Abstract
Nucleotide availability is crucial for DNA replication and repair; however, the coordinating mechanisms in vivo remain unclear. Here, we show that the circadian clock in the liver controls the activity of the pentose phosphate pathway (PPP) to support de novo nucleotide biosynthesis for DNA synthesis demands. We demonstrate that disrupting the hepatic clock by genetic manipulation or mistimed feeding impairs PPP activity in male mice, leading to nucleotide imbalance. Such defects not only elicit DNA replication stress to limit liver regeneration after resection but also allow genotoxin-induced hepatocyte senescence and STING signalling-dependent inflammation. Mechanistically, the molecular clock activator BMAL1 synergizes with hypoxia-inducible factor-1α (HIF-1α) to regulate the transcription of the PPP rate-limiting enzyme glucose-6-phosphate dehydrogenase (G6PD), which is enhanced during liver regeneration. Overexpressing G6PD restores the compromised regenerative capacity of the BMAL1- or HIF-1α-deficient liver. Moreover, boosting G6PD expression genetically or through preoperative intermittent fasting potently facilitates liver repair in normal mice. Hence, our findings highlight the physiological importance of the hepatic clock and suggest a promising pro-regenerative strategy.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout







Similar content being viewed by others
Data availability
The time-series microarray data for the postoperative mouse livers25 have been deposited in the Gene Expression Omnibus (GEO) under accession number GSE215216. All other supporting data are included in this paper. Source data are provided with this paper.
Code availability
No custom code was used in this study.
References
Welsh, D. K., Takahashi, J. S. & Kay, S. A. Suprachiasmatic nucleus: cell autonomy and network properties. Annu. Rev. Physiol. 72, 551–577 (2010).
Izumo, M. et al. Differential effects of light and feeding on circadian organization of peripheral clocks in a forebrain Bmal1 mutant. eLife 3, e04617 (2014).
Damiola, F. et al. Restricted feeding uncouples circadian oscillators in peripheral tissues from the central pacemaker in the suprachiasmatic nucleus. Genes Dev. 14, 2950–2961 (2000).
Reinke, H. & Asher, G. Crosstalk between metabolism and circadian clocks. Nat. Rev. Mol. Cell Biol. 20, 227–241 (2019).
Stokkan, K. A., Yamazaki, S., Tei, H., Sakaki, Y. & Menaker, M. Entrainment of the circadian clock in the liver by feeding. Science 291, 490–493 (2001).
Asher, G. et al. Poly(ADP-ribose) polymerase 1 participates in the phase entrainment of circadian clocks to feeding. Cell 142, 943–953 (2010).
Vollmers, C. et al. Time of feeding and the intrinsic circadian clock drive rhythms in hepatic gene expression. Proc. Natl Acad. Sci. USA 106, 21453–21458 (2009).
Mukherji, A., Kobiita, A. & Chambon, P. Shifting the feeding of mice to the rest phase creates metabolic alterations, which, on their own, shift the peripheral circadian clocks by 12 hours. Proc. Natl Acad. Sci. USA 112, E6683–E6690 (2015).
Hatori, M. et al. Time-restricted feeding without reducing caloric intake prevents metabolic diseases in mice fed a high-fat diet. Cell Metab. 15, 848–860 (2012).
Koronowski, K. B. et al. Defining the independence of the liver circadian clock. Cell 177, 1448–1462 (2019).
Manella, G. et al. The liver-clock coordinates rhythmicity of peripheral tissues in response to feeding. Nat. Metab. 3, 829–842 (2021).
Matsuo, T. et al. Control mechanism of the circadian clock for timing of cell division in vivo. Science 302, 255–259 (2003).
Gréchez-Cassiau, A., Rayet, B., Guillaumond, F., Teboul, M. & Delaunay, F. The circadian clock component BMAL1 is a critical regulator of p21WAF1/CIP1 expression and hepatocyte proliferation. J. Biol. Chem. 283, 4535–4542 (2008).
Farshadi, E. et al. The positive circadian regulators CLOCK and BMAL1 control G2/M cell cycle transition through cyclin B1. Cell Cycle 18, 16–33 (2019).
