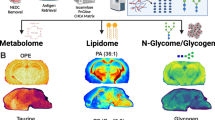
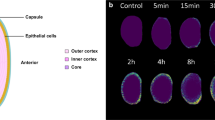
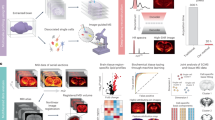
Abstract
High-resolution spatial imaging is transforming our understanding of foundational biology. Spatial metabolomics is an emerging field that enables the dissection of the complex metabolic landscape and heterogeneity from a thin tissue section. Currently, spatial metabolism highlights the remarkable complexity in two-dimensional (2D) space and is poised to be extended into the three-dimensional (3D) world of biology. Here we introduce MetaVision3D, a pipeline driven by computer vision, a branch of artificial intelligence focusing on image workflow, for the transformation of serial 2D MALDI mass spectrometry imaging sections into a high-resolution 3D spatial metabolome. Our framework uses advanced algorithms for image registration, normalization and interpolation to enable the integration of serial 2D tissue sections, thereby generating a comprehensive 3D model of unique diverse metabolites across host tissues at submesoscale. As a proof of principle, MetaVision3D was utilized to generate the mouse brain 3D metabolome atlas of normal and diseased animals (available at https://metavision3d.rc.ufl.edu) as an interactive online database and web server to further advance brain metabolism and related research.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to the full article PDF.
USD 39.95
Prices may be subject to local taxes which are calculated during checkout






Similar content being viewed by others
Data availability
The 3D atlas of the brain metabolome and downloadable files are available at https://metavision3d.rc.ufl.edu. Raw data in .csv format are available via Zenodo at https://doi.org/10.5281/zenodo.14212426 (ref. 47) and in imzML format are available at https://sunlabresources.rc.ufl.edu. Tutorials on how to use the online atlas and ImageJ are available at https://www.youtube.com/channel/UC5tUZiZHCjS33Un4yxfIWdw. Source data are provided with this paper.
Code availability
Python code for the entire MetaVision3D framework and test dataset is available via GitHub at https://github.com/XinBiostats/MetaVision3D and via Zenodo at https://doi.org/10.5281/zenodo.14213133 (ref. 48).
References
Moses, L. & Pachter, L. Museum of spatial transcriptomics. Nat. Methods 19, 534–546 (2022).
Rao, A., Barkley, D., França, G. S. & Yanai, I. Exploring tissue architecture using spatial transcriptomics. Nature 596, 211–220 (2021).
Bendall, S. C. et al. Single-cell mass cytometry of differential immune and drug responses across a human hematopoietic continuum. Science 332, 687–696 (2011).
Angelo, M. et al. Multiplexed ion beam imaging of human breast tumors. Nat. Med. 20, 436–442 (2014).
Rappez, L. et al. SpaceM reveals metabolic states of single cells. Nat. Methods 18, 799–805 (2021).
Alexandrov, T. Spatial metabolomics: from a niche field towards a driver of innovation. Nat. Metab. 5, 1443–1445 (2023).
Wang, L. et al. Spatially resolved isotope tracing reveals tissue metabolic activity. Nat. Methods 19, 223–230 (2022).
Schwaiger-Haber, M. et al. Using mass spectrometry imaging to map fluxes quantitatively in the tumor ecosystem. Nat. Commun. 14, 2876 (2023).
Zhang, H. et al. Single-cell lipidomics enabled by dual-polarity ionization and ion mobility-mass spectrometry imaging. Nat. Commun. 14, 5185 (2023).
Li, Z. et al. Single-cell lipidomics with high structural specificity by mass spectrometry. Nat. Commun. 12, 2869 (2021).
Young, L. E. et al. In situ mass spectrometry imaging reveals heterogeneous glycogen stores in human normal and cancerous tissues. EMBO Mol. Med. 14, e16029 (2022).
Sun, R. C. et al. Brain glycogen serves as a critical glucosamine cache required for protein glycosylation. Cell Metab. 33, 1404–1417 (2021).
Harrison, A. C. et al. Spatial metabolome lipidome and glycome from a single brain section. Preprint at bioRxiv https://doi.org/10.1101/2023.07.22.550155 (2023).
