Abstract
Hematologic adverse events are common dose-limiting toxicities in drug development. Classical animal models for preclinical safety assessment of immunotherapies are often limited due to insufficient cross-reactivity with non-human homologous proteins, immune system differences, and ethical considerations. Therefore, we evaluate a human bone marrow (BM) microphysiological system (MPS) for its ability to predict expected hematopoietic liabilities of immunotherapeutics. The BM-MPS consists of a closed microfluidic circuit containing a ceramic scaffold covered with human mesenchymal stromal cells and populated with human BM-derived CD34+ cells in chemically defined growth factor-enriched media. The model supports on-chip differentiation of erythroid, myeloid and NK cells from CD34+ cells over 31 days. The hematopoietic lineage balance and output is responsive to pro-inflammatory factors and cytokines. Treatment with a transferrin receptor-targeting IgG1 antibody results in inhibition of on-chip erythropoiesis. The immunocompetence of the chip is established by the addition of peripheral blood T cells in a fully autologous setup. Treatment with T cell bispecific antibodies induces T cell activation and target cell killing consistent with expected on-target off-tumor toxicities. In conclusion, this study provides a proof-of-concept that this BM-MPS is applicable for in vitro hematopoietic safety profiling of immunotherapeutics.
Similar content being viewed by others
Introduction
In recent decades, drug development for many diseases has evolved from small molecules to biologics. Biologics include various forms of recombinant proteins, including monoclonal antibodies, cytokines and hormones. The market share of biologics of overall global pharmaceutical sales has grown from 18% in 2010 to over 30% in 2021 reaching a total market share of $343 billion in 20211. Biologics target a wide range of diseases, including cancer (solid tumors and hematological malignancies), autoimmune and inflammatory conditions (e.g., rheumatoid arthritis, psoriasis), infectious diseases (e.g., COVID-19, Ebola, anthrax), and neurological disorders (e.g., Alzheimer’s). They also address metabolic and endocrine disorders such as diabetes, rare genetic diseases like β-thalassemia and spinal muscular atrophy, hematological disorders (e.g., neutropenia and anemia), and cardiovascular diseases1.
Although biologics tend to be more target-specific than small molecules, they can still cause adverse drug reactions (ADRs). These ADRs include hematologic toxicities such as thrombocytopenia, anemia, neutropenia, and monocytopenia2, as described for antibody drug conjugates, such as Gemtuzumab ozogamicin that frequently induces severe thrombocytopenia by targeting CD33-positive cells and impacting bone marrow progenitors3 and immune checkpoint inhibitors, such as Magrolimab, an anti-CD47 antibody, that has been associated with moderate to severe anemia caused by hemophagocytosis triggered through its binding to CD47 on red blood cells4. The underlying mechanisms of these ADRs are often complex and may include unexpected on- and off-target effects, immune responses and the induction of local or systemic cytokine release. Complex human microphysiological systems (MPS) to predict these toxicities are being developed because they offer intriguing opportunities to solve challenges of classical animal models such as (i) providing human-relevant data, aiming at improving the translational accuracy of preclinical predictions, (ii) providing mechanistic insights through detailed studies at the cellular and molecular levels, and (iii) reducing animal use by providing early dose-response data, addressing ethical concerns and resource limitations. By leveraging these advantages, in vitro models can streamline the dose-finding process, making preclinical drug development more efficient and ethically responsible. An overview of MPS modelling the human bone marrow can be found in the review by Rosalem et al.5. However, the majority of these models are not suitable for modeling large molecule-induced hematologic ADRs because they are either based on animal cells6,7, lack the hematopoietic component8, mimic only a specific pathology9,10,11,12, or include only single cell types such as megakaryocytes13.
Recent examples of human MPS bone marrow models show promise for overcoming these limitations and allowing the study of hematologic ADRs in vitro14,15,16. Nelson et al. and Glaser et al. developed co-culture systems of osteoblasts, mesenchymal stromal cells (MSCs), CD34+ hematopoietic stem and progenitor cells (HSPCs), and endothelial cells to mimic the endosteal and perivascular niche in vitro. Glaser et al. used doxorubicin and G-CSF and showed decreased and increased release of granulocytes from the model, respectively. Chou et al. developed a co-culture system of MSC, HSPCs and endothelial cells that was cultured for 28 days. Drug exposure was performed with an Aurora B kinase inhibitor and 5-FU to induce myelosuppression.
While those studies have demonstrated the potential of microphysiological systems (MPSs) to model hematopoiesis, biologics have not been tested in those systems and they were lacking features such as fully autologous immunocompetence or robust multilineage hematopoietic differentiation, which are required for the safety evaluation of biologics.
The HUMIMIC Chip2 bone marrow model was originally developed for the long-term in vitro culture of human hematopoietic stem cells17 and was subsequently applied for lineage-specific toxicity testing of small molecules18. Here, we further developed and characterized the model, providing:
-
1.
A fully autologous setup, incorporating MSCs, CD34+ hematopoietic progenitors, and peripheral blood T cells.
-
2.
Support for multilineage differentiation (erythroid, myeloid, NK cells) over a prolonged culture period (31 days).
-
3.
The ability to model immune interactions and hematopoietic imbalances induced by immunotherapies.
The aim of this study was the development of a human bone marrow microphysiological system suitable for the assessment and investigation of hematopoietic adverse events induced by immunotherapeutics. By addressing the clinical need for reliable preclinical safety assessment tools, this study seeks to establish the BM-MPS as a robust platform for hematopoietic safety profiling in drug development.
Results
Stromal compartment maintained osteogenic phenotype during chip culture
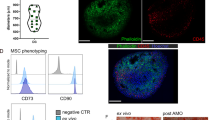
An in vitro model of human bone marrow in the HUMIMIC Chip2 system (Fig. 1A) was established based on the model previously published by Sieber et al.17. A zirconium oxide scaffold was seeded with 500,000 primary human bone marrow-derived MSCs and 40,000 primary human bone marrow-derived CD34+ HSPCs (Fig. 1B). The primary human BM-derived MSCs were expanded for 7 to 15 days under animal-content-free 2D culture conditions, then seeded onto the ceramics and cultured for 10 days under static conditions before HSPCs were seeded into the scaffold, the scaffold was transferred into the chip and the dynamic on-chip co-culture was conducted for up to 31 days (Fig. 1B).
A Exploded view of the HUMIMIC Chip2 showing the three structural layers. B 2D view of the HUMIMIC Chip2 microfluidics: the outer compartment houses the bone marrow model; the inner compartment is used as a medium reservoir and as a sampling point for hematopoietic cells. C Calcein-AM RedOrange staining (red) of vital mesenchymal stromal cells populating the ceramic, scale is 250 µm. D The top 20 enriched GO BP terms in MSC samples from static and dynamic cultivation in MSC and HSC cultivation medium. Only GO BP terms from a list of GO biological processes significantly enriched among osteocyte transcriptome signature genes according to Youlten et al.21 were considered. E Dimensionality reduction of the gene expression profiles using PCA. The PCA is computed based on the top 500 most variable genes using TMM-normalized logCPM. Arrows indicate time course of the experiment. F GO biological processes enrichment analysis for differential expressed genes in different MSC culture conditions. Signatures correspond to rows and indications/contrasts to columns. The color shows the median log2 fold-changes of the genes in the signatures. Significance level is indicated with asterisks: *** 1% FDR, ** 5% FDR, *10% FDR.
After ten days of pre-culture, MSCs formed an interconnected, viable cellular network that completely covered the visible ceramic scaffold surface and spanned the pores of the spongy scaffold structure (Fig. 1C). Previous work indicated that the culture on the zirconium oxide scaffold induced osteogenesis and upregulation of HSC niche factors in MSCs, which was stable over four weeks in an HSPC medium containing thrombopoietin (TPO) and FLT-3L, supporting the maintenance of HSPCs on the chip17. Here, the differentiation of HSPCs into mature blood cells required the addition of a new mixture of cytokines and growth factors to the HSPC medium. To confirm that the favorable MSC phenotype on the scaffolds was maintained in the new medium and to further characterize the stromal culture compartment, MSCs cultured in different media and conditions were harvested from the ceramic and analyzed by bulk RNA sequencing. The scaffolds were either cultivated under static (24-well plate) or dynamic (on chip) conditions in MSC medium or HSPC medium for up to 24 days.
The samples clustered primarily by cultivation medium and time (Fig. 1E and Supplementary Fig. S1A). Unbiased tissue identity analysis using the BioQC method19 showed an enrichment for various stromal tissues including mesenchymal stem cells and osteoblasts (Supplementary Fig. S1B). The analysis of selected mesenchymal tissue signatures indicated that the prolonged usage of stromal cell culture medium induced an endothelial cell signature while the HSPC medium induced an adipose tissue signature besides the prominent osteoblast signature present in both media (Supplementary Fig. S1C). Since the strong osteoblast signature was maintained, an additional adipocyte signature was not considered alarming given that bone marrow adipose tissue is an important constituent of the healthy human bone marrow20.
Targeted analysis of GO biological process terms for osteocytes21 showed activity of pathways associated with bone and cartilage formation, extracellular matrix collagen deposition and regulation of proliferation under all culture conditions (Fig. 1D). Biological processes observed to be significantly downregulated in the ceramic scaffold over time were DNA replication and cell cycle progression. At the same time, multiple processes including cytoplasmic translation, ATP synthesis coupled electron transport and oxidative phosphorylation were significantly enhanced, indicating a high energy demand and activity despite significantly reduced proliferation, for example due to ECM production (Fig. 1F).
The cultivation of stromal cells in the HSPC medium in comparison to stromal cell medium resulted in a significant induction of processes associated with antigen processing and presentation, including pathways associated with lysosome functionality. Alternatively, the induction of those processes could be indicative of a contamination of macrophages in the original stromal cell population. However, unbiased tissue identity analysis did not indicate the presence of macrophages (Supplementary Fig. S1B) and a flow cytometric analysis of expanded stromal cells for the markers CD14 and CD206 did not reveal a macrophage subpopulation (Supplementary Fig. S2).
No differences were observed between static and dynamic cultures in terms of stromal cell identity, which was expected, since stromal cells are mainly found inside of the ceramic scaffold pores and mostly shielded from direct fluid flow-induced shear stress (Fig. 1F).
On-chip lineage differentiation of CD34+ cells into erythroid, myeloid and NK-cells could be stimulated with different growth factor cocktails over 31 days
The dynamic co-cultures were maintained for up to 31 days with media changes every 2–3 days. Various combinations of the growth factors TPO, Flt-3 ligand (FLT-3L), stem cell factor (SCF), interleukin-3 (IL-3), granulocyte-macrophage colony-stimulating factor (GM-CSF), granulocyte colony-stimulating factor (G-CSF), erythropoietin (EPO), interleukin-7 (IL-7), and interleukin-15 (IL-15)) were added to the medium to stimulate the differentiation of HSPCs into hematopoietic lineages. Non-adherent cells were transported by the medium flow from the ceramic scaffold compartment to the medium compartment and back. Cells accumulating in the medium compartment were harvested for counting and flow cytometric analysis on days 10, 17, 24, and 31 of the assays (Fig. 2A and Supplementary Figs. S8 and S9). At the end of the assays, non-adherent cells were washed out of the ceramic scaffold and were analyzed separately by flow cytometry.