Wang, H. et al. Time-restricted feeding shifts the skin circadian clock and alters UVB-induced DNA damage. Cell Rep. 20, 1061–1072 (2017).
Zada, D. et al. Parp1 promotes sleep, which enhances DNA repair in neurons. Mol. Cell 81, 4979–4993 (2021).
Zada, D., Bronshtein, I., Lerer-Goldshtein, T., Garini, Y. & Appelbaum, L. Sleep increases chromosome dynamics to enable reduction of accumulating DNA damage in single neurons. Nat. Commun. 10, 895 (2019).
Bellesi, M., Bushey, D., Chini, M., Tononi, G. & Cirelli, C. Contribution of sleep to the repair of neuronal DNA double-strand breaks: evidence from flies and mice. Sci. Rep. 6, 36804 (2016).
Forey, R. et al. Mec1 is activated at the onset of normal S phase by low-dNTP pools impeding DNA replication. Mol. Cell 78, 396–410 (2020).
Rudd, S. G., Valerie, N. C. K. & Helleday, T. Pathways controlling dNTP pools to maintain genome stability. DNA Repair (Amst.) 44, 193–204 (2016).
Fustin, J.-M. et al. Rhythmic nucleotide synthesis in the liver: temporal segregation of metabolites. Cell Rep. 1, 341–349 (2012).
Caldez, M. J. et al. Metabolic remodeling during liver regeneration. Dev. Cell 47, 425–438 (2018).
Su, A. I., Guidotti, L. G., Pezacki, J. P., Chisari, F. V. & Schultz, P. G. Gene expression during the priming phase of liver regeneration after partial hepatectomy in mice. Proc. Natl Acad. Sci. USA 99, 11181–11186 (2002).
Böhm, F., Köhler, U. A., Speicher, T. & Werner, S. Regulation of liver regeneration by growth factors and cytokines. EMBO Mol. Med. 2, 294–305 (2010).
Keys, H. R. & Knouse, K. A. Genome-scale CRISPR screening in a single mouse liver. Cell Genom. 2, 100217 (2022).
Eguchi, A., Wree, A. & Feldstein, A. E. Biomarkers of liver cell death. J. Hepatol. 60, 1063–1074 (2014).
Bravo, R., Frank, R., Blundell, P. A. & Macdonald-Bravo, H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-δ. Nature 326, 515–517 (1987).
Takhaveev, V. et al. Temporal segregation of biosynthetic processes is responsible for metabolic oscillations during the budding yeast cell cycle. Nat. Metab. 5, 294–313 (2023).
Angus, S. P. et al. Retinoblastoma tumor suppressor targets dNTP metabolism to regulate DNA replication. J. Biol. Chem. 277, 44376–44384 (2002).
Gaucher, J., Montellier, E. & Sassone-Corsi, P. Molecular cogs: interplay between circadian clock and cell cycle. Trends Cell Biol. 28, 368–379 (2018).
Liu, Z. et al. SIRT7 couples light-driven body temperature cues to hepatic circadian phase coherence and gluconeogenesis. Nat. Metab. 1, 1141–1156 (2019).
Newberry, E. P. et al. Altered hepatic triglyceride content after partial hepatectomy without impaired liver regeneration in multiple murine genetic models. Hepatology 48, 1097–1105 (2008).
Lamia, K. A., Storch, K.-F. & Weitz, C. J. Physiological significance of a peripheral tissue circadian clock. Proc. Natl Acad. Sci. USA 105, 15172–15177 (2008).
Walsby-Tickle, J. et al. Anion-exchange chromatography mass spectrometry provides extensive coverage of primary metabolic pathways revealing altered metabolism in IDH1 mutant cells. Commun. Biol. 3, 247 (2020).
Thaben, P. F. & Westermark, P. O. Detecting rhythms in time series with RAIN. J. Biol. Rhythms 29, 391–400 (2014).
Peek, C. B. et al. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science 342, 1243417 (2013).
Michalopoulos, G. K. & Bhushan, B. Liver regeneration: biological and pathological mechanisms and implications. Nat. Rev. Gastroenterol. Hepatol. 18, 40–55 (2021).
