Abstract
Precision oncology has emerged as a promising strategy for treating high-grade gliomas, yet its clinical impact has been disappointing, with over 300 clinical trials on targeted therapies failing to yield substantial improvements in patient outcomes. Current approaches primarily focus on static, marker-driven tumor features, which capture only a small portion of the complex biology that governs therapeutic responses. Functional precision oncology (FPO) offers a complementary approach, enhancing treatment selection in a personalized manner by dynamically testing patient-derived tumor cells against a range of available therapeutic agents. Here this review examines both historical and contemporary treatment strategies for high-grade gliomas and explores underlying reasons for the limited success of multiple precision oncology initiatives. We demonstrate how the incorporation of FPO in the armamentarium of glioma therapies may address these challenges and outline its proposed role as well as the practical considerations in utilizing FPO for clinical decision-making in patients with glioma.
Similar content being viewed by others
Introduction
The success of targeted therapies ushered in the era of precision oncology, a field focused on identifying and targeting cancer-specific alterations to guide more effective treatments. Pioneering therapies such as imatinib and trastuzumab, which revolutionized the management of chronic myeloid leukemia and HER2-positive breast cancer, respectively, laid the foundation for targeted approaches. Since then, the field has expanded substantially, fueled by large-scale cancer genomic projects aimed at uncovering actionable targets for therapeutic intervention1.
While numerous success stories illustrate the transformative potential of precision oncology, it is key to realize that these successes are exceptions rather than the rule. The effectiveness of targeted therapies often varies markedly depending on the cancer type and its underlying biological context. Glioblastoma (GBM), the most aggressive form of high-grade glioma (HGG), was the first cancer to be molecularly characterized by The Cancer Genome Atlas (TCGA)2. This landmark effort has since transformed glioma classification, culminating in the World Health Organization (WHO) 2021 classification, which integrates molecular diagnostics into the taxonomy of central nervous system (CNS) tumors3. Over the past two decades, more than 300 clinical trials exploring targeted therapies and 50 trials investigating immunotherapy have failed to show significant improvements in patient outcomes4,5. Efforts have focused on targeting genetic alterations implicated in HGG pathogenesis, including EGFR overexpression (evaluated through receptor tyrosine kinase inhibitors and ADC against EGFRvIII), PTEN loss (addressed by PI3K pathway inhibitors) and tumor angiogenesis (targeted by antiangiogenic agents such as bevacizumab). While a new wave of immuno-oncology studies is emerging, past immuno-oncology strategies, both as monotherapies and in combination therapies, have failed to establish a new standard of care (SOC)5. As a result, HGG continues to rank among the malignancies with the highest failure rates in cancer treatment.
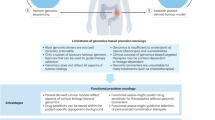
Despite extensive molecular characterization, it is increasingly evident that measurable genomic data represents only a fraction of the intricate biology driving therapeutic responses in HGG. This highlights the need to better understand the complexity of HGG, in particular the dynamic interplay of factors such as cellular state plasticity, microenvironmental interactions and epigenetic modifications that collectively shape tumor behavior6. Crucially, an incomplete understanding of all the factors influencing therapeutic outcomes lends support to the inadequacy of current genomic-centric precision oncology approaches and clinical trials over the past two decades7.
More recently, functional precision oncology (FPO) has emerged as a complementary strategy that bridges the gap between static genomic data and dynamic tumor behavior by directly testing a patient’s tumor cells against therapeutic agents. This real-time assessment of drug efficacy provides a more objective basis for identifying effective treatments beyond genetic probabilities, and is especially valuable in cases such as HGG, where mutation-based approaches have failed to reliably predict clinical outcomes. By expanding the focus beyond targetable mutations alone, FPO enhances current precision oncology strategies, offering a more personalized and comprehensive evaluation of drug responses8. In practice, FPO workflows begin with the collection of patient-derived specimens to generate ex vivo models, such as 2D cells, 3D organoids or tissue explants, which are rapidly screened against a panel of drugs to quantify tumor cell responses using functional assays that measure viability, growth inhibition or apoptosis. The findings are subsequently integrated with molecular and clinical data, often through multidisciplinary tumor boards, to inform individualized treatment recommendations within clinically relevant timeframes. Studies in hematological malignancies and solid tumors, including HGG, have demonstrated the feasibility and predictive power of FPO, with clinical correlations and successful therapeutic outcomes8,9,10,11.
In this review, we provide a comprehensive overview of existing approaches in managing HGGs, outline three key pillars fundamental to precision oncology strategies and explore the unique biological context of HGG that may have limited the success of previous trials. Finally, we discuss how FPO offers a promising and innovative approach to address the shortcomings of genomics-based approaches, paving the way for more effective and personalized treatment strategies.
The current treatment paradigm for HGGs: past trials, future challenges
Historically, HGG referred to a heterogeneous group of gliomas defined based on histopathological features supported by ancillary tissue-based tests (for example, immunohistochemistry), encompassing grade 3 and 4 tumors. More recently, the WHO 2021 classification advanced the role of molecular diagnostics in CNS tumor classification, dividing adult HGG into four subtypes: grade 3 oligodendroglioma (1p/19 co-deleted, IDH-mutant), grade 3 IDH-mutant astrocytoma, grade 4 IDH-mutant astrocytoma and grade 4 IDH wild-type GBM3. HGG carries a median overall survival (OS) of 14 months and a median progression-free survival (PFS) of 7 months, with tumor recurrence observed in over 90% of patients8,9. Unfortunately, no established SOC treatment exists at recurrence, for which patients have a median OS of 6.2 months10.
Timeline of FDA-approved systemic therapies in HGG
There have been very few FDA-approved therapies for HGG (Fig. 1). Most approved therapies are traditional cytotoxic therapies belonging to the DNA alkylating class, including temozolomide (TMZ), lomustine and carmustine (both intravenous and wafer implants). Exceptions to this include bevacizumab and tumor-treating field (TTF) devices. In this review, we focus on treatment regimens used for high-grade astrocytic gliomas, which constitute the majority of HGGs in adults.
A timeline illustrating the evolution of FDA-approved cancer therapies for HGG, categorized by their application in primary or recurrent disease. The modalities are color coded to illustrate their evolution, transitioning from chemotherapy (blue) to targeted therapies (pink) and device-based treatments (green). Key clinical trials that occurred during the corresponding periods are also shown.
Maximal safe resection followed by radiotherapy remains the cornerstone of treatment for HGG, with its role established through studies dating back to the 1970s11. Before the introduction of TMZ, systemic therapy was predominantly based on cytotoxic agents such as lomustine and carmustine. These agents continue to be widely used for the management of recurrent HGG, although a universally accepted SOC treatment remains elusive12. The efficacy of lomustine was first demonstrated as part of the regimen comprising of procarbazine, lomustine, and vincristine (PCV), during which it was combined with procarbazine and vincristine following radiotherapy in newly diagnosed HGG13. Lomustine received FDA approval in 1976 for the treatment of primary and recurrent HGG; while its efficacy remains largely limited to tumors with O6-methylguanine DNA methyltransferase (MGMT) promoter methylation, it continues to be used in the recurrent setting regardless of MGMT status14. Similarly, carmustine demonstrated a survival benefit in two pivotal phase III trials, which showed that the addition of carmustine to surgery and radiotherapy improved survival, with a greater proportion of patients surviving beyond 18 months. Carmustine was subsequently approved by the FDA in 1977 for the treatment of recurrent HGG15,16.
The landscape of HGG management shifted notably in 2005, when Stupp et al. demonstrated superior survival outcomes with the addition of TMZ to radiotherapy in the first-line treatment of HGG9. This survival benefit was considered a therapeutic milestone for systemic therapy in HGG management and established the Stupp protocol as the new SOC. Since then, the evolving molecular understanding of HGG has led to countless trials evaluating chemotherapy, targeted therapy and immunotherapy, but none has succeeded in replacing the Stupp protocol.
