Abstract
Systemic lupus erythematosus (SLE) is an autoimmune disease driven by autoantibody production. Lupus nephritis (LN), a severe SLE complication, is primarily caused by renal autoantibodies. Long-lived plasma cells (LLPCs), the main producers of these autoantibodies, are especially elevated in the kidney of LN patients, particularly in refractory or recurrent cases. However, the cause of increased LLPCs in LN kidneys remains unknown. This study uses an LN mouse model and combines single-cell RNA sequencing with spatial transcriptomics, finding that kidney-resident Cxcl9high macrophages and their secreted chemokine Cxcl9 significantly rise with disease progression. This increase in Cxcl9 attracts Cxcr3+ plasmablasts in peripheral blood into the kidneys, where they differentiate into LLPCs and produce autoantibodies. Based on these findings, this study suggests that Cxcl9high macrophages are the inducing factor causing the increase of LLPCs in LN kidneys and may be a potential therapeutic target for LN.

Similar content being viewed by others
Introduction
Systemic Lupus Erythematosus (SLE) is an autoimmune disease characterized by autoantibody production by plasma cells (PCs). Lupus nephritis (LN) is the most severe complication of SLE, potentially leading to renal failure1,2.
PCs primarily consist of plasmablasts, short-lived plasma cells (SLPCs), and long-lived plasma cells (LLPCs). Plasmablasts, mainly located in secondary lymphoid organs and bone marrow, can differentiate into either LLPCs or SLPCs. After antigen stimulation, plasmablasts and SLPCs trigger a transient and timely immune response before undergoing rapid apoptosis, while LLPCs can persist for several years, secreting approximately 80% of serum antibodies3,4,5.
Studies indicate a significant increase in LLPCs in the kidneys of LN mice, which are the primary site for autoantibody production (50% in kidneys, 20% in spleen, 30% in bone marrow)6. Patients with refractory and relapsed LN have more LLPCs in their kidneys than untreated, newly diagnosed patients7. Therefore, LLPCs within the kidneys are critical in the pathogenesis of LN, especially in refractory and relapsed cases. But why LLPCs increase in LN kidney and where does they come from is unclear.
Tertiary lymphoid structures (TLS) resemble lymphoid follicles in secondary lymphoid organs like the spleen and lymph nodes8. TLS have been identified in the target tissues of autoimmune diseases, including the kidneys (lupus nephritis), salivary glands (Sjögren’s syndrome), and synovial joints (rheumatoid arthritis)9.
TLS presence in the kidneys of LN patients correlates with an increased disease activity, severe histological findings, and poorer renal prognosis10. About 10% of LN patients with kidney biopsies showed TLS with typical features of germinal center11. So, TLS might be the origin site of LLPC. However, A recent study found that B cells in the kidneys of LN patients cannot differentiate into LLPC10,12.
In the present study, we combined single-cell RNA sequencing (scRNA-seq) with spatial transcriptomics (ST) to comprehensively analyze the origin, inducing factors, and spatial distribution of LLPCs in the kidneys of LN mice model. We discovered that the number of Cxcl9 high macrophages in the LN kidneys gradually increased, and they secreted a large amount of chemokine Cxcl9, which recruited Cxcr3+ plasmablasts from the circulation into the kidneys, where plasmablasts further differentiated into LLPCs. Therefore, our results indicate that Cxcl9high macrophages may a core factor to cause the increase of LLPCs in the kidneys of LN mice.
Results
The immune response increased as renal function declined during LN progression
MRL/lpr mice serve as a common model for spontaneous LN, displaying renal lesions similar to those in humans13. To simulate the pathological grades of human LN, we grouped the MRL/lpr and control MRL/MPJ mice by age: 4, 8, 12, 16, 20, and 24 weeks and analyzed the corresponding pathological features (Fig. 1A).
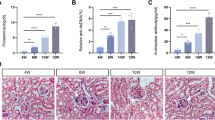
A Schematic representation of experimental grouping. The animals were divided into six groups based on age: 4 weeks (4w), 8 weeks (8w), 12 weeks (12w), 16 weeks (16w), 20 weeks (20w), 24 weeks (24w). Sac: sacrifice. B 24-h proteinuria levels were measured in six groups of mice: Con (control group, MRL/MPJ) and LN (LN group, MRL/lpr). C Serum ANA (antinuclear antibody) levels in the six groups of mice. D PAS staining, immunofluorescence staining (IgG, IgM, C3), and electron microscopy results of glomeruli from 4w LN, 12w LN, and 20w LN mice, with red arrows indicating immune complexes deposited in different parts of the glomerulus. E H&E staining, immunohistochemical CD45 staining, and Masson’s trichrome staining were used to observe the infiltration of interstitial immune cells in different parts of the renal cortex and medulla and the degree of fibrosis in the renal cortex of mice, with black arrows indicating immune cells infiltrated in different parts of the kidneys. Data are presented as mean ± SD. (n = 6). *p < 0.05, **p < 0.01, ***p < 0.001; #p < 0.05, ##p < 0.01, ###p < 0.001; * represents the significant degree of difference between the LN and Con groups. # represents the significance of the differences among the six groups of LN mice. PAS scale bar: 10 μm, immunofluorescence scale bar: 50 μm, transmission electron microscopy scale bar: 2 μm, H&E scale bar: 50 μm, CD45 scale bar: 20 μm, Masson scale bar: 50 μm.
The results indicated that the survival rate of the mice decreased with age (Supplementary Fig. 1A), while 24-h proteinuria, urinary protein-to-creatinine ratio (UPCR) (Fig. 1B and Supplementary Fig. 1B), serum creatinine, serum urea nitrogen (Supplementary Fig. 1C, D), serum ANA, anti-dsDNA antibody, spleen coefficient (Fig. 1C and Supplementary Fig. 1E, F), and kidney coefficient (Supplementary Fig. 1G) increased gradually with age.
PAS, immunofluorescence, and transmission electron microscopy results showed that glomerular injury, IgG, IgM, C3, and immune complexes deposition increased with age (Fig. 1D and Supplementary Fig. 2A). Renal interstitial inflammatory cell infiltration and tubular atrophy are closely linked to the prognosis of LN, and are important markers for its progression to uremia14. The hematoxylin and eosin (H&E) staining and CD45 immunohistochemistry indicated that the number of interstitial immune cells increased in kidneys of LN mice with age. Masson staining revealed significant renal interstitial fibrosis in LN mice from 20 weeks of age (Fig. 1E and Supplementary Fig. 2B).
The above renal pathology feature of LN mice in different ages are listed in Supplementary Table 1. According 2016 revised ISN/RPS renal pathological classification criteria and NIH LN activity and chronicity scoring15: the 4-weeks-old mice showed no disease; the kidney lesions of 8-weeks-old LN mice were similar to human LN class II; At 12 weeks and 16–20 weeks, the renal pathology in the mice corresponded to human LN class III and LN class IV, respectively; At 24 weeks, they resembled human LN class VI. Finally, based on survival rates and kidney injury severity, we selected the 4-weeks LN (4w LN), 12-weeks LN (12w LN), and 20-weeks (20w LN) mice for the following experiments.
Changes in the cell profile in LN kidney tissues
We identified 87,906 cells in the kidneys of 4w, 12w, and 20w LN mice using the 10× Genomics scRNA-seq platform, consisting of 80% CD45+ immune cells and 20% CD45- kidney-resident cells. The cell counts were 27,888 from 4w LN, 29,496 from 12w LN, and 30,522 from 20w LN (Fig. 2A).
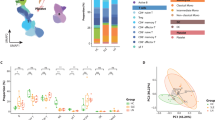
A Experimental flowchart. B UMAP plot illustrating cell clusters derived from combined single-cell data (87,906 cells) from the kidneys of three groups of LN mice, with each dot representing an individual cell and each color indicating a distinct cell subgroup. C Violin plot displaying the expression levels of typical marker genes for each cell subgroups. D Heatmap depicting the expression levels of the top 10 DEGs across each cell subgroups. E UMAP illustrating the distribution of cell clusters by sample grouping after integrating the single-cell data from the kidneys of the three groups of LN mice. F Circular plots showing the distribution ratio of each cell subgroups in the kidneys of the three groups for LN mice.
Cell annotation identified 11 subgroups in LN kidneys based on unbiased clustering and typical lineage markers, including immune cells (T lymphocytes (T), B lymphocytes (B), plasma cells (PC), dendritic cells (DC), natural killer cells (NK), macrophages (Mφ), innate lymphoid cells (ILC), and neutrophils (NEUT)) and non-immune populations (renal tubular epithelial cells (TEC), endothelial cells (EC), and pericytes (PEC)) (Fig. 2B). Typical lineage markers for each cell subgroup are shown in Fig. 2C. The heatmap illustrates the expression levels of the top 10 differentially expressed genes (DEGs) in each subgroup (Fig. 2D), with the gene list in Supplementary Table 2. We found that the proportion of the infiltrating plasma cells in the kidneys of LN mice is positively correlated with LN progression (Fig. 2E, F). The proportions of 11 cell subgroups were detailed in Supplementary Table 3.
