Abstract
Circular RNAs are an unusual class of single-stranded RNAs whose ends are covalently linked via back-splicing. Due to their versatility, the need to express circular RNAs in vivo and in vitro has increased. Efforts have been made to efficiently and precisely synthesize circular RNAs. However, a review on the optimization of the processes of circular RNA design, synthesis, and delivery is lacking. Our review highlights the multifaceted aspects considered when producing optimal circular RNAs and summarizes the available options for each step of exogenous circular RNA design and synthesis, including circularization strategies. Additionally, this review describes several potential applications of circular RNAs.

Similar content being viewed by others
Introduction
Circular RNAs (circRNAs) are RNA molecules whose 5′-ends are covalently linked to their 3′-ends, a structure achieved through back-splicing1,2,3,4,5. Back-splicing is a process in which a downstream 3′-splice donor is joined to a 5′-splice acceptor that is positioned upstream of the donor. Due to their unique structure, circRNAs were discovered with low expression levels in normal and neoplastic mammalian cells more than three decades ago1 but were initially thought to be a byproduct of erroneous splicing. However, later studies revealed that circRNAs are widely expressed not only in mammalian species2 but also in a diverse range of other organisms, including other vertebrates6, worms3, flies7, plants8, fungi and protists4, and lower eukaryotes5. High-throughput transcriptome sequencing has revealed that ≥10% of the genes expressed in mammalian cells and tissues can produce circRNAs, further establishing the widespread presence of circRNAs9. The biological and clinical importance and potential of circRNAs have been increasingly highlighted in many fields. Numerous studies have reported the expression of thousands of circRNAs under normal and abnormal conditions10. CircRNAs are associated with stress-related11 and immune-related responses12 and have been implicated in many human illnesses, including cancer and neurodegenerative diseases13,14.
CircRNAs act as efficient platforms for the expression of functional molecules15,16. Especially during the coronavirus disease 2019 pandemic, circRNAs that undergo internal ribosome entry site (IRES)-mediated translation were highlighted as potential mRNA vaccine candidates. Although circRNA-encoded peptides need to be carefully validated17,18, it is possible to design and synthesize circRNAs that exhibit robust and stable protein synthesis ability16,19,20,21. Wesselhoeft et al. reported that circRNAs produced 9- and 1.5-fold more proteins than unmodified and modified nucleoside linear RNAs, respectively, with 1.7- to 2.4-fold longer half-lives than linear RNAs in human cells20. That same group later demonstrated that nanoparticle delivery and in vivo translation of synthetic circRNAs were feasible21. A recent study showed that circRNA vaccines could protect mice22 and macaques15 against different variants of SARS-CoV-2, with improved efficacy and similar immunogenicity relative to linear RNA vaccines. These promising applications have been noted by industry, with Merck & Co. being one of the important investors. Merck agreed to spend up to $3.75 billion on Orna Therapeutics, Inc., a new startup aiming to develop medicines from synthetic circRNAs23. Laronde, another group with a similar goal, had raised $440 million by 2021.
Considering these trends, determining the optimal methods for exogenous circRNA synthesis and delivery, as well as for specific expression in desired tissues, is of utmost importance. In this review, we describe how each step in the expression of exogenous circRNAs can be optimized, along with the strengths and limitations of possible options. Furthermore, we discuss the potential of circRNAs as effector molecules in therapeutic applications.
Exogenous circRNA synthesis
To express exogenous circRNAs, one should first decide whether to generate circRNAs via in vitro transcription (IVT), circularization, and delivery of the RNA or to inject a DNA construct and generate circRNAs via in vivo transcription. There are various protocols for RNA circularization and in vivo transcription or IVT that lead to different choices for the expression of exogenous circRNAs (Table 1). Here, we describe validated trials for the expression of synthetic circRNAs in vivo and in vitro.
Generation of circRNAs by in vivo transcription
Earlier studies used naturally occurring introns from highly expressed circRNAs to ensure robust transcription and circularization of circRNAs19,24,25,26,27,28 (Fig. 1a). However, these constructs produced a mixture of RNA forms, i.e., circRNAs and linear RNAs, suggesting that the constructs must be optimized to robustly transcribe the intended linear RNAs and induce their circularization via back-splicing in vivo.
a Endogenous circularization methods that have been used or mimicked. b A highly efficient circularization method that leaves a scar sequence in the product. c RNAs have been designed to enable the effective circularization of scarless circRNAs. d The sequence of interest has been permuted; thus, it resembles exonic sequences in the T4 td gene. The yellow and orange boxes indicate natural introns and ribozymes, respectively. Sequences that bring the splice sites together are shown as cyan boxes. Genes of interest are represented as green boxes. Promoters and SV40 terminators are shown as gray circles and red hexagons, respectively. DNA constructs are depicted with black lines, and RNA transcripts are depicted with gray lines. RNA transcription is indicated with blue arrows. FL flag, NLS nuclear localization signal, PUF PUF binding motif, DIM dimerization domain, FPG split GFP, H homology arm, E1 and E2 exonic sequences, CDS coding sequence, L ligation sequence, GOI gene of interest.
As a growing number of cis- and trans-acting factors involved in back-splicing have been identified, studies aimed at ectopically expressing circRNAs using these factors have been performed (Table 2). Researchers have simulated natural introns by inserting intronic complementary sequences (ICSs) that bring splice sites together24,29,30,31,32 (Fig. 1b). Qi et al. described the engineering of circRNA regulators, which combine circRNA vectors with the RNA-binding motifs of homodimerizing RNA-binding proteins (RBPs) and nuclear localization signals33 (Fig. 1b). Introduction of the RNA-binding domains from PUM1 paired with those from ZBTB18, HNRNPA1, and PRKAR1A resulted in expression levels similar to those of ICSs without affecting linear RNA expression.