A Time schedule of the co-culture assay in the Chip2 with weekly sampling points for flow cytometry analysis and media exchanges every 2–3 days. B Sampled cell counts of erythroid cells (CD45−, CD33−, CD71+ and/or CD235+), granulocytes (CD34−, CD33+, CD15+, CD14−) and NK-cells (CD45+, CD33−, CD56+) from the medium circulation at day 10, 17, 24, and 31. Mean cell count values +/− SD of three CD34+ cell donors (large symbols) and three chips each (small symbols) are shown. Development of cell counts in the microfluidic chip was modeled using a non-linear regression with a 4-parameter variable slope sigmoidal curve fit. For comparison of total cell counts in between differentiation media, log transformed total count data (sum of all produced cells of the respective lineage over time) were analyzed and compared with a repeated measures one-way ANOVA and Dunnett’s multiple comparisons test against the mixed lineage condition (N = 3 donors, DFn = 3, DFd = 6, F(erythroid) = 243.9, F(Granulocytes) = 156.3, F(NK-cells) = 17.59). Statistical significance was defined as ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05 and ns (p > 0.05). C Representative flow cytometry plots depicting the maturation of CD45-CD33- erythroid cells on the chip over time in erythroid differentiation medium. Cells were sampled weekly from the medium reservoir and at the end of the assay (day 31) from the ceramic scaffold. D Representative flow cytometry plots depicting the maturation of granulocytes on the chip over time in myeloid differentiation medium. Cells were sampled weekly from the medium reservoir and at the end of the assay (day 31) from the ceramic scaffold.
The growth factor cocktails induced differentiation of the intended lineages over time. Addition of EPO and IL3 to the basic medium containing TPO, FLT-3L, and SCF induced erythroid differentiation (red lines in Fig. 2B). Addition of IL3, G-CSF, and GM-CSF induced granulocyte differentiation (blue lines in Fig. 2B), and addition of IL-7 and IL-15 induced NK cell differentiation (orange lines in Fig. 2B). Without these specific factors, only small numbers of these cell types were observed. The complete mixture of growth factors induced the differentiation of erythroid cells, granulocytes and NK cells simultaneously with a similar potency as in the single lineage differentiation media (green lines in Fig. 2B). Marked donor-to-donor differences were observed for NK cell maturation. While the erythroid and myeloid output over time was comparable across 3 donors, one donor (Donor 3) showed a much higher absolute and relative output of CD56 + NK cells compared to the other two donors (Supplementary Fig. S3).
The maturation of CD45 negative erythroid cells progressed over time, with cells maturing from blast-forming units (BFU-E, CD71low CD235−), through colony-forming units (CFU-E, CD71high CD235−), early erythroblasts (CD71high CD235+), late erythroblasts (CD71low CD235+) to orthochromatic erythroblasts (CD71− CD235+)22,23. The orthochromatic erythroblast stage was reached after 31 days and was present in the circulation as well as inside of the ceramic scaffold (Fig. 2C). The maturation of CD15+ granulocytes in the myeloid culture medium progressed from CD13high CD16− promyelocytes, to CD13low CD16− myelocytes, to CD13low CD16+ metamyelocytes, to CD13 + CD16+ neutrophils24. Efficient differentiation up to the metamyelocyte stage was observed, while only low numbers of CD13high neutrophils were detected (Fig. 2D). In addition, the numbers of CD34 + HSPC, CD14+ monocytes and CD41+ megakaryocytes were monitored in the different cell culture media (Supplementary Fig. S4A). The number of CD34+ HSPCs in circulation decreased until day 17 but remained stable thereafter, regardless of the growth factors applied. Monocytes developed in all conditions, with higher numbers in the IL3-containing media (Supplementary Fig. S4A). The monocyte population could be further subdivided into classical (CD16-), intermediate (CD13high CD16+) and non-classical (CD13low CD16+) monocytes (Supplementary Fig. S4B, C), with intermediate monocytes being CD14high and non-classical monocytes being CD14low (Supplementary Fig. S4D), as described previously25. Consistent with the known role of IL3 in megakaryocyte development26, megakaryocytes developed in the erythroid, myeloid, and mixed lineage media, but not in the lymphoid medium. (Supplementary Fig. S4A).
Despite constant flow, the majority of the cells on day 31 were located in the ceramic compartment of the chip. The relative distribution of cells between the ceramic scaffold and the circulation at the end of the assay differed between the media compositions. For example, in the erythroid medium, the fraction of erythroid cells in the ceramic scaffold was significantly higher than in the circulating cell pool, while granulocytes in the granulocyte medium were more equally distributed in the two compartments (Supplementary Fig. S4E).
Expression of CD71 and CD10 in the CD34+ cell starting cell population correlated with the donor’s capacity to generate erythroid and NK-cells in vitro
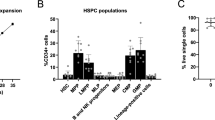
In order to investigate the donor-to-donor differences in lineage outputs, CD34+ cells from the 13 donors used in the experiments were characterized right after thawing by flow cytometry using the markers CD34+, CD38, CD71, CD33, and CD10. All donors had CD34+ purity greater than 95% (Fig. 3A). The frequencies of individual progenitor subpopulations differed between the donors. The largest standard deviation was observed for the CD34 + CD10+ subpopulation, followed by the CD34 + CD38−, CD34 + CD71−, and CD34 + CD33+ subpopulations (Fig. 3B). We correlated these initial subfractions with the output of erythroid cells, granulocytes and NK cells at day 31 of the assay by non-linear regression assuming exponential growth behavior. The size of the CD10+ fraction of CD34+ cells correlated well with the number of CD56 + NK cells at day 31 of the assay (Fig. 3C), and the CD71 expression of the initial population of CD34+ cells correlated well with the number of CD45− erythroid cells at day 31 of the assay (Fig. 3D). Only a weak correlation was observed between the CD33+ expression of the initial CD34+ cell population and the CD15+ granulocyte count at day 31 of the assay (Fig. 3E).
A Composition of starting material, percentages of CD38-Lin- stem cells, CD38+Lin- progenitor cells, CD71+ erythroid progenitor cells, CD33+ myeloid progenitor cells, CD10+ lymphoid progenitor cells and of CD34- differentiated cells are shown for 13 donors used throughout experiments. B Percentage of starting cell population of CD38+progenitor cells, CD71+ erythroid progenitor cells, CD33+ myeloid progenitor cells, CD10+ lymphoid progenitor cells. Values of 13 donors are plotted with mean and standard deviation. C Non-linear regression of CD10+ cells at day 0 of the assay and CD56 + NK-cell output at day 31 assuming exponential growth behavior. Individual NK-counts of three donors (N = 3) with three replicates (n = 3) are shown. D Non-linear regression of CD71+ cells at day 0 of the assay and CD45- erythroid cell output at day 31 assuming exponential growth behavior. Individual erythroid cell counts of five donors (N = 5) with three replicates (n = 3) are shown. E Non-linear regression of CD33+ cells at day 0 of the assay and CD14+ granulocyte output at day 31 assuming exponential growth behavior. Individual granulocyte counts of five donors (N = 5) with three replicates (n = 3) are shown.
Inflammatory factors induced reversible imbalances between erythroid and myeloid lineage output
Inflammatory factors can alter the proliferation and lineage fate of bone marrow HSPCs in acute and chronic disease27, often leading to a bias toward enhanced myelopoiesis and reduced erythroid and lymphoid lineage output28,29. To test how differentiation patterns in the BM-MPS respond to the addition of inflammatory factors, the BM-MPS were established in a mixed lineage medium and treated from day 7 with either interleukin-1β (IL-1β), lipopolysaccharides (LPS) or interferon-γ (IFN-γ) for 10 days, followed by a 2-week recovery period.
IL-1β has been shown to stimulate myelopoiesis while inhibiting erythropoiesis and lymphopoiesis. The addition of 10 ng/mL IL-1β to the BM-MPS for 10 days resulted in a potent inhibition of erythropoiesis with a significantly reduced number of erythroid cells past the BFU-E stage in circulation during the treatment phase (Fig. 4A). Erythroid differentiation was arrested at the CFU-E stage (Fig. 4B), but progressed after removal of IL-1β (Supplementary Fig. S5). The number of erythroid cells fully recovered to control levels within the 2-week recovery period (Fig. 4A). Application of IL-1β also impaired NK cell differentiation for two donors. The third donor failed to develop measurable levels of NK cells on chip under control conditions. While the number of circulating NK cells returned to baseline levels during the 2-week recovery period, the number of NK cells in the ceramic scaffold remained reduced at the end of the assay compared to untreated control circuits (Fig. 4C). The total number of circulating monocytes was transiently decreased by ~40% after treatment, and the numbers in the ceramic scaffold remained significantly reduced on day 31 compared to untreated controls (Supplementary Fig. S6B). However, a subpopulation of non-classical circulating monocytes increased significantly and remained significantly elevated during the recovery phase (Fig. 4E). Granulopoiesis was also increased upon IL-1β treatment, and the number of granulocyte lineage cells in the circulation remained significantly elevated during the recovery phase. (Fig. 4G). In addition, granulopoiesis was accelerated, with cells reaching the metamyelocyte stage by day 17, when this differentiation stage had not yet been reached in the untreated population (Fig. 4H). The number of megakaryocytes initially decreased significantly by 75% after IL-1β treatment, but strongly increased during the recovery phase, reaching 50–100% elevated levels compared to the untreated control in the circulation and in the ceramic scaffold on day 31 (Supplementary Fig. S6D). Inflammation-associated cytokines IL-23, G-CSF, TNF-α, MCP-1, and IL-6 in the cell culture medium were all significantly induced within 6 h of IL-1β treatment and remained highly elevated throughout the treatment. With the exception of IL-6, cytokine levels returned to baseline levels within the recovery period (Fig. 4I). Taken together, IL-1β treatment of BM-MPS induced effects on hematopoiesis consistent with those reported in the literature.
