Abstract
Adipose tissue (AT) is a complex connective tissue with a high relative proportion of adipocytes, which are specialized cells with the ability to store lipids in large droplets. AT is found in multiple discrete depots throughout the body, where it serves as the primary repository for excess calories. In addition, AT has an important role in functions as diverse as insulation, immunity and regulation of metabolic homeostasis. The Human Cell Atlas Adipose Bionetwork was established to support the generation of single-cell atlases of human AT as well as the development of unified approaches and consensus for cell annotation. Here, we provide a first roadmap from this bionetwork, including our suggested cell annotations for humans and mice, with the aim of describing the state of the field and providing guidelines for the production, analysis, interpretation and presentation of AT single-cell data.
Similar content being viewed by others
Main
The Human Cell Atlas (HCA) is a global consortium of researchers working to develop a catalogue of all human cell types throughout development (https://www.humancellatlas.org/). The HCA comprises a series of bionetworks focused on specific tissues, organs or developmental pathways. These bionetworks are responsible for identifying and addressing critical issues that face researchers in that area, and for helping the HCA generate reference atlases of the particular tissue, organ or pathway. Towards that end, the HCA Adipose Bionetwork will be involved in generating the Adipose Atlas 1.0, which will include (a) the integration of multiple single-nucleus RNA-sequencing (snRNA-seq) datasets from different healthy human AT depots; and (b) the development of unified approaches and consensus on cell annotation. To support this work, the bionetwork held a series of virtual meetings open to all researchers in the field aiming to agree on guidelines in four areas of importance for single-cell analyses of ATs, that is, nomenclature for the anatomical localization of adipose depots, nomenclature for cell annotation, and experimental and computational best practices. In this Review, we provide a roadmap of AT at single-cell resolution based on these meetings.
AT: a multifocal, heterogeneous and dynamic organ
Once thought to be an inert, cellularly bland storage reservoir for energy in the form of triacylglycerol, AT is now recognized to be a multifocal, complex, heterogeneous and dynamic organ that controls metabolic homeostasis via both endocrine and paracrine mechanisms. Unlike most organs, AT is multifocal, found both in discrete depots as well as scattered throughout other organs and tissues. AT is complex because it contains a multitude of cell types and subtypes, including adipocytes, fibroblastic stromal cells (many of which represent adipocyte progenitor cells), immune cells, Schwann cells and vascular cells. AT is heterogeneous because its cellular composition varies substantially between and within depots. AT is dynamic because its composition changes during development1,2,3 and with perturbation, such as after changes in nutrition, physical activity or ambient temperature4,5,6. The transcriptional plasticity of AT has been studied using various methods, including bulk RNA-seq and quantitative PCR of whole tissue. More recently, single-cell transcriptome profiling has enabled the cellular and molecular plasticity of AT to be resolved at a single-cell level7. Defining ‘normal AT’ in terms of its component cellular subtypes and linking them to function is a critical unmet milestone towards a complete understanding of metabolic homeostasis and disease.
Dysfunctional AT is associated with a variety of cardiometabolic diseases. A clear example is obesity, a condition in which excess adiposity significantly increases the risk of type 2 diabetes (T2D), metabolic dysfunction-associated steatotic liver disease and cardiovascular disease. But AT is not only associated with disease when in excess; lack of AT, either generally or limited to specific regions, also confers risk for severe T2D, metabolic dysfunction-associated steatotic liver disease and atherosclerosis. Individuals with either inherited or acquired forms of lipodystrophy store excess lipids in non-adipose organs, such as liver, muscle and pancreatic islets, contributing to insulin resistance and reduced insulin secretion8. Many gene expression changes in whole AT have been associated with cardiometabolic disease, but a true understanding of this relationship will depend on the ability to determine molecular and cellular changes at the level of single cells. For example, some subpopulations of macrophages are more likely to promote insulin resistance than others9, and certain adipocyte subtypes are more tightly associated with T2D than others5,10.
Anatomical location of adipose depots
Most AT is found in discrete depots in stereotypical locations throughout the body, although scattered adipocytes can be found in various tissues and organs. Because the cellular composition, molecular attributes and associations with disease of AT vary depending on the depot, it is critically important that researchers share a common language for describing where their samples originate. Broadly speaking, AT is divided into two major groups: subcutaneous adipose tissue (SAT) and visceral adipose tissue (VAT), which have different associations with cardiometabolic diseases11,12. Excess VAT (stereotypically seen in men, thus the older term ‘android’ obesity) is associated with a relatively high risk of T2D, insulin resistance and atherosclerosis13. Conversely, excess SAT, especially in the gluteofemoral region (stereotypically seen in women, thus the older term ‘gynoid’ obesity), is associated with a relatively lower risk of metabolic dysfunction11,14. This simple dichotomy obscures several critical issues: (1) most individuals with obesity exhibit a mixed pattern of visceral and subcutaneous adiposity; (2) there are many depots within the broad category of VAT or SAT, each possessing distinct compositions and functions; and (3) there are many other depots of biological interest that become enlarged in obesity and that cannot be categorized within the duality of VAT versus SAT.
Another distinction can be made between white adipose tissue (WAT) and brown adipose tissue (BAT). The latter contains brown adipocytes that express high levels of genes for mitochondrial biogenesis, oxidative phosphorylation and heat production (thermogenesis) via futile cycling. The best-studied thermogenic pathway involves uncoupling protein 1 (UCP1), although futile calcium and creatine cycling can also generate heat15,16. Thermogenic adipocytes can also be found in white fat depots and are variously referred to as beige17,18 or BRITE19 adipocytes; these cells are also rich in mitochondria and express UCP1. In rodents and other small mammals, brown adipocytes are found in a cluster of small, dedicated BAT depots in the interscapular area, while beige adipocytes emerge in WAT depots in response to specific stimuli, such as cold exposure. Brown and beige adipocytes have different developmental origins and express overlapping but distinct sets of marker genes20,21. In humans, BAT was thought to exist only in the newborn period, but recent studies show that it can also be found in adults in the deep cervical, supraclavicular, paravertebral and perirenal regions22,23,24,25,26.
There have been few systematic efforts to catalogue and characterize the different adipose depots in any species27,28. In part, the diffuse nature of some adipocytes in different tissues and organs may make a truly comprehensive list of depots difficult to compile. This challenge is compounded by the fact that there is generally little correlation between depots across species. For example, the depot most often used by adipose researchers to represent VAT in mice is the perigonadal depot (typically the epididymal AT of the male mouse), which has no precise analogue in humans29. Conversely, one of the most important visceral depots in humans, the omental AT, is rudimentary in mice. This issue goes beyond mice and men. For example, the hump of the camel is mainly composed of AT and represents one of the largest depots in that animal, yet it is absent in nearly all other mammalian species. Despite these constraints, here, we present a compendium of generally accepted adipose depots in humans and mice (Fig. 1 and Table 1).
Human adipose depots
Subcutaneous adipose depots
Most studies have not drawn major distinctions between SAT at different locations; however, it is becoming increasingly clear that there are substantial regional differences between SAT depots. For example, abdominal SAT has different attributes and confers a different metabolic risk profile than gluteofemoral SAT30. Such differences may be explained by larger, yet more insulin-sensitive adipocytes in the thigh31, higher retention of lipids in gluteofemoral adipocytes14, differences in cell composition between abdominal and gluteofemoral depots32 and/or variations in the differentiation capacity of their progenitors33.
Furthermore, both truncal and gluteofemoral SAT are separated into two planes, called deep and superficial, by a discrete subcutaneous fascia (termed Scarpa’s fascia in the abdominal region)34. This fascial plane is discontinuous, and in some parts of the body, the deep SAT can dominate. Deep and superficial truncal SAT exhibit different microanatomy and may have distinct metabolic functions and disease associations35,36,37.
Craniofacial AT provides another example of the differences found within the broad category of SAT. Not only does craniofacial AT exhibit different metabolic characteristics than SAT found elsewhere in the body, but there are actually several compartments within the category of craniofacial AT (for example, buccal AT), each of which may have its own molecular and functional characteristics38. We recommend that all studies (single-cell or otherwise) using SAT precisely define the site and depth from which it was collected.
Dermal AT has also been proposed as a separate depot. In mice, a thin muscular layer called the panniculus carnosus separates the dermal AT from SAT, but humans have only vestigial remnants of this layer in scattered areas39. Therefore, it is unclear whether the subcutaneous and dermal AT represent distinct depots in humans. However, small islands of adipocytes can be found in the human dermis, especially around hair follicles40,41.
Visceral adipose depots
Mesenteric and omental
The mesenteric and omental depots together make up most of the human VAT. The mesentery is a membranous fold that extends from the posterior peritoneum and suspends the loops of the large and small bowel within the peritoneal cavity. The AT that accumulates within the mesentery (mesenteric adipose tissue (MAT)) also contains lymphoid tissue in the form of AT-associated lymphoid clusters, which are milky, spot-like structures that grossly resemble fat. In inflammatory bowel disease, MAT can infiltrate the intestinal wall, a phenomenon called ‘creeping fat’, which is closely associated with disease pathogenesis42. MAT is sometimes named by the specific intestinal segment that it connects (for example, mesocolic, mesosigmoid and mesorectal AT), and including this information in the reporting metadata would be of considerable value given the recognized heterogeneity of MAT adjacent to specific intestinal segments43. The omentum is a large, apron-like fold from the visceral peritoneum that extends downward from the stomach. Similarly to MAT, omental AT contains milky spots, which are lymphoid aggregates analogous to fat-associated lymphoid clusters44 with a rich vasculature. As the dominant sites of fat storage in obesity, MAT and omental AT are highly correlated with adverse cardiometabolic outcomes.
Perirenal and retroperitoneal
Perirenal adipose tissue (PRAT), which covers the kidneys and adrenal glands, is in the retroperitoneal space; as such, PRAT is not technically ‘visceral’, although it is commonly viewed as VAT. PRAT lies between the renal capsule and renal sinus, although the AT within the renal sinus itself is considered perivascular (see below), given its proximity to the renal vasculature. Similar to other VAT depots, PRAT is associated with obesity and cardiovascular disease. Interestingly, PRAT contains numerous thermogenic brown or beige adipocytes45,46. Retroperitoneal AT is found in the retroperitoneal space that is not within the anatomical limits described for PRAT.
Epiploic
Epiploic AT, also known as the appendices epiploicae, consists of two rows of 50–100 lobules attached to the outside of the colon. This rarely studied depot has recently been shown to have a unique mRNA and protein expression profile relative to other visceral depots47.
Periprostatic
As the name implies, periprostatic AT surrounds the prostate gland, and is a critical part of the tumour microenvironment in prostate cancer48.
Pericardial and epicardial
There has been some confusion regarding the depots around the heart. The AT inside the pericardial sac and adherent to the myocardium should be called epicardial AT, while the term pericardial AT should be reserved for adipose depots that are exterior to the pericardium (Fig. 1a). One sometimes finds the term ‘paracardial’ AT in the literature; this term has no clear meaning and should be abandoned. Epicardial AT, which can make up 20% of gross cardiac weight, increases with obesity, and is associated with coronary artery disease and atrial fibrillation in humans and non-human primates49,50.
Mediastinal
Roughly 70% of intrathoracic AT in humans is not found in the pericardial or epicardial depots—this depot is called mediastinal AT. Mediastinal AT has some brown-like characteristics51. Some mediastinal AT is associated with the thymus, especially as it undergoes involution with ageing; adipocytes become a dominant component of the thymus by middle age52. Although the origin and function of these adipocytes remain incompletely understood, it has been suggested that thymic AT accumulation may contribute to age-related decline in adaptive immune function.
Other adipose depots
There are also AT depots located outside the primary subcutaneous and visceral regions, each with distinct anatomical locations and metabolic roles.
Bone marrow
AT occupies up to 70% of the marrow cavity, mostly in the so-called ‘yellow marrow’, although there are adipocytes in the red marrow as well. Bone marrow adipose tissue has several unique features, not least of which is the paradoxical expansion in the face of severe calorie restriction and weight loss53,54.