Zhang, Z. et al. Serine catabolism generates liver NADPH and supports hepatic lipogenesis. Nat. Metab. 3, 1608–1620 (2021).
Lewis, C. A. et al. Tracing compartmentalized NADPH metabolism in the cytosol and mitochondria of mammalian cells. Mol. Cell 55, 253–263 (2014).
Fan, J. et al. Quantitative flux analysis reveals folate-dependent NADPH production. Nature 510, 298–302 (2014); erratum 513, 574 (2014).
Dröge, W. Free radicals in the physiological control of cell function. Physiol. Rev. 82, 47–95 (2002).
Beyer, T. A. et al. Impaired liver regeneration in Nrf2 knockout mice: role of ROS-mediated insulin/IGF-1 resistance. EMBO J. 27, 212–223 (2008).
Humpton, T. J. et al. p53-mediated redox control promotes liver regeneration and maintains liver function in response to CCl4. Cell Death Differ. 29, 514–526 (2022).
Diehl, F. F. et al. Nucleotide imbalance decouples cell growth from cell proliferation. Nat. Cell Biol. 24, 1252–1264 (2022).
Donne, R. et al. Replication stress triggered by nucleotide pool imbalance drives DNA damage and cGAS–STING pathway activation in NAFLD. Dev. Cell 57, 1728–1741 (2022).
Köhler, E., Barrach, H.-J. & Neubert, D. Inhibition of NADP dependent oxidoreductases by the 6-aminonicotinamide analogue of NADP. FEBS Lett. 6, 225–228 (1970).
Clark, P. M. et al. Positron emission tomography probe demonstrates a striking concentration of ribose salvage in the liver. Proc. Natl Acad. Sci. USA 111, E2866–E2874 (2014).
Saldivar, J. C., Cortez, D. & Cimprich, K. A. The essential kinase ATR: ensuring faithful duplication of a challenging genome. Nat. Rev. Mol. Cell Biol. 18, 622–636 (2017).
Hu, H. et al. Long-term expansion of functional mouse and human hepatocytes as 3D organoids. Cell 175, 1591–1606 (2018).
Babalola, A. O., Beetlestone, J. G. & Luzzatto, L. Genetic variants of human erythrocyte glucose-6-phosphate dehydrogenase. Kinetic and thermodynamic parameters of variants A, B, and A—in relation to quaternary structure. J. Biol. Chem. 251, 2993–3002 (1976).
Stokes, K. et al. The circadian clock gene BMAL1 coordinates intestinal regeneration. Cell. Mol. Gastroenterol. Hepatol. 4, 95–114 (2017).
King, D. P. et al. Positional cloning of the mouse circadian clock gene. Cell 89, 641–653 (1997).
Wu, Y. et al. Reciprocal regulation between the circadian clock and hypoxia signaling at the genome level in mammals. Cell Metab. 25, 73–85 (2017).
Peek, C. B. et al. Circadian clock interaction with HIF1α mediates oxygenic metabolism and anaerobic glycolysis in skeletal muscle. Cell Metab. 25, 86–92 (2017).
Tajima, T. et al. HIF-1α is necessary to support gluconeogenesis during liver regeneration. Biochem. Biophys. Res. Commun. 387, 789–794 (2009).
Kinouchi, K. et al. Fasting imparts a switch to alternative daily pathways in liver and muscle. Cell Rep. 25, 3299–3314 (2018).
Sarac, T. P. et al. Preoperative fasting improves survival after 90% hepatectomy. Arch. Surg. 129, 729–733 (1994).
Serger, E. et al. The gut metabolite indole-3 propionate promotes nerve regeneration and repair. Nature 607, 585–592 (2022).
Patterson, R. E. & Sears, D. D. Metabolic effects of intermittent fasting. Annu. Rev. Nutr. 37, 371–393 (2017).
Longo, V. D. & Panda, S. Fasting, circadian rhythms, and time-restricted feeding in healthy lifespan. Cell Metab. 23, 1048–1059 (2016).