Sampled cell counts are plotted relative to the untreated control condition of the same donor and day. Mean relative cell count values +/− s.e.m of three CD34+ cell donors (large symbols) and two to three chips (small symbols) each are shown. Statistical comparison of means was performed on log-transformed count data of the treatment phase (day 10 + day 17), the recovery phase (day 24 + day 31) and the ceramic scaffold by a repeated measures two-way ANOVA plus Dunnett’s post hoc test with the untreated condition of each day as control condition (N = 3 donors, DFn = 6, DFd = 12, F(ery) = 16.79, F(NC Mono) = 4.867, F(Granu) = 11.89). Statistical significance was defined as ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05 and ns (p > 0.05). A Sampled relative cell counts of erythroid cell in circulation (left plot) and in the ceramic scaffold at the end of cultivation (right plot). B Exemplary flow cytometry dot plot (CD71 vs CD235) of one circuit treated with IL-1β in comparison to one untreated circuit at day 17. C Sampled relative cell counts of NK cells in circulation (left plot) and in the ceramic scaffold at the end of cultivation (right plot). One donor did not produce NK-cells so only values from two CD34+ cell donors are shown. D Exemplary flow cytometry dot plot of lymphoid cell populations (CD56 vs CD10) of one circuit treated with IFN-γ in comparison to one untreated circuit at day 31. E Sampled cell counts of non-classical monocytes in circulation (left plot) and in the ceramic scaffold at the end of cultivation (right plot). F Exemplary flow cytometry dot plot of granulocytes (CD16 vs CD13) of one circuit treated with LPS in comparison to one untreated circuit at day 24. G Sampled cell counts of granulocytes in circulation (left plot) and in the ceramic scaffold at the end of cultivation (right plot). H Exemplary flow cytometry dot plot of monocytes (CD13 vs CD16) of one circuit treated with IL-1β in comparison to one circuit from the same donor without treatment at day 17 of the assay. I Concentrations of the inflammatory factors IL-23, G-CSF, TNFa, MCP-1, IL-6, and IL-1β in the cell culture medium over time. For comparison of cytokine concentration between groups, area under curve values were analyzed and compared with a repeated measures one-way ANOVA and Dunnett’s multiple comparisons test (N = 3 donors, DFn = 3, DFd = 6, F(IL-23) = 303.7, F(G-CSF) = 593.5, F(TNFa) = 42.65, F(MCP1) = 160.3, F(IL-6) = 114.8. F(IL-1β) = 1876).
LPS has been shown to bind directly to several hematopoietic cells including HSCs through toll-like receptors, inducing myeloid skewing and the production of pro-inflammatory cytokines including IL-1β, IL-6, IFNs, and TNF-α29. Consistent with IL-1β being a downstream effector molecule of LPS, the response to 10 µg/ml LPS in BM-MPS was comparable to the response to IL-1β treatment, including a significant increase in granulocytes and non-classical monocytes during the recovery phase (Fig. 4E, G) and significantly reduced numbers of monocytes in the ceramic scaffold (Supplementary Fig. S6B). Non-classical monocytes formed a distinct cell population that was not present in untreated control samples at day 24 (Fig. 4F). In contrast to IL-1β treatment, HSPC numbers did not recover during the recovery phase under LPS treatment. HSPC numbers remained significantly reduced in the circulation and in the ceramic scaffold at the end of the assay on day 31 (Supplementary Fig. S6C). LPS caused a low level of IL-1β release and a similar but lower cytokine release pattern as observed with IL-1β treatment, however, not only IL-6 but also MCP1 and IL-1β levels remained elevated during the recovery period (Fig. 4I).
IFN-γ plays a critical role in immune responses, including macrophage activation and modulation of antigen presentation. In hematopoiesis, IFN-γ has been shown to have both stimulatory and inhibitory effects, depending on the context and specific cell lineages involved28,30. While inhibition of erythropoiesis is a well-established effect of IFN-γ, opposing effects on the myeloid lineage have been reported, such as skewing of myeloerythroid differentiation toward myelopoiesis by MSC-derived IL-631 or disruption of multipotent progenitor differentiation into myeloid progenitors in mice upon constitutive low-level elevation of IFN-γ32.
Treatment of the BM-MPS with 10 ng/mL IFN-γ for 10 days resulted in a significant decrease in total hematopoietic cell counts in the circulation of the BM-MPS (Supplementary Fig. S6A). Erythroid and megakaryocytic lineage counts decreased significantly by ~50% in circulation on culture days 17 and 24, respectively, but partially recovered during the 2-week recovery period on day 31, both in circulation and within the ceramic scaffold (Fig. 4A and Supplementary Fig. S6D). Non-classical monocyte and granulocyte lineage counts also decreased upon treatment and were significantly lower than in the control condition during the recovery period (Fig. 4E, G). A significant 50% decrease in the number of CD34+ HSPCs was also observed during the recovery phase. In contrast to the other lineages, the numbers of NK cells from two donors increased after IFN-γ treatment compared to untreated control chips (Fig. 4C, D). IFN-γ did not induce the release of any of the pro-inflammatory cytokines tested, including IL-6, indicating that the observed effects were directly caused by IFN-γ and potentially explaining the lack of myelopoiesis induction often described in other studies (Fig. 4I). Thus, the effects of IFN-γ in the BM-MPS reproduced previously described inhibitory effects on HSPC proliferation and differentiation.
Taken together, the data indicated that physiologically relevant effects of haematopoietically active cytokines can be recapitulated in the BM-MPS and that the system could serve as a tool to investigate cytokine-mediated effects on human hematopoiesis.
A transferrin receptor-binding antibody induced stage-dependent suppression of erythroid progenitor cells
Antibodies targeting the transferrin receptor (CD71) are in clinical development for targeted drug delivery to the brain and as anticancer agents33,34. CD71 is responsible for the uptake of transferrin-bound iron into cells, and is therefore strongly expressed in the erythroid lineage but also in other proliferating progenitor cell populations of the human bone marrow35. In preclinical studies with CD71-engaging antibodies, a decrease in circulating reticulocytes and anemia has been observed due to antibody-dependent cell-mediated and complement-mediated effects via Fcγ receptor binding36,37,38.
An antibody with a wild-type Fc region and a C-terminal TfR1-binding moiety (TfR1-FcWT), which does not interfere with the binding of transferrin to TfR1, was applied to the BM-MPS at 120 µg/ml in a mixed lineage medium driving erythropoiesis and myelopoiesis for 14 days. The effects were compared to a control antibody which had an Fc region (TfR-FcSilent) that was effector-silenced by a PG-LALA mutation (Fig. 5A). Compared to the untreated control and the TfR-FcSilent treatment, the TfR-FcWT treatment resulted in a significantly reduced number of erythroid lineage cells on the chip after two weeks (Fig. 5B). Analysis of erythroid subpopulations showed significant and strong hits on BFU-E, CFU-E and erythroblast populations, which express high levels of CD71 (Fig. 5C).
A Time schedule of 14-day treatment with the TfR-FcWT and TfR-FcSilent at a concentration of 120 µg/mL in the Chip2 system with weekly sampling points for flow cytometry analysis and media exchanges every 2–3 days. B–E Relative counts and mean +/− s.e.m. of B: erythroid cells, C: erythroid subpopulations, D: myeloid cells, and E: myeloid subpopulations from multiple CD34+ donors after 14 days of treatment with a wildtype transferrin-binder antibody (TfR-FcWT) and a Fc-silenced transferrin-binder isotype control antibody (TfR-FcSilent). TfR-FcSilent (N = 7 donors, n = 2–3 chips) and TfR-FcWT (N = 6 donors, n = 3 chips) are normalized to the untreated control (N = 8 donors, n = 3 chips) of the same donor in the same experiment. Statistical comparison of means was performed on log transformed cell count data with a repeated measures mixed effects model REML (N = 8 donors, DFn=2, DFd = 12, F(ery) = 19.79, F(BFU-E) = 17.21, F(CFU-E) = 49.44,F(EE) = 38.12, F(LE) = 11.69, F(R) = 11.56, F(myelo) = 7.686, F(MB) = 1.194, F(PMC) = 3.747, F(MC) = 5.270, F(MM) = 10.28, F(Neu) = 4.822, F(Mono) = 2.484)) and Tukey’s multiple comparisons test (B, D) or Dunnett’s multiple comparisons test (C, E) F Relative concentrations as mean +/− SD of the inflammatory factors MCP-1, G-CSF and IL-8 in the cell culture medium at day 19, 21, 24 and 31. Concentrations of three technical replicates (chip circuits) were averaged and normalized to the concentration at day 17 before treatment. For comparison of cytokine concentration between groups, area under curve values of raw concentration data was analyzed and compared with a repeated measures one-way ANOVA and Dunnett’s multiple comparisons test (MCP1: N = 5, DFn = 2, DFd = 8, F = 8.375; G-CSF/IL-8: N = 3, DFn = 2, DFd = 4, F(G-CSF) = 10.81, F(IL-8) = 16.86.) Statistical significance was defined as ****p < 0.0001, ***p < 0.001, **p < 0.01, *p < 0.05, and ns (p > 0.05).
Total myeloid cell counts were slightly reduced after two weeks of treatment with TfR-FcSilent and TfR-FcWT antibodies (Fig. 5D). Analysis of myeloid subpopulations showed that TfR-FcWT induced a significant decrease in myelocytes, metamyelocytes and neutrophils, but not in myeloblasts, promyelocytes or monocytes. (Fig. 5E).
In addition to the cellular effects, TfR-FcWT treatment induced a minor but significant release of the myeloid effector cell-derived pro-inflammatory cytokines MCP-1, G-CSF and IL-8 in the culture (Fig. 5F). Levels of several other cytokines, including IL-23, M-CSF, Osteopontin, IL-1b, IL-6, and TNF-a, remained stable after treatment with the TfR-FcWT antibody (Supplementary Fig. S7).
Taken together, these data demonstrate that clinically relevant side effects of TfR-binding therapeutic antibodies can be recapitulated with the BM-MPS and support an Fcγ receptor-mediated mechanism involving myeloid effector cells.
Allogeneic CD3 + T cells were activated by T-cell bispecific antibodies on chip and showed pronounced cytotoxicity
T cell bispecific antibodies are a cancer immunotherapy drug modality that simultaneously bind to surface antigens on tumor cells and on T cells, thereby engaging and activating the T cells to kill the tumor cells. TCBs target tumor-associated antigens (TAAs). However, most TAAs are also present on healthy tissue cells, and TCB treatment has been observed to result in dose-dependent on-target off-tumor toxicities39. To make the BM-MPS immunocompetent (iBM-MPS) for the prediction of on-target off-tumor toxicities of TCBs, cryopreserved allogeneic blood-derived pan-CD3 + T cells were added to the BM-MPS on culture day 17. First, the effect of different seeding numbers of T cells and the effect of T cell pre-activation (CD3/CD28) or stimulation (IL-2) on T cell survival and responsiveness in the iBM-MPS were investigated.
The cells were harvested from the chips after 4 days (day 21) and 7 days (day 24) of co-culture and analyzed by flow cytometry (Supplementary Figs. S10, S12, and S13). The recovery rate of unstimulated T cells after 7 days of BM-MPS co-culture was ~80% independent of the presence of IL-2. Pre-activation induced T cell proliferation on-chip, resulting in recovery rates of 130%–320% after seven days. The proliferation of pre-activated T cells on-chip was more pronounced in the presence of IL-2 or when lower numbers were seeded into the chip. (Fig. 6A).