Epineural/perineural
Some of the larger nerves have adipose depots that adhere to the outer nerve sheath, or epineurium. The mass of some neural-associated adipose depots can increase after nerve injury, and leptin from epineural adipocytes enhances neuronal regeneration and recovery55.
Mammary
AT makes up approximately 90% of the mass of the breast in non-lactating women. Adipocytes in the breast have an important role in development of the ductal epithelium and contribute to tumorigenesis and metastasis of breast cancer56.
Perithyroidal
There is AT adherent to the surface of the thyroid gland, which can be a site of local invasion in some cases of thyroid carcinoma57.
Perivascular
As the name implies, perivascular adipose tissue (PVAT) encases many blood vessels, especially arteries. By consensus, vascular biologists have agreed that PVAT extends to a distance of one luminal width at the site under examination, or within 2 cm of any vessel >2 cm in diameter58. PVAT has been shown to have robust interactions with the encased vessel and may be involved in such conditions as hypertension and atherosclerosis58.
Retro-orbital
AT provides a cushion for the eye in its socket. In some people, the adipose stromal and progenitor cells (ASPCs) within this retro-orbital (sometimes called periorbital) AT express antigens that make the depot expand in the setting of Graves’ disease, causing ophthalmopathy59.
Intra-articular
AT also provides an important cushioning function within joints. A recent mixed single-cell RNA-sequencing (scRNA-seq)/snRNA-seq study of the intra-articular AT of the knee (that is, the infrapatellar fat pad) demonstrated a developmental relationship between adipocytes here and the synovial lining, and suggested paracrine interactions that may promote osteoarthritis60.
Intermuscular/intramuscular
AT that is located under the deep fascia and between major muscle bundles is called intermuscular AT61. Some investigators make a distinction between intermuscular AT and adipose depots located within or between muscle fibres (sometimes called intramuscular AT)62, although a functional, molecular or developmental basis for this distinction is not entirely clear. Most of the literature on intermuscular AT emerges from the livestock industry, as a measure of meat quality63. Nonetheless, in humans there are clear associations between intermuscular AT mass, obesity and insulin resistance61. Importantly, lipid deposition within myocytes (intramyocellular triacylglycerol) is distinct from AT.
Structural adipose depots of the heel and fingers
Some parts of the body use adipocytes as a mechanical cushion, such as the heel and the tips of the fingers and toes. This AT is encased in tough septated connective tissue and is relatively inert metabolically64.
Brown (and beige) adipose depots
Unlike rodents, most adult humans at room temperature have little constitutively active thermogenic AT. In general, women have more thermogenic AT than men, and the amount diminishes with age65. Many people respond to cold exposure by increasing the amount of 18F-fluorodeoxyglucose (18F-FDG) positron emission tomography-positive thermogenic AT22,23,24,25. There are also substantial interindividual differences in BAT volume that may in part be genetic in origin. Interestingly, there is a clear association between the amount of brown and beige AT (as determined by scanning) and metabolic health, with reduced rates of T2D, dyslipidaemia, coronary artery disease, cerebrovascular disease, congestive heart failure and hypertension observed in individuals with greater 18F-FDG uptake65. Depots with the highest 18F-FDG uptake are found in the cervical, supraclavicular, axillary, intercostal, mediastinal, ventral spinal and perirenal areas27 (Table 1). In addition to the depots defined in adults, infants have classical BAT in the interscapular depot and around the heart and great vessels66, but much of this is lost during childhood. There is ongoing debate about whether the other human thermogenic depots represent brown, beige, some other type of adipocyte or a mixed population. Marker studies have been contradictory and/or inconclusive, highlighting the complexity of human thermogenic AT and the need for clarity and consensus around nomenclature, cellular annotation and experimental approaches20,21,67,68,69.
Mouse adipose depots
There are many similarities between mouse and human AT, but also profound differences. Some of the most obvious differences relate to the specific depots, as highlighted below, but there are also important molecular and functional differences between mouse and human adipocytes. Table 1 lists the mouse adipose depots that have been described to date.
Subcutaneous adipose depots
Mouse SAT exists in more anatomically defined locations than human SAT. In mice, the major accessible subcutaneous depot is the inguinal fat pad, and virtually all studies that look at mouse SAT focus on this depot. This inguinal depot contains a large lymph node that demarcates two distinct lobes, denoted the dorsolumbar and inguinal lobes70. In addition to its distinct structure, inguinal AT differs markedly from human SAT in two key aspects. First, the inguinal depot is a major site of thermogenesis in cold-exposed mice, as it houses a large number of inducible beige adipocytes. Importantly, these beige cells are not distributed evenly throughout the depot70. Second, the inguinal depot also contains a substantial portion of the mammary ducts, which cannot be easily separated physically from the rest of the AT. Other white subcutaneous depots in the mouse are found in the anterior axillary region, the triceps area, the popliteal fossa, and encasing and infiltrating the interscapular BAT (see below)28,71.
Mice have a thin muscular layer below the dermis called the panniculus carnosus; adipocytes located between the panniculus carnosus and the dermis are called dermal adipocytes (Fig. 1b). By contrast, subcutaneous adipose depots lie below the panniculus carnosus. Mouse dermal adipocytes have a different developmental origin than SAT, and have been implicated in hair growth and cycling, wound healing and antibacterial defence40,72.
Visceral adipose depots
Mouse visceral depots also differ substantially from their human counterparts. Mice have a rudimentary omentum and store relatively little fat in their mesenteric depot. Instead, the perigonadal depots (epididymal in males and periovarian in females) are the major sites of intraperitoneal fat storage. Recently, new adipose depots affiliated with the perigonadal AT were described in mice, referred to as lymph node-cervical adipose tissue in female mice and lymph node-prostatic adipose tissue in male mice73. Finally, mice have only a small amount of epicardial AT74, which becomes more evident in obese animals.
Other adipose depots
Similarly to humans, mice also have bone marrow adipose tissue, perirenal, retroperitoneal, perivascular, intra-articular and intramuscular/intermuscular AT (Fig. 1b).
Brown (and beige) adipose depots
Mice have distinct BAT depots that maintain their identity regardless of ambient temperature75; the best studied of these is the interscapular BAT, but there are small cervical and axillary depots that share this trait. In addition, almost all ‘white’ depots of the mouse can undergo browning to some degree in the face of a cold challenge. This is most often studied in the inguinal SAT, but most other depots have at least some abilities to remodel to a more thermogenic state28. Accordingly, it is imperative that studies looking at AT composition in mice are rigorous about reporting ambient temperature.
Definition and annotation of adipose cell types
The rise of single-cell sequencing technologies has revealed a much greater cellular heterogeneity of most tissues, including ATs, than previously appreciated. This has led to confusion regarding the precise definition and annotation of distinct cell types, subtypes and cell states. The concept of ‘cell type’ refers to a distinct category of cells that share fundamental characteristics, functions and lineage. In single-cell data, a cell type is recognized as a distinct cluster of cells sharing overall marker gene expression and transcriptional networks. A cell type may be composed of many different cellular ‘subtypes’, which are specific populations within a cell type having specific characteristics and known or inferred functions. In single-cell data, cellular subtypes can be identified as distinct subclusters of a cell type. Finally, a cell state is the specific physiological or functional condition of a cell at a given moment. The condition can be dynamic and change in response to environmental cues, developmental signals or metabolic states. In single-cell data, cell states will often appear as less distinct subclusters of a cell (sub)type. The context-dependent and dynamic nature of cell states makes them more difficult to classify than cell types. Moreover, as they often occur as a continuum of states, their classification is highly sensitive to the choice of computational strategy and the resolution imposed. For now, we propose focusing on the annotation of adipose cell types and subtypes in human and mouse tissues based on transcriptomic markers (Tables 2 and 3), acknowledging the inherent limitations of this approach. For example, as marker genes are rarely unique to one cellular subtype, accurate annotation of closely related cellular subtypes often requires a combination of multiple marker genes. Moreover, although subtype markers are useful for distinguishing between subtypes within a specific cell type, they may not be effective in differentiating these subtypes from other cell types present in the tissue. Therefore, the use of subtype markers is often limited to identifying subtypes within one or a few specific cell types, rather than across the entire tissue. Importantly, proper annotation of cellular subtypes and states will require inclusion of other modalities such as proteomics, metabolomics, and eventually isolation and functional characterization. Moreover, for cell (sub)types that are also abundant in other tissues, such as vascular cells and immune cells, nomenclature will eventually have to be aligned between tissues, as clarity with respect to functional characteristics is reached.
Here we provide the nomenclature and characteristics of the adipose cell types and subtypes agreed upon in the HCA Adipose Bionetwork.
Adipocytes
Adipocytes are the parenchymal cells of AT. They are large, lipid-filled cells with the primary function of storing excess calories mainly in the form of triacylglycerol. Furthermore, adipocytes act as endocrine cells that communicate systemically to inform other tissues about the state of their lipid stores, and regulate a large number of physiological functions, including appetite, glucose homeostasis, coagulation, immune activity and blood pressure76. The large size and fragile nature of adipocytes mean that they are not amenable to conventional flow-sorting and microfluidic devices. Thus, single-cell analyses of AT rely largely on snRNA-seq techniques7 (Fig. 2).
AT samples are collected from participants with informed consent, and metadata, including demographic and clinical data, are recorded. For snRNA-seq, AT can be snap frozen, allowing nuclei to be extracted from frozen samples. For scRNA-seq, fresh tissue is preferred, but fresh-frozen tissue can also be used if dissociated into single-cell suspensions before freezing. The samples are then processed to isolate cells or nuclei through enzymatic digestion or mechanical dissociation. Sample barcoding for multiplexing can be used to pool multiple samples in a single sequencing run, reducing costs and increasing throughput. Flow cytometry can be used to further sort and enrich specific cell populations before sequencing, or to remove low-quality nuclei or cells. Finally, the prepared samples can be loaded onto, for example, the 10x Chromium Controller, which uses advanced microfluidics to partition individual cells or nuclei into droplets, each containing a unique barcode for downstream sequencing and analysis. SVF, stromal vascular fraction; UMAP, uniform manifold approximation and projection.
Adipocytes are easily identifiable in snRNA-seq data by their expression of adipocyte-specific markers such as ADIPOQ and PLIN1. Brown and beige adipocyte subtypes can be distinguished from white adipocytes by their high and moderate expression of UCP1, CIDEA, PPARGC1A, PRDM16 and CKMT2 (refs. 77,78,79). Several additional markers of brown and beige adipocytes have been reported; however, some of these have turned out to be positional markers that do not distinguish between white, beige and brown80. In addition to the white, brown and beige subtypes, several lines of evidence indicate that adipocytes from different depots, in addition to their positional markers, also have slightly different properties and transcriptional profiles, which may justify classification as different subtypes. Moreover, different adipocyte cell states, potentially depending on nutritional status, have been reported in both mice and humans3,5,77,78,81,82,83. However, the exact classification and characteristics of these cell states remains obscure and controversial.
ASPCs
The stromal cells of AT are a heterogeneous group of cells generally distinguished by high expression of PDGFRA and DCN84,85. These cells have been called adipose stromal cells, adipose-derived stromal (or stem) cells, adipose progenitor cells and fibro-adipogenic progenitors, among dozens of other designations7,86. We recommend the term ASPCs, as this term captures both the supportive (stromal) and differentiative (progenitor) roles played by these cells.
Several subpopulations of ASPCs have been identified, and although the exact taxonomy of these cells is still being worked out, some consensus has been reached. DPP4, for example, marks ASPCs that have the capacity to differentiate into adipocytes, but which are the farthest removed from the mature cell87,88. ASPCs that express mature adipocyte marker genes, such as PPARG and CD36, represent late preadipocytes3. Interestingly, subpopulations of ASPCs with the ability to inhibit in vitro adipogenesis, called adipogenesis-regulatory (Areg) cells, have been highlighted in some studies89,90,91. The relative proportions of different ASPCs may depend on depot, age, body weight, metabolic health, sex and ethnicity. The developmental trajectories of different progenitor cells can be inferred computationally by trajectory analyses3,88; however, detailed insight into the plasticity and lineage relationships between ASPC subtypes will ultimately require the development of new in vitro and in vivo models.
