Abstract
Discovery of novel antibiotics is crucial to counteract bacterial resistance spread. Aiming to expand the available arsenal of last-resort glycopeptide antibiotics (GPAs), we mined the actinobacterial genomes of Pseudonocardiales. We thus identified a biosynthetic gene cluster (BGC) encoding for a GPA with a novel peptide scaffold, not fitting into the existing classification of GPA types. By cultivating the producer strain, Actinokineospora auranticolor DSM 44650, an antibiotic complex—named kineomicins (Kmc)—was identified and characterized by microbiological assays, LC-MS, and MS/MS analyses. A comprehensive model for Kmc biosynthesis was then proposed by a thorough investigation of kineomicin BGC (knm). The structure of the main complex congener (KmcB), resolved by NMR spectroscopy, proved to be unique. Finally, the remarkably high antibiotic production rate, up to >1 g L−1 Kmc in benchtop bioreactor, indicated A. auranticolor as a natural GPA overproducer, holding promise as a potential host for heterologous expression of GPA BGCs.
Similar content being viewed by others
Introduction
Glycopeptide antibiotics (GPAs) constitute a diverse and still expanding group of naturally occurring non-ribosomally synthesized peptide antibiotics1,2,3,4. These antibiotics are produced by Gram-positive bacteria found in soil, belonging to actinobacterial orders such as Kitasatosporales, Micromonosporales, Streptosporangiales, and Pseudonocardiales1. GPAs have proven clinically effective in treating infections caused by multidrug-resistant (MDR) Gram-positive pathogens in both children and adults5,6. Notable examples include vancomycin and teicoplanin, which are natural GPAs in clinical use, derived from Amycolatopsis spp. (order Pseudonocardiales)7,8 and Actinoplanes teichomyceticus ATCC 311219 (order Micromonosporales), respectively. Additionally, semi-synthetic GPAs such as dalbavancin, oritavancin, and telavancin10 have been developed from A40926 (produced by Nonomuraea gerenzanensis ATCC 3972711, order Streptosporangiales), chloroeremomycin (Kibdelosporangium aridum A8284612, order Pseudonocardiales), and vancomycin, respectively, and more recently approved for clinical application.
The story of this antibiotic class began with the discovery of vancomycin, ristocetin, and actinoidin in the 1950s13,14,15. The clinical success of vancomycin brought GPAs into the spotlight, leading to various bioactivity-guided screening programs of environmental isolates of actinomycetes1. During the 1980s, numerous and structurally diverse natural sourced GPAs were isolated, and teicoplanin16 entered the clinics. It was meantime discovered that GPAs bind to the d-alanyl-d-alanine (d-Ala-d-Ala) termini of nascent peptidoglycan, hindering cell wall biosynthesis in Gram-positive bacteria17. The common feature of these GPAs was recognized as a sidechain cross-linked peptide core consisting of seven amino acids, whose structural variations led to the first (and still valid) classification (dated 1999) into five structural types1. GPAs of Types I-IV are considered glycopeptides sensu stricto, and they are also named dalbaheptides18, whereas Type V comprises non-glycosylated antibiotics1. Type I compounds (e.g., vancomycin, balhimycin, chloroeremomycin19) contain aromatic amino acids in positions AA2, 4-7, and aliphatic amino acids in positions AA1 and AA3 (Supplementary Fig. 1). Cross-linking occurs between aryl rings at AA7 (denoted as A) and AA5 (B), AA6 (C) and AA4 (D), and AA4 and AA2 (E) in Type I GPAs (Supplementary Fig. 1). In Type II GPAs, all amino acids are aromatic, with aryl rings introduced at AA3 (F) and AA1 (G), although F and G are not cross-linked (e.g., avoparcin, keratinimicin20) (Supplementary Fig. 1). Type III GPAs display cross-linking between aromatic AA1 and AA3 (e.g., ristocetin21) (Supplementary Fig. 1). Type IV GPAs (e.g., teicoplanin, A4092622) differ from Type III by carrying an aliphatic side chain (Supplementary Fig. 1). Type V compounds (e.g., complestatin, kistamicin23) feature a tryptophan residue in position AA2 and display more variable cross-linking patterns (Supplementary Fig. 1).
At the beginning of the 21st century, the development of DNA sequencing technologies enabled the identification of large biosynthetic gene clusters (BGCs) that encode GPA biosynthesis as in the case of chloroeremomycin24, complestatin25, teicoplanin26, and A4092627. An extensive investigation of the roles of different genes and enzymes in GPA biosynthesis followed and led to the understanding of its main steps, i.e., the aglycone synthesis28,29,30, the biosynthesis of non-proteinogenic amino acids31,32,33 and aminosugars34, as well as of the tailoring reactions involved in GPA diversification35,36,37. Shortly after, the onset of the “genomic” era allowed for the complete sequencing of the genomes of GPA producers or of new isolates, facilitating the identification of novel BGCs and promoting the discovery of structurally novel compounds8,21,38,39. Actually, we are in a “postgenomic” era, in which phylogenomic tools are being applied to identify novel GPA BGCs and investigate their evolution40,41,42,43,44. Novel gene-engineering tools are being utilized to design heterologous hosts for GPA production and manipulate native producer strains40,45,46, and multidisciplinary approaches are being employed to unravel the intricate details of non-ribosomal peptide synthetase (NRPS) structure and functions23,47,48,49, with the essential support of advanced analytical chemistry techniques20. In the light of this progress, the 1999-dated back classification of GPAs based on their chemical structure has been recently widened including diverse novel members of Type V GPAs discovered in Streptomyces spp.40,42,43 (order Kitasatosporales), whose peptide core might extend up to 10 amino acids, exhibiting variable cross-linking patterns40,42,43. Furthermore, these Type V GPAs inhibit the growth of Gram-positive bacteria by a novel mode of action, i.e., impeding cell wall turnover through the inhibition of autolysins42,43. A recent extensive evolutionary reconstruction has thus recommended categorizing Type V antibiotics as a novel class of glycopeptide-related peptides (GRPs), suggesting that the term GPA should be limited to Type I-IV dalbaheptides18,50.
In this context, our discovery strategy was searching for novel GPAs with unconventional peptide scaffolds through comparative genomic analysis, exploiting such actinobacterial orders as Micromonosporales, Streptosporangiales, and Pseudonocardiales that are less represented (in terms of number of isolates and of genome sequences available) than the well-studied streptomycetes (order Kitasatosporales)51. In this study, we focused on Pseudonocardiales, that were demonstrated to be a quite prolific source of GPA BGCs and corresponding compounds40,41,52. Pseudonocardiales produce: Type I GPAs as vancomycin and chloroeremomycin (vide supra), as well as the model GPA balhimycin (this last produced by Amycolatopsis balhimycina DSM 5908)35; Type II GPAs, such as avoparcin (from Amycolatopsis coloradensis DSM 44225)53, and the recently discovered keratinimicin (from Amycolatopsis keratiniphila NRRL B-24117)20; Type III GPAs (with multiple Amycolatopsis strains described as producers of ristocetin)21,39,54; and Type IV GPAs (e.g., teicoplanin-like GP1416 from Amycolatopsis sp. WAC 01416)40. Although the vast majority of these GPA producers belong to the Amycolatopsis55 genus, our recent research indicated that GPA producers could be found among lesser-known genera of Pseudonocardiales, such as Actinokineospora52.
Thanks to the comparative analysis of the available Pseudonocardiales genomes, in this work we reported the identification of two GPA BGCs with unusual NRPS properties, respectively in the genomes of Actinokineospora auranticolor YU 961-1 (=DSM 44650) and of Umezawaea endophytica DSM 103496. Intriguingly, in both cases the specificity of the NRPS module 3 (M3) adenylation (A)-domains could not be unambiguously predicted by bioinformatics tools. Further experimental investigation of A. auranticolor DSM 44650 confirmed that this strain actively produces an unknown GPA complex. Surprisingly, we discovered that GPA productivity in A. auranticolor DSM 44650 was in the unexpected range of 0.7–1.3 g L−1 in the commonly used R5 medium. Consequently, we isolated this bioactive GPA complex and characterized it using liquid chromatography-mass spectrometry (LC-MS) and high-resolution tandem mass spectrometry (MS/MS). The structure of the main congener was resolved using nuclear magnetic resonance (NMR) spectroscopy, revealing a novel aglycone type consisting of an aromatic AA1 and an aliphatic AA3. GPA complex produced in A. auranticolor DSM 44650 was named kineomicins. Elucidated structure of kineomicins appears to be the first example of a different type of GPAs, transitional between Type I and Type II. Further investigations allowed us to characterize the corresponding biosynthetic gene cluster, knm, and to propose an integrated model for kineomicins biosynthesis.
Results and discussion
Harnessing the potential of Pseudonocardiales spp. to produce GPAs with novel peptide scaffolds
At the time of our research (2023), GenBank contained up to 600 genomic records for Pseudonocardiales spp., which were available either as complete assemblies or as drafts of different quality (https://www.ncbi.nlm.nih.gov/assembly/?term=pseudonocardiales). We screened these genomic records using MultiGeneBlast56 with the sequences of bbr, oxyA, oxyB, and oxyC from the balhimycin BGC (bal) as query-probes. Nucleic acid sequences containing co-localized BLAST hits were further subjected to antiSMASH 7.057 to identify BGCs. As a result, we identified 40 genomic records potentially encoding for GPA BGCs (Supplementary Table 1) which might be attributed to 35 different strains belonging to Pseudonocardiales order. Some of these BGCs were already known and described previously41,52. Notably, in addition to several species of Amycolatopsis (carrying BGCs for Types I-IV GPAs, Supplementary Table 1) and two species of Kibdelosporangium (carrying BGCs for Type I chloroeremomycin and for an unknown Type II GPA), we found that poorly known genera as Actinokineospora, Kutzneria, and Umezawaea might produce GPAs as well, further expanding the range of putatively GPA-producing Pseudonocardiales spp. Phylogenetic relationships of these strains are illustrated by Supplementary Fig. 2.
We then placed the 35 identified BGCs into a phylogenetic framework by building a multi-locus phylogeny (MLP) (Fig. 1). Concatenated sequences of a conserved set of proteins encoded in each GPA BGC from Pseudonocardiales spp. described to date were used, including: an ABC-transporter (orthologue of Tba coded in bal58); the crosslinking oxygenases that install A-B, C-O-D, and D-O-E crosslinks in type I-IV GPAs (orthologues of OxyA, OxyB, and OxyC coded in bal19); and the β-hydroxytyrosine (Bht), 4-hydroxyphenylglycine (Hpg), and 3,5-dihydroxyphenylglycine (Dpg) biosynthetic enzymes (orthologues of Bhp, BpsD, HmaS, DpgA, DpgB, and DpgC coded in bal32,33,59,60) (Supplementary FASTA File 1). This concatenation was possible for 33 out of the 35 investigated BGCs, since the sequences for two of them (Supplementary Table 1) were incomplete and missing the corresponding genes.