A significant increase in plasmablasts and LLPCs in kidneys of LN mice
To further characterize LLPCs in LN mice kidneys, we performed secondary cluster analysis by merging identified B cells and plasma cells. We categorized these cells into 8 subtypes: Naive B cells, memory B cells (MBC), transitional plasma cells (Trans PC1-4), and mature plasma cells (PC1 and PC2) (Fig. 3A). The gene expression patterns of Trans PC4, PC1, and PC2 matched those of plasmablasts, SLPCs, and LLPCs from previous studies16,17.
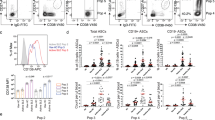
A UMAP representation of 8 distinct subtypes of B cells and plasma cells (PCs) in the kidneys of three groups of LN mice. Each dot represents an individual cell, with color coding indicating different cell types. B–D Nomenclature for the 8 cell subtypes. E Proportions of 8 various subtypes in the kidneys of the three groups of LN mice. F Flow cytometry analysis of CD45+ immune cells, B cells, Trans PC4, PC1, and PC2 in the kidneys of the three groups of LN mice. Data are presented as mean ± SD. (n = 6), *p < 0.05, **p < 0.01, ***p < 0.001.
B cells highly express Ptprc (B220), Cd19, major histocompatibility complex class II (MHC II) molecules (Cd74, H2-Ab1, H2-Ea, H2-Eb1, H2-Aa, H2-Ob, H2-DMb2, H2-DMa), and chemokine receptors (Cxcr4, Cxcr5)18.
B cell clusters are divided into Naive B cells and MBC based on cell differentiation and maturation. Naive B cells express key transcription factors for early B cell development, including Ebf1, Pax5, Cd19, Cd79a, Cd79b, Vpreb3, Bank1, Ms4a1, Iglc2 or Igkc, Ighm, and Ighd19,20,21, along with follicular B cell-related genes like Fcer2a and Ccr7. MBC express mature B cell genes such as H2-Aa, H2-Ab1, Ighg1, Ighg2b, and Ighg2c, as well as memory B cell-related genes like Cd38, Nt5e, Pdcd1lg2, and Cd8022,23. They also show high expression of B1 cell-related genes like Zbtb20 and Plac819,24 (Fig. 3B, C). In addition, MBC cells express a higher level of Igha, which may be due to the presence of more autoantigens in LN mice that can bind to MBCs with high Igha expression.
Mature plasma cells, including PC1 (SLPCs) and PC2 (LLPCs), express high levels of plasma cell-related genes (Sdc1 (CD138), Prdm1, Xbp1, Jchain, Epcam)25,26. PC1 (SLPCs) have low expression of MHC II and chemokine receptor genes, while PC2 (LLPCs) show minimal expression of these genes16 (Fig. 3D).
Transitional plasma cells, including Trans PC1, Trans PC2, Trans PC3, and Trans PC4 (plasmablasts), represent distinct populations during the differentiation of B cells into plasma cells, each expressing different levels of genes related to mature B and plasma cells. The proliferation-related genes Pcna and Mki67 are highly expressed in Trans PC4, while the other transitional plasma cell types and PC1 (SLPCs) show low expression. B cells and PC2 (LLPCs) basically did not express the two genes (Fig. 3D).
scRNA-seq data were used to analyzed the percentage of 8 cell subtypes in three groups of LN mice, revealing that the proportions of Trans PC4 (plasmablasts) and PC2 (LLPCs) increased as disease progressed (Fig. 3E). Flow cytometry further validated that the proportions of total immune cells, B cells, Trans PC4 (plasmablasts), PC1 (SLPCs), and PC2 (LLPCs) also rose with LN progression (Fig. 3F). The flow cytometry gating strategy is listed in Supplementary Fig. 3.
Significantly enhanced in antibody synthesis and secretion of plasmablasts and LLPCs in the kidneys of LN mice
The top 5 DEGs in Naive B cells, MBC, Trans PC1, Trans PC2, Trans PC3, Trans PC4 (plasmablasts), PC1 (SLPCs), and PC2 (LLPCs) are listed in Fig. 4A. Analysis of highly expressed differentially genes (Log2 FC > 0.26) across 8 cell subtypes using KEGG and GO functional enrichment tool revealed that Naive B cells and MBC are mainly linked to local immune defense, the pathways include B cell receptor signaling pathway, chemokine-mediated signaling pathway, lysosome and antigen processing and presentation. Trans PC1, Trans PC2, and Trans PC3 are mainly involved in chemotaxis activities, such as neutrophil chemotaxis, monocyte chemotaxis, eosinophil chemotaxis, and chemokine-mediated signaling pathway. The enriched pathways of Trans PC4 (plasmablasts) focus on cell proliferation and division, including DNA replication, G1/S transition of mitotic cell cycle, cell division. However, PC1 (SLPCs) and PC2 (LLPCs) are linked to immunoglobulin synthesis and secretion, involving protein export, protein processing in endoplasmic reticulum and endoplasmic reticulum unfolded protein response (Fig. 4B, C).
A The top 5 highly expressed genes in Naive B cells, MBC (memory B cells), Trans PC1, Trans PC2, Trans PC3, Trans PC4, PC1, and PC2. B Heatmap showing KEGG enriched pathways of 8 cell subtypes. C Heatmap showing GO enriched pathways of 8 cell subtypes. Heatmap was performed using the OmicStudio tools at https://www.omicstudio.cn. D–K Dot plots showed significant differences in enrichment of the main enriched pathways among three groups of the eight types of cells. KEGG/GO legend = Log10 (Q.value), legend_max = 0 legend_min = −6, log_tip = 1e-6; Size: Rich.Factor; Color: Log10 (Q.value)/20. PB (plasmablasts), SLPCs (Short-lived plasma cells), LLPCs (Long-lived plasma cells). G Enzyme-linked immunosorbent spot (ELISPOT) assay was used to detect the number of IgG-secreting cells in the kidneys of LN mice. Data are presented as mean ± SD. (n = 6), *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we compared the enrichment functions of 8 cell subtypes at various disease stages. The results show that, unlike MBC, the enrichment level of related pathways for Naive B cells decreased with disease advancement, indicating limited involvement of these cells in LN progression (Fig. 4D, E, G). Meanwhile, the enrichment significance of the main pathways involved in antibody production and secretion in Trans PC1, Trans PC3, and Trans PC4 (plasmablasts) gradually increased with LN progression.
Interestingly, we found that the enrichment significance of DNA replication and cell cycle pathways in Trans PC4 (plasmablasts) exhibits a markedly upward trend, suggesting that Trans PC4 (plasmablasts) is extremely active during LN progression (Fig. 4F, H, I). Additionally, the primary enriched pathways for PC1 (SLPCs) and PC2 (LLPCs) involve protein processing in the endoplasmic reticulum and protein export. Their enrichment significance increases with LN progression, highlighting their crucial role in local antibody production and secretion within the kidneys (Fig. 4J, K).
We then used Enzyme-Linked Immunospot Assay to measure the number of IgG-secreting cells in the kidneys of three groups of lupus mice, which showed a gradual increase in these cells with LN progression (Fig. 4G).
The results above indicate that, in addition to Naive B cells and Trans PC2, the MBC, Trans PC1, Trans PC3, Trans PC4 (plasmablasts), PC1 (SLPCs), and PC2 (LLPCs) all actively participate in the progression of LN.
Circulating plasmablasts are attracted to the LN kidneys and differentiate into LLPCs
Plasmablasts, precursors of LLPCs, express chemokine receptors like CXCR3 and CXCR4, allowing them to migrate to survival niches in the bone marrow and inflamed tissues from the spleen and lymph nodes along the corresponding chemokine concentration gradients. Subsequently, plasmablasts differentiate into LLPCs, which do not express chemokine receptors27,28,29,30,31,32. However, this study found that Naive B cells and MBCs are present in the kidneys of LN mice. Thus, LLPCs may originate from circulating plasmablasts or differentiate locally from Naive B cells and MBCs within the kidneys.
Cell division and differentiation are interrelated processes vital for multicellular organism development. Cell differentiation typically depends on cell proliferation (cell division)33,34. Therefore, we conducted a comprehensive analysis of the expression of cell cycle- and apoptosis-related genes in the 8 cell subtypes in three groups of LN mice. We also analyzed the cell differentiation trajectories of these cells.
The results showed that among the 8 cell subtypes, Trans PC4 (plasmablasts) was the only cell population that highly expressed the genes related to various stages of the cell cycle, such as the G1 phase (Ccnd2, Ccne1, Ccne2, Cdk2, Cdk4, Cdk6), the S phase (Ccna2, Cdk1, Cdk2), and the G2/M phase (Ccnb1, Ccnb2, Cdk1) (Fig. 5A). Moreover, with LN progression, gene expression levels related to the S and G2/M phases significantly rise in Trans PC4 (plasmablasts) (Fig. 5A, B). In contrast, Naive B cells and PC2 (LLPCs) basically do not express the cell cycle-related genes. MBC, Trans PC1, Trans PC2, Trans PC3, and PC1 (SLPCs) highly express Ccnd2 and Cdkn2a, while the expression levels of other cell cycle-related genes remain low during LN progression. This indicates that Trans PC4 (plasmablasts) can continue to differentiate in the LN kidneys, while other subtypes have very limited proliferation and differentiation capabilities.