Mesothelial cells
The mesothelium consists of a layer of epithelial cells that lines the visceral cavities and organs. From the perspective of AT, mesothelial cells are only found in intraperitoneal (for example, omental and mesenteric AT) and intrathoracic (for example, epicardial) depots. These cells are not per se adipogenic92,93 and can be distinguished from ASPCs by the expression of markers such as MSLN and KRT19 (ref. 5). There is conflicting evidence about the role that mesothelial cells play in AT. Although the data indicate that mouse mesothelial cells cannot become adipocytes, single-cell studies have continued to identify a subset of mesothelial cells that share markers with ASPCs and exhibit the potential to transition between the mesothelial and mesenchymal state93,94, suggesting that there is a relationship between these cells that is not yet fully understood.
Immune cells
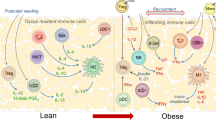
The AT immune compartment identified by expression of CD45 (PTPRC) has representation from all major immune cell types with markers summarized in Tables 2 and 3 (refs. 95,96,97,98,99). Single-cell analyses corroborate experimental studies showing that macrophages, dendritic cells (DCs), monocytes and T lymphocytes are the most abundant immune cells in AT, while B cells, natural killer (NK) cells, mast cells and innate lymphoid cells (ILCs) are comparatively less represented within the immune cell population. There is a disconnect between experimental evidence of adipose neutrophil and eosinophil populations in AT and single-cell studies that show a paucity of these cell types100,101. This disconnect may be due to known limitations in RNA yields from neutrophils in single-cell experiments102,102. Furthermore, blood contamination in highly vascularized AT biopsy samples may lead to increased proportions of blood immune cells such as lymphocytes and monocytes103,104. Distinguishing blood and tissue-resident immune cells in AT is important, as resident AT cells have distinct molecular profiles compared to circulating blood immune cells (for example, regulatory T (Treg) cells)105,106. However, a clear separation of blood and tissue-resident immune cells may be difficult, as monocytes have been reported to have altered profiles as they transit through AT107.
The proportions and features of immune cells in AT differ based on body weight, age, sex and anatomical location. The diversity of AT immune cells and their role in obesity and its complications have been extensively reviewed elsewhere9,108,109,110. Multiple molecular and functional subtypes exist within each broad immune cell type, and a full catalogue of all immune types may require enrichment of certain populations before single-cell profiling93. For example, AT macrophages can be categorized into various subpopulations, such as perivascular macrophages (PVMs), characterized by LYVE1 expression107,111; lipid-associated macrophages (LAMs), marked by CD9 and TREM2 expression112,113; and non-perivascular macrophages (non-PVMs), which include sympathetic nerve-associated macrophages expressing MAOA104,114,115,116. Importantly, the molecular profiles of LAMs and PVMs are seen in other tissue/disease contexts117,118, suggesting convergence of signals leading to specific macrophage activation profiles.
Vascular cells
Vascular cells can be classified into three main types: endothelial cells (ECs), which line the interior of blood vessels; smooth muscle cells (SMCs), which regulate vessel diameter and blood flow; and pericytes, which support and stabilize capillaries and venules.
Blood endothelium
Adipose ECs, like those in other tissues, comprise several functionally and transcriptionally distinct subtypes, which express common markers such as PECAM1 and CDH5 in humans and mice119,120. The subtypes include arterial ECs, expressing markers such as GJA5 and NEBL; venous ECs, expressing ACKR1; and capillary ECs, expressing RBP7, BTNL9 and CA4 (ref. 121). ECs exhibit remarkable plasticity, responding dynamically to changes in their surrounding environment. Integrative data analysis has revealed several additional potential subpopulations of ECs. Some of these subpopulations exhibit hybrid transcriptional profiles, with gene signatures resembling those of adipose monocytes or macrophages, suggesting a possible immunomodulatory role122.
Lymphatic endothelium
Lymphatic vessels, lined by lymphatic endothelial cells, are integral to maintaining fluid balance and conducting immune surveillance within AT. Lymphatic endothelial cells are particularly prevalent in VAT compared to SAT or BAT123. These cells are identifiable by their expression of PROX1 and LYVE1. However, it is important to note that LYVE1 also serves as a marker for specific subsets of macrophages and, therefore, should not be used by itself to identify lymphatic endothelial cells.
Pericytes and SMCs
Pericytes and SMCs, both classified as mural cells, have key roles in forming the structure of blood vessel walls. Pericytes are mainly located around small blood vessels, including capillaries and microvessels, while SMCs are the predominant cells in the walls of arterioles and arteries. Although both cell types express PDGFRB, SMCs in both humans and mice can be further distinguished by their expression of MYH11 and MYOCD, whereas pericytes selectively express STEAP4 (refs. 5,122).
Schwann cells
Schwann cells are glial cells that coat the axon and essential components of the peripheral nerves that innervate AT. They have been captured in some integrated single-cell atlases of AT in humans and mice and are characterized by markers such as XKR4 and MPZ124.
Challenges in AT collection and processing
Proper collection and handling of AT samples are critical to preserve cellular or nuclear integrity and minimize experimental artefacts in single-cell analyses. Factors such as exact anatomical location, collection technique and processing protocol need to be thoroughly considered and accurately documented to ensure the generation of reliable and consistent single-cell data (Fig. 2 and Box 1).
Adipose tissue sampling
Several methods exist for the collection of AT, each potentially influencing downstream analyses. Here we briefly summarize our collective recommendation for AT sampling and storage. Although these recommendations are informed by current scientific understanding and expert insights of the Adipose Biological Network, some aspects may require further investigation and stronger experimental evidence to achieve optimal sampling and storage practices.
The workflow begins with aligning reads to the genome using, for example, Cell Ranger. Ambient RNA is removed using tools such as CellBender, SoupX or DecontX and doublets are identified and removed using scDblFinder, scdx, DoubletFinder or scrublet. Cells and genes are filtered on the basis of QC metrics. Data are then combined and integrated using Seurat or Scanpy, with integration methods such as Harmony, canonical correlation analysis (CCA) and reciprocal principal component analysis (RPCA). Clusters and marker genes are identified using Seurat, Scanpy or Liger. Data can be mapped to reference datasets using Azimuth or CellTypist. Additional analyses include trajectory analysis with slingshot or monocole3, differential expression analysis using pseudobulk, and prediction of cell–cell interactions with CellphoneDB or Cellchat. Listed programmes are suggestions and not exhaustive.
The choice of collection method is usually guided by the location of the depots and should be properly documented (Box 1), as it can help account for potential confounding effects, when analysing the data. In humans, SAT biopsy samples can be acquired using minimally invasive techniques such as needle aspiration, although surgical procedures are typically used to obtain VAT due to its deep location. Most elective intra-abdominal operations are now performed laparoscopically, which limits the amount of tissue that can be collected (approximately 50 mg). However, larger amounts of PVAT can be collected during open heart surgery. An often-overlooked aspect is the impact of surgical procedures on AT125, where factors such as prolonged preoperative fasting, exposure to lower ambient temperatures in the operating room, and the choice of tissue collection methods (for example, aspiration needle, electrocautery or scalpel) may influence gene expression in AT. Furthermore, the administration of oral or topical anaesthetics may also affect gene expression signatures in AT biopsy samples. For isolation of mouse AT, the mouse is typically euthanized using inhalational anaesthetics (for example, isoflurane) in combination with cervical dislocation or a lethal dose of injectable anaesthetics (for example, ketamine). Perfusion can be performed to flush out blood from the AT before harvest, which could be particularly useful when studying tissue-resident immune compartments103,104.
After isolation of AT, it is often necessary to remove excess blood or moisture. However, rinsing the tissue with water or PBS may cause damage during subsequent freezing, as formation of ice crystals can disrupt cell membranes and intracellular structures126. To avoid this, the tissue can be cleaned using clean laboratory wipes. Prompt handling of biopsy material is critical to minimize RNA degradation, and when immediate processing is not possible, the tissue should be frozen down as soon as possible after collection127. The most used method is snap freezing in liquid nitrogen; however, for some downstream assays this method can be problematic, because liquid nitrogen boils in contact with warm tissues and forms a vapour barrier acting as an insulator. As a result, the inner core of the tissue sample may freeze slowly and unevenly. To improve preservation of cellular structures for downstream assays, one can immerse the tissue in pre-cooled isopentane or freeze down tissue pieces in slow-freezing medium containing dimethylsulfoxide128. Additionally, sample integrity may be affected by the storage condition and duration. Although storage at −80 °C may be sufficient for most downstream applications, cryotanks with controlled temperature stability may offer advantages for preservation of nuclear integrity.
Preparation of AT for single-cell/single-nuclei sequencing analyses
AT presents unique challenges for single-cell sequencing technologies, particularly due to the fragile nature of adipocytes. scRNA-seq approaches have been widely used to profile cells originating from the stromal vascular fraction, such as ASPCs, ECs and immune cells1,32,81,83,88,90,97,98,113,129,130,131,132,133. However, capturing all cell types in AT requires snRNA-seq3,5,82,132,134,135,136 (Fig. 2). The pros and cons of scRNA-seq versus snRNA-seq for AT studies have been discussed in detail elsewhere7,132,134, and the method of choice depends heavily on the specific research question. For example, snRNA-seq is necessary to map the transition from preadipocytes to fully mature adipocytes during adipogenesis3,132.
Single-cell analyses
Various methods have been used for dissociating AT for single-cell analyses, including enzymatic, mechanical and automated methods81,97,127,137,138,139. Careful consideration is needed regarding differences in the types and proportions of subpopulations that are recovered across these methods, particularly when integrating and comparing various datasets. Gentle dissociation methods are preferred for maintenance of cellular integrity, whereas harsher techniques may be required for enriching certain cell populations, such as vascular cells, thereby potentially compromising the integrity of other cell types. Several single-cell studies of populations from AT use fluorescence-activated cell sorting (FACS) to enrich specific subpopulations or rare cell types93,98,113,129,130,133,140. In these studies, it is important to consider the potential introduction of bias in the cellular fractions being analysed as well as potential effects of FACS on cell integrity and features. In all instances, meticulous quality-control (QC) measures are required to guarantee optimal cell quality during sample preparation.
Single-nucleus analyses
Nuclei are fragile structures that can be easily damaged during isolation, necessitating careful handling and the use of appropriate methods and buffers to maintain their integrity137. Furthermore, the use of low-protein-binding tubes and pipette tips during the nuclei extraction process is strongly recommended to minimize contamination from oil droplets and residual tissue fragments. Importantly, contamination with cytoplasmic components, organelles and debris from ruptured cells can compromise the purity of the isolated nuclei and impact subsequent applications. Pulverization of the tissue is occasionally preferred to achieve a more homogeneous nuclei preparation by effectively releasing cells embedded in the extracellular matrix.
However, in recent years, automated systems, such as the gentle MACS Dissociator, have gained popularity for isolating high-quality nuclei from AT127. These systems combine mechanical agitation and enzymatic digestion, offering standardized and reproducible dissociation that enhances experimental consistency. Regardless of the approach, customizing dissociation protocols to accommodate structural variations in AT, such as those seen in obesity, might be needed to ensure sufficient yield of high-quality and representative nuclei for downstream analyses137. A major challenge with snRNA-seq is that the enclosed RNA becomes increasingly susceptible to RNases, resulting in rapid RNA degradation. A recent study, however, showed that using a combination of vanadyl ribonucleoside complex and recombinant RNase inhibitors during nuclei isolation from AT preserves RNA integrity and nuclear structure, resulting in high-quality snRNA-seq data with high unique molecular identifier (UMI) counts and minimal ambient RNA contamination141. Another strategy that has been shown to increase the quality of nuclei and reduce ambient RNA load for snRNA-seq analyses is fluorescence-activated nuclear sorting127. However, sorting leads to loss of nuclei, which could introduce compositional biases as well as challenges in the analysis of biopsy samples with limited material.