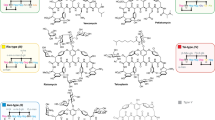
On the left side, the MLP of the 33 GPA BGCs selected from the genomic records of Pseudonocardiales spp. (see Supplementary Table 1) is represented: concatenated protein sequences of central biosynthetic enzymes encoded in each BGC (see main text) were used to build the unrooted maximum-likelihood phylogenetic tree (bootstrap values of branch support analysis were obtained from 500 replicates). The names of the GPAs are reported under the species previously known to produce them. To the right, the predicted domain architecture of each NRPS (where the non-ribosomal codes are given) is reported, as well as the schematic representation of monooxygenases and of the other tailoring enzymes taking part in post NRPS biosynthetic steps, coded within the corresponding BGCs (see Supplementary Table 2).
In the further analyses, the organization of NRPSs encoded within each BGC, their A-domain specificities, and the presence of different cross-linking oxygenases, were compared by extrapolating this information from the genomic data and correlating it with phylogeny (Fig. 1, Supplementary Table 2). Then, the repertoire of the tailoring genes (i.e., halogenases, methyltransferases, glycosyltransferases, sulfotransferases, and acyltransferases) found in each BGC (Supplementary Table 2) was analyzed. The amino acid composition of each peptide scaffold, together with their cross-linking patterns, were predicted, and each BGC was assigned to the currently known GPA Types. The result was that well-separated clades of the tree coherently grouped the BGCs coding for the biosynthesis of structurally similar GPAs (Fig. 1).
Clade A (Fig. 1) included the BGCs from the already known Type III GPA ristocetin producers (Amycolatopsis lurida NRRL 2430, Amycolatopsis japonica MG417-CF17, Amycolatopsis sp. MJM2582, and Amycolatopsis sp. TNS10621,39,54, further denoted as ris BGCs) and all the BGCs predicted by us or other authors41 that appeared identical to those of the known producers (Supplementary Fig. 3). Coherently, these BGCs lacked genes for halogenases, since the corresponding GPAs are not halogenated (Supplementary Table 2). The sister clade B (Fig. 1) grouped two BGCs identical in terms of genetic organization (Supplementary Fig. 3) that code for the biosynthesis of teicoplanin-like acylated GPAs: the product from one of them (known as GP1416) was previously identified from Amycolatopsis sp. WAC 0141640. Clade C (Fig. 1) contained three BGCs respectively from the azureomycin producer Amycolatopsis azurea DSM 4385461 and from two other Amycolatopsis spp. still not known to produce GPAs. Of note, the structure of azureomycin is still unresolved. These BGCs seemed identical in terms of genetic organization (Supplementary Fig. 3). Their predicted products would be Type III GPAs, although the repertoire of glycosyltransferases (GTFs) coded by their BGCs is more limited than in ris BGCs, and halogenase genes are present (Supplementary Table 2). Clade D (Fig. 1) included the known producer (Am. keratiniphila ssp. nogabecina FH 1893) of the Type II GPA nogabecin and two additional identical BGCs from two other Amycolatopsis spp. (Supplementary Fig. 3). Interestingly, the BGC sequence for the Type II GPA avoparcin from Am. coloradensis DSM 44225 appeared as an outgroup to clades A, B, and C. Unlike nogabecin-like Type II GPAs, avoparcin carries Hpg instead of Phe in AA3 position1, indicating its different origin and its unrelatedness to clade D Type II GPA BGCs (confirmed by the different genetic organization, Supplementary Fig. 3). Clade E grouped the vancomycin-like BGCs coding for Type I GPAs including vancomycin and norvancomycin (Fig. 1 and Supplementary Fig. 3), the BGCs for decaplanin from Amycolatopsis decaplanina DSM 4459462, and for dimethylvancomycin from Amycolatopsis sp. WAC 041698.
The last two clades of the tree, F and G, were much more sparsely populated and included many putatively novel BGCs. In clade F, two BGCs were present (Fig. 1), likely coding for Type III GPAs, but different from those grouped in clades A and C due to their repertoire of tailoring genes (Supplementary Table 2, Supplementary Fig. 3). Notably, one of these BGCs was found in the genome of Kutzneria sp. CA-10326063, although members of this genus were not previously known to produce GPAs. The second was unusual GPA BGC of Amycolatopsis bartoniae CGMCC 4.7679 (Supplementary Fig. 3)64. Clade G grouped BGCs for GPAs of different types (Fig. 1 and Supplementary Fig. 3), including well known BGCs as the ones encoding for Type I balhimycin (bal) and chloroeremomycin BGC (cem). Sequences from bal formed a subclade with sequences from the Amycolatopsis vastitatis H5 BGC, which, on the basis of our analyses, likely codes for the biosynthesis of a yet unknown Type IV GPA. At the same time, sequences from cem grouped together with sequences from Kibdelosporangium philippinense ATCC 49844 BGC, which putatively codes for the biosynthesis of a novel Type II GPA, carrying a Hpg residue in the AA3 position.
Finally, the last G subclade included the BGCs from Actinokineospora auranticolor YU 961-1 (=DSM 44650) and Umezawaea endophytica DSM 103496, and for both of them we found impossible to predict the specificity of the A-domain from NRPS M3. The non-ribosomal code of the M3 A-domain from U. endophytica DSM 103496—DAYMWGIVCK—was never reported before and it shared the highest similarity with the code (DAYLWGGVFK) found in the A-domain of KtzE (ABV56585) from kutzneride biosynthesis65, which is specific for incorporating a unit of 2-(1-methylcyclopropyl)-d-glycine65. In the case of A. auranticolor YU 961-1, the code of the M3 A-domain (DAFMWGIVIK) was also previously unknown and shared the highest similarity to the code (DAFWWGGVFK) found in the M3 A-domain of ecumicin biosynthesis NRPS (AIW58892)66 from Nonomuraea sp. MJM5123, where it was found specific for l-allo-isoleucine66. Thus, the GPA NRPSs from U. endophytica DSM 103496 and A. auranticolor YU 961-1 represent the unique cases so far described, where the A-domain of M1 recognizes the aromatic amino acid (Hpg), while the M3 A-domain likely recognizes an aliphatic amino acid. To the best of our knowledge, such recognition pattern is unprecedented among the biosynthetic pathways of known GPAs, and the corresponding compounds do not actually fit into the existing classification of GPAs types1, thus requiring its update.
Driven by the novelty of the predicted chemical structures, our efforts were then devoted to the investigation of putative GPA production in A. auranticolor YU 961-1 and U. endophytica DSM 103496 with the goal to purify and characterize these yet-unknown compounds. In this paper, we focused on A. auranticolor YU 961-1 (=DSM 44650), as our preliminary studies demonstrated that this strain produced the putative GPA at high level in standard cultivation conditions (see below). GPA production in U. endophytica DSM 103496 will be instead investigated in future.
GPA production in A. auranticolor DSM 44650
To the best of our knowledge, A. auranticolor DSM 44650 was never tested before for GPA production67. Therefore, we grew the strain at Erlenmeyer flask scale in a range of vegetative and production media using Bacillus subtilis HB095068 as an indicator strain. The latter strain might serve as an antimicrobial assay system in which lipid II binders, including GPAs, induce the chromogenic conversion of X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) at the edge of the growth inhibition zone69,70. Through this assay, we discarded several media (VSP71, TM172, FM271, YMPG73, ISP274, GYM Streptomyces medium75, GYC76, and SG77) since no GPA-related antimicrobial activity (or only a weak one) was detectable. On a contrary, A. auranticolor DSM 44650 gave a clearly green-blue edged antimicrobial inhibition halo when cultivated in TSB78, Lysogeny broth (Miller), SAM79, and liquid R5 medium78 (Supplementary Fig. 4). This last one, which was previously used for the routine production of balhimycin35,80,81 and ristocetin21, seemed to allow the highest production of the putative antibiotic, and was, therefore, selected for the subsequent cultivations of A. auranticolor DSM 44650 at flask and bioreactor scale (Fig. 2a, Supplementary Fig. 5, Supplementary Data 1).
a, b Time course of A. auranticolor DSM 44650 growth (a) and production of Kmc congeners (b) in R5 medium at 500 mL Erlenmeyer flasks-scale. pH (circles, a), residual glucose (squares, a), dry weight (rhombi, a) Kmc total production and complex composition by PDA-HPLC analyses (b) were monitored every 24 h. Results provided are mean values of seven independent experiments ± standard deviations. In panel (b) all minor components of Kmc complex are grouped as “other”. c, d HPLC chromatograms (λ = 236 nm detection wavelength, see the methods section for details) of a culture extract from A. auranticolor DSM 44650 cultivated for 336 h in R5 medium (c) and of a partially purified fraction from the same extract following d-Ala-d-Ala affinity chromatography (d). In both panels, arrows indicate major (KmcA, KmcB, KmcC, and KmcD) and minor peaks attributable to the different congeners of Kmc complex.
A. auranticolor DSM 44650 grew in R5-containing flasks, reaching the maximum biomass production (48.6 ± 3.2 g L−1 in dry weight) after 192 h of cultivation (Fig. 2a, Supplementary Data 1). Glucose was slowly consumed during the microbial growth, being completely depleted after 264 h from the inoculum, whereas pH tended to remain stable in the 7.0-7.5 interval for the entire cultivation time (Fig. 2a, Supplementary Data 1). Putative GPA production was detected by the photodiode-array (PDA) HPLC of day-by-day culture extracts (Fig. 2b, Supplementary Data 1). Starting from 24 h of cultivation, several peaks at various retention times (tR) and showing UV spectra with high similarity to those from GPA standards (e.g., vancomycin and teicoplanin), became detectable (Fig. 2c), indicating that A. auranticolor DSM 44650 produces, as in the case of many other GPAs1, a complex of structurally related congeners. We named this putative GPA complex kineomicins (from the name of the producing strain Actinokineospora, Kmc). Four major peaks were identified (named KmcA-KmcD), with KmcB being the most abundant congener, together with other structurally related minor components (Fig. 2b, c). Kmc production started during the exponential growth phase but reached its maximum during the stationary phase, then remaining almost stable till the end of cultivation—336 h after the inoculum (Fig. 2b, Supplementary Data 1). A slightly different growth curve and production time course were observed at the level of 3-L bench bioreactor cultivation, where biomass accumulation followed a diauxic profile (Supplementary Fig. 5a) and Kmc production continued to increase up to the end of the cultivation at 336 h (Supplementary Fig. 5b). Notwithstanding these differences, the same kineomicin complex composition, with four major peaks (KmcA-KmcD) and various other minor congeners, was highlighted by PDA-HPLC analyses also at bioreactor level (Supplementary Fig. 5b, c). The total Kmc productivity (calculated as the sum of the major and minor congeners detectable by PDA-HPLC) indicated a maximum productivity of 685 ± 95 mg L−1 after 288 h of cultivation at flask level (Fig. 2b, Supplementary Data 1) and of 1320 ± 280 mg L−1 at bioreactor scale after 336 h (Supplementary Fig. 5b).