A Dot plot shows the expression levels of cell cycle-related genes in Naive B cell, MBC (memory B cells), Trans PC1, Trans PC2, Trans PC3, Trans PC4, PC1, and PC2. B Violin plot shows the expression levels of genes (Ccna2 and Cdk1) representing DNA synthesis and division (S and G2/M phases) in eight subtypes of cells in the kidneys. C Dot plot shows the expression levels of cell apoptosis-related genes in eight subtype cells in the kidneys. D Violin plot shows the expression levels of apoptosis-executing gene (Casp3) and representative anti-apoptotic gene (Bcl2) in eight subtypes of cells in the kidneys. E Pseudotime visualization of cell differentiation trajectories for Trans PC4, PC1, and PC2. F Pseudotime differentiation trajectories of Trans PC4, PC1, and PC2. G Differences in the differentiation trajectories of Trans PC4, PC1, and PC2 in three groups of LN mice. H Dynamic expression levels of the top six genes in pseudotime differentiation trajectories. I GSEA revealed the differences in enrichment of oxidative phosphorylation and glycolytic metabolic pathways in Trans PC4, PC1, and PC2. J The dot plot illustrates the expression levels of chemokine receptors in Trans PC4, PC1, and PC2, and it lists the corresponding chemokines for the four chemokine receptor genes that are highly expressed in these three cell subtypes. K The protein expression of Cxcl9 in the kidney of LN was measured using ELISA. (n = 6). L Flow cytometry was used to detect the proportion of CD45+ CD138+ B220+ Brdu+ Cxcr3+ Trans PC in the kidneys. (n = 3). Data are expressed as mean ± SD; one-way ANOVA was used for comparisons of three or more groups. *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we analyzed the expression levels of pro-apoptotic, anti-apoptotic, and pyroptosis genes across 8 cell subtypes35. Trans PC4 (plasmablasts) showed high expression of apoptotic pathway genes such as Bax, Bak1, Bid, Casp2, and Casp3, while Naive B cells and PC2 (LLPCs) had markedly lower expression levels. MBC, Trans PC1, Trans PC2, Trans PC3, and PC1 (SLPCs) exhibited variable expression levels of these genes (Fig. 5C, D). These results further indicate that Trans PC4 (plasmablasts) is highly active during LN progression and exhibits a rapid turnover rate. Meanwhile, Naive B cells are in a highly quiescent state, and PC2 (LLPCs) are also in a largely inert state.
Additionally, we analyzed the differentiation trajectories of Trans PC4 (plasmablasts), PC1 (SLPCs), and PC2 (LLPCs). The results showed that Trans PC4 (plasmablasts) can differentiate into both PC1 (SLPCs) and PC2 (LLPCs) along a continuous trajectory. The above findings indicate that PC1 (SLPCs) have limited proliferative and differentiation capabilities. Thus, we speculate that intermediate cells may exist during the differentiation of Trans PC4 (plasmablasts) into PC2 (LLPCs), with gene expression similar to that of PC1 (SLPCs), but represent only a transient stage in the differentiation process of PC2 (LLPCs). In contrast, the infiltrating PC1 (SLPCs) in the kidney are mature plasma cells that are completely independent of PC2 (LLPCs), and there is no differentiation or maturation sequence between PC1 (SLPCs) and PC2 (LLPCs) (Fig. 5E–G). The differentiation trajectories result also confirmed that the expression levels of cell proliferation and differentiation-related genes H2ac24, H2ac8, Mki67, and Tmsb4x gradually decreased during the differentiation process from Trans PC4 (plasmablasts) into PC2 (LLPCs) (Fig. 5H).
Subsequently, we analyzed the differences in enrichment of glycolysis and oxidative phosphorylation pathways, two major cellular metabolic pathways, in Trans PC4 (plasmablasts), PC1 (SLPCs), and PC2 (LLPCs). The results indicate that, for both pathways, Trans PC4 exhibited the highest enrichment levels, while PC2 (LLPCs) demonstrated the lowest enrichment levels (Fig. 5I). These results further indicate that Trans PC4 (plasmablasts) is highly active during LN progression.
Therefore, rather than B lymphocyte being the origin cells, Trans PC4 (plasmablasts) are the cells of origin that differentiate into PC2 (LLPCs) within the kidneys of LN mice.
Cxcl9 binds to its receptor Cxcr3 on plasmablasts, attracting them into the kidneys and differentiation into LLPCs
The above results show that the B lymphocyte have very limited proliferation and differentiation capabilities. This raises another important question: where do the Trans PC4 (plasmablasts) that infiltrate the kidneys come from? Previous studies have suggested that chemokine genes, such as Cxcl9, Cxcl10, and Cxcl11, are highly expressed in the kidneys of LN mice31. Therefore, these chemokines may attract plasmablasts expressing their corresponding receptors into the kidneys.
To confirm this hypothesis, we analyzed the gene expression levels of all chemokine receptors that can be identified in scRNA-seq data for Trans PC4, PC1, and PC236. The results indicated that, in Trans PC4 (plasmablasts), four chemokine receptors—Cxcr3, Cxcr4, Ccr7, and Ccrl2—showed high expression levels (Fig. 5J and Supplementary Fig. 4). Subsequently, we measured the protein concentration of all ligands corresponding to these four receptors in the kidneys using ELISA kits, including Cxcl4, Cxcl9, Cxcl10, Cxcl11, Cxcl12, Cxcl13, Ccl19, and Ccl21. Only Cxcl9 was significantly elevated during LN progression (Fig. 5K and Supplementary Fig. 5). Additionally, flow cytometry results confirmed that the proportion of Cxcr3+ Trans PC4 in the kidneys, which binds to Cxcl9, gradually increased throughout the progression of LN (Fig. 5L). The flow cytometry gating strategy was listed in Supplementary Fig. 6.
In summary, we found that the production of chemokine Cxcl9 is elevated in the kidneys of LN mice with disease progression, which attracts circulating Trans PC4 (plasmablasts) that express Cxcr3 into the kidneys.
Kidney-resident Cxcl9high macrophages recruit circulating plasmablast into kidneys by secreting Cxcl9
Studies have indicated that the chemokine Cxcl9 can be secreted by antigen-presenting cells, including DC and macrophages, as well as some tumor cells37. However, the specific cell types in the kidneys of LN mice that secrete Cxcl9 are still unknown. To investigate the source of Cxcl9, we first analyzed the expression levels of Cxcl9 in 11 cell subtypes. The results showed that Cxcl9 is highly expressed in macrophages and has low expression in DC and PEC (Fig. 6A).
A The dot plot illustrating the gene expression levels of Cxcl9 across 11 renal cell subgroups. B The relative proportions of macrophages (Mφ), dendritic cells (DC), and pericytes (PEC) in the kidneys of LN. (Mφ/DC/PEC)/Total kidney cells (%) = 1.25 × (Mφ/DC/PEC accounted for in single-cell enriched populations) × (Proportion of immune cells among total kidney cells). (n = 3). C UMAP visualization illustrating the regrouping of renal macrophage cell clusters. D The dot plot illustrates the gene expression level of Cxcl9 across 12 distinct macrophage cell clusters. E The dot plot illustrates the expression level of related genes that have been previously identified to differentiate monocyte-derived macrophages from tissue-resident macrophages across 12 distinct macrophage cell clusters. F UMAP visualization of five macrophage subtypes in the kidney. G The heatmap displays the top 10 differentially expressed genes across five macrophage subtypes. H The dot plot was utilized to compare the enrichment significance of KEGG and GO enriched pathways in Cxcl9high macrophages across three groups of LN. I The relative proportions of five macrophage subtypes among three groups in the kidney of lupus mice. Relative percentage of five macrophage subtypes in kidney = log10 (Proportion of Subtypes macrophages in total cells of macrophages) × Percentage of (macrophages)/Total kidney cells (%). J Flow cytometry was used to detect the proportion of total macrophages and Cxcl9high macrophages in the kidney of three groups of lupus mice. (n = 6). All data are expressed as mean ± SD; one-way ANOVA was used for comparisons of three or more groups. *p < 0.05, **p < 0.01, ***p < 0.001.
Next, we compared the relative proportions of macrophages, DC, and PEC in the kidneys of three groups of LN mice. When compared to the other two cell types, the proportion of macrophages significantly increased with LN progression (Fig. 6B). These findings suggest that macrophages are the primary cells responsible for Cxcl9 production.
