Abstract
Dentin, the main component of dental hard tissues, is produced by differentiated odontoblasts. How odontoblast differentiation is regulated remains understudied. Here, we screen that the expression of membrane-associated RING finger protein 2 (March2) is the highest among all March family members, with an increasing trend during odontoblast differentiation. In mouse incisors and molars, MARCH2 is moderately expressed in the undifferentiated dental papilla cells and strongly expressed in the odontoblasts. Knockdown and overexpression experiments demonstrate that MARCH2 inhibits odontoblastic differentiation of mouse dental papilla cells (mDPCs). Additionally, both March2 deficient mice and mice with odontoblast specific knockdown of March2 exhibit the phenotype of increased dentin thickness, accelerated dentin deposition as well as elevated expression levels of odontoblast markers compared with control littermates. Therefore, MARCH2 plays an inhibitory role in odontoblast differentiation. Mechanistically, MARCH2 interacts with protein tyrosine phosphatase receptor delta (PTPRD) and facilitates its K27-linked polyubiquitination and subsequent degradation, which is dependent on the ligase activity of MARCH2. The presence of MARCH2 promotes the translocation of PTPRD from the cell membrane to the lysosome, thereby enhancing its degradation via the lysosomal pathway. Further experiments show that knockdown of endogenous Ptprd impairs odontoblastic differentiation of mDPCs. Ptprd and March2 double knockdown in mDPCs apparently reversed the enhanced odontoblastic differentiation by knockdown of March2 alone, indicating that MARCH2 inhibits odontoblastic differentiation by promoting PTPRD degradation. This study unveils a novel mechanism where an E3 ubiquitin ligase regulates odontoblast differentiation through post-translational modification of a membrane protein, highlighting a promising direction for future exploration.
Similar content being viewed by others
Introduction
As the main constituent of dental hard tissues, dentin supports the outer enamel and protects the inner dental pulp. At the bell stage of tooth development, the mesenchymal cells adjacent to the dental epithelium differentiate into odontoblasts and initiate dentin production. Abnormalities of odontoblast differentiation can lead to defects in dentin formation.1
Membrane proteins are essential components of the cell membrane and play pivotal roles in various physiological processes.2 Notably, some membrane proteins participate in the differentiation processes of diverse tissue or cell types,3,4 including odontoblast differentiation.5 Ubiquitination, a form of post-translational modification, modulates the conformation, stability, and localization of membrane proteins, thereby influencing their function.6,7 The membrane-associated RING-CH (MARCH) E3 ubiquitin ligases are a family of RING-type E3 ligases, comprising 11 members, named membrane-associated RING finger protein 1 (MARCH1) to MARCH11.8 All members of the family except MARCH7 and MARCH10 contain at least one transmembrane domain and are located at the cell membranes or organelle membranes,9 and function through controlling the stability and activity of various cellular membrane proteins.10 Members of the MARCH family play critical roles in many developmental processes. For instance, MARCH1 mediates the removal of major histocompatibility complex class II (MHCII) and cluster of differentiation 86 (CD86) on dendritic cells residing in the lymph nodes, thereby promoting the differentiation of Type 2T helper cells.11 MARCH2 targets Xenopus dishevelled (DSH) for ubiquitin-mediated degradation, counteracting Wnt signaling and thus controlling proper head development.8 MARCH10 is involved in spermiogenesis by regulating the assembly and maintenance of the flagella in developing spermatids.12 However, the role of MARCH family members in odontoblast differentiation and dentinogenesis as well as the membrane proteins targeted by them remain unclear.
In this study, we show that MARCH2 exhibits the highest expression level among all MARCH family members during odontoblastic differentiation of mouse dental papilla cells (mDPCs). Knockdown and overexpression experiments reveal that MARCH2 inhibits odontoblastic differentiation of mDPCs. Using March2 deficient mice and mice with odontoblast specific knockdown of March2, we demonstrate that in mouse molars and incisors, MARCH2 exerts an inhibitory effect on dentinogenesis and odontoblast differentiation. Mechanistically, MARCH2 targets the membrane protein, protein tyrosine phosphatase receptor delta (PTPRD) for polyubiquitination, which promotes subsequent lysosomal degradation of PTPRD.
Results
MARCH2 is expressed during odontoblast differentiation
To screen the expression levels of MARCH family members during odontoblastic differentiation, real-time quantitative polymerase chain reaction (RT-qPCR) was conducted using mDPCs cultured in differentiation medium for 0 day (d), 3 d and 5 d. The results revealed that, among all MARCH members, the expression of March2 was the highest with an increasing trend during odontoblastic differentiation (Supplementary Fig. S1a, b). Next, Western blot (WB) analysis was performed. Similarly, the results showed a gradual increase of MARCH2 protein during this process (Supplementary Fig. S1c, d). Meanwhile, immunocytochemistry (ICC) also revealed that the expression of MARCH2 increased with odontoblastic differentiation, and MARCH2 was present in the subcellular compartments of the cytoplasm and cell membrane but not in the nucleus (Supplementary Fig. S1e, f). Analysis of single-cell sequencing data from tooth samples at postnatal day 3.5 (PN3.5)13 showed that March2 exhibited the highest average mRNA level within the odontoblast subpopulation among all dental populations (Supplementary Fig. S2a–d).
To further explore the in vivo expression of MARCH2, immunohistochemistry (IHC) was performed on PN2 mouse incisors which encompass all stages of odontoblast differentiation within a single section (Fig. 1a a’). The results showed that MARCH2 exhibited a moderate level in the undifferentiated dental papilla cells (DPCs) and polarizing odontoblasts (Fig. 1a b’, c’), and an intense level in the secretory and mature odontoblasts (Fig. 1a d’, e’). It was noted that MARCH2 was extensively distributed except the nucleus compartment, consistent with its reported subcellular localization at endosomes, lysosomes and the cell membrane.14 Meanwhile, IHC of MARCH2 was also performed in mouse molars. The results showed that MARCH2 was moderately expressed in the undifferentiated DPCs and strongly expressed in the odontoblasts at embryonic day 18.5 (E18.5), PN3, PN7, and PN10 (Fig. 1b a’–d’).
The expression pattern of MARCH2 during odontoblast differentiation in vivo. a Immunohistochemistry (IHC) of MARCH2 in postnatal day 2 (PN2) mouse incisor. b’, c’, d’, and e’ are enlarged views of the rectangles in a’. Red arrows and red dashed lines indicate dental papilla cells or odontoblasts. b IHC of MARCH2 in mouse molars from embryonic day 18.5 (E18.5) to PN10. a”-d” and a”’-d”’ are enlarged views of the rectangles in a’-d’. Red arrows and red dashed lines mark the odontoblasts. Black arrows mark the dental papilla cells. The black dotted lines represent the boundaries between pre-dentin and odontoblast layer. Od odontoblasts, DPC dental papilla cell, Pd pre-dentin. Except for the labeled scale bars, all others are 5 µm
In addition, ameloblasts were also positive for MARCH2 (Fig. 1a, b), but no positive signals were observed in the negative controls (Supplementary Fig. S3).