Computational issues related to the analysis of AT single-cell data
Comprehensive guides on best practices for the computational processing of single-cell data in general are available142,143. Here we describe the major issues, with emphasis on how they relate to the analysis of AT data142,143 (Fig. 3).
As described above, most AT studies use snRNA-seq because of its ability to simultaneously capture information on all cells, including mature adipocytes. The use of nuclear data requires stricter QC cut-offs for mitochondrial reads than are commonly used in scRNA-seq, as these should not be present in nuclear preparation144. Fairly strict cut-offs for ribosomal reads should also be used. Additionally, it is important not to discard reads that map to intronic regions, as these can constitute up to 50% of the reads in a snRNA-seq dataset, compared to ~10% in a scRNA-seq experiment145. Because the cell membrane is destroyed to isolate nuclei, snRNA-seq data tend to be more susceptible to artefacts from ambient RNA than scRNA-seq. Although this should be addressed experimentally by using extra washes and/or FACS sorting, computational methods can also be used to clean and adjust ambient reads from datasets142.
Doublet removal is a major challenge in analysing AT datasets (Fig. 3). Several algorithms that detect ‘within-sample’ doublets are available in both Python and R146, and at least one of these should be applied to all datasets. Additionally, if samples are multiplexed, demultiplexing using genetic or hashtag identifiers can also detect sample-to-sample doublets. After an initial round of doublet removal, we recommend annotating cells with the ‘doublet score’ calculated by the detection software and plotting this along with other QC metrics such as UMI number and mitochondrial percentage; cells with higher doublet scores will often cluster together and can be removed. Users should be mindful that overly stringent doublet removal might eliminate transition states or other bona fide cell types. If a cluster is suspected to represent a novel cell type or state, this can be supported by the presence of unique markers that are not present in either of the cell types that make up the putative ‘doublet’, as well as by staining for markers in tissue sections.
Benchmarking analysis comparing ASPC gene expression in whole-cell and single-nuclear datasets has shown that similar populations are retrieved using both methods, although there is less RNA diversity in nuclear data145. This can affect the accuracy of quantifying transcript abundances and the ability to detect low abundance transcripts. Because there is a higher ratio of unspliced-to-spliced transcripts in the nucleus compared to the whole cell, there is increased length bias in transcript detection for snRNA-seq compared to scRNA-seq, leading to, for example, enrichment of long noncoding RNAs in snRNA-seq145. Additionally, because of the differences in spliced RNA content in the nucleus versus the whole cell, certain analytical approaches, such as trajectory and velocity analyses, which rely on the ratio of spliced-versus-unspliced RNA, may require adaptation.
Most single-cell AT datasets have been obtained using the droplet-based 10x Genomics platform3,5,60,82,97,98,133,136,147 and some initial integration efforts have been made124,134,148. More recently, however, datasets acquired using SMART-seq technology have been published, offering full-length and more comprehensive transcript coverage that can serve as a complementary resource for characterizing transcript variants of interest associated with AT149. However, this comes at the expense of reduced capacity to detect rare cellular subpopulations due to the ~10-fold reduction in numbers of profiled cells compared to 10x technology32,132,135. It is important to note that the platforms used may limit the computational tools available to clean the data. For example, SMART-seq-based methods do not automatically sequence ‘empty’ wells, which are typically required to estimate and remove ambient RNA.
Accurate classification and annotation of cell types in AT are crucial for QC and downstream analysis. Several automated correlation-based annotation tools, such as Azimuth150 and CellTypist151 are effective for annotating datasets that are not expected to contain novel cell types. These methods are built on existing reference datasets5,134, which limits their use for benchmarking previously uncharacterized tissues and annotating rare cell types. Similarly, supervised classification-based methods such as CellAssign152 and Garnett153 use deep learning to annotate cells, which also requires good-quality reference single-cell datasets containing all expected cell types.
These tools can be useful for datasets in which background noise or batch effects make other classification methods problematic. Most other methods rely on marker gene expression to annotate each cell, which has the advantage of not needing a reference dataset and, therefore, more easily allowing for the discovery of novel cell types. However, the expression of many genes can vary considerably due to the inherent heterogeneity across different samples, potentially leading to variable annotations of the same cell type. This inconsistency in cell-type labelling across different datasets can hinder comparative analyses and data integration efforts, underscoring the need for the standardized naming conventions that we are working towards as a network.
One of the goals of scRNA-seq and snRNA-seq experiments is the identification of new cell types or subtypes, and any new cell type identified in a dataset should be rigorously validated, both computationally and experimentally. A common pitfall is the annotation of a doublet population as a novel cell population. Groups should, therefore, ensure that any observed novel population expresses marker genes that are specific to that population and should validate these marker genes using complementary techniques such as immunohistochemistry, immunocytochemistry and/or flow cytometry.
Integration of data is an important step in creating a usable dataset and is especially important in the development of an atlas based on data from multiple laboratories using different protocols. Multiple variables must be taken into consideration when integrating, most notably the platform and method (for example, nuclear versus whole cell). Differences in cohort composition, batches, tissue lysis and single-nucleus/cell isolation method, library preparation, sequencing platform, sequencing depth and RNA diversity in differing methodologies can lead to difficulties in harmonizing data. Various integration methods can be explored to assess their impact on the results154; however, when comparing two highly distinct datasets, it may be essential to use techniques such as reference mapping150,155, which enables the comparison of cells across datasets while preserving the unique analysis of the component datasets.
Because a fully integrated adipose atlas will contain data from multiple depots, there is concern that data integration may inadvertently overcorrect true biological differences between depots. To preserve depot-specific features in an individual study, when multiple depot samples (for example, SAT and VAT) are available from the same individual, they should be aggregated at the individual level before integrating across different individuals. Efforts should be made to collect data for every depot in at least three laboratories to help identify the differences between datasets that could be lab-versus-depot driven.
Similarly, data integration for comparisons of variables across individuals can lead to overcorrection of biological differences. One way to address this issue is by combining multiple samples into the same run and computationally demultiplexing using genotype information156,157, or by multiplexing using barcoded antibodies127 and using run information as the integrating variable. Pseudobulking, that is, averaging/aggregating expression profiles across biological replicates142,158, effectively reduces over-integration artefacts but sacrifices information that might be gleaned from intra-sample variation. Another approach is to extend deep learning algorithms often used in the analysis of single-cell transcriptomes159 to explicitly model donor identity and intra-donor variation160.
Incorporation of bulk RNA-seq data
Single-cell/single-nucleus analyses are still performed on relatively small numbers of samples, although translating the knowledge gained by these approaches to the clinic requires validation in large (or larger) cohorts. snRNA-seq data can help identify novel putative molecular markers that can then be tested individually using dedicated approaches for validation (PCR of specific cell-type markers, as an example). Complementarily, deconvolution algorithms can be used to deduce cell-type proportions from bulk RNA-seq161, some of which are designed to use gene expression derived from snRNA-seq data to provide reliable estimates of cell-type composition of human AT162. As with annotation, the success of these techniques relies on a robust adipose reference atlas and a consensus definition of cell types.
Adipose Atlas 1.0
Choosing datasets
The primary objective of Adipose Atlas 1.0 is to create a comprehensive and inclusive representation of the major AT depots of the human body. On the basis of availability, we will include high-quality datasets from abdominal visceral, abdominal subcutaneous, and superficial and deep neck AT, ensuring representation of both WAT and BAT and independent replicates from different laboratories. The atlas will ideally include samples from both sexes across a wide range of ages. In addition, we will include diverse ethnic and racial backgrounds, when possible, although currently available datasets are somewhat limited in this regard. This information will be included in the metadata, allowing users to easily split and analyse the dataset on the basis of these key variables. We will further strive to include individuals across a broad spectrum of body mass index (BMI) categories to ensure that the atlas captures the heterogeneity of AT in different metabolic states. Pronounced differences have been observed between male and female AT163, which imposes an important analytical challenge to seamlessly integrate datasets across both sexes. Whenever possible, data on lifestyle and social determinants of health, such as diet, activity, occupation, socioeconomic status, income, education and domestic status164, will also be included for humans.
To ensure comprehensive representation of all cell types in the tissue, we will include both snRNA-seq and scRNA-seq data, despite the integration challenges this may pose. Such challenges arise from variations in the mRNA profiles between cells and nuclei, spliced-versus-unspliced RNA content, differences in isolation techniques for snRNA-seq and scRNA-seq, and disparities in the capture efficiency and sensitivity of the two approaches. We will only include studies analysing a meaningful number of cells or nuclei, as a robust sample size is essential to mitigate potential batch effects. The initial focus will be on datasets that do not involve enrichment of specific cellular subpopulations. Given that most data have been generated using the 10x Genomics platform, we will begin by integrating these datasets, with plans to incorporate technologies offering full transcript coverage at a later stage. Although we recognize the growing interest in spatial and multi-omics approaches, the initial atlas will concentrate solely on RNA-seq data.
Metadata—required and suggested
An important task of the HCA Adipose Bionetwork is to ensure that the data are accessible and useful for groups with a wide range of interests. In this regard, we will collect comprehensive metadata for AT samples involved in the HCA integration efforts. It is, therefore, important to develop clear guidelines for reporting these metadata. This includes the exact anatomical location of the biopsy (Box 1). Currently, there is no consensus on the optimal biopsy site within a depot, such as the omentum, which exhibits considerable regional structural variability. Providing more detailed information on the biopsy site can aid data integration and comparison and offer a more comprehensive overview of subpopulations within the different adipose depots. In addition, specifics should be provided on sample acquisition, including the method of sample collection, storage and handling.
In addition to providing detailed information on the sample itself, individual-level metadata focused on physiological and metabolic parameters relevant to AT will ideally also be collected for these studies. These metadata would include anthropometric data, such as age, sex, weight, height, BMI, waist, waist–hip ratio and body fat percentage (with method specified), as well as information on disease states (for example, T2D, polycystic ovarian syndrome and cardiovascular disease), a comprehensive medication list and relevant blood work such as fasting glucose and insulin as well as lipid panels.
Information on the conditions at the time of sample collection such as the feeding status of the donor (duration of fast) at the time of biopsy acquisition, ambient temperature at the time of collection, and time of day and season should also be collected. If the study allows for collection of peripheral blood that can be banked along with the samples, it will allow for queries of factors such as circulating leptin and adiponectin levels as hypotheses arise. Although it is unlikely that all recommended metadata will be available for all studies, efforts should be made to collect whatever is possible during study design. When Adipose Atlas 1.0 is created, we will include the metadata that are available and make a note of which studies the metadata come from to assist with interpretation of the datasets.
Data sharing
As groups generate ever-larger sequencing datasets, one question is how best to share data with the community. Although sharing raw FASTQ files complies with the data sharing policies of many journals, these are often too large and computationally onerous for less bioinformatically inclined groups to query for genes and cell types of interest. Therefore, data should ideally be shared both as raw FASTQ files and as processed data matrices and/or data objects such as RDS files. The HCA Data Repository (https://data.humancellatlas.org/contribute/) offers a platform for sharing FASTQ files and associated metadata for HCA datasets. Additionally, the HCA operates the Cell Annotation Platform (https://celltype.info/), which stores matrices and cell annotation metadata in a specialized portal that enables collaborative data annotation within the HCA community. Other similar platforms for querying single-cell datasets online include CZI’s CellxGene (https://cellxgene.cziscience.com/) and the Broad Institute’s Single Cell Portal (https://singlecell.broadinstitute.org/single_cell/). The Adipose Biological Network will actively be using these platforms for our atlases to ensure comprehensive data sharing and accessibility within the scientific community.