Macrophages represent a heterogeneous population. To identify the specific macrophage subtype that secretes Cxcl9, we conducted a secondary cluster analysis of macrophages. Using UMAP clustering, we divided the macrophages into 12 distinct clusters (Fig. 6C). Among the 12 clusters, Cxcl9 was highly expressed only in cluster 10. The expression level of Cxcl9 in cluster 10 was significantly increased in 12w LN and 20w LN compared with 4w LN (Fig. 6D). Then, we identified five macrophage subtypes based on the characteristic marker genes of monocyte-derived macrophages and tissue-resident macrophages (Fig. 6E)38,39,40,41. These subtypes include Cxcl9high macrophages (cluster 10), C1q+ macrophages (clusters 0, 1, 6, 9), Plac8+ macrophages (clusters 2, 5, 8), TCR+ macrophages (clusters 3, 6, 9), and Cd83+ macrophages (clusters 4, 7) (Fig. 6F). Cxcl9high macrophages and C1q+ macrophages exhibited high expression levels of tissue-resident macrophage marker genes, including Cx3cr1, Cd81, Cd74, C1qa, and C1qc. In contrast, Plac8+ macrophages and TCR+ macrophages demonstrated elevated levels of monocyte-derived macrophage-related genes, such as Ly6c1, Ly6c2, and Ccr2. However, Cd83+ macrophages showed relatively lower expression of genes associated with both types of macrophages.
An analysis of the top 10 DEGs in the five macrophage subtypes showed that Cxcl9high macrophages highly express chemokine genes, such as Cxcl9, Ccl8, Cxcl10, Cxcl13, and Ccl12, indicating a strong chemotactic capability42 (Fig. 6G). We conducted KEGG and GO functional enrichment analysis of the Cxcl9high macrophages. The results showed that the functional enrichment of these cells was primarily related to defense, chemotaxis, adhesion, and pro-inflammatory responses (Fig. 6H).
Next, we compared the relative proportions of five macrophage subtypes in the kidneys of three groups of LN mice and found that the proportions of these cells increased consistently during LN progression (Fig. 6I). Flow cytometry also confirmed these results (Fig. 6J). The flow cytometry gating strategy was listed in Supplementary Fig. 7.
Taken together, these results indicate that the Cxcl9high macrophages are the primary source of increased Cxcl9 production in the kidneys of LN mice.
Spatial distribution of macrophages and plasma cells in the kidneys of LN mice
Our results indicate that the interaction between macrophages and plasma cells in the kidneys of lupus mice is positively correlated with LN progression. The tissue-specific microenvironment and spatial architecture critically influence cell interactions. We used ST to investigate the distribution of these cell subgroups.
The ST map includes numerous 55 µm diameter spots, each exhibiting distinct gene expression patterns. We identified 11,505 spots in the renal pathological sections of 4w LN, 12w LN, and 20w LN mice. Unbiased clustering and canonical lineage markers revealed 9 distinct cell subgroups: Glomerulus (GLO), Proximal Tubule (PT), Loop of Henle (LOH), Distal Tubule/Connecting Tubule/Collecting Duct (DT/CNT/CD), Transitional Epithelium (TE), Artery/Immune Cell (ART/IC), Immune Cell (IC), and Adipose Tissue (AT) (Fig. 7A, B).
A UMAP visualization of 11,505 spots in the kidney of LN. B UMAP visualization of cell subgroups in the kidneys of LN. C Representative genes used for annotating spatial transcriptomic cell maps in the kidneys of LN, along with the corresponding gene expression levels of immune cells in each cell subgroup. D H&E staining profiles and corresponding spatial transcriptome profiles of the kidneys of LN, with small black arrows indicating the distribution of immune cells in the kidneys. Glomerulus (GLO), Proximal Tubule (PT), Loop of Henle (LOH), Distal Tubule/Connecting Tubule/Collecting Duct (DT/CNT/CD), Transitional Epithelium (TE), Artery/Immune Cell (ART/IC), Immune Cell (IC), and Adipose Tissue (AT).
Due to differences in enzymatic hydrolysis timing between glomeruli and renal tubulointerstitium during the preparation of single-cell suspensions, we obtained gene expression data for tubulointerstitial and immune cells, but did not obtain data for glomerular cells in our scRNA-seq data. The scRNA-seq data identified Cxcl9high macrophages as the primary contributors to Cxcl9 production in the kidneys of LN mice. However, it cannot be ruled out that cells within the GLO, such as podocytes and mesangial cells, may also secrete Cxcl9 as LN progresses. Therefore, we utilized ST data to analyze the gene expression levels of Cxcl9 in the glomeruli. The results indicated that the expression level of Cxcl9 in the glomeruli was extremely low (Fig. 7C). This further confirms that Cxcl9high macrophages are the primary producers of Cxcl9 in the kidneys during LN progression.
We integrated the 9 cell subgroups identified by spot annotation with H&E pathological sections. Compared to 4w LN, results showed that the number of immune cells in the kidneys of 12w LN significantly increased, distributed along the direction of arterial blood flow, around the renal artery of the anterior and posterior branches and arcuate arteries, mainly distributed in the renal hilum, renal medulla, and corticomedullary junction, and almost did not involve the renal cortex. As disease progressed, immune cells in the kidneys of 20w LN increased and were widely distributed, not only around the renal hilum and the renal medulla, but also scattered in the renal cortex (Fig. 7D).
To determine the spatial distribution of Trans PC4 (plasmablasts) and Cxcl9high macrophages in the kidneys of LN mice, we performed co-localization analysis. We co-localized five marker genes (Cd68, Cxcl9, Jchain, Cxcr3, Mki67) representing plasmablasts and Cxcl9high macrophages. Results showed no co-localized spots in 4w LN, but an increase was observed in 12w and 20w LN. In 12w LN, spots were mainly around the medullary arteries, while in 20w LN, they were scattered around the renal medulla and cortical blood vessels. Single-gene expression analysis indicated that macrophages with high Cd68 expression were dispersed in the kidneys of all three groups, increasing with LN progression. Conversely, plasma cell-related spots with high Jchain expression were scarce in 4w LN kidneys but increased with disease progression, primarily located in the renal interstitium and displaying a perivascular morphology, and nearly absent in the glomeruli (Fig. 8A). The results of immunofluorescence staining confirmed the above findings (Fig. 8B).
A The spatial transcriptomic co-localization map of five representative genes (Cd68, Cxcl9, Jchain, Cxcr3, and Mki67) for Trans PC4 and Cxcl9high macrophages. B The results of multicolor immunofluorescence demonstrated the distribution of macrophages and plasma cells within the kidney of LN. The red arrow indicates the arteries, the white arrow highlights the infiltrating immune cells surrounding the glomeruli. The scales for the immunofluorescence staining as follows: Merge 2×: 500 μm, Merge 12×: 100 μm, DAPI: 100 μm, CD45: 100 μm, CD68: 100 μm, CD138: 100 μm, and Merge 20×: 50 μm.
Taken together, these results indicate that Cxcl9high macrophages are located adjacent to Cxcr3+Trans PC4 (plasmablasts) in the kidneys of LN mice. As LN progresses, the distribution of these cells gradually spreads from the renal medulla to the cortex, following the direction of renal arterial blood flow.
Anti-Cxcl9 antibody showed a curative efficacy in LN mice
Our studies revealed that Cxcl9high macrophages are a core factor inducing renal damage in LN. Targeting Cxcl9high macrophages or neutralizing Cxcl9 within the kidneys may thus alleviate renal damage in LN mice.
To verify this hypothesis, starting from 12 weeks of age, LN mice were intraperitoneally injected with anti-Cxcl9 antibody or matched irrelevant isotype control antibody every two weeks. Treatment efficacy was evaluated after 4 injections (Fig. 9A). The results showed that, compared with the control group, the UPCR and serum creatinine levels were significantly reduced in the anti-Cxcl9 antibody-injected group (Fig. 9B, C). Renal histopathological results demonstrated a decreased infiltration of immune cells in the renal interstitium and a mild glomerular damage in the anti-Cxcl9 antibody-injected group (Fig. 9D).
A Schematic representation of experimental grouping. The animals were divided into two groups: anti-mouse Cxcl9 antibody treatment group (Cxcl9 Ab): MRL/lpr mice (n = 6) were intraperitoneal injected with anti-mouse Cxcl9 antibody; Cxcl9 isotype-matched irrelevant antibody treatment group (Irr Ab): MRL/lpr mice (n = 6) were intraperitoneal injected with Cxcl9 isotype-matched irrelevant antibody. i.p. intraperitoneal inject: Sac: sacrifice. B Urinary protein/creatinine ratio (UPCR) in the two groups of MRL/lpr mice. C Serum creatinine levels in the two groups of MRL/lpr mice. D H&E staining were used to observe the infiltration of interstitial immune cells in different parts of the renal cortex and medulla for the two groups of MRL/lpr mice; PAS staining results were used to observe the degree of lesion in the glomerulus. H&E scale bar: 50 μm, PAS scale bar: 10 μm. E Flow cytometry was used to detect the proportion of CD45+ CD138+ CD20+ Brdu+ Cxcr3+ Trans PC in the kidneys. (n = 3). F Flow cytometry was used to detect the proportion of CD45+ CD138+ B220+ Cxcr3+ Trans PC in the peripheral blood. (n = 3). Data are expressed as mean ± SD; t-test was used for comparisons of the two groups. *p < 0.05, **p < 0.01, ***p < 0.001.