Initial attempt to purify Kmc from culture extracts using the d-Ala-d-Ala affinity chromatography indicated that the four main congeners bound to the functionalized resin, which mimics the canonical GPA target in the bacterial cell wall82, confirming their dalbaheptide mode of action (Fig. 2d). This preparation was also tested for its antimicrobial activity, measuring minimum inhibitory concentrations (MICs) and minimal bactericidal concentrations (MBCs)83 against a collection of clinically relevant Gram-positive and Gram-negative bacteria, in parallel with vancomycin and teicoplanin (Table 1). Kmc showed the typical GPA antimicrobial spectrum, targeting Gram-positive bacteria (staphylococci and enterococci), but not the Gram-negative Escherichia coli ATCC 35218 and Moraxella catarrhalis ATCC 3293. Kmc, albeit tested as a still not completely purified extract, was particularly effective versus staphylococci, including methicillin-resistant Staphylococcus aureus ATCC 43300 (MRSA), and the clinical isolates Staphylococcus epidermidis strain 4 and Staphylococcus haemolyticus 3902, when compared to the pure standards of vancomycin and teicoplanin. On enterococci, the antimicrobial activity of Kmc was overall lower than on staphylococci, being, as vancomycin, ineffective on the clinical isolate with VanA phenotype Enterococcus faecalis 9160188401-EF-34 and on VanB-type E. faecalis ATCC 51299, but showing MBCs lower than those of vancomycin and teicoplanin towards E. faecalis ATCC 29212 (Table 1).
These results confirm that A. auranticolor DSM 44650 synthetizes a GPA complex with a promising antimicrobial activity and produces it at a remarkable level. For example, balhimycin productivity in the model Am. balhimycina DSM 5908 (cultivated in R5 and other media) ranged from ca.100 to 250 mg L−1 84,85,86. Ristocetin production in wild type strains reached around 100 mg L−1 54. In non-Pseudonocardiales GPA producers, antibiotic production levels in optimized production media may vary in the range of ca. 200–300 mg L−1 (e.g., teicoplanin production in Act. teichomyceticus ATCC 31121 or A40926 production in N. gerenzanensis ATCC 39727)46,72. Only GPA overproducers, generated through gene-engineering or classical mutagenesis and selection, have the potential to yield antibiotics in grams per liter range87,88,89. In this context, A. auranticolor DSM 44650 stands out as a unique naturally tailored GPA overproducer and for this reason it might also be considered in future as an alternative platform for the production of heterologous GPAs.
Characterization of kineomicin complex and structure elucidation of kineomicin B
Production of Kmc complex in A. auranticolor DSM 44650 culture extracts was then investigated by HPLC-ESI-HRMS detecting a set of structurally related ions (Supplementary Fig. 6, Supplementary Table 3). [M + H]+ of KmcA-KmcD were m/z 1698.5786, 1712.5990, 1729.5773, and 1713.5826, respectively, and corresponded to the four major congeners. In addition, a group of thirteen minor components were identified by LC-MS (Supplementary Fig. 6, Supplementary Table 3). The most abundant KmcB congener (m/z of 1712.5990) displayed characteristics consistent with the presence of one chlorine atom, as discerned from the HR-MS isotope distribution pattern20, a typical modification found in GPAs. KmcB was then purified from the producing cultures of A. auranticolor DSM 44650 cultivated in R5 medium for 288–336 h, by d-Ala-d-Ala affinity chromatography90 followed by reverse phase HPLC (Supplementary Fig. 7), yielding 6.1 mg of a white solid. Data interpretation from 1D/2D NMR spectra revealed that KmcB is composed of seven amino acids (as other Type I-IV dalbaheptides) with discernible δ (1Hα) values at 4.32, 4.61, 4.05, 5.54, 4.46, 4.21, and 4.34 ppm (Supplementary Fig. 8, Supplementary Table 4)20. Further investigation by means of 1H-13C HSQC, 1H-1H COSY, 1H-1H TOCSY, and 1H-13C HMBC (Supplementary Figs. 9, 10, 11, and 12, respectively) identified these amino acids as 4-hydroxyphenylglycine (1Hpg), β-hydroxytyrosine (2Bht), isoleucine (3Ile), two cross-linked Hpg residues (4Hpg-5Hpg), β-OH-3-Cl-tyrosine (6Bht), and one crosslinked 3,5-dihydroxyphenylglycine (7Dpg). Notably, the cross-links occur between 5Hpg and 7Dpg, via a carbon-carbon bond, and between 2Bht and 4Hpg, and 4Hpg and 6Bht, mediated by aryl ether bonds, as confirmed by HSQC, HMBC, and NOESY spectra (Supplementary Figs. 9, 12, and 13, respectively). Moreover, four glycosyl residues were identified by 2D-NMR analysis: α-mannose, α-ristosamine, and a β-glucosyl-α-ristosaminyl disaccharide attached to residues 7Dpg, 6Bht, and 4Hpg, respectively (Supplementary Fig. 14a), thus completing the 2D structure elucidation of the novel GPA KmcB (Fig. 3a, b, and Supplementary Fig. 15). The molecular formula of C80H94ClN9O31 deduced from the NMR experiments is in agreement with the molecular formula determined by HPLC-ESI-HRMS measurements for KmcB (Supplementary Fig. 16a), and the resulting structure of KmcB is also in full agreement with MS/MS data (Supplementary Fig. 16b)90.
a Key COSY/NOESY/TOCSY and (b) HMBC correlations are highlighted in the structure of KmcB. c Structures of KmcA-C with varying substitution patterns and assignment of the chiral centers. d 1H-13C HSQC section of KmcB with chemical shift ranges from 5.10–5.23 ppm (1H) and 90.5–99.0 ppm (13C) (DMSO-d6, 298 K). The cross peaks belonging to the anomeric carbons within the mannose moieties in both KmcB (blue) and minor KmcB’ (red) are annotated.
Confirming what highlighted by genomic analyses and phylogenetics, KmcB is therefore a peculiar GPA with a novel heptapeptide aglycone type containing an aromatic amino acid (Hpg) in AA1 and an aliphatic (isoleucine) in AA3. Such configuration is transitional between Type I vancomycin-type dalbaheptides (both AA1 and AA3 aliphatic) and Type II dalbaheptides such as avoparcin and keratinimicin (AA1 and AA3 aromatic).
Despite this, it remained to establish the absolute configuration of 3Ile in KmcB, that was conclusively established utilizing the advanced Marfey’s method. Following hydrolysis (6 N HCl, 110 °C, 12 h) of KmcB, the resultant mixture underwent d/l-FDLA derivatization and subsequent analysis via HPLC-ESI-HRMS mass spectrometry. The obtained data unequivocally indicated the presence of 3Ile residue in the l-configuration (Supplementary Fig. 17)91. Therefore, we can conclude that the novel non-ribosomal code of Kmc M3 A-domain—DAFMWGIVIK—is specific for l-Ile. Finally, utilizing bioinformatics-based A-domain specificity prediction, the assignment of chiral centers for the other amino acids was performed57, thus finalizing the proposed structure of KmcB, as illustrated in Fig. 3c.
Interestingly, in the series of consecutive NMR measurements of KmcB, an additional set of signals was observed (Supplementary Fig. 18a). The analysis of these spectra facilitated the elucidation of a second compound (termed KmcB’). Notably, its chemical shifts closely resemble those of KmcB, with the exception of an alteration of 5.6 ppm in the chemical shift of the anomeric carbon of mannose (Supplementary Table 4), indicating a β-configurated mannose in KmcB’ (Fig. 3d and Supplementary Fig. 14b)92,93. As in the 1H-NMR spectra the peaks corresponding to KmcB did not show any significant decrease (Supplementary Fig. 18b), we explained this observation with a solubility effect or dimerization/monomerization during HPLC purification and analytics94, rather than an isomerization during sample handling steps (e.g., rotary evaporation or lyophilization). This conclusion was also supported by the retention time detected of a new peak with the same MS patterns of KmcB (Supplementary Fig. 7b).
Although the structure of the three other main congeners of Kmc complex (KmcA, C, and D) was not accessed with NMR-spectrometry, further analysis of LC-MS and MS/MS data allowed us to speculate about the structures of KmcA and KmcC. Unfortunately, the quality of the MS/MS spectra for KmcD was not sufficient for its structure determination, due to low abundance. Thus, we assumed that KmcA differs from KmcB by carrying l-Val residue at AA3 (Supplementary Table 3, Supplementary Figs. 16 and 19), considering that the mass of KmcA is 14 Da smaller than that of KmcB, and such loss of a methyl group could not be apparently attributed to any other reaction during the biosynthesis of Kmc (see below). Moreover, it is highly likely that the other major congener KmcC differs from KmcB (Supplementary Table 3, Supplementary Figs. 16 and 20) by carrying a d-rhamnose residue attached to AA4 d-glucose instead of l-ristosamine.
Kineomicin BGC and biosynthesis model
In parallel to the structure elucidation of KmcB, we analyzed the organization of the Kmc BGC (named knm), assigning the functions to knm genes in Kmc biosynthesis. As in the other known GPA BGCs2, knm genes encode for the biosynthesis of the antibiotic scaffold including the supply of non-proteinogenic amino acids and the tailoring reactions, for the self-resistance protecting the producer during antibiotic production, for the cluster-situated regulatory genes, and for the export of the antibiotic. knm consists of 41 ORFs, the majority of them having orthologues in other Pseudonocardiales GPA BGCs, as exemplified by a comparison with the model bal and ris BGCs54,95 (Table 2, Fig. 4a). Searching for clues to explain the natural high production levels of Kmc, we also completely sequenced and assembled the full genome of A. auranticolor DSM 44650 (yielding a single circular 8,488 kbp chromosome). In addition, by semi-quantitative reverse transcription-PCR (RT-PCR), we investigated the operon structure of knm and compared it with those of bal and ris (from Amycolatopsis sp. TNS106), which were the only ones so far described at the transcriptomic level from Pseudonocardiales spp.54,95. knm consists of 8 operons (O1-8) and 6 monogenic transcriptional units (Fig. 4b). Integrating all the data obtained from in silico analysis, transcriptional studies, and analytical chemistry, we thus build a streamlined model for Kmc biosynthesis, moving from genes to product, that is presented below.
a knm consists of 41 ORFs (resembling bal and ris BGCs, but lacking genes for methyltransferases). It is organized into 8 operons and 6 monogenic transcriptional units, according to (b) the results of semi-quantitative RT-PCR analysis showing the expression (or absence thereof) of all intergenic regions (35, except for the ones where neighboring genes lie on opposite strands and cannot be co-expressed) within knm. RNA was extracted from Kmc producing culture cultivated in R5 medium for 96 h, processed into cDNA, and used as a template for PCRs; chromosome DNA (gDNA) served as a positive control for each reaction, while non-treated RNA as a negative control; electrophoregrams represent the average results of three independent experiments.
Resistance
knm contains a complete set of GPA resistance (van) genes52. This includes orthologues of vanHAX genes—knm1-3, a vanRS two-component regulatory system – knm4-5, and an orthologue of vanY—knm6 (Table 2, Fig. 4a). GPA resistance genes are organized as O1 (knm1-2-3 – vanHAX) and O2 (knm4-5 – vanRS), but knm6 (vanY) exists as a single gene (Fig. 4b). Thus, knm combines the features of bal (carrying the vanRS operon and the vanY gene within the BGC) and of GPA BGCs from other Pseudonocardiales spp. (carrying only the vanHAX operon within the BGC, e.g., ris)82 (Fig. 4a). If these genes are all expressed, they could probably confer a highly self-resistant phenotype to the producer strain which may contribute to its surprisingly elevated antibiotic productivity. The pattern of GPA resistance in A. auranticolor DSM 44650 would merit further investigation.