Ethical and societal considerations
As spatial and single-cell technologies advance, consideration must be given to data privacy and ethical use of the detailed molecular data. Ensuring participant privacy and consent is paramount. Raw FASTQ level data can be used to identify a participant165, and some of the metadata described above may also be specific enough to identify an individual participant. In some regions, rules are already in place that restrict the sharing of raw data and metadata reporting with specific demographic information. The HCA data ingestion process is designed to comply with these laws, and groups that prefer to share their raw data through controlled-access databases can do so via the HCA Data Repository or dbGaP, which is operated by the National Center for Biotechnology Information. Groups that cannot share specific demographic information due to concerns about participant privacy should bin these values and report the appropriate range for each individual (for example, a 49-year-old individual could be reported as being aged 45–50).
To avoid bias and ensure that findings are broadly applicable, it is important to promote equity in research by ensuring access to advanced technologies and inclusion of diverse populations in AT research. When possible, researchers should recruit individuals from a wide demographic range. One of the missions of the HCA is to collect data from researchers around the world, ensuring diversity in both participants and the researchers themselves.
Atlas 2.0 and beyond
Although the HCA Adipose Atlas 1.0 is focused on currently available snRNA-seq datasets from healthy individuals, we anticipate that future versions of the Adipose Atlas will include more diverse datasets, comprising different adipose depots, genetically diverse cohorts and a variety of physiological and pathological conditions. Other modalities, such as spatial transcriptomics, and single-cell epigenomics, proteomics and metabolomics will also feature in later atlases. This should also include cellular trajectories, including modelling of adipogenesis166,167,168,169,170 and activation of immune cells.
Moreover, the human atlas will become more functional and interpretable when comparable atlases are developed for other species, especially for emerging model organisms171 and species of agricultural importance172, which will also help to evaluate cross-species similarities and differences. Furthermore, adipocytes are not necessarily confined to specific adipose depots but can also be found in a variety of other tissues. As snRNA-seq datasets emerge for more tissues, such as breast173 and skin174,175, we will learn more about adipocytes residing in these tissues. Finally, as mentioned previously, all of these huge datasets need to be accessible widely across the data sharing options suggested above to allow for exploring and querying single-cell datasets.
Metabolic and disease states
Most studies to date have used tissue biopsy samples obtained in the fasting state. However, AT is highly responsive to the dynamic hormonal and metabolic signals in the postprandial state, and future studies should also probe the impact of metabolic state and nutrition on the adipose transcriptome and cellular composition. Recent longitudinal snRNA-seq studies have addressed the plasticity of human AT in response to short-term and long-term weight loss176,177, revealing an obesogenic memory that may be driven by stable epigenetic changes in certain AT cell types177. These findings warrant further investigation at the single-cell level, which could be incorporated into future versions of the atlases. AT also needs to be studied across the entire life course, as some studies have suggested that AT may be a sentinel tissue for changes associated with ageing135,178. Furthermore, AT dysfunction is one of the main factors contributing to variability in metabolic disease risk, including T2D, cardiovascular disease, polycystic ovarian syndrome and other endocrine and reproductive disorders. Although the initial version of the adipose atlas will focus on tissue from healthy individuals across a wide range of BMIs, one of the intended uses of this atlas is to enable comparison with AT during disease progression. To this end, future expansions of the atlas will incorporate recent single-cell studies that have uncovered associations between specific AT cell types and the severity of certain metabolic disease94. In addition to metabolic diseases, future atlases might encompass data from liposarcoma samples, which are malignancies of mesenchymal origin with elements of adipocytic differentiation and unique gene signatures compared to normal AT179.
Emerging technologies
Many new technologies are being utilized to query tissues and organs at single-cell resolution, including proteomics, metabolomics and epigenomics coupled with spatial mapping and profiling of cell types. Advances in single-cell epigenomics have enabled the mapping of accessible genomic regions at single-nucleus resolution using the assay for transposase-accessible chromatin with sequencing (ATAC-seq)1,180,181,182, despite persistent technical challenges with frozen human AT. DNA methylation can also be assessed at the single-cell level, allowing exploration of how environmental factors such as diet or obesity influence epigenetic states in cellular subpopulations183. Furthermore, future advances in single-cell long-read sequencing are expected to enhance chromatin accessibility and DNA methylation profiling, enabling simultaneous profiling of single-nucleotide polymorphisms, large-scale genetic alterations and epigenetic modifications184.
Some studies have already explored the transcriptional profile of AT using spatial approaches10,134,185,186, although most of these studies lack single-cell resolution. New sequencing-based and bioimaging-based technologies now allow the study of spatial cell organization at single-cell and subcellular levels, enhancing our understanding of how spatial relationships between cell types in the adipose niche affect tissue function187.
Integrating various single-cell data modalities holds the potential to enhance our understanding of how different molecular layers interact in AT biology. For example, combining snRNA-seq and single-nucleus ATAC-seq to detect both the transcriptome and epigenome within the same cell is a powerful approach for inferring gene regulatory networks and causal transcription factors controlling AT function1. Additionally, scRNA-seq has also recently been combined with cellular indexing of transcriptomes and epitopes by sequencing (CITE-seq) to interrogate immunometabolic adaptations in mouse AT during weight loss188.
Efforts to integrate various multi-omics data types from the same cell still face considerable technological and computational challenges, including issues related to data alignment, synchronization and batch effects. Future efforts by members of the HCA and the Adipose Bionetwork will, therefore, also focus on developing improved techniques for simultaneous or sequential multi-omics measurements, as well as sophisticated computational tools for data integration and analysis.
Conclusion
Over the past few decades, AT has emerged as a central player in mammalian metabolism, with important roles in normal physiology and in disease. As a complex, heterogeneous, multifocal and dynamic tissue, AT needs to be understood at single-cell resolution. Despite the technical challenges associated with this fatty tissue, the field has made substantial progress by generating comprehensive cellular atlases of AT, identifying novel cell types and states, and determining the physical and functional relationships between cell types. The adipose research community has begun to establish standards for depot and cellular nomenclature, and for experimental and computational best practices. Going forward, there will be considerable efforts throughout the HCA Bionetwork system and within the Adipose Bionetwork to harmonize data and achieve consensus on cell annotation. As a bionetwork, we hope to establish resources that will enable data mining by experts and non-experts alike to usher in a new paradigm for AT research.
References
Rondini, E. A., Ramseyer, V. D., Burl, R. B., Pique-Regi, R. & Granneman, J. G. Single cell functional genomics reveals plasticity of subcutaneous white adipose tissue (WAT) during early postnatal development. Mol. Metab. 53, 101307 (2021).
Holman, C. D. et al. Aging impairs cold-induced beige adipogenesis and adipocyte metabolic reprogramming. eLife https://doi.org/10.7554/eLife.87756 (2024).
Sarvari, A. K. et al. Plasticity of epididymal adipose tissue in response to diet-induced obesity at single-nucleus resolution. Cell Metab. 33, 437–453 (2021). This snRNA-seq study uncovers the adaptive responses of epididymal AT to diet-induced obesity in mice, providing insights into cellular heterogeneity and adipogenic trajectories.
Yang, J. et al. Single-cell dissection of the obesity-exercise axis in adipose-muscle tissues implies a critical role for mesenchymal stem cells. Cell Metab. 34, 1578–1593 (2022).
Emont, M. P. et al. A single-cell atlas of human and mouse white adipose tissue. Nature 603, 926–933 (2022). This study presents a comprehensive single-cell atlas of subcutaneous and visceral WAT in mouse and human across a range of body weights.
Lundgren, P. et al. A subpopulation of lipogenic brown adipocytes drives thermogenic memory. Nat. Metab. 5, 1691–1705 (2023).
Maniyadath, B., Zhang, Q., Gupta, R. K. & Mandrup, S. Adipose tissue at single-cell resolution. Cell Metab. 35, 386–413 (2023). This review provides a comprehensive overview of mouse and human AT biology at single-cell resolution.
Garg, A. Acquired and inherited lipodystrophies. N. Engl. J. Med. 350, 1220–1234 (2004).
Chavakis, T., Alexaki, V. I. & Ferrante, A. W. Jr. Macrophage function in adipose tissue homeostasis and metabolic inflammation. Nat. Immunol. 24, 757–766 (2023).
Backdahl, J. et al. Spatial mapping reveals human adipocyte subpopulations with distinct sensitivities to insulin. Cell Metab. 33, 1869–1882 (2021). This study combines scRNA-seq and spatial mapping to identify distinct human adipocyte subpopulations with varying levels of insulin responsiveness.
Fox, C. S. et al. Abdominal visceral and subcutaneous adipose tissue compartments: association with metabolic risk factors in the Framingham Heart Study. Circulation 116, 39–48 (2007).
Gupta, O. T. & Gupta, R. K. The expanding problem of regional adiposity: revisiting a 1985 diabetes classic by Ohlson et al. Diabetes 73, 649–652 (2024).
Neeland, I. J. et al. Visceral and ectopic fat, atherosclerosis, and cardiometabolic disease: a position statement. Lancet Diabetes Endocrinol. 7, 715–725 (2019).
Karpe, F. & Pinnick, K. E. Biology of upper-body and lower-body adipose tissue–link to whole-body phenotypes. Nat. Rev. Endocrinol. 11, 90–100 (2015).
Cohen, P. & Kajimura, S. The cellular and functional complexity of thermogenic fat. Nat. Rev. Mol. Cell Biol. 22, 393–409 (2021).
Sharma, A. K., Khandelwal, R. & Wolfrum, C. Futile lipid cycling: from biochemistry to physiology. Nat. Metab. 6, 808–824 (2024).
Ishibashi, J. & Seale, P. Medicine. Beige can be slimming. Science 328, 1113–1114 (2010).
Seale, P. et al. Prdm16 determines the thermogenic program of subcutaneous white adipose tissue in mice. J. Clin. Invest. 121, 96–105 (2011).
Petrovic, N. et al. Chronic peroxisome proliferator-activated receptor gamma (PPARγ) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. J. Biol. Chem. 285, 7153–7164 (2010).
Wu, J. et al. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell 150, 366–376 (2012).
Cypess, A. M. et al. Anatomical localization, gene expression profiling and functional characterization of adult human neck brown fat. Nat. Med. 19, 635–639 (2013).
Virtanen, K. A. et al. Functional brown adipose tissue in healthy adults. N. Engl. J. Med. 360, 1518–1525 (2009).
Cypess, A. M. et al. Identification and importance of brown adipose tissue in adult humans. N. Engl. J. Med. 360, 1509–1517 (2009).
van Marken Lichtenbelt, W. D. et al. Cold-activated brown adipose tissue in healthy men. N. Engl. J. Med. 360, 1500–1508 (2009).
Saito, M. et al. High incidence of metabolically active brown adipose tissue in healthy adult humans: effects of cold exposure and adiposity. Diabetes 58, 1526–1531 (2009).
Zingaretti, M. C. et al. The presence of UCP1 demonstrates that metabolically active adipose tissue in the neck of adult humans truly represents brown adipose tissue. FASEB J. 23, 3113–3120 (2009).
Zhang, F. et al. An adipose tissue atlas: an image-guided identification of human-like BAT and beige depots in rodents. Cell Metab. 27, 252–262 (2018).
de Jong, J. M., Larsson, O., Cannon, B. & Nedergaard, J. A stringent validation of mouse adipose tissue identity markers. Am. J. Physiol. Endocrinol. Metab. 308, E1085–E1105 (2015).
Borgeson, E., Boucher, J. & Hagberg, C. E. Of mice and men: pinpointing species differences in adipose tissue biology. Front Cell Dev. Biol. 10, 1003118 (2022).
Manolopoulos, K. N., Karpe, F. & Frayn, K. N. Gluteofemoral body fat as a determinant of metabolic health. Int. J. Obes. 34, 949–959 (2010).
Casey, B. A., Kohrt, W. M., Schwartz, R. S. & Van Pelt, R. E. Subcutaneous adipose tissue insulin resistance is associated with visceral adiposity in postmenopausal women. Obesity 22, 1458–1463 (2014).
Divoux, A. et al. Distinct subpopulations of human subcutaneous adipose tissue precursor cells revealed by single-cell RNA sequencing. Am. J. Physiol. Cell Physiol. 326, C1248–C1261 (2024).