Furthermore, flow cytometry results indicated a significant reduction in Cxcr3+ Trans PC (plasmablasts) in kidneys (Fig. 9E) and a significant increase in Cxcr3+ Trans PC (plasmablasts) in the peripheral circulation in the anti-Cxcl9 antibody-injected group when compared with the control group (Fig. 9F). The gating strategy for flow cytometry is provided in Supplementary Fig. 8. These results suggest that anti-Cxcl9 antibody can inhibit the migration of Cxcr3+ Trans PC (plasmablasts) from the peripheral circulation into kidneys, thereby ameliorating LN.
Discussion
Current research on LLPCs in LN faces a major obstacle: the difficulty of obtaining adequate kidney specimens from LN patients. Plasma cells make up 0.1–5% of the total cell population in mouse bone marrow or spleen, with only 10–20% classified as LLPCs, which makes them extremely rare43. Besides, plasma cell proportions in peripheral tissues are significantly lower than in the bone marrow and spleen. Human renal biopsy samples, about 1.5 cm long and 1 mm in diameter, pose challenges in obtaining enough LLPCs for detailed studies. MRL/lpr mice serve as a common animal model for LN, exhibiting renal lesions similar to those seen in humans44.
This study analyzed peripheral blood, urine, and renal pathological parameters in MRL/lpr mice of various ages. According to the human LN renal pathology score (ISN/RPS, NIH), the lupus mice at 4 weeks of age did not exhibit any signs of disease and displayed normal renal pathology. In contrast, 12-week- and 20-week-old lupus mice exhibited renal damage similar to human LN class III and IV, respectively. Thus, we selected these age groups to model human LN pathogenesis and explore LLPCs in the kidneys during disease progression.
Similar to the heterogeneity in bone marrow B cells and plasma cells45, we found that the B cells and plasma cells in the kidneys of lupus mice also represent heterogeneous populations. We identified two types of B cells: naive B cells and MBC; four types of transitional plasma cells: Trans PC1, Trans PC2, Trans PC3, and Trans PC4; and two types of mature plasma cells: PC1 and PC2.
Naive B cells express Ighm and Ighd, indicating that they have not undergone antigen stimulation or immunoglobulin class switching46. They may migrate from the bone marrow to peripheral organs, such as the kidneys and liver, to form tissue-resident immune cells during the fetal stage, aiding local immune surveillance47. MBC highly express Ighg, Igha, and MHC class II molecules, indicating their presence in the kidneys as memory cells after the primary immune response48. The low expression of proliferation- and cell cycle-related genes in Naive B cells and MBC suggests that the capacity of these two cell subtypes to proliferate and differentiate within the kidneys is extremely lower.
Trans PC4 is a specific type of mature plasma cell precursor25, and some studies call them plasmablasts16,49. Among the 8 B cell and plasma cell subtypes, only Trans PC4 demonstrated high expression levels of genes related to proliferation and the cell cycle, and therefore, they were the only cell subtype capable of further proliferation and differentiation within the kidneys. PC2, namely LLPCs, are a type of cell that lacks proliferative capacity and does not express chemokine receptors. They can only differentiate from Trans PC4 (plasmablasts) in the kidneys of LN mice. Notably, one of our intriguing findings is that LLPCs can also be observed in the kidneys of 4w LN. We hypothesize that LLPCs in 4w LN may represent non-autoantibody-producing cells that reside in the kidneys during the embryonic or fetal stages of the mice, serving a role in the kidneys as surveillance and protection.
Plasmablasts, originating from secondary lymphoid organs, express chemokine receptors like CXCR3 and CXCR4, as shown in previous studies50,51. They can migrate from the spleen and lymph nodes to the bone marrow and inflammatory sites along chemokine gradients. Our research indicates that B cells in the kidneys of LN mice lack the ability to proliferate and differentiate. Trans PC4 (plasmablasts) is absent in the kidneys of 4w LN, but their infiltration number increases as LN progresses, suggesting they may translocate from peripheral circulation. Previous studies indicated that the gene expression level of various chemokines (Cxcl9, Cxcl10, Cxcl11, Cxcl12, Cxcl13) are elevated in the kidneys of LN mice. However, protein levels of these chemokines in the kidneys were not examined31,49,52. This study revealed that the protein expression level of the chemokines only for Cxcl9 in mice kidneys significantly increased during LN progression. Additionally, our study not only found that the protein and gene expression levels of Cxcl9 are increased in the kidneys of LN mice, but also revealed that Cxcl9 is secreted by a new identified subtype of macrophages, Cxcl9high macrophages.
The GLO is the primary site of immune complex deposition, and previous studies have shown that LLPCs in the kidney are the primary producers of autoantibodies in LN6. However, this study found that plasma cells are mainly located in the renal interstitium and are nearly absent from the GLO. Therefore, we speculate that the kidney, like the bone marrow, is another favorable niche for the survival of LLPCs in LN mice. Due to the limited spatial scope of the bone marrow53, the inflammatory kidneys can provide a larger space for the survival of LLPCs. Once LLPCs reside in the kidneys, they may produce more tissue-specific autoantibodies and have a stronger ability to attack the kidneys. These antibodies do not directly affect the interstitium of the kidneys, and may deposit in the GLO as circulating antibodies after entering the bloodstream. However, these conjectures need to be further confirmed by follow-up studies.
In summary, previous studies indicate an increase in the number of LLPCs in the kidneys of patients with LN, particularly in refractory or recurrent cases. However, the origin, inducing factors, and spatial distribution of these cells in the kidneys have not been further explored. In this study, we combined scRNA-seq and ST to comprehensively explore these questions. We found that Cxcl9high macrophages are an inducing factor for LLPCs and they are positively correlated with the progression of LN. So, targeting Cxcl9high macrophages during the early stages of LN is vital for preventing the migration of Cxcr3 + plasmablasts into the kidneys and reducing the accumulation of LLPCs in the kidneys of LN. Therefore, our study provides a robust theoretical foundation for the future development of targeted therapeutic drugs targeting Cxcl9high macrophages and LLPCs in the kidneys for the treatment of LN patients, especially those with refractory or recurrent conditions.
Our study has some limitations: (1) We used only one animal model of LN, necessitating validation in other LN models and patient renal biopsy specimens; (2) Future research should create Cxcl9 gene knockout models to clarify the link between Cxcl9 and the disease progression of LN.
Methods
Animal models and treatment
Total 63 MRL/lpr mice (LN, lupus nephritis model) and 51 female MRL/MPJ mice (Con, LN control model) were utilized in this study. Mice were obtained from the Shanghai SLAC Laboratory Animal Company Limited (Shanghai, China). Mice were housed in individually ventilated cages under specific pathogen-free conditions with a 12-h light/dark cycle at the Medical Experimental Animal Center of the Chinese PLA General Hospital. Mice used were 4 weeks old, female animals were used. Firstly, when evaluating the survival rate of mice, mice were divided into 2 groups: Control (Con/MRL/MPJ mice, n = 15), Lupus nephritis (LN / MRL/lpr mice, n = 15). Secondly, the mice (n = 72; Con n = 36, LN n = 36) were randomly divided into 6 groups using the random number table. They were euthanized (By intraperitoneal injection of sodium pentobarbital at a dose of 50 mg/kg) at various time points: 4 weeks (4w Con n = 6, 4w LN n = 6), 8 weeks (8w Con n = 6, 8w LN n = 6), 12 weeks (12w Con n = 6, 12w LN n = 6), 16 weeks (16w Con n = 6, 16w LN n = 6), 20 weeks (20w Con n = 6, 20w LN n = 6), and 24 weeks (24w Con n = 6, 24w LN n = 6). Finally, for Cxcl9 blocking assay, the MRL/lpr mice were randomly divided into 2 groups: anti-Cxcl9 group (n = 6) and Irr Ab group (n = 6). The anti-Cxcl9 group was treated with 10 µg/g anti-mouse Cxcl9 antibody (supplier name: BioXcell, catalog number: BE0309). The Irr Ab group was treated with the irrelevant isotype-matched antibody (polyclonal Armenian hamster IgG) (supplier name: BioXcell, catalog number: BE0091). The treatment was administered via intraperitoneal injection biweekly, starting at 12 weeks of age, for a total of 4 times. Mice in the 4w LN, 12w LN, 20w LN, anti-Cxcl9 group, and Irr Ab group were administered drinking water containing BrdU (0.8 mg/mL; supplier name: Sigma, catalog number: # B5002) for 14 days prior to euthanasia. Estimating the sample size based on the resource equation (The basic expression of the resource equation is: E = N − B – T. E: Error degrees of freedom, a key indicator for measuring the power of statistical tests. It is generally recommended to range from 10 to 20 (this range is considered to balance test power and resource consumption); N: Total number of subjects/observations. B: Number of blocks (if there are no blocks in the experimental design, such as a completely random design, then B = 0). T: Number of treatment groups (including the control group): When E is set to 10 and at least 2 groups are involved, we calculated that each group of mice should have at least 6 individuals. We excluded from the analysis animals, which did not present signs on LN (The concentration of autoantibodies in the serum). All animals received equal care, and animal caregivers were blinded. We have complied with all relevant ethical regulations for animal use. Animal experiments were performed in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals and approved by the Institutional Animal Care and Use Committee of the Chinese PLA General Hospital (No. 2023-X19-24).