Mean values +/− SD of two to five circuits (n = 2–5) of one CD34+ cell donor (N = 1) are shown if not specified otherwise. A Number of sampled T cells from the chip at day 21 and day 24 in the presence of absence of IL-2 relative to the amount of seeded naive or preactivated T cells at day 17. B Percentage of naive or preactivated CD4+ and CD8 + T cells that express activation markers CD69, PD-1, CD25, and HLA-DR before chip culture (day 17) and after 7 days cultivation on chip without additional stimulation (d24) in the presence of absence of IL-2. C–E Sampled HSPC (C), erythroid cell counts (D) and granulocytes (E) at day 24 in circuits without T cells and in circuits with naive or preactivated T cells in the presence or absence of IL-2. F Time schedule of 7 day treatment with ESK-1-like TCB and an isotype control TCB at a concentration of 1 µg/mL in the Chip2 system with cell sampling for flow cytometry analysis after four and seven days after treatment start. G Sampled CD3 negative non-T cell counts from circulation and the ceramic scaffold at day 24 in circuits treatedwith the ITC-TCB or ESK1-TCB. Counts are normalized to the mean of untreated control circuits. Circuitsthat were supplemented with IL-2 are marked by squares, circuits without IL-2 supplementation with circles. H Expression of activation markers CD69, PD-1, CD25, and HLA-DR on naive CD4+ and CD8 + T cells at day 21 and day 24 treated with the ITC-TCB or ESK1-TCB.
Flow cytometry was used to investigate the effects of the BM-MPS co-culture on the activation status of T cells. Initially, as expected, the levels of activation markers (HLA-R, CD25, CD69, PD-1) on the T cell surface were low. The pre-activation of T cells induced a pronounced upregulation of HLA-DR and CD25 on both CD4+ and CD8 + T cells. After BM-MPS culture, all activation markers increased on both CD4 and CD8 T cells. The activation level on the chip was higher in pre-activated T cells compared to naive T cells, but was independent of seeding number and IL-2 supplementation (Fig. 6B).
The addition of T cells affected the lineage output of erythroid cells and HSPCs. With the addition of pre-activated T cells, the HSPC output from BM-MPS was reduced by ~50% (Fig. 6C). Furthermore, the number of erythroid lineage cells was reduced by ~50% upon addition of naive T cells and by ~75% upon addition of pre-activated T cells (Fig. 6D). No pronounced differences were observed in the numbers of T cells, HSPCs, erythroid cells, and myeloid cells between circuits treated with or without IL-2 (Fig. 6C–E).
As a biological proof of concept that TCBs can induce T cell-mediated cytotoxicity against target cells in the BM-MPS, the ESK1-like TCB was used to treat BM-MPS. The ESK1-like TCB non-specifically targets HLA-A2, a common MHC class 1 receptor isotype in humans. HSCs, MSCs and T cells on the chip were all derived from HLA-A2 positive human donors. Concurrent with the addition of T cells to the MPS, treatment with ESK1-like TCB and isotype control TCB (ITC-TCB) was initiated (Fig. 6F). After four and seven days of treatment, non-adherent cells were harvested from the chip and analyzed by flow cytometry. Hematopoietic cell counts were reduced by approximately 70% after 7 days of ESK1-like TCB treatment, regardless of the number of initially added T cells, pre-activation status, or addition of IL-2 (Fig. 6G). Furthermore, ESK1-TCB promoted a strong and significant induction of T cell activation markers on naive CD4+ and CD8 + T cells, as evidenced by upregulation of the activation markers PD-1, HLA-DR, CD69 and CD25. The activation level did not increase further from day 4 to day 7 on the chip (Fig. 6H).
For further experiments we decided to work with naive T cells and to limit the treatment time with TCBs to 4 days.
Application of autologous T-cells reduced unspecific activation while showing T-cell bispecific antibody induced lineage-specific cytotoxicity
To eliminate confounding effects of potential alloreactivity in the iBM-MPS, a fully autologous model was established. HSPCs, MSCs, and T lymphocytes were isolated from the same HLA-A2+ human donor and assembled in an iBM-MPS assay setup (Fig. 7A).
Mean values +/− s.e.m. of two chips (n = 2, small symbols) of three CD34+ cell donors (N = 3, large symbols) are shown if not specified otherwise. A Schematic showing isolation of CD34+ hematopoietic progenitor cells, mesenchymal stromal cells and T-lymphocytes from one donor. This process was performed for three donors in total. Cells were then used to build autologous models from all three cell types in the HUMIMIC Chip2. B Time schedule of experiment with addition of 400,000 naive autologous T cells at day 17 and subsequent 4-day treatment with ESK-1-like TCB, CD33-TCB, Nivolumab and an isotype control TCB at a concentration of 1 µg/mL. C LDH concentrations in circulation over time in circuits without T cells and in circuits including T cells with and without test antibody treatment. Dotted line visualized the treatment start point at day 17 of the assay. Area under curve LDH concentration values was analyzed with a repeated measures one-way ANOVA and Dunnett’s multiple comparisons test (N = 3 donors, DFn = 5, DFd = 10, F = 44.26). D Activation status of CD4+ and CD8 + T cells in the chip after four days (day 21 of the assay) with and without test antibody treatment. Expression of CD69 on CD4+ and CD8 + T cells was compared with a repeated measures two-way ANOVA and Dunnett’s multiple comparisons test (N = 3 donors, DFn = 5, DFd = 20, F = 265.7). E–H Sampled living cells (E), CD3 + T cell (F), erythroid cell (G) and myeloid (H) counts from circulation and the ceramic scaffold on day 21. Counts are normalized to the mean of untreated control circuits. Raw count data was compared by repeated measures one-way ANOVA and Dunnett’s multiple comparisons test (N = 3 donors, DFn = 4, DFd = 8, F(living) = 6.903, F(T cells) = 41.65, F(ery) = 14.33), F(myelo) = 2.547). I Sampled myeloid subpopulation cell counts from circulation and the ceramic scaffold at day 21 treated with CD33-TCB. Counts are normalized to the mean of untreated control circuits. Mean cell counts of myeloid subpopulations (I) in untreated and CD33-TCB treated circuits were compared with multiple ratio paired t-tests (N = 3 donors, DF = 2, t(MB) = 2.228, t(PMC) = 0.8779, t(MC) = 6.964), t(MM/Neu) = 1.182, t(Mono) = 2.813).Statistical significance was defined as ****p < 0.0001, ***p < 0.001, **p < 0.01, and ns (p > 0.05).
T cells were added to the dynamic bone marrow cultures on day 17. Concurrently, treatment with ESK1-like TCB40, a CD33 TCB, nivolumab and ITC-TCB was initiated. CD33-directed bispecific antibodies are currently under investigation in clinical trials for acute myeloid leukemia41. CD33-TCBs have been shown to have significant on-target off-tumor side effects due to the broad expression of CD33 on healthy mature myeloid cells and myeloid progenitor cells42,43. While HLA-A2 was broadly expressed on all cells in the chip, including all hematopoietic lineages, MSCs and T cells, CD33 was expressed only on mature myeloid lineage cells and the common myeloid progenitors that give rise to both the myeloid and erythroid lineages. Nivolumab is a human immune checkpoint inhibitor blocking the programmed cell death protein 1 (PD-1) receptor on T cells, thereby enhancing the immune system’s response against tumor cells44. It was used as a negative control clinical antibody in this study, since we did not expect any short term adverse effects on hematopoiesis based on clinical experience.
After four days of treatment, the non-adherent cells were harvested from the chip and analyzed by flow cytometry and the supernatants were sampled to investigate LDH release (Fig. 7B, and Supplementary Figs. S11–13). LDH levels increased significantly in cultures treated with ESK1-like and non-significantly upon CD33-TCB treatment, indicating T cell-induced cell death. As expected, due to the higher numbers of target-expressing cells, LDH release was more pronounced after ESK1-TCB treatment compared to CD33-TCB treatment. No differences in LDH levels were observed between chips cultured with and without T cells and after treatment with the control antibodies (Fig. 7C).
The baseline activation status of T cells in the autologous setup on day 0 was similar to that of T cells in the allogeneic setup. T cell activation was induced by ESK1-like TCB and CD33-TCB, as evidenced by the significant upregulation of the early and late T cell activation markers on CD4+ and CD8 + T cells (Fig. 7D). Interestingly, the overall activation status of naïve, untreated T cells under control conditions in the autologous setup Fig. 7D) remained lower than in the allogeneic setup (Fig. 6B, H).
After 4 days of treatment, the total living cell counts were reduced by ~40% by ESK1-like TCB and by ~25% by CD33 TCB (Fig. 7E). The T cell counts recovered from the chip on day 4 in all treatment groups except for the ESK1-like TCB were in the range of ~60% of the initially seeded counts. After treatment with the ESK1-like TCB, the proportion of T cells recovered from the chip significantly decreased to ~30% of the initially seeded number, indicating that T cells were also targeting each other upon ESK1-like TCB treatment (Fig. 7F).
Both ESK1-like TCB and CD33 TCB induced a significant reduction of erythroid lineage cells (Fig. 7G), however, only CD33-TCB treatment showed a trend towards reducing total myeloid cell counts (Fig. 7H). While the effect on the total myeloid cell population was not statistically significant, a closer examination of myeloid subpopulations revealed a significant reduction of myelocyte counts (Fig. 7I).
Taken together, these data provide proof-of-principle that this BM-MPS could serve as an adaptable tool for predicting and investigating clinically relevant safety liabilities of biologics such as cytokines and immune-engaging antibodies.
Discussion
Here we report the generation of a chemically defined and adaptable human bone marrow microphysiological system with hematopoietic and stromal compartments that supported functional in vitro myelo-erythroid and lymphoid differentiation and maintenance of CD34+ HSPCs for 31 days.
Supporting the differentiation of erythroid, myeloid, and NK cells over 31 days while maintaining a stable pool of HSPCs represents a significant challenge due to the complex and dynamic requirements of hematopoiesis. Commonly used semi-solid cultures, such as methylcellulose-based CFU assays, are limited to endpoint analyses and poor standardization and do not allow continuous monitoring or modeling of dynamic processes, such as drug recovery45.
Liquid culture systems without methylcellulose like the HALO ATP assay46, overcame many of the prior limitations but failed to maintain the hematopoietic lineage balance and typically resulted in rapid differentiation and exhaustion of HSPCs. These high-throughput screening systems were not able to recapitulate functional aspects of in vivo cellular microenvironments. Consequently the pharmaceutical industry is exploring novel advanced methods to overcome these limitations47 such as microphysiological systems14,15,16 or mimicking stem cell niches in vitro48.
We set out to develop a human bone marrow model that mimics key aspects of the bone marrow niche, including 3D architecture, cell-cell interactions, and dynamic perfusion. These features enabled long-term maintenance of hematopoiesis, preserved the HSPC pool regardless of cytokine addition, and allowed the study of inflammatory and drug-induced hematopoietic changes in real time. These capabilities are particularly valuable for pre-clinical safety profiling of biologics, where the functional co-culture and interaction of immune cells and target cells as well as the ability to monitor recovery after drug removal are critical.
Setting up such a model in a microfluidic organ-on-a-chip context instead of a 2D static environment is setting the stage for further advancements such as the development of a multi-organ setup connecting the bone marrow model with vasculature, liver, or secondary lymphoid tissue compartments.
