-stained E. coli genome DNA. The blue fluorescence in Ec-AB4* and Ec-AB3* appeared evenly dispersed throughout the cells, in contrast to Ec-B3*4 and Ec-A3*4 that had blue fluorescence condensed into more restricted nucleoid space (Fig. 3c). The significant condensation of chromosomal DNA observed in Ec-B3*4 and Ec-A3*4 cells may be linked to the protected DNA fragments of ~70 bp from the MNase assay, which were present only in Ec-B3*4 and Ec-A3*4 cells (Fig. 3b).
For Ec-AB4* and Ec-AB3*, we observed that the red fluorescence from mCherry tethered to H3 or H4, had a dispersed distribution being inverse to the density of blue fluorescence representing DAPI-stained E. coli genome DNA (Fig. 3c). In these two cases, the labeled histone proteins not co-localizing with chromosomal DNA is consistent with the MNase-digestion assay results (Fig. 3b), which further confirmed the necessity of H3 and H4 for in vivo assembly of nucleosome in E. coli. On the other hand, for Ec-B3*4 and Ec-A3*4 in which condensed chromosomal DNA was observed, mCherry-labeled H3 was found co-localizing with chromosomal DNA in nucleoid (Fig. 3c). In conjunction with the protected DNA bands of ~70 bp in Ec-B3*4 and Ec-A3*4 (Fig. 3b), these data suggest the presence of a smaller-size subnucleosomal structure, distinctly different from the nucleosome that had the characteristic 147 bp DNA fragments from MNase digestion. This structure was found only in Ec-B3*4 and Ec-A3*4 cells, which suggested their shared histones, H3 and H4, are required for its formation. Conversely, Ec-AB4* and Ec-AB3* did not form this structure when either H3 or H4 was missing in the makeup. This smaller-size structure was found to have a profile closely resembling that of subnucleosomal particles previously reported as (H3-H4)2 tetrasomes that were reconstituted in vitro42,43.
Taking together, by subtracting one histone at a time we further proved the in vivo formation of eukaryotic nucleosomes in E. coli required all core histones. Intriguingly, through the process we observed, for the first time, the in vivo formation of stable subnucleosomal particles, the (H3-H4)2-like structure, in a bacterium, which caused condensation of chromosomal DNA in vivo, and protected genome DNA from in situ MNase digestion.
In vivo assembly of (H3-H4)-only tetrasomes and octasomes in E. coli
We were intrigued by the observation of in vivo formation of subnucleosomal structure in E. coli when either H2A or H2B was subtracted from the core nucleosome. To test our hypothesis that the subnucleosomal structure is (H3-H4) 2 tetrasome, we designed and generated three new constructs, Ec-3*4, Ec-3*, and Ec-4*, which expressed both H3 and H4, H3-only, or, H4-only, respectively (Fig. 4a). Note that mCherry was tethered to C-terminus of H3 for Ec-3*4 and Ec-3*, or that of H4 in case of Ec-4*.
a Strain constructs containing different histone components. The symbols ‘+’ and ‘−’ denote the presence and absence of corresponding histone gene, whereas ‘+m’ indicates the histone gene is fused with a mCherry gene. b DNA fragmentation profiles of five strains: Ec-pET29a/21a, Ec-AB*34, Ec-3*4, Ec-3*, Ec-4* using ecMNase assay. The green arrow marks the (H3-H4)2 tetrasome or (H3-H4)4 octasome, while the red arrow marks mononucleosome particle. M, DNA standards. The complete gel pieces are provided in Supplementary Fig. 6. c Confocal images of three strains Ec-3*4, Ec-3*, and Ec-4* using a Leica TCS SP8 STED 3× confocal laser scanning microscope. DAPI was used to stain E. coli DNA, and the ‘fire’ pseudocolor mode of ImageJ was applied to the DAPI images. BF, bright field. DAPI, mCherry and BF images are shown for the same field of cells. Scale bars = 2 μm.
Using optimized conditions for MNase digestion assay, we observed protected DNA bands at size of 70 bp and of ~130 bp in Ec-3*4, but not for Ec-3* or Ec-4* (Fig. 4b; Supplementary Fig. 4). These protected DNA fragments were notably different from those of the core nucleosomes (canonical octasomes) in Ec-AB*34. These results suggested that histones H3 and H4 were sufficient for formation of the subnucleosomal particles in vivo in E. coli. While the 70 bp protected DNA band were consistent with that of (H3-H4)2 tetrasome, the ~130 bp DNA fragments were consistent with that of (H3-H4)4 octasome27. The (H3-H4)4 octasomes were found to be wrapped by ∼130 bp of DNA segment, which were recently reported to assemble in vitro under a specified condition of high particle/DNA ratio26,27,44. So, using the specified conditions we were able to successfully assemble both the (H3-H4)2 tetrasomes and (H3-H4)4 octasomes in vivo in a bacterium.