MARCH2 suppresses odontoblastic differentiation of mDPCs in vitro
To investigate the role of MARCH2 in odontoblastic differentiation of mDPCs, knockdown experiments were performed using small interfering RNAs (siRNAs), which result in a significant reduction of March2 mRNA as assessed by RT-qPCR (Fig. 2a). The levels of odontoblast marker genes including dentin matrix protein 1 (Dmp1), dentin sialophosphoprotein (Dspp) and collagen type I alpha 1 chain (Col1a1) were significantly increased in the March2 knockdown cells after culture in differentiation medium as shown by RT-qPCR (Fig. 2b). In accordance, WB analysis verified that the protein levels of DMP1, DSPP and collagen type I (COL I) were elevated in the March2 knockdown group (Fig. 2c; Supplementary Fig. S4a). Alkaline phosphatase (ALP) staining and alizarin red S (ARS) staining showed that both ALP activity (Fig. 2d) and mineralized nodule formation (Fig. 2e; Supplementary Fig. S4b) were increased upon March2 knockdown.
Knockdown of March2 promotes the odontoblastic differentiation of mouse dental papilla cells (mDPCs) in vitro. a The knockdown efficiency of two March2 small interfering RNAs (siRNAs) in mDPCs was assessed by real-time quantitative PCR (RT-qPCR) (n = 4). b The effects of March2 knockdown on mRNA levels of Dmp1, Dspp and Col1a1 after differentiation induction of mDPCs for 5 days (5 d) (n = 3). c The effects of March2 knockdown on protein levels of DMP1, DSPP and COL I after differentiation induction of mDPCs for 5 d. d The effects of March2 knockdown on the alkaline phosphatase (ALP) activity detected by ALP staining. e The effects of March2 knockdown on the formation of mineralized nodule assessed by Alizarin Red S (ARS) staining. Scr Scramble siRNA, D differentiation induction. The statistical difference was analyzed by one-way ANOVA with Tukey’s post hoc test (a, b), where *P < 0.05; ***P < 0.001; ****P < 0.000 1
Next, to further confirm its role, MARCH2 was overexpressed in mDPCs. The results showed that, after an increase of MARCH2 protein by 13.5-fold (Supplementary Fig. S5a), mRNA and protein levels of DMP1, DSPP and COL I (Supplementary Fig. S5b–d) as well as ALP activity (Supplementary Fig. S5e) and calcified nodule formation (Supplementary Fig. S5f, g) were all downregulated compared with the control group.
Therefore, both the knockdown and overexpression experiments support that MARCH2 exerts an inhibitory role in the odontoblastic differentiation of mDPCs.
March2 deletion leads to increased dentin formation in vivo
To observe the in vivo function of MARCH2 in dentin formation, March2 deficient mice were generated using CRISPR/Cas9 technology (Supplementary Fig. S6a). Sanger sequencing of the March2 deficient mice revealed a 7 bp deletion in exon 3, leading to a frameshift and premature termination of translation of MARCH2 protein (Supplementary Fig. S6b). The successful inactivation of March2 was further confirmed by IHC analysis (Supplementary Fig. S6c). Hematoxylin and eosin (H&E) staining revealed that the widths of dentin in March2 deficient mice were significantly increased compared with those in control littermates at PN10, PN14 and PN21 (Fig. 3a; Supplementary Fig. S7a). Additionally, double-calcein labeling experiments suggested an accelerated rate of dentin deposition in March2 deficient mice (Fig. 3b; Supplementary Fig. S7b). Micro-computed tomography (micro-CT) confirmed increased dentin thickness in the molars of March2 deficient mice (Fig. 3c). Three-dimensional (3D) reconstruction analysis of PN14 and PN21 molars revealed increased dentin volume in March2 deficient mice, while the enamel volume remained comparable to that of control littermates (Fig. 3d; Supplementary Fig. S7c). Micro-CT analysis of incisors also revealed increased dentin thickness in March2 deficient mice compared with control littermates (Supplementary Fig. S8a). Furthermore, double-calcein labeling experiments indicated accelerated dentin deposition rate in the incisors of March2 deficient mice (Supplementary Fig. S8b, c). These findings indicate that MARCH2 inhibits dentinogenesis in vivo.
March2 deficient mice exhibit enhanced odontoblast differentiation and dentinogenesis. a Hematoxylin and Eosin (H&E) staining of molars from control littermates and March2 deficient mice at PN10 (a’, b’), PN14 (c’, d’) and PN21 (e’, f’). The white dotted lines represent the boundaries between dentin and odontoblast layer. The white arrows represent the widths of dentin. a”-f” are magnified views of the black box in a’-f’. Scale bar, 100 μm. b Calcein labeling was performed to compare the dentin deposition rate between control littermates and March2 deficient mice. The white arrows indicate the dentin deposited in the period between drug administrations. Scale bar, 10 μm. c Micro-computed tomography (micro-CT) images of the mandibular molars from control littermates and March2 deficient mice at PN10 (a’, b’), PN14 (c’, d’) and PN21 (e’, f’). The black lines delineate the interface between the dentin and enamel. Scale bar, 100 μm. d Three-dimensional (3D) reconstruction of the first mandibular molars from control littermates and March2 deficient mice at PN14 and PN21. Scale bar, 100 μm. e Immunofluorescent (IF) staining of the odontoblast markers DMP1 (a’, a”), DSPP (b’, b”) and COL I (c’, c”) in molars of control littermates (a’, b’, c’) and March2 deficient (a”, b”, c”) mice at PN14. Scale bar, 10 μm. CTR control littermates, March2 KO March2 deficient, D dentin, Od odontoblasts
To verify the in vivo effects of MARCH2 on the differentiation of odontoblasts, the cell type responsible for dentin formation, immunofluorescence (IF) was performed to assess the expression levels of the odontoblast markers including DMP1, DSPP and COL I. The results demonstrated that all these markers were upregulated in the odontoblast layer of March2 deficient mice at PN14 (Fig. 3e; Supplementary Fig. S7d). Therefore, deletion of March2 leads to enhanced dentinogenesis and odontoblast differentiation in mouse molars.
Mice with specific knockdown of March2 in the odontoblast layer recapitulates the dentin phenotype of March2 deficient mice
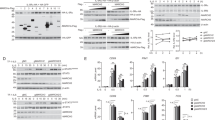
Systemic March2 inactivation may introduce confounding effects from other tissues. To achieve specific knockdown of March2 in the odontoblast layer, we constructed an adeno-associated virus serotype 6 (AAV6) vector expressing a short hairpin RNA (shRNA) targeting March2 under the control of the mouse Dmp1 promoter (designated as AAV6-shMarch2), which was locally injected into the mandibular molar germs (Fig. 4a) as previously reported.1 This vector also includes a GFP reporter to enable tracking of transduction efficiency. 2 days after injection, GFP was detected in the odontoblast layer but not other cell types of AAV6-shMarch2-injected molars (Fig. 4b) and the percentage of GFP-positive odontoblasts reached 83% (Supplementary Fig. S9a). IF analysis further showed reduced MARCH2 expression within the odontoblast layer, whereas its expression in the dental epithelium and other cells remained unchanged (Fig. 4c), indicating successful specific knockdown of March2 in the odontoblast layer. H&E staining confirmed that the widths of dentin in AAV6-shMarch2 group were increased compared with those in AAV6-Scramble shRNA (AAV6-shScr) group at PN10, PN14 and PN21 (Fig. 4d; Supplementary Fig. S9b). Micro-CT analysis revealed that AAV6-shMarch2-injected molars exhibited increased dentin thickness (Fig. 4e). 3D reconstruction of PN14 and PN21 molars demonstrated increase of dentin volume in these mice, while enamel volume remained similar to that of AAV6-shScr-injected control molars (Fig. 4f; Supplementary Fig. S9c). Moreover, IF showed upregulated expression of DMP1, DSPP and COL I in the odontoblast layer of AAV6-shMarch2 group at PN14 (Fig. 4g; Supplementary Fig. S9d). Together, these results indicate that specific knockdown of March2 in the odontoblast layer enhances odontoblast differentiation and dentinogenesis.