In the model described here, the stromal cells kept an osteoblast phenotype and key osteogenic pathway expression over 24 days in HSPC medium. Induction of osteoblast differentiation by culture on hydroxyapatite-coated zirconium oxide scaffolds has already been shown previously49, and the RNA sequencing data overall indicate that osteoblast maturation was present in the model. General effects of prolonged culture such as reduced proliferation and increased metabolism were present in both stromal as well as hematopoietic cell culture medium. Hematopoietic culture medium induced processes associated with antigen processing and presentation in stromal cells. Antigen processing and presentation via MHC class I molecules by MSCs can be induced upon stimulation with cytokines such as IFN-γ50. Taken together, the data indicated that the stromal compartment of the chip provided a viable and functional support system for HSPC maintenance and differentiation.
In combination, the co-culture with stromal cells and the addition of specific cytokines to the medium supported the maintenance of HSPCs as well as their differentiation and maturation into granulocytes, erythrocytes, megakaryocytes, NK cells and monocytes. Compared to in vivo hematopoiesis in the human bone marrow, monopoiesis was overrepresented, while granulopoiesis, erythropoiesis and especially lymphopoiesis were underrepresented in the BM-MPS with full differentiation medium51.
The setup was used to model changes in hematopoiesis upon exposure to inflammatory cytokines and LPS. After ten days of exposure to pro-inflammatory factors, multiple changes in lineage differentiation were observed that were in line with previously reported clinical manifestations of acute and chronic inflammation52, including increased granulopoiesis and impaired erythropoiesis. The inflammatory phenotype was reinforced by the responsiveness of the BM-MPS-resident cells, which differentiated into inflammation-associated phenotypes such as non-classical monocytes25 and released additional pro-inflammatory cytokines into the system. Non-classical monocytes appeared upon treatment with LPS and IL-1β. Under inflammatory conditions, they have been described as the primary producers of TNF-α, IL-1β, and IL-625, which were all found strongly increased in the BM-MPS in their presence. In addition, G-CSF was released upon treatment with IL-1β and LPS, in line with increased numbers and accelerated maturation of granulocytes in the BM-MPS53. The observed effects were mostly reversible within two weeks after removal of the treatments, demonstrating the ability of the model to recover from treatment, and indicating that the BM-MPS may be suited to model recovery scenarios. The BM-MPS was further evaluated for its applicability in nonclinical safety assessment of complex large molecules. First, the effects of TfR1-binding antibodies with and without a functional Fcγ receptor binding site were investigated. Transferrin receptor (TfR1) binding antibodies have been developed for applications like targeted delivery at the blood-brain barrier or as anticancer agents33,54. Hematologic toxicities are well known on-target immune-related ADRs of TfR-binding antibodies due to high TfR levels on erythroid progenitor cells36. Mechanistically, the Fcγ receptor (FcγR) binding site in the Fc region of the antibodies is engaging FcγR-expressing effector cells of the immune system such as macrophages, T and NK cells, triggering antibody-dependent cellular cytotoxicity or phagocytosis38. In mouse studies, a murine wild-type Fc TfR-BACE1 bispecific antibody caused reversible hematotoxic effects, while an Fc-silenced analog did not36. The wild-type antibody affected erythroid progenitor cells but not mature erythrocytes. Similar toxicities were observed in rhesus monkeys37, again circumventable by attenuating the antibody’s Fc effector function34. Those hematopoietic effects of anti-TfR1 antibodies with and without Fc effector function were recapitulated in the BM-MPS, where a wild-type TfR1-binding antibody evoked pronounced effects, while a LALA-PG silenced TfR1-binding antibody55 did not induce effects on erythroid lineage differentiation. The most affected differentiation stages were the CFU-E and early erythroblast stages, which expressed the highest levels of TfR1.
Next, we investigated whether the BM-MPS could be applied as a tool for preclinical safety assessment of TCBs. The preclinical prediction of on-target and off-target56 ADRs of TCBs is often not possible in classical animal models due to species-specific differences in target protein epitopes, in immune system components and in antigen processing pathways. Hence, novel approach methodologies (NAMs) such as human microphysiological systems and computational models are increasingly used to better predict human responses. As a proof of concept, we showed here that two different TCBs were causing the biologically expected T cell mediated cytotoxic effects on hematopoietic lineages in an immunocompetent BM-MPS which contained either allogeneic or autologous T lymphocytes. ESK1, a TCB developed to target a specific peptide of an intracellular protein (WT1) presented on the MHC Class I molecule HLA-A2, was discontinued due to a lack of specificity for the WT1 peptide57,58,59. It was therefore expected to induce T cell-mediated cytotoxicity to all HLA-A2-positive cells in the iBM-MPS. Indeed, ESK1 activated allogeneic T cells in the iBM-MPS and caused significant hematopoietic cell killing over the course of seven days, in line with recently published data from co-cultures of human liver spheroids, alveolar cells, and intestinal organoids with PBMCs56. Interestingly, in a fully autologous iBM-MPS, the T cells had lower levels of activation markers and the cytotoxicity was reduced compared to the allogeneic setup, suggesting that alloreactivity may have amplified T cell cytotoxicity in the allogeneic setup. This amplification should not be neglected as it may affect the estimation of safety thresholds or therapeutic windows if such assays are used for in vitro therapeutic window modeling or dose finding calculations. CD33-targeting T cell engagers are in clinical development for AML. CD33 is widely expressed in myeloid cells. However, in addition to the myeloid lineage, CD33 is expressed in common myeloid precursor cells (CMPs) and early erythroid precursor cells (BFU-E and CFU-E stages)60, hence, CD33-TCB induced significant cytotoxicity in the myeloid and erythroid lineages.
There are several limitations that remain to be solved in the future. While the BM-MPS supports differentiation and maturation of multiple hematopoietic lineages, the lymphopoiesis, particularly B and early T lymphocyte development, was missing. In the absence of a B cell compartment, the safety assessment for therapies targeting B lymphocytes is not possible. Although the iBM-MPS incorporated peripheral blood T cells, the lack of B cells, secondary lymphoid tissues and vasculature restricted its ability to fully mimic immune responses. This limitation reduced the system’s applicability for predicting immune-related ADRs that require complex immunological processes such as systemic cytokine release or autoantibody production. Furthermore, the composition of the stromal compartment does not fully recapitulate the heterogeneity of the bone marrow niche, lacking for example endothelial cells or adipocytes with regulatory roles in hematopoiesis.
The BM-MPS is well-suited for testing biologics that are directly targeting and directly modulating hematopoietic and immune cells, including monoclonal antibodies, bispecific antibodies, and cytokines. The presence of the expected target cells and the expected levels of target protein expression should be confirmed as a first step, before further testing. Furthermore, the BM-MPS could be valuable for evaluating off-target effects of T cell-engaging therapies, inflammatory mediators, and cell-based products like CAR-T cells. However, it is less suitable for biologics that rely on interactions with secondary lymphoid organs (e.g., vaccines), require a fully functional vascular system (e.g., anti-angiogenic therapies), target non-hematopoietic cell types, or, in case of immune checkpoint inhibitors, present with rare and late onset hematological toxicities of uncertain mechanism, most likely involving systemic immune dysregulation and autoantibody production.
We conclude that this human iBM-MPS could be a valuable tool for the safety profiling and optimization of complex large molecules during pre-clinical development, as well as for the elucidation of biological mechanisms of action. It could be used for the head-to-head comparison of candidate molecules against clinical benchmarks at multiple drug concentrations and at a higher throughput compared to animal models, thereby facilitating the identification of effective dose ranges and potential toxicities more efficiently than in vivo models.
Validation of new approach methodologies (NAMs) such as organ-on-a chip is an absolute necessity. It is essential to base the validation of these system on comparison with clinical data instead of comparing it with animal models, since the goal is to enhance the overall methodology to predict human responses rather than to directly replace animal testing. A common approach is to validate these systems on legacy data of compounds that showed safety or efficacy issues that were not predicted in preclinical phases. However, this approach is challenging since these datasets are often not published or otherwise accessible61. We therefore propose a validation concept where the most promising NAMs are applied in parallel with ongoing (pre-)clinical studies to receive an unbiased validation.
Methods
Order of work packages
The studies were carried out over a timeframe of four years. The data generated in the first year are not part of the dataset presented here; the system was set up including the establishment of cell sources, readouts and optimization of culture and medium parameters. In the second year, several experiments investigating different media compositions (Fig. 2) and first experiments with a TfR-binding antibody were performed. In the third year, three experiments with TfR-binding antibodies (Fig. 5) and two experiments with T-cell coculture and testing of bispecific antibodies (Figs. 6 and 7) were carried out in an alternating schedule. In the fourth year, experiments investigating the effects of inflammatory factors (Fig. 4) and an additional experiment with TfR-binding antibodies (Fig. 5) were performed. Data characterizing the CD34+ starting population was collected throughout all studies (Fig. 3). The experiment for the analysis of MSC identity by RNA sequencing was carried out in the second year (Fig. 1).
Preculture of mesenchymal stromal cells
Cryopreserved human bone marrow stromal cells (Stemcell Technologies, Vancouver, BC, Canada) were thawed and initially seeded at a density of 1750 cells/cm2 in T175 cell culture flasks in Mesencult-ACF Plus medium (Stemcell Technologies, Vancouver, BC, Canada). Medium was changed every 2–3 days until the cells reached 80–90% confluence. Cells were passaged using 0.05% trypsin/0.44 mM EDTA (Corning, Corning, NY, USA) and reseeded at a density of 2500 cells/cm2 into T175 cell culture flasks. Cells were grown to 80–90% confluence and then used to seed ceramic scaffolds.
One day before seeding, the hydroxyapatite-coated zirconium oxide scaffolds (Zellwerk GmbH, Berlin, Germany) were transferred to a 48-well plate and each scaffold was washed with 1 mL PBS. ACF cell attachment substrate from the Mesencult-ACF Plus Kit (Stemcell Technologies, Vancouver, BC, Canada) was diluted 1:300 in PBS. The scaffolds were incubated in 1 mL of the prepared coating solution for 2 h at 37 °C or overnight at 4 °C. The next day, MSC were passaged in monolayer culture and a MSC seeding solution was prepared in Mesencult-ACF Plus medium at a cell density of 3.33 × 106 cells/mL. The coating solution was aspirated from the ceramic scaffolds, the scaffolds were washed once with 1 ml PBS and transferred to 96-well flat-bottomed ULA plates (Corning, Corning, NY, USA) using sterile forceps. To seed MSCs onto the ceramic scaffolds, 150 µL of cell seeding solution was added to each scaffold/well and incubated for 5 h. After 5 h, the medium was removed from the top of the ceramic scaffolds and the ceramic scaffolds were transferred to 24-well ULA plates. Scaffolds seeded with MSCs were cultured for 10 days with media changes every 2 to 3 days until seeding with CD34+ bone marrow cells.