Meganck et al. sought to increase the circularization efficiency of natural introns by partially deleting ZKSCAN1 and HIPK3 introns with inverted ALU elements19. In a more recent study, a circPVT1 backbone was used because shorter exonic sequences did not circularize efficiently with the widely used ZKSCAN1 introns34. In 2021, the same group generated a series of insertions and deletions in the upstream and downstream introns of the model to investigate the effect of the distance between the ALU elements and the splice junction35. These experiments revealed that truncating the upstream and downstream introns to bring the ALU elements closer to the splice junction enhanced circRNA and protein expression by up to fivefold. The authors stated that systems with synthetic introns have multiple advantages compared to the Tornado system36, which utilizes the “Twister” ribozyme, whose splicing leaves RtcB-compatible reactive RNA ends, because the synthetic introns can be further improved, and their short lengths enable researchers to package the system into recombinant adeno-associated viral (AAV) vectors, allowing long-term gene expression in a wide range of tissues37,38. Controlling the amount of circRNA expressed in cells can be difficult, as transcription and circularization may be affected by endogenous factors.
In vitro RNA circularization
Another common method for inducing RNA circularization involves the use of a permuted intron‒exon system (PIE), which comprises exonic sequences flanked by group I self-splicing introns39. This method enables the expression of desired circRNAs both in vitro and in vivo. Anabaena pre-tRNA introns and T4 bacteriophage td gene introns are widely used with some modifications20 (Fig. 1c). Litke et al. devised the Tornado system, which utilizes ubiquitously expressed, tRNA precursor-ligating, RtcB-compatible 5ʹ and 3ʹ ends and “Twister” ribozymes36,40 (Fig. 1c). Further studies have shown that the Tornado system can robustly express stable circRNAs in vivo and that these circRNAs play designated biological roles41. However, the caveat of the Tornado system is that ribozyme-based circularization leaves an unintended exonic sequence, called a “scar”, in the resulting circRNAs. Generating circRNAs based on group I introns inevitably leaves 80–180 nt long sequences derived from the two adjacent exons (commonly referred to as E1 and E2) in the products. These scars introduce undesired sequences into the final product, and these sequences elicit immune responses or have unexpected effects on the experimental results.
Strategies for generating “scarless” circRNAs
To overcome the scar issue, Rausch et al. screened for possible exon‒intron pairs necessary for the self-splicing of T4 td introns and suggested permuting desired exonic sequences to ensure that the 5ʹ- and 3ʹ-termini resemble the E2 and E1 exonic sequences of the T4 td gene42 (Fig. 1d). The authors demonstrated that their constructs could easily produce circRNAs without T4 exon sequences in vitro. Unlike transfection of the Tornado system, transfection of the modified scarless system did not induce an immune response21,43. Efforts to identify methods for synthesizing scarless circRNAs are ongoing and represent an active field of research. Zuo et al. devised a novel strategy, termed Clean-PIE, that could be applied in vivo and in vitro using permuted T4 td introns44. The authors concealed the E1 and E2 sequences necessary for the splicing reaction in the ORF of their construct and optimized the variable parts of the E1 and E2 sequences. In another study by Wang et al., group II introns were used to produce scarless circRNAs in vitro45. In this study, the exon-binding site in the D1 domain of group II introns was modified; therefore, it could bind to the circular exon for self-splicing. This backbone is not universally applicable because the sequence of circular exons differs among genes; thus, the D1 sequence should be modified differently for different genes.
Another group adopted the group I intron of Tetrahymena, a trans-splicing ribozyme that enables the efficient circularization of RNAs without scars. Lee et al. concatenated the target sequence (5ʹ-NNNNNU-3ʹ) recognized by the Tetrahymena intron at the 3ʹ end of the gene of interest and the intron itself, allowing end-to-end self-targeting and splicing to occur46. The results indicated that this system could induce more robust expression of circRNAs than the PIE method, although the authors stated that self-circularization was effective only in vitro. They investigated whether this system could generate multimeric circRNAs via intermolecular splicing and concluded that this was unlikely. The authors recommended that only a single target site be present to achieve precise splicing. Similarly, Cui et al. devised a construct with a backbone and flanking antisense sequences to aid in the self-splicing of Tetrahymena thermophila introns47. The efficiency was approximately 80% both in vitro and in vivo. They synthesized circFOXO3 using this method and found that the product could be utilized to regulate various cellular phenotypes, such as proliferation, migration, and apoptosis, in prostate cancer cells.
Generation of circRNAs with chemicals or enzymes
CircRNAs can be generated in vitro from linear precursors via reactions catalyzed by chemicals or enzymes48. Generally, RNA synthesis using chemicals results in the production of short oligomers (~50–70 nt). Therefore, an additional step of ligating several RNAs is required to synthesize larger molecules. Another challenge in the chemical circularization of RNA is that the concentration of the linear precursor should be low to prevent its oligomerization, which leads to low throughput. This method requires preorientation of the two reactive ends, which may be performed using a linear or hairpin helper oligonucleotide or a splint.
Several enzymes can be used for intramolecular ligation; the most commonly used are T4 DNA ligase and T4 RNA ligases 1 and 2. Specifically, T4 RNA ligase produces large amounts of homogenous and pure circRNAs49.
Although circularization by chemical reactions has many disadvantages, each method has its strengths and limitations, and the method should be chosen based on several characteristics of the circRNA product of interest (i.e., in vivo or in vitro production, natural or modified nucleotides, and construct size)48. The length of the sequence of interest could limit the choice of synthesis method owing to the difficulties in synthesizing large molecules using chemical methods and the PIE system39. In fact, PIE system does not work if there are long (1.1 kb) intervening regions between the splice sites20. Additionally, long RNAs tend to be less efficiently circularized and are more prone to nicking when magnesium ions are present during and after IVT20. Chemical- or enzyme-based methods produce circRNAs only in vitro, whereas methods based on ribozymes can robustly generate circRNAs both in vitro and in vivo. Therefore, ribozyme-based methods are frequently used to express endogenous RNA sequences.
Design of messenger circRNA vectors
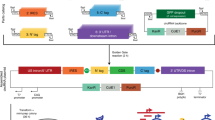
Synthesizing “messenger circRNAs” that encode polypeptides requires the design of circRNA vectors that consider cis-acting factors to robustly express the desired protein (Fig. 2). The choice of the IRES, 5ʹ- and 3ʹ-untranslated regions (UTRs), and coding region can affect the translation efficiency50.
Each factor to be considered is presented for each region of the vector of interest. Spacers were added to lessen the structural hindrance between the IRES and the gene of interest or the splice junction. Adding motifs for RBPs that enhance translation is beneficial. Several IRESs with stronger activity than the canonically used IRESs and additional codon optimization can result in faster and more abundant protein production.
Vector topology is crucial for IRES-mediated translation, as the IRES is a structural element that recruits ribosomes. It is important to ensure that the sequences flanking the IRES do not interfere with IRES activity by forming complex secondary structures. In this case, the addition of spacers to separate each secondary structure can facilitate translation. One study reported that the addition of spacers to attenuate the structural hindrances caused by IRESs can improve translation20. A more recent study concluded that placing spacers of 50 nt in length between the splicing scar of T4 td introns and the IRES resulted in the most robust translation50. Furthermore, Liu et al. showed that RNA duplexes in circRNAs may activate degradation by PKR51. Thus, it is important to design the overall sequence to minimize the formation of RNA duplexes.
The IRES of encephalomyocarditis virus is the most commonly used IRES owing to its robust and nonspecific expression, which does not require many IRES trans-acting factors19,20,35,52. However, an extensive comparison of various IRESs revealed that an IRES from coxsackievirus B3 (CVB3) was the most efficient across several cell lines (HEK293, HeLa, A549, and Min6)20. Further investigation revealed that the IRES of Echovirus 29 had a stronger translation signal than the IRES of CVB344,53. A later study investigated a wide range of viral IRESs to optimize circRNA translation; the authors concluded that IRESs of human rhinovirus B and enterovirus B species could drive strong translation and further elucidated that the translation efficiency of viral IRESs could be further improved by the insertion of eukaryotic translation initiation factor (eIF) G4-associated aptamers50. Moreover, random sequences were generated in this study to screen for IRES activity, and several sequences with strong translation-driving power were identified50.
UTR sequences are known to regulate multiple aspects of RNA translation, post-transcriptional regulation, and RNA stability54. UTRs harbor many sequences and structural elements that positively or negatively affect translation. One of the well-characterized examples is the binding site for poly(A)-binding proteins (PABPs) in the 5ʹ UTR, which aids in the binding of eIFs55; in addition, a highly structured 5ʹ-UTR is known to attenuate translation efficiency56. Including poly(A)20 or poly(AC)44 sequences in the construct improved translational strength and reduced immunogenicity16. Chen et al. noted that adding PABP motifs and an aptamer sequence that recruits eIF4G increased the translation of the circular reporter50. A few 3ʹUTRs of linear mRNAs, such as that of human β-globin, have been shown to enhance protein production57. Most of the 3ʹUTRs that tend to drive efficient translation of linear RNAs, except for the 3ʹUTR of human α-globin 2, do not seem to do so for circRNAs50.
Codon triplets are recognized by tRNAs during translation, and it has long been debated whether codon usage and the abundance of tRNAs can affect translation efficiency and speed58. The kinetics of translation are crucial for proper protein folding and translation elongation59,60; therefore, optimizing codons for the same amino acid may promote effective protein production. This field of study has not been rigorously explored; however, it was shown that eliminating unfavorable base-pairing interactions between the adjacent ends of an IRES and a coding sequence can further facilitate circRNA translation50.
Delivery of synthesized circRNA or DNA constructs
During IVT-mediated circRNA synthesis, the resulting molecules must be rigorously purified. Purification by gel extraction or size-exclusion high-performance liquid chromatography is necessary because Anabaena introns or rare circular concatenations are resistant to degradation by RNase R20. The solid-phase DNA probe method61, in which a DNA probe is designed to hybridize the back-splice junction of the desired product62, can also be used to purify circRNAs from total RNA.
The size of the construct and the required targeting specificity can influence the choice of delivery vehicle. CircRNAs can be efficiently delivered using lipids21, gold nanoparticles63, AAV vectors19, lentiviral vectors, exosomes64, and transposons65. Recent advances in nanodrug delivery have suggested that nanoparticles may increase target specificity66. Although each method has distinct limitations and strengths, the toxicity of gold nanoparticles is under debate. Exosomes may be more biocompatible than nanoparticles but require complex manufacturing processes. For the delivery of naked circRNAs, optimization of the solvent may result in greater cellular uptake, as shown in a study by Yang et al., in which the use of Ringer’s solution resulted in the highest reported uptake at tumor sites53.
Intracellular regulation of circRNA expression
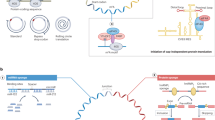
Endogenous circular RNAs exhibit tightly regulated expression, are stable with a long half-life, and are resistant to RNA decay mechanisms. The level of circRNAs is affected by numerous factors in multiple steps; therefore, several factors need to be considered to achieve stable expression (Fig. 3).
Regulation of back-splicing
Back-splicing efficiency is a combination of numerous factors at multiple levels, including chromatin states and sequence context in exons and flanking introns25,67 (Fig. 3a). At the epigenomic level, several histone modifications, including H3K4me1, H3K36me3, H3K79me2, and H4K20me1, affect circRNA biogenesis68. However, some exons are more preferentially processed by circRNAs than others, and the more back-splicing that occurs on an exon, the more exon skipping occurs during forward splicing24,69. However, exon skipping does not guarantee the inclusion of the exon in a circRNA; therefore, an additional level of regulation is required for exon circularization69. There are a few reported cases of Schizosaccharomyces pombe in which circRNA biogenesis occurs independently of cis or trans elements5, despite various cis- or trans-acting factors having been reported to facilitate or hinder circRNA production. ICSs are among the most important cis-acting elements, although their importance varies among species10. ICSs can be either inverted repeats9,25 or nonrepetitive elements24,70. In human fibroblasts, the vast majority (88%) of ICSs contain ALU repeats9.
In addition to cis-acting factors, RBPs can promote or disrupt exon circularization (Table 2). Quaking binds to flanking introns and forms a homodimer, bringing the splice sites together27. In contrast, A-to-I editing protein (ADAR1) can destabilize RNA pairs necessary for back-splicing via A-to-I editing71. However, the role of RBPs in circRNA biogenesis requires further investigation, as the effect of RBPs may vary depending on the type of circRNA and cell line. For example, HNRNPL increases the expression of the circmCherry reporter in HeLa cells32; however, a study of HNRNPL-knockdown LNCaP cells showed that endogenous circRNAs were downregulated rather than upregulated72 (Table 2). Similarly, knockdown of Slu7 resulted in circRNA enrichment in DL1 cells73 and depletion of circmCherry in HeLa cells32.
Back-splicing is performed by the spliceosome machinery and involves canonical splice sites in most cases (99%); therefore, it competes with forward splicing, although forward splicing is >100-fold more efficient74,75. Thus, the inhibition of forward splicing may be important for promoting back-splicing. Ablation of some core factors of the spliceosomal complex and treatment with a splicing inhibitor allows more back-splicing events to occur73,76,77. Despite the expected competition between forward and back-splicing, early genome-wide studies have shown that the levels of circular and linear isoforms are not fully correlated with each other7,27,59,78,79, although researchers have made further efforts to define the efficiency of back-splicing in terms of the circular-to-linear ratio (CLR). The CLR is the ratio of mapped sequencing reads that support back-splicing to those that support forward splicing59. Although the CLR is known to be <1% for most human loci, some circRNAs are robustly expressed and sometimes accumulate to levels that exceed those of their corresponding linear forms7,59,78. However, the precise mechanism underlying the regulation of back splicing efficiency remains to be elucidated.
Context specificity of circRNA expression
CircRNAs are known to show expression patterns that are strongly specific for certain biological conditions and independent of those of linear isoforms, increasing the difficulty of understanding the control of circRNA expression27,59,80,81. A recent study using 90 human tissue transcriptomes revealed that 36–75% of alternative back-splicing events are tissue-specific82. Investigation of tissue-wide circRNA profiles revealed that different brain compartments, such as the olfactory bulb, prefrontal cortex, hippocampus, and cerebellum, have the greatest number of tissue-specific circRNAs59. By comparison, the heart, liver, and muscle have the lowest number of tissue-specific circRNAs83. These characteristics are not limited to endogenous circRNAs. Advances in the engineering of synthetic circRNAs have revealed similar tissue- and cell-type specificities for exogenous circRNAs. Injecting AAV vectors carrying sequences encoding the circular form of green fluorescent protein into mice resulted in different transduction rates across tissues19, although AAV vectors are known to broadly express encoded sequences without any tissue preference84.
CircRNA specificity extends beyond the tissue or cell type level to include cell-to-cell variations and distinct subcellular localization patterns (Fig. 3c). Single-cell studies have reinforced the idea that circRNA profiles vary from cell to cell85,86,87. Little is known about the subcellular localization of circRNAs; however, exonic circRNAs are localized mostly in the cytoplasm, whereas those with intronic sequences primarily remain in the nucleus2,5,88,89. The nuclear export of some circRNAs appears to be mediated by their length-dependent association with UAP56 or URH49, which are RNA helicases that recruit the REF adapter protein to RNAs90. Another study showed that YTHDC1, an m6A reader protein, mediates the nuclear export of circNSUN2 via m6A modification; this was the first report of an association between m6A and circRNA translocation91. In a more recent study, circRNA representation in the nuclear, cytoplasmic, mitochondrial, ribosomal, cytosolic, and exosomal fractions of HepG2 cells was systematically examined92. The results indicated that circRNAs in different compartments had different characteristics regarding length and G/C content. In neurons, some circRNAs have been shown to localize to synapses59,78; however, the elements that dictate this localization are unknown34. Several studies have examined functional mitochondrial circRNAs and revealed that their intracellular expression levels are altered under stress conditions93,94,95,96. Overall, these results suggested that circRNA expression is tightly regulated at different subcellular locations. Data from continued efforts to investigate the subcellular localization of circRNAs have been integrated into platforms for the visual presentation of localization information97.
Regulation of the intracellular levels of circRNAs
One prominent feature that distinguishes circRNAs is their marked stability. Researchers have found that the half-lives of circRNAs are, on average, two- to fourfold longer than those of linear mRNAs and sometimes as much as 10-fold longer98. This difference results mainly from the absence of 5ʹ- and 3ʹ-terminal nucleotides that can be attacked by exonucleases, which block the degradation of circRNAs under normal or stressful conditions. During viral infection, RNase L is activated via an unknown mechanism and globally degrades circRNAs associated with PKR as part of the innate immune response51 (Fig. 3d). Park et al. identified RNase P and MRP as circRNA-degrading agents that interact with YTHDF2 and HRSP12, two proteins that recognize circRNAs with m6A modifications and a GGUUC motif99. Drosophila GW182, a key component of P-bodies, and its human homologs TNRC6A/TNRC6B/TNRC6C participate in circRNA decay through an AGO2-independent mechanism, and their depletion substantially increases the steady-state levels of cytoplasmic circRNAs100. Considering that these mechanisms function sequentially, it is likely that circRNA isoforms are subjected to decay via different mechanisms, possibly leading to the enrichment of circRNAs related to stress responses.
Under normal conditions, approximately one-third of human circRNAs are predicted to be highly structured, and their degradation is globally regulated by UPF1 and G3BP1 via structure-mediated RNA decay (SRD)101. G3BP1 selectively binds to highly structured circRNAs and is a determining factor in SRD. SRD targets appear to be preferentially excluded from stress granules, where UPF1 and G3BP1 localize after stress-inducing treatment. Taken together, these results indicate that circRNA decay mechanisms vary between normal and stressful conditions. None of the factors mentioned above exclusively target circRNAs; thus, it is likely that there are additional unknown pathways responsible for the regulation of circRNA steady-state levels102.
Studies using human cell lines have suggested that circRNAs can be actively exported103 (Fig. 3e). Several reports have shown that circRNAs are enriched in extracellular vesicles103,104, in the circulation and urine105, and in exosomes secreted by various cell lines106,107. Additionally, circRNAs with a 5ʹ-GMWGVWGRAG-3ʹ motif were found to be selectively packaged into exosomes92. However, the exact mechanism of circRNA secretion and its effect on donor and recipient cells remain unknown108.
Immunogenicity of exogenous circRNAs
The immunogenicity of engineered circRNAs remains controversial. Chen et al. showed that transfection of circRNAs using a PIE system containing the T4 td gene intron triggered the expression of several immune genes, whereas transfection of a circRNA generated with the ZKSCAN1 intron did not43. Subsequently, they observed that m6A modification could act as a molecular marker for “self” circRNAs109. Another study using the Anabaena intron reported that the resulting circRNA did not elicit an immune response20. This was later challenged by a more recent study, which concluded that circRNAs produced by group I introns are immunogenic, possibly due to the intron “scars” that remain in the final product110. This inconsistency could be a result of differences in the methods used to test immunogenicity or the type of linear RNA used for comparison since the above studies all used distinct methods to evaluate the immunogenicity of a circRNA111. However, the immunogenicity of vector-carrying circRNAs has not been discussed in detail. To date, the transduction of circRNAs via AAV vectors or lentiviruses has shown negligible immune activation ability.
Application of engineered circRNAs
Recently, several studies have used circRNA technology to investigate or control cellular processes and immune responses (Fig. 4) 21,36,43,51,109. Circular mRNA vaccines showed efficient protection against SARS-CoV-2 infection (Fig. 4a)15,22. Furthermore, circRNAs aid in reducing the effective vector dose for gene therapy applications because the expression levels of their protein products are likely to increase over time19.
CircRNAs have also been used in DNA editing and RNA regulation. Two groups used ADAR with a circular guide RNA for in vivo and in vitro RNA editing (Fig. 4b, c)112,113. Several siRNA mimics114, RNA dumbbells115, and aptamers36 have been shown to perform robustly, with improved stability in the circular form (Fig. 4d–f).
Conclusion
CircRNAs have demonstrated potential as molecules for next-generation vaccines and therapeutics. Herein, we presented several options that could be chosen and aspects that could be considered when developing a platform for circRNAs. A rational sequence design that guarantees the maximal cellular level of a circRNA or a desired protein is of pivotal importance. One could also adopt an adequate delivery method and enhance cellular uptake by optimizing the solution. CircRNAs can be expressed differently in different tissues and cell types, and efforts should be made to minimize the immunogenicity of circRNAs.
Our aim was to show how the process for exogenous circRNA synthesis can be modified to be more efficient and suitable for circRNA synthesis. By considering each step of the application of a circRNA, we believe that one can accomplish the desired results with maximum potential. Ultimately, these steps will become standard procedures for the industrial synthesis of circRNAs.
References
Nigro, J. M. et al. Scrambled exons. Cell 64, 607–613 (1991).
Salzman, J., Gawad, C., Wang, P. L., Lacayo, N. & Brown, P. O. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS ONE 7, e30733 (2012).
Memczak, S. et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495, 333–338 (2013).
Wang, P. L. et al. Circular RNA is expressed across the eukaryotic tree of life. PLoS ONE 9, e90859 (2014).
Barrett, S. P., Wang, P. L. & Salzman, J. Circular RNA biogenesis can proceed through an exon-containing lariat precursor. Elife 4, e07540 (2015).
Wu, W., Ji, P. & Zhao, F. CircAtlas: an integrated resource of one million highly accurate circular RNAs from 1070 vertebrate transcriptomes. Genome Biol. 21, 101 (2020).
Salzman, J., Chen, R. E., Olsen, M. N., Wang, P. L. & Brown, P. O. Cell-type specific features of circular RNA expression. PLoS Genet. 9, e1003777 (2013).
Lu, T. et al. Transcriptome–wide investigation of circular RNAs in rice. RNA 21, 2076–2087 (2015).
Jeck, W. R. et al. Circular RNAs are abundant, conserved & associated with ALU repeats. RNA 19, 141–157 (2013).
Dong, R., Ma, X. K., Chen, L. L. & Yang, L. Increased complexity of circRNA expression during species evolution. RNA Biol. 14, 1064–1074 (2017).
Fischer, J. W. & Leung, A. K. CircRNAs: a regulator of cellular stress. Crit. Rev. Biochem. Mol. Biol. 52, 220–233 (2017).
Yan, L. & Chen, Y. G. Circular RNAs in immune response and viral infection. Trends Biochem. Sci. 45, 1022–1034 (2020).
Kim, E., Kim, Y. K. & Lee, S. V. Emerging functions of circular RNA in aging. Trends Genet. 37, 819–829 (2021).
Wang, Y. et al. Circular RNAs in human cancer. Mol. Cancer 16, 25 (2017).
Qu, L. et al. Circular RNA vaccines against SARS-CoV-2 and emerging variants. Cell 185, 1728–1744.e1716 (2022).
Kameda, S., Ohno, H. & Saito, H. Synthetic circular RNA switches and circuits that control protein expression in mammalian cells. Nucleic Acids Res. 51, e24 (2023).
Ho-Xuan, H. et al. Comprehensive analysis of translation from overexpressed circular RNAs reveals pervasive translation from linear transcripts. Nucleic Acids Res. 48, 10368–10382 (2020).
Nielsen, A. F. et al. Best practice standards for circular RNA research. Nat. Methods 19, 1208–1220 (2022).
Meganck, R. M. et al. Tissue-dependent expression and translation of circular RNAs with recombinant AAV vectors in vivo. Mol. Ther. Nucleic Acids 13, 89–98 (2018).
Wesselhoeft, R. A., Kowalski, P. S. & Anderson, D. G. Engineering circular RNA for potent and stable translation in eukaryotic cells. Nat. Commun. 9, 2629 (2018).
Wesselhoeft, R. A. et al. RNA circularization diminishes immunogenicity and can extend translation duration in vivo. Mol. Cell. 74, 508–520.e504 (2019).
Seephetdee, C. et al. A circular mRNA vaccine prototype producing VFLIP-X spike confers a broad neutralization of SARS-CoV-2 variants by mouse sera. Antivir. Res. 204, 105370 (2022).
Shen, H. et al. Circular RNAs: characteristics, biogenesis, mechanisms, and functions in liver cancer. J. Hematol. Oncol. 14, 134 (2021).
Zhang, X. O. et al. Complementary sequence-mediated exon circularization. Cell 159, 134–147 (2014).
Liang, D. & Wilusz, J. E. Short intronic repeat sequences facilitate circular RNA production. Genes Dev. 28, 2233–2247 (2014).
Kramer, M. C. et al. Combinatorial control of Drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins. Genes Dev. 29, 2168–2182 (2015).
Conn, S. J. The RNA binding protein quaking regulates formation of circRNAs. Cell 160, 1125–1134 (2015).
Xu, J. Z. et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 10, 175 (2019).
Hansen, T. B. et al. Natural RNA circles function as efficient microRNA sponges. Nature 495, 384–388 (2013).
Wang, Y. & Wang, Z. Efficient backsplicing produces translatable circular mRNAs. RNA 21, 172–179 (2015).
Pamudurti, N. R. et al. Translation of CircRNAs. Mol. Cell. 66, 9–21.e27 (2017).
Li, X. et al. Coordinated circRNA biogenesis and function with NF90/NF110 in viral infection. Mol. Cell. 67, 214–227.e217 (2017).
Qi, Y. et al. Engineering circular RNA regulators to specifically promote circular RNA production. Theranostics 11, 7322–7336 (2021).
Ron, M. & Ulitsky, I. Context-specific effects of sequence elements on subcellular localization of linear and circular RNAs. Nat. Commun. 13, 2481 (2022).
Meganck, R. M. et al. Engineering highly efficient backsplicing and translation of synthetic circRNAs. Mol. Ther. Nucleic Acids 23, 821–834 (2021).
Litke, J. L. & Jaffrey, S. R. Highly efficient expression of circular RNA aptamers in cells using autocatalytic transcripts. Nat. Biotechnol. 37, 667–675 (2019).
Wang, D., Tai, P. W. L. & Gao, G. Adeno-associated virus vector as a platform for gene therapy delivery. Nat. Rev. Drug Discov. 18, 358–378 (2019).
Li, C. & Samulski, R. J. Engineering adeno-associated virus vectors for gene therapy. Nat. Rev. Genet. 21, 255–272 (2020).
Puttaraju, M. & Been, M. D. Group I permuted intron-exon (PIE) sequences self-splice to produce circular exons. Nucleic Acids Res. 20, 5357–5364 (1992).
Roth, A. et al. A widespread self-cleaving ribozyme class is revealed by bioinformatics. Nat. Chem. Biol. 10, 56–60 (2014).
Schreiner, S., Didio, A., Hung, L. H. & Bindereif, A. Design and application of circular RNAs with protein–sponge function. Nucleic Acids Res. 48, 12326–12335 (2020).
Rausch, J. W. et al. Characterizing and circumventing sequence restrictions for synthesis of circular RNA in vitro. Nucleic Acids Res. 49, e35 (2021).
Chen, Y. G. et al. Sensing self and foreign circular RNAs by intron identity. Mol. Cell. 67, 228–238.e225 (2017).
Zonghao, Q. H. et al. Clean-PIE: a novel strategy for efficiently constructing precise circRNA with thoroughly minimized immunogenicity to direct potent and durable protein expression. https://www.biorxiv.org/content/10.1101/2022.06.20.496777v1 (2022).
Chuyun, C. et al. A flexible, efficient, and scalable platform to produce circular RNAs as new therapeutics. https://www.biorxiv.org/content/10.1101/2022.05.31.494115v2 (2022).
Lee, K. H. et al. Efficient circular RNA engineering by end–to–end self–targeting and splicing reaction using Tetrahymena group I intron ribozyme. Mol. Ther. Nucleic Acids 33, 587–598 (2023).
Cui, J. et al. A precise and efficient circular RNA synthesis system based on a ribozyme derived from Tetrahymena thermophila. Nucleic Acids Res. 51, e78 (2023).
Muller, S. & Appel, B. In vitro circularization of RNA. RNA Biol. 14, 1018–1027 (2017).
Beaudry, D. & Perreault, J. P. An efficient strategy for the synthesis of circular RNA molecules. Nucleic Acids Res. 23, 3064–3066 (1995).
Chen, R. et al. Engineering circular RNA for enhanced protein production. Nat. Biotechnol. 41, 262–272 (2023).
Liu, C. X. et al. Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177, 865–880.e821 (2019).
Yang, Y. et al. Extensive translation of circular RNAs driven by N(6)-methyladenosine. Cell. Res. 27, 626–641 (2017).
Yang, J. et al. Intratumoral delivered novel circular mRNA encoding cytokines for immune modulation and cancer therapy. Mol. Ther. Nucleic Acids 30, 184–197 (2022).
Mignone, F., Gissi, C., Liuni, S. & Pesole, G. Untranslated regions of mRNAs. Genome Biol. 3, REVIEWS0004 (2002).
Mangus, D. A., Evans, M. C. & Jacobson, A. Poly(A)-binding proteins: multifunctional scaffolds for the post-transcriptional control of gene expression. Genome Biol. 4, 223 (2003).
Leppek, K., Das, R. & Barna, M. Functional 5ʹʹUTR mRNA structures in eukaryotic translation regulation and how to find them. Nat. Rev. Mol. Cell Biol. 19, 158–174 (2018).
Jiang, Y., Xu, X. S. & Russell, J. E. A nucleolin-binding 3ʹʹ untranslated region element stabilizes beta–globin mRNA in vivo. Mol. Cell Biol. 26, 2419–2429 (2006).
Gingold, H. & Pilpel, Y. Determinants of translation efficiency and accuracy. Mol. Syst. Biol. 7, 481 (2011).
Rybak-Wolf, A. et al. Circular RNAs in the mammalian brain are highly abundant, conserved, and dynamically expressed. Mol. Cell. 58, 870–885 (2015).
Kim, S. J. et al. Protein folding. Translational tuning optimizes nascent protein folding in cells. Science 348, 444–448 (2015).
Suzuki, T., Suzuki, T., Wada, T., Saigo, K. & Watanabe, K. Taurine as a constituent of mitochondrial tRNAs: new insights into the functions of taurine and human mitochondrial diseases. EMBO J. 21, 6581–6589 (2002).
Umekage, S. & Kikuchi, Y. In vitro and in vivo production and purification of circular RNA aptamer. J. Biotechnol. 139, 265–272 (2009).
Zeng, Y. et al. A circular RNA binds to and activates AKT phosphorylation and nuclear localization reducing apoptosis and enhancing cardiac repair. Theranostics 7, 3842–3855 (2017).
Yang, L. et al. Extracellular vesicle-mediated delivery of circular RNA SCMH1 promotes functional recovery in rodent and nonhuman primate ischemic stroke models. Circulation 142, 556–574 (2020).
Mecozzi, N. et al. Genetic tools for the stable overexpression of circular RNAs. RNA Biol. 19, 353–363 (2022).
Dancy, J. G. et al. Decreased nonspecific adhesivity, receptor-targeted therapeutic nanoparticles for primary and metastatic breast cancer. Sci. Adv. 6, eaax3931 (2020).
Ashwal-Fluss, R. et al. circRNA biogenesis competes with pre-mRNA splicing. Mol. Cell. 56, 55–66 (2014).
Zhang, M. et al. Revealing epigenetic factors of circRNA expression by machine learning in various cellular contexts. iScience 23, 101842 (2020).
Kelly, S., Greenman, C., Cook, P. R. & Papantonis, A. Exon skipping is correlated with exon circularization. J. Mol. Biol. 427, 2414–2417 (2015).
Starke, S. et al. Exon circularization requires canonical splice signals. Cell Rep. 10, 103–111 (2015).
Ivanov, A. et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 10, 170–177 (2015).
Fei, T. et al. Genome-wide CRISPR screen identifies HNRNPL as a prostate cancer dependency regulating RNA splicing. Proc. Natl Acad. Sci. USA 114, E5207–E5215 (2017).
Liang, D. et al. The output of protein-coding genes shifts to circular RNAs when the pre-mRNA processing machinery is limiting. Mol. Cell. 68, 940–954.e943 (2017).
Zhang, Y. et al. The biogenesis of nascent circular RNAs. Cell Rep. 15, 611–624 (2016).
Vo, J. N. et al. The landscape of circular RNA in cancer. Cell 176, 869–881.e813 (2019).
Li, X. et al. A unified mechanism for intron and exon definition and back-splicing. Nature 573, 375–380 (2019).
Wang, M., Hou, J., Muller-McNicoll, M., Chen, W. & Schuman, E. M. Long and repeat-rich intronic sequences favor circular RNA formation under conditions of reduced spliceosome activity. iScience 20, 237–247 (2019).
You, X. et al. Neural circular RNAs are derived from synaptic genes and regulated by development and plasticity. Nat. Neurosci. 18, 603–610 (2015).
Zhang, X. O. et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26, 1277–1287 (2016).
Westholm, J. O. et al. Genome-wide analysis of Drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep. 9, 1966–1980 (2014).
Gruner, H., Cortes-Lopez, M., Cooper, D. A., Bauer, M. & Miura, P. CircRNA accumulation in the aging mouse brain. Sci. Rep. 6, 38907 (2016).
Zhang, P. et al. Comprehensive identification of alternative back-splicing in human tissue transcriptomes. Nucleic Acids Res. 48, 1779–1789 (2020).
Mahmoudi, E. & Cairns, M. J. Circular RNAs are temporospatially regulated throughout development and ageing in the rat. Sci. Rep. 9, 2564 (2019).
Zincarelli, C., Soltys, S., Rengo, G. & Rabinowitz, J. E. Analysis of AAV serotypes 1-9 mediated gene expression and tropism in mice after systemic injection. Mol. Ther. 16, 1073–1080 (2008).
Fan, X. et al. Single-cell RNA-seq transcriptome analysis of linear and circular RNAs in mouse preimplantation embryos. Genome Biol. 16, 148 (2015).
Zhong, C., Yu, S., Han, M., Chen, J. & Ning, K. Heterogeneous circRNA expression profiles and regulatory functions among HEK293T single cells. Sci. Rep. 7, 14393 (2017).
Wu, W., Zhang, J., Cao, X., Cai, Z. & Zhao, F. Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 13, 3242 (2022).
Li, Z. et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat. Struct. Mol. Biol. 22, 256–264 (2015).
Veno, M. T. et al. Spatio-temporal regulation of circular RNA expression during porcine embryonic brain development. Genome Biol. 16, 245 (2015).
Huang, C., Liang, D., Tatomer, D. C. & Wilusz, J. E. A length-dependent evolutionarily conserved pathway controls nuclear export of circular RNAs. Genes Dev. 32, 639–644 (2018).
Chen, R. X. et al. N(6)–methyladenosine modification of circNSUN2 facilitates cytoplasmic export and stabilizes HMGA2 to promote colorectal liver metastasis. Nat. Commun. 10, 4695 (2019).
Zhang, J. et al. Circular RNA profiling provides insights into their subcellular distribution and molecular characteristics in HepG2 cells. RNA Biol. 16, 220–232 (2019).