Li, X. & Heyer, W.-D. Homologous recombination in DNA repair and DNA damage tolerance. Cell Res. 18, 99–113 (2008).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999).
Li, T. & Chen, Z. J. The cGAS–cGAMP–STING pathway connects DNA damage to inflammation, senescence, and cancer. J. Exp. Med. 215, 1287–1299 (2018).
Hong, Z. et al. STING inhibitors target the cyclic dinucleotide binding pocket. Proc. Natl Acad. Sci. USA 118, e2105465118 (2021).
Eckel-Mahan, K. L. et al. Coordination of the transcriptome and metabolome by the circadian clock. Proc. Natl Acad. Sci. USA 109, 5541–5546 (2012).
Weglarz, T. C. & Sandgren, E. P. Timing of hepatocyte entry into DNA synthesis after partial hepatectomy is cell autonomous. Proc. Natl Acad. Sci. USA 97, 12595–12600 (2000).
Adamovich, Y., Ladeuix, B., Golik, M., Koeners, M. P. & Asher, G. Rhythmic oxygen levels reset circadian clocks through HIF1α. Cell Metab. 25, 93–101 (2017).
Rey, G. et al. The pentose phosphate pathway regulates the circadian clock. Cell Metab. 24, 462–473 (2016).
Patel, J. H., Ong, D. J., Williams, C. R., Callies, L. K. & Wills, A. E. Elevated pentose phosphate pathway flux supports appendage regeneration. Cell Rep. 41, 111552 (2022).
Cheng, J. et al. TRIM21 and PHLDA3 negatively regulate the crosstalk between the PI3K/AKT pathway and PPP metabolism. Nat. Commun. 11, 1880 (2020).
Li, Q. et al. Rac1 activates non-oxidative pentose phosphate pathway to induce chemoresistance of breast cancer. Nat. Commun. 11, 1456 (2020).
Sarkar, A. et al. Intermittent fasting induces rapid hepatocyte proliferation to restore the hepatostat in the mouse liver. eLife 12, e82311 (2023).
Delahunty, T. J. & Rubinstein, D. Accumulation and release of triglycerides by rat liver following partial hepatectomy. J. Lipid Res. 11, 536–543 (1970).
Zhan, C. et al. Preoperative short-term fasting protects liver injury in patients undergoing hepatectomy. Ann. Transl. Med. 6, 449 (2018).
Mitchell, C. & Willenbring, H. A reproducible and well-tolerated method for 2/3 partial hepatectomy in mice. Nat. Protoc. 3, 1167–1170 (2008).
Gil, J., Bernard, D., Martínez, D. & Beach, D. Polycomb CBX7 has a unifying role in cellular lifespan. Nat. Cell Biol. 6, 67–72 (2004); erratum 24, 1445 (2022).
Meng, F. et al. Synergy between SIRT1 and SIRT6 helps recognize DNA breaks and potentiates the DNA damage response and repair in humans and mice. eLife 9, e55828 (2020).
Peng, L. et al. Hippo-signaling-controlled MHC class I antigen processing and presentation pathway potentiates antitumor immunity. Cell Rep. 43, 114003 (2024).
Acknowledgements
We thank all members of the laboratory of the late P. Sassone-Corsi (University of California, Irvine) for insightful discussions and advice, as well as J. Tamanini (Shenzhen University and ET Editing) for editing the paper before submission. This work was supported by grants from the National Key Research and Development Program of China (2022YFC2303200, 2023YFA1801900 to M.Q.); the National Natural Science Foundation of China (82022025, 91949124 and 82271602 to M.Q.; 32071259 to Z.W.; 81972894 to C.S.); the Innovation and Entrepreneurship (Shuangchuang) Program of Jiangsu Province (2023); the Natural Science Foundation of Chongqing (CSTB2024NSCQ-MSX1152); the Project Program of State Key Laboratory of Natural Medicines, China Pharmaceutical University (SKLNMZZ2024JS39); and the ‘Double First-Class’ University Project of China Pharmaceutical University (CPUQNJC22). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the paper.