Chip preparation and seeding of CD34+ bone marrow cells
One day before the start of the chip experiment, HUMIMIC Chip2 (TissUse GmbH, Berlin, Germany) were prepared. The chips were opened, washed once with PBS + 1% penicillin/streptomycin (P/S) and filled with 400 µL PBS + 1% P/S per culture compartment. The Chip2 were connected to the control units and pumped at 0.45 Hz and +/− 300 mbar. On the first day of BM chip cultures, human bone marrow CD34+ cells (Stemcell Technologies, Vancouver, BC, Canada or Lonza, Basel, Switzerland) were thawed according to the supplier’s instructions. For each scaffold, 40,000 CD34+ cells were added in 150 µl StemSpan AOF (Stemcell Technologies, Vancouver, BC, Canada) supplemented with 10 ng/mL TPO, 25 ng/mL FLT-3L, 20 ng/mL SCF (all Peprotech, Cranbury, NJ, USA), 100 µg/mL holo-transferrin (Sigma Aldrich, St. Louis, MO, USA), 100 IU/mL penicillin + 100 µg/mL streptomycin and 5 µg/mL gentamicin (both Corning, Corning, NY, USA) (HSC basal medium). The HSC seeding solution was adjusted to 2.67 × 105 live cells/mL. Before adding the CD34+ cells to the pre-seeded scaffolds, the scaffolds were transferred from the 24-well ULA plates to sterile 96-well ULA plates. 150 µl of the CD34+ cell suspension was carefully added to the top of each scaffold and incubated at 37 °C for 5 h. After 2.5 h of incubation, the plate was inverted to ensure even distribution of the CD34+ cells in the ceramic.
In the meantime, the HUMIMIC Chip2 was prepared for insertion of the co-culture constructs (ceramic scaffold + MSCs + CD34+ cells) by aspirating the PBS in the culture compartments and adding HSC basal medium plus additional cytokines depending on the experiment. 400 µl of the respective medium was added to each inner culture compartment and 300 µl to each outer culture compartment of each chip circuit. The chips were then incubated until the co-culture constructs were inserted.
For analysis of lineage differentiation capacity, 10 ng/mL IL-3 (Miltenyi Biotec, Bergisch Gladbach, Germany), 20 ng/mL GM-CSF and 5 ng/mL GSF (both Peprotech, Cranbury, NJ, USA) were added to induce myeloid differentiation, 50 ng/mL IL-7 and 50 ng/mL IL-7 and 50 ng/mL IL-15 (both Miltenyi Biotec, Bergisch Gladbach, Germany) were added to induce lymphoid differentiation, 10 ng/mL IL-3 (Miltenyi Biotec, Bergisch Gladbach, Germany), 10 ng/mL EPO (Peprotech, Cranbury, NJ, USA) was added to induce erythroid differentiation, and a mixture of all growth factors was used to induce broad lineage differentiation. EPO concentration was increased to 50 ng/mL starting on day 10 of the experiment.
For the experiment investigating the influence of temporary application of inflammatory factors, the HSC basal medium was further supplemented with 10 ng/mL IL-3, 50 ng/mL IL-7, 50 ng/mL IL-15 (all Miltenyi Biotec, Bergisch Gladbach, Germany), 20 ng/mL GM-CSF and 50 ng/mL EPO (both Peprotech, Cranbury, NJ, USA). EPO concentration was increased to 250 ng/mL starting on day 10 of the experiment.
For the TfR binding antibody experiments and the T cell experiments, the HSC basal medium was further supplemented with 10 ng/mL IL-3 (Miltenyi Biotec, Bergisch Gladbach, Germany), 20 ng/mL GM-CSF, and 10 ng/mL EPO (both Peprotech, Cranbury, NJ, USA). The EPO concentration was increased to 50 ng/mL starting on day 10 of the experiment. During the establishment of the T cell co-culture, some groups were additionally supplemented with 10 ng/mL IL-2 (Biolegend, San Diego, CA, USA) starting on day 17.
Chip cultivation and cell sampling
Cell culture medium was changed on Monday, Wednesday, and Friday during the course of the assay. Each day, 200 µL were removed from the outer ceramic compartment and the inner medium compartment and transferred to 1.5 mL centrifuge tubes. The compartments were then refilled with 200 µL of the respective cell culture medium. Cell culture supernatants were used for LDH measurement and the remaining volume was stored in deep well assay plates at −80 °C.
Starting on day 10 of the assay, cells were harvested weekly for flow cytometric analysis. Instead of taking 200 µL from each compartment, 400 µL were taken from the medium compartment with resuspension and transferred to 1.5 mL centrifugation tubes to remove the circulating cell fraction from the chip. After all chips were finished, the sample cell suspension was centrifuged at 300 g for 5 min. The supernatants were used as described above and the cell pellet was resuspended in PBS + 3% FCS (flow cytometry (FC) buffer).
On the last day of cultivation, cells from the ceramic scaffold were also sampled. After sampling the circulating cell fraction as described above, the ceramic scaffold was transferred to a 24-well plate containing 1.2 mL PBS plus 0.6% (w/v) BSA and 1 mM EDTA (PBE buffer). The ceramic scaffold was thoroughly washed by pipetting and then incubated in a second well containing 1.2 mL PBE buffer for 10 min at room temperature. After this incubation period, the ceramic scaffold was washed a second time by pipetting. Both wash fractions, the remaining medium in the ceramic scaffold and the remaining cells in the ceramic scaffold compartment of the Chip2 were pooled and the cells were resuspended in FC buffer. This additional cell sample is referred to as the ceramic cell sample.
T cell handling
Cryopreserved human peripheral blood (PB) T cells (Lonza, Basel, Switzerland) were cultured for 1 week (with preactivation) or 1 day (without preactivation) before addition to the chip culture. Thawed cells were resuspended in IMDM + 10% FCS and centrifuged at 300 g for 10 min.
Pre-activated T cells were then resuspended in ImmunoCult-XF T cell expansion medium supplemented with 2.5 mL ImmunoCult Human CD3/CD28 T cell activator (both from Stemcell Technologies, Vancouver, BC, Canada) and 10 ng/mL IL-2 (Biolegend, San Diego, CA, USA) to adjust the live cell concentration to 106 cells/mL and seeded at 4 mL/well in 6-well ULA plates (d0). After three days (d3), cells were harvested again, centrifuged at 400 g for 5 min, and the live cell concentration was adjusted to 105 cells/mL and seeded at 4 mL/well in 6-well ULA plates. After two days (d5), cells were harvested again, centrifuged at 400 g for 5 min and the live cell concentration was adjusted to 2 × 105 cells/mL and seeded at 4 mL/well in 6-well ULA plates. On day 7 after thawing, the cells were again counted and used for seeding in the chip system.
T cells without pre-activation were resuspended in ImmunoCult-XF T Cell Expansion Medium (Stemcell Technologies, Vancouver, BC, Canada) and counted. The concentration of live cells was adjusted to 106 cells/mL and seeded at 4 mL/well in 6-well ultra-low attachment plates. One day after thawing, the cells were counted again and used for seeding in the chip system.
Compound application
Treatment antibodies or cytokines were added during medium preparation. On the first treatment day, 2× concentrations were prepared to account for the 50% medium exchange rate. All subsequent exchanges were made with 1× concentration of treatment. All antibodies used in the study were provided by Roche (Table 1). Antibodies were shipped on dry ice in 1.5 ml tubes and stored at −80 °C prior to the start of the experiment. Antibody stock solutions were added directly to the prepared cell culture medium. If direct dilution was not possible due to a high dilution factor >1000, a predilution of the antibody in the respective cell culture medium was prepared.
Cell counting
To measure cell number and viability, cells were counted using an automated cell counter (ChemoMetec, Allerod, Denmark) running the NucleoView NC-200 Revision 1.17 software. After extraction of hematopoietic cells from each circuit of the BM chips, the total volume of the extracted cell suspension was measured with a 1000 µL pipette and recorded. Total cell count, live cell count and viability were measured and used as the basis for further calculations with the data from the flow cytometry analysis.
Flow cytometry analysis
The cell suspension from the cell counting procedure was divided into 96-well U-bottom plates depending on the total number of flow cytometry staining panels, e.g., divided into two plates for staining two panels or three plates for staining three separate panels. Splitting was done either by volume (e.g., 100 µL of cell suspension per plate) or by cell number (e.g., 100,000 cells per plate). All remaining cells from each sample were pooled and this cell suspension was used for single staining controls and fluorescence minus one (FMO) controls. All centrifugation steps were performed at 300 g for 5 min at 4 °C.
Plates were centrifuged and cell pellets were resuspended in 50 µl of 5% TruStain FcX (Biolegend, San Diego, CA, USA) in FC buffer and incubated for 20 min at 4 °C. 150 µL FACS buffer per well was added to all plates and the plates were centrifuged. The supernatants were discarded and the cell pellets were resuspended in 50 µL of the respective antibody staining solution and incubated for 20 min at 4 °C. Antibody staining solutions were freshly prepared on each day of flow cytometry by diluting the antibodies in a 1:1 mixture of BD Horizon Brilliant Stain Buffer (BD, East Rutherford, NJ, USA) and FC Buffer. The staining procedure was stopped by adding 150 µL of PBS to each staining sample followed by centrifugation. The supernatant was discarded and cells from the single staining controls and the viability dye FMO were resuspended in 150 µL PBS and stored at 4 °C until further processing. Cells from the full panel staining, the viability dye single staining control, and the remaining FMO controls were resuspended in 50 µL of viability dye, diluted 1:400 in PBS, and incubated at 4 °C for 20 min. Depending on the panel, Zombie Yellow (405 nm excitation) or Zombie Green (488 nm excitation) dye (both Biolegend, San Diego, CA, USA) was used as a viability stain. The cells were then washed once with 150 µL FC buffer and resuspended in 100 µL Fluorofix fixation buffer (Biolegend, San Diego, CA, USA). The cells were fixed for 30 min at 4 °C. Then 100 µL FC buffer was added and the plates were centrifuged for 5 min at 600 × g at 4 °C. Samples were resuspended in 200 µL FACS buffer and stored overnight until assayed.
The next day, the unstained control as well as the individual staining of each marker were used to set the flow cytometer voltage settings of the measurement, before measuring all FMO control staining as well as the full panel staining on a MACSQuant 16 analyzer (Miltenyi Biotec, Bergisch Gladbach, Germany) running the MACSQuantify Version 2.13 software. Analysis of flow cytometry data and gating was performed in FlowJo v10.8.0 software.
Panels and antibodies that have been used are shown in Tables 2, 3:
LDH measurement
To monitor cell death and/or cell proliferation in the chip cultures, LDH concentrations were measured using the LDH IFCC kit (Thermo Fisher Diagnostics) on an Indiko Plus chemical analyzer (Thermo Fisher Scientific, Waltham, MA, USA). The results of the photometric analysis are automatically calculated by the Indiko Plus based on a two-point calibration with enzyme calibrator serum (eCal) and 0.9% NaCl, which is performed every 10 days or when the reagent batch is changed. A quality control is performed using abnormal (Abtrol) and normal (Nortrol) human control serum (Thermo Fisher Scientific) before starting measurements.