Analysis of serum and urine samples
The urinary protein concentration was determined by urine protein test kit (supplier name: Nanjing Jiancheng Bioengineering Institute, catalog number: C035-2-1). The creatinine concentration was measured using the Cayman Urinary Creatinine Kit (supplier name: Cayman, catalog number: 500701). The UPCR and 24-h proteinuria were calculated to assess protein excretion in mouse urine. Serum creatinine and urea nitrogen levels were measured using an automatic biochemical analyzer. Serum anti-dsDNA (supplier name: Alpha Diagnostic Intl, catalog number: 5120, lot number: 240822K4) and antinuclear antibodies (ANA) (supplier name: Alpha Diagnostic Intl, catalog number: 5210, lot number: 240822K5) were quantified using an ELISA kit.
Evaluation of kidney coefficient and spleen coefficient
The body weight of the mice was recorded prior to euthanasia, and the organs were subsequently weighed. Organ coefficients were calculated using the following formulas: kidney coefficient = (100 × kidney weight)/body weight (g/100 g); spleen coefficient = (100 × spleen weight)/body weight (g/100 g)54.
Pathological examination and immunological staining
Kidneys were fixed in 4% formaldehyde, embedded in paraffin, sectioned to a thickness of 2 μm, and subsequently stained using standard protocols with periodic acid-Schiff (PAS), Masson’s trichrome stain, or H&E. Additionally, 1 mm³ of renal tissue was fixed in 2.5% glutaraldehyde, embedded in acetone, and sectioned to a thickness of 100 nm, followed by staining with uranyl acetate and lead citrate. The steps of immunohistochemistry mainly include the following: dewaxing and hydration of renal paraffin sections, microwave antigen retrieval, elimination of peroxidase activity, serum blocking of non-specific staining, incubation with primary and secondary antibodies, and final DAB color development. The steps of immunofluorescence staining mainly include the following: first, blocking frozen sections of kidney tissue with goat serum, then incubating them with primary and secondary antibodies in the dark, and finally observing after DAPI staining of cell nuclei. The primary antibodies utilized were as follows: IgG (1:500, supplier name: abcam, catalog number: ab6785), IgM (1:1000, supplier name: abcam, catalog number: ab150121), C3 (1:200, supplier name: abcam, catalog number: ab11862), and anti-CD45 (1:100, supplier name: abcam, catalog number: ab10558, clone name: Rabbit polyclonal, lot number: 1072771-1), anti-CD68 (1:100, supplier name: abcam, catalog number: ab283654, clone name: EPR23917-164, lot number: 1008461-36), anti-CD138 (1:1500, supplier name: abcam, catalog number: ab128936, clone name: EPR6454, lot number: 1068804-2). All the above test results were evaluated according 2016 revised ISN/RPS renal pathological classification criteria and NIH LN activity and chronicity scoring system. The percentage of immunofluorescent area for IgG, IgM, C3 were calculated using ImageJ software. The percentage of CD45-positive area and collagen deposition with Masson-positive area were calculated using ImageJ software.
Enzyme-linked immunosorbent assay
The protein levels of Cxcl4 (supplier name: RayBiotech, catalog number: EML-PF4-1, lot number: 0617248026), Cxcl9 (supplier name: RayBiotech, catalog number: EML-MIG-1, lot number: 0812240485), Cxcl10 (supplier name: RayBiotech, catalog number: EML-CRG2-1, lot number: 0401240568), Cxcl11 (supplier name: RayBiotech, catalog number: EML-ITAC-1, lot number: 0308240510), Cxcl12 (supplier name: RayBiotech, catalog number: ELM-SDF1a-1, lot number: 032724232), Cxcl13 (supplier name: RayBiotech, catalog number: ELM-BLC-1, lot number: 0408248010), Ccl19 (supplier name: RayBiotech, catalog number: ELM-MIP3b-1, lot number: 0617247043), and Ccl21 (supplier name: RayBiotech, catalog number: ELM-6Ckine-1, lot number: 0617242001) were measured using ELISA kits. Kidneys (4w LN, 12w LN, 20w LN) were lysed. The protein concentration of kidney was assayed using a BCA protein assay kit (Thermo Fisher Scientific, MA, USA). According to the experimental protocol, standards or samples (50 μg of protein) were added to the wells of each enzyme-labeled plate, and absorbance was measured at 450 nm for each well.
Flow cytometry
Freshly surgically resected kidney samples (4w LN, 12w LN, 20w LN) were first minced and enzymatically digested (1 ml of 0.25 mg/mL LiberaseTM (supplier name: Roche, catalog number: 5401119001) + 100 μL of 100 U/mL Dnase I (supplier name: Solarbio, catalog number: D8071)) in a 37 °C laboratory water bath for 30 min to obtain tissue lysis solution. Subsequently, the tissue lysis solution will be filtered through 100 μm and 40 μm cell filters, respectively to obtain single-cell suspensions. Collect 100 μL peripheral blood samples from mice and store them in anticoagulant tubes. The peripheral blood and single-cell suspensions of kidney were prepared and stained successively with the Zombie Aqua™ viability dye (supplier name: BioLegend, catalog number: 423101), cell-surface antibodies, Foxp3/Transcription Factor Staining Buffer Set (supplier name: eBioscience, catalog number: 00-5523-00), and intracellular antibodies. Data were acquired using a Beckman DXFLEX cytometer. Analysis was conducted with BD FlowJo™ v. 10.0.7 software. The following antibodies were utilized: PE/Dazzle™ 594 anti-mouse CD45 Antibody (supplier name: BioLegend, catalog number: 103146, clone name: 30-F11, lot number: B407721), Brilliant Violet 421™ anti-mouse CD138 (Syndecan-1) Antibody (supplier name: BioLegend, catalog number: 142523, clone name: 281-2, lot number: B387829), APC anti-mouse/human CD45R/B220 Antibody (supplier name: BioLegend, catalog number: 103211, clone name: RA3-6B2, lot number: B379792), FITC anti-BrdU Antibody (supplier name: BioLegend, catalog number: 364104, clone name: 3D4, lot number: B377768), Brilliant Violet 650™ anti-mouse CD183 (CXCR3) Antibody (supplier name: BioLegend, catalog number: 126531, clone name: CXCR3-173, lot number: B379381), FITC anti-mouse CD45 Antibody (supplier name: BioLegend, catalog number: 103108, clone name: 30-F11, lot number: B399569), PerCP/Cyanine5.5 anti-mouse/human CD11b Antibody (supplier name: BioLegend, catalog number: 101227, clone name: M1/70, lot number: B375903), Brilliant Violet 421™ anti-mouse F4/80 Antibody (supplier name: BioLegend, catalog number: 123137, clone name: BM8, lot number: B402385), PE anti-mouse CXCL9 (MIG) Antibody (supplier name: BioLegend, catalog number: 515604, clone name: MIG-2F5.5, lot number: B413041).
ELISPOT
To detect total IgG-secreting plasma cells, ELISPOT plates (supplier name: MabTech, catalog number: 3654-EP-10) were coated overnight at 4 °C with anti-mouse IgG (supplier name: MabTech, catalog number: 3825-2A) antibodies. Single-cell suspensions of kidney were added to saturated ELISPOT plates at a concentration of 2.5 × 105 cells per 100 µL in six replicates and incubated overnight at 37 °C in a humidified incubator with 5% CO2 and culture medium. Antibody-forming cells were detected using anti-IgG biotin, streptavidin-ALP, and BCIP/NBT. The plates were analyzed with an Mabtech ELISPOT reader following the manufacturer’s instructions.
Single-cell RNA sequencing by Chromium 10× genomics
Single cells isolated from the kidneys (kidneys were perfused by Saline at 4 °C) of 4w LN, 12w LN, and 20w LN mice, were stained with Zombie NIR viability dye (supplier name: BioLegend, catalog number: 423106). CD45+ and CD45− live cells were sorted using a Cytek Aurora cell sorter. FITC anti-mouse CD45 Antibody (supplier name: BioLegend, catalog number: 103108, clone name: 30-F11, lot number: B399569) was utilized. The CD45+ immune cells and CD45- cells were then mixed in a ratio of 4:1 and pooled together. The purity of each cell type reached 90%. The proportion of cell types are listed in the Table S3. Cells were resuspended to a final concentration of 700–1200 cells/μL, with over 80% viability as determined by a Countstar Rigel S2 Automated Fluorescence Cell Counter (Shanghai Ruiyu Biotechnology Co., Ltd.). The 3′ sequencing library was constructed using the 10× Genomics Chromium Next GEM Single-Cell 3′ Reagent Kit v3.1 (Dual Index) according to the manufacturer’s protocols. In summary, isolated single cells were encapsulated and barcoded using a Chromium Controller (10× Genomics, Pleasanton, CA). After reverse transcription and emulsion breaking, the resulting cDNA was purified and amplified, followed by sample index attachment. Libraries were sequenced on an Illumina NovaSeq 6000 platform using 150 paired-end reads at a minimum depth of 20,000 reads per cell.