Author information
Authors and Affiliations
Contributions
M.Q., Z.W. and B.L. conceived of and designed the research and wrote the paper. L.P. and S.X. performed most experiments, with help from T.W., M.Y., Y.D., X.M., S.L., C.Y. and X.Z., and analysed the data. H.H., Z.L. and J.S. provided technical assistance and/or suggestions for experiments. C.S. and C.W. reviewed and edited the paper.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Jean-Michel Fustin, Meritxell Huch and Tara Teslaa for their contribution to the peer review of this work. Primary Handling Editor: Yanina-Yasmin Pesch, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
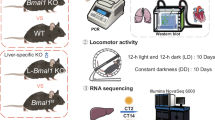
Extended Data Fig. 1 Expression profiles of the central clock genes under different experimental conditions.
a, qRT-PCR showing expression of Cry1, Per2, and Dbp in C57BL/6 J mouse livers at the indicated time points after PH. b. Heatmap showing differential expression of genes related to the circadian clock and cell cycle during liver regeneration. Original data were derived from GSE215216 dataset. c-e. The expression patterns of the core clock genes in the livers of WT and LBKO mice (c), or WT mice fed AL or DRF (d,e), determined by qRT-PCR. For a,c-e, n = 3 mice per group. Data are mean ± s.e.m. P-values were calculated using two-way ANOVA with Sidak’s test (a). Rhythmicity analysis was performed using JTK_CYCLE as shown in Supplementary Table 1.
Extended Data Fig. 2 Circadian clock disruption limits hepatocyte proliferation after resection.
a,b, Representative IHC images (a) and quantification (b) of Ki67-positive staining cells in AL, DRF and LBKO mice with sham- or PH- operation. More than three fields for each of three independent samples were counted. Scale bar, 100 μm. c-g, Graphs showing the expression of Bmal1 (c), the liver-body weight ratio (d), the contents of ALT (e) and AST (f), and representative IHC images and quantification of Ki-67 staining (g) in the livers on day 4 post-PH. Male adult Bmal1f/f mice were infected with control AAV8 (AAV-NC) or AAV8-TbgCre (AAV-Cre) and then subjected to PH. Five fields for each of three independent samples were counted. Scale bar, 100 μm. h-j, Graphs showing the liver-body weight ratio (h) and the contents of ALT (i) and AST (j) in mice which fed with DRF for 10 days were then subjected to PH. The samples were collected on day 4 post-PH. The red line indicates the mean value of DRF-D4 group. k, qPCR analysis showing the pattern of Pcna mRNA expression in the livers of WT, LBKO and DRF mice at various ZTs. For c,k, n = 3 mice per group. For d-f,h-j, n = 5 mice per group. Data are mean ± s.e.m. P-values were calculated using one-way ANOVA with Dunnett’s test (b) and unpaired two-tailed Student’s t-test (c-j). For k, JTK_CYCLE was applied to determine rhythmicity. P < 0.05 was considered to be rhythmic.
Extended Data Fig. 3 Bmal1 depletion alters glucose metabolic flux, while G6pdx deficiency attenuates hepatocyte proliferation.
a-c, Examples of rhythmic metabolites generated from the PPP. 6-PG, 6-phosphogluconate (a); R5P, ribose-5-phosphate (b); S7P, sedoheptulose 7-phosphate (c). * P < 0.05, ** P < 0.01, *** P < 0.001. d,e, Mass isotopomer distribution of nucleotides (d) and lactate (e) derived from 13C-labeled glucose in primary hepatocytes from WT and LBKO mice. n = 4 independent biological samples. ND, not detected. f-h, Flow cytometric analysis (f) and quantification (g) of 2-NBDG intake, and measurement of lactate contents (h) in primary hepatocytes from WT and LBKO mice. n = 3 independent biological replicates. i,j, The mRNA expression of glycolytic genes Ldha and Pdk1 in hepatocytes from WT and LBKO mice with or without overexpressing G6pdx (i) and those from WT mice infected with AAV-shNC or AAV-shG6pdx (j). k,l, Representative IF images (k) and quantification (l) of staining with anti-Ki-67 and anti-G6pdx antibodies for the livers of AAV8-shNC or AAV8-shG6pdx infected mice on day 4 post-PH. Seven fluorescent fields for each of three samples were counted. Scale bar, 20 μm. m, qRT-PCR showing the mRNA levels of G6pdx in the livers of mice infected with AAV-shNC or AAV-shG6pdx for four weeks. For a-c,i,j,m, n = 3 mice per group. Data are mean ± s.e.m. P-values were calculated using two-way ANOVA with Sidak’s test (a-c,e,i), unpaired two-tailed Student’s t-test (g,h,l,m) and multiple unpaired two-tailed Student’s t-test (j).