Confirmation of stromal cell purity by flow cytometry
To confirm stromal cell purity, stromal cells were expanded until the day when they were seeded onto ceramic scaffolds. Cells were stained by flow cytometry as described before with the following set of antibodies: CD45-BV605 (Biolegend, 368524), CD14-PE (Biolegend, 325606), CD206-FITC (Biolegend, 321103). The cell population were subsequently analyzed for presence of CD45+ hematopoietic cells and in that population, presence of CD14 + CD206− monocytes and CD14 + CD206+ macrophages.
RNA-sequencing of stromal cells grown on ceramic scaffolds
To analyze stromal cell identity, ceramic scaffolds with stromal cells were cultured for 24 days under static conditions in 24-well plates in Mesencult-ACF Plus medium (Stemcell Technologies, Vancouver, BC, Canada) or StemSpan AOF (Stemcell Technologies) supplemented with 10 ng/mL TPO, 25 ng/mL FLT-3L, 20 ng/mL SCF, 20 ng/mL GM-CSF (all Peprotech, Cranbury, NJ, USA), 10 ng/mL IL-3 (Miltenyi Biotec, Bergisch Gladbach, Germany), 100 µg/mL holo-transferrin (Sigma Aldrich, St. Louis, MO, USA), 100 IU/mL penicillin + 100 µg/mL streptomycin and 5 µg/mL gentamicin (both Corning, Corning, NY, USA). In addition, ceramic scaffolds were seeded with stromal cells for 24 days in the HUMIMIC Chip2 under in StemSpan AOF with the same supplements. After 24 days, the scaffolds were washed with PBS and total RNA was extracted using the NucleoSpin 8 RNA Core Kit (Macherey-Nagel, Düren, Germany). RNA was quantified by spectrophotometric analysis using the NanoDrop 2000 UV–vis instrument (Thermo Fisher Scientific) and quality controlled by capillary electrophoresis on the 2100 Bioanalyzer Instrument (Agilent Technologies, Santa Clara, CA, USA) using the RNA 6000 Nano Kit (Agilent Technologies).
Paired-end (PE) sequencing libraries were prepared from 100 ng of RNA using the TruSeq Stranded mRNA Sample Preparation Kit (Illumina, San Diego, CA, USA) according to the manufacturer’s instructions. Sequencing libraries were quantified using the Qubit dsDNA High Sensitivity Kit (Thermo Fisher Scientific) on Qubit 4.0 Fluorometer (Thermo Fisher Scientific) and quality control was performed on a 2100 Bioanalyzer Instrument (Agilent Technologies) using the DNA High Sensitivity Kit (Agilent Technologies). PE Libraries were normalized and multiplexed into a single sequencing library pool. The library pool was sequenced on a NovaSeq6000 sequencer (Illumina) for 2 × 50 cycles using a NovaSeq6000 S1 100 cycles Reagent Kit (Illumina). As a sequencing control 1% of a PhiX control library (Illumina) was spiked into the library pool. The obtained sequencing depths were ranging between ∼30 and 81 million PE reads with an average depth of ∼55 million PE reads.
The RNA-Seq data processing involved mapping the reads to the human reference genome (hg38) using the STAR algorithm62. Gene-level read counts were collated and normalized to account for variations in library size and composition. The analysis was restricted to genes expressed in at least three samples, with a minimum read count per million (CPM) greater than one.
Differential gene expression analysis was conducted using the voom/limma framework63. To identify enriched gene signatures and pathways among the differentially expressed genes, we applied the CAMERA method, a competitive gene set test that considers inter-gene correlations64. We defined the entire set of protein-coding genes as the background. CAMERA was then employed to analyze the ranked differentially expressed genes for each contrast, using the standard inter-gene correlation factor of 0.01.
Cytokine quantification
Media supernatant samples for cytokine quantification were collected during media exchange as described above. In the experiment testing the effects of inflammatory factors, 55 µL were collected from the outer compartment without volume replacement 6 h after application of the inflammatory factors. Sampled media supernatants were stored at −80 °C until cytokine quantification.
Medium supernatants were analyzed for cytokine levels using a Luminex Human Discovery Assay (R&D Systems) according to the manufacturer’s protocol. Customized cytokine panels were used. A panel detecting CCL2/MCP-1, TNF-α, LAP (TGF-β), IP-10, IL-7, IL-6, IL-4, IL-12, IL-10, IL-1β, and IFN-γ was used for the TfR binding antibody experiments, and a panel detecting CCL2/MCP-1, G-CSF, IL-1β, IL-6, IL-8/CXCL8, IL-23, M-CSF, Osteopontin/OPN, and TNF-α was used for the inflammatory factor experiments. Samples were analyzed on a Bioplex200 instrument (BioRad # 171000201) running the Bio-Plex Manager 6.0 software including standard curves for all cytokines. The concentration of the samples was calculated in the BioPlex acquisition software.
Statistics and reproducibility
Results are presented as mean ± s.e.m. or ± SD as indicated in the figure legends. All data were initially stored and processed in Microsoft excel 2019. Statistical analyses were performed using GraphPad Prism 10 (GraphPad Software Inc.). Different levels of significance (p-values) are indicated by asterisks in each figure (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001).
Development of cell counts in the microfluidic chip was analyzed using a non-linear regression with a 4-parameter variable slope sigmoidal curve fit. This model best represents the observed cell proliferation characteristics within the microcircuit chip. Initially, cell numbers in the medium reservoir exhibit a lag phase while cells undergo differentiation and proliferation within the ceramic scaffold. This is followed by an exponential growth phase as cells proliferate rapidly. Finally, the growth enters a plateau phase as the system reaches maximal growth capacity.
For each experiment the total number of replicates was limited to 24 chips equaling 48 technical replicates. Test groups were splitted evenly among the available replicates. All experiments were carried out with multiple CD34+ donors when feasible. We did not perform power analysis prior to initiation of the study.
Figure legends indicate the number of biological samples (N) of individual CD34+ cell donors and the number of (technical) pseudoreplicates (n) of individual independent chips. All experiments use a matched-samples design, where the same set of donors is used for all treatment groups. Each donor cell was exposed to multiple treatment conditions, ensuring that the samples are inherently matched across these conditions. This design allows us to control for inter-donor variability and focus on the within-donor effects of treatments.
Multiple chips (n = 2–3) were used as pseudoreplicates for each donor. For statistical comparison of cell count data, the means of these pseudoreplicates were calculated and log-transformed before statistical tests were performed to stabilize the variance and normalize the distribution. For analysis of cytokine concentrations in the cell culture medium, area under the curve values were calculated, averaged across pseudoreplicates, and compared.
Statistical comparison of means was performed using multiple ratio paired t-tests (Fig. 7I), repeated measures one- or two-way ANOVA (Figs. 2B, 4, 5F and 7C–H), or a repeated measures mixed effects model REML (Fig. 5B–E) when not all treatment conditions were tested in all experiments. Because the experimental design relies on matching rather than repeated measures, we assumed sphericity of the data as violations are unlikely. Following repeated measures ANOVA, Dunnett’s multiple comparison test was used to compare each treatment group with the untreated control group, and Tukey’s test was used for pairwise comparisons among all treatment groups to control for type I error.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the results of this study are available in the article and in the Supplementary Information. Raw data sets of flow cytometry count data and cytokine concentration data are made available in the Supplementary data file. The single cell RNA sequencing datasets generated in this study have been deposited in GEO database under accession code GSE281012. Extra data are available from the corresponding author upon reasonable request.
Bispecific antibodies used in the study: CD33tcb-D04, TfR-FcSilent and TfR-FcWT were developed by F. Hoffmann-La Roche AG and are subject to patent applications and therefore cannot be made available to the scientific community at this time.
References
Walsh, G. & Walsh, E. Biopharmaceutical benchmarks 2022. Nat. Biotechnol. 40, 1722–1760 (2022).
Everds, N. E. & Tarrant, J. M. Unexpected hematologic effects of biotherapeutics in nonclinical species and in humans. Toxicol. Pathol. 41, 280–302 (2013).
Zhu, Y., Liu, K., Wang, K. & Zhu, H. Treatment-related adverse events of antibody–drug conjugates in clinical trials: a systematic review and meta-analysis. Cancer 129, 283–295 (2023).
Kroll, M. H., Rojas-Hernandez, C. & Yee, C. Hematologic complications of immune checkpoint inhibitors. Blood 139, 3594–3604 (2022).
Rosalem, G. S. et al. Microfluidics and organ-on-a-chip technologies: a systematic review of the methods used to mimic bone marrow. PLoS ONE 15 https://doi.org/10.1371/journal.pone.0243840 (2020).
Sharipol, A., Lesch, M. L., Soto, C. A. & Frisch, B. J. Bone marrow microenvironment-on-chip for culture of functional hematopoietic stem cells. Front. Bioeng. Biotechnol. 10, 1–15 (2022).
Torisawa, Y. et al. Bone marrow–on–a–chip replicates hematopoietic niche physiology in vitro. Nat. Methods 11, 663–669 (2014).
Marturano-Kruik, A. et al. Human bone perivascular niche-on-a-chip for studying metastatic colonization. Proc. Natl. Acad. Sci. USA 115, 1256–1261 (2018).
Bruce, A. et al. Three-dimensional microfluidic tri-culture model of the bone marrow microenvironment for study of acute lymphoblastic leukemia. PLoS ONE 10, e0140506 (2015).
McAleer, C. W. et al. Multi-organ system for the evaluation of efficacy and off-target toxicity of anticancer therapeutics. Sci. Transl. Med. 11, https://doi.org/10.1126/scitranslmed.aav1386 (2019).
Zhang, W., Lee, W. Y., Siegel, D. S., Tolias, P. & Zilberberg, J. Patient-specific 3D microfluidic tissue model for multiple myeloma. Tissue Eng. Part C. Methods 20, 663–670 (2014).
Houshmand, M. et al. Mimicking the acute myeloid leukemia niche for molecular study and drug screening. Tissue Eng. Part C. Methods 23, 72–85 (2017).
Thon, J. N. et al. Platelet bioreactor-on-a-chip. Blood 124, 1857–1867 (2014).
Nelson, M. R. et al. A multi-niche microvascularized human bone marrow (hBM) on-a-chip elucidates key roles of the endosteal niche in hBM physiology. Biomaterials 270, 120683 (2021).
Glaser, D. E. et al. Organ-on-a-chip model of vascularized human bone marrow niches. Biomaterials 280, 121245 (2022).
Chou, D. B. et al. On-chip recapitulation of clinical bone marrow toxicities and patient-specific pathophysiology. Nat. Biomed. Eng. 4, 394–406 (2020).
Sieber, S. et al. Bone marrow-on-a-chip: long-term culture of human haematopoietic stem cells in a three-dimensional microfluidic environment. J. Tissue Eng. Regen. Med. 12, 479–489 (2018).
Cairns, J. et al. Optimal experimental design for efficient toxicity testing in microphysiological systems: a bone marrow application. Front. Pharmacol. 14, https://doi.org/10.3389/fphar.2023.1142581 (2023).
Zhang, C., Zhang, B., Lin, L.-L. & Zhao, S. Evaluation and comparison of computational tools for RNA-seq isoform quantification. BMC Genomics 18, 583 (2017).