We demultiplexed cell barcodes using the Cell Ranger software pipeline (version 7.1.0) provided by 10× Genomics. We mapped the reads to the Ensembl GRCm38/mm10 reference genome (v105) using the STAR aligner and down-sampled the reads as necessary to generate normalized aggregate data across samples, resulting in a matrix of gene counts versus cells. Doublets were detected using the R package DoubletFinder (v2.0.3) and were subsequently removed from the analysis. The unique molecular identifier (UMI) count matrix was processed using the R package Seurat (version 4.1.2)55. We filtered out cells with gene counts of fewer than 500 to eliminate low-quality cells and potential multiple captures. Additionally, we removed low-quality cells exhibiting more than 25% mitochondrial gene expression. A total of 87,906 single cells were included in the downstream analysis after applying the quality control criteria. The library size was normalized using the Normalize Data function in Seurat to obtain normalized counts. Specifically, the global scaling normalization method gene expression measurements for each cell relative to total expression, multiplies by a scaling factor (default 10,000), and then log-transforms the result.
The top variable genes in single cells were identified using the method described by Macosko et al. 56. The variable genes were identified using the “FindVariableFeatures” function in Seurat (selection.method = “vst”,nfeatures = 2000). Principal component analysis (PCA) was conducted to reduce dimensionality through the “RunPCA” function in Seurat. Graph-based clustering was executed using the “FindClusters” function in Seurat, based on the gene expression profiles of the cells. Total kidney cell populations, as well as B cell and plasma cell populations, were all clustered at a resolution of 0.5. The macrophage populations were clustered at a resolution of 0.8. The cells were visualized using the two-dimensional Unified Manifold Approximation and Projection (UMAP) algorithm with the “RunUMAP” function in Seurat. We used the “FindAllMarkers” function in Seurat (test.use = bimod) to identify marker genes for each cluster. Gene expression was visualized using dot plots, violin plots, heatmaps, and volcano plots, generated by the “FeaturePlot()” and “VlnPlot()” functions in the Seurat package. Differentially expressed genes (DEGs) were identified using the “FindMarkers” function in Seurat (test.use = bimod). P < 0.01 and |log2 fold change| > 0.26 were established as thresholds for significant differential expression. Gene Ontology (GO) enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analyses of DEGs were conducted using hypergeometric distribution-based R software. All single-cell data analyses were performed using the OmicStudio tools developed by LC-BIO Co., Ltd. (Hangzhou, China) at https://www.omicstudio.cn/cell.
Spatial transcriptomics by Chromium 10× genomics
Three-micrometer formalin-fixed paraffin-embedded (FFPE) sections from three samples (4w LN, 12 W LN, 20 W LN) were placed on the designated capture areas of the immunocytochemistry slides. After incubating for 2 h at 42 °C and allowing them to dry at room temperature, the slides were further dried for 3 h at 60 °C. Hematoxylin (Dako, Part number S330930-2) and Eosin (Sigma-Aldrich, Product number HT110216) were then added for H&E staining. Approximately 100 µL of 85% Glycerol (Thermo Fisher, Catalog number 15514011) was added, and a coverslip was applied before proceeding with tissue imaging. To remove the coverslip, a beaker filled with Milli-Q water was used.
The Visium slides were placed into a cassette. Add 100 µL of 0.1 N HCl (Sigma-Aldrich, Product Number H1758) to each well and incubate for 15 min at 42 °C. After incubation, remove the HCl from each well, add the decrosslinking buffer, and incubate at 95 °C for 1 h. Immediately proceed with the pre-hybridization step according to the Visium Spatial Gene Expression for FFPE Reagent Kit (10× Genomics, User Guide CG000407 Rev C, Product Number 1000339). Add 100 µL of the pre-hybridization mix to each well and incubate for 15 min at room temperature. At the end of the incubation, remove the pre-hybridization mix and add 100 µL of the hybridization mix. Incubate the Visium slide with the hybridization mix at 50 °C overnight.
For the remainder of the library preparation process, which includes probe ligation, probe release and extension, probe elution, and FFPE library construction, we followed the user guide for the “Visium Spatial Gene Expression for FFPE Reagent Kit” (10× Genomics, User Guide CG000407 Rev C, Product Number 1000339). The completed libraries were sequenced on the NovaSeq 6000 (Illumina). The lengths of Read 1 and Read 2 were 28 base pairs and 91 base pairs, respectively.
Data analysis
Each sequenced ST library was processed and aligned to the mm10 mouse reference genome using Space Ranger software (version 2.0.0) from 10× Genomics. UMI counts were summarized for each spot. To differentiate tissue-overlapping spots from the background, these spots were identified based on the images. Only barcodes associated with the tissue overlaying spots were retained, resulting in the generation of filtered UMI count matrices. We also manually excluded spots that were not covered by tissue but were detected by Space Ranger, and further filtered the UMI count matrices. After importing the data into R, the filtered UMI count matrix was analyzed using the R package Seurat (version 4.1.0). Regularized negative binomial regression (SCTransform) was employed to normalize the UMI count matrices. Three slices were merged for a comprehensive analysis. A total of 3000 highly variable genes were identified in each matrix, and the union of these genes was designated as the set of highly variable genes for the merged dataset. This set was subsequently utilized in PCA to reduce dimensionality. Based on the elbow plot, we decided to use the first 30 principal components for clustering analysis. Uniform Manifold Approximation and Projection (UMAP) dimensionality reduction was conducted using these 30 principal components as input to visualize the spots. All spatial data analysis was performed using the OmicStudio tools created by LC-BIO Co., Ltd (HangZhou, China) at https://www.omicstudio.cn/cell.
Statistics and reproducibility
The statistical analysis and graphing were performed using SPSS 22.0 and GraphPad Prism 9.0 software. All measurements were taken from distinct samples; sample sizes were listed in every experiment, and each experiment was measured repeatedly three times. For quantitative data between two groups, the Mann–Whitney U test or t-test was used, depending on whether the data followed a normal distribution. For comparing quantitative data among multiple groups, analysis of variance (ANOVA) or the Kruskal–Wallis test was employed, based on whether the data variance was homogeneous and follow normal distribution. When a statistically significant difference was detected among different groups, pairwise comparisons were conducted using the least-significant difference test or Tamhane T2 test. The statistical tests used one-side. Data are presented as mean ± SD. tatistical significance is indicated as follows: no significance; *p < 0.05, **p < 0.01, ***p < 0.001; #p < 0.05, ##p < 0.01, ###p < 0.001.
Data availability
All the data are available in the main text, supplementary materials and supplementary data (Supplementary Data 1–9 and the caption for Supplementary Data 1–9). scRNA-seq data and spatial transcriptomic data that support the findings of this study are openly available in the Genome Sequence Archive at https://ngdc.cncb.ac.cn/gsa/s/Jcm71pYO, reference number CRA027691. All other data are available from the corresponding author (or other sources, as applicable) on reasonable request.
Code availability
The website and the detailed methods for analyzing scRNA-seq data and spatial transcriptomic data are described in detail in the “Methods” section.
References
Parikh, S. V., Almaani, S., Brodsky, S. & Rovin, B. H. Update on lupus nephritis: core curriculum 2020. Am. J. Kidney Dis. 76, 265–281 (2020).
Contreras Martin, G. N. Lupus nephritis treatment strategies. J. Rheumatol. 51, 549–550 (2024).
Glaros, V. & Kreslavsky, T. Putting a stamp on plasma cells’ age certificate. Immunity 56, 1434–1436 (2023).
Fairfax, K. A., Kallies, A., Nutt, S. L. & Tarlinton, D. M. Plasma cell development: from B-cell subsets to long-term survival niches. Semin. Immunol. 20, 49–58 (2008).
Calame, K. L., Lin, K. I. & Tunyaplin, C. Regulatory mechanisms that determine the development and function of plasma cells. Annu. Rev. Immunol. 21, 205–230 (2003).
Espeli, M. et al. Local renal autoantibody production in lupus nephritis. J. Am. Soc. Nephrol. 22, 296–305 (2011).
Crickx, E. et al. Molecular signatures of kidney antibody-secreting cells in lupus patients with active nephritis upon immunosuppressive therapy. Arthritis Rheumatol. 73, 1461–1466 (2021).
Zhao, L. et al. Tertiary lymphoid structures in diseases: immune mechanisms and therapeutic advances. Signal Transduct. Target Ther. 9, 225 (2024).
Wang, M. et al. Tertiary lymphoid structures as local perpetuators of organ-specific immune injury: implication for lupus nephritis. Front. Immunol. 14, 1204777 (2023).
Yoshikawa, T. & Yanagita, M. Single-cell analysis provides new insights into the roles of tertiary lymphoid structures and immune cell infiltration in kidney injury and chronic kidney disease. Am. J. Pathol. 195, 40–54 (2025).
Chang, A. et al. In situ B cell-mediated immune responses and tubulointerstitial inflammation in human lupus nephritis. J. Immunol. 186, 1849–1860 (2011).
Arazi, A. et al. The immune cell landscape in kidneys of patients with lupus nephritis. Nat. Immunol. 20, 902–914 (2019).