Hauner, H. & Entenmann, G. Regional variation of adipose differentiation in cultured stromal-vascular cells from the abdominal and femoral adipose tissue of obese women. Int. J. Obes. 15, 121–126 (1991).
Markman, B. & Barton, F. E. Jr. Anatomy of the subcutaneous tissue of the trunk and lower extremity. Plast. Reconstr. Surg. 80, 248–254 (1987).
Marinou, K. et al. Structural and functional properties of deep abdominal subcutaneous adipose tissue explain its association with insulin resistance and cardiovascular risk in men. Diabetes Care 37, 821–829 (2014).
Tordjman, J. et al. Structural and inflammatory heterogeneity in subcutaneous adipose tissue: relation with liver histopathology in morbid obesity. J. Hepatol. 56, 1152–1158 (2012).
Golan, R. et al. Abdominal superficial subcutaneous fat: a putative distinct protective fat subdepot in type 2 diabetes. Diabetes Care 35, 640–647 (2012).
Wollina, U., Wetzker, R., Abdel-Naser, M. B. & Kruglikov, I. L. Role of adipose tissue in facial aging. Clin. Inter. Aging 12, 2069–2076 (2017).
Naldaiz-Gastesi, N., Bahri, O. A., Lopez de Munain, A., McCullagh, K. J. A. & Izeta, A. The panniculus carnosus muscle: an evolutionary enigma at the intersection of distinct research fields. J. Anat. 233, 275–288 (2018).
Kruglikov, I. L. & Scherer, P. E. Dermal adipocytes: from irrelevance to metabolic targets? Trends Endocrinol. Metab. 27, 1–10 (2016).
Nicu, C. et al. A guide to studying human dermal adipocytes in situ. Exp. Dermatol. 27, 589–602 (2018).
Eder, P., Adler, M., Dobrowolska, A., Kamhieh-Milz, J. & Witowski, J. The role of adipose tissue in the pathogenesis and therapeutic outcomes of inflammatory bowel disease. Cells https://doi.org/10.3390/cells8060628 (2019).
Zhang, H. et al. Characteristics of mesenteric adipose tissue attached to different intestinal segments and their roles in immune regulation. Am. J. Physiol. Gastrointest. Liver Physiol. 322, G310–G326 (2022).
Meza-Perez, S. & Randall, T. D. Immunological functions of the omentum. Trends Immunol. 38, 526–536 (2017).
Grigoras, A. et al. Perirenal adipose tissue-current knowledge and future opportunities. J. Clin. Med. https://doi.org/10.3390/jcm10061291 (2021).
Jespersen, N. Z. et al. Heterogeneity in the perirenal region of humans suggests presence of dormant brown adipose tissue that contains brown fat precursor cells. Mol. Metab. 24, 30–43 (2019).
Krieg, L. et al. Multiomics reveal unique signatures of human epiploic adipose tissue related to systemic insulin resistance. Gut 71, 2179–2193 (2022).
Cao, H. et al. Periprostatic adipose tissue: a new perspective for diagnosing and treating prostate cancer. J. Cancer 15, 204–217 (2024).
Corradi, D. et al. The ventricular epicardial fat is related to the myocardial mass in normal, ischemic and hypertrophic hearts. Cardiovasc. Pathol. 13, 313–316 (2004).
Willar, B., Tran, K. V. & Fitzgibbons, T. P. Epicardial adipocytes in the pathogenesis of atrial fibrillation: an update on basic and translational studies. Front. Endocrinol. 14, 1154824 (2023).
Cheung, L. et al. Human mediastinal adipose tissue displays certain characteristics of brown fat. Nutr. Diabetes 3, e66 (2013).
Dixit, V. D. Thymic fatness and approaches to enhance thymopoietic fitness in aging. Curr. Opin. Immunol. 22, 521–528 (2010).
Scheller, E. L., Cawthorn, W. P., Burr, A. A., Horowitz, M. C. & MacDougald, O. A. Marrow adipose tissue: trimming the fat. Trends Endocrinol. Metab. 27, 392–403 (2016).
Suchacki, K. J., Cawthorn, W. P. & Rosen, C. J. Bone marrow adipose tissue: formation, function and regulation. Curr. Opin. Pharmacol. 28, 50–56 (2016).
Sundaram, V. K. et al. Adipo-glial signaling mediates metabolic adaptation in peripheral nerve regeneration. Cell Metab. 35, 2136–2152 (2023).
Kothari, C., Diorio, C. & Durocher, F. The importance of breast adipose tissue in breast cancer. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21165760 (2020).
Xu, B. & Ghossein, R. A. Crucial parameters in thyroid carcinoma reporting - challenges, controversies and clinical implications. Histopathology 72, 32–39 (2018).
Antoniades, C. et al. Perivascular adipose tissue as a source of therapeutic targets and clinical biomarkers. Eur. Heart J. 44, 3827–3844 (2023).
Khoo, T. K. & Bahn, R. S. Pathogenesis of Graves’ ophthalmopathy: the role of autoantibodies. Thyroid 17, 1013–1018 (2007).
Tang, S. et al. Single-cell atlas of human infrapatellar fat pad and synovium implicates APOE signaling in osteoarthritis pathology. Sci. Transl. Med. 16, eadf4590 (2024).
Goodpaster, B. H., Bergman, B. C., Brennan, A. M. & Sparks, L. M. Intermuscular adipose tissue in metabolic disease. Nat. Rev. Endocrinol. 19, 285–298 (2023).
Ogawa, M., Lester, R., Akima, H. & Gorgey, A. S. Quantification of intermuscular and intramuscular adipose tissue using magnetic resonance imaging after neurodegenerative disorders. Neural Regen. Res. 12, 2100–2105 (2017).
Ford, H., Liu, Q., Fu, X. & Strieder-Barboza, C. White adipose tissue heterogeneity in the single-cell era: from mice and humans to cattle. Biology https://doi.org/10.3390/biology12101289 (2023).
Pettenuzzo, S. et al. Mechanical behaviour of plantar adipose tissue: from experimental tests to constitutive analysis. Bioengineering https://doi.org/10.3390/bioengineering11010042 (2023).
Becher, T. et al. Brown adipose tissue is associated with cardiometabolic health. Nat. Med. 27, 58–65 (2021).
Lidell, M. E. et al. Evidence for two types of brown adipose tissue in humans. Nat. Med. 19, 631–634 (2013).
Pino, M. F., Divoux, A., Simmonds, A. V., Smith, S. R. & Sparks, L. M. Investigating the effects of Orexin-A on thermogenesis in human deep neck brown adipose tissue. Int. J. Obes. 41, 1646–1653 (2017).
Jespersen, N. Z. et al. A classical brown adipose tissue mRNA signature partly overlaps with brite in the supraclavicular region of adult humans. Cell Metab. 17, 798–805 (2013).
Xue, R. et al. Clonal analyses and gene profiling identify genetic biomarkers of the thermogenic potential of human brown and white preadipocytes. Nat. Med. 21, 760–768 (2015).
Chi, J. et al. Three-dimensional adipose tissue imaging reveals regional variation in beige fat biogenesis and PRDM16-dependent sympathetic neurite density. Cell Metab. 27, 226–236 (2018).
Bagchi, D. P. & MacDougald, O. A. Identification and dissection of diverse mouse adipose depots. J. Vis. Exp. https://doi.org/10.3791/59499 (2019).
Chen, S. X., Zhang, L. J. & Gallo, R. L. Dermal white adipose tissue: a newly recognized layer of skin innate defense. J. Invest. Dermatol. 139, 1002–1009 (2019).
Hu, L. et al. Characterization of a novel adipose tissue located between abdominal lymph nodes and cervix/prostate in mice. Am. J. Physiol. Endocrinol. Metab. 327, E134–E144 (2024).
Yamaguchi, Y. et al. Adipogenesis and epicardial adipose tissue: a novel fate of the epicardium induced by mesenchymal transformation and PPARγ activation. Proc. Natl Acad. Sci. USA 112, 2070–2075 (2015).
Roh, H. C. et al. Warming induces significant reprogramming of beige, but not brown, adipocyte cellular identity. Cell Metab. 27, 1121–1137 (2018).
Hagberg, C. E. & Spalding, K. L. White adipocyte dysfunction and obesity-associated pathologies in humans. Nat. Rev. Mol. Cell Biol. 25, 270–289 (2024).
Sakers, A., De Siqueira, M. K., Seale, P. & Villanueva, C. J. Adipose-tissue plasticity in health and disease. Cell 185, 419–446 (2022).
Sun, W., Modica, S., Dong, H. & Wolfrum, C. Plasticity and heterogeneity of thermogenic adipose tissue. Nat. Metab. 3, 751–761 (2021).
Abdul Majeed, S. et al. COBL, MKX and MYOC are potential regulators of brown adipose tissue development associated with obesity-related metabolic dysfunction in children. Int. J. Mol. Sci. https://doi.org/10.3390/ijms24043085 (2023).
Pilkington, A. C., Paz, H. A. & Wankhade, U. D. Beige adipose tissue identification and marker specificity-overview. Front. Endocrinol. 12, 599134 (2021).
Rajbhandari, P. et al. Single cell analysis reveals immune cell-adipocyte crosstalk regulating the transcription of thermogenic adipocytes. eLife https://doi.org/10.7554/eLife.49501 (2019).
Sun, W. et al. snRNA-seq reveals a subpopulation of adipocytes that regulates thermogenesis. Nature 587, 98–102 (2020). This study identifies a rare, temperature-induced subpopulation of adipocytes capable of regulating the thermogenic capacity of neighbouring adipocytes.
Lazarescu, O. et al. Human subcutaneous and visceral adipocyte atlases uncover classical and nonclassical adipocytes and depot-specific patterns. Nat. Genet. https://doi.org/10.1038/s41588-024-02048-3 (2025).
Burl, R. B. et al. Deconstructing adipogenesis induced by beta3-adrenergic receptor activation with single-cell expression profiling. Cell Metab. 28, 300–309 (2018).
Lee, Y. H., Petkova, A. P., Mottillo, E. P. & Granneman, J. G. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metab. 15, 480–491 (2012).
Cannavino, J. & Gupta, R. K. Mesenchymal stromal cells as conductors of adipose tissue remodeling. Genes Dev. 37, 781–800 (2023).
Stefkovich, M., Traynor, S., Cheng, L., Merrick, D. & Seale, P. Dpp4+ interstitial progenitor cells contribute to basal and high fat diet-induced adipogenesis. Mol. Metab. 54, 101357 (2021).
Merrick, D. et al. Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science https://doi.org/10.1126/science.aav2501 (2019). This study identifies a hierarchical structure of mesenchymal progenitor cells in mouse AT with DPP4+ multipotent interstitial progenitors serving as the starting point.
Dong, H. et al. Identification of a regulatory pathway inhibiting adipogenesis via RSPO2. Nat. Metab. 4, 90–105 (2022).
Schwalie, P. C. et al. A stromal cell population that inhibits adipogenesis in mammalian fat depots. Nature 559, 103–108 (2018). This study identifies Areg cells in mice, which exhibit paracrine inhibitory effects on adipogenesis.
Zachara, M. et al. Mammalian adipogenesis regulator (Areg) cells use retinoic acid signalling to be non- and anti-adipogenic in age-dependent manner. EMBO J. 41, e108206 (2022).
Westcott, G. P. et al. Mesothelial cells are not a source of adipocytes in mice. Cell Rep. 36, 109388 (2021).
Ferrero, R. et al. A human omentum-specific mesothelial-like stromal population inhibits adipogenesis through IGFBP2 secretion. Cell Metab. https://doi.org/10.1016/j.cmet.2024.04.017 (2024). This review represents an initial effort to compare and annotate adipocyte stem and progenitor cells across multiple single-cell studies.
Reinisch, I. et al. Unveiling adipose populations linked to metabolic health in obesity. Cell Metab. 37, 640–655 (2025). This study presents an AT single-cell atlas of metabolically healthy and unhealthy obesity, linking mesothelial cells to VAT dysfunction and metabolic disease.
