Abstract
The balance between mitochondrial calcium (mCa2+) uptake and efflux is essential for ATP production and cellular homeostasis. The mitochondrial sodium-calcium exchanger, NCLX, is a critical route of mCa2+ efflux in excitable tissues, such as the heart and brain, and animal models support NCLX as a promising therapeutic target to limit pathogenic mCa2+ overload. However, the mechanisms that regulate NCLX activity are largely unknown. Using proximity biotinylation proteomic screening, we identify the mitochondrial inner membrane protein TMEM65 as an NCLX binding partner that enhances sodium (Na+)-dependent mCa2+ efflux. Mechanistically, acute pharmacological NCLX inhibition or genetic deletion of NCLX ablates the TMEM65-dependent increase in mCa2+ efflux, and loss-of-function studies show that TMEM65 is required for Na+-dependent mCa2+ efflux. In line with these findings, knockdown of Tmem65 in mice promotes mCa2+ overload in the heart and skeletal muscle and impairs both cardiac and neuromuscular function. Collectively, our results show that loss of TMEM65 function in excitable tissue disrupts NCLX-dependent mCa2+ efflux, causing pathogenic mCa2+ overload, cell death, and organ-level dysfunction. These findings demonstrate the essential role of TMEM65 in regulating NCLX-dependent mCa2+ efflux and suggest modulation of TMEM65 as a therapeutic strategy for a variety of diseases.
This is a preview of subscription content, access via your institution
Access options
Access Nature and 54 other Nature Portfolio journals
Get Nature+, our best-value online-access subscription
$32.99 / 30 days
cancel any time
Subscribe to this journal
Receive 12 digital issues and online access to articles
$119.00 per year
only $9.92 per issue
Buy this article
- Purchase on SpringerLink
- Instant access to full article PDF
Prices may be subject to local taxes which are calculated during checkout




Similar content being viewed by others
Data availability
The MS proteomics data for the proximity biotinylation screen were uploaded to the ProteomeXchange Consortium via MassIVE under accession nos. PXD045834 and MSV000093004, respectively. The public datasets used in this study include the 2018 release (IMPI-2018-Q2) of the IMPI database (https://www.mrc-mbu.cam.ac.uk/research-resources-and-facilities/impi), and GTEx Analysis V8 RNA-seq data (GTEx_Analysis_2017-06-05_v8_RNASeQCv1.1.9_gene_median_tpm.gct.gz) available via the GTEx Portal (https://www.gtexportal.org/home/downloads/adult-gtex/bulk_tissue_expression). Source data are provided with this paper.
References
Palty, R. et al. NCLX is an essential component of mitochondrial Na+/Ca2+ exchange. Proc. Natl Acad. Sci. USA 107, 436–441 (2010).
Palty, R. et al. Lithium-calcium exchange is mediated by a distinct potassium-independent sodium-calcium exchanger. J. Biol. Chem. 279, 25234–25240 (2004).
Luongo, T. S. et al. The mitochondrial Na+/Ca2+ exchanger is essential for Ca2+ homeostasis and viability. Nature 545, 93–97 (2017).
Jadiya, P. et al. Neuronal loss of NCLX-dependent mitochondrial calcium efflux mediates age-associated cognitive decline. iScience 26, 106296 (2023).
Garbincius, J. F. et al. Enhanced NCLX-dependent mitochondrial Ca2+ efflux attenuates pathological remodeling in heart failure. J. Mol. Cell. Cardiol. 167, 52–66 (2022).
Jadiya, P. et al. Impaired mitochondrial calcium efflux contributes to disease progression in models of Alzheimer’s disease. Nat. Commun. 10, 3885 (2019).
Pathak, T. et al. Dichotomous role of the human mitochondrial Na+/Ca2+/Li+ exchanger NCLX in colorectal cancer growth and metastasis. eLife 9, e59686 (2020).
Ruknudin, A. M. & Lakatta, E. G. The regulation of the Na/Ca exchanger and plasmalemmal Ca2+ ATPase by other proteins. Ann. N. Y. Acad. Sci. 1099, 86–102 (2007).
Brustovetsky, T., Pellman, J. J., Yang, X. F., Khanna, R. & Brustovetsky, N. Collapsin response mediator protein 2 (CRMP2) interacts with N-methyl-d-aspartate (NMDA) receptor and Na+/Ca2+ exchanger and regulates their functional activity. J. Biol. Chem. 289, 7470–7482 (2014).