Extended Data Fig. 4 Redox imbalance in hepatic clock-disturbed mice after PH.
a, The rhythmicity of NADPH content in the livers from WT and LBKO mice at various ZTs. n = 3 mice per group. b,c, Representative IF images (b) and quantification (c) of DHE staining of liver tissues of AL, DRF, and LBKO mice after sham or PH. Scale bar, 20 μm. d,e, Representative IHC images (d) and quantification (e) of 8-OhdG staining for AL, DRF and LBKO mice with sham- or PH- operation. Scale bar, 100 μm. More than three fields for each of three samples were counted. Data are mean ± s.e.m. P-values were calculated using one-way ANOVA with Dunnett’s test (c,e). For a, JTK_CYCLE was applied to determine rhythmicity. P < 0.05 was considered to be rhythmic.
Extended Data Fig. 5 Bmal1-regulating nucleotide pool is required for cell proliferation and survival.
a, Measurement of the nucleoside/nucleotide contents by LC-MS/MS in WT mice with or without shG6pdx via the AAV8 system. b, Examples of oscillating nucleosides/nucleotides from the metabolome of WT and Bmal1-null mouse livers. The processing data were included in Supplementary Table 3. Box plots extend from the 25th and 75th percentile, with a horizontal line representing the median value, and the whiskers indicate the min and max values. c, Measurement of the contents of nucleotides in the livers from WT and LBKO mice before and after PH by LC-MS/MS. d, Representative images of hepatic organoids cultured in Matrigel assessed by bright field microscopy, with quantification of the numbers of hep-organoids formed from 104 cells. Primary hepatocytes from LBKO mice were treated with or without a combination of NAC and NTPs during the 14-day organoid culture. n = 3 independent experiments. e, Immunoblotting showing the protein levels of Bmal1 and G6pdx in primary hepatocytes, which were isolated from Bmal1f/f mouse livers and then treated with AAV-Cre. f-h, Graphs showing representative images (f) of hepatic organoids cultured in Matrigel and quantification of the numbers of hep-organoids formed from 104 cells (g) and the mean diameter (h). Primary hepatocytes, as pretreated in e, were subjected to 3D culture with or without a combination of NAC and dNTPs (N + N) for 14 days. n = 3 independent experiments. Scale bar, 50 μm. For a,c, n = 3 or 4 mice per group. Data are mean ± s.e.m. P-values were calculated using multiple unpaired two-tailed Student’s t-test (a), one-way ANOVA with Dunnett’s test (c,g,h) and two-tailed unpaired t-test (d).
Extended Data Fig. 6 Bmal1 depletion has a less pronounced effect on the expression of other PPP-related genes.
a, Diagram outlining the oxidative and non-oxidative branches of the PPP in glucose metabolism. b-f, qRT-PCR showing oscillations of the PPP-related genes in AL, DRF, and LBKO mouse livers at various ZTs, including Pgd (b), Tkt (c), Rpe (d), Taldo1 (e), and Rpia (f). The arrows indicate the ZT point at which the expression of the indicated gene peaked in the livers of AL and DRF groups (b,c). g, qRT-PCR showing the mRNA levels of the PPP-related genes in Hepa1-6 cells expressing shNC or shBmal1. h, Immunoblotting showing the protein levels of Bmal1 and G6pdx in the livers from Bmal1f/f mice with AAV-NC or AAV-TbgCre injection. i, qRT-PCR showing the mRNA levels of G6pdx in the indicated tissues from WT and Bmal1-null mice. j, Representative immunofluorescence images of WT and Bmal1-null mouse intestines stained with the indicated antibodies. Nuclei were stained with DAPI. Scale bar, 15 μm. k, Quantification of the intensity of Ki67-positive staining shown in j. Four fluorescent fields for each of three independent samples were counted. For b-f,g-i, n = 3 mice per group. Data are mean ± s.e.m. P-values were calculated using multiple unpaired two-tailed Student’s t-test (g,i) and unpaired two-tailed Student’s t-test (k). For b-f, JTK_CYCLE was applied to determine rhythmicity as shown in Supplementary Table 4. P < 0.05 was considered to be rhythmic.