Pathway specific regulation and export
Like all other BGCs from Pseudonocardiales spp. (Supplementary Fig. 3), knm contains a single gene for the StrR-like pathway-specific regulator – knm7R (Table 2, Fig. 4a). Similar to the orthologue from ris BGC, knm7R is a monogenic transcription unit (Fig. 4a, b)54. The ABC transporter, coded by knm9, shares 77% of its amino acid sequence identity with the balhimycin exporter Tba, whose function was proved experimentally58, and it is most likely responsible for the export of Kmc. knm9 is co-transcribed with knmA-B NRPS genes (forming O3, Fig. 4b); in bal and ris, knm9 orthologues were also shown to be co-transcribed with NRPS genes (Fig. 4a)54,95.
Tyrosine supply
The Kmc aglycone is mainly composed of aromatic amino acids, including two residues of Bht, three Hpg residues, and one Dpg residue. Their biosynthesis requires tyrosine either as a direct or indirect precursor84,86. To meet the high demand for tyrosine, knm carries two genes of the shikimate pathway enzymes: knm8 and knm13, coding for prephenate dehydrogenase (Pdh) and for 3-deoxy-d-arabinoheptulosonate 7-phosphate (Dahp) synthase, respectively (Table 2, Figs. 4b and 5). Notably, while Knm8 is an orthologue of Pdh from bal84, Knm13 is an orthologue of Tei14* from teicoplanin BGC22 (and not of Dahpsec from bal). Knm13 and Tei14* belong to a different evolutionary lineage than Dahpsec within the Superfamily I of Dahp synthases, PRK08763 and PRK0926196, respectively. Peculiarly, such properties of knm8 and knm13 are also reflected in the positions of genes within the BGC. knm8 is located just downstream the cluster-situated regulatory gene and forms a separate transcriptional unit (like its orthologues in bal and ris, Fig. 4a, b), while knm13 is located between the monooxygenase genes (within the O5 operon) contrary to the bal and ris BGCs, where Dahp synthase gene is found on the 3’ boundary of the BGC (Fig. 4a). In addition to knm13, we found that the genome of A. auranticolor DSM 44650 carries two genes for PRK08763-like Dahp synthases (V5P93_005139 and V5P93_004074, Fig. 5), while only one of them (dahpprim) was previously reported in the genome of Am. balhimycina DSM 590886. Moreover, the genome of A. auranticolor DSM 44650 carries two genes for Superfamily II of Dahp synthases (V5P93_002217 and V5P93_006080, Fig. 5), while only one of them (A3CE_RS0135135) was found in the genome of Am. balhimycina DSM 5908 (A3CE_RS0135135)86. On the basis of these analyses, we might speculate that the extended repertoire of Dahp synthase genes found in the genome of A. auranticolor DSM 44650 contributes to its unusually high Kmc production rate.
Biosynthesis of Bht, Hpg and Dpg
The knm BGC contains the typical set of genes for the biosynthesis of non-proteinogenic amino acids found in Pseudonocardiales spp. GPA BGCs (Table 2, Fig. 4a). However, the gene for DpgD enoyl-CoA hydratase, which serves as an accessory enzyme to DpgB, is absent in knm, resembling the situation observed in pekiskomycin BGCs8. The Bht and Hpg biosynthetic genes (knm22-23-24-25-26) constitute the O7, although knm21 (4-hydroxyphenylglycine transaminase, Hpgt) exists as monogenic transcriptional unit (Fig. 4a, b). In ris and bal BGCs, Bht and Hpg biosynthetic genes also belong to one operon, and both carry Hpgt genes as a separate transcriptional unit. In knm, Dpg biosynthesis genes (knm33-34-35) belong to the O8, where they are co-transcribed with some other biosynthetic genes (discussed below), resembling the organization of Dpg genes in bal95, whereas in ris, the corresponding genes form a separate operon54.
Biosynthesis of the aglycone
The features of the NRPS coded within knm were analyzed in previous sections. The module/domain organizations of KnmA-D corresponded to the experimentally resolved structure of KmcB (Figs. 1 and 3). Nevertheless, some of the Kmc congeners (KmcA, vide supra) likely carry l-Val residue at AA3 position of the aglycone (Supplementary Fig. 19). This evidence might suggest a certain promiscuity in the substrate specificity of the M3 A-domain in Kmc biosynthesis NRPS, or of the corresponding C-domain. Unusual structure of Kmc aglycone raised some additional questions about the evolutionary route of the NRPS. Previous works demonstrated that aliphatic amino acids in AA1 and AA3 positions of Type I GPAs appeared as a result of gradual evolution of M1 A-domain substrate specificity and due to larger recombination events exchanging M3, respectively44. However, it is unclear which of these two routes was followed by Kmc M3. To address this, we reconstructed the phylogeny of 363 A-domains and 312 C-domains coming from GPA NRPSs from 52 BGCs (Supplementary Table 5). As a result, sequence of the M3 A-domain of Kmc grouped with the counterpart from U. endophytica DSM 103496 and with the phenylalanine (Phe)-specific A-domains from Type II GPA NRPSs (from Am. keratiniphila ssp. nogabecina FH 1893, Amycolatopsis sp. EV170708-02-1, and Amycolatopsis sp. QT-25), altogether belonging to a larger clade with the sequences of Dpg-specific M3 A-domains (Supplementary Fig. 21). Unfortunately, the reconstruction lacked the M3 A-domain sequence of Type II GPA avoparcin BGC from Am. coloradensis DSM 44225 (Hpg-specific). A similar picture was obtained for the M3 C-domain phylogeny (Supplementary Fig. 22). Thus, according to the reconstructed phylogenies, Ile-specific M3 A-domain of Kmc NRPS, Phe-specific M3 A-domains of Type II GPAs, and Dpg-specific M3 A-domains of other GPAs share immediate common ancestor.
To reflect these findings and fit Kmc into the existing classification of GPAs, we propose herein to separate Type II into two subtypes: Type IIa and Type IIb. In this way, GPAs with aromatic AA1 and aliphatic AA3 (first example here being Kmc) would belong to Type IIa, while the GPAs with both AA1 and AA3 aromatic will form Type IIb.
The knm BGC encodes three cross-linking oxygenases: Knm11, Knm12, and Knm14. These oxygenases are orthologues to OxyA, OxyB, and OxyC of balhimycin biosynthesis, respectively. They install D-O-E, C-O-D, and A-B crosslinks in the Kmc aglycone. Additionally, a single knm-encoded halogenase (Knm15) adds chlorine to AA6 Bht of the aglycone. The substrate specificity of Knm15 is the same as that of Pek27 (pekiskomycin halogenase8), and both proteins are related (Supplementary Table 6 and Supplementary Fig. 23).
The NRPS genes belong to the operons O3 and O4; knmA-B are co-transcribed with knm9 encoding for the ABC transporter (see above), and knmC-D are co-transcribed with knm10 (MbtH, NRPS chaperone protein) (Fig. 4a, b). O5 includes the genes knm11-12-13-14-15, coding for cross-linking oxygenases, Dahp synthase, and halogenase. Such organization shares some features with what observed in both bal (cross-linking oxygenase genes form a separate operon with halogenase gene, but all NRPS genes are co-transcribed) and ris (NRPS genes are divided into two separate operons, but the latter is extended with cross-linking oxygenase genes).
Ristosamine biosynthesis and glycosylation
knm contains a complete set of genes required for ristosamine biosynthesis (knm28-32, Table 2, Fig. 4a), three genes for GT1-family glycosyltransferases (GTFs) (knm16-18), and one gene for a GT39-family GTF (knm20) (Table 2, Fig. 4a). Since GT39-GTFs are mannosyltransferases97, Knm20 is most likely responsible for the addition of the d-mannose residue to AA7 Dpg (similar to ORF22 in ristocetin biosynthesis39). Instead, to assign the functions to the three GT1-GTFs, we included them in a phylogenetic tree published earlier98. We found (Supplementary Table 7 and Supplementary Fig. 24) that Knm17 groups together with the GTFs installing d-glucose at AA4, while Knm16 groups with those that install an amino sugar at AA6. Thus, Knm17 most likely adds d-glucose at AA4 Hpg and Knm16 l-ristosamine at AA6 Bht (Fig. 5). This leaves Knm18 (grouping together with GTFs that add a second sugar residue to d-glucose at AA4, Supplementary Fig. 24) responsible for the addition of a second l-ristosamine residue to AA4 d-glucose in KmcB biosynthesis. As stated previously, at least some Kmc congeners (e.g., KmcC) likely carry a d-rhamnose instead of l-ristosamine attached to AA4 d-glucose (Supplementary Fig. 20). Since no other GT1-GTF genes were found in the genome of A. auranticolor DSM 44650, Knm18 most likely has an extended specificity to sugar donors, which has been also observed in keratinimicin biosynthesis20.
The GT1-GTF genes (knm16-17-18) are co-transcribed as O6, while the mannosyltransferase gene (knm20) and the GlcNAc-deacetylase gene (knm19) form separate monogenic transcriptional units. Ristosamine biosynthesis enzymes (knm28-29-30-31-32) belong to the last operon (O8, the largest in the BGC), also including knm27 (coding for Na+/H+-antiporter), knm33-34-35 (Dpg biosynthesis enzymes, see above), as well as two genes with unclear function (knm36-37), apparently encoding proteins not relevant to GPA biosynthesis.
The biosynthetic model (Fig. 5) proposed from in silico analysis of knm and A. auranticolor DSM 44650 genome indicates that Kmc is produced following a scheme which is overall similar to the other GPAs produced by Pseudonocardiales spp. Anyhow, the notable differences which make Kmc unique are the following. First, the structural elucidation of the main congener KmcB demonstrated that it is a novel chemical structure (proposed as a model for the new chemical Type IIa of GPAs), with the heptapeptide aglycone containing Hpg in AA1 and l-Ile in AA3, this last incorporated due to the previously unknown code of the Kmc NRPS M3 A-domain, DAFMWGIVIK. Secondly, Kmc is produced as a complex of an unusually high numbers of congeners (at least 17 detected by LC-MS analyses, including the four major ones KmcA-D), demonstrating a high flexibility of the GPA biosynthetic machine of A. auranticolor DSM 44650. These congeners, in analogy with other GPA complexes20,99, can be produced as intermediates during the biosynthesis, for example lacking some sugar residues/chlorine atoms added by post NRPS reactions, as reported in teicoplanin overproducing strains100 and/or as degradation/detoxification products during GPA secretion (as in the case of recombinant strains producing increased amount of A40926101,102). A third peculiarity of Kmc, is, in fact, the elevated level of its production by the wild type strain: as a consequence, tailoring reactions might easily become bottleneck steps along the biosynthesis (thus leading to the observed heterogeneity of the complex), as well as detoxification reactions may be needed to sustain such high antibiotic productivity. A. auranticolor DSM 44650 genome analysis revealed that this naturally high producing phenotype is likely due to the expanded repertoire of Dahp synthase genes, which guarantee the aromatic amino acid supply for the aglycone synthesis, making this strain an interesting model for the heterologous production of GPA antibiotics. Finally, although further NMR studies are needed to confirm the structure of KmcA, if our prediction combining mass spectrometry data with in silico analysis of the BGC is correct, this specific congener might be produced due to the intriguing promiscuity of the M3 A-domain in the Kmc NRPS (supposed to introduce l-Val in KmcA instead if l-Ile in KmcB). To our knowledge, that would be the first case, in which a structural variation in a congener is generated during the assembling of the heptapeptide scaffold instead of being due to the peripheral tailoring reactions occurring on the already formed peptide103. The promiscuity of Kmc NRPS can open novel perspectives for reengineering the GPA aglycone assembling, generating a set of new variants. To this purpose, our future efforts will be devoted to the purification, structural elucidation and biological activity testing of the different congeners of Kmc, with the aim to identify the more active ones and eventually modulate their production by adapting cultivation conditions and/or engineering the BGC.