Pulina, M. V., Rizzuto, R., Brini, M. & Carafoli, E. Inhibitory interaction of the plasma membrane Na+/Ca2+ exchangers with the 14-3-3 proteins. J. Biol. Chem. 281, 19645–19654 (2006).
Garbincius, J. F. & Elrod, J. W. Mitochondrial calcium exchange in physiology and disease. Physiol. Rev. 102, 893–992 (2022).
Zhang, Y. et al. Loss of TMEM65 causes mitochondrial disease mediated by mitochondrial calcium. Preprint at bioRxiv https://doi.org/10.1101/2022.08.02.502535 (2023).
Vetralla, M. et al. TMEM65-dependent Ca2+ extrusion safeguards mitochondrial homeostasis. Preprint at bioRxiv https://doi.org/10.1101/2023.10.10.561661 (2023).
Cai, X. & Lytton, J. Molecular cloning of a sixth member of the K+-dependent Na+/Ca2+ exchanger gene family, NCKX6. J. Biol. Chem. 279, 5867–5876 (2004).
Stavsky, A. et al. Aberrant activity of mitochondrial NCLX is linked to impaired synaptic transmission and is associated with mental retardation. Commun. Biol. 4, 666 (2021).
Roux, K. J., Kim, D. I., Raida, M. & Burke, B. A promiscuous biotin ligase fusion protein identifies proximal and interacting proteins in mammalian cells. J. Cell Biol. 196, 801–810 (2012).
Kim, D. I. et al. An improved smaller biotin ligase for BioID proximity labeling. Mol. Biol. Cell. 27, 1188–1196 (2016).
Nishimura, N., Gotoh, T., Oike, Y. & Yano, M. TMEM65 is a mitochondrial inner-membrane protein. PeerJ 2, e349 (2014).
Nazli, A. et al. A mutation in the TMEM65 gene results in mitochondrial myopathy with severe neurological manifestations. Eur. J. Hum. Genet. 25, 744–751 (2017).
Logan, C. V. et al. Loss-of-function mutations in MICU1 cause a brain and muscle disorder linked to primary alterations in mitochondrial calcium signaling. Nat. Genet. 46, 188–193 (2014).
Wilton, K. M. et al. Developmental brain abnormalities and acute encephalopathy in a patient with myopathy with extrapyramidal signs secondary to pathogenic variants in MICU1. JIMD Rep. 53, 22–28 (2020).
Musa, S. et al. A Middle Eastern founder mutation expands the genotypic and phenotypic spectrum of mitochondrial MICU1 deficiency: a report of 13 patients. JIMD Rep. 43, 79–83 (2019).
Liu, J. C. et al. MICU1 serves as a molecular gatekeeper to prevent in vivo mitochondrial calcium overload. Cell Rep. 16, 1561–1573 (2016).
Rudolf, R., Mongillo, M., Magalhaes, P. J. & Pozzan, T. In vivo monitoring of Ca2+ uptake into mitochondria of mouse skeletal muscle during contraction. J. Cell Biol. 166, 527–536 (2004).
Lambert, J. P. et al. MCUB regulates the molecular composition of the mitochondrial calcium uniporter channel to limit mitochondrial calcium overload during stress. Circulation 140, 1720–1733 (2019).
Roy, S., Dey, K., Hershfinkel, M., Ohana, E. & Sekler, I. Identification of residues that control Li+ versus Na+ dependent Ca2+ exchange at the transport site of the mitochondrial NCLX. Biochim. Biophys. Acta Mol. Cell Res. 1864, 997–1008 (2017).
Refaeli, B., Giladi, M., Hiller, R. & Khananshvili, D. Structure-based engineering of lithium-transport capacity in an archaeal sodium-calcium exchanger. Biochemistry 55, 1673–1676 (2016).
Goldman, A., Harper, S. & Speicher, D. W. Detection of proteins on blot membranes. Curr. Protoc. Protein Sci. 86, 10.8.1–10.8.11 (2016).
Aguet, F. et al. The GTEx Consortium atlas of genetic regulatory effects across human tissues. Science 369, 1318–1330 (2020).
Lombardi, A. A. et al. Mitochondrial calcium exchange links metabolism with the epigenome to control cellular differentiation. Nat. Commun. 10, 4509 (2019).
Mallilankaraman, K. et al. MICU1 is an essential gatekeeper for MCU-mediated mitochondrial Ca2+ uptake that regulates cell survival. Cell 151, 630–644 (2012).