Bevacizumab is a monoclonal antibody targeting vascular endothelial growth factor, inhibiting angiogenesis thereby restricting tumor vascularization. Several phase II and III trials have explored the inclusion of bevacizumab to chemoradiation with TMZ in the front-line treatment of HGG, including the phase III RTOG 0825 (NCT00884741) and phase III AVAGlio (NCT00943826). Despite showing a modest prolongation in PFS, these trials did not demonstrate a significant improvement in OS17. In 2006, a phase II study (NCT00345163) assessed the efficacy of bevacizumab, both as a monotherapy and in combination with irinotecan, for patients with recurrent HGG. The promising results from this study led to FDA approval of bevacizumab in 2009 as salvage therapy for recurrent HGG18. Subsequently, several other trials explored the use of bevacizumab in combination with various chemotherapy agents17. While the combination of lomustine and bevacizumab appeared promising in the phase II BELOB trial (NCT01290939) and the phase III EORTC 26101 trial (NCT01290939), these trials failed to meet their primary endpoint of an OS benefit19. Nonetheless, bevacizumab and lomustine remain the most commonly used systemic treatments for recurrent HGG so far19.
In recent years, TTFs, a noninvasive anti-cancer modality utilizing low-intensity, intermediate-frequency alternating electric fields, has become more widely accessible. The EF-11 trial (NCT00379470) in 2006 demonstrated the efficacy of TTFs in patients with recurrent HGG, leading to FDA approval in 2011 for its use in recurrent tumors. Subsequently, the EF-14 trial (NCT00916409) in 2009 showed that combining TTFs with maintenance therapy significantly improved OS in newly diagnosed patients, resulting in FDA approval in 2017. This pivotal study marked the first advancement in over a decade to extend OS for newly diagnosed HGG patients since the addition of TMZ to radiotherapy20. However, real-world uptake remains limited, with only 18–20% of eligible patients receiving TTF in recent multicenter cohorts, with optimal usage (≥75% daily, ≥18 h/day) achieved by only a minority21,22. Barriers include device burden, cost and quality-of-life considerations, contributing to variable adoption despite guideline endorsement21,23.
Emerging therapeutic modalities in HGG
In recent years, emerging targeted therapies and immunotherapy approaches, supported by robust preclinical data, have been extensively investigated in HGG, as reviewed previously4. Despite substantial monetary investments and their representation in a large proportion of clinical trials, these therapies have largely failed to demonstrate meaningful improvements in survival outcomes4,5. Among the numerous strategies evaluated, bevacizumab remains the only FDA-approved targeted therapy for HGG. Here, we discuss the development of a few key therapies.
Targeted therapy
EGFR amplification is observed in approximately 50% of HGGs, with the EGFRvIII variant the most common alteration. So far, approximately 40 ongoing or completed trials have explored various strategies targeting EGFR, including small-molecule tyrosine kinase inhibitors, monoclonal antibodies, antibody–drug conjugates (ADC) and vaccines. However, despite the perceived importance of EGFR as a target in other cancers, such as non-small cell lung cancer and colorectal cancer, EGFR-targeted trials have largely failed to provide clinical benefit in HGG24. Several phase II trials investigating gefitinib and erlotinib showed no clinical efficacy in patients with newly diagnosed HGG, regardless of EGFR amplification or mutation status, attributable to the limited brain penetrance of these compounds24,25,26,27,28. While limited brain penetrance may contribute, emerging evidence suggests that context-specific resistance mechanisms may play a larger role. Interestingly, newer brain-penetrant EGFR inhibitors such as osimertinib, while effective in EGFR-mutant non-small cell lung cancer brain metastases29,30, have not demonstrated meaningful benefit in HGG, with reports showing rapid progression on the drug31,32,33. More recently, the fourth-generation EGFR inhibitor, silevertinib, is being investigated in an ongoing phase I/II trial (NCT05256290) for patients with recurrent GBM harboring EGFR alterations, with results currently pending34. Other strategies evaluating monoclonal antibodies, as well as the ADC, depatuxizumab mafodotin, have similarly yielded disappointing results, showing no improvement in clinical outcomes35. Several factors may underlie this unexpected discrepancy between preclinical promise and clinical failure, underscoring the unique challenges of targeting EGFR in HGGs.
Recent studies have demonstrated that while EGFRvIII is present in primary tumors, it is often undetectable in nearly half of recurrent tumors following SOC treatment. The loss of EGFRvIII expression in recurrent HGG is hypothesized to be driven by clonal selection and tumoral heterogeneity, where treatment pressures favor tumor subpopulations without EGFRvIII expression36. This suggests that tumor recurrence may not be dependent on EGFRvIII, raising important questions about its role in glioma biology. While EGFRvIII may be a critical driver in primary tumor initiation and early progression, it may no longer play a central role in the mechanisms driving treatment failure and tumor recurrence36,37 However, despite EGFRvIII loss, EGFR expression is rarely lost at recurrence, suggesting alternative resistance mechanisms, with the transition of EGFR from chromosomal to extrachromosomal DNA playing a potential role38. In addition, as gliomas are rarely dependent on a single gene or pathway, EGFR and EGFRvIII are likely to represent just one component of a broader, highly interconnected signaling network39. This complexity explains the limited clinical efficacy of therapies targeting individual molecular alterations, as such approaches fail to address the adaptive and multifaceted nature of HGG biology4.
The PI3K/AKT/mTOR pathway, a hallmark of HGG associated with poorer outcomes, has been the focus of numerous strategies supported by strong preclinical evidence40. Initial optimism arose from phase III trials showing that the mTOR inhibitor everolimus was able to reduce the volume of subependymal giant cell astrocytomas41. However, mTOR inhibitors in HGG have largely failed to demonstrate meaningful benefit42. These failures may stem from compensatory AKT activation via unregulated mTORC2 signaling and the absence of reliable predictive biomarkers to identify treatment-responsive patients. In the recently completed phase II/III GBM AGILE trial (NCT03970447), paxalisib, a potent oral selective brain-penetrant small-molecule PI3K/mTOR inhibitor, demonstrated a promising 3.8-month OS improvement compared to SOC treatment in the MGMT unmethylated subpopulation of newly diagnosed GBM, suggesting potential benefit in selected patients42. This has led to the design of an anticipated phase III first-line study of paxalisib, in addition to the ongoing studies of the drug in recurrent gliomas (NCT05009992 and NCT05183204)43,44.
BRAFV600E mutations, which are present in 1–8% of HGGs, and up to 20% of cases in patients under 30 years of age, have emerged as potentially actionable mutation in HGG45. Several case reports have highlighted rapid and occasionally complete responses to BRAF inhibitors (BRAFi) and BRAF–MEK inhibitor combinations, a rarity seen in the setting of HGGs46. This has driven ongoing research, with larger basket trials reporting similar encouraging outcomes. Notably, the phase II ROAR trial (NCT02034110) demonstrated a 33% objective response rate in 31 patients with HGG, including three complete responses47. While further studies are needed to understand the long-term clinical course of BRAFi in HGG, a key limitation of BRAFi is the transient nature of their efficacy, with tumor recurrence developing over time. This phenomenon has been well-documented in cancers such as melanoma, where reactivation of the MAPK pathway or activation of alternative survival pathways drives tumor regrowth despite continued BRAFi treatment. Similarly, this effect has been reported in HGGs with the acquisition of mutations in the RAS/MAPK pathway and histologic transformation to gliosarcoma48. Nonetheless, the promising initial response observed in some patients with HGG underscores the therapeutic potential of BRAFi.