Vicari, M. et al. Spatial multimodal analysis of transcriptomes and metabolomes in tissues. Nat. Biotechnol. 42, 1046–1050 (2024).
Luo, L. et al. Spatial metabolomics reveals skeletal myofiber subtypes. Sci. Adv. 9, eadd0455 (2023).
He, Y. et al. Prosaposin maintains lipid homeostasis in dopamine neurons and counteracts experimental parkinsonism in rodents. Nat. Commun. 14, 5804 (2023).
Conroy, L. R. et al. Spatial metabolomics reveals glycogen as an actionable target for pulmonary fibrosis. Nat. Commun. 14, 2759 (2023).
Xia, C.-R., Cao, Z.-J., Tu, X.-M. & Gao, G. Spatial-linked alignment tool (SLAT) for aligning heterogenous slices. Nat. Commun. 14, 7236 (2023).
Zeira, R., Land, M., Strzalkowski, A. & Raphael, B. J. Alignment and integration of spatial transcriptomics data. Nat. Methods 19, 567–575 (2022).
Zhou, X., Dong, K. & Zhang, S. Integrating spatial transcriptomics data across different conditions, technologies and developmental stages. Nat. Comput. Sci. 3, 894–906 (2023).
Andersson, M., Groseclose, M. R., Deutch, A. Y. & Caprioli, R. M. Imaging mass spectrometry of proteins and peptides: 3D volume reconstruction. Nat. Methods 5, 101–108 (2008).
Trede, D. et al. Exploring three-dimensional matrix-assisted laser desorption/ionization imaging mass spectrometry data: three-dimensional spatial segmentation of mouse kidney. Anal. Chem. 84, 6079–6087 (2012).
Giordano, S. et al. 3D mass spectrometry imaging reveals a very heterogeneous drug distribution in tumors. Sci. Rep. 6, 37027 (2016).
Alexandrov, T. Spatial metabolomics and imaging mass spectrometry in the age of artificial intelligence. Annu. Rev. Biomed. Data Sci. 3, 61–87 (2020).
Cai, K. et al. Magnetic resonance imaging of glutamate. Nat. Med. 18, 302–306 (2012).
Evangelidis, G. D. & Psarakis, E. Z. Parametric image alignment using enhanced correlation coefficient maximization. IEEE Trans. Pattern Anal. Mach. Intell. 30, 1858–1865 (2008).
Brunet, D., Vrscay, E. R. & Wang, Z. On the mathematical properties of the structural similarity index. IEEE Trans. Image Process. 21, 1488–1499 (2011).
Wang, Z. & Bovik, A. C. Mean squared error: love it or leave it? A new look at signal fidelity measures. IEEE Signal Process. Mag. 26, 98–117 (2009).
Schneider, C. A., Rasband, W. S. & Eliceiri, K. W. NIH Image to ImageJ: 25 years of image analysis. Nat. Methods 9, 671–675 (2012).
Eimer, W. A. & Vassar, R. Neuron loss in the 5XFAD mouse model of Alzheimer’s disease correlates with intraneuronal Aβ 42 accumulation and caspase-3 activation. Mol. Neurodegen. 8, 1–12 (2013).
Leon-Astudillo, C. et al. Current avenues of gene therapy in Pompe disease. Curr. Opin. Neurol. 36, 464–473 (2023).
Andersen, J. V. et al. Hippocampal disruptions of synaptic and astrocyte metabolism are primary events of early amyloid pathology in the 5xFAD mouse model of Alzheimer’s disease. Cell Death Dis. 12, 954 (2021).
Ullman, J. C. et al. Small-molecule inhibition of glycogen synthase 1 for the treatment of Pompe disease and other glycogen storage disorders. Sci. Transl. Med. 16, eadf1691 (2024).
Kaya, I. et al. Spatial lipidomics reveals region and long chain base specific accumulations of monosialogangliosides in amyloid plaques in familial Alzheimer’s disease mice (5xFAD) brain. ACS Chem. Neurosci. 11, 14–24 (2020).