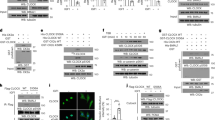
Extended Data Fig. 7 G6pdx expression is regulated by Bmal1 and Hif-1α, which is required for facilitating liver regeneration.
a, Schematic diagram depicting the promoter region of murine G6pdx gene. Expanded fragments show the DNA sequence of the E-box in the P1 region and the E’-box in the P2 region. b, Measurement of the relative luciferase activity in Hepa1-6 cells transfected with the indicated expression plasmids. c,d, qRT-PCR showing G6pdx expression in Hepa1-6 cells expressing shNC, shBmal1, or shClock (c) and immortalized WT and Clock mutant MEFs (d). Delta 19 (Δ19), deletion of exon 19 in the Clock gene. e, Measurement of the activity of P2-driven luciferase reporter in Hepa1-6 cells expressing shNC, shBmal1, or shHif-1α. f, Representative images of IF staining showing the G6pdx expression in the livers. WT and LBKO mice were infected with AAV8-GFP or AAV8-G6pdx virus for three weeks. Scale bar, 15 μm. g, Representative IHC images and quantification of Ki67-positive cells in the livers of postoperative mice treated as in f. The red line indicates the mean value of the AL group. Scale bar, 100 μm. h, Graphs showing the liver-body weight ratio and the contents of ALT and AST in DRF mice that were injected with AAV8-GFP or AAV8-Flag-G6pdx. The samples were collected on day 4 post-PH. i,j, Graphs showing Ki-67 staining (i) and the indicated protein levels (j) in mice treated as in Fig. 6b. Scale bar, 100 μm. k-m, Identification of G6pdx overexpression in WT mice after AAV8 injection, as measured by immunoblotting (k), qRT-PCR (l) and enzymatic activity assay (m). n, Measurement of the nucleoside/nucleotide contents in WT mice with or without G6pdx overexpression by LC-MS/MS. For g,i, Five fields for each of three samples were counted. For b-e, n = 3 independent biological samples. For h, n = 6 mice per group. For l-n, n = 3 mice per group. Data are mean ± s.e.m. P-values were calculated using two-way ANOVA with Tukey’s test (b) or Sidak’s test (c), unpaired two-tailed Student’s t-test (d,g,h,l,m), one-way ANOVA with Dunnett’s test (e,i) and multiple unpaired two-tailed Student’s t-test (n).
Extended Data Fig. 8 Changes to the feeding schedule alter G6pdx expression and affect hepatocyte proliferation.
a-c, Immunoblotting showing the indicated protein levels in Hepa1-6 cells cultured in glucose-free medium (a), and in the livers from mice fasted for 2 days (b) or AAV-infected (c). d, Representative IHC images and quantification of Ki67-positive cells from mice with the indicated treatment. More than three fields for each of three independent samples were counted. Scale bar, 100 μm. e,f, The mRNA levels (e) and enzymatic activity (f) of G6pdx in the livers of AL- or 10-day IF- mice. g, qRT-PCR showing the expression of other PPP related genes in the livers of mice fed AL or IF. h, Quantification of the ratio of phosphorylated (p)-Bmal1 to total Bmal1 from the blots as shown in Fig. 6h. i, Immunoblotting showing the levels of the indicated proteins in chromatin fraction. j,k, Representative IHC images (j) and quantification (k) of Ki67-positive cells from mice with the indicated treatment. Five fields for each of three independent samples were counted. For e-h, n = 3 or 4 mice per group. Data are mean ± s.e.m. P-values were calculated using two-way ANOVA with Tukey’s test (d), unpaired two-tailed Student’s t-test (e,f,h) and multiple unpaired two-tailed Student’s t-test (g,k).