We further investigated the in vivo distribution of (H3-H4)2 tetrasomes and (H3-H4)4 octasomes by staining with DAPI and analyzing with confocal microscopy. We found their formation in Ec-3*4 caused chromosomal DNA to condense in nucleoid, whereas in Ec-3* and Ec-4* genome DNA was more dispersed (Fig. 4c). Furthermore, there was distinct contrast of mCherry-labeled histones in their distribution. While in Ec-3*4 cells mCherry-labeled H3 was co-localized with blue fluorescence that represents DAPI-stained E. coli genome DNA, the mCherry-labeled histones had a distribution being inverse to the density of blue fluorescence in Ec-3* and Ec-4* cells (Fig. 4c).
Taking together, we for the first time assembled the (H3-H4)2 tetrasomes and (H3-H4)4 octasomes in vivo in a bacterium, corroborated by co-localization of these non-canonical nucleosome particles with the E. coli chromosomal DNA. Like the canonical core nucleosome, the (H3-H4)2 tetrasome and (H3-H4)4 octasome were capable of causing condensation of bacterial genome DNA in vivo, and protecting genome DNA from in situ MNase digestion.
In vivo assembly of complete eukaryotic nucleosomes with addition of histone H1
Histone H1 is known to bind to linker DNA between nucleosomes and promote compaction of nucleosome arrays into chromatin fibers, the higher-order chromosome structure5,45. H1 is associated with increased nucleosome repeat length and the compaction of chromatin and mitotic chromosomes in eukaryotes46,47. To attempt in vivo assembly of the complete eukaryotic nucleosome that included the canonical octasome and H1 histone, we generated the xenopus histone H1-expressing construct, pET21a-H1 (Fig. 5a). Histone H1 was co-expressed alongside H2A, H2B, H3, and H4, in strain Ec-AB341, and was evaluated by SDS-PAGE analysis. The expression of H1 as well as H2A, H2B, H3, and H4, was found primarily in the soluble fraction of the Ec-AB341 lysate (Fig. 5b). The expression level of H1 was lower than that of the core histones, as indicated by the molar ratio of H1 to H2A, which was approximately 0.74 (Methods).
a A schematic of polycistronic construct, pET21a-H1, for expression of xenopus histone H1 (B4) in E. coli. b Protein expression profiles in strains Ec-pET29a/21a and Ec-AB341. tag-H1, 31.00 kDa; tag-H2A, 18.15 kDa; H2B, 13.64 kDa; H3, 15.07 kDa; tag-H4, 13.31 kDa. c DNA fragmentation profiles of strains Ec-AB34 and Ec-AB341 using ecMNase assay. Arrows mark mono-, and di- nucleosome bands, respectively. M, DNA standards. The complete gel pieces are provided in Supplementary Fig. 6. d Length distribution of over-digestion ecMNase-seq reads from strains Ec-AB34 and Ec-AB341. Source data are provided as Supplementary Data 1. e Thin-section transmission electron photomicrographs of three strains: Ec-pET29a, Ec-AB34, and Ec-AB341 using a H7650 electron microscope. Scale bars = 500 nm.
To determine whether histone H1 can form the complete eukaryotic nucleosome with other histones in E. coli, we performed the ecMNase assay on Ec-AB341, using Ec-AB34 as control. As predicted, the protected DNA bands generated for mono-nucleosomes in Ec-AB341 had a larger size (~165 bp) than that of Ec-AB34 (~147 bp) (Fig. 5c; Supplementary Figs. 3, 5). In addition, the vague signal of di-nucleosomes in Ec-AB341 also appeared larger than that in Ec-AB34. To examine H1 histone-binding to space DNA and its association with the core-nucleosome, we performed over-digestion in ecMNase assay for both Ec-AB341 and Ec-AB34 (Methods). Ec-AB34 had 147 and 137 bp protected DNA bands, whereas Ec-AB341 exhibited 165 and 147 bp protected fragments (Supplementary Fig. 5). The size of the digested products for Ec-AB341 and Ec-AB34 was determined using ecMNase-seq analysis, which showed corresponding peaks (Fig. 5d). Notably, the 137 bp fragments in Ec-AB34 were similar to previously reported products from internal cleavage of core-nucleosomes48. The 165 bp fragments from Ec-AB341 represent the complete nucleosome-protected genome DNA49,50. The addition of H1 histone in Ec-AB341 appeared to make genome DNA more resistant to MNase cleavage than the core nucleosomes in Ec-AB34. Furthermore, we examine the cell thin sections from Ec-AB341, Ec-AB34, and Ec-pET29a using transmission electron microscopy (TEM), which showed an equally, if not more condensed nucleoid in Ec-AB341 than in Ec-AB34 (Fig. 5e). Given all the evidence from ecMNase, ecMNase-seq, and TEM, these results demonstrated the in vivo assembly of complete eukaryotic nucleosomes in E. coli with the addition of H1 histone.
Discussion
The eukaryotic nucleosome, a complex of eight histone proteins wrapped around with DNA, plays a central role in organizing and compacting genome DNA, as well as regulating genome access in eukaryotic cells. The nucleosome is a distinctive hallmark of eukaryotes in chromosome organization, distinguishing them from bacteria that lack these features. Since the discovery of the nucleosome in eukaryotic cells in 197451, many heterogeneous nucleosome variants, canonical and non-canonical, were also reported. Many belong to the transient states of nucleosome assembly or disassembly, which were only constituted in vitro under specified conditions. In this study, by developing an in vivo assembly system of the nucleosomes in a model bacterium E. coli that has not encountered the nucleosomes, we reconstituted and investigated the properties of the nucleosomes and its non-canonical variants in a ‘living’ bacterium. This ‘naive’ system may also facilitate study on the emergence of eukaryotic nucleosomes that is hypothesized in the theory of eukaryogenesis52. Remarkably, we accidentally assembled the (H3-H4)2 tetrasomes and (H3-H4)4 octasomes in E. coli, marking the in vivo reconstitution of these nucleosome variant structures in a living organism.