In vivo knockdown of March2 in odontoblasts leads to increased odontoblast differentiation and dentin formation in mice. a Schematic diagram showing time points for AAV injection and tissue collection. Mice were injected with AAV6-shScr or AAV6-shMarch2 at PN1. Samples were collected at PN3 to assess the transduction efficiency, and harvested at PN10, PN14 and PN21 to evaluate the phenotype. b Mandibles from PN3 mice after injection with AAV6-shMarch2 or PBS were processed for frozen sectioning, and GFP fluorescence was examined. c IF staining of MARCH2 in molars of PN3 mice in AAV6-shScr or AAV6-shMarch2 group. d HE staining of molars in the AAV6-shScr and AAV6-shMarch2 group at PN10 (a’, b’), PN14 (c’, d’) and PN21 (e’, f’). a”-f” are magnified views of the black boxes in a’-f’. The white dotted lines in b, c, d represent the boundaries between dentin and the odontoblast layer. The white arrows in d represent the widths of dentin. e Micro-CT images of the mandibular molars at PN10 (a’, b’), PN14 (c’, d’) and PN21 (e’, f’) after AAV6-shScr or AAV6-shMarch2 injection. The black lines delineate the interface between the dentin and enamel. f 3D reconstruction of the first mandibular molars from mice at PN14 and PN21 after following injection with AAV6-shScr or AAV6-shMarch2. g IF staining of the odontoblast markers DMP1 (a’, a”), DSPP (b’, b”) and COL I (c’, c”) in molars of PN14 mice of AAV6-shScr (a’, b’, c’) or AAV6-shMarch2 (a”, b”, c”) group. Ep epithelium, Od odontoblasts, DPC dental papilla cells, D dentin. Scale bar, 100 μm
MARCH2 polyubiquitinates PTPRD and promotes its lysosomal degradation
To explore the underlying mechanisms by which MARCH2 influences odontoblast differentiation, the bioinformatic platforms including UbiBrowser (http://ubibrowser.bio-it.cn/) and BioGrid (https://www.biogrid.org.au/) were used to analyze potential targets of MARCH2. After screening, PTPRD, a member of the Leukocyte Common Antigen-Related Receptor Protein Tyrosine Phosphatase (LAR-RPTP) family, attracted our attention as a potential candidate target of MARCH2. As a membrane protein, PTPRD has been found to be involved in diverse developmental processes, including neurogenesis,15 gliogenesis,3 and adipogenesis.16
To confirm whether PTPRD acted as the substrate of MARCH2 during odontoblast differentiation, several lines of experiments were conducted. Firstly, the physical interaction between MARCH2 and PTPRD, which is the prerequisite for PTPRD as the substrate of MARCH2, was analyzed in cultured cell types using co-immunoprecipitation (co-IP) assays. The results showed that overexpressed MARCH2 and PTPRD were able to pull down each other in human embryonic kidney 293 T (HEK293T) cells (Supplementary Fig. S10a, b). Furthermore, MARCH2 was found to interact with endogenous PTPRD in mDPCs (Supplementary Fig. S10c). Meanwhile, in situ proximity ligation assay (PLA) demonstrated the physical proximity between the overexpressed MARCH2 and endogenous PTPRD in mDPCs (Supplementary Fig. S10d). Ubiquitination modification includes polyubiquitination and monoubiquitination according to the ubiquitin chains.17 Co-IP assays showed that wild-type MARCH2 increased the polyubiquitination of PTPRD but its ligase-inactive mutants (March2 C64S, C67S, and C80S) did not (Fig. 5a). Meanwhile, knockdown of March2 in mDPCs reduced the ubiquitination of endogenous PTPRD as assessed by in situ PLA (Fig. 5b). Therefore, MARCH2 polyunbiquitinates PTPRD.
MARCH2 promotes the polyubiquitination of PTPRD and its subsequent degradation. a Wild-type March2-Myc or its ligase-inactive mutants (C64S, C67S, and C80S) were overexpressed together with Ptprd-Flag and HA-ubiquitin in human embryonic kidney 293T (HEK293T) cells. Co-immunoprecipitation (Co-IP) assays show that MARCH2 ubiquitinated PTPRD dependent of its ligase activity. b The level of ubiquitinated PTPRD is reduced following March2 knockdown in mDPCs, as demonstrated by in situ proximity ligation assay (PLA) with anti-PTPRD and anti-ubiquitin antibodies. Red punctate fluorescence indicates the physical proximity of two proteins. c Cycloheximide (CHX) chase assays demonstrate that overexpressed MARCH2 decreases the half-life of endogenous PTPRD protein in cultured mDPCs. Cells were treated with CHX for 0, 2, 5 and 8 h (h) before harvest for WB analysis. d Quantitative analysis of the relative PTPRD levels at different time points in (c), suggesting that MARCH2 promotes the degradation of PTPRD. e Wild-type March2-Myc or its ligase-inactive mutants (C64S, C67S, and C80S) were overexpressed in mDPCs. WB analysis shows that wild-type MARCH2 reduced the endogenous protein level of PTPRD but its ligase-inactive mutants did not show this effect. f IF staining shows a higher level of PTPRD protein in the molars of March2 deficient mice at PN10 and PN14. The white dashed line represents the junction between odontoblasts and dentin. Scr Scramble siRNA, CTR control littermates, March2 KO March2 deficient, Od odontoblasts. Scale bar, 10 μm
Polyubiquitination leads to different outcomes for the substrates, with protein degradation being the most frequent.18 Therefore, we examined whether MARCH2 targets PTPRD for degradation. Our results showed that MARCH2 overexpression did not affect the mRNA level of Ptprd, ruling out the possibility of transcriptional regulation of PTPRD by MARCH2. (Supplementary Fig. S11). Cycloheximide (CHX, a protein synthesis inhibitor) chase assays showed that MARCH2 overexpression indeed increased the degradation velocity of PTPRD in mDPCs (Fig. 5c, d). Additionally, wild-type MARCH2, but not its ligase-inactive mutants, promoted the degradation of endogenous PTPRD (Fig. 5e; Supplementary Fig. S12a), suggesting that the increased degradation of PTPRD caused by MARCH2 is dependent on the ligase activity of MARCH2. Consistently, IF showed that PTPRD protein level was elevated in the odontoblasts of March2 deficient mice at PN10 and PN14 (Fig. 5f; Supplementary Fig. S12b). To define ubiquitination type of PTPRD mediated by MARCH2, we co-transfected PTPRD with ubiquitin mutants that contain only a single lysine residue. This analysis revealed that MARCH2 specifically promoted K27-linked polyubiquitination of PTPRD. By cotransfection of PTPRD with ubiquitin mutants where a single lysine residue was substituted with arginine, we found that MARCH2 could not promote the polyubiquitination of PTPRD only when K27 was mutated to arginine (Supplementary Fig. S13). Therefore, MARCH2 facilitates the K27-linked polyubiquitination of PTPRD and its subsequent degradation.