In conclusion, in this work we discovered a completely new GPA, that demonstrates yet another example of the pharmacophore-retaining evolutionary diversification of the ancestral teicoplanin-like aglycone44. These results open further insights into the chemical diversity of this clinically relevant antibiotic class and pave the way for developing in future new derivatives active on emerging resistant pathogens.
Methods
Bacterial strains and resources used in the work
Bacterial strains, used in the work, are listed in the Supplementary Table 8. Chemical substances, enzymes, cultivation media components, etc. are listed in the Supplementary Table 9.
Cultivation conditions of A. auranticolor DSM 44650
A. auranticolor DSM 44650 was routinely cultivated on solid YMPG medium73. Glycerol stocks of A. auranticolor DSM 44650, purchased from Leibniz Institute DSMZ (German Collection of Microorganisms and Cell Cultures), were prepared by growing the strain first in 50 mL of Tryptic Soy Broth (TSB) medium (g L−1 distilled water: 17 tryptone, 3 soya peptone, 5 NaCl, 2.5 K2HPO4, 2.5 dextrose, pH corrected to 7.3 before sterilization) in 300 mL Erlenmeyer flasks with ca. 10 glass beads (ø5 mm). After a 72 h incubation on a rotary shaker at 30 °C and 200 rotations per minute (rpm), 5 mL of the culture were transferred into 100 mL of the liquid version of YMPG medium (g L−1 distilled water: 2 yeast extract, 10 malt extract, 2 bacto peptone, 10 g glucose, 2 g KH2PO4, and 1 g MgSO4 × 7H2O, pH corrected to 7.3 before sterilization)73 in 500 mL flasks with ca. 10 glass beads (ø5 mm), then incubated for 96 h as above. Glycerol stocks were then prepared dividing the culture in 2-mL aliquots, stored at −80 °C with 10% v/v glycerol.
Cultivation of A. auranticolor DSM 44650 for Kmc production was initiated by inoculating one glycerol stock in 50 mL of SGC2 medium (g L−1 distilled water: 5 CaCO3, 3 cane molasses, 5 casein hydrolysate, 30 soluble starch, 15 soya peptone, 15 dextrose, pH corrected to 7.0 before sterilization) in 300 mL Erlenmeyer flasks with ca. 10 glass beads (ø5 mm). After 72 h of incubation at 30 °C and 200 rpm, a 5% v/v inoculum was performed, transferring 5 mL of the culture into 100 mL of R5 medium78 (g L−1 distilled water: 103 sucrose, 0.25 K2SO4, 10.12 MgCl2 × 6 H2O, 10 glucose, 0.1 casein hydrolysate, 5.73 TES buffer, 5 yeast extract, 2 mL trace element solution, 10 mL KH2PO4 solution at 0.5% w/v, 4 mL CaCl2 × 2 H2O solution at 5 M, 15 mL l-proline solution at 20% w/v, 7 mL NaOH 1 N) in 500 mL Erlenmeyer flasks with ca. 10 glass beads (ø5 mm). The cultivation continued at 30 °C and 200 rpm for 336 h. Samples were collected every 24 h to estimate biomass accumulation (dry weight), pH (with a pH meter), glucose consumption (with Diastix sticks, Bayer AG, Leverkusen, Germany), and Kmc production (see below PDA-HPLC). For the cultivation in bioreactor, the culture of A. auranticolor DSM 44650 grown in SGC2 medium was used to inoculate at 5% v/v a 3-L P-100 Applikon glass reactor (height 250 mm, ø130 mm) equipped with an AD1030 Biocontroller and AD1032 motor, and containing 2 L of R5 medium, supplemented with 200 µL of Hodag antifoam (Hodag Chemical Corporation, Chicago, IL, US). Fermentation was carried out at 30 °C, with stirring at 400 rpm with Rushton blades and 2 L min−1 aeration rate. Dissolved oxygen (measured as % pO2) was monitored using an Ingold polarographic oxygen electrode, while pH values were monitored using a pH meter. Foam production was controlled by adding Hodag antifoam through an antifoam sensor.
PDA-HPLC analyses
A. auranticolor DSM 44650 cultures were extracted by correcting their pH to 12.0 using NaOH 10 N, then centrifuged at 13,400 rpm for 15 min. Chromatographic analyses were performed with a VWR Hitachi diode array L-2455 HPLC system, with detection at 236 nm. 50 µL of each sample were injected onto a 5-µm-particle size KromaPhase 100 C18 4.6 × 250 mm HPLC column. Elution was done at a flow rate of 0.95 mL min−1, with a linear gradient from 5% v/v to 30% v/v of Phase B in 25 min. Phase A was 0.1% v/v phosphoric acid, while Phase B was 100% v/v HPLC-grade acetonitrile. Kmc production was quantified taking into consideration the areas of all major (KmcA-KmcD) and minor congeners identified by the UV spectrum, and using 50 µL of 50 µg mL−1 vancomycin as internal standard.
LC-MS and MS/MS analyses
LC-MS and MS/MS analyses were conducted using an LTQ-Orbitrap XL hybrid ion trap-orbitrap mass spectrometer (Thermo Fisher Scientific GmbH, Bremen, Germany) coupled with an analytical HPLC 1290 Infinity system (Agilent Technologies, Waldbronn, Germany). To analyze the culture extracts and crude extracts, an HPLC column (Poroshell 120, EC-C18, 50 × 2.1 mm, 2.7 µm, Agilent Technologies, Waldbronn, Germany) enabled the sample separation and was eluted by a linear gradient using water with 0.1% v/v formic acid as Phase A and acetonitrile with 0.1% v/v formic acid as Phase B. The separation started with an isocratic elution at 5% v/v B for 4 min, followed by a linear gradient from 5 to 10% v/v B over 4 min, from 10–15% v/v B over 5 min, from 15 to 100% v/v B over 2 min. The column was re-equilibrated with 5% v/v B for an additional 3 min. The injection volume was 5 µL and the flow rate was set to 0.5 mL min−1. The drawing and ejection speed were set to 100 μL min−1 and 400 μL min−1, respectively. The ESI source parameters were set as follows: product ion spectra were recorded in data-dependent acquisition (DDA) mode with a mass range from m/z 180 to m/z 2000 (MS1: FTMS, normal, resolution = 60,000, full, positive; MS2: FTMS, normal, resolution = 30,000, positive). An auxiliary gas flow of 10 units, capillary temperature of 270 °C, capillary voltage of 1 V, sheath gas flow of 45 units, and source voltage of 4000 V were used. The parameter for the DDA mode was set as follows: activation type: CID, minimum signal required: 10,000, isolation width: m/z 2.00, normalized collision energy: 35.0, default charge state: 2, activation Q: 0.250, and activation time: 30 ms. The dynamic exclusion enabled was set as follows: repeat count: 3, repeat duration: 30 s, exclusion list size: 50, and exclusion duration: 180 s. To analyze the pure compound (KmcB), the above gradient was optimized as it started isocratically with 5% v/v B for 4 min, followed by a linear gradient from 5 to 100% v/v B over 5 min. The column was re-equilibrated with 5% v/v B for an additional 3 min. The mass range was from m/z 500 to m/z 2000. For MS/MS fragmentation, the product ions were formed by in-source surface-induced dissociation (SID: 70 V). The mass range spanned from m/z 100 to m/z 2000. Alternatively, the two most intensive precursors per MS1 were selected for subsequent collision-induced dissociation (CID: 35 V). The MS and MS/MS data derived from the LTQ-Orbitrap XL were acquired with Xcalibur 2.2 (Thermo Fisher Scientific GmbH, Bremen, Germany), displayed and analyzed with Freestyle 1.8 SP2 (Thermo Fisher Scientific GmbH, Bremen, Germany).
d-Ala-d-Ala based affinity chromatography
Activation of 5 mL HiTrap NHS-activated HP affinity columns (GE Healthcare) and ligand binding was conducted as described before90 with slight modifications. Briefly, the resin was activated with 30 mL of 1 mM HCl, followed by injection of 200 mM d-Ala-d-Ala dipeptide, dissolved into 5 mL of coupling buffer (0.2 M NaHCO3, 0.5 M NaCl, pH 8.3). After 30 min incubation, the resin was washed with 0.5 M ethanolamine hydrochloride, 0.5 M NaCl (pH 8.3, 30 mL – Buffer A), followed by 0.1 M sodium acetate, 0.5 mM NaCl (pH 4.0, 30 mL – Buffer B), and again 30 mL of Buffer A. After 30 min incubation, the resin was washed with three cycles of Buffer B, Buffer A, and Buffer B. Culture extracts were prepared as described above and their pH brought back to 8.0 using HCl 6 N. They were then filtered with a cut-off of 0.22 µm and loaded onto d-Ala-d-Ala column at a flow rate of 0.5 mL min−1. After extensive washing with coupling buffer, the bound GPA was eluted with 0.1 M ammonium hydroxide and the eluate was lyophilized to obtain a crude pre-fractionated GPA mixture.
Purification of KmcB
An Agilent 1100 system consisting of a G1312A binary pump, a G1315D diode array detector (DAD), a G1316A column compartment, a G1329A automatic liquid sampler (ALS) and a G1364C analytical fraction collector (FC) was used for KmcB purification. The analytical HPLC chromatograms were acquired and displayed with Agilent ChemStation for LC 3D systems B.03.02 (Agilent Technologies, Waldbronn, Germany). For purification, a partially purified crude extract (1.973 g) obtained by d-Ala-d-Ala based affinity chromatography was resuspended in MQ-H2O to give a final concentration at 80 mg mL−1, followed by centrifugation at 4000 rpm and 4 °C for 10 min (Eppendorf® Centrifuge 5810 R). The separation of the supernatant obtained was carried out using a Fortis Diphenyl column (150 × 4.6 mm, 5 μm, Fortis Technologies Ltd., Cheshire, UK), which was eluted by a linear gradient using water with 0.1% v/v formic acid as Phase A and methanol with 0.1% v/v formic acid as Phase B. The gradient was started with an isocratic elution with 5% v/v B for 2 min, followed by a linear gradient from 5–10% v/v B over 10 min, from 10–35% v/v B over 2 min. The column was washed with 100% v/v B for 1.5 min and re-equilibrated with 5% v/v B for an additional 4.5 min. The injection volume was 60 µL. The flow rate and UV monitoring were set to 1.0 mL min−1 and 236 nm, respectively. All fractions containing KmcB were pooled. 6.1 mg of pure KmcB was obtained after evaporation and lyophilization.