Matsushita, Y., Ono, W. & Ono, N. Toward marrow adipocytes: adipogenic trajectory of the bone marrow stromal cell lineage. Front. Endocrinol. (Lausanne). 13, https://doi.org/10.3389/fendo.2022.882297 (2022).
Youlten, S. E. et al. Osteocyte transcriptome mapping identifies a molecular landscape controlling skeletal homeostasis and susceptibility to skeletal disease. Nat. Commun. 12, 2444 (2021).
Migliaccio, A. R., Masselli, E., Varricchio, L. & Whitsett, C. Ex-vivo expansion of red blood cells: how real for transfusion in humans?. Blood Rev. 26, 81–95 (2012).
Chen, K. et al. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc. Natl. Acad. Sci. USA 106, 17413–17418 (2009).
Aanei, C. M., Picot, T., Tavernier, E., Guyotat, D. & Campos Catafal, L. Diagnostic utility of flow cytometry in myelodysplastic syndromes. Front. Oncol. 6, https://doi.org/10.3389/fonc.2016.00161 (2016).
Mukherjee, R. et al. Non-Classical monocytes display inflammatory features: validation in sepsis and systemic lupus erythematous. Sci. Rep. 5, 13886 (2015).
Dolzhanskiy, A., Hirst, J., Basch, R. S. & Karpatkin, S. Complementary and antagonistic effects of IL-3 in the early development of human megakaryocytes in culture. Br. J. Haematol. 100, 415–426 (1998).
Xu, Y., Murphy, A. J. & Fleetwood, A. J. Hematopoietic progenitors and the bone marrow niche shape the inflammatory response and contribute to chronic disease. Int. J. Mol. Sci. 23, 2234 (2022).
Najafi, S., Ghanavat, M., Shahrabi, S., Gatavizadeh, Z. & Saki, N. The effect of inflammatory factors and their inhibitors on the hematopoietic stem cells fate. Cell Biol. Int. 45, 900–912 (2021).
Hormaechea-Agulla, D., Le, D. T. & King, K. Y. Common sources of inflammation and their impact on hematopoietic stem cell biology. Curr. Stem Cell Rep. 6, 96–107 (2020).
Morales-Mantilla, D. E. & King, K. Y. The role of interferon-gamma in hematopoietic stem cell development, homeostasis, and disease. Curr. Stem Cell Rep. 4, 264–271 (2018).
Chou, D. B. et al. Stromal-derived IL-6 alters the balance of myeloerythroid progenitors during Toxoplasma gondii infection. J. Leukoc. Biol. 92, 123–131 (2012).
Lin, F. et al. IFN-γ causes aplastic anemia by altering hematopoietic stem/progenitor cell composition and disrupting lineage differentiation. Blood 124, 3699–3708 (2014).
Candelaria, P. V., Leoh, L. S., Penichet, M. L. & Daniels-Wells, T. R. Antibodies targeting the transferrin receptor 1 (TfR1) as direct anti-cancer agents. Front. Immunol. 12, https://doi.org/10.3389/fimmu.2021.607692 (2021).
Yu, Y. J. et al. Therapeutic bispecific antibodies cross the blood-brain barrier in nonhuman primates. Sci. Transl. Med. 6, https://doi.org/10.1126/scitranslmed.3009835 (2014).
Dinh, H. Q. et al. Coexpression of CD71 and CD117 identifies an early unipotent neutrophil progenitor population in human bone marrow. Immunity 53, 319–334.e6 (2020).
Couch, J. A. et al. Addressing safety liabilities of TfR bispecific antibodies that cross the blood-brain barrier. Sci. Transl. Med. 5, https://doi.org/10.1126/scitranslmed.3005338 (2013).
Pardridge, W. M., Boado, R. J., Patrick, D. J., Ka-Wai Hui, E. & Lu, J. Z. Blood-Brain barrier transport, plasma pharmacokinetics, and neuropathology following chronic treatment of the rhesus monkey with a brain penetrating humanized monoclonal antibody against the human transferrin receptor. Mol. Pharm. 15, 5207–5216 (2018).
Lo, M. et al. Effector-attenuating substitutions that maintain antibody stability and reduce toxicity in mice. J. Biol. Chem. 292, 3900–3908 (2017).
Saber, H., Del Valle, P., Ricks, T. K. & Leighton, J. K. An FDA oncology analysis of CD3 bispecific constructs and first-in-human dose selection. Regul. Toxicol. Pharmacol. 90, 144–152 (2017).
Kroll, K. T. et al. Immune-infiltrated kidney organoid-on-chip model for assessing T cell bispecific antibodies. Proc. Natl. Acad. Sci. USA 120, https://doi.org/10.1073/pnas.2305322120 (2023).
Clark, M. C. & Stein, A. CD33 directed bispecific antibodies in acute myeloid leukemia. Best. Pract. Res. Clin. Haematol. 33, 101224 (2020).
Walter, R. B., Appelbaum, F. R., Estey, E. H. & Bernstein, I. D. Acute myeloid leukemia stem cells and CD33-targeted immunotherapy. Blood 119, 6198–6208 (2012).
Liu, Y. et al. CD33-directed immunotherapy with third-generation chimeric antigen receptor T cells and gemtuzumab ozogamicin in intact and CD33-edited acute myeloid leukemia and hematopoietic stem and progenitor cells. Int. J. Cancer 150, 1141–1155 (2022).
Chaudhari, P. B. Nivolumab – Pearls of evidence. Indian J. Med. Paediatr. Oncol. 38, 520–525 (2017).
Pamphilon, D. et al. Current practices and prospects for standardization of the hematopoietic colony-forming unit assay: a report by the cellular therapy team of the Biomedical Excellence for Safer Transfusion (BEST) Collaborative. Cytotherapy 15, 255–262 (2013).
Olaharski, A. J. et al. In vitro to in vivo concordance of a high throughput assay of bone marrow toxicity across a diverse set of drug candidates. Toxicol. Lett. 188, 98–103 (2009).
Ewart, L. et al. Application of microphysiological systems to enhance safety assessment in drug discovery. Annu. Rev. Pharmacol. Toxicol. 58, 65–82 (2018).
Donnelly, H., Salmeron-Sanchez, M. & Dalby, M. J. Designing stem cell niches for differentiation and self-renewal. J. R. Soc. Interface 15, 20180388 (2018).
Suck, K. et al. Cultivation of MC3T3-E1 cells on a newly developed material (Sponceram®) using a rotating bed system bioreactor. J. Biomed. Mater. Res. Part A 80A, 268–275 (2007).
François, M. et al. Mesenchymal stromal cells cross-present soluble exogenous antigens as part of their antigen-presenting cell properties. Blood 114, 2632–2638 (2009).
Sakamoto, C. et al. Automated enumeration of cellular composition in bone marrow aspirate with the CELL-DYN 4000TM automated hematology analyzer. Acta Haematol. 101, 130–134 (1999).
Ganz, T. Anemia of inflammation. N. Engl. J. Med. 381, 1148–1157 (2019).
Martin, K. R., Wong, H. L., Witko-Sarsat, V. & Wicks, I. P. G-CSF – A double edge sword in neutrophil mediated immunity. Semin. Immunol. 54, 101516 (2021).
Moos, T., Gudbergsson, J. M. & Johnsen, K. B. Transport of transferrin receptor-targeted antibodies through the blood-brain barrier for drug delivery to the brain. 527–549; https://doi.org/10.1007/978-3-030-88773-5_17 (2022).
Schlothauer, T. et al. Novel human IgG1 and IgG4 Fc-engineered antibodies with completely abolished immune effector functions. Protein Eng. Des. Sel. 29, 457–466 (2016).
Marrer-Berger, E. et al. The physiological interactome of TCR-like antibody therapeutics in human tissues. Nat. Commun. 15, 3271 (2024).
Dao, T. et al. Therapeutic bispecific T-cell engager antibody targeting the intracellular oncoprotein WT1. Nat. Biotechnol. 33, 1079–1086 (2015).
Gejman, R. S. et al. Identification of the targets of t-cell receptor therapeutic agents and cells by use of a high-throughput genetic platform. Cancer Immunol. Res. 8, 672–684 (2020).
Ataie, N. et al. Structure of a TCR-mimic antibody with target predicts Pharmacogenetics. J. Mol. Biol. 428, 194–205 (2016).
Brashem-Stein, C. et al. Ontogeny of hematopoietic stem cell development: reciprocal expression of CD33 and a novel molecule by maturing myeloid and erythroid progenitors. Blood 82, 792–799 (1993).
Shamliyan, T. A. & Kane, R. L. Availability of results from clinical research: failing policy efforts. J. Epidemiol. Glob. Health 4, https://doi.org/10.1016/j.jegh.2013.08.002 (2013).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Law, C. W., Chen, Y., Shi, W. & Smyth, G. K. voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014).
Wu, D. & Smyth, G. K. Camera: a competitive gene set test accounting for inter-gene correlation. Nucleic Acids Res. 40, e133 (2012).
Acknowledgements
We thank Adrian Roth, Thomas Singer, Ekaterina Breous-Nystrom, Estelle Marrer-Berger, Reyk Horland and Uwe Marx for their supervision of the research cooperation in the joint steering committee.
This study was funded by the ROADS program of the Innovation Partnerships at Roche.
Author information
Authors and Affiliations
Contributions
L.K., L.J., D.S. and A.W. conceived, designed, and performed experiments, and analyzed and interpreted the data. L.K. and D.S. co-wrote the manuscript. L.K. generated the figures. T.P.T. and S.F. performed experiments and analyzed the data. S.I.J. and J.N. supplied test molecules and contributed to discussion. N.J. designed experiments, provided critical review and conclusions for TfR1-related results. R.M. designed experiments, provided critical review and conclusions for TfR1-related results and reviewed the manuscript. I.C. performed the RNA QC, library preparation and sequencing, and contributed to the manuscript preparation. D.M. performed data curation, analysis and plotting of bulk RNA Seq data.
Corresponding author
Ethics declarations
Competing interests
The authors declare the following competing interests: L.J., S.F., I.C., S.I.J., N.J., R.M., D.M., J.N., and D.S. are paid employees of Roche at the time of writing this manuscript. L.K., T.P.T., and A.W. are paid employees of TissUse GmbH at the time of writing this manuscript and received grants and non-financial or other support from F. Hoffmann-La Roche during the conduct of the study.
Peer review
Peer review information
Communications Biology thanks Benjamin J. Frisch, Yu-Suke Torisawa, and Petek Korkusuz for their contribution to the peer review of this work. Primary Handling Editor: Johannes Stortz. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Koenig, L., Juglair, L., Tao, T.P. et al. A microfluidic bone marrow chip for the safety profiling of biologics in pre-clinical drug development. Commun Biol 8, 754 (2025). https://doi.org/10.1038/s42003-025-08137-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08137-1
This article is cited by
-
Human preclinical multiple myeloma in vitro models for disease modeling and therapy screening
Journal of Biological Engineering (2025)