More recently, IDH1/2 mutations, present in up to 80% of low-grade gliomas (LGGs) and associated with favorable prognoses, have been investigated as therapeutic targets in gliomas. The phase III INDIGO trial (NCT04164901) demonstrated that vorasidenib, an IDH1/2 inhibitor, significantly improved median PFS (27.7 months versus 11.1 months with placebo) in patients with IDH1- or IDH2-mutant LGGs. Following these results, vorasidenib received FDA approval in 2024 for the treatment of IDH mutant LGGs. While these results are promising for LGGs, the role of IDH inhibitors in HGG remains uncertain with further trials necessary to define their role49.
It remains clear that genomics-driven strategies have failed to deliver the anticipated clinical success in HGGs. Nevertheless, precision oncology remains a promising strategy; however, realizing its full potential requires acknowledgement of its core principles and context-specific limitations. Ultimately, therapeutic efficacy depends not on a treatment’s intrinsic potency but on how well it aligns with the biological landscape it is applied to—a critical consideration that must guide future therapeutic development.
Factors limiting the success of targeted therapies
Precision oncology was founded on the premise of identifying genetic or molecular changes that distinguish tumors from normal tissues, thereby enabling the selection of efficient treatment specific to the tumor. Fundamentally, the success of precision oncology depends on three key factors: identifying targetable variants, developing drugs that can effectively target these variants and ensuring the effective delivery of these targeted therapies. Challenges at any of these stages can hinder the success of targeted treatments. We discuss these factors in the context of HGG.
Identification of targetable variants
Large-scale cancer genomics initiatives, such as TCGA, International Cancer Genome Consortium (ICGC), and Genomics Evidence Neoplasia Information Exchange (GENIE) have facilitated the analysis of thousands of tumor genomes to identify targetable variants50. However, the identification of clinically actionable variants in HGG remains highly challenging. A major challenge in this process is the differentiation between true ‘driver mutations’, which promote cancer, from a vast majority of ‘passenger’ mutations, which accumulate without causal relevance to oncogenesis. This is further complicated by the fact that certain passenger mutations, although seemingly noncontributory in isolation, may act as ‘conditional’ driver mutations in the presence of other mutations51,52,53. Furthermore, current computational algorithms, which are principally guided by mutational frequencies, may overlook low-penetrance genetic events, thereby missing rare but potentially actionable drivers54. More importantly, even after identifying potential targets, our understanding of these variants often remains limited as the discovery of new variants outpaces our ability to classify them effectively54. Together, these challenges underscore the difficulty of reliably identifying targetable genomic variants in HGG.
Consequently, alternative approaches such as transcriptomic profiling have been pursued to better capture the dynamic nature of tumor biology. In HGGs, a landmark study by Verhaak et al. introduced gene expression-based classification of HGG into classical, mesenchymal and proneural subtypes55. Building on this, Neftel et al. demonstrated the existence of four cellular states—NPC-like (neural progenitor like), OPC-like (oligodendrocyte progenitor like), AC-like (Astrocyte like) and MES-like (mesenchymal like)—while highlighting the dynamic plasticity between states and the potential for a single cell to generate all four states56. Remarkably, a single tumor often contains a mixture of these states, with 60% of gliomas presenting with two or three different subtypes within the same tumor mass. Glioma stem-cells (GSCs) add further complexity, representing the apex cell population in HGG, underpinning phenotypic diversity, therapeutic resistance and recurrence57. Through self-renewal and multipotency, they regenerate diverse cellular lineages following treatment, while evading cytotoxic stress via enhanced DNA damage repair, metabolic adaptation, quiescence and activation of signaling networks56,58. The cellular diversity of GSCs, spanning distinct molecular subtypes and existing along gradients of transcriptional states with unique vulnerabilities, further exemplifies the difficulty of defining a single targetable vulnerability in HGG59.
Ability to target oncogenic drivers
Second, there remains a discordance in our ability to translate identified genetic or molecular targets into clinically relevant drugs. Approximately 60% of small-molecule drug discovery projects fail owing to identified targets considered ‘undruggable’. Prominent examples include RAS, MYC and fusion transcription factors, which are challenging to target owing to extensive protein–protein interaction interfaces and the absence of deep protein-binding sites. Loss-of-function mutations are particularly challenging to treat therapeutically, with frequently affected genes in HGG, such as NF1, RB1, TP53, PTEN and ATRX, remaining largely uncharted territory60.
In addition, delivering therapeutic agents to the brain is notoriously challenging owing to the combined effects of the blood–brain barrier (BBB) and the brain–tumor barrier. The BBB, a neurovascular construct regulating the passage of substances into healthy brain tissue, limits the penetration of over 98% of small drug molecules, with those that do cross often being expelled by efflux pumps such as P-glycoprotein. The brain–tumor barrier, driven by the secretion of factors such as vascular endothelial growth factor and hepatocyte growth factor, forms an abnormal vascular structure with heterogeneous permeability, creating hypoxic regions that impede drug delivery and promote resistance. These stringent barriers have contributed to the high failure rate of drug candidates during early development stages, with FDA approval rates for CNS drugs notably lower than those for non-CNS drugs61. Understanding this barrier, recent approaches have been developed to overcome the BBB and enhance the bioavailability and efficacy of therapeutic agents, including nanoparticles, hyperosmolar therapy, convection-enhanced delivery and magnetic resonance-guided focused ultrasound62.
Clinical efficacy of targets
Finally, it is imperative to note that only a small subset of precision oncology strategies have resulted in meaningful changes to cancer management, with the clinical impact of targeted therapy varying considerably across cancer types63.
Basket trials, designed to evaluate the efficacy of targeted therapies across multiple cancer types sharing the same actionable target, have become a widely employed strategy to evaluate the effectiveness of molecular-targeted therapies for oncogene-defined subsets of cancers. This approach offers the advantage of simultaneously evaluating therapies for multiple tumor types, including rare cancers, potentially accelerating the drug development process. However, clinical trial outcomes revealed significant limitations in patient–treatment matching and drug accessibility. The phase II SHIVA trial (NCT01771458) found that although 30–50% of patients harbored only driver mutations, only 26% received matched treatments64. Similarly, in the phase II NCI-MATCH trial (NCT02465060), 38% of patients were matched to targeted therapies, but only 18% received treatment65. A pooled analysis of basket trials involving 1100 patients across 33 different cancers found that targeted therapy guided by biomarkers associated with driver mutations achieved only a 25% response rate, highlighting the limitations of static biomarker-driven approaches63. Beyond these low matching rates, none of the trials demonstrated a significant improvement in PFS63. Consistent with these findings, a recent large-scale real-world meta-analysis reported that genomic profiling-guided therapy was delivered to 15.6% of patients with metastatic solid tumors, achieving an objective response rate of 23.9% and modest gains in PFS and OS, although the overall proportion of patients deriving benefit remained limited66.
These findings emphasize a central challenge in the practical application of precision oncology: even when genomic profiling successfully identifies actionable mutations, the clinical utility of this information remains heavily constrained by the availability of effective agents and the low proportion of patients who ultimately receive matched therapy. While continued innovation in the development of pharmacological targets is crucial, more can be done to optimize the use of existing agents that may work beyond genomically matched indications, allowing us to maximize the potential of current therapies.
Therapeutic responses are shaped by the confluence of intricate tumor–microenvironment interactions, epigenetic changes, phenotypic plasticity and factors far exceeding our current understanding or measurement capabilities67. Biomarker-driven strategies simplify these complexities by reducing gene and molecular interactions to binary terms—such as the presence or absence of a specific mutation—and measurable parameters focused solely on targets under investigation. These approaches assume that shortlisted parameters alone are sufficient to predict clinical outcomes, disregarding the broader biological influences that govern treatment response68. In reality, gene products rarely function in isolation, as discussed previously, but operate within highly interconnected networks where parallel systems can compensate for missing components39. Correspondingly, precision oncology has advanced toward integrative multiomics, combining genomic, transcriptomic, proteomic, metabolomic and epigenomic data, offering a panoramic view of tumor biology69. Recent integrative multiomics studies on HGG tumors have comprehensively characterized the disease, stratifying patients into distinct molecular subtypes, each with its own unique predicted susceptibilities70,71,72,73. While these approaches have deepened our understanding of the disease, current strategies are largely retrospective or predictive in nature, and prospective evidence demonstrating that multiomics data can directly inform clinical decision-making remains limited. Furthermore, the generation of vast, multidimensional datasets poses substantial translational challenges, particularly in analyzing such complex information in a clinically relevant manner that directly captures how these alterations converge to determine therapy response and can be readily leveraged by clinicians69.