Hernandez, J. D. et al. Linking adipose tissue eosinophils, IL-4, and leptin in human obesity and insulin resistance. JCI Insight https://doi.org/10.1172/jci.insight.170772 (2024).
Muir, L. A. et al. Human CD206+ macrophages associate with diabetes and adipose tissue lymphoid clusters. JCI Insight https://doi.org/10.1172/jci.insight.146563 (2022).
Vijay, J. et al. Single-cell analysis of human adipose tissue identifies depot and disease specific cell types. Nat. Metab. 2, 97–109 (2020). The scRNA-seq study highlights depot-specific and disease-specific cellular characteristics in different ATs in humans, including a distinct population of adipogenic progenitors found exclusively in VAT.
Hildreth, A. D. et al. Single-cell sequencing of human white adipose tissue identifies new cell states in health and obesity. Nat. Immunol. 22, 639–653 (2021). This study investigates the diverse immune cell landscape in healthy lean and obese human WAT at single-cell resolution.
Li, J. H., Hepworth, M. R. & O’Sullivan, T. E. Regulation of systemic metabolism by tissue-resident immune cell circuits. Immunity 56, 1168–1186 (2023).
Wu, D. et al. Eosinophils sustain adipose alternatively activated macrophages associated with glucose homeostasis. Science 332, 243–247 (2011).
Talukdar, S. et al. Neutrophils mediate insulin resistance in mice fed a high-fat diet through secreted elastase. Nat. Med. 18, 1407–1412 (2012).
Salcher, S. et al. High-resolution single-cell atlas reveals diversity and plasticity of tissue-resident neutrophils in non-small cell lung cancer. Cancer Cell 40, 1503–1520 (2022).
Goldberg, E. L. et al. Ketogenesis activates metabolically protective gammadelta T cells in visceral adipose tissue. Nat. Metab. 2, 50–61 (2020).
Gonzalez-Hurtado, E. et al. Nerve-associated macrophages control adipose homeostasis across lifespan and restrain age-related inflammation. Preprint at bioRxiv https://doi.org/10.1101/2024.10.12.618004 (2024).
Li, C. et al. TCR transgenic mice reveal stepwise, multi-site acquisition of the distinctive fat-Treg phenotype. Cell 174, 285–299 (2018).
Munoz-Rojas, A. R., Wang, G., Benoist, C. & Mathis, D. Adipose-tissue regulatory T cells are a consortium of subtypes that evolves with age and diet. Proc. Natl Acad. Sci. USA 121, e2320602121 (2024).
Silva, H. M. et al. Vasculature-associated fat macrophages readily adapt to inflammatory and metabolic challenges. J. Exp. Med. 216, 786–806 (2019).
Man, K., Kallies, A. & Vasanthakumar, A. Resident and migratory adipose immune cells control systemic metabolism and thermogenesis. Cell Mol. Immunol. 19, 421–431 (2022).
Weinstock, A., Moura Silva, H., Moore, K. J., Schmidt, A. M. & Fisher, E. A. Leukocyte heterogeneity in adipose tissue, including in obesity. Circ. Res. 126, 1590–1612 (2020).
Jacks, R. D. & Lumeng, C. N. Macrophage and T cell networks in adipose tissue. Nat. Rev. Endocrinol. 20, 50–61 (2024).
Chakarov, S. et al. Two distinct interstitial macrophage populations coexist across tissues in specific subtissular niches. Science https://doi.org/10.1126/science.aau0964 (2019).
Hill, D. A. et al. Distinct macrophage populations direct inflammatory versus physiological changes in adipose tissue. Proc. Natl Acad. Sci. USA 115, E5096–E5105 (2018).
Jaitin, D. A. et al. Lipid-associated macrophages control metabolic homeostasis in a Trem2-dependent manner. Cell 178, 686–698 (2019). This study describes a subset of Trem2-positive lipid-associated macrophages in ATs required to maintain proper adipocyte function and systemic metabolic homeostasis.
Pirzgalska, R. M. et al. Sympathetic neuron-associated macrophages contribute to obesity by importing and metabolizing norepinephrine. Nat. Med. 23, 1309–1318 (2017).
Camell, C. D. et al. Inflammasome-driven catecholamine catabolism in macrophages blunts lipolysis during ageing. Nature 550, 119–123 (2017).
Felix, I. et al. Single-cell proteomics reveals the defined heterogeneity of resident macrophages in white adipose tissue. Front. Immunol. 12, 719979 (2021).
Zhou, L. et al. Hepatic danger signaling triggers TREM2+ macrophage induction and drives steatohepatitis via MS4A7-dependent inflammasome activation. Sci. Transl. Med. 16, eadk1866 (2024).
Nugent, A. A. et al. TREM2 regulates microglial cholesterol metabolism upon chronic phagocytic challenge. Neuron 105, 837–854 (2020).
Kalucka, J. et al. Single-cell transcriptome atlas of murine endothelial cells. Cell 180, 764–779 (2020).
Bondareva, O. et al. Single-cell profiling of vascular endothelial cells reveals progressive organ-specific vulnerabilities during obesity. Nat. Metab. 4, 1591–1610 (2022).
AlZaim, I., de Rooij, L., Sheikh, B. N., Borgeson, E. & Kalucka, J. The evolving functions of the vasculature in regulating adipose tissue biology in health and obesity. Nat. Rev. Endocrinol. 19, 691–707 (2023).
AlZaim, I. et al. Defining the vascular niche of human adipose tissue across metabolic conditions. Preprint at bioRxiv https://doi.org/10.1101/2024.09.22.610444 (2024).
AlZaim, I., Festa, J. & Kalucka, J. Adipose tissue lymphatic endothelial cells: revisited functions in the modulation of adipose biology. Curr. Opin. Physiol. 34, 100675 (2023).
Moller, A. F. & Madsen, J. G. S. JOINTLY: interpretable joint clustering of single-cell transcriptomes. Nat. Commun. 14, 8473 (2023).
Kremen, J. et al. Increased subcutaneous and epicardial adipose tissue production of proinflammatory cytokines in cardiac surgery patients: possible role in postoperative insulin resistance. J. Clin. Endocrinol. Metab. 91, 4620–4627 (2006).
Karow, A. M. Jr. & Webb, W. R. Tissue freezing. A theory for injury and survival. Cryobiology 2, 99–108 (1965).
Gulko, A. et al. Protocol for flow cytometry-assisted single-nucleus RNA sequencing of human and mouse adipose tissue with sample multiplexing. STAR Protoc. 5, 102893 (2024).
Fujiwara, S., Baek, S., Varticovski, L., Kim, S. & Hager, G. L. High quality ATAC-seq data recovered from cryopreserved breast cell lines and tissue. Sci. Rep. 9, 516 (2019).
Hepler, C. et al. Identification of functionally distinct fibro-inflammatory and adipogenic stromal subpopulations in visceral adipose tissue of adult mice. eLife https://doi.org/10.7554/eLife.39636 (2018).
Cho, D. S., Lee, B. & Doles, J. D. Refining the adipose progenitor cell landscape in healthy and obese visceral adipose tissue using single-cell gene expression profiling. Life Sci. Alliance https://doi.org/10.26508/lsa.201900561 (2019).
Nahmgoong, H. et al. Distinct properties of adipose stem cell subpopulations determine fat depot-specific characteristics. Cell Metab. 34, 458–472 (2022).
Whytock, K. L. et al. Single cell full-length transcriptome of human subcutaneous adipose tissue reveals unique and heterogeneous cell populations. iScience 25, 104772 (2022).
Angueira, A. R. et al. Defining the lineage of thermogenic perivascular adipose tissue. Nat. Metab. 3, 469–484 (2021).
Massier, L. et al. An integrated single cell and spatial transcriptomic map of human white adipose tissue. Nat. Commun. 14, 1438 (2023). This study represents an initial effort to develop a comprehensive integrated single-cell atlas of human WAT.
Whytock, K. L. et al. Aging human abdominal subcutaneous white adipose tissue at single cell resolution. Aging Cell https://doi.org/10.1111/acel.14287 (2024).
Wang, T. et al. Single-nucleus transcriptomics identifies separate classes of UCP1 and futile cycle adipocytes. Cell Metab. 36, 2130–2145 (2024).
Van Hauwaert, E. L. et al. Isolation of nuclei from mouse white adipose tissues for single-nucleus genomics. STAR Protoc. 2, 100612 (2021).
Whytock, K. L. et al. Isolation of nuclei from frozen human subcutaneous adipose tissue for full-length single-nuclei transcriptional profiling. STAR Protoc. 4, 102054 (2023).
Elingaard-Larsen, L. O. et al. Isolation of nuclei from human intermuscular adipose tissue and downstream single-nuclei RNA sequencing. J. Vis. Exp. https://doi.org/10.3791/66784 (2024).
Han, S. M. et al. Unique adipose tissue invariant natural killer T cell subpopulations control adipocyte turnover in mice. Nat. Commun. 14, 8512 (2023).
So, J. et al. Robust single-nucleus RNA sequencing reveals depot-specific cell population dynamics in adipose tissue remodeling during obesity. eLife https://doi.org/10.7554/eLife.97981 (2025).
Heumos, L. et al. Best practices for single-cell analysis across modalities. Nat. Rev. Genet. 24, 550–572 (2023). This recommendation summarizes benchmarking studies of unimodal and multimodal single-cell analysis, providing comprehensive best-practice workflows for the most common analysis steps.
Andrews, T. S., Kiselev, V. Y., McCarthy, D. & Hemberg, M. Tutorial: guidelines for the computational analysis of single-cell RNA sequencing data. Nat. Protoc. 16, 1–9 (2021).
Denisenko, E. et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 21, 130 (2020).
Gupta, A. et al. Characterization of transcript enrichment and detection bias in single-nucleus RNA-seq for mapping of distinct human adipocyte lineages. Genome Res. 32, 242–257 (2022).
Xi, N. M. & Li, J. J. Benchmarking computational doublet-detection methods for single-cell RNA sequencing data. Cell Syst. 12, 176–194 (2021).
Shamsi, F. et al. Vascular smooth muscle-derived Trpv1+ progenitors are a source of cold-induced thermogenic adipocytes. Nat. Metab. 3, 485–495 (2021).
Ferrero, R., Rainer, P. & Deplancke, B. Toward a consensus view of mammalian adipocyte stem and progenitor cell heterogeneity. Trends Cell Biol. 30, 937–950 (2020).
Engelhard, C. A., Khani, S., Derdak, S., Bilban, M. & Kornfeld, J. W. Nanopore sequencing unveils the complexity of the cold-activated murine brown adipose tissue transcriptome. iScience 26, 107190 (2023).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587 (2021).
Dominguez Conde, C. et al. Cross-tissue immune cell analysis reveals tissue-specific features in humans. Science 376, eabl5197 (2022).
Zhang, A. W. et al. Probabilistic cell-type assignment of single-cell RNA-seq for tumor microenvironment profiling. Nat. Methods 16, 1007–1015 (2019).
Pliner, H. A., Shendure, J. & Trapnell, C. Supervised classification enables rapid annotation of cell atlases. Nat. Methods 16, 983–986 (2019).
Luecken, M. D. et al. Benchmarking atlas-level data integration in single-cell genomics. Nat. Methods 19, 41–50 (2022).
Kang, J. B. et al. Efficient and precise single-cell reference atlas mapping with Symphony. Nat. Commun. 12, 5890 (2021).
Kang, H. M. et al. Multiplexed droplet single-cell RNA-sequencing using natural genetic variation. Nat. Biotechnol. 36, 89–94 (2018).
Huang, Y., McCarthy, D. J. & Stegle, O. Vireo: Bayesian demultiplexing of pooled single-cell RNA-seq data without genotype reference. Genome Biol. 20, 273 (2019).
Squair, J. W. et al. Confronting false discoveries in single-cell differential expression. Nat. Commun. 12, 5692 (2021).