Antón-Bolaños, N., Espinosa, A. & López-Bendito, G. Developmental interactions between thalamus and cortex: a true love reciprocal story. Curr. Opin. Neurobiol. 52, 33–41 (2018).
Yao, Z. et al. A taxonomy of transcriptomic cell types across the isocortex and hippocampal formation. Cell 184, 3222–3241.e3226 (2020).
Bohne, P., Schwarz, M. K., Herlitze, S. & Mark, M. D. A new projection from the deep cerebellar nuclei to the hippocampus via the ventrolateral and laterodorsal thalamus in mice. Front. Neural Circuits 13, 51 (2019).
Yu, W. J. & Krook-Magnuson, E. Cognitive collaborations: bidirectional functional connectivity between the cerebellum and the hippocampus. Front. Syst. Neurosci. 9, 177 (2015).
Watson, T. C. et al. Anatomical and physiological foundations of cerebello-hippocampal interaction. eLife 8, e41896 (2018).
Oh, S. W. et al. A mesoscale connectome of the mouse brain. Nature 508, 207–214 (2014).
Nasiriavanaki, M. et al. High-resolution photoacoustic tomography of resting-state functional connectivity in the mouse brain. Proc. Natl Acad. Sci. USA 111, 21–26 (2013).
Shi, H. et al. Spatial atlas of the mouse central nervous system at molecular resolution. Nature 622, 552–561 (2023).
Nunes, J. B. et al. Integration of mass cytometry and mass spectrometry imaging for spatially resolved single-cell metabolic profiling. Nat. Methods 21, 1796–1800 (2024).
Xia, J. & Wishart, D. S. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics 55, 14.10.1–14.10.91 (2016).
Jewison, T. et al. SMPDB 2.0: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 42, D478–D484 (2014).
Molenaar, M. R. et al. LION/web: a web-based ontology enrichment tool for lipidomic data analysis. Gigascience 8, giz061 (2019).
Ma, X. MetaVision3D: automated framework for the generation of spatial metabolome atlas in 3D | MALDI data. Zenodo https://doi.org/10.5281/zenodo.14212426 (2024).
Ma, X. XinBiostats/MetaVision3D: version 1.0.0. Zenodo https://doi.org/10.5281/zenodo.14213133 (2024).
Acknowledgements
This study was supported by National Institutes of Health (NIH) grants R01AG066653, R01CA266004, R01AG078702, 1R01CA288696 and RM1NS133593 to R.C.S., R35NS116824 to M.S.G., R35GM142701 to L.C., T32HL134621 to H.A.C. and 1R01NS127280 to N.N.Y.
Author information
Authors and Affiliations
Contributions
Conceptualization, R.C.S. Methodology, R.C.S.,L.C., N.N.Y., X.M., B.E.C.Z., H.K.R.G., C.J.S., T.M. and R.A.R. Investigation, X.M., C.J.S., T.M., R.A.R., H.A.C., B.E.C.Z., H.K.R.G., T.R.H., P.K.D., L.W., S.N.B., M.E.M., C.W.V.K., L.C., N.N.Y. and M.S.G. Writing—original draft, R.C.S. Writing—review and editing, R.C.S., X.M., C.J.S., T.M., R.A.R., M.E.M., C.W.V.K., M.S.G. and L.C. Funding acquisition, R.C.S. and M.S.G. Resources, R.C.S. Supervision, L.C., M.S.G. and R.C.S.
Corresponding authors
Ethics declarations
Competing interests
R.C.S. has research support and received consultancy fees from Maze Therapeutics. R.C.S. is a member of the Medical Advisory Board for Little Warrior Foundation. M.S.G. received research support and research compounds from Maze Therapeutics, Valerion Therapeutics and Ionis Pharmaceuticals. M.S.G. also received a consultancy fee from Maze Therapeutics, PTC Therapeutics and the Glut1-Deficiency Syndrome Foundation. The other authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Juan Bolaños and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Alfredo Giménez-Cassina, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 2D MALDI heatmap images of serial mouse brain sagittal sections.
a. Schematic of five serial sagittal cut brain sections from the medial plane (left). Heatmap images for the spatial distribution of the free fatty acid (FFA) 20:4 in 2D for all five serial brain sections tested for alignment. b. Schematic of serial sagittal cut brain sections from the medial to posterior brain with different section sizes (left). Heatmap images for the spatial distribution of the free fatty acid (FFA) 20:4 in 2D for all six serial brain sections with different sizes tested for alignment.