Zhao, Q. et al. Targeting mitochondria-located circRNA SCAR alleviates NASH via reducing mROS output. Cell 183, 76–93.e22 (2020).
Wu, Z. et al. Mitochondrial genome-derived circRNA mc-COX2 functions as an oncogene in chronic lymphocytic leukemia. Mol. Ther. Nucleic Acids 20, 801–811 (2020).
Liu, X. et al. Identification of mecciRNAs and their roles in the mitochondrial entry of proteins. Sci. China Life Sci. 63, 1429–1449 (2020).
Gong, W. et al. Nuclear genome–derived circular RNA circPUM1 localizes in mitochondria and regulates oxidative phosphorylation in esophageal squamous cell carcinoma. Signal Transduct. Target. Ther. 7, 40 (2022).
Lin, Y. C. et al. CircVIS: a platform for circRNA visual presentation. BMC Genom. 22, 921 (2022).
Enuka, Y. et al. Circular RNAs are long–lived and display only minimal early alterations in response to a growth factor. Nucleic Acids Res. 44, 1370–1383 (2016).
Park, O. H. et al. Endoribonucleolytic cleavage of m(6)A-containing RNAs by RNase P/MRP complex. Mol. Cell. 74, 494–507.e498 (2019).
Jia, R., Xiao, M. S., Li, Z., Shan, G. & Huang, C. Defining an evolutionarily conserved role of GW182 in circular RNA degradation. Cell Discov. 5, 45 (2019).
Fischer, J. W., Busa, V. F., Shao, Y. & Leung, A. K. L. Structure-mediated RNA decay by UPF1 and G3BP1. Mol. Cell. 78, 70–84.e76 (2020).
Chen, L. L. The expanding regulatory mechanisms and cellular functions of circular RNAs. Nat. Rev. Mol. Cell Biol. 21, 475–490 (2020).
Lasda, E. & Parker, R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PLoS ONE 11, e0148407 (2016).
Preusser, C. et al. Selective release of circRNAs in platelet-derived extracellular vesicles. J. Extracell. Vesicles. 7, 1424473 (2018).
Wang, Y. et al. Exosomal circRNAs: biogenesis, effect and application in human diseases. Mol. Cancer 18, 116 (2019).
Li, Y. et al. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 25, 981–984 (2015).
Dou, Y. et al. Circular RNAs are down–regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci. Rep. 6, 37982 (2016).
Patop, I. L., Wust, S. & Kadener, S. Past, present, and future of circRNAs. EMBO J. 38, e100836 (2019).
Chen, Y. G. et al. N6-Methyladenosine modification controls circular RNA immunity. Mol. Cell. 76, 96–109.e109 (2019).
Liu, C. X. et al. RNA circles with minimized immunogenicity as potent PKR inhibitors. Mol. Cell. 82, 420–434.e426 (2022).
Tai, J. & Chen, Y. G. Differences in the immunogenicity of engineered circular RNAs. J. Mol. Cell Biol. 15, mjad002 (2023).
Katrekar, D. et al. Efficient in vitro and in vivo RNA editing via recruitment of endogenous ADARs using circular guide RNAs. Nat. Biotechnol. 40, 938–945 (2022).
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
Jahns, H. et al. Small circular interfering RNAs (sciRNAs) as a potent therapeutic platform for gene-silencing. Nucleic Acids Res. 49, 10250–10264 (2021).
Abe, N. et al. Synthesis, structure, and biological activity of dumbbell-shaped nanocircular RNAs for RNA interference. Bioconjugate Chem. 22, 2082–2092 (2011).
Aktas, T. et al. DHX9 suppresses RNA processing defects originating from the Alu invasion of the human genome. Nature 544, 115–119 (2017).
Yu, C. Y. et al. The circular RNA circBIRC6 participates in the molecular circuitry controlling human pluripotency. Nat. Commun. 8, 1149 (2017).
Errichelli, L. et al. FUS affects circular RNA expression in murine embryonic stem cell–derived motor neurons. Nat. Commun. 8, 14741 (2017).
Ho, J. S. et al. HNRNPM controls circRNA biogenesis and splicing fidelity to sustain cancer cell fitness. Elife 10, e59654 (2021).
Okholm, T. L. H. et al. Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 12, 112 (2020).
Knupp, D., Cooper, D. A., Saito, Y., Darnell, R. B. & Miura, P. NOVA2 regulates neural circRNA biogenesis. Nucleic Acids Res. 49, 6849–6862 (2021).
Khan, M. A. et al. RBM20 regulates circular RNA production from the titin gene. Circ. Res. 119, 996–1003 (2016).
Stagsted, L. V. W., OʹLeary, E. T., Ebbesen, K. K. & Hansen, T. B. The RNA-binding protein SFPQ preserves long-intron splicing and regulates circRNA biogenesis in mammals. Elife 10, e63088 (2021).
Acknowledgements
We thank all BIG lab members for their helpful discussions. This work was supported by the Basic Science Research Program and the Bio & Medical Development Program through the National Research Foundation, funded by the Ministry of Science and ICT [2021R1A2C3005835, 2022M3E5F1018502, 2022M3A9I2082294, RS-2023-00207840, 2023R1A6C101A009].
Author information
Authors and Affiliations
Contributions
SWC and JWN contributed to the design and writing of the manuscript and JWN conceived the idea.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Choi, SW., Nam, JW. Optimal design of synthetic circular RNAs. Exp Mol Med 56, 1281–1292 (2024). https://doi.org/10.1038/s12276-024-01251-w
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s12276-024-01251-w
This article is cited by
-
Scarless circular mRNA-based CAR-T cell therapy elicits superior antitumor efficacy
Signal Transduction and Targeted Therapy (2025)
-
Nucleic acid aptamers in orthopedic diseases: promising therapeutic agents for bone disorders
Bone Research (2025)
-
Regulatory RNA: from molecular insights to therapeutic frontiers
Experimental & Molecular Medicine (2024)