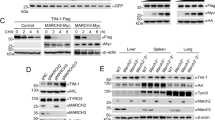
To investigate the degradation pathway of PTPRD, we used inhibitors of proteasomal and lysosomal pathways and cultured for different time intervals. The results showed that application of Chloroquine (CQ, a lysosome inhibitor) but not MG132 (a proteasome inhibitor) attenuated MARCH2-induced PTPRD degradation, suggesting that MARCH2 triggered lysosomal rather than proteasomal degradation of PTPRD (Fig. 6a, b). When PTPRD alone was overexpressed, it was predominantly present on the cell membrane (Fig. 6c a’–c’). However, when MARCH2 was concurrently overexpressed, PTPRD was progressively translocated to the cytoplasm and exhibited colocalization with lysosome (Fig. 6c d’–f’), indicating that MARCH2 promotes the translocation of PTPRD from the cell membrane to the intracellular lysosomes. These findings demonstrated that MARCH2 facilitates K27-linked polyubiquitination of PTPRD, thereby directing its degradation through the lysosomal pathway.
MARCH2 promotes the lysosomal degradation of PTPRD. a Chloroquine (CQ), a lysosome inhibitor, suppresses the degradation of endogenous PTPRD by overexpressed MARCH2 in mDPCs, while MG132, a proteasome inhibitor, does not. Cells were cultured with CHX and the inhibitors for up to 8 h before harvest as indicated, with MG132 and CQ administered at working concentrations of 40 nM and 10 mol/L, respectively. b Quantitative analysis of the relative PTPRD levels at different time points in (a). c MARCH2 facilitates the translocation of PTPRD from the cell membrane to intracellular area where PTPRD shows co-localization with lysosomes. Henrietta Lacks (Hela) cells were transfected with Ptprd-Flag plasmid alone or together with March2-Myc plasmid, and cultured for 16 h, 18 h or 20 h. The cells were incubated with LysoTracker Red (200 mol/L) for 1 h prior to collection. After collection, cells were fixed with 4% paraformaldehyde, and then stained with mouse anti-FLAG and rabbit anti-MYC antibodies. PTPRD and MARCH2 were visualized using Alexa Fluor 488- and Alexa Fluor 647-conjugated secondary antibodies, respectively. Scale bar, 10 μm
PTPRD is expressed in the odontoblast layer and promotes the odontoblastic differentiation of mDPCs in vitro
To unveil the role of PTPRD during odontoblast differentiation, the expression of PTPRD in mouse teeth was first investigated. IHC showed that PTPRD was expressed in the odontoblast layer of both incisors and molars in mice (Supplementary Fig. S14). Ptprd was then knocked down in mDPCs using two siRNAs which gave rise to over 70% reduction of Ptprd mRNA as shown by RT-qPCR (Supplementary Fig. S15a). Compared with the control group transfected with scramble siRNA, the mRNA levels of Dmp1, Dspp and Col1a1 were significantly reduced in the Ptprd knockdown group after culture in differentiation medium (Supplementary Fig. S15b). Consistently, WB analysis demonstrated reduced protein levels of DMP1, DSPP and COL I (Supplementary Fig. S15c, d). Additionally, ALP and ARS staining revealed decreased ALP activity (Supplementary Fig. S15e) and reduced formation of mineralized nodules (Supplementary Fig. S15f, g) following Ptprd knockdown. These results indicate that PTPRD plays a positive role in the odontoblastic differentiation of mDPCs.
MARCH2 inhibits odontoblastic differentiation via PTPRD
To determine whether PTPRD was the key mediator of the role of MARCH2 during odontoblast differentiation, simultaneous knockdown of March2 and Ptprd in mDPCs was performed. March2 knockdown led to upregulation of both mRNA and protein levels of DMP1, DSPP and COL I, which were remarkably reversed by concurrent knockdown of Ptprd shown by RT-qPCR and WB analysis (Fig. 7a, b; Supplementary Fig. S16a). Additionally, concurrent knockdown of Ptprd partially reversed the alterations in ALP activity (Fig. 7c) and mineralized nodules formation (Fig. 7d; Supplementary Fig. S16b) induced by March2 knockdown. These findings suggest that MARCH2 inhibits odontoblastic differentiation through PTPRD.
Ptprd knockdown reverses the enhanced odontoblastic differentiation of mDPCs by March2 deficiency. a RT-qPCR shows the mRNA levels of Dmp1, Dspp and Col1a1 in mDPCs cultured in the differentiation medium for 5 d following transfection with siRNAs targeting March2 and Ptprd each or both (n = 3). b WB analysis shows the protein levels of DMP1, DSPP and COL I in mDPCs cultured for 5 d in the differentiation medium following transfection with March2 siRNA or Ptprd siRNA or both. c The increase in ALP activity in differentiation-induced cells due to March2 knockdown is partially reversed by the simultaneous knockdown of Ptprd and March2. d ARS staining indicates that March2 knockdown promotes mineralized nodule formation in differentiation-induced cells, an effect that is abolished by the simultaneous knockdown of Ptprd and March2. D, differentiation induction. The statistical difference was analyzed by one-way ANOVA with Dunnett’s post hoc test (a), where ns denotes not significant; *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
E3 ligases target a broad spectrum of substrates to regulate fundamental developmental processes, including cell differentiation.19 In this study, we identified MARCH2, a membrane-associated E3 ligase, plays a negative regulatory role in odontoblast differentiation and dentinogenesis through mediating the polyubiquitination and lysosomal degradation of the membrane protein PTPRD. Several other E3 ubiquitin ligases including smad ubiquitination regulatory factor 1 (SMURF1), murine double minute 2 (MDM2), WW domain-containing E3 ubiquitin protein ligase 2 (WWP2) and carboxyl-terminus of Hsc70 interacting protein (CHIP) have been reported to play pivotal roles in modulating odontoblast differentiation and dentinogenesis.20,21,22,23,24,25 However, the substrates of these E3 ligases are all cytoplasmic or nuclear proteins, different from the present study with a membrane protein as the substrate of the E3 ubiquitin ligase MARCH2.