Extended Data Fig. 2 Normalization of serial brain sections for 3D construction using MetaNorm3D.
a. Representative intensity distribution of PIP 38:4 among all 79 serial sections of the mouse sagittal hemi-brain. b. Representative intensity distribution of PIP 38:4 after slide normalization (see method) among all 79 serial sections of the mouse sagittal hemi-brain.
Extended Data Fig. 3 2D MALDI heatmap images of serial mouse brain sagittal sections before and after normalization.
a. Heatmap images for the spatial distribution of PIP 38:4 in 2D for all 79 serial brain sections for 3D construction before and after normalization. Insert sections represents tissue sections with disparities in total abundance are zoomed in below. b. Heatmap images for the spatial distribution of PI 36:4 in 2D for all 79 serial brain sections for 3D construction before and after normalization. Insert sections represents tissue sections with disparities in total abundance are zoomed in below.
Extended Data Fig. 4 MetaImpute3D to fill in imperfections during sample handling.
a. Heatmap images for the spatial distribution of PE 44:10 in 2D for all 79 serial brain sections for 3D after normalization, but before and after imputation of gaps in tissue. A representative tissue section after imputation is highlighted in the insert.
Extended Data Fig. 5 MetaInterp3D to fill in imperfections during sample handling.
a. Representatives heatmap images for the spatial distribution of PI 36:2 in 2D for all 79 real and 158 phantom serial brain sections for 3D construction. b. same as a, except phantom sections are highlighted in green. c. Statistical measures of alignment and fit quality measured by enhanced correlation coefficient (ECC), structural similarity index measure (SSIM), and mean squared error (MSE) after interpolation (316 section). The box-and-whisker plot in panel d shows the median (line), interquartile range (box), and variability (whiskers extending to the maximum and minimum data points) n = 857 (11 different lipid class across 79 brain section) before interpolation and n = 2571 (11 different lipid class across 237 brain section).
Extended Data Fig. 6 Additional examples of spatial metabolome atlas of by MetaVision3D.
a. Front cross section and top-down view of fine brain anatomical structures of mouse brain cerebellum and hippocampal region in manual fit and after MetaVision3D framework. b and c. MetaVision3D rendering of PI 38:5 and PS 40:6 in 3D space. Top down (transverse) and front cross section (coronal) views are designated as 1, 2 or 3 shown on the 2D side view image. 3D rendering is visualized by ImageJ using both projection mode and 3D fill of regions with high intensity of both features.
Extended Data Fig. 7 Reproducibility of 3D lipid distribution in independent WT mouse brains.
3D rendered views of two independent WT mouse brains (Brain 1 and Brain 2) showing the spatial distribution of three lipid species: PS 36:2, PI 38:5, and PS 40:6. The consistent patterns across the two brains demonstrate the reproducibility of lipid localization in the hippocampus and surrounding regions. 3D renderings are produced by MetaVision website.
Extended Data Fig. 8 3D distribution of representative lipids highlighting anatomical regions in the mouse brain.
Representative images showing the spatial distribution of specific lipid species in mouse brain regions. (Left) Nissl-stained sagittal brain section of the same plane from the Allen Brain Atlas and Allen Reference Atlas, – Mouse Brain, Allen Mouse Brain Atlas, mouse.brain-map.org and atlas.brain-map.org with regions of interest highlighted in red, including the isocortex, hindbrain, cerebellar molecular layer, hippocampal region, ventricular system, and cerebral nuclei. (Middle) Sagittal plane images obtained using 2D MALDI imaging from our study, illustrating the relative abundance and localization of the indicated lipid species. (Right) 3D visualization of lipid distributions, highlighting the top 1% highest intensity pixels to resolve their spatial organization. Lipid species are annotated above each row: PE 38:6 [M-H]⁻, PE 36:2 [M-H]⁻, PS 44:12 [M-H]⁻, PE 38:4 [M-H]⁻, PI 40:4 [M-H]⁻, and LPE 20:4 [M-H]⁻. Intensity scales range from minimum (purple) to maximum (yellow/red) as depicted in the bottom scale bar.