NMR spectroscopy
1D- and 2D-NMR spectra were acquired on a Bruker Avance III 700 MHz spectrometer (700 MHz for 1H, and 176 MHz for 13C) with a 5 mm TXI probe (Bruker, Karlsruhe, Germany) at 298 K. The 1H and 13C NMR chemical shifts were referenced to the solvent peaks at 2.50 ppm (1H) and 39.5 ppm (13C) for DMSO when DMSO-d6 was utilized as solvent. TopSpin 3.5 (Bruker, Karlsruhe, Germany) was used for data acquisition and TopSpin 4.1.4 (Bruker, Karlsruhe, Germany) for data processing.
Advanced Marfey’s amino analysis
KmcB (0.35 mg) was hydrolyzed with stirring in 6 N HCl (200 μL) at 110 °C for 12 h. The acid hydrolysates were frozen at −80 °C followed by lyophilization. The dry acid hydrolysates were resuspended in H2O (50 μL; analytical grade) and equally divided into two portions A (25 μL) and B (25 μL). Portion A was treated with 1 M NaHCO3 (10 μL) and then with Marfey’s reagent l-FDLA (50 μL of a 10 mg mL−1 solution in acetone), and the mixture was stirred at 37 °C for 1 h. The reaction was quenched with 1 N HCl (10 μL) and diluted with MeOH (105 μL) up to a final volume of 200 μL. Portion B was treated following the similar protocol but with l-FDLA (25 μL of a 10 mg mL−1 solution in acetone) and d-FDLA (25 μL of a 10 mg mL−1 solution in acetone). Authentic standards of l-Ile, l-allo-Ile, and d-Ile were treated with l-FDLA and d/l-FDLA as described above and yielded the l-FDLA and d-FDLA derivatives of standards91. Sample analysis was carried out with the HPLC-ESI-HRMS mass spectrometer as described above but in negative ionization mode. The column was eluted isocratically at 5% v/v B for 5 min, followed by a linear gradient from 5–50% v/v B over 40 min, from 50–100% v/v B over 5 min. The column was re-equilibrated with 5% v/v B for an additional 5 min. The injection volume was 5 µL and the flow rate was set to 0.5 mL min−1. The detection mass range was set between m/z 100 to m/z 1000.
Minimum Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC) determination
MICs and MBCs of Kmc, vancomycin, and teicoplanin towards a panel of Gram-positive and Gram-negative bacteria were determined following the guidelines of the Clinical and Laboratory Standards Institute83, with broth dilution method in Müller Hinton Broth, cation adjusted (MHB2). Escherichia coli ATCC 35218, Moraxella catarrhalis ATCC 3293, Staphylococcus aureus ATCC 6538 P (MSSA), Staphylococcus aureus ATCC 25923 (MSSA), Staphylococcus aureus ATCC 43300 (MRSA), Enterococcus faecalis ATCC 29212, and Enterococcus faecalis ATCC 51299 (VanB phenotype) were obtained from the American Type Culture Collection (ATCC). Enterococcus faecalis 9160188401-EF-34 (VanA phenotype) and Staphylococcus epidermidis strain 4 are clinical isolates, kindly provided by Laboratorio Microbiologia Clinica—Ospedale di Circolo, Varese, Italy. Staphylococcus haemolyticus 3902 is a teicoplanin-resistant clinical isolate104, received from FIIRV (Fondazione Istituto Insubrico Ricerca per la Vita), Gerenzano Varese, Italy. For long-term preservation, bacterial cultures were stored at –80 °C in 10% v/v glycerol.
For MIC determination, 50 µL of a suspension of bacterial cells in exponential growth phase (at 1 × 106 colony forming unit (CFU) mL−1 concentration) in MHB2 were inoculated in 96-well plates, together with 50 µL of GPA (from 0 to 128 µg mL−1 final concentration). Plates were then incubated for 20 h at 37 °C and 100 rpm. MICs were expressed as the minimal concentration of antibiotic at which no turbidity could be detected. For MBC determination, the content of the wells in which no growth was observed, was spread in 48-well plates containing 900 µL of Müller Hinton Agar, incubated at 37 °C for 24 h. MBCs were defined as the lowest concentration of antibiotic at which no growth could be seen. All experiments were repeated at least in triplicate.
Isolation of genomic DNA from A. auranticolor DSM 44650
Genomic DNA, further used as a positive control in semi-quantitative RT-PCR analysis, was isolated from A. auranticolor DSM 44650 using the Kirby procedure78. For the genome sequencing, genomic DNA of A. auranticolor DSM 44650 was extracted using the NucleoSpin® Microbial DNA kit (MACHEREY-NAGEL GmbH & Co. KG) according to the supplier’s protocol.
RNA isolation from A. auranticolor DSM 44650, cDNA synthesis, and semi-quantitative RT-PCR
RNA was isolated from vegetative mycelium of A. auranticolor DSM 44650 obtained from three independent Kmc producing cultures cultivated for 96 h in R5 medium. RNAqueous Phenol-free total RNA isolation kit (Invitrogen, Thermo Fisher Scientific, Waltham, USA) was used for RNA isolation according to the protocol of supplier, but omitting the on-column DNase treatment step. Isolated RNA samples (contaminated with DNA) were further subjected to first DNase treatment (1 h at 37 °C), using 1 U of DNase I, RNase-free (Thermo Fisher Scientific, Waltham, USA) per 10 µL of solution. After first DNase treatment, RNA concentration and quality were assessed using DeNovix DS-11 FX+ (DeNovix Inc., Wilmington, USA), and 50 µg of RNA were subjected to a second DNase treatment with 8 U of DNase I, RNase-free (Thermo Fisher Scientific, Waltham, USA) for 2 h at 37 °C. To test if the second DNase treatment removed all residual DNA contamination, 1 µL of the mixture was used as a template for PCR with HrdB_Akin_F/R (Supplementary Table 10) to amplify the 253 bp internal region of hrdB (RNA polymerase sigma factor gene, V5P93_005973); in case the results of PCR were negative, DNAse was inactivated by adding 10% v/v EDTA (50 mM) and incubating it at 65 °C for 10 min, otherwise additional DNase treatment step was performed. Concentration and quality of DNA-free RNA sample was assessed using DeNovix DS-11 FX+ (DeNovix Inc., Wilmington, USA), and 5 µg of RNA were subjected to the cDNA synthesis step using RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher Scientific, Waltham, USA) according to the supplier’s protocol. 1 µL of cDNA obtained in the previous step was used as a template for the PCR reaction with Q5 High-Fidelity DNA Polymerase (New England Biolabs, Ipswich, USA) and oligonucleotide primers listed in Supplementary Table 10. This reaction was set in parallel with the positive control reaction (with 100 ng of gDNA as a template) and a negative control reaction (with 1 µL of DNA-free RNA sample). Obtained amplicons were visualized by horizontal 2% w/v agarose (Thermo Fisher Scientific, Waltham, USA) gel-electrophoresis using GeneRuler 1 kb DNA Ladder (Thermo Fisher Scientific, Waltham, USA).
Sequencing and complete assembly of A. auranticolor DSM 44650 genome
Long and short DNA reads were generated by Nanopore and Illumina sequencing, respectively. For library preparation, the TruSeq DNA PCR-free high-throughput library prep kit (Illumina) and the SQK-LSK112 sequencing kit (Oxford Nanopore Technologies [ONT]) were used without prior shearing of the DNA. To generate the short reads, a 2 × 300-nucleotide run (MiSeq reagent kit v3, 600 cycles) was executed. The long reads were generated on a GridION platform using a R9.4.1 flow cell. Base calling and demultiplexing were performed using guppy v6.3.8.0 with the super-accurate basecalling model. Assemblies were done using flye v.2.9105 for the Nanopore long read data and Newbler v2.8106 for the Illumina short read data. After polishing of the flye-based assembly using pilon v1.22107 using bowtie2108 for mapping, the respective flye and Newbler assemblies were combined in consed v28.0109. The resulting single contig representing the circular genome was annotated using the PGAP pipeline110,111. All raw sequencing data are available via BioProject ID PRJNA1076217. The annotated genome is accessible via GenBank ID CP154825.
Details of in silico analysis
MultiGeneBlast56 was used to search annotated Pseudonocardiales spp. genomes, available in GenBank, for co-localized homologs of bbr, oxyA, oxyB, and oxyC from balhimycin BGC (Y16952.3) under the default parameters (except “maximum distance between genes in locus (kb)” parameter, that was set to 34 kbp – distance between bbr and oxyC). antiSMASH 7.057 was used for the automated annotation of GPA BGCs, prediction of the module/domain organization of NRPSs, and non-ribosomal codes. Mega 11.0.13112 package was used for the phylogenetic reconstructions. Geneious 4.8.5113 was used for the oligonucleotide primer design and routine analysis of nucleic acid and amino acid sequences.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All unique reagents generated in this study are available from the corresponding authors upon reasonable request. The availability of kineomicins may be limited to a small batch and might require preparation. The published article includes all biological data generated during this study. Complete genome assembly of A. auranticolor DSM 44650 was deposited to the GenBank under the CP154825 accession number. All mass spectrometry data, obtained in this work, were deposited in the MassIVE mass spectrometry data repository (MSV000097194 dataset).
References
Nicolaou, K. C., Boddy, C. N. C., Bräse, S. & Winssinger, N. Chemistry, biology, and medicine of the glycopeptide antibiotics. Angew. Chem. - Int. Ed. 38, 2096–2152 (1999).
Yushchuk, O. & Ostash, B. Glycopeptide antibiotics: genetics, chemistry, and new screening approaches. In Natural Products from Actinomycetes (eds. Rai, R. V. & Bai, J. A.) (Springer, 2022).
Yim, G., Thaker, M. N., Koteva, K. & Wright, G. Glycopeptide antibiotic biosynthesis. J. Antibiot. (Tokyo). 67, 31–41 (2014).
Hansen, M. H., Stegmann, E. & Cryle, M. J. Beyond vancomycin: recent advances in the modification, reengineering, production and discovery of improved glycopeptide antibiotics to tackle multidrug-resistant bacteria. Curr. Opin. Biotechnol. 77, 102767 (2022).
Van Bambeke, F. Lipoglycopeptide antibacterial agents in gram-positive infections: a comparative review. Drugs 75, 2073–2095 (2015).
Van Bambeke, F. Glycopeptides in clinical development: pharmacological profile and clinical perspectives. Curr. Opin. Pharmacol. 4, 471–478 (2004).
Xu, L. et al. Complete genome sequence and comparative genomic analyses of the vancomycin-producing Amycolatopsis orientalis. BMC Genomics 15, 1–18 (2014).
Thaker, M. N. et al. Identifying producers of antibacterial compounds by screening for antibiotic resistance. Nat. Biotechnol. 31, 922–927 (2013).
Bardone, M. R., Paternoster, M. & Coronelli, C. Teichomycins, new antibiotics from Actinoplanes teichomyceticus nov. sp. II. Extraction and chemical characterization. J. Antibiot. (Tokyo). 31, 170–177 (1978).
Van Groesen, E., Innocenti, P. & Martin, N. I. Recent advances in the development of semisynthetic glycopeptide antibiotics: 2014-2022. ACS Infect. Dis. 8, 1381–1407 (2022).