Among all E3 ubiquitin ligases localized to cellular and organelle membranes, MARCH family members comprise a substantial proportion.8 Based on our preliminary experiments, MARCH2 was screened out since its expression was the highest among all MARCH family members and increased during in vitro odontoblastic differentiation of mDPCs. IHC analysis of incisors at PN2 and molars at E18.5, PN3, PN7 and PN10 revealed that the expression of MARCH2 was moderate in the undifferentiated DPCs and strong in the odontoblasts in vivo. The expression pattern of MARCH2 suggests a possible role of it during odontoblast differentiation.
MARCH2 has been reported to be involved in various physiological processes including vesicular trafficking26,27 and autophagy,28,29 as well as in several pathological processes, such as myocardial ischemia30 and allergic airway inflammation.9 It is also involved in the developmental process of the Xenopus head.8 To investigate the role of MARCH2 during the odontoblastic differentiation of mDPCs, a series of knockdown and overexpression experiments were performed in mDPCs. The results of these in vitro experiments collectively demonstrated an inhibitory role of MARCH2 during this process. Furthermore, March2 deficient mice and mice with odontoblast specific knockdown of March2 exhibited increased dentin thickness, accelerated dentin deposition rate as well as elevated expression levels of odontoblast markers in both incisors and molars compared with control mice, consistent with the findings of in vitro experiments. The high expression of MARCH2 in odontoblasts appears paradoxical given its inhibitory role in odontoblast differentiation. However, we speculate that MARCH2 act as a negative regulatory mechanism to restrain pro-differentiation signals, thereby preventing excessive odontoblast differentiation and maintaining balanced dentin formation. A similar mechanism is seen in osteoblasts, where the E3 ligase SMURF1 targets runt-related transcription factor 2 (RUNX2) for degradation, thereby delaying osteoblast differentiation and limiting excessive bone formation.31
Based on phylogenetic analysis, the MARCH family is classified into four subgroups, with MARCH2 and MARCH3 sharing significant similarity and being placed in the same subgroup. Previous studies have demonstrated that MARCH2 and MARCH3 exhibit redundant roles in regulating allergic airway inflammation.9 However, during odontoblastic differentiation, March3 is hardly expressed in the dental papilla (Supplementary Figs. S1a, b, S2e), thereby ruling out a compensatory role of it for MARCH2 in dentinogenesis.
As a member of the MARCH family, MARCH2 typically targets substrates localized at the cell membrane and organelle membranes, promoting their ubiquitination and subsequent degradation.9,27,28,30 This study revealed that MARCH2 interacted with PTPRD, a membrane protein, and promoted its polyubiquitination and degradation dependent of the ubiquitin ligase activity of MARCH2. March2 deficient mice exhibited elevated PTPRD protein level, supporting that the degradation of PTPRD is mediated by MARCH2. Further experiments confirmed that MARCH2 promoted the K27-linked polyubiquitylation of PTPRD. Therefore, MARCH2 targets PTPRD for the K27-linked polyubiquitination and subsequent degradation. Likewise, MARCH2 has been reported to interact with interleukin-5 receptor alpha (IL-5Rα), mediating its K27-linked polyubiquitination and subsequent degradation.9 Notably, the residual ubiquitination of PTPRD observed in March2-Myc-untransfected groups may be attributed to functional redundancy between MARCH2 and other E3 ubiquitin ligases. Ubiquitination mediated protein degradation has been found to occur through proteasomal and/or lysosomal pathways.32 Application of CQ, rather than MG132, rescued MARCH2-induced degradation of endogenous PTPRD, indicating that MARCH2 promotes PTPRD degradation through the lysosomal pathway. Additionally, we found that overexpressed MARCH2 facilitated translocation of PTPRD from cell membrane to intracellular area where PTPRD showed colocalization with lysosome, supporting the above finding of MARCH2-mediated lysosomal degradation of PTPRD.
Multiple receptor proteins have been found to be involved in the process of odontoblast differentiation.33,34,35 PTPRD, as a member of the LAR-RPTP family, is localized on the cell membrane and influences signal transduction by mediating tryosine residues dephosphorylation of intracellular proteins.36 In this study, knockdown of Ptprd impaired the mRNA and protein levels of DMP1, DSPP and COL I, as well as ALP activity and mineralized nodule formation ability, revealing that PTPRD was able to promote the odontoblastic differentiation of mDPCs. Previous studies have shown that PTPRD facilitates the differentiation of mesenchymal progenitor cell into adipocytes by modulating FYN tyrosine phosphorylation levels.16 Furthermore, PTPRD has been reported to specifically dephosphorylate glycogen synthase kinase 3 beta (GSK3β) at tyrosine 216,37 a modification known to reduce the kinase activity of GSK3β.38 Notably, inhibition of GSK3β activity, which leads to increased intracellular β-catenin levels, has been shown to promote the differentiation of dental pulp stem cells into odontoblasts.39 Thus, we speculate that PTPRD may promote odontoblast differentiation by dephosphorylating GSK3β at tyrosine 216, which result in cytoplasmic accumulation of β-catenin, activation of Wnt/β-catenin signaling, and upregulation of differentiation-related genes such as Dmp1 and Dspp. Moreover, double knockdown of March2 and Ptprd remarkably reversed the facilitating effects by knockdown of March2 alone, indicating that the inhibitory role of MARCH2 in odontoblastic differentiation was mainly mediated through the ubiquitination and subsequent degradation of PTPRD.
Among all E3 ubiquitin ligases involved in tooth development, SMURF1 was the first to be identified.20 In recent years, research on E3 ubiquitin ligases in tooth development has increased considerably. In the nucleus, MDM2 and WWP2 monoubquitinate distal-less homeobox 3 (DLX3) and kruppel-like factor 5 (KLF5), respectively, enhancing their transcriptional regulatory activities on the Dmp1 and Dspp genes.21,23 Additionally, WWP2 promotes phosphatase and tensin homolog (PTEN) degradation through polyubiquitination, thereby relieving its inhibitory effect on KLF5.24 Conditional knockout of Mdm2 in the odontoblast layer or global knockout of Wwp2 in mice results in molars with thinner dentin and shorter roots.22,24 Another E3 ubiquitin ligase, CHIP promote the polyubiquitination and subsequent degradation of DLX3, thus inhibiting odontoblast differentiation. Notably, Chip-deficient mice exhibit increased dentin formation.25 Our study shows that MARCH2 suppresses odontoblast differentiation and dentin formation. Mechanistically, PTPRD, a membrane protein, is able to promote the odontoblastic differentiation of mDPCs. MARCH2 suppresses odontoblast differentiation through mediating the K27-linked polyubiquitination and subsequent lysosomal degradation of PTPRD (Fig. 8).
Our findings demonstrate an important mechanism by which the differentiation of odontoblasts is delicately regulated at the membrane protein level through ubiquitination modification. Given that MARCH2 destabilizes PTPRD and consequently suppresses odontoblast differentiation, modulation of this axis may represent a promising strategy for dentin regeneration. For instance, pharmacological inhibition of MARCH2 activity or stabilization of PTPRD could potentially enhance odontoblast differentiation, offering therapeutic benefit in conditions such as dental pulp injury or dentin caries. Future studies are warranted to explore the feasibility and safety of targeting the MARCH2-PTPRD axis in dentin tissue engineering and regenerative dentin therapies.