Our work showed the core nucleosomes assembled in E coli cells had many features resembling those found in their eukaryotic hosts. The mono-nucleosome bands had a length of ~147 bp when analyzed with ecMNase assay (Fig. 1a, Supplementary Fig. 2), identical to those of nucleosomes from eukaryotic hosts. On the other hand, the di-nucleosome bands are weak and smaller than 300 bp, below the fragment length of those native di-nucleosomes53,54. The weak intensity of the di-nucleosome bands suggests that only a portion of histones assembled into nucleosomes, given the short period allowed for nucleosome assembly after IPTG induction. The smaller size of the di-nucleosome fragments, below 300 bp, can be explained by the collapse of linker space between neighboring core-nucleosomes due to the lack of histone H1 in the E. coli system. In agreement, we later showed that histone H1 was associated with increased nucleosome repeat length in experiments when H1 was co-expressed alongside H2A, H2B, H3, and H4 - the mono-nucleosome repeat was extended by 18 bp to ~165 bp in ecMNase assay (Fig. 5c, d; Supplementary Figs. 3, 5). Notably, the 165 bp fragment length is consistent to the repeat length of complete nucleosomes in native eukaryotic hosts49,50.
To visualize the eukaryotic nucleosomes formed in E. coli cells, we took two technical approaches: 1) labeling histones with a fluorescent protein, namely mCherry, and observing them using laser scanning confocal microscopy; and 2) observing the nucleosomes in E. coli nucleoid using transmission electron microscopy. Compared to the control expressing free-moving mCherry that was broadly distributed within E. coli cells, the mCherry-labeled histones were co-localized with the bacterial chromosomal DNA in the nucleoids (Fig. 1d), corroborating the formation of nucleosome complexes in E. coli. Furthermore, the co-localization of mCherry-labeled histones and chromosomal DNA was abolished when either H3 or H4 was subtracted from the core nucleosome constructs (Fig. 3b). Furthermore, not unexpectedly, the formation of eukaryotic nucleosomes in vivo caused condensation of E. coli nucleoids, likely due to the chromosomal DNA condensed from formation of nucleosome complex, which was revealed in more details by transmission electron microscopy (TEM) (Figs. 1d and 2b). As a reference, it was estimated in previous studies the condensation ratio for genome DNA from formation of nucleosomes complexes was ~7-fold4,55. E. coli chromosome was organized into topologically independent domains and within each domain there exist large unfolded loops of DNA56,57,58. It was known nucleosomes themselves generate negative supercoiled DNA41,59, and the enhanced wrapping of supercoiled DNA around nucleosomes drove toward higher DNA superhelical density in the nucleosome-forming E. coli, resulting in condensed nucleoids. The nucleosome-forming E. coli maintaining a folding structure of high superhelical density, to certain degree, may reflect how the genome DNA is constrained in eukaryotic chromatin.
In eukaryotes, the (H3-H4)2 tetrasome is an initial stage in nucleosome assembly, followed by deposition of two H2A-H2B dimers individually in a process mediated by histone chaperones in vivo14,60. However, the transient stage was only stably observed in in vitro studies, where the (H3-H4)2 tetrasomes were associated with ∼70 bp DNA segment. The non-canonical (H3-H4)4 octasomes were only found by in vitro experiments, wrapped in ∼130 bp DNA with a lefthanded ramp of 1.5 turns27,43. An AFM study showed the (H3-H4)2 tetrasomes had a hairpin-like appearance, whereas the (H3-H4)4 octasomes exhibited a round-shaped structure26,44. Notably, both the (H3-H4)2 tetrasomes and (H3-H4)4 octasomes were only reconstituted in vitro under specialized conditions. The former formed at low histone protein/DNA ratio (1:1) via continuous salt-gradient dialysis, whereas the latter was obtained at higher histone protein/DNA ratio (2.2:1)27,61. We completed the assembly of (H3-H4)2 tetrasomes and (H3-H4)4 octasomes for the first time in a bacterium, indicating that stable (H3-H4)2 tetrasomes and (H3-H4)4 octasomes can exit in vivo. In presence of H2A-H2B dimer molecules, assembly moved forwards canonical octasome stage with fewer (H3-H4)2 tetrasomes remaining, whereas the assembly was largely stalled at (H3-H4)2 tetrasome stage when either H2A or H2B was subtracted (Fig. 3b). The results indicated that the (H3-H4)4 octasomes cannot compete with the canonical octasomes, which is illustrated in a tilted balance model (Fig. 6).