Collectively, these challenges underscore the need for strategies that bridge the complex and dynamic molecular networks with functional therapeutic insights. FPO has emerged as a promising strategy to address this gap by directly linking functional responses of biological systems with actionable therapeutic implications, evaluating tumor cell behavior in its entirety rather than focusing on discrete molecular alterations39.
FPO: the bridge between genotype and phenotype
FPO represents a transformative strategy that carries the potential to bridge the gap between molecular characterization and actionable therapeutic insights. Functional approaches that involve dynamic perturbations of tumor cells and the measurement of their responses capture the complexities and functional dependencies of cancer cells74. By harnessing the potential of existing drug libraries, FPO broadens the scope of treatment options available to patients, extending beyond molecular alterations and standard protocols. Prioritizing functional outputs over purely mechanistic understanding, FPO aligns with the need for clinically translatable strategies capable of addressing the heterogeneity and adaptability of malignancies, an approach that has shown promise in both pediatric and adult cancers75. This is especially noteworthy in the context of hard-to-treat, highly heterogeneous malignancies such as gliomas and sarcomas, and rare cancers, where limited molecular insights and a scarcity of effective treatment options pose considerable challenges to clinical management76. While FPO may be a relatively recent addition to the precision oncology landscape, its conceptual roots lie in foundational pharmacological practices: selecting therapeutic targets based on cellular responses, a principle through which virtually every chemotherapy agent used in oncology was derived from. This same principle is now applied at an individual patient level, allowing for personalized treatment strategies tailored to the specific functional characteristics of each patient’s cancer.
FPO workflows commence with the collection of patient-derived samples, such as tumor tissue or blood, which are utilized to generate patient-derived models, including organoids, explants or spheroids. These models are then employed for drug screening and functional assays to inform personalized therapeutic strategies. This approach may be used to predict patient responses to SOC therapies, enabling improved patient stratification, while also expanding to a broader drug repository to facilitate the repurposing of chemotherapy and targeted agents (Fig. 2). A central assumption underpinning the success of this strategy is the ability of these models to faithfully retain the molecular and phenotypic fidelity of their parental tumor, which is crucial for accurate clinical extrapolation. Patient-derived xenograft (PDX) models have been shown to faithfully recapitulate tumor features with high fidelity, accurately reflecting the biological characteristics and therapeutic responses observed in the original patient tumors77. In PDX models of HGGs, in addition to genomic, morphological and pathological similarities, patient-specific responses to TMZ and radiotherapy as part of the SOC treatment were successfully replicated78. Similarly, patient-derived organoids have demonstrated strong genotypic and phenotypic concordance with their parental tumors, including histological features, cellular diversity, gene expression and mutational profiles of corresponding parental tumors79. A growing area of research has focused on enriching these models for GSC populations, given their rarity and tendency to be missed by bulk genomic or transcriptomic analyses80. Enrichment of GSC populations has been successfully achieved through the establishment of patient-derived gliomaspheres, which serve as in vitro models that selectively expand stem-like cell populations from tumor samples for downstream functional assays81,82. Such enrichment enables direct interrogation of the drug sensitivities and adaptive resistance mechanisms of the very cells that drive recurrence, thereby guiding the development of more effective, patient-tailored therapies. Emerging platforms, such as microfluidics and engineered microenvironments aim to further recapitulate molecular and mechanical cues in human tissues83. Although the overarching concept of the various patient-derived models are the same, model-specific drawbacks and challenges that are cancer specific exist that need to be considered when selecting the appropriate model84.
Patient-derived HGG samples can be obtained from primary or recurrent tumors through patient biopsies or resections. Tumor cells are isolated and processed to generate patient-derived tumor models, including organoids, tumorspheres or xenografts. These models undergo high-throughput drug screening to identify effective, personalized drug combinations, facilitating the development of individualized therapeutic strategies for patients with HGG.
While FPO approaches are still emerging, they have demonstrated notable success across multiple cancer types in both pediatric and adult populations (Tables 1 and 2). In pediatric patients with relapsed or refractory malignancies, a prospective observational study (NCT03860376) demonstrated that FPO-derived treatments significantly improved PFS by more than 1.3-fold in 83% of pediatric cases compared to prior treatments85. Encouraged by these findings, ongoing trials are expanding this approach to larger cohorts of children (NCT05857969) and adults (NCT06024603)85. In adults, EXALT-1, a phase IIb trial (NCT03096821), was the first precision oncology trial to use functional assays to guide the identification of personalized therapies86. At a median follow-up of 23.9 months, 54% of patients demonstrated a more than 1.3-fold improvement in PFS compared to prior therapies, with 40% achieving exceptional responses86. Notably, 23% of patients remained progression-free at 12 months after FPO-guided therapy, significantly outperforming the 5% progression-free rate observed with previous treatments86. This work forms the basis for the recently initiated prospective randomized trial EXALT-2 (NCT04470947)87. Since then, several other trials have been initiated across both hematological malignancies and solid tumors, albeit considerably fewer in the latter75,88,89.
In HGG, four ongoing FPO trials are employing ex vivo approaches in guiding the management of primary and recurrent tumors (Table 2). Among them, the phase II EVIDENT trial (NCT05231655), part of a pan-cancer study on solid tumors, and the phase II 3D-PREDICT trial (NCT03561207) employ functional testing in treatment-naive HGG to predict response to SOC treatment. Preliminary results from the 3D-PREDICT study demonstrated the ability to identify TMZ responders irrespective of MGMT methylation status, with the identification of patients who responded to TMZ despite being MGMT unmethylated. Test-predicted responders had a median OS postsurgery of 11.6 months compared to 5.9 months for test-predicted nonresponders. With functional data provided within 7–10 days of tissue receipt, patients with HGG whose tumors are predicted not to respond favorably to TMZ could be preferentially directed to clinical trials or managed in ways that might offer greater clinical benefit90. Currently, MGMT promoter methylation status is the primary metric used to predict responses to TMZ clinically. However, its limitations are well-documented, as both methylated patients who respond poorly to therapy, and unmethylated patients who respond favorably have been observed in numerous studies. Consequently, while MGMT methylation status may serve as a predictor of response, its impact on clinical decision-making regarding TMZ administration remains limited91. Its role is also limited owing to the lack of approved alternative cytotoxic therapies in the first-line setting. These findings highlight the importance of complementing static biomarkers such as MGMT methylation status with functional drug studies to improve the selection of therapeutic targets for patients. Other ongoing trials include the phase Ia/IIb trial (NCT05532397) for recurrent HGG, which utilizes the quadratic phenotypic optimization platform (QPOP), a computational analytic platform designed to identify top ranking drug combinations and the phase II ATTRACT trial (NCT06512311), which similarly employs ex vivo drug sensitivity testing to personalize treatment strategies for patients92,93,94.