Materials and methods
Animals
All animal experiments were performed following the guidelines of the Animal Ethics Committee of Wuhan University (BZ20230063). March2 deficient (March2 KO) mice were kindly provided by Professor Hong-Bing Shu from Wuhan University. All mice were housed in a specific pathogen-free (SPF) animal facility at the Center for Animal Experiment, Wuhan University. Housing conditions were strictly controlled, with a temperature of 20–24 °C, humidity of 40%–70%, and a 12-h light/dark cycle. No more than four mice were kept per cage. Comprehensive information on the experimental mice is shown in Supplementary Table 1.
Genotyping and tissue preparation
March2 heterozygous mice were interbred, and the offspring underwent toe amputation on PN7. The toe tissue was subjected to heating in 200 μL 50 mol/L NaOH for 30 min (min), followed by adding 20 μL 1 mol/L Tris-HCl (pH 8.0) (Beyotime). Genotyping was performed using Polymerase Chain Reaction (PCR) analysis with primers as below: forward, 5′-GGACGTCTGGAGCTGGGCTG-3′; reverse, 5′-AGGGAGGGGCTTGCAGACCT-3′ (March2). The target gene sequences were amplified by PCR using 2× Hieff PCR Master Mix (Yeasen). The resulting products were separated using 1% agarose gel through electrophoresis. Then the gel was excised under ultraviolet light for subsequent Sanger sequencing.
The mandibles of March2 deficient and littermate control mice were harvested and fixed in 4% paraformaldehyde (PFA) at 4 °C overnight, which were also applied to Kunming mice at different ages.
Construction of AAV vectors and in vivo injection
A previous study has reported that AAV6 exhibits strong tropism for odontoblast-like cells (OLCs).1 We constructed an AAV6 vector expressing a shRNA targeting March2 under the control of a mouse Dmp1 promoter, which consists of 2 600 base pairs upstream of the transcription start site (TSS) and the first exon.40 A GFP reporter was included to enable tracking of transduction efficiency. The shRNA sequences are listed in Supplementary Table 2. All AAVs were obtained from Tsingke Biotechnology (Wuhan, China)
In vivo injections were performed using a 10 μL microsyringe with a 32-gauge needle via the submandibular approach as previously reported.1 2 μL of AAV6-shMarch2 (2 × 1012 vg/mL) was injected into wild-type C57BL/6 mice at PN1; littermate controls received 2 μL of AAV6-shScr (2 × 1012 vg/mL) under the same conditions. To verify the in vivo transduction efficacy of AAV6-shMarch2, mandibles were collected at PN3. To investigate the functional impact of March2 knockdown in odontoblasts on dentin formation, mandibles were harvested at PN10, PN14 and PN21.
Cell culture and transfection
To obtain mDPCs, mandibles were harvested from mice at E16.5 and the tooth germs were isolated under a stereomicroscope (Nikon). The dental mesenchyme was collected and digested with 0.25% trypsin (Hyclone) before being cultured in plates. The mDPCs, HEK293T cells, and Henrietta Lacks (Hela) cells were cultured in Dulbecco’s Modified Eagle’s Medium (DMEM; HyClone) containing 10% fetal bovine serum (FBS; Gibco) and 100 µ/mL penicillin/streptomycin (Hyclone). The cells were cultured in a humidified incubator at 37 °C with 5% CO2. mDPCs were cultured in differentiation medium (DM) with 10 mmol/L β-glycerophosphate, 50 µg/mL ascorbic acid, and 100 mol/L dexamethasone to induce odontoblastic differentiation.
siRNAs utilized for in vitro knockdown experiments were obtained from GenePharma and siRNA sequences are listed in the Supplementary Table 2. pCMV-HA-Ub plasmids were obtained from Miaoling. pLOV-CMV-March2-Flag; pcDNA3.1-March2-myc, C64S, C67S, and C80S; pRK-HA-K6R, K11R, K27R, K29R, K33R, K48R and K63R; pRK-HA-K6O, K11O, K27O, K29O, K33O, K48O and K63O were generously provided by Professor Hong-Bing Shu from Wuhan University. pcDNA3.1-Ptprd were provided by BaoLi Wang from Tianjin Medical University. pcDNA3.1-Ptprd-Flag was constructed using the Mut Express II Fast Mutagenesis Kit V2 (Vazyme). The aforementioned siRNAs and plasmids were transfected into mDPCs using the Lipofectamine 3000 Transfection Reagent (Invitrogen) following the manufacturer’s protocol, while the plasmids were transfected into HEK293T cells and Hela cells using the Lipo8000 Transfection Reagent (Beyotime) following the manufacturer’s protocol.
H&E staining, IHC, IF, and ICC
The mandibles were decalcified by 10% Ethylene Diamine Tetraacetic Acid (EDTA), dehydrated, and embedded in paraffin. Sections at 4-µm thickness were prepared. For frozen sectioning, mandibles were fixed in 4% paraformaldehyde. Following decalcification, the samples were infiltrated sequentially with 15% and 30% sucrose at 4 °C, embedded in optimal cutting temperature (OCT) compound (Tissue-Tek, Japan), and cryosectioned at a thickness of 10 µm using a CM1950 cryostat (Leica Biosciences, Germany).
For H&E staining, tissue slides were dewaxed, rehydrated through graded alcohols, and stained with hematoxylin and eosin.
For IHC, after dewaxing and rehydration, tissue slides underwent antigen retrieval using an enzyme digestion solution (Maxin). The slides were blocked with normal serum and incubated with primary antibodies at 4 °C overnight, as listed in Supplementary Table 3. In the case of IHC, subsequent steps were performed according to the immunohistochemistry kit (Maxin) protocol. Visualization was achieved using Diaminobenzidine (DAB; Maxin), and the nuclei were counterstained with hematoxylin before mounting with neutral gum. For tissue IF, the samples were incubated with fluorescent secondary antibodies (Abbkine) at 37 °C for 1 h (h), counterstained using DAPI-containing mounting medium (ZSGB-Bio), and finally visualized under a Leica DM4000B microscope (Leica).
For ICC, mDPCs and Hela cells were cultured on cover glasses (Nest). To label intracellular lysosomes, Hela cells were treated with 200 nmol/L LysoTracker Red DND-99 (Thermo Fisher) for 1 h before sample collection. Next, the cells were fixed with 4% PFA, permeabilized using 0.1% Triton X-100, blocked with 2.5% BSA, and incubated with the primary antibody at 4 °C overnight. Subsequent steps were carried out the same as those for tissue IF. The images were captured using the FluoView FV1200 (Olympus).
ALP staining and ARS staining
For ALP staining, mDPCs were cultured with growth medium (GM) or DM for 4 days (d) and then stained with the BCIP/NBT Alkaline Phosphatase Kit (Beyotime) according to the manufacturer’s instruction.
For ARS staining, mDPCs were cultured with either GM or DM for 5 d, then fixed and stained with 1% Alizarin Red S (Alladin). The stained cells were then eluted at 37 °C overnight with 10% cetylpyridinium chloride (CPC), and the absorbance of the eluate at 562 nm was measured for quantification.