Luongo, T. S. et al. The mitochondrial calcium uniporter matches energetic supply with cardiac workload during stress and modulates permeability transition. Cell Rep. 12, 23–34 (2015).
Garbincius, J. F. et al. MCU gain-and loss-of-function models define the duality of mitochondrial calcium uptake in heart failure. Preprint at bioRxiv https://doi.org/10.1101/2023.04.17.537222 (2023).
Frezza, C., Cipolat, S. & Scorrano, L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2, 287–295 (2007).
Toye, A. A. et al. A genetic and physiological study of impaired glucose homeostasis control in C57BL/6J mice. Diabetologia 48, 675–686 (2005).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Aartsma-Rus, A. & van Putten, M. Assessing functional performance in the mdx mouse model. J. Vis. Exp. https://doi.org/10.3791/51303 (2014).
Briguet, A., Courdier-Fruh, I., Foster, M., Meier, T. & Magyar, J. P. Histological parameters for the quantitative assessment of muscular dystrophy in the mdx-mouse. Neuromuscul. Disord. 14, 675–682 (2004).
Acknowledgements
We thank X. Cui and the Genome Engineering and iPSC Center at Washington University in St. Louis for gRNA validation and genotyping services; X. Hua and the Fox Chase Cancer Center Transgenic Mouse Facility for assistance in generating the Nclx-3×Flag mouse line; and M. Andrake of the Fox Chase Cancer Center Molecular Modeling Facility for assistance with in silico modelling of protein–protein interactions. The research was supported by the National Institutes of Health (nos. T32HL091804 and F32HL151146 to J.F.G.; T32HL091804 and F30AG082407 to H.M.C.; DK120876 to D.T.; and R01NS121379, P01HL147841, P01HL134608 and R01HL142271 to J.W.E.), a Frances Velay Fellowship (to A.E.S.) and the American Heart Association (no. 24CDA1268083 to J.F.G.; no. 20EIA35320226 to J.W.E.).
Author information
Authors and Affiliations
Contributions
Conceptualization: J.F.G. and J.W.E. Investigation: J.F.G., O.S., H.M.C., C.C.-F., A.S.M., A.D.M., A.E.S., D.Y.K., J.J.D., A.S.W., E.K.M., M.P.L., A.N.H. and D.T. Formal analysis: J.F.G., H.M.C., C.C.-F., D.T. and J.W.E. Data curation: J.F.G. and J.W.E. Figure preparation: J.F.G. Writing—original draft preparation: J.F.G. Writing—review and editing: J.F.G. and J.W.E. All authors approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Metabolism thanks Elizabeth Jonas and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Alfredo Giménez-Cassina, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 Validation of mitochondrial localization of NCLX-BioID2-HA fusion protein and TMEM65.
a, Western blots showing progressive digestion of mitochondria from AC16 cardiomyocytes expressing NCLX-BioID2HA with increasing concentrations of trypsin, +/− increasing concentrations of digitonin to disrupt the inner mitochondrial membrane. WCL, whole cell lysate; Cyto, cytosolic fraction. CypD, cyclophilin D. O.M.M., outer mitochondrial membrane; I.M.M., inner mitochondrial membrane. Data are representative of 3 independent experiments. b, Immunofluorescence staining of AC16 cardiomyocytes transiently transfected with a plasmid encoding TMEM65-Myc-FLAG or empty vector control, corresponding to experiment shown in Fig. 1f. Data are representative of 2 independent experiments.
Extended Data Fig. 2 Effects of TMEM65 overexpression or deletion on mCa2+ and cytosolic Ca2+ handling and expression of mCa2+-handling proteins.