In addition to the identification of personalized drug targets, functional drug screens have played a pivotal role in accelerating therapeutic discovery. A notable example is the identification of panobinostat as a potential therapy for pediatric diffuse midline gliomas following a drug screen of 83 compounds. These findings have progressed to several phase I clinical trials (NCT02717455, NCT03566199 and NCT03632317)95. Similarly, in adult HGGs, functional screens across 132 neuroactive drugs revealed that the antidepressant vortioxetine synergizes with current SOC chemotherapies in vivo96. Furthermore, functional screens also provide valuable mechanistic insights, elucidating the mechanisms of action of existing approved drugs, thereby optimizing their clinical use. For instance, the lack of efficacy of BRAFi in BRAFV600E-mutated colorectal cancers, despite their success in melanoma with the same mutation, was clarified through functional studies, which identified feedback activation of EGFR in colorectal tumors97. Together, these examples highlight the potential of FPO both in driving therapeutic innovation and in refining the clinical application of existing agents through deeper mechanistic understanding of tumor biology.
Delivering drug sensitivity profiles within clinically relevant timelines remains a crucial hurdle, requiring efficient clinic-to-bench workflows and rapid generation of results. Achieving this demands close collaboration between neurosurgeons, oncologists, pathologists and scientists. Ideally, laboratories conducting FPO analysis should be located near major hospitals performing brain tumor resections to ensure timely access to high-quality tumor samples. A major obstacle in HGG is the acquisition of sufficient, high-quality tumor material for analysis, which often requires invasive surgical resections or biopsies. This limitation is exacerbated in deep-seated or recurrent tumors, where less than 10% of recurrent HGGs are eligible for resection. In addition, factors such as tumor necrosis and preoperative steroid use frequently impede successful ex vivo expansion of tumor cells, with these challenges particularly pronounced in the context of recurrent tumors84. Despite these obstacles, studies by Jacob et al. have demonstrated the feasibility of generating and biobanking patient-derived HGG organoids, achieving an overall success rate of 91.4%79. However, success rates vary depending on the tumor subtype, with lower success rates observed for IDH1-mutant tumors (66.7%) and recurrent tumors79. Most recently, a 2025 study indicated a 66.5% success rate for establishing patient-derived HGG organoids98, while other studies have reported success rates of 80–90% in HGG PDX models78.
Another key challenge of current models is their inability to replicate key aspects of the tumor microenvironment necessary for evaluating specific therapies. Therapeutic agents such as bevacizumab, which target angiogenesis cannot be effectively tested in many current models owing to their inability to replicate the vascular architecture required for assessing such treatments. Similarly, immunotherapies require advanced 3D culture systems capable of replicating immune–tumor interactions, as traditional models, including 2D systems and basic tumorspheres models lack the complexity needed to evaluate these therapies effectively. The development of advanced platforms, including microfluidic systems, cerebral organoids and humanized PDX models, has broadened the range of therapies99. In HGG, preclinical studies have demonstrated the feasibility of co-culturing organoids with chimeric antigen receptor T cells for therapeutic testing84. Notably, ongoing evaluations in six patients involve real-time parallel assays using the same therapeutics administered to patients, providing prospective insights into treatment responses100,101.
Although FPO theoretically enables the screening of an extensive range of therapeutic compounds, its practical application remains constrained by challenges such as limited tumor sample availability and the labor-intensive nature of comprehensive drug screening. In addition, with combinatorial strategies gaining prominence in oncology, the identification of effective drug combinations has become a priority in translational research. However, large-scale studies have shown that true synergies are uncommon and challenging to detect102,103. Development of biology-guided screening approaches that incorporate molecular context and mechanistic insights have shown to further improve the likelihood of uncovering clinically meaningful combinations from high-throughput screening strategies104. An emerging opportunity is to integrate these approaches with AI-based models, such as DeepSynergy and COSTAR, which could enable the assimilation of complex datasets and improve the prediction of synergistic drug pairs96,105. Such integration would build on the reciprocal and interdependent nature of genomics and FPO, with each providing critical insights that enhance the effectiveness of the other.
While FPO holds great potential to transform HGG management through personalized, biology-driven therapeutic strategies, it remains in its early stages and several challenges must be addressed before it can be widely adopted in clinical practice. Current evidence is largely derived from single-arm trials in highly selected patient populations, requiring cautious interpretation. The lack of randomized, prospective trials means that the true clinical benefit of FPO has yet to be definitively established, with future efforts focused on validating FPO within structured clinical trial frameworks. Furthermore, the utility of ex vivo drug platforms must be approached with nuance as they may demonstrate responses to targeted therapies even in the absence of the corresponding molecular target, such as BRAFi showing activity in tumors lacking BRAF mutations106. These discrepancies highlight the complexity of tumor biology, suggesting the need for a more comprehensive approach that integrates ex vivo results with genomic, transcriptomic and proteomic data, where cross-validation may enhance the accuracy of tumor vulnerability assessments and improve treatment selection (Fig. 3).
Genomic and functional approaches provide distinct but complementary insights for guiding treatment. Genomics enables rapid, evidence-based identification of actionable mutations, but may overlook nongenomic vulnerabilities. Functional testing, though more technically demanding, captures tumor dynamics and drug responses beyond what is detectable through molecular profiling.
It is imperative to acknowledge that while FPO approaches, based on N-of-1 trial designs offer many advantages, they are not suitable or necessary for all cancers107. These trials are resource intensive, requiring substantial time, effort and financial investment. Their personalized nature complicates scalability, making integration into routine clinical practice challenging. Furthermore, regulatory frameworks and evidence-based guidelines, designed for population-level efficacy, provide limited support and few incentives for such individualized approaches. The selection of treatment strategies and trial design should be guided by the unique characteristics of the cancer in question, ensuring alignment with its biological complexity and therapeutic challenges. For cancers with well-defined SOCs that are effective for the majority of patients, population-based trials may remain the most practical approach. By contrast, rare, refractory or highly heterogeneous cancers that lack an established SOC or perform poorly under existing treatments may benefit from N-of-1 trials107. In this context, personalized treatment strategies enabled by FPO-based N-of-1 trials present a promising alternative for HGGs, offering hope where conventional approaches have consistently fallen short.
Conclusion and future perspectives
In summary, despite two decades of effort, precision oncology in HGG has fallen short of delivering transformative clinical outcomes. FPO holds promise as a complementary approach, moving beyond molecular profiling to incorporate patient-specific models that capture the unique biology of each tumor. Advancing the scalability of patient-derived models and integrating them with high-throughput drug screening and molecular profiling are crucial for positioning FPO as a foundational approach for both therapeutic development and personalized patient treatment. A critical challenge remains in translating these insights into clinical workflows. Achieving this will require coordinated efforts among multidisciplinary teams, including oncologists, pathologists, bioinformaticians and laboratory scientists, to streamline sample processing, ensure robust data interpretation and deliver individualized treatment strategies.
Availability of data and materials
Data for this Review were identified through searches of PubMed, MEDLINE, ClinicalTrials.gov and references from relevant articles using the terms ‘functional precision oncology’, ‘high-grade glioma’, ‘patient-derived models’, ‘drug screening’, ‘clinical trials’ and ‘genomics’. Abstracts and meeting reports were included only when directly related to published work. Only articles published in English between 1975 and April 2025 were considered.
References
Tsimberidou, A. M., Fountzilas, E., Nikanjam, M. & Kurzrock, R. Review of precision cancer medicine: Evolution of the treatment paradigm. Cancer Treat. Rev. 86, 102019 (2020).
The Cancer Genome Atlas Research Network Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature 455, 1061–1068 (2008).
Louis, D. N. et al. The 2021 WHO Classification of Tumors of the Central Nervous System: a summary. Neuro Oncol. 23, 1231–1251 (2021).
Cruz Da Silva, E., Mercier, M. C., Etienne-Selloum, N., Dontenwill, M. & Choulier, L. A systematic review of glioblastoma-targeted therapies in phases II, III, IV clinical trials. Cancers https://doi.org/10.3390/cancers13081795 (2021).
Sferruzza, G. et al. A systematic review of immunotherapy in high-grade glioma: learning from the past to shape future perspectives. Neurol. Sci. 45, 2561–2578 (2024).