RT-qPCR analysis
Total RNAs were extracted from cells using Trizol reagent (Takara), and cDNAs were synthesized from 1 μg RNAs with the ABScript III RT Master Mix (Abclonal). The reaction system was prepared according to the instruction provided with the ChamQ SYBR qPCR Master Mix (Yeason) for RT-qPCR program. PCR primers are listed in the Supplementary Table 2.
WB analysis
Cells were lysed using NP40 (Beyotime) and Cocktail (MedChemExpress), and the lysate was then sonicated. After centrifugation, the protein supernatant was collected and mixed with 5× SDS-PAGE Sample Loading Buffer (Biosharp), then heated at 95 °C. Protein separation was performed by SDS-PAGE gel, followed by transfer to a polyvinylidene difluoride membrane (Millipore). The membrane was incubated with 5% non-fat milk in TBS containing 0.1% Tween 20 (Biosharp) at room temperature for 1 h, then with primary antibodies at 4 °C overnight, as listed in Supplementary Table 3. Afterwards, the membrane was incubated with secondary antibodies for 1 h at room temperature, followed by chemiluminescent detection using Western Bright ECL (Advansta) and Super ECL Detection Reagent (Yeasen). Protein bands were visualized using Dual-color near-infrared fluorescence imaging system (LI-COR). ImageJ was used to quantify band intensities.
Co-IP assay
Cells were lysed with NP40 and Cocktail, and then sonicated. After centrifugation, the supernatant was collected. Ten percent of the supernatant was reserved as the input, and 1 mg cell lysate was incubated with anti-FLAG antibody, anti-MYC antibody, or nonimmune IgG under rotation at 4 °C overnight. Subsequently, 40 µL of Protein A/G magnetic beads (Bimake) were added and incubated at 4 °C for 1 h under rotation. The beads were next washed for five times with lysis buffer and resuspended in 80 µL of 1× SDS buffer, and heated at 95 °C. The samples were then analyzed by WB analysis.
Micro-CT
Micro-CT scanning was performed using a SkyScan 1276 (Bruker) with an effective pixel size of 10 μm, at 50 kV voltage, 114 μA current, and a 14 min exposure time across 180 rotational steps. The two-dimensional image slices were reconstructed using the CTAn software, and quantitative analyses were conducted with Mimics Research 21.0.
Double-Calcein labeling
March2 deficient mice and control littermates at PN5 were intraperitoneally injected with 20 mg/kg Calcein (2.5 mg/mL in 2% NaHCO3 solution) (Sigma) with a second injection at PN10. At PN14, the mice were euthanized, and the mandibles were collected. After fixation and washes, the samples were sent to Servicebio company for tissue dehydration, embedding, and sectioning. Slices were observed under the Aperio digital pathology (Leica).
In situ PLA assay
The detailed procedure for in situ PLA has been previously reported.41,42 Briefly, cells on cover glasses were treated with 0.1% Triton X-100, and then blocked using reagents from the Duolink In Situ PLA Probe (Sigma). Then the cells were incubated with primary antibodies at 4 °C overnight. Next, the PLA probe solution was applied and incubated at 37 °C for 1 h. Amplification process was completed using the Duolink In Situ Red Detection Reagents (Sigma). The slices were subsequently mounted with DAPI-containing mounting medium and imaged using the FluoView FV1200.
Bioinformatics analysis
The PN3.5 molar single-cell sequencing data used for single-cell analysis were retrieved from the GEO database (No. GSE189381).13 The single-cell data underwent quality control using the Seurat 4 and were normalized accordingly. Umap was employed for dimensionality reduction, followed by unsupervised clustering based on gene expression patterns. Classical marker genes were utilized to identify different cell subpopulations.
Statistical analysis
All experiments were conducted independently at least three times. Statistical analyses were carried out using GraphPad Prism software (version 8.0). Normality of the data was assessed using the Shapiro–Wilk test, and homogeneity of variances was evaluated with Levene’s test. The differences between two groups were analyzed by using Student’s t-test, and three or more groups were analyzed by one-way ANOVA with Tukey’s post hoc test or Dunnett’s post hoc test, with P-value < 0.05 considered statistically significant (*P < 0.05; **P < 0.01; ***P < 0.001; ****P < 0.000 1).
Change history
16 January 2026
The Acknowledgment section has been added to the article.
References
Fu, J. et al. AAV6-mediated gene therapy prevents developmental dentin defects in a dentinogenesis imperfecta type III mouse model. Hum. Gene Ther. 34, 567–577 (2023).
Jelokhani-Niaraki, M. Membrane proteins: structure, function and motion. Int. J. Mol. Sci. 24, 5 (2023).
Cornejo, F., Franchini, N., Cortes, B. I., Elgueta, D. & Cancino, G. I. Neural conditional ablation of the protein tyrosine phosphatase receptor Delta PTPRD impairs gliogenesis in the developing mouse brain cortex. Front. Cell. Dev. Biol. 12, 13 (2024).
Yue, R., Zhou, B. O., Shimada, I. S., Zhao, Z. Y. & Morrison, S. J. Leptin receptor promotes adipogenesis and reduces osteogenesis by regulating mesenchymal stromal cells in adult bone marrow. Cell Stem Cell 18, 782–796 (2016).
Fu, Y. et al. Identification of GPI-anchored protein LYPD1 as an essential factor for odontoblast differentiation in tooth development. J. Biol. Chem. 299, 17 (2023).
Patwardhan, A., Cheng, N. & Trejo, J. Post-translational modifications of G protein-coupled receptors control cellular signaling dynamics in space and time. Pharmacol. Rev. 73, 120–151 (2021).
Reiche, J. & Huber, O. Post-translational modifications of tight junction transmembrane proteins and their direct effect on barrier function. Biochim. Biophys. Acta Biomembr. 1862, 11 (2020).
Lee, H. et al. Head formation requires Dishevelled degradation that is mediated by March2 in concert with Dapper1. Development 145, 13 (2018).
Zeng, L. W. et al. The membrane-associated ubiquitin ligases MARCH2 and MARCH3 target IL-5 receptor alpha to negatively regulate eosinophilic airway inflammation. Cell. Mol. Immunol. 19, 1117–1129 (2022).
Liu, X. M. et al. MARCH8 inhibits influenza A virus infection by targeting viral M2 protein for ubiquitination-dependent degradation in lysosomes. Nat. Commun. 12, 13 (2021).
Castellanos, C. A. et al. Lymph node-resident dendritic cells drive TH2 cell development involving MARCH1. Sci. Immunol. 6, 17 (2021).
Iyengar, P. V., Hirota, T., Hirose, S. & Nakamura, N. Membrane-associated RING-CH 10 (MARCH10 Protein) is a microtubule-associated E3 ubiquitin ligase of the spermatid flagella. J. Biol. Chem. 286, 39082–39090 (2011).
Jing, J. J. et al. Spatiotemporal single-cell regulatory atlas reveals neural crest lineage diversification and cellular function during tooth morphogenesis. Nat. Commun. 13, 14 (2022).