The dynamic process of nucleosome assembly is initiated with action of (H3-H4)2-tetramer being deposited onto a DNA template, creating a tetrasome platform that resembles their archaeal ancestors. The subsequent loading of H2A-H2B dimers leads to formation of the canonical octasome (eukaryotic core nucleosome). In the absence of H2A-H2B dimer, the non-canonical (H3-H4)4 octasomes may form, which represent a ‘fossil complex’ that marks the intermediate before emergence of the eukaryotic nucleosomes. The H2A-H2B dimers showed a greater tendency to bind (H3-H4)2 tetrasomes to form the eukaryotic core nucleosome, tilting evolution to eukaryotic structures.
It is known that archaeal histone-like proteins had the capacity to form multimeric particles in archaea28,62. As histones H3 and H4 share structure-fold with archaeal histone-like proteins, the (H3-H4)2 tetrasomes and (H3-H4)4 octasomes are likely inherited multimeric structures from their archaeal ancestors, which as ‘fossil complex’, possibly mark the intermediates in the progressive development of the eukaryotic nucleosomes. The fossil complex structures would dissipate when new partners, i.e., H2A-H2B dimers, emerged in evolution -- our study thus may have visited the early stage of eukaryogenesis (Fig. 6). Notably, previous study found the presence of H2A-H2B dimers acted as a molecular ‘cap’ by binding to the so-called sticky regions on either side of the (H3-H4)-tetramer, which prevented their self-aggregation from growing into larger complexes63. The observed properties of H2A-H2B dimers were important for tilting balance for evolution towards eukaryotic nucleosome structures. Also in the process, eukaryotic histones evolved extended terminal tails and posttranslational modifications, and new capability for histone-based epigenetic mechanisms. Whether the fossil complex, (H3-H4)4 octasome, exists naturally in eukaryotic cells has not been definitively answered by our results or the previous study in yeast cells27, and needs to be further explored.
In summary, we have developed an in vivo assembly system of eukaryotic nucleosomes in a model bacterium, E. coli. We successfully reconstituted in vivo in a bacterial system, the nucleosome, and its non-canonical variants, like (H3-H4)2 tetrasomes and (H3-H4)4 octasomes. These non-canonical nucleosomes, the so called ‘fossil complex’, likely inherit the multimeric structures from their ancestors. The in vivo formation of the different nucleosome structures in a bacterium supports the in vitro reconstitution results, and also provides a unique platform for studying the properties of the nucleosome, and its non-canonical variants. This synthetic biology approach to assemble the nucleosome and its variants in a ‘naive’ system may also present a opportunity to enhance our understanding on the important question of how the eukaryotic nucleosomes evolved through the process of eukaryogenesis.
Methods
Plasmids and histone-expression constructs
The plasmids and construction of various histone-expression vectors are as described (Supplementary Table 1; Supplementary Methods). The oligonucleotides used in vector construction are listed in Supplementary Table 3. Polymerase chain reaction (PCR) was performed using Taq (Thermo Fisher Scientific) or KOD FX DNA polymerase (TOYOBO). Plasmids and chromosomal DNA were extracted using the Plasmid Mini Kit I and Gel Extraction Kit from OMEGA. Cloning was performed using either restriction endonucleases and T4 DNA ligase (New England Biolabs) or the ClonExpress® II One Step Cloning Kit (Vazyme). E. coli strain DH5α was used for the purpose of molecular cloning.
Strains and expression of histones in bacterial culture
E. coli strain Rossetta (DE3) was transformed via heat-shocked method to construct various histone-expressing strains in this study (Supplementary Table 2). All strains were grown in LB medium at 37 °C with agitation (220 rpm) unless otherwise indicated. For induced histone expression, a single colony of histone-expression strains was inoculated into 3 mL LB media containing appropriate antibiotics, i.e., kanamycin (50 µg/mL), chloramphenicol (25 µg/mL) or carbenicillin (50 µg/mL). This starter culture was grown overnight at 37 °C, 220 rpm before being amplified to 20 mL culture of the same media and grown at 37 °C, 220 rpm, to exponential phase (OD600 = 0.6). Then, the expression of various histones was induced with addition of 400 µM IPTG and harvested for assay when the OD600 reached 1.2.
SDS-PAGE analysis of histone protein expression
A volume of 16.7 mL E. coli culture (OD600 ~ 1.2) after induction with IPTG were harvested by centrifugation (5000 rpm for 5 min at 4 °C). The cells were re-suspended in 2 mL lysis buffer (50 mM Tris-HCl, pH 8.0, 500 mM NaCl, 10 mM 2-mercaptoethanol, 5% glycerol and 0.1 mM proteinase inhibitor (PMSF)). Cells were lysed, and genomic DNA was fragmented to obtain soluble fractions using a Scientz sonication system (Ningbo Scientz Biotechnology CO. LTD, Ningbo, China) in an ice-water bath. For total protein analysis, 80 µL of total cell lysate were mixed with 20 µL 5× SDS-PAGE Protein Loading Buffer.
100 µL of total cell lysate were centrifuged at 15,000 rpm for 15 min at 4 °C to separate soluble proteins from cell pellet. The supernatant was treated with 20 µL 5× SDS-PAGE Protein Loading Buffer (#20315ES05, Yeasen, Shanghai). The pellet was suspended in 80 µL lysis buffer and treated with 20 µL 5× SDS-PAGE Protein Loading Buffer.
Samples for supernatant, pellet and total lysate (representing roughly 1.0 OD600 bacterial culture) were incubated at 95 °C for 10 min before centrifuged at 15,000 rpm for 30 min at 4 °C. Samples were loaded on SDS-PAGE (15% Bis-Tris) for electrophoresis, and protein contents were revealed using Coomassie Blue Fast Staining Solution (P0017, Beyotime, Shanghai).
We used molar ratio of H1 to H2A to characterize the relative expression levels of histone H1 in comparison to core histones. To calculate the molar ratio of H1 to H2A, we used the formula (below):
The grayscale values of the protein bands corresponding to histone H1 and H2A on SDS-PAGE gel depicted in Fig. 5b were measured using ImageJ software.
In situ micrococcal nuclease digestion assay for E. coli (ecMNase)
The micrococcal nuclease (MNase) digestion of genome DNA was previously used to analyze nucleosome formation in eukaryotes, which displayed a unique pattern of genomic DNA fragmentation due to nucleosome protection35,36. To make it work with the nucleosome-forming E. coli cells, we developed the in situ MNase digestion assay to work with E. coli cells (ecMNase) by generating protoplasts from bacterial cells and applying MNase to them (Supplementary Fig. 1d). A volume of 16.7 mL bacterial culture (OD600 ~ 1.2) were harvested by centrifugation (5000 rpm for 5 min at 4 °C). The pelleted cells were washed with 1 mL chilled buffer B (25 mM Tris-HCl pH 8.0, and 456 mM sucrose). Following centrifugation at 5000 rpm for 5 min at 4 °C, bacterial cells were incubated in 2 mL buffer A (25 mM Tris-HCl pH 8.0, 456 mM sucrose, 0.042% EDTA pH 7.0, and 0.21 mg/mL lysozyme) for 17 min at 25 °C to generate protoplasts. The protoplasts were pelleted (5000 rpm for 5 min at 4 °C) and washed with buffer B for three times. Then the protoplasts were re-suspended in 2.0 mL MNase buffer (1.0 M sorbitol, 50 mM NaCl, 1 mM CaCl2, 10 mM Tris-HCl pH 7.4, 5.0 mM MgCl2, 1.0 mM beta-mercaptoethanol, 0.5 mM spermidine and 0.1% Nonidet NP-40). Aliquots of 250 µL protoplast suspension (representing 2.5 OD of bacterial culture) were treated with 200 unit/mL MNase (N863776-10KU, Maklin, Shanghai) at 37 °C for different length of time, before reactions were stopped by addition of 12.5 µL 0.5 M EDTA to each reaction. For prolonged MNase digestion, 280, 320, 360 and 400 unit/mL MNase were used, respectively, to treat protoplast suspension for 12 min at 37 °C. Each aliquot was further treated with 0.5% SDS, 100 µg/mL ribonuclease A, and 200 µg/mL proteinase K at 37 °C for 1 h, before DNA was precipitated with ethanol and re-suspended in 20 µL distilled water. The DNA fragmentation profiles were assessed by electrophoresis (2% agarose gel in 1× TAE). For the prolonged MNase digestion assay, the DNA fragments were analyzed with 6% agarose gel in 1× TAE (Supplementary Fig. 5).
Fluorescence imaging analysis of various nucleosome-forming bacteria
To visualize the nucleosome complex formation in different strains, 1.43 mL of bacterial cultures (OD600 ~ 1.2) treated IPTG were harvested by centrifugation (5000 rpm for 3 min at 4 °C). Pellets were washed with PBS buffer (0.137 M Sodium chloride, 0.0027 M Potassium Chloride, 0.01 M Sodium Phosphate Dibasic and 0.0018 M Potassium Phosphate Monobasic, pH 7.4), and stained with 50 µg/mL DAPI (4’,6-diamidino-2-phenylindole, Sigma) for 10 minutes on ice. 1 µL of stained cell- suspension was spread onto pre-prepared 1% agarose pad on slides. Cells were visualized and images were taken using Leica TCS SP8 STED 3× confocal laser scanning microscope with a HC PL APO CS2 100×/1.30 oil objective and a HyD1 detector. The excitation and emission wavelengths were 405 nm and 430–480 nm for DAPI, and were 561 nm and 592-671 nm for mCherrry, respectively. The bright field (BF) was visualized with the PMT detector. Images were analyzed using the Leica Application suite X and ImageJ 1.53e.
Transmission electron microscope (TEM) imaging analysis of various nucleosome-forming bacteria
A volume of 83 mL bacterial culture (OD600 ~ 1.2) treated with IPTG were harvested by centrifugation (5000 rpm for 5 min at 4 °C). The pellets were washed twice with the glutaraldehyde solution (2.5% [vol/vol] in 0.1 M phosphate buffer, pH 7.2) and fixed with the glutaraldehyde solution for 24 h at 4 °C. Fixed cells were washed with 0.1 M phosphate buffered saline (PBS, pH 7.2) several times and then postfixed in 1% osmium tetroxide in 0.1 M PBS for 2 h at 4 °C. Next, the cells were dehydrated in a series of ethanol solution (30, 50, 70, 80, 90, 95, and 100%) for 15 min each, and then 100% acetone (30 min each; 3 times). For resin penetration, dehydrated cells were incubated in a mixture of acetone and epoxy resin (3:1) for 2 h, of an equal volume of acetone and resin (1:1) for 2 h, and of acetone and epoxy resin (1:3) for 2 h, before treated with pure epoxy resin 3 times for 8 h each. Finally, the samples were embedded in Epon 812 resin and allowed to polymerize at 35 °C and 45 °C for 12 h successively. The epoxy resin-embedded cells were cured in a 60 °C oven for 2 days. The cured blocks were cut on an ultramicrotome with glass knives, and 70 nm-thick sections were placed on copper grids and stained with uranyl acetate and lead citrate solution. The sections were observed and imaged with a H7650 electron microscope (HITACHI, Tokyo) operated at 80 kv.
ecMNase sequencing (ecMNase-seq) of the nucleosome-forming E. coli
A volume of 16.7 mL bacterial culture (OD600 ~ 1.2) were harvested and treated with ecMNase assay using the digestion conditions as described above. Gel slices containing the mono-nucleosome products (approximately 100 to 180 bp) were obtained, and DNA fragments were extracted for sequencing using Illumina Hiseq X Ten instrument according to manufacturer’s instructions (Illumina, San Diego, CA). Note a reduced number of PCR cycles (~4 cycles) were used for library construction to ensure minimum redundancy of ecMNase-seq data. Illumina sequencing was performed by Genewiz (Suzhou, China), using a 2 × 150 paired-end (PE) configuration. Paired-end MNase-seq reads were first trimmed for adapter sequences using Cutadapt (version 2.7)64 (parameters: -a AGATCGGAAGAGCACACGTCTGAACTCCAGTCAC, -A AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGT), and then merged using BBmerge (version 38.26)65 with default parameters. The merged reads were analyzed for length distribution using R.
DNA size detection using Agilent 2100
To analyze the size of the DNA fragments produced from stains Ec-AB34, Ec-AB341, and Ec-34* in the ecMNase experiments, the samples was detected using the Agilent directly to capillary electrophoresis analysis using Agilent 2100 bioanalyzer and DNA10000 kit without running agarose gel first according to the protocals, performed by Genewiz (Suzhou, China).
Statistics and reproducibility
For growth profiles, we performed 4 replicates of curves to ensure the reproducibility of the results, and the data were presented as mean ± sem.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
MNase-seq data are available on NCBI under BioProject ID PRJNA1088616. Source data are provided in this paper. Datasets used and analyzed are available in Supplementary Data 1. All the original gels images are included as Supplementary Fig. 6. in the Supplementary Information. All other data will be available from the corresponding authors upon reasonable request.
References
Luger, K., Mäder, A. W., Richmond, R. K., Sargent, D. F. & Richmond, T. J. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389, 251–260 (1997).
Richmond, T. J., Finch, J. T., Rushton, B., Rhodes, D. & Klug, A. Structure of the nucleosome core particle at 7 A resolution. Nature 311, 532–537 (1984).
Annunziato, A. DNA packaging: nucleosomes and chromatin. Nat. Educ. 1, 26 (2008).
Cutter, A. R. & Hayes, J. J. A brief review of nucleosome structure. FEBS Lett. 589, 2914–2922 (2015).
Fan, Y. et al. Histone H1 depletion in mammals alters global chromatin structure but causes specific changes in gene regulation. Cell 123, 1199–1212 (2005).
Allan, J., Mitchell, T., Harborne, N., Bohm, L. & Crane-Robinson, C. Roles of H1 domains in determining higher order chromatin structure and H1 location. J. Mol. Biol. 187, 591–601 (1986).
Hansen, J. C. Conformational dynamics of the chromatin fiber in solution: determinants, mechanisms, and functions. Annu Rev. Biophys. Biomol. Struct. 31, 361–392 (2002).
Klemm, S. L., Shipony, Z. & Greenleaf, W. J. Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019).
Kornberg, R. D. & Lorch, Y. Primary Role of the Nucleosome. Mol. Cell 79, 371–375 (2020).
Millán-Zambrano, G., Burton, A., Bannister, A. J. & Schneider, R. Histone post-translational modifications - cause and consequence of genome function. Nat. Rev. Genet. 23, 563–580 (2022).
Kurumizaka, H., Kujirai, T. & Takizawa, Y. Contributions of Histone Variants in Nucleosome Structure and Function. J. Mol. Biol. 433, 166678 (2021).
Tropberger, P. et al. Regulation of transcription through acetylation of H3K122 on the lateral surface of the histone octamer. Cell 152, 859–872 (2013).
Venkatesh, S. & Workman, J. L. Histone exchange, chromatin structure and the regulation of transcription. Nat. Rev. Mol. Cell Biol. 16, 178–189 (2015).
Verreault, A. De novo nucleosome assembly: new pieces in an old puzzle. Genes. Dev. 14, 1430–1438 (2000).
Mello, J. A. & Almouzni, G. The ins and outs of nucleosome assembly. Curr. Opin. Genet. Dev. 11, 136–141 (2001).
Kireeva, M. L. et al. Nucleosome remodeling induced by RNA polymerase II: loss of the H2A/H2B dimer during transcription. Mol. Cell 9, 541–552 (2002).
Bina-Stein, M. & Simpson, R. T. Specific folding and contraction of DNA by histones H3 and H4. Cell 11, 609–618 (1977).
Ruiz-Carrillo, A. & Jorcano, J. L. An octamer of core histones in solution: central role of the H3-H4 tetramer in the self-assembly. Biochemistry 18, 760–768 (1979).
Arimura, Y., Tachiwana, H., Oda, T., Sato, M. & Kurumizaka, H. Structural analysis of the hexasome, lacking one histone H2A/H2B dimer from the conventional nucleosome. Biochemistry 51, 3302–3309 (2012).
Ehara, H., Kujirai, T., Shirouzu, M., Kurumizaka, H. & Sekine, S. I. Structural basis of nucleosome disassembly and reassembly by RNAPII elongation complex with FACT. Science 377, eabp9466 (2022).
Kato, D. et al. Crystal structure of the overlapping dinucleosome composed of hexasome and octasome. Science 356, 205–208 (2017).
Matsumoto, A. et al. Structural Studies of Overlapping Dinucleosomes in Solution. Biophys. J. 118, 2209–2219 (2020).
Kulaeva, O. I., Hsieh, F. K. & Studitsky, V. M. RNA polymerase complexes cooperate to relieve the nucleosomal barrier and evict histones. Proc. Natl Acad. Sci. USA 107, 11325–11330 (2010).
Moss, T., Stephens, R. M., Crane-Robinson, C. & Bradbury, E. M. A nucleosome-like structure containing DNA and the arginine-rich histones H3 and H4. Nucleic Acids Res. 4, 2477–2486 (1977).
Simon, R. H., Camerini-Otero, R. D. & Felsenfeld, G. An octamer of histones H3 and H4 forms a compact complex with DNA of nucleosome size. Nucleic Acids Res. 5, 4805–4818 (1978).
Zou, T. et al. Direct Observation of H3-H4 Octasome by High-Speed AFM. Chemistry 24, 15998–16002 (2018).
Nozawa, K. et al. Cryo-electron microscopy structure of the H3-H4 octasome: A nucleosome-like particle without histones H2A and H2B. Proc. Natl Acad. Sci. USA 119, e2206542119 (2022).
Mattiroli, F. et al. Structure of histone-based chromatin in Archaea. Science 357, 609–612 (2017).
Henneman, B., van Emmerik, C., van Ingen, H. & Dame, R. T. Structure and function of archaeal histones. PLoS Genet. 14, e1007582 (2018).
Talbert, P. B., Meers, M. P. & Henikoff, S. Old cogs, new tricks: the evolution of gene expression in a chromatin context. Nat. Rev. Genet. 20, 283–297 (2019).
Bowerman, S., Wereszczynski, J. & Luger, K. Archaeal chromatin ‘slinkies’ are inherently dynamic complexes with deflected DNA wrapping pathways. Elife 10, e65587 (2021).
Rojec, M., Hocher, A., Stevens, K. M., Merkenschlager, M. & Warnecke, T. Chromatinization of Escherichia coli with archaeal histones. Elife 8, e49038 (2019).
Shim, Y., Duan, M. R., Chen, X., Smerdon, M. J. & Min, J. H. Polycistronic coexpression and nondenaturing purification of histone octamers. Anal. Biochem 427, 190–192 (2012).
Bell, O., Tiwari, V. K., Thomä, N. H. & Schübeler, D. Determinants and dynamics of genome accessibility. Nat. Rev. Genet. 12, 554–564 (2011).
Wei, G., Hu, G., Cui, K. & Zhao, K. Genome-wide mapping of nucleosome occupancy, histone modifications, and gene expression using next-generation sequencing technology. Methods Enzymol. 513, 297–313 (2012).
Mieczkowski, J. et al. MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016).
Schwartz, U. et al. Characterizing the nuclease accessibility of DNA in human cells to map higher order structures of chromatin. Nucleic Acids Res. 47, 1239–1254 (2019).
Chereji, R. V. et al. Genome-wide profiling of nucleosome sensitivity and chromatin accessibility in Drosophila melanogaster. Nucleic Acids Res. 44, 1036–1051 (2016).
Bosco, N. et al. KaryoCreate: A CRISPR-based technology to study chromosome-specific aneuploidy by targeting human centromeres. Cell 186, 1985–2001.e1919 (2023).
Wery, M., Woldringh, C. L. & Rouviere-Yaniv, J. HU-GFP and DAPI co-localize on the Escherichia coli nucleoid. Biochimie 83, 193–200 (2001).
Arents, G., Burlingame, R. W., Wang, B. C., Love, W. E. & Moudrianakis, E. N. The nucleosomal core histone octamer at 3.1 A resolution: a tripartite protein assembly and a left-handed superhelix. Proc. Natl Acad. Sci. USA 88, 10148–10152 (1991).
Oudet, P. et al. Nucleosome structure I: all four histones, H2A, H2B, H3, and H4, are required to form a nucleosome, but an H3-H4 subnucleosomal particle is formed with H3-H4 alone. Cold Spring Harb. Symp. Quant. Biol. 42, 287–300 (1978).
Bowman, A., Ward, R., El-Mkami, H., Owen-Hughes, T. & Norman, D. G. Probing the (H3-H4)2 histone tetramer structure using pulsed EPR spectroscopy combined with site-directed spin labelling. Nucleic Acids Res. 38, 695–707 (2010).
Lavelle, C. & Prunell, A. Chromatin polymorphism and the nucleosome superfamily: a genealogy. Cell Cycle 6, 2113–2119 (2007).
Thoma, F., Koller, T. & Klug, A. Involvement of histone H1 in the organization of the nucleosome and of the salt-dependent superstructures of chromatin. J. Cell Biol. 83, 403–427 (1979).
Freedman, B. S. & Heald, R. Functional comparison of H1 histones in Xenopus reveals isoform-specific regulation by Cdk1 and RanGTP. Curr. Biol. 20, 1048–1052 (2010).
Happel, N. & Doenecke, D. Histone H1 and its isoforms: contribution to chromatin structure and function. Gene 431, 1–12 (2009).
Cole, H. A., Ocampo, J., Iben, J. R., Chereji, R. V. & Clark, D. J. Heavy transcription of yeast genes correlates with differential loss of histone H2B relative to H4 and queued RNA polymerases. Nucleic Acids Res. 42, 12512–12522 (2014).
Yuan, G. C. et al. Genome-scale identification of nucleosome positions in S. cerevisiae. Science 309, 626–630 (2005).
Cole, H. A. et al. Novel nucleosomal particles containing core histones and linker DNA but no histone H1. Nucleic Acids Res. 44, 573–581 (2016).
Olins, A. L. & Olins, D. E. Spheroid chromatin units (v bodies). Science 183, 330–332 (1974).
Brunk, C. F. & Martin, W. F. Archaeal Histone Contributions to the Origin of Eukaryotes. Trends Microbiol. 27, 703–714 (2019).
Kent, N. A., Adams, S., Moorhouse, A. & Paszkiewicz, K. Chromatin particle spectrum analysis: a method for comparative chromatin structure analysis using paired-end mode next-generation DNA sequencing. Nucleic Acids Res. 39, e26 (2011).
Truong, D. M. & Boeke, J. D. Resetting the Yeast Epigenome with Human Nucleosomes. Cell 171, 1508–1519.e1513 (2017).
Gross, D. S., Chowdhary, S., Anandhakumar, J. & Kainth, A. S. Chromatin. Curr. Biol. 25, R1158–R1163 (2015).
Worcel, A. & Burgi, E. On the structure of the folded chromosome of Escherichia coli. J. Mol. Biol. 71, 127–147 (1972).
Sinden, R. R. & Pettijohn, D. E. Chromosomes in living Escherichia coli cells are segregated into domains of supercoiling. Proc. Natl Acad. Sci. USA 78, 224–228 (1981).
Griffith, J. D. Visualization of prokaryotic DNA in a regularly condensed chromatin-like fiber. Proc. Natl Acad. Sci. USA 73, 563–567 (1976).
Baranello, L., Levens, D., Gupta, A. & Kouzine, F. The importance of being supercoiled: how DNA mechanics regulate dynamic processes. Biochim Biophys. Acta 1819, 632–638 (2012).
Böhm, V. et al. Nucleosome accessibility governed by the dimer/tetramer interface. Nucleic Acids Res. 39, 3093–3102 (2011).
Dyer, P. N. et al. Reconstitution of nucleosome core particles from recombinant histones and DNA. Methods Enzymol. 375, 23–44 (2004).
Pereira, S. L., Grayling, R. A., Lurz, R. & Reeve, J. N. Archaeal nucleosomes. Proc. Natl Acad. Sci. USA 94, 12633–12637 (1997).
Baxevanis, A. D., Godfrey, J. E. & Moudrianakis, E. N. Associative behavior of the histone (H3-H4)2 tetramer: dependence on ionic environment. Biochemistry 30, 8817–8823 (1991).
Kechin, A., Boyarskikh, U., Kel, A. & Filipenko, M. cutPrimers: A New Tool for Accurate Cutting of Primers from Reads of Targeted Next Generation Sequencing. J. Comput Biol. 24, 1138–1143 (2017).
Bushnell, B., Rood, J. & Singer, E. BBMerge - Accurate paired shotgun read merging via overlap. PLoS One 12, e0185056 (2017).
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China (32393971, 92451303, 32270719, and 32200093), and the Strategic Projects of Chinese Academy of Sciences (KFJ-BRP-009-002). We thank Wenjuan Cai at the core facility of the Center for Excellence in Molecular Plant Sciences (CEMPS) for assistance with confocal microscopy imaging experiments.
Author information
Authors and Affiliations
Contributions
X.L. and X.J. conceived the project, and wrote the manuscript. X.Z., X.J., N.Z., J.G., K.Z., P.C., and X.C. performed experiments, analyzed data, and/or prepared the manuscript. G.Z., and B.C.Y. advised on the study, and/or helped with data analysis.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Jeffrey Hayes, Yoshifumi Nishimura and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Joanna Timmins and David Favero.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhou, X., Zhang, N., Gong, J. et al. In vivo assembly of complete eukaryotic nucleosomes and (H3-H4)-only non-canonical nucleosomal particles in the model bacterium Escherichia coli. Commun Biol 7, 1510 (2024). https://doi.org/10.1038/s42003-024-07211-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-024-07211-4