a–f, Densitometric quantifications of western blots shown in Fig. 2a. (n = 3 biological replicates/group; P-values vs. control by 1-way ANOVA with Dunnett’s post-hoc test). g, Western blots for pyruvate dehydrogenase (PDH) phosphorylation status in control and stable TMEM65-Myc-FLAG overexpression lines (TMEM65-OE). Data include 3 biological replicates/group. h, Densitometric quantification of PDH phosphorylation normalized to total PDH. (n = 3 biological replicates/group; P-values vs. control by 1-way ANOVA with Dunnett’s post-hoc test). i, mCa2+ uptake rate for permeabilized cell experiments shown in Fig. 2b. (n = 4 for empty vector control, 3 for TMEM65-OE. P-values by 2-way ANOVA with Sidak’s post-hoc test, as indicated). j, Western blotting of human AC16 cardiomyocytes transduced for 24 hours with adenovirus encoding untagged human TMEM65. Both full-length TMEM65 (~25kD) and mature TMEM65 following cleavage of the mitochondrial targeting sequence (~20kD) were detected. MOI, multiplicity of infection. Dashed line is to aid in visualizing separate conditions. Similar results were obtained after 48 hours and after 72 hours of transduction. k, Pooled traces for AC16 cardiomyocytes transduced with adenovirus encoding LacZ (Ad-LacZ) or human TMEM65 (Ad-TMEM65), showing extramitochondrial bath Ca2+ (Fura-FF). 4 million cells were permeabilized with digitonin (Dg) in the presence of thapsigargin (Tg), then administered a single 10 µM Ca2+ bolus, with subsequent additions of the mitochondrial calcium uniporter inhibitor Ru360 and FCCP as indicated. (n = 3/group). Inset shows initial mCa2+ efflux phase. mCa2+ efflux rate over the first 25 seconds following addition of Ru360 (l); % of matrix Ca2+ effluxed post-Ru360 (m); and % net mCa2+ uptake following 10 µM Ca2+ bolus (n). (n = 3 assay runs/genotype; P-values by unpaired two-tailed t-test). o, Western blotting of mouse C2C12 myoblasts transduced for 24 hours with adenovirus encoding human TMEM65. Data include 3 biological replicates/group. p, Pooled traces for 2 million mouse C2C12 skeletal myoblasts with acute transduction of Ad-LacZ or Ad-TMEM65, in the presence or absence of the NCLX inhibitor CGP-37157 (CGP), showing extramitochondrial bath Ca2+ (Fura-FF) as in (k) (n = 3/group). mCa2+ efflux rate over the first 25 seconds following addition of Ru360 (q), % of matrix Ca2+ effluxed post-Ru360 (r); % net mCa2+ uptake following the 10 µM Ca2+ bolus (s), and initial mCa2+ uptake rate for the first 25 seconds following the Ca2+ peak (t). (n = 3 assay runs/group. P-values by 2-way ANOVA with Sidak’s post-hoc test, as indicated). Time to 50% decay (u) and half rise time (v) of KCl-evoked cytosolic Ca2+ transients as measured by Fluo-4 fluorescence in intact AC16 cardiomyocytes for experiment shown in Fig. 2j. (n = 9 cells/group. P-values by unpaired two-tailed t-test). w, Peak amplitude of KCl-evoked cytosolic Ca2+ transients for experiment shown in Fig. 2j. (n = 9 cells/group. P-value by two-tailed Mann-Whitney test). x-b’, Densitometric quantifications of western blots shown in Fig. 2n. (n = 3 biological replicates/group; P-values vs. WT by 1-way ANOVA with Dunnett’s post-hoc test). Data are presented as mean ± SEM.
Extended Data Fig. 3 Effects of NCLX deletion on TMEM65 expression, validation of Nclx-3xFLAG mouse, and size exclusion chromatography standards.
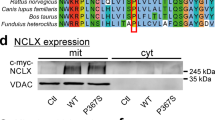
a, Western blots for TMEM65 expression in Nclxfl/fl and Nclx−/− mouse embryonic fibroblasts, assessed 7 days after cre-mediated Nclx deletion. b, Densitometric quantification of western blots shown in (a). (n = 3 biological replicates/group; P-value by unpaired two-tailed t-test). Data are presented as mean ± SEM. c, Chromatogram of sequencing confirming proper in-frame insertion of DNA encoding the NCLX-3xFLAG epitope tag prior to the stop codon of the endogenous mouse Nclx gene. d, Genotyping gel showing pups heterozygous (+/−) or homozygous (+/+) for the 3xFLAG knock-in. Data are representative of >10 litters. e, Western blots validating solubilization and specific detection of NCLX-3xFLAG protein (arrow) in isolated adult mouse heart mitochondria. Amido black staining for total protein is shown as a loading control. f, Absorbance at 280 nm for gel filtration standards separated by size-exclusion chromatography on 1 MD column. g, Standard curve of elution fraction vs. known molecular weight for gel filtration standards separated by size-exclusion chromatography on 1 MD column. The equation shows the fit of the standards to a one-phase exponential decay. h, Absorbance at 280 nm for gel filtration standards separated by size-exclusion chromatography on 600 kD column. i, Standard curve of elution fraction vs. known molecular weight for gel filtration standards separated by size-exclusion chromatography on 600 kD column. The equation shows the fit of the standards to a one-phase exponential decay.