Lin, H., Li, S. & Shu, H. B. The membrane-associated MARCH E3 ligase family: emerging roles in immune regulation. Front. Immunol. 10, 10 (2019).
Tomita, H. et al. The protein tyrosine phosphatase receptor delta regulates developmental neurogenesis. Cell Rep. 30, 215 (2020).
Chen, M. et al. Metastasis suppressor 1 interacts with protein tyrosine phosphatase receptor-δ to regulate adipogenesis. Faseb J. 37, 13 (2023).
Liu, Y. C., Penninger, J. & Karin, M. Immunity by ubiquitylation: a reversible process of modification. Nat. Rev. Immunol. 5, 941–952 (2005).
Cheng, J. Y. et al. Linear polyubiquitylation of Gli protein regulates its protein stability and facilitates tumor growth in colorectal cancer. Cell Death Discov. 10, 14 (2024).
Rape, M. Ubiquitylation at the crossroads of development and disease. Nat. Rev. Mol. Cell Biol. 19, 59–70 (2018).
Lee, D. S. et al. Crosstalk between nuclear factor I-C and transforming growth factor-β1 signaling regulates odontoblast differentiation and homeostasis. PLoS ONE 6, 12 (2011).
Zheng, H., Yang, G., Fu, J., Chen, Z. & Yuan, G. Mdm2 promotes odontoblast-like differentiation by ubiquitinating Dlx3 and p53. J. Dent. Res. 99, 320–328 (2020).
Zheng, H., Fu, J., Chen, Z., Yang, G. & Yuan, G. Dlx3 ubiquitination by nuclear Mdm2 is essential for dentinogenesis in mice. J. Dent. Res. 101, 1064–1074 (2022).
Fu, J. et al. WWP2 promotes odontoblastic differentiation by monoubiquitinating KLF5. J. Dent. Res. 100, 432–439 (2021).
Fu, J. et al. A WWP2-PTEN-KLF5 signaling axis regulates odontoblast differentiation and dentinogenesis in mice. J. Biol. Chem. 298, 12 (2022).
Zheng, H. W. et al. CHIP inhibits odontoblast differentiation through promoting DLX3 polyubiquitylation and degradation. Development 150, 13 (2023).
Nakamura, N., Fukuda, H., Kato, A. & Hirose, S. MARCH-II is a syntaxin-6-binding protein involved in endosomal trafficking. Mol. Biol. Cell 16, 1696–1710 (2005).
Yoo, W., Cho, E. B., Kim, S. & Yoon, J. B. The E3 ubiquitin ligase MARCH2 regulates ERGIC3-dependent trafficking of secretory proteins. J. Biol. Chem. 294, 10900–10912 (2019).
Xia, D. et al. MARCH2 regulates autophagy by promoting CFTR ubiquitination and degradation and PIK3CA-AKT-MTOR signaling. Autophagy 12, 1614–1630 (2016).
Li, H. L. et al. Linc00707 regulates autophagy and promotes the progression of triple negative breast cancer by activation of PI3K/AKT/mTOR pathway. Cell Death Discov. 10, https://doi.org/10.1038/s41420-024-01906-7 (2024)
Liu, S. L. et al. The E3 ubiquitin ligase MARCH2 protects against myocardial ischemia-reperfusion injury through inhibiting pyroptosis via negative regulation of PGAM5/MAVS/NLRP3 axis. Cell Discov. 10, 23 (2024).
Shimazu, J., Wei, J. W. & Karsenty, G. Smurf1 inhibits osteoblast differentiation, bone formation, and glucose homeostasis through serine 148. Cell Rep. 15, 27–35 (2016).
Dikic, I. Annual Review of Biochemistry, Vol. 86 (ed Kornberg, R. D.) 193–224 (Annual Reviews, 2017).
Chmilewsky, F. et al. C5L2 regulates DMP1 expression during odontoblastic differentiation. J. Dent. Res. 98, 597–604 (2019).
Miyazaki, A. et al. Coordination of WNT signaling and ciliogenesis during odontogenesis by piezo type mechanosensitive ion channel component 1. Sci. Rep. 9, 13 (2019).
Mohamed, F. F., Ge, C., Binrayes, A. & Franceschi, R. T. The role of discoidin domain receptor 2 in tooth development. J. Dent. Res. 99, 214–222 (2020).
Cornejo, F., Cortes, B. I., Findlay, G. M. & Cancino, G. I. LAR Receptor tyrosine phosphatase family in healthy and diseased brain. Front. Cell. Dev. Biol. 9, 21 (2021).
Henderson, I. M. et al. Substrate-selective positive allosteric modulation of PTPRD’s phosphatase by flavonols. Biochem. Pharmacol. 202, https://doi.org/10.1016/j.bcp.2022.115109 (2022).
Shimasaki, T. et al. Glycogen synthase kinase 3β inhibition sensitizes pancreatic cancer cells to gemcitabine. J. Gastroenterol. 47, 321–333 (2012).
Rajasekar, V., Abdalla, M. M., Basbrain, M. S., Neelakantan, P. & Yiu, C. K. Y. Odontogenic differentiation of dental pulp stem cells by glycogen synthase kinase-3β inhibitory peptides. Stem Cell Res. Therapy 16, https://doi.org/10.1186/s13287-025-04150-7 (2025).
Chen, Z., Xie, H., Yuan, J., Lan, Y. & Xie, Z. Kruppel-like factor 6 promotes odontoblastic differentiation through regulating the expression of dentine sialophosphoprotein and dentine matrix protein 1 genes. Int. Endod. J. 54, 572–584 (2021).
Bagchi, S., Fredriksson, R. & Wallen-Mackenzie, A. In situ proximity ligation assay (PLA). Methods Mol. Biol. 1318, 149–159 (2015).
Xu, X. M. et al. Dentin sialoprotein acts as an angiogenic factor through association with the membrane receptor endoglin. J. Biol. Chem. 301, https://doi.org/10.1016/j.jbc.2025.108279 (2025)
Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (82370913, 82170914 to Guohua Yuan and 82270947 to Guobin Yang); the National Natural Science Foundation of China Key Program (82230029 to Z. C); the Fundamental Research Funds for the Central Universities (2042022dx0003), and the Innovation Project of Municipal Science and Technology Bureau (2023020201010170) to Guohua Yuan. We thank Prof. Hong-bing Shu from Wuhan University for providing the March2 deficient mice. We thank Prof. Baoli Wang from Tianjin Medical University for providing the Ptprd plasmid.
Author information
Authors and Affiliations
Contributions
H.F. contributed to data acquisition, analysis and interpretation, and drafted the manuscript. J.N. contributed to data acquisition, analysis, and critically revised the manuscript. Z.C. and G.Y. (Guobin Yang) contributed to conception, and critically revised the manuscript. G.Y. (Guohua Yuan) contributed to the conception, design, and critically revised the manuscript. All authors gave their final approval and agreed to be accountable for all aspects of the work.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feng, H., Niu, J., Chen, Z. et al. MARCH2 suppresses odontoblast differentiation by polyubiquitinating PTPRD. Int J Oral Sci 18, 5 (2026). https://doi.org/10.1038/s41368-025-00407-2
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41368-025-00407-2