Gayoso, A. et al. A Python library for probabilistic analysis of single-cell omics data. Nat. Biotechnol. 40, 163–166 (2022).
Boyeau, P. et al. Deep generative modeling of sample-level heterogeneity in single-cell genomics. Preprint at bioRxiv https://doi.org/10.1101/2022.10.04.510898 (2024).
Nguyen, H., Nguyen, H., Tran, D., Draghici, S. & Nguyen, T. Fourteen years of cellular deconvolution: methodology, applications, technical evaluation and outstanding challenges. Nucleic Acids Res. 52, 4761–4783 (2024).
Sorek, G. et al. sNucConv: a bulk RNA-seq deconvolution method trained on single-nucleus RNA-seq data to estimate cell-type composition of human adipose tissues. iScience 27, 110368 (2024).
Palmer, B. F. & Clegg, D. J. The sexual dimorphism of obesity. Mol. Cell. Endocrinol. 402, 113–119 (2015).
Ferreira, S. R. G., Macotela, Y., Velloso, L. A. & Mori, M. A. Determinants of obesity in Latin America. Nat. Metab. 6, 409–432 (2024).
Walker, C. R. et al. Private information leakage from single-cell count matrices. Cell https://doi.org/10.1016/j.cell.2024.09.012 (2024).
Truong, D. D. et al. Mapping the single-cell differentiation landscape of osteosarcoma. Clin. Cancer Res. 30, 3259–3272 (2024).
Gupta, A. et al. Mapping the transcriptional landscape of human white and brown adipogenesis using single-nuclei RNA-seq. Mol. Metab. 74, 101746 (2023).
Yang Loureiro, Z. et al. Wnt signaling preserves progenitor cell multipotency during adipose tissue development. Nat. Metab. 5, 1014–1028 (2023).
Palani, N. P. et al. Adipogenic and SWAT cells separate from a common progenitor in human brown and white adipose depots. Nat. Metab. 5, 996–1013 (2023).
Li, J. et al. Single-cell transcriptome dataset of human and mouse in vitro adipogenesis models. Sci. Data 10, 387 (2023).
Han, L. et al. Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature 604, 723–731 (2022).
Michelotti, T. C. et al. Single-nuclei analysis reveals depot-specific transcriptional heterogeneity and depot-specific cell types in adipose tissue of dairy cows. Front. Cell Dev. Biol. 10, 1025240 (2022).
Bhat-Nakshatri, P. et al. Single-nucleus chromatin accessibility and transcriptomic map of breast tissues of women of diverse genetic ancestry. Nat. Med. https://doi.org/10.1038/s41591-024-03011-9 (2024).
Zhu, R. et al. Updated skin transcriptomic atlas depicted by reciprocal contribution of single-nucleus RNA sequencing and single-cell RNA sequencing. J. Dermatol Sci. 111, 22–31 (2023).
Almet, A. A. et al. A roadmap for a consensus human skin cell atlas and single-cell data standardization. J. Invest. Dermatol. 143, 1667–1677 (2023).
Loft, A. et al. Single cell-resolved transcriptional dynamics of human subcutaneous adipose tissue during lifestyle- and bariatric surgery-induced weight loss. Preprint at bioRxiv https://doi.org/10.1101/2025.01.30.634294 (2025).
Hinte, L. C. et al. Adipose tissue retains an epigenetic memory of obesity after weight loss. Nature 636, 457–465 (2024). This snRNA-seq study demonstrates that both human and mouse ATs retain cellular transcriptional changes after weight loss, suggesting the existence of an obesogenic memory in certain adipogenic cell types.
Gao, H. et al. Age-induced reduction in human lipolysis: a potential role for adipocyte noradrenaline degradation. Cell Metab. 32, 1–3 (2020).
Bevill, S. M. et al. Impact of supraphysiologic MDM2 expression on chromatin networks and therapeutic responses in sarcoma. Cell Genom. 3, 100321 (2023).
Buenrostro, J. D., Giresi, P. G., Zaba, L. C., Chang, H. Y. & Greenleaf, W. J. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013).
Zhang, K. et al. A single-cell atlas of chromatin accessibility in the human genome. Cell 184, 5985–6001 (2021).
Lee, S. et al. Remodeling of gene regulatory networks underlying thermogenic stimuli-induced adipose beiging. Commun. Biol. 5, 584 (2022).
Gu, H. et al. Smart-RRBS for single-cell methylome and transcriptome analysis. Nat. Protoc. 16, 4004–4030 (2021).
Gupta, P., O’Neill, H., Wolvetang, E. J., Chatterjee, A. & Gupta, I. Advances in single-cell long-read sequencing technologies. NAR Genom. Bioinform. 6, lqae047 (2024).
Nigro, P. et al. Exercise training remodels inguinal white adipose tissue through adaptations in innervation, vascularization, and the extracellular matrix. Cell Rep. 42, 112392 (2023).
Stansbury, C. M. et al. A lipid-associated macrophage lineage rewires the spatial landscape of adipose tissue in early obesity. JCI Insight https://doi.org/10.1172/jci.insight.171701 (2023).
Vandereyken, K., Sifrim, A., Thienpont, B. & Voet, T. Methods and applications for single-cell and spatial multi-omics. Nat. Rev. Genet. 24, 494–515 (2023).
Cottam, M. A., Caslin, H. L., Winn, N. C. & Hasty, A. H. Multiomics reveals persistence of obesity-associated immune cell phenotypes in adipose tissue during weight loss and weight regain in mice. Nat. Commun. 13, 2950 (2022).
Strieder-Barboza, C. et al. Single-nuclei transcriptome of human AT reveals metabolically distinct depot-specific adipose progenitor subpopulations. Preprint at bioRxiv https://doi.org/10.1101/2022.06.29.496888 (2022).
Peters, H. et al. Cell and transcriptomic diversity of infrapatellar fat pad during knee osteoarthritis. Ann. Rheum. Dis. https://doi.org/10.1136/ard-2024-225928 (2024).
Niu, R. Z. et al. Single-cell transcriptome unveils unique transcriptomic signatures of human organ-specific endothelial cells. Basic Res. Cardiol. 119, 973–999 (2024).
Boesch, M. et al. Adipose tissue macrophage dysfunction is associated with a breach of vascular integrity in NASH. J. Hepatol. 80, 397–408 (2024).
Weinstock, A. et al. Single-cell RNA sequencing of visceral adipose tissue leukocytes reveals that caloric restriction following obesity promotes the accumulation of a distinct macrophage population with features of phagocytic cells. Immunometabolism https://doi.org/10.20900/immunometab20190008 (2019).
Song, A. et al. Low- and high-thermogenic brown adipocyte subpopulations coexist in murine adipose tissue. J. Clin. Invest. 130, 247–257 (2020).
Rondini, E. A. & Granneman, J. G. Single cell approaches to address adipose tissue stromal cell heterogeneity. Biochem. J. 477, 583–600 (2020).
Burl, R. B., Rondini, E. A., Wei, H., Pique-Regi, R. & Granneman, J. G. Deconstructing cold-induced brown adipocyte neogenesis in mice. eLife https://doi.org/10.7554/eLife.80167 (2022).
Henriques, F. et al. Single-cell RNA profiling reveals adipocyte to macrophage signaling sufficient to enhance thermogenesis. Cell Rep. 32, 107998 (2020).
Spallanzani, R. G. et al. Distinct immunocyte-promoting and adipocyte-generating stromal components coordinate adipose tissue immune and metabolic tenors. Sci. Immunol. https://doi.org/10.1126/sciimmunol.aaw3658 (2019).
Zhang, Q. et al. Distinct functional properties of murine perinatal and adult adipose progenitor subpopulations. Nat. Metab. 4, 1055–1070 (2022).
LaMarche, N. M. et al. Distinct iNKT cell populations use IFNγ or ER stress-induced IL-10 to control adipose tissue homeostasis. Cell Metab. 32, 243–258 (2020).
Acknowledgements
As an initial initiative, members of the HCA Adipose Bionetwork formed subcommittees focused on ‘anatomical location’, ‘cell annotation’, ‘experimental best practices’ and ‘computational best practices’ to develop recommendations for the entire adipose research community. All listed authors are members of the HCA Adipose Bionetwork and contributed actively, through participation in these subcommittees and/or during the manuscript editing process. Further, a special thanks goes to the following members of the HCA Adipose Bionetwork: A. R. Peltonen, A. Streets, A. L. Hunter, A. Körner, A. Rudich, C. Sarathy, D. Li, E. G. Klinggaard, E. Pimenta, F. Zhao, H. Dobrzynski, J. Stephens, J. L. Rinker, J. Nie, K. Delgado, K. Warmink, L. van Gijn, L. Qin, M. N. Khan, Q. Yao, R. Rydbirk, R. Gualtierotti, T. Tong, Y. Zhu and Z. Y. Loureiro. This work was supported by The Danish National Research Foundation to the Center for Functional Genomics and Tissue Plasticity (ATLAS; DNRF 141, to S.M. and A.L.), the Lundbeck Foundation (R413-2022-471 to A.L.), Sygeforsikring ‘danmark’ (2021-0297 to S.M.), and the National Institutes of Health (K01 DK134806 to M.P.E., RC2 DK116691 to E.D.R.). This publication is part of the HCA (www.humancellatlas.org/publications/).
Author information
Authors and Affiliations
Contributions
HCA Adipose Bionetwork co-coordinators: A.L., M.P.E., S.M. and E.D.R. HCA Adipose Bionetwork subcommittee members: A.D., A.G., B.M., B.D., B.L., C. Lumeng, C.M., C.W., C.S.-B., C. Li, E.S.-V., F.S., H.C., J.Z., J.K., J.J., K.L.W., L.M.S., L.A.M., L.M., M.A.M., M.B., M.-E.P., M.R., N.M., R.K.G., S.C., T.E.O., V.E., Y.T. and Y.-H.T. Writing of manuscript: A.L., M.P.E., S.M. and E.D.R. Editing of manuscript: A. Weinstock, A.D., A.G., A. Wagner, A.V.H., B.M., B.D., B.L., C.S., C. Lumeng, C.D., C.M., C.W., C.S.-B., C. Li, D.D.T., D.A.B., E.S-.V., E.E.K., E.Y-.L., F.S., H.X.H., H.C., J.Z., J.K., J.A.L., J.A.S., J.J., K.L.W., K.D.D., L.M.S., L.A.M., L.F., L.M., L.R.S., M.D.B., M.G.J., M.A.M., M.B., M.J.W., M.-E.P., M.D.L., M.B., M.R., N.H., N.L.S., N.M., P.G., R.K.G., R.M., S.P., S.C., S.T., S.D., S.F.S., T.Z., T.M.N., T.E.O., V.E., W.W., Y.T. and Y.-H.T.
Corresponding authors
Ethics declarations
Competing interests
M.B. received honoraria as a consultant and speaker from Amgen, AstraZeneca, Bayer, Boehringer Ingelheim, Daiichi Sankyo, Eli Lilly, Novo Nordisk, Novartis and Sanofi.
Peer review
Peer review information
Nature Metabolism thanks Prashant Rajbhandari, James G. Granneman and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editor: Revati Dewal, in collaboration with the Nature Metabolism team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Loft, A., Emont, M.P., Weinstock, A. et al. Towards a consensus atlas of human and mouse adipose tissue at single-cell resolution. Nat Metab 7, 875–894 (2025). https://doi.org/10.1038/s42255-025-01296-9
Received:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s42255-025-01296-9
This article is cited by
-
Bridging the variant-to-function gap in type 2 diabetes: advances and challenges
Diabetologia (2026)
-
Depot-dependent effects of subclinical ketosis on visceral and subcutaneous adipose tissue transcriptional cellular diversity in dairy cows
Journal of Animal Science and Biotechnology (2025)
-
Weight loss alters adipose tissue beyond just reducing fat
Nature (2025)
-
Adipose tissue ageing: implications for metabolic health and lifespan
Nature Reviews Endocrinology (2025)