Extended Data Fig. 9 Comparative analysis of lipid and metabolite distribution in WT, 5xFAD, and Gaa-/- mouse brains.
2D slice and 3D rendered views showing the distribution of specific lipids (LPE 22:4, PE 38:5, LPE 20:0, PE 38:4) and metabolites (ADP) across different mouse models: WT, 5xFAD (Alzheimer’s disease model), and Gaa-/-. The left panel compares WT and 5xFAD brains, while the right panel compares WT and Gaa-/- brains. The color scales indicate the relative intensity of the lipid/metabolite signals, illustrating differences in spatial distribution between the genotypes. These comparisons highlight alterations in brain metabolism and lipid composition associated with Alzheimer’s disease and GAA deficiency.
Extended Data Fig. 10 Pathway enrichment analysis of neuronal layers of the cerebellum in WT and 5xFAD mouse brains.
a. 2D slice and 3D rendered views of WT and 5xFAD mouse brains displaying the distribution of phosphatidylinositol (PI) 38:5 in the hippocampus and cerebellum. b. The excitatory neuronal bodies of the cerebellum from a Nissl-stained sagittal brain section from the Allen Brain Atlas and Allen Reference Atlas, – Mouse Brain, Allen Mouse Brain Atlas, mouse.brain-map.org and atlas.brain-map.org (left) and pixels corresponding to these regions were extracted (right) for targeted analysis. c. Pathway enrichment analysis was performed on metabolomics (left) and lipidomics (right) datasets extracted from these pixels. Metabolomics pathway enrichment was conducted using MetaboAnalyst using the hypergeometric test and adjusted for multiple comparison (bonferroni correction) highlighting key pathways such as the transfer of acetyl groups into mitochondria, the Warburg effect, and the citric acid cycle based on -log10(pvalue). Lipid ontology enrichment using the one-sided Kolmogorov-Smirnov test and adjusted for multiple comparison (bonferroni correction) was performed using Lipid Ontology (LION) pathway analysis, identifying significant lipid categories related to bilayer thickness and lateral diffusion properties based on -log10(pvalue).
Supplementary information
Supplementary Video 1
Two-dimensional serial slide transition from first to last section by manual fit and MetaVision3D. Examples are LPE 16:1, PS 38:4, PS 40:6 and PS 44:12.
Supplementary Video 2
Front or coronal view of plane transition of PS 44:12 after Metavision3D rendering. ImageJ is used for visualization.
Supplementary Video 3
Top-down or transverse view of plane transition of PS 44:12 after Metavision3D rendering. ImageJ is used for visualization.
Supplementary Video 4
Metavision3D rendering of PS 44:12 in projection mode generated in ImageJ.
Supplementary Video 5
Front or coronal view of plane transition of PS 38:6 after Metavision3D rendering. ImageJ is used for visualization.
Supplementary Video 6
Top-down or transverse view of plane transition of PS 38:6 after Metavision3D rendering. ImageJ is used for visualization.
Supplementary Video 7
Metavision3D rendering of PS 38:6 in projection mode generated in ImageJ.
Supplementary Video 8
Front or coronal view of plane transition of PS 40:6 after Metavision3D rendering. ImageJ is used for visualization.
Supplementary Video 9
Top-down or transverse view of plane transition of PS 40:6 after Metavision3D rendering. ImageJ is used for visualization.
Supplementary Video 10
Metavision3D rendering of PS 40:6 in projection mode generated in ImageJ.
Source data
Source Data Fig. 2
Statistical source data.
Source Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ma, X., Shedlock, C.J., Medina, T. et al. AI-driven framework to map the brain metabolome in three dimensions. Nat Metab 7, 842–853 (2025). https://doi.org/10.1038/s42255-025-01242-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42255-025-01242-9