Goldstein, B. P. et al. A40926, a new glycopeptide antibiotic with anti-Neisseria activity. Antimicrob. Agents Chemother. 31, 1961–1966 (1987).
Hamill, R. L., Mabe, J. A., Mahoney, D. F., Nakatsukasa, W. M., Yao, R. C. A82846 antibiotics (U. S. Patent No. US5312738A). U.S. Patent and Trademark Office. https://patents.google.com/patent/US5312738A/en. (1987)
Levine, D. P. Vancomycin: a history. Clin. Infect. Dis. 42, S5–S12 (2006).
Levine, J. F. Vancomycin: a review. Med. Clin. North Am. 71, 1135–1145 (1987).
Grundy, W. E. et al. Ristocetin, microbiologic properties. Antibiot. Annu. 1956, 687–692 (1956).
Cavalleri, B., Pagani, H., Volpe, G., Selva, E. & Parenti, F. A-16686, a new antibiotic from Actinoplanes: I. Fermentation, isolation and preliminary physico-chemical characteristics. J. Antibiot. (Tokyo). 37, 309–317 (1984).
Williams, D. H. The glycopeptide story – how to kill the deadly ‘superbugs. Nat. Prod. Rep. 13, 469–477 (1996).
Parenti, F. & Cavalleri, B. Proposal to name the vancomycin-ristocetin like glycopeptides as dalbaheptides. J. Antibiot. (Tokyo). 42, 1882–1883 (1989).
Stegmann, E. et al. Genetic analysis of the balhimycin (vancomycin-type) oxygenase genes. J. Biotechnol. 124, 640–653 (2006).
Xu, F. et al. A genetics-free method for high-throughput discovery of cryptic microbial metabolites. Nat. Chem. Biol. 15, 161–168 (2019).
Spohn, M. et al. Overproduction of ristomycin a by activation of a silent gene cluster in Amycolatopsis japonicum MG417-CF17. Antimicrob. Agents Chemother. 58, 6185–6196 (2014).
Yushchuk, O., Ostash, B., Truman, A. W., Marinelli, F. & Fedorenko, V. Teicoplanin biosynthesis: unraveling the interplay of structural, regulatory, and resistance genes. Appl. Microbiol. Biotechnol. 104, 3279–3291 (2020).
Greule, A. et al. Kistamicin biosynthesis reveals the biosynthetic requirements for production of highly crosslinked glycopeptide antibiotics. Nat. Commun. 10, 2613 (2019).
van Wageningen, A. M. et al. Sequencing and analysis of genes involved in the biosynthesis a vancomycin group antibiotic. Chem. Biol. 5, 155–162 (1998).
Chiu, H. T. et al. Molecular cloning and sequence analysis of the complestatin biosynthetic gene cluster. Proc. Natl Acad. Sci. USA. 98, 8548–8553 (2001).
Li, T. L. et al. Biosynthetic gene cluster of the glycopeptide antibiotic teicoplanin: characterization of two glycosyltransferases and the key acyltransferase. Chem. Biol. 11, 107–119 (2004).
Sosio, M., Stinchi, S., Beltrametti, F., Lazzarini, A. & Donadio, S. The gene cluster for the biosynthesis of the glycopeptide antibiotic A40926 by Nonomuraea species. Chem. Biol. 10, 541–549 (2003).
Recktenwald, J. et al. Nonribosomal biosynthesis of vancomycin-type antibiotics: a heptapeptide backbone and eight peptide synthetase modules. Microbiology 148, 1105–1118 (2002).
Woithe, K. et al. Oxidative phenol coupling reactions catalyzed by OxyB: a cytochrome P450 from the vancomycin producing organism. Implications for vancomycin biosynthesis. J. Am. Chem. Soc. 129, 6887–6895 (2007).
Hubbard, B. K. & Walsh, C. T. Vancomycin assembly: nature’s way. Angew. Chem. Int. Ed. Engl. 42, 730–765 (2003).
Sandercock, A. M. et al. Biosynthesis of the di-meta-hydroxyphenylglycine constituent of the vancomycin-group antibiotic chloroeremomycin. Chem. Commun. 2001, 1252–1253 (2001)
Pfeifer, V. et al. A polyketide synthase in glycopeptide biosynthesis. The biosynthesis of the non-proteinogenic amino acid (S)-3,5-dihydroxyphenylglycine. J. Biol. Chem. 276, 38370–38377 (2001).
Puk, O. et al. Biosynthesis of chloro-β-hydroxytyrosine, a nonproteinogenic amino acid of the peptidic backbone of glycopeptide antibiotics. J. Bacteriol. 186, 6093–6100 (2004).
Kirkpatrick, P. N. et al. Characterisation of a sugar epimerase enzyme involved in the biosynthesis of a vancomycin-group antibiotic. Chem. Commun. 17, 1565–1566 (2000).
Pelzer, S. et al. Identification and analysis of the balhimycin biosynthetic gene cluster and its use for manipulating glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908. Antimicrob. Agents Chemother. 43, 1565–1573 (1999).
Losey, H. C. et al. Tandem action of glycosyltransferases in the maturation of vancomycin and teicoplanin aglycones: тovel glycopeptides. Biochemistry 40, 4745–4755 (2001).
Losey, H. C. et al. Incorporation of glucose analogs by GtfE and GtfD from the vancomycin biosynthetic pathway to generate variant glycopeptides. Chem. Biol. 9, 1305–1314 (2002).
Yim, G. et al. Harnessing the synthetic capabilities of glycopeptide antibiotic tailoring enzymes: сharacterization of the UK-68,597 biosynthetic cluster. ChemBioChem. 15, 2613–2623 (2014).
Truman, A. W. et al. Antibiotic resistance mechanisms inform discovery: identification and characterization of a novel Amycolatopsis strain producing ristocetin. Antimicrob. Agents Chemother. 58, 5687–5695 (2014).
Xu, M. et al. GPAHex – a synthetic biology platform for type IV–V glycopeptide antibiotic production and discovery. Nat. Commun. 11, 5232 (2020).
Waglechner, N., McArthur, A. G. & Wright, G. D. Phylogenetic reconciliation reveals the natural history of glycopeptide antibiotic biosynthesis and resistance. Nat. Microbiol. 4, 1862–1871 (2019).
Culp, E. J. et al. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature 578, 582–587 (2020).
Xu, M. et al. Phylogeny-informed synthetic biology reveals unprecedented structural novelty in type V glycopeptide antibiotics. ACS Cent. Sci. 8, 615–626 (2022).
Hansen, M. H. et al. Resurrecting ancestral antibiotics: unveiling the origins of modern lipid II targeting glycopeptides. Nat. Commun. 14, 7842 (2023).
Yue, X., Xia, T., Wang, S., Dong, H. & Li, Y. Highly efficient genome editing in N. gerenzanensis using an inducible CRISPR/Cas9–RecA system. Biotechnol. Lett. 42, 1699–1706 (2020).
Yushchuk, O. et al. New molecular tools for regulation and improvement of A40926 glycopeptide antibiotic production in Nonomuraea gerenzanensis ATCC 39727. Front. Microbiol. https://doi.org/10.3389/fmicb.2020.00008 (2020).
Haslinger, K., Peschke, M., Brieke, C., Maximowitsch, E. & Cryle, M. J. X-domain of peptide synthetases recruits oxygenases crucial for glycopeptide biosynthesis. Nature 521, 105–109 (2015).
Peschke, M., Haslinger, K., Brieke, C., Reinstein, J. & Cryle, M. J. Regulation of the P450 oxygenation cascade involved in glycopeptide antibiotic biosynthesis. J. Am. Chem. Soc. 138, 6746–6753 (2016).
Schoppet, M., Tailhades, J., Kulkarni, K. & Cryle, M. J. Precursor manipulation in glycopeptide antibiotic biosynthesis: are β-amino acids compatible with the oxidative cyclization cascade? J. Org. Chem. 83, 7206–7214 (2018).
Gavriilidou, A. et al. Phylogenetic distance and structural diversity directing a reclassification of glycopeptide antibiotics. bioRxiv https://doi.org/10.1101/2023.02.10.526856 (2023)
Nouioui, I. et al. Genome-based taxonomic classification of the phylum actinobacteria. Front. Microbiol. 9, 1–119 (2018).
Andreo-Vidal, A., Binda, E., Fedorenko, V., Marinelli, F. & Yushchuk, O. Genomic insights into the distribution and phylogeny of glycopeptide resistance determinants within the Actinobacteria phylum. Antibiotics 10, 1533 (2021).
Kunstmann, M. P., Mitscher, L. A., Porter, J. N., Shay, A. J. & Darken, M. A. LL-AV290, a new antibiotic. I. Fermentation, isolation, and characterization. Antimicrob. Agents Chemother. 8, 242–245 (1968).
Liu, K. et al. Enhancing ristomycin a production by overexpression of ParB-like StrR family regulators controlling the biosynthesis genes. Appl. Environ. Microbiol. 87, 1–19 (2021).
Adamek, M. et al. Comparative genomics reveals phylogenetic distribution patterns of secondary metabolites in Amycolatopsis species. BMC Genomics 19, 426 (2018).
Medema, M. H., Takano, E. & Breitling, R. Detecting sequence homology at the gene cluster level with MultiGeneBLAST. Mol. Biol. Evol. 30, 1218–1223 (2013).
Blin, K. et al. AntiSMASH 7.0: new and improved predictions for detection, regulation, chemical structures and visualisation. Nucleic Acids Res. 51, W46–W50 (2023).
Menges, R., Muth, G., Wohlleben, W. & Stegmann, E. The ABC transporter Tba of Amycolatopsis balhimycina is required for efficient export of the glycopeptide antibiotic balhimycin. Appl. Microbiol. Biotechnol. 77, 125–134 (2007).
Mulyani, S. et al. The thioesterase Bhp is involved in the formation of β-hydroxytyrosine during balhimycin biosynthesis in Amycolatopsis balhimycina. ChemBioChem. 11, 266–271 (2010).
Stegmann, E., Frasch, H. J. & Wohlleben, W. Glycopeptide biosynthesis in the context of basic cellular functions. Curr. Opin. Microbiol. 13, 595–602 (2010).
Omura, S. et al. Studies on bacterial cell wall inhibitors: VII. Azureomycins A and B, new antibiotics produced by Pseudonocardia azurea nov. sp. Taxonomy of the producing organism, isolation, characterization and biological properties. J. Antibiot. (Tokyo). 32, 985–994 (1979).
Kaur, N., Kumar, S., Bala, M., Raghava, G. P. S. & Mayilraj, S. Draft genome sequence of Amycolatopsis decaplanina strain DSM 44594T. Genome Announc 1, e0013813 (2013).
Jørgensen, T. S. et al. Complete genome sequence of the rare actinobacterium Kutzneria sp. strain CA-103260. Microbiol. Resour. Announc. 10, e0049921 (2021).
Stepanyshyn, A. et al. Complete genome assembly of Amycolatopsis bartoniae DSM 45807T allows the characterization of a novel glycopeptide biosynthetic gene cluster. 15, 1651 (2024).
Fujimori, D. G. et al. Cloning and characterization of the biosynthetic gene cluster for kutznerides. Proc. Natl Acad. Sci. USA 104, 16498–16503 (2007).