Marine, J. C., Dawson, S. J. & Dawson, M. A. Non-genetic mechanisms of therapeutic resistance in cancer. Nat. Rev. Cancer 20, 743–756 (2020).
Comba, A. et al. Uncovering spatiotemporal heterogeneity of high-grade gliomas: from disease biology to therapeutic implications. Front. Oncol. 11, 703764 (2021).
Ostrom, Q. T. et al. CBTRUS statistical report: primary brain and other central nervous system tumors diagnosed in the United States in 2015–2019. Neuro Oncol. 24, v1–v95 (2022).
Stupp, R. et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. N. Engl. J. Med. 352, 987–996 (2005).
Knisely, J. P. S. & Fine, H. A. Reirradiation for recurrent glioblastoma: what we know and what we do not. J. Clin. Oncol. 41, 1183–1188 (2023).
Walker, M. D. et al. Evaluation of BCNU and/or radiotherapy in the treatment of anaplastic gliomas. A cooperative clinical trial. J. Neurosurg. 49, 333–343 (1978).
Weller, M. & Le Rhun, E. How did lomustine become standard of care in recurrent glioblastoma? Cancer Treat. Rev. 87, 102029 (2020).
Gutin, P. H. et al. Phase II study of procarbazine, CCNU, and vincristine combination chemotherapy in the treatment of malignant brain tumors. Cancer 35, 1398–1404 (1975).
Hochberg, F. H., Linggood, R., Wolfson, L., Baker, W. H. & Kornblith, P. Quality and duration of survival in glioblastoma multiforme. Combined surgical, radiation, and lomustine therapy. JAMA 241, 1016–1018 (1979).
Walker, M. D. et al. Randomized comparisons of radiotherapy and nitrosoureas for the treatment of malignant glioma after surgery. N. Engl. J. Med. 303, 1323–1329 (1980).
Green, S. B. et al. Comparisons of carmustine, procarbazine, and high-dose methylprednisolone as additions to surgery and radiotherapy for the treatment of malignant glioma. Cancer Treat. Rep. 67, 121–132 (1983).
Sferruzza, G., Malcangi, M., Bosco, L. & Finocchiaro, G. Reassessing the efficacy of bevacizumab in newly diagnosed glioblastoma: a systematic review and external pseudodata-based analysis. Neurooncol. Adv. 6, vdad174 (2024).
Friedman, H. S. et al. Bevacizumab alone and in combination with irinotecan in recurrent glioblastoma. J. Clin. Oncol. 27, 4733–4740 (2009).
Vaz-Salgado, M. A. et al. Recurrent glioblastoma: a review of the treatment options. Cancers https://doi.org/10.3390/cancers15174279 (2023).
Khagi, S. et al. Recent advances in tumor treating fields (TTFields) therapy for glioblastoma. Oncologist https://doi.org/10.1093/oncolo/oyae227 (2025).
Crompton, D. J. et al. Tumor treating fields utilization and efficacy for glioblastoma at a large multicenter academic practice. J Clin Neurosci 138, 111395 (2025).
Riegel, D. C., Bureau, B. L., Conlon, P., Chavez, G. & Connelly, J. M. Long-term survival, patterns of progression, and patterns of use for patients with newly diagnosed glioblastoma treated with or without Tumor Treating Fields (TTFields) in a real-world setting. J. Neurooncol. 173, 49–57 (2025).
Kumthekar, P. et al. A qualitative evaluation of factors influencing tumor treating fields (TTFields) therapy decision making among brain tumor patients and physicians. BMC Cancer 24, 527 (2024).
Eskilsson, E. et al. EGFR heterogeneity and implications for therapeutic intervention in glioblastoma. Neuro Oncol. 20, 743–752 (2018).
Agarwal, S., Sane, R., Gallardo, J. L., Ohlfest, J. R. & Elmquist, W. F. Distribution of gefitinib to the brain is limited by P-glycoprotein (ABCB1) and breast cancer resistance protein (ABCG2)-mediated active efflux. J. Pharmacol. Exp. Ther. 334, 147–155 (2010).
Togashi, Y. et al. Cerebrospinal fluid concentration of gefitinib and erlotinib in patients with non-small cell lung cancer. Cancer Chemother. Pharmacol. 70, 399–405 (2012).
Zhao, J. et al. Cerebrospinal fluid concentrations of gefitinib in patients with lung adenocarcinoma. Clin. Lung Cancer 14, 188–193 (2013).
Wen, P. Y. et al. Phase I/II study of erlotinib and temsirolimus for patients with recurrent malignant gliomas: North American Brain Tumor Consortium trial 04-02. Neuro Oncol. 16, 567–578 (2014).
Reungwetwattana, T. et al. CNS response to osimertinib versus standard epidermal growth factor receptor tyrosine kinase inhibitors in patients with untreated EGFR-mutated advanced non-small-cell lung cancer. J. Clin. Oncol. 36, 3290–3297 (2018).
Soria, J. C. et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med 378, 113–125 (2018).
Villanacci, F. et al. Resistance of recurrent epidermal growth factor receptor-altered glioblastoma to anti-epidermal growth factor receptor targeted therapy with osimertinib. Oncologist https://doi.org/10.1093/oncolo/oyaf146 (2025).
Boongird, A. et al. Glioblastoma with novel EGFR mutations (T790M and exon 20 insertion) yet unresponsive to osimertinib: a case report. Genes Chromosomes Cancer 62, 423–429 (2023).
Makhlin, I. et al. Clinical activity of the EGFR tyrosine kinase inhibitor osimertinib in EGFR-mutant glioblastoma. CNS Oncol. 8, CNS43 (2019).
Johnson, M. L. et al. A phase 1 study to assess BDTX-1535, an oral EGFR inhibitor, in patients with glioblastoma or non-small-cell lung cancer. J. Clin. Oncol. 41, TPS9156–TPS9156 (2023).
Lassman, A. B. et al. Depatuxizumab mafodotin in EGFR-amplified newly diagnosed glioblastoma: a phase III randomized clinical trial. Neuro Oncol. 25, 339–350 (2023).
van den Bent, M. J. et al. Changes in the EGFR amplification and EGFRvIII expression between paired primary and recurrent glioblastomas. Neuro Oncol. 17, 935–941 (2015).
Sampson, J. H. et al. Immunologic escape after prolonged progression-free survival with epidermal growth factor receptor variant III peptide vaccination in patients with newly diagnosed glioblastoma. J. Clin. Oncol. 28, 4722–4729 (2010).
Niclou, S. P. Gauging heterogeneity in primary versus recurrent glioblastoma. Neuro Oncol. 17, 907–909 (2015).
Oda, K., Matsuoka, Y., Funahashi, A. & Kitano, H. A comprehensive pathway map of epidermal growth factor receptor signaling. Mol. Syst. Biol. https://doi.org/10.1038/msb4100014 (2005).
Li, X. et al. PI3K/Akt/mTOR signaling pathway and targeted therapy for glioblastoma. Oncotarget 7, 33440–33450 (2016).
Franz, D. N. et al. Efficacy and safety of everolimus for subependymal giant cell astrocytomas associated with tuberous sclerosis complex (EXIST-1): a multicentre, randomised, placebo-controlled phase 3 trial. Lancet 381, 125–132 (2013).
Akhavan, D., Cloughesy, T. F. & Mischel, P. S. mTOR signaling in glioblastoma: lessons learned from bench to bedside. Neuro Oncol. 12, 882–889 (2010).
Jackson, E. R. et al. ONC201 in combination with paxalisib for the treatment of H3K27-altered diffuse midline glioma. Cancer Res. 83, 2421–2437 (2023).
Noch, E. K. et al. Insulin feedback is a targetable resistance mechanism of PI3K inhibition in glioblastoma. Neuro Oncol. 25, 2165–2176 (2023).