Extended Data Fig. 4 Validation of expression and mitochondrial localization for TMEM65 mutant constructs.
a, Sequence alignments of C-terminal Myc-FLAG-tagged wild-type (WT) and mutant human TMEM65 constructs encoded by TMEM65 adenoviruses, corresponding to schematic shown in Fig. 3k. (TM = transmembrane helix). b, c, Western blots of AC16 cardiomyocytes transduced with adenovirus encoding control LacZ, WT TMEM65-Myc-FLAG, or mutant TMEM65-Myc-FLAG. Data include 3 biological replicates/group. d, Immunofluorescence staining of AC16 cardiomyocytes transduced with adenovirus encoding control LacZ, WT TMEM65-Myc-FLAG, or mutant TMEM65-Myc-FLAG. TOM20 served as a mitochondrial marker. Images are representative of 2 independent replicates/group. Cterm., C-terminus.
Extended Data Fig. 5 Phenotypic consequences of altered TMEM65 expression.
a, Representative images showing WGA labelling (green) and nuclear staining with DAPI (blue) in cross sections of the gastrocnemius muscle at 24 weeks of age. scr., scrambled. b, Total mass of the gastrocnemius + soleus + plantaris muscle, dissected as a unit, normalized to total body mass at 24 weeks of age. Open symbols represent male mice, filled symbols represent female mice throughout. (n = 10 mice + scrambled shRNA, 9 mice + Tmem65 shRNA. P-value by unpaired two-tailed t-test with Welch’s correction). Left ventricular end diastolic dimension (LVEDD) (c) and left ventricular end systolic dimension (LVESD) (d) as measured by echocardiography in mice at 6 weeks of age (n = 11 mice + scrambled shRNA, 7 mice + Tmem65 shRNA. P-values by unpaired two-tailed t-test). e, Pooled traces showing extramitochondrial bath Ca2+ (Fura-FF) and mitochondrial membrane potential (JC-1) in permeabilized WT and TMEM65−/− AC16 cardiomyocytes in response to repeated additions of 5 µM Ca2+ (arrows). Red arrows indicate the Ca2+ bolus typically triggering permeability transition in each genotype. (n = 3 replicates/genotype). f, Pooled traces showing extramitochondrial bath Ca2+ (Fura-FF) and mitochondrial membrane potential (JC-1) in permeabilized control or TMEM65-OE AC16 cardiomyocytes in response to repeated additions of 10 µM Ca2+ (arrows). Red arrow indicates Ca2+ bolus triggering permeability transition in control cells. (n = 4 replicates for empty vector controls, 5 replicates for TMEM65-OE). Data are presented as mean ± SEM.
Supplementary information
Supplementary Information
Supplementary Table 1.
Supplementary Video 1
Representative normal motor phenotype in a 24-week-old mouse injected as neonate with AA9-scrambled shRNA.
Supplementary Video 2
Representative motor phenotype in a 24-week-old mouse injected as neonate with AAV9-Tmem65 shRNA.
Source data
Source Data Fig. 1
Statistical source data.
Source Data Fig. 1
Full-length immunoblots.
Source Data Fig. 2
Statistical source data.
Source Data Fig. 2
Full-length immunoblots.
Source Data Fig. 3
Statistical source data.
Source Data Fig. 3
Full-length immunoblots.
Source Data Fig. 4
Statistical source data.
Source Data Fig. 4
Full-length immunoblots.
Source Data Extended Data Fig. 1
Full-length immunoblots.
Source Data Extended Data Fig. 2
Statistical source data.
Source Data Extended Data Fig. 2
Full-length immunoblots.
Source Data Extended Data Fig. 3
Statistical source data.
Source Data Extended Data Fig. 3
Full-length immunoblots.
Source Data Extended Data Fig. 4
Full-length immunoblots.
Source Data Extended Data Fig. 5
Statistical source data.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Garbincius, J.F., Salik, O., Cohen, H.M. et al. TMEM65 regulates and is required for NCLX-dependent mitochondrial calcium efflux. Nat Metab 7, 714–729 (2025). https://doi.org/10.1038/s42255-025-01250-9
Received:
Accepted:
Published:
Issue date:
DOI: https://doi.org/10.1038/s42255-025-01250-9
This article is cited by
-
TMEM65 joins the mitochondrial Ca2+ club
Nature Metabolism (2025)