Extended Data Fig. 9 Accumulated DNA damage by Bmal1 depletion triggers senescence and inflammation.
a, Clonogenic survival experiments showing the colony formation capacity of NIH3T3 cells expressing shNC or shBmal1 after treatment with the indicated doses of CPT with or without dNTPs. b, Representative images and quantification of IHC staining for the indicated markers of livers from 6-month-old WT and whole-body Bmal1 knockout (BKO) mice. More than three fields for each of three samples were counted. c, Representative IF images of primary MEFs at passage 5 stained with the anti-γH2ax antibody and quantification of the number of γH2ax foci in individual cells. n = 53 cells counted for each condition taken from three independent experiments. Scale bar, 15 μm. d, qRT-PCR showing the expression of Bmal1, G6pdx, and the senescent markers P21 and P16, in shNC or shBmal1 MEFs at passage 5. e, Representative IHC images in mouse liver sections stained with anti-F4/80 antibody and optical density-based quantification. WT and LBKO mice with AAV8-GFP or AAV8-G6pdx were subjected to paraquat (20 mg kg−1) treatment and euthanized after 5 days. More than three fields for each of three independent samples were counted. Scale bar, 100 μm. f, qRT-PCR showing the expression of the indicated SASP genes in shNC- or shBmal1-expressing primary MEFs at passage 5. g, Immunoblotting showing the indicated protein levels in the liver lysates of WT or LBKO mice treated with 20 mg kg−1 paraquat and then with 0 or 5 mg kg−1 SN-011 treatment for 2 days before sacrifice. h-j, qRT-PCR showing the expression of the indicated genes (h,j) and representative images of IHC staining with the anti-F4/80 antibody (i) of livers from mice as treated in g. Scale bar, 100 μm. For a,d,f, n = 3 independent biological samples. For h,j, n = 3 mice per group. Data are mean ± s.e.m. P-values were calculated using RM two-way ANOVA (a), unpaired two-tailed Student’s t-test (b), two-way ANOVA with Sidak’s test (d,f,h,j) and one-way ANOVA with Dunnett’s test (e).
Extended Data Fig. 10 A working model.
The proposal model depicting the hepatic clock controls the oscillation of pentose phosphate pathway through cooperating with HIF-1α, that is crucial for liver recovery from damage. Glu, Glucose; senescence-associated secretory phenotype, SASP. Created by figdraw.com.
Supplementary information
Supplementary Information
Supplementary Figs. 1–6 and unprocessed blots for Supplementary Fig. 6.
Supplementary Tables
Supplementary Tables 1–5. Rhythmicity analysis, primers and metabolome dataset.
Supplementary Data
Statistical source data for supplementary figures.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 5
Statistical source data.
Source Data Fig. 6
Statistical source data.
Source Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 1
Statistical source data.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 4
Statistical source data.
Source Data Extended Data Fig. 5
Statistical source data.
Source Data Extended Data Fig. 6
Statistical source data.
Source Data Extended Data Fig. 7
Statistical source data.
Source Data Extended Data Fig. 8
Statistical source data.
Source Data Extended Data Fig. 9
Statistical source data.
Source Data Figs. 1 and 4–7 and Extended Data Figs. 5–9
Unprocessed blots.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Peng, L., Xiang, S., Wang, T. et al. The hepatic clock synergizes with HIF-1α to regulate nucleotide availability during liver damage repair. Nat Metab 7, 148–165 (2025). https://doi.org/10.1038/s42255-024-01184-8
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42255-024-01184-8
This article is cited by
-
M6A methyltransferase METTL3 promotes glucose metabolism hub gene expression and induces metabolic dysfunction-associated steatotic liver disease (MASLD)
BMC Genomics (2025)
-
Unlocking the therapeutic potential of the STING signaling pathway in anti-tumor treatment
Clinical and Experimental Medicine (2025)