Schindler, G. et al. Analysis of BRAF V600E mutation in 1,320 nervous system tumors reveals high mutation frequencies in pleomorphic xanthoastrocytoma, ganglioglioma and extra-cerebellar pilocytic astrocytoma. Acta Neuropathol. 121, 397–405 (2011).
Arbour, G. et al. Upfront BRAF/MEK inhibitors for treatment of high-grade glioma: a case report and review of the literature. Neurooncol. Adv. 4, vdac174 (2022).
Wen, P. Y. et al. Dabrafenib plus trametinib in patients with BRAF(V600E)-mutant low-grade and high-grade glioma (ROAR): a multicentre, open-label, single-arm, phase 2, basket trial. Lancet Oncol. 23, 53–64 (2022).
Villanueva, J., Vultur, A. & Herlyn, M. Resistance to BRAF inhibitors: unraveling mechanisms and future treatment options. Cancer Res. 71, 7137–7140 (2011).
Mellinghoff, I. K. et al. Vorasidenib in IDH1- or IDH2-mutant low-grade glioma. N. Engl. J. Med. 389, 589–601 (2023).
Tegally, H. et al. Discovering novel driver mutations from pan-cancer analysis of mutational and gene expression profiles. PLoS ONE 15, e0242780 (2020).
Kumar, S. et al. Passenger mutations in more than 2,500 cancer genomes: overall molecular functional impact and consequences. Cell 180, 915–927 e916 (2020).
Saito, Y. et al. Landscape and function of multiple mutations within individual oncogenes. Nature 582, 95–99 (2020).
Ostroverkhova, D., Przytycka, T. M. & Panchenko, A. R. Cancer driver mutations: predictions and reality. Trends Mol. Med. 29, 554–566 (2023).
Bostrom, M. & Larsson, E. Somatic mutation distribution across tumour cohorts provides a signal for positive selection in cancer. Nat. Commun. 13, 7023 (2022).
Verhaak, R. G. et al. Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell 17, 98–110 (2010).
Neftel, C. et al. An integrative model of cellular states, plasticity, and genetics for glioblastoma. Cell 178, 835–849 e821 (2019).
Prager, B. C., Bhargava, S., Mahadev, V., Hubert, C. G. & Rich, J. N. Glioblastoma stem cells: driving resilience through chaos. Trends Cancer 6, 223–235 (2020).
Bao, S. et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 444, 756–760 (2006).
MacLeod, G. et al. Fitness screens map state-specific glioblastoma stem cell vulnerabilities. Cancer Res. 84, 3967–3983 (2024).
Xie, X. et al. Recent advances in targeting the “undruggable” proteins: from drug discovery to clinical trials. Signal Transduct. Target. Ther. 8, 335 (2023).
Gribkoff, V. K. & Kaczmarek, L. K. The need for new approaches in CNS drug discovery: why drugs have failed, and what can be done to improve outcomes. Neuropharmacology 120, 11–19 (2017).
Giantini-Larsen, A. M. et al. Therapeutic manipulation and bypass of the blood-brain barrier: powerful tools in glioma treatment. Neurooncol. Adv. 7, vdae201 (2025).
Hazim, A. & Prasad, V. A pooled analysis of published, basket trials in cancer medicine. Eur. J. Cancer 101, 244–250 (2018).
Woodcock, J. & LaVange, L. M. Master protocols to study multiple therapies, multiple diseases, or both. N. Engl. J. Med. 377, 62–70 (2017).
Corrigendum to: The Molecular Analysis for Therapy Choice (NCI-MATCH) Trial: lessons for genomic trial design. J. Natl Cancer Inst. 114, 325, (2022).
Zerdes, I. et al. Comprehensive genome profiling for treatment decisions in patients with metastatic tumors: real-world evidence meta-analysis and registry data implementation. J. Natl Cancer Inst. 117, 1117–1124 (2025).
Gantner, B. N. et al. Metabolism and epigenetics: drivers of tumor cell plasticity and treatment outcomes. Trends Cancer 10, 992–1008 (2024).
Duval, M. X. The inadequacy of the reductionist approach in discovering new therapeutic agents against complex diseases. Exp. Biol. Med. 243, 1004–1013 (2018).
Akhoundova, D. & Rubin, M. A. Clinical application of advanced multi-omics tumor profiling: ahaping precision oncology of the future. Cancer Cell 40, 920–938 (2022).
Ding, Y. et al. Comprehensive multi-omics and machine learning framework for glioma subtyping and precision therapeutics. Sci. Rep. 15, 24874 (2025).
Migliozzi, S. et al. Integrative multi-omics networks identify PKCdelta and DNA-PK as master kinases of glioblastoma subtypes and guide targeted cancer therapy. Nat. Cancer 4, 181–202 (2023).
Kim, K. H. et al. Integrated proteogenomic characterization of glioblastoma evolution. Cancer Cell 42, 358–377 e358 (2024).
Wang, L. B. et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell 39, 509–528 e520 (2021).
Letai, A. Functional precision cancer medicine-moving beyond pure genomics. Nat. Med. 23, 1028–1035 (2017).
Letai, A., Bhola, P. & Welm, A. L. Functional precision oncology: testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell 40, 26–35 (2022).
Pauli, C. et al. A functional precision oncology approach to identify treatment strategies for myxofibrosarcoma patients. Mol. Cancer Res. 20, 244–252 (2022).
Woo, X. Y. et al. Conservation of copy number profiles during engraftment and passaging of patient-derived cancer xenografts. Nat. Genet. 53, 86–99 (2021).
Joo, K. M. et al. Patient-specific orthotopic glioblastoma xenograft models recapitulate the histopathology and biology of human glioblastomas in situ. Cell Rep. 3, 260–273 (2013).
Jacob, F. et al. A patient-derived glioblastoma organoid model and biobank recapitulates inter- and intra-tumoral heterogeneity. Cell 180, 188–204 e122 (2020).
Singh, S. K. et al. Identification of human brain tumour initiating cells. Nature 432, 396–401 (2004).
Galli, R. et al. Isolation and characterization of tumorigenic, stem-like neural precursors from human glioblastoma. Cancer Res. 64, 7011–7021 (2004).
Lee, J. et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell 9, 391–403 (2006).
Logun, M., Zhao, W., Mao, L. & Karumbaiah, L. Microfluidics in malignant glioma research and precision medicine. Adv. Biosyst. https://doi.org/10.1002/adbi.201700221 (2018).
Jacob, F., Ming, G. L. & Song, H. Generation and biobanking of patient-derived glioblastoma organoids and their application in CAR T cell testing. Nat. Protoc. 15, 4000–4033 (2020).
Acanda De La Rocha, A. M. et al. Feasibility of functional precision medicine for guiding treatment of relapsed or refractory pediatric cancers. Nat. Med. 30, 990–1000 (2024).
Kornauth, C. et al. Functional precision medicine provides clinical benefit in advanced aggressive hematologic cancers and identifies exceptional responders. Cancer Discov. 12, 372–387 (2022).
Kazianka, L. et al. Functional precision medicine vs genomics vs clinical experience: feasibility results from the multicentric, prospective, randomized controlled Exalt-2 Trial. Blood 142, 5006–5006 (2023).
Acanda de la Rocha, A. M., Berlow, N. E. & Azzam, D. J. Functional precision medicine: the future of cancer care. Trends Mol. Med. https://doi.org/10.1016/j.molmed.2024.10.015 (2024).
van Renterghem, A. W. J., van de Haar, J. & Voest, E. E. Functional precision oncology using patient-derived assays: bridging genotype and phenotype. Nat. Rev. Clin. Oncol. 20, 305–317 (2023).
Ledford, A. et al. Functional prediction of response to therapy prior to therapeutic intervention is associated with improved survival in patients with high-grade glioma. Sci. Rep. 14, 19474 (2024).
Hegi, M. E. & Stupp, R. Withholding temozolomide in glioblastoma patients with unmethylated MGMT promoter-still a dilemma? Neuro Oncol. 17, 1425–1427 (2015).
Toh, T. B. et al. Evaluation of ex vivo drug combination optimization platform in recurrent high grade astrocytic glioma: An interventional, non-randomized, open-label trial protocol. PLoS ONE 19, e0307818 (2024).
Berghoff, A. S. et al. Personalized targeted glioblastoma therapies by ex vivo drug screening: Advanced brain tumor therapy clinical trial (ATTRACT). J. Clin. Oncol. 43, TPS2097–TPS2097 (2025).
Berghoff, A. S. et al. Personalized targeted glioblastoma therapies by ex vivo drug screening: study protocol of the Advanced brain Tumor TheRApy Clinical Trial (ATTRACT). Neurooncol. Adv. 7, vdaf056 (2025).
Monje, M. et al. Phase I trial of panobinostat in children with diffuse intrinsic pontine glioma: a report from the Pediatric Brain Tumor Consortium (PBTC-047). Neuro Oncol. 25, 2262–2272 (2023).
Lee, S. et al. High-throughput identification of repurposable neuroactive drugs with potent anti-glioblastoma activity. Nat. Med. 30, 3196–3208 (2024).
Prahallad, A. et al. Unresponsiveness of colon cancer to BRAF(V600E) inhibition through feedback activation of EGFR. Nature 483, 100–103 (2012).
Souberan, A. et al. Brain tumoroids: treatment prediction and drug development for brain tumors with fast, reproducible, and easy-to-use personalized models. Neuro Oncol. 27, 415–429 (2025).
Moysidou, C. M., Barberio, C. & Owens, R. M. Advances in engineering human tissue models. Front. Bioeng. Biotechnol. 8, 620962 (2020).
Bagley, S. J. et al. Intrathecal bivalent CAR T cells targeting EGFR and IL13Ralpha2 in recurrent glioblastoma: phase 1 trial interim results. Nat. Med. 30, 1320–1329 (2024).
Logun, M. et al. Patient-derived glioblastoma organoids as real-time avatars for assessing responses to clinical CAR-T cell therapy. Cell Stem Cell https://doi.org/10.1016/j.stem.2024.11.010 (2024).
O’Neil, J. et al. An unbiased oncology compound screen to identify novel combination strategies. Mol. Cancer Ther. 15, 1155–1162 (2016).
Jaaks, P. et al. Effective drug combinations in breast, colon and pancreatic cancer cells. Nature 603, 166–173 (2022).
Ariey-Bonnet, J. et al. Combination drug screen targeting glioblastoma core vulnerabilities reveals pharmacological synergisms. EBioMedicine 95, 104752 (2023).
Preuer, K. et al. DeepSynergy: predicting anti-cancer drug synergy with deep learning. Bioinformatics 34, 1538–1546 (2018).
Kohtamaki, L. et al. High-throughput ex vivo drug testing identifies potential drugs and drug combinations for NRAS-positive malignant melanoma. Transl. Oncol. 15, 101290 (2022).
Gouda, M. A., Buschhorn, L., Schneeweiss, A., Wahida, A. & Subbiah, V. N-of-1 trials in cancer drug development. Cancer Discov. 13, 1301–1309 (2023).
Labots, M. et al. Selection of protein kinase inhibitors based on tumor tissue kinase activity profiles in patients with refractory solid malignancies: an interventional molecular profiling study. Oncologist 23, 1135–e1118 (2018).
Mathonnet, M. et al. ONCOGRAM: study protocol for the evaluation of therapeutic response and survival of metastatic colorectal cancer patients treated according to the guidelines of a chemosensitivity assay, the Oncogramme(R). Trials 22, 556 (2021).
Lau, L. M. S. et al. Precision-guided treatment in high-risk pediatric cancers. Nat. Med. 30, 1913–1922 (2024).
Coffey, D. G. et al. High-throughput drug screening and multi-omic analysis to guide individualized treatment for multiple myeloma. JCO Precis. Oncol. https://doi.org/10.1200/PO.20.00442 (2021).
Kazianka, L. et al. Comparing functional and genomic-based precision medicine in blood cancer patients. Hemasphere 9, e70129 (2025).
Diehl, A., Harris, W. P. & King, G. T. Trial in progress: A feasibility study of multi-platform profiling using biospecimens from patients with resected biliary tract cancer. J. Clin. Oncol. 39, TPS359–TPS359 (2021).
King, G. et al. 114P Prospective longitudinal tumor-informed ctDNA in resectable biliary tract cancers. Ann. Oncol.34, S225 (2023).
Chan, S. P. Y. et al. Functional combinatorial precision medicine for predicting and optimizing soft tissue sarcoma treatments. npj Precis. Oncol. 9, 83 (2025).
Sun, H. et al. Prediction of clinical precision chemotherapy by patient-derived 3D bioprinting models of colorectal cancer and its liver metastases. Adv. Sci. 11, e2304460 (2024).
Varinelli, L. et al. Colorectal carcinoma peritoneal metastases-derived organoids: results and perspective of a model for tailoring hyperthermic intraperitoneal chemotherapy from bench-to-bedside. J. Exp. Clin. Cancer Res. 43, 132 (2024).
Jaulin, F. et al. 12P ORGANOTREAT-01: a pioneer phase I/II multicenter trial of organoid-driven precision medicine for refractory colorectal cancer. Ann. Oncol. 36, S9–S10 (2025).
Wang, H., He, M., Zhong, L. & Zou, D. EPV191/# 327 Drug screening of patient-derived organoids from ovarian cancer culture to personalized therapy, an exploratory research. Int. J. Gynecol. Cancer 31, A105–A106 (2021).
Vaklavas, C. et al. TOWARDS study: patient-derived xenograft engraftment predicts poor survival in patients with newly diagnosed triple-negative breast cancer. JCO Precis. Oncol. 8, e2300724 (2024).
Seiler, R. et al. Guidance of adjuvant instillation in intermediate-risk non-muscle invasive bladder cancer by drug screens in patient derived organoids: a single center, open-label, phase II trial. BMC Urol. 23, 89 (2023).
Berlow, N. et al. Clinical utility and accessibility of functional precision medicine for relapsed/refractory pediatric and adult cancers. J. Clin. Oncol. 42, 1551–1551 (2024).
Rahman, M. M. et al. 2302P Efficacy assessment of targeted and immunotherapies for personalised treatment of melanoma using 2D and 3D ex-vivo assays. Ann. Oncol. 34, S1177 (2023).
Acknowledgements
All figures were created with BioRender.com. T.B.T. acknowledges funding from the WisDM Seed Fund (WisDM/Seed/002/2021, NUS) and Tahir-Surgery Education and Research Funding, NUSMed Department of Surgery. A.L.A.W. acknowledges funding from the National University Health System (NUHS) Seed Fund (NUHSRO/2021/052/RO5+6/Seed-Mar/01).
Author information
Authors and Affiliations
Contributions
L.G.S.H. wrote the manuscript and prepared the figures. D.K.H.T., Y.L.A., D.H., T.B.T. and A.L.A.W. edited the manuscript and figures. T.B.T. and A.L.A.W. conceptualized, provided guidance and supervision. All authors have read and agree with the final version of this manuscript.
Corresponding author
Ethics declarations
Competing interests
D.H. is a scientific co-founder and shareholder of KYAN Technologies, which has licensed intellectual property pertaining to AI-based oncology drug development and functional precision medicine.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Goh, L.S.H., Thng, D.K.H., Ang, Y.L.E. et al. The functional imperative in high-grade glioma. Exp Mol Med (2026). https://doi.org/10.1038/s12276-025-01614-x
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s12276-025-01614-x