Lemay, S., Mao, C. & Singh, A. K. Cytokine gene expression in the MRL/lpr model of lupus nephritis. Kidney Int. 50, 85–93 (1996).
Rao, D. A., Arazi, A., Wofsy, D. & Diamond, B. Design and application of single-cell RNA sequencing to study kidney immune cells in lupus nephritis. Nat. Rev. Nephrol. 16, 238–250 (2020).
Bajema, I. M. et al. Revision of the International Society of Nephrology/Renal Pathology Society classification for lupus nephritis: clarification of definitions, and modified National Institutes of Health activity and chronicity indices. Kidney Int 93, 789–796 (2018).
Oracki, S. A., Walker, J. A., Hibbs, M. L., Corcoran, L. M. & Tarlinton, D. M. Plasma cell development and survival. Immunol. Rev. 237, 140–159 (2010).
Liu, X., Yao, J., Zhao, Y., Wang, J. & Qi, H. Heterogeneous plasma cells and long-lived subsets in response to immunization, autoantigen and microbiota. Nat. Immunol. 23, 1564–1576 (2022).
Wilkinson, S. T. et al. Partial plasma cell differentiation as a mechanism of lost major histocompatibility complex class II expression in diffuse large B-cell lymphoma. Blood 119, 1459–1467 (2012).
Morgan, D. & Tergaonkar, V. Unraveling B cell trajectories at single cell resolution. Trends Immunol. 43, 210–229 (2022).
Wang, Y., Liu, J., Burrows, P. D. & Wang, J. Y. B cell development and maturation. Adv. Exp. Med. Biol. 1254, 1–22 (2020).
Carsetti, R., Rosado, M. M. & Wardmann, H. Peripheral development of B cells in mouse and man. Immunol. Rev. 197, 179–191 (2004).
Good-Jacobson, K. L. Strength in diversity: phenotypic, functional, and molecular heterogeneity within the memory B cell repertoire. Immunol. Rev. 284, 67–78 (2018).
Cancro, M. P. & Tomayko, M. M. Memory B cells and plasma cells: the differentiative continuum of humoral immunity. Immunol. Rev. 303, 72–82 (2021).
Luo, Y. et al. Single-cell genomics identifies distinct B1 cell developmental pathways and reveals aging-related changes in the B-cell receptor repertoire. Cell Biosci. 12, 57 (2022).
Manakkat Vijay, G. K. & Singh, H. Cell fate dynamics and genomic programming of plasma cell precursors. Immunol. Rev. 303, 62–71 (2021).
Shaffer, A. L. et al. XBP1, downstream of Blimp-1, expands the secretory apparatus and other organelles, and increases protein synthesis in plasma cell differentiation. Immunity 21, 81–93 (2004).
Manz, R. A., Hauser, A. E., Hiepe, F. & Radbruch, A. Maintenance of serum antibody levels. Annu. Rev. Immunol. 23, 367–386 (2005).
Slifka, M. K., Matloubian, M. & Ahmed, R. Bone marrow is a major site of long-term antibody production after acute viral infection. J. Virol. 69, 1895–1902 (1995).
Hiepe, F. et al. Long-lived autoreactive plasma cells drive persistent autoimmune inflammation. Nat. Rev. Rheumatol. 7, 170–178 (2011).
Nutt, S. L., Hodgkin, P. D., Tarlinton, D. M. & Corcoran, L. M. The generation of antibody-secreting plasma cells. Nat. Rev. Immunol. 15, 160–171 (2015).
Lacotte, S. et al. Early differentiated CD138(high) MHCII+ IgG+ plasma cells express CXCR3 and localize into inflamed kidneys of lupus mice. PLoS ONE 8, e58140 (2013).
Wang, A. et al. CXCR4/CXCL12 hyperexpression plays a pivotal role in the pathogenesis of lupus. J. Immunol. 182, 4448–4458 (2009).
Liu, L., Michowski, W., Kolodziejczyk, A. & Sicinski, P. The cell cycle in stem cell proliferation, pluripotency and differentiation. Nat. Cell Biol. 21, 1060–1067 (2019).
Wiggins, K. J. & Scharer, C. D. Roadmap to a plasma cell: epigenetic and transcriptional cues that guide B cell differentiation. Immunol. Rev. 300, 54–64 (2021).
Yuan, J. & Ofengeim, D. A guide to cell death pathways. Nat. Rev. Mol. Cell Biol. 25, 379–395 (2024).
Zlotnik, A. & Yoshie, O. The chemokine superfamily revisited. Immunity 36, 705–716 (2012).
Reschke, R. & Gajewski, T. F. CXCL9 and CXCL10 bring the heat to tumors. Sci. Immunol. 7, eabq6509 (2022).
Yao, W. et al. Single cell RNA sequencing identifies a unique inflammatory macrophage subset as a druggable target for alleviating acute kidney injury. Adv. Sci. 9, e2103675 (2022).
Zimmerman, K. A. et al. Single-cell RNA sequencing identifies candidate renal resident macrophage gene expression signatures across species. J. Am. Soc. Nephrol. 30, 767–781 (2019).
Gentek, R., Molawi, K. & Sieweke, M. H. Tissue macrophage identity and self-renewal. Immunol. Rev. 262, 56–73 (2014).
Vendrov, A. E. et al. Cardiomyocyte NOX4 regulates resident macrophage-mediated inflammation and diastolic dysfunction in stress cardiomyopathy. Redox Biol. 67, 102937 (2023).
Cappuyns, S. et al. PD-1(-) CD45RA(+) effector-memory CD8 T cells and CXCL10(+) macrophages are associated with response to atezolizumab plus bevacizumab in advanced hepatocellular carcinoma. Nat. Commun. 14, 7825 (2023).
Fooksman, D. R., Jing, Z. & Park, R. New insights into the ontogeny, diversity, maturation and survival of long-lived plasma cells. Nat. Rev. Immunol. 24, 461–470 (2024).
Andrews, B. S. et al. Spontaneous murine lupus-like syndromes. Clinical and immunopathological manifestations in several strains. J. Exp. Med. 148, 1198–1215 (1978).
Duan, M. et al. Understanding heterogeneity of human bone marrow plasma cell maturation and survival pathways by single-cell analyses. Cell Rep. 42, 112682 (2023).
Korzhenevich, J., Janowska, I., van der Burg, M. & Rizzi, M. Human and mouse early B cell development: so similar but so different. Immunol. Lett. 261, 1–12 (2023).
Zhao, J. et al. Single-cell RNA sequencing reveals the heterogeneity of liver-resident immune cells in human. Cell Discov. 6, 22 (2020).
Glaros, V. et al. Limited access to antigen drives generation of early B cell memory while restraining the plasmablast response. Immunity 54, 2005–2023.e2010 (2021).
Hiepe, F. & Radbruch, A. Plasma cells as an innovative target in autoimmune disease with renal manifestations. Nat. Rev. Nephrol. 12, 232–240 (2016).
Cyster, J. G. Homing of antibody secreting cells. Immunol. Rev. 194, 48–60 (2003).
Hoyer, B. F., Mumtaz, I. M., Yoshida, T., Hiepe, F. & Radbruch, A. How to cope with pathogenic long-lived plasma cells in autoimmune diseases. Ann. Rheum. Dis. 67, iii87–iii89 (2008).
Moreth, K. et al. The proteoglycan biglycan regulates expression of the B cell chemoattractant CXCL13 and aggravates murine lupus nephritis. J. Clin. Invest. 120, 4251–4272 (2010).
Radbruch, A. et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat. Rev. Immunol. 6, 741–750 (2006).
Zhou, C., Bai, X., Yang, Y., Shi, M. & Bai, X. Y. Single-cell sequencing informs that mesenchymal stem cell alleviates renal injury through regulating kidney regional immunity in lupus nephritis. Stem Cells Dev. 32, 465–483 (2023).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Acknowledgements
This work was supported by the following funding: National Key Research and Development Program of China 2022YFC3602000 (P.L.), National Key Research and Development Program of China 2020YFA0113004 (X.Y.B.)
Author information
Authors and Affiliations
Contributions
Xue-Yuan Bai, Ping Li, and Xiangmei Chen obtained the research funding and is the principal investigator. Xue-Yuan Bai and Jing Zhao conceived and designed the study. Jing Zhao, Xinlong Bai, and Cheng Zhou performed the experiments. Qing Ouyang and Qinggang Li guided the experiment. Yingjie Zhang, Xiao Zhang, Xumin Zheng, and Chaofan Li participated in mouse models experiments. Wanjun Shen performed the transmission electron microscopy analysis. Jing Zhao analyzed and interpreted the data. Jing Zhao drafted the manuscript. Xue-Yuan Bai revised and edited the manuscript. Xue-Yuan Bai, Xiangmei Chen, and Guangyan Cai supervised the study.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks Kristin Fenton and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Dario Ummarino.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhao, J., Bai, X., Zhou, C. et al. Cxcl9high macrophages recruit circulating Cxcr3+ plasmablasts into kidneys to promote pathogenesis of lupus nephritis mice. Commun Biol 8, 1446 (2025). https://doi.org/10.1038/s42003-025-08852-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42003-025-08852-9