Gao, W. et al. Discovery and characterization of the tuberculosis drug lead ecumicin. Org. Lett. 16, 6044–6047 (2014).
Otoguro, M. et al. Numerical phenetic and phylogenetic analyses of Actinokineospora isolates, with a description of Actinokineospora auranticolor sp. nov. and Actinokineospora enzanensis sp. nov. Actinomycetologica 15, 30–39 (2001).
Mascher, T., Zimmer, S. L., Smith, T. A. & Helmann, J. D. Antibiotic-inducible promoter regulated by the cell envelope stress-sensing two-component system LiaRS of Bacillus subtilis. Antimicrob. Agents Chemother. 48, 2888–2896 (2004).
Mascher, T., Margulis, N. G., Wang, T., Ye, R. W. & Helmann, J. D. Cell wall stress responses in Bacillus subtilis: the regulatory network of the bacitracin stimulon. Mol. Microbiol. 50, 1591–1604 (2003).
Andreo-Vidal, A., Yushchuk, O., Marinelli, F. & Binda, E. Cross-talking of pathway-specific regulators in glycopeptide antibiotics (teicoplanin and A40926) production. Antibiotics 12, 641 (2023).
Marcone, G. L., Binda, E., Carrano, L., Bibb, M. & Marinelli, F. Relationship between glycopeptide production and resistance in the actinomycete Nonomuraea sp. ATCC 39727. Antimicrob. Agents Chemother. 58, 5191–5201 (2014).
Taurino, C., Frattini, L., Marcone, G. L., Gastaldo, L. & Marinelli, F. Actinoplanes teichomyceticus ATCC 31121 as a cell factory for producing teicoplanin. Microb. Cell Fact. 10, 82 (2011).
Su, W. & Beuchat, L. R. Combined effect of growth medium, age of cells and phase of sporulation on heat resistance and recovery of Hansenula anomala. Mycopathologia 87, 129–134 (1984).
Shirling, E. B. & Gottlieb, D. Method for characterization of Streptomyces species. Int. J. Syst. Bacteriol. 16, 313–340 (1966).
Koshla, O. et al. Gene miaA for post-transcriptional modification of tRNAXXA is important for morphological and metabolic differentiation in Streptomyces. Mol. Microbiol. 112, 249–265 (2019).
Zahoor, T., Siddique, F. & Farooq, U. Isolation and characterization of vinegar culture (Acetobacter aceti) from indigenous sources. Br. Food J. 108, 429–439 (2006).
Rebets, Y. et al. Production of landomycins in Streptomyces globisporus 1912 and S. cyanogenus S136 is regulated by genes encoding putative transcriptional activators. FEMS Microbiol. Lett. 222, 149–153 (2003).
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F. & Hopwood, D. A. Practical Streptomyces Genetics (John Innes Foundation, 2000).
Neu, J. M. & Wright, G. D. Inhibition of sporulation, glycopeptide antibiotic production and resistance in Streptomyces toyocaensis NRRL 15009 by protein kinase inhibitors. FEMS Microbiol. Lett. 199, 15–20 (2001).
Puk, O. et al. Glycopeptide biosynthesis in Amycolatopsis mediterranei DSM5908: function of a halogenase and a haloperoxidase/perhydrolase. Chem. Biol. 9, 225–235 (2002).
Schäberle, T. F. et al. Self-resistance and cell wall composition in the glycopeptide producer Amycolatopsis balhimycina. Antimicrob. Agents Chemother. 55, 4283–4289 (2011).
Yushchuk, O., Binda, E. & Marinelli, F. Glycopeptide antibiotic resistance genes: distribution and function in the producer actinomycetes. Front. Microbiol. https://doi.org/10.3389/fmicb.2020.01173 (2020).
Wayne, P. Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Third Informational Supplement. CLSI Document M100-S23. Performance Standards for Antimicrobial Susceptibility Testing. https://clsi.org/shop/standards/m100/ (2013).
Thykaer, J. et al. Increased glycopeptide production after overexpression of shikimate pathway genes being part of the balhimycin biosynthetic gene cluster. Metab. Eng. 12, 455–461 (2010).
Nadkarni, S. R. et al. Balhimycin, a new glycopeptide antibiotic produced by Amycolatopsis sp. Y-86,21022. Taxonomy, production, isolation and biological activity. J. Antibiot. (Tokyo). 47, 334–341 (1994).
Goldfinger, V. et al. Metabolic engineering of the shikimate pathway in Amycolatopsis strains for optimized glycopeptide antibiotic production. Metab. Eng. 78, 84–92 (2023).
Jung, H. M., Kim, S. Y., Moon, H. J., Oh, D. K. & Lee, J. K. Optimization of culture conditions and scale-up to pilot and plant scales for vancomycin production by Amycolatopsis orientalis. Appl. Microbiol. Biotechnol. 77, 789–795 (2007).
Horbal, L. et al. The pathway-specific regulatory genes, tei15* and tei16*, are the master switches of teicoplanin production in Actinoplanes teichomyceticus. Appl. Microbiol. Biotechnol. 98, 9295–9309 (2014).
Ramoni, G. et al. New avoparcin-like molecules from the avoparcin producer Amycolatopsis coloradensis ATCC 53629. Fermentation 8, 1–11 (2022).
Yushchuk, O. et al. Genomic-led discovery of a novel glycopeptide antibiotic by Nonomuraea coxensis DSM 45129. ACS Chem. Biol. 16, 915–928 (2021).
Wu, Y. et al. Fuscasins A-D, cycloheptapeptides from the marine sponge Phakellia fusca. J. Nat. Prod. 82, 970–979 (2019).
Jonker, H. R. A. et al. NMR spectroscopic characterization of the C-mannose conformation in a thrombospondin repeat using a selective labeling approach. Angew. Chem. Int. Ed. Engl. 59, 20659–20665 (2020).
Deslauriers, R., Jarrell, H. C., Byrd, R. A. & Smith, I. C. Observation by 13C NMR of metabolites in differentiating amoeba. Trehalose storage in encysted Acanthamoeba castellanii. FEBS Lett. 118, 185–190 (1980).
Gerhard, U., Mackay, J. P., Maplestone, R. A. & Williams, D. H. The role of the sugar and chlorine substituents in the dimerization of vancomycin antibiotics. J. Am. Chem. Soc. 115, 232–237 (1993).
Shawky, R. M. et al. The border sequence of the balhimycin biosynthesis gene cluster from Amycolatopsis balhimycina contains bbr, encoding a StrR-like pathway-specific regulator. J. Mol. Microbiol. Biotechnol. 13, 76–88 (2007).
Lu, S. et al. CDD/SPARCLE: The conserved domain database in 2020. Nucleic Acids Res. 48, D265–D268 (2020).
Drula, E. et al. The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res. 50, D571–D577 (2022).
Yushchuk, O., Binda, E., Fedorenko, V. & Marinelli, F. Occurrence of vanHAX and related genes beyond the Actinobacteria phylum. Genes (Basel). 13, 1960 (2022).
Debono, M. et al. Actaplanin, new glycopeptide antibiotics produced by Actinoplanes missouriensis the isolation and preliminary chemical characterization of actaplanin. J. Antibiot. (Tokyo). 37, 85–95 (1984).
Yushchuk, O. et al. Characterization of the post-assembly line tailoring processes in teicoplanin biosynthesis. ACS Chem. Biol. 11, 2254–2264 (2016).
Alt, S. et al. Toward single-peak dalbavancin analogs through biology and chemistry. ACS Chem. Biol. 14, 356–360 (2019).
Sosio, M., Canavesi, A., Stinchi, S. & Donadio, S. Improved production of A40926 by Nonomuraea sp. through deletion of a pathway-specific acetyltransferase. Appl. Microbiol. Biotechnol. 87, 1633–1638 (2010).
James, R. C., Pierce, J. G., Okano, A., Xie, J. & Boger, D. L. Redesign of glycopeptide antibiotics: back to the future. ACS Chem. Biol. 7, 797–804 (2012).
Beltrametti, F., Lazzarini, A., Brunati, C., Selva, E. & Marinelli, F. Production of demannosyl-A40926 by a Nonomuraea sp. ATCC 39727 mutant strain. J. Antibiot. (Tokyo). 56, 310–313 (2003).
Kolmogorov, M., Yuan, J., Lin, Y. & Pevzner, P. A. Assembly of long, error-prone reads using repeat graphs. Nat. Biotechnol. 37, 540–546 (2019).
Miller, J. R., Koren, S. & Sutton, G. Assembly algorithms for next-generation sequencing data. Genomics 95, 315–327 (2010).
Walker, B. J. et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9, e112963 (2014).
Langmead, B. & Salzberg, S. L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Gordon, D. & Green, P. Consed: A graphical editor for next-generation sequencing. Bioinformatics 29, 2936–2937 (2013).
Li, W. et al. RefSeq: expanding the prokaryotic genome annotation pipeline reach with protein family model curation. Nucleic Acids Res. 49, D1020–D1028 (2021).
Tatusova, T. et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 44, 6614–6624 (2016).
Tamura, K., Stecher, G. & Kumar, S. MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027 (2021).
Kearse, M. et al. Geneious basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649 (2012).
Acknowledgements
This work was supported by Humboldt Research Fellowship for Postdocs from Alexander von Humboldt Foundation and Ministry of Education and Science of Ukraine BG-14E grant (to O.Y.); by Fondo per il Programma Nazionale di Ricerca e Progetti di Rilevante Interesse Nazionale (PRIN) for the project DiGlycAn—project number 2022J7W7LW (to F.B. and F.M.); by the University of Insubria grant Fondo di Ateneo per la Ricerca 2022 and 2023 (to F.M. and F.B.); by the University of Insubria Starting grant for young researchers 2022 and 2023 (to F.B.); and by the Deutsche Forschungsgemeinschaft—RTG 2473 ‘Bioactive Peptides’, project number 392923329 (to L.Z. and R.D.S.). We would like to thank Dr. Elisa Binda from University of Insubria for her help with fermentation at bioreactor scale, and Dr. Enrico Selva for the enthusiastic support and the invaluable suggestions for kineomicins purification. O.Y. would like to thank all defenders of Ukraine, who made working on this manuscript possible. L.Z. gratefully acknowledges the China Scholarship Council (CSC) for a granted Ph.D. scholarship. L.B. is a Ph.D. student of the “Life Sciences and Biotechnology” course at University of Insubria.
Author information
Authors and Affiliations
Contributions
O.Y., F.M., and R.D.S.—conceptualization; O.Y., F.B., L.Z., E.B., and L.B.—investigation; C.R.-R., T.B., and J.K.—software; O.Y., L.Z., C.R.-R.—formal analysis; F.B., L.Z., and R.D.S.—methodology; O.Y., F.B., L.Z., F.M., and R.D.S.—writing, original draft; F.M. and R.D.S.—supervision and funding acquisition.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Chemistry thanks Max Cryle and the other, anonymous, reviewers for their contribution to the peer review of this work. Peer review reports are available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yushchuk, O., Berini, F., Zhong, L. et al. A rare peptide scaffold in kineomicins, the glycopeptide antibiotics produced by Actinokineospora auranticolor DSM 44650. Commun Chem 8, 134 (2025). https://doi.org/10.1038/s42004-025-01534-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s42004-025-01534-x