Abstract
Peroxisome proliferator-activated receptor γ (PPARγ), which is expressed in a variety of malignancies, governs biological functions through transcriptional programs. Defining the molecular mechanisms governing the selection of canonical versus non-canonical PPARγ binding sequences may provide the opportunity to design regulators with distinct functions and side effects. Acetylation at K268/293 in mouse Pparγ2 participates in the regulation of adipose tissue differentiation, and the conserved lysine residues (K154/155) in mouse Pparγ1 governs lipogenesis in breast cancer cells. Herein, the PPARγ1 acetylated residues K154/155 were shown to be essential for oncogenic ErbB2 driven breast cancer growth and mammary tumor stem cell expansion in vivo. The induction of transcriptional modules governing growth factor signaling, lipogenesis, cellular apoptosis, and stem cell expansion were dependent upon K154/155. The acetylation status of the K154/155 residues determined the selection of genome-wide DNA binding sites, altering the selection from canonical to non-canonical (C/EBP) DNA sequence-specific binding. The gene signature reflecting the acetylation-dependent genomic occupancy in lipogenesis provided predictive value in survival outcomes of ErbB2+ breast cancer. The Pparγ1 acetylation site is critical for ErbB2-induced breast cancer tumor growth and may represent a relevant target for therapeutic coextinction.
Similar content being viewed by others
Introduction
The activation of oncogenic signals, downregulation of tumor suppressor pathways, metabolic changes and alterations in the tumor microenvironment, including immune cells, adipocytes, and tumor-associated fibroblasts, contribute to tumor onset and progression [1,2,3]. Identifying endogenous target genes governing tumor metabolism and inflammation is essential to provide a rational approach to extinguishing multiple pathways activated in cancer. Upregulation of lipid metabolism occurs in breast tumor epithelium [4, 5]. Enhanced synthesis or uptake of lipids contributes to rapid cancer cell growth and tumor formation. Increased fatty acid synthesis occurs in cancers, and lipogenesis is essential for tumor growth [6]. Increased lipogenesis and mevalonate pathway activation are supported by enhanced expression of the enzymes belonging to these pathways, regulated by the sterol regulatory element-binding proteins (SREBPs), the liver X receptors, (LXRα and LXRβ) and PPARγ [5, 7]. The induction of lipid metabolism occurs early in “normal” breast epithelium of women who subsequently develop breast cancer, associated with the upstream regulator PPARγ. Understanding the mechanisms governing tumor lipogenesis may identify therapeutic vulnerabilities. For example, mTORC1 increases SREBP activity and cancer cells driven by the AKT/mTOR pathway become reliant on de novo lipogenesis [8].
HER2/neu overexpression in breast cancer confers a lipogenic phenotype [9, 10]. ErbB2 expressing breast cancers represent ~25% of human breast cancers and respond to treatment with Her2 inhibitors, including Herceptin and the dual tyrosine kinase inhibitor lapatinib [11]. Although the introduction of anti-Her2 therapies has led to dramatic improvements in survival, nearly all patients with metastatic Her2-positive breast cancer will progress on treatment suggesting the importance of developing coextinction approaches targeting multiple pathways. The resistance mechanisms to anti-Her2 therapy include alterations in receptor tyrosine kinases, local immune cell infiltration, expression of the nuclear receptor (NR) ERα, and cyclin D1/Cdk4 activity. Recent studies identified KEGG pathway enrichment of Peroxisome proliferator-activated receptor γ (PPARγ) signaling in lapatinib-resistant breast cancer [12].
PPARγ is a NR superfamily member that regulates diverse biological functions, including lipogenesis and differentiation, inflammation, insulin sensitivity, cellular proliferation and autophagy [13,14,15,16]. Evidence for PPARγ as a tumor growth inhibitor includes the detection of heterozygous PPARγ mutations in colon cancer and the finding that PPARγ agonists reduce tumorigenesis in murine models [17,18,19]. In contrast, several lines of evidence suggest PPARγ augments growth, as PPARγ ligands increased gastrointestinal polyp number in the Apc mouse model of familial adenomatosis [20], and mammary tumor growth [21]. Furthermore, Cre mediated deletion of Pparγ1 reduced ErbB2-induced mammary tumorigenesis in transgenic mice [22].
PPARγ regulates much of its known functions through binding DNA, either at canonical or non-canonical binding sequences. The canonical DNA binding properties of PPARγ are determined by a DNA binding domain (DBD) together with the ligand binding domain or the hinge region. The DBD of PPARγ forms a heterodimer with the DBD of RXR in a head-to-tail orientation, which binds consensus DNA elements, known as DR1 sites (a direct repeat of the AGGTCA element separated by a single nucleotide). Chromatin immunoprecipitation (ChIP) of PPARγ/RXRα coupled with whole genome tiling identified enrichment of the consensus DR1 binding motif in mouse 3T3-L1 adipocytes [23]. PPARγ binds additional DNA cis elements associated with other transcription factors (TFs), including CCAAT/enhancer-binding proteins (C/EBPs), NFκB and AP-1 proteins, to promote non-canonical signaling [24,25,26].
Although Cre based Pparγ1 gene deletion in mammary tumor oncomice showed Pparγ1 participates in the onset and progression of ErbB2-induced mammary tumorigenesis [22], the molecular mechanisms and the post-translational modifications of PPARγ governing Pparγ1 tumorigenic function remained to be determined. Post-translational modifications of PPARγ include phosphorylation [27,28,29], sumoylation [24], acetylation [30], and O-GlcNAcylation [24, 28, 31]. Ligand-dependent sumoylation of PPARγ2 at lysine residue K77 governs the transcription of inflammatory response genes in mouse macrophages [24, 28]. Acetylation of NRs was initially shown to occur at a conserved lysine motif shared amongst evolutionarily related NRs [32, 33]. The action of PPARγ is mediated via two isoforms, the widely expressed PPARγ1, and the adipose tissue-restricted PPARγ2. PPARγ is acetylated at multiple residues, including K268/293 in mouse Pparγ2 [30] and the conserved lysine residues (K154/155) in mouse Pparγ1 [7]. Substitution of these lysines to generate residues that cannot be acetylated, either arginine (R) [34] or glutamine (Q) [7] revealed the importance of this modification as the murine Pparγ1 K154/155Q and Pparγ1 K154/155A were defective in lipogenesis in tissue culture [7]. The K268/293R substitution in murine Pparγ2 reduced the conversion of white to brown fat gene expression [30] and K268/293R acetylation dead mutant mice were protected from visceral adiposity [34].
The role of Pparγ1 in the growth of mammary adenocarcinoma and the function of the Pparγ1 acetylation site in breast tumor growth was not previously known. Herein, we defined the role of the PPARγ1 acetylation site in breast cancer growth in immune-deficient mice using distinct breast cancer cell lines (MCF10-Ha-Ras, MCF10A-NeuT). Furthermore, using ChIP and ChIP-Seq we show that the acetylated residues of Pparγ1 contribute to altered preference of cis-element binding in chromatin to augment Pparγ non-canonical binding (C/EBP proteins). The Pparγ1 acetylation site governs Pparγ-mediated tumor growth, stem cell expansion and autophagy.
Materials and methods
Mouse tumor models
All animal experiments were approved by the Institutional Animal Care and Use Committee of Thomas Jefferson University, protocol number 782A. All methods were performed in accordance with the relevant guidelines and regulations. Animals were housed and maintained at Thomas Jefferson University in a pathogen-free barrier facility under National Institutes of Health (NIH) guidelines. 12-week-old Female NCr nu/nu (NCI, Bethesda, MD) mice received 1 × 106 of MCF10A-NeuT or MCF10A-Ras cells stably expressing PPARγ1 WT, PPARγ1 K154/155Q, PPARγ1 K77R or its vector control suspended in 50 μL of Dulbecco PBS lacking calcium and magnesium (DPBS) and 50 μL of BD Matrigel Basement Membrane Matrix (BD Biosciences) by subcutaneous injection at one dorsal flank. The injection was performed using 27.5-gauge needle. Transgenic mice encoding tamoxifen inducible CRE recombinase with the genotypes of Pparγ1FLOX/FLOX ROSA26mTmG/CRE-ERT2 or Pparγ1WT/WT/ROSA26mTmG/CRE-ERT2 were described previously [22].
Antibodies
All Antibodies for Western blotting and IHC were previously described [7, 35] and were purchased from commercial sources. BNIP3 (ANa40) was from Abcam, BNIP3L (GTX28399) was from GeneTex. SCD1 (M38), LC3A (D50G8 XP), Cleaved Caspase-3 (Asp175) and p-Rb (Ser780) were from Cell Signaling. BECN1 (H-300), Cyclin D1 (DCS-6), SREBP1 (H-160), SREBP2 (H-164), FASN (H-300), PPARγ (H-100), PPARγ (E-8), Bcl-XL (H-5) and total Rb (C-15) were from Santa Cruz. Vinculin (SPM227) was from Novus. FLAG (M2) was from Sigma-Aldrich. GDI was RTG Sol (Gaithersburg, MD).
Cell culture, plasmid DNA, and transfection
The HEK293T and MCF-10A cell line were initially purchased from ATCC. The early passages of the cells were stored. MCF10A-NeuT, and MCF10A-Ha-Ras cell lines, the expression plasmids encoding 3XFLAG-PPARγ1 and mutants (K154/155Q and K77R), the MCF10A-NeuT cells stably expressing PPARγ1 WT, PPARγ1 K154/155Q and PPARγ1 K77R of MSCV-IRES-GFP and the MCF10A-Ha-Ras cells stably expressing PPARγ1 WT, PPARγ1 K154/155Q and vector control of pCDH-CMV-MCS-EF1-RFP were established as described [7]. The cells thawed from low passage stocks were used within 1 month of the initial thaw. During the experiments, the morphology of all cell lines was checked under phase contrast microscope routinely. The mycoplasma contamination was determined with Hoechst 33258 staining under high magnification fluorescent microscope routinely and/or with ATCC Universal Mycoplasma Detection Kit.
Mammosphere assays were conducted as previously described [36, 37]. Markers for breast cancer stem cells using CD24-CD44+ [38,39,40] was conducted by FACS analysis of isogenic breast cell lines (MCF10A-NeuT-vector, MCF10A-NeuT-PPARγ1 and MCF10A-NeuT-PPARγ1 K154/155Q) as described [36, 41]. Aldehyde dehydrogenase (ALDH) activity was detected with ALDEFLUOR Kit (STEMCELL Technologies) as described previously [36, 42].
Oil Red-O staining and Triglycerides-Glo assay
Oil red-O staining for neutral lipid was described previously [7]. Triglycerides-Glo assay was conducted according to manufacturer’s instruction [43]. 5000 cells per well were plated into 96-well plate for 24 h.
ChIP assays and ChIP-Seq analysis
ChIP assays [44, 45] and ChIP-Seq analysis [46] were conducted as described. Raw ChIP-seq reads were aligned to the hg38 reference genome using the Bowtie v.2 algorithm. Only uniquely aligned reads were retained for subsequent analyses. Peak calling of individual ChIP-seq replicates was performed with MACS2 with default parameters [47]. Consensus peaks were identified by intersecting MACS2 peaks obtained from each sample using bedtools intersect (v.2.25.0) with minimum overlap >0.6. MCF10A PPARγ1 WT and PPARγ1 K154/155Q consensus peaks were than intersected with bedtools intersect (-v, minimum overlap = 1 bp) to obtain a list of uniquely-bound peaks in PPARγ1 WT and PPARγ1 K154/155Q. Merged bigwig tracks for visualization were created from merged bam files from all replicates using the bamCoverage function with scaling factor normalization and heatmaps and average profiles were plotted with deepTools2 [48]. The gene annotations of the peaks were ascribed using ChIP-enrich [49]. The HOMER motif discovery suite (v.4.10) was used for motif analysis, using random, matched regions as background. For CEBP sites enriched at K154/155Q mutant only peak we used the Homer motif CEBP(bZIP)/ThioMac-CEBPb-ChIP-Seq (GSE21512)/Homer. Motifs were ranked by log P value from hypergeometric enrichment calculations (or binomial) to determine motif enrichment. Observed/expected fold change enrichment analyses were performed using GAT [50].
Gene expression analysis
Total RNA was extracted from frozen MCF10A-NeuT PPARγ1 wildtype and MCF10A-NeuT PPARγ1 K154/155Q mutant murine xenograft tumor tissues using the RNeasy Kit (Qiagen). cDNA was generated using the Super- Script II Reverse Transcriptase Kit according to the manufacturer’s instructions. RNA quality was determined by an Agilent 2100 bioanalyzer. Probe synthesis and hybridization to Affymetrix gene chips, Human Exon 1.0 ST Array (Affymetrix, Santa Clara, CA), were performed according to the manufacturer’s manual. Chips were scanned on an Affymetrix Gene Chip Scanner 3000, using Command Console Software. Background correction and normalization were done using Iterative plier 16 with GeneSpring V12.0 software (Agilent, Palo Alto, CA, USA). 2-fold (p < 0.05) differentially expressed gene lists were generated and loaded to David Functional Annotation Tool (https://david.ncifcrf.gov/home.jsp) for Gene Ontology (GO), and Kyoto Encyclopedia of Genes and Genomes (KEGG) [51] pathway analysis. Further downstream pathway exploration was performed by Preranked Gene Set Enrichment Analysis (GSEA) (https://www.gsea-msigdb.org/gsea/index.jsp) based on log2 fold change between PPARγ1 WT and K154/155Q mutant and groups across the cell types using the Hallmark pathway database.
Breast cancer patients’ data set and statistical analysis
Breast cancer microarray datasets previously compiled from the public repositories of Gene Expression Omnibus (GEO) and ArrayExpress were used to evaluate GPASS expression in the context of clinical samples. The data were obtained from public databases and were de-identified; therefore, ethical approval was not required. Differential gene expression among these sample subsets was evaluated using a 2-tailed Student t-test. These studies were extended into a larger cohort of patients to examine GPASS expression in breast cancer genetic subtypes and its correlation with outcome. Breast cancer microarray datasets from the public repository GEO, The European Genome-phenome Archive (EGA), and The Cancer Genome Atlas were used to evaluate GPASS association with patient survival. The most reliable probe set for each gene was selected using JetSet. Analysis of GPASS expression was then evaluated by assigning patients into two cohorts based on the high or low expression of the GPASS. The intersection of the gene expression data with Raw ChIP-seq reads aligned to the hg19 reference genome, was calculated. The correlation between survival and the averaged gene signature magnitude among these sample subsets was evaluated using Cox proportional hazards regression. Kaplan–Meier analysis was used to visualize survival trends within the sample subsets.
Statistical significance of differences in means was determined with two-tailed Student’s t-tests.
Public datasets
MCF7 CEBPB ChIP-seq data sets were downloaded from ENCODE ENCSR000BSR https://www.encodeproject.org/experiments/ENCSR000BSR/.
Results
The PPARγ1 K154/155 is required for the induction of tumor growth
The 3DZY PDB structure PMID:19043829) of hPPARγ1 (green) with DNA double helix (blue) and RXR (purple) [52] places the acetylation sites lysine 154 and 155 in proximity to the consensus DNA binding site (Fig. 1A). To determine whether PPARγ1 increased cellular growth, we employed tissue culture and immune-deficient mouse xenograft tumor models. In prior mass spectrometry analysis, PPARγ1 was shown to be acetylated at nine lysine residues, including a conserved lysine motif at K154/155. To determine the role of the PPARγ1 Κ154/155 acetylation site in cellular growth, MCF10A-NeuT cells were transfected with PPARγ1 WT, an acetylation site mutant (K154/155Q mutant) or a control vector. In addition, a comparison was made with a mutation of another residue (K77R), as this lysine residue is required for PPARγ1 sumoylation and was previously shown to enhance PPARγ1 transactivity [53, 54]. Both PPARγ1 WT and the PPARγ1 K77R mutant transduced cells showed increased cell proliferation. In contrast, the PPARγ1 K154/155Q mutant failed to induce cell proliferation (Fig. 1B). The lack of growth induction by the K154/155Q mutant was not due to altered subcellular distribution. Similar distribution for the PPARγ1 WT and PPARγ1 K154/155Q were identified in the nuclear and cytoplasmic compartments (Fig. S1A, B). Endogenous levels of PPARγ1 in MCF10A increased upon oncogenic transformation (Fig. S1C) but were very low compared with the abundance of PPARγ expressed from the introduced vectors (Fig. S1D, E). The lack of growth induction by the K154/155Q mutant was not due to reduced levels of the PPARγ1 K154/155Q protein compared with PPARγ1 WT as higher levels of protein were identified by Western blot in the MCF10A-NeuT cells (Fig. S1D, E). Quantitative analysis of lipid production in MCF10A-NeuT transduced with either PPARγ Wt or PPARγ mutants showed a 3- to 4-fold induction of Oil-Red O staining with PPARγ WT, with the PPARγ1 K154/155R acetylation mimic or PPARγ1 K77R, but a ~90% reduction in Oil Red O staining with the PPARγ1 mutant of K154/155Q, or K154/155A (Fig. S2A,B), consistent with our prior studies [7]. Intracellular triglycerides were increased in PPARγ Wt vs K154/155Q (2.31 ± 0.11 vs 0.42 ± 0.06 nmol/5000 cells, n = 4, p < 0.001) (Fig. S2C). MCF10A NeuT-PPARγ1 and MCF10A NeuT-PPARγ1-K77R cells were enriched for S phase, which was reduced ~90% by PPARγ1 mutants of K154/155Q, and K154/155A (Fig. S2D,E).
A the 3DZY PDB structure of hPPARγ1 (green) with DNA double helix (blue) and RXR (purple) in cartoon representation, NCOA2 peptides are displayed in light pink, and a small molecule ligand in cyan. The two lysines receiving the acetylations fall into a loop of one of the PPARγ zinc fingers and are depicted in red stick representation. The inset picture zooms in on the two lysines. Zinc ions coordinated by the zinc finger-type DNA-binding domains are displayed as blue spheres, and the cysteins coordinating the zinc atoms in PPARγ are depicted in green stick representation. B MCF10A-NeuT cells transduced with retrovirus expressing PPARγ1 WT or PPARγ1 mutants were seeded at the same density, and the cell growth curve plotted over a period of 4 days. C MCF10A-NeuT cells were transduced with PPARγ1 WT or PPARγ1 mutants as indicated. 5 × 106 cells were implanted into the mammary fat pad of immune-deficient nude mice (n ≥ 12 each group). Tumor growth was measured every 3 days by digital caliper, and tumor volume was calculated. D Tumors from (C) were weighed at the time of sacrifice and the mean tumor weight was calculated and graphically represented. Data are mean ± SEM. E MCF10A-Ha-Ras cells were transduced with PPARγ1 WT or PPARγ1 mutants as indicated. 5 × 106 cells were implanted into the mammary fat pad of immune-deficient nude mice (n = 10 each group). Tumor growth was monitored by measuring tumor size every 3 days. F Tumors from (E) were weighed at the time of sacrifice and the mean tumor weight was calculated and graphically represented. Data are mean ± SEM. *P < 0.05, **P < 0.01, and ***P < 0.001.
MCF10A-NeuT cells stably expressing PPARγ1 WT, PPARγ1 K154/155Q or vector control were injected into the mammary fat pad of nude mice. The MCF10A-NeuT cells transduced with the empty vector failed to populate in the mammary fat pad, as previously described [55], whereas the PPARγ1 WT was sufficient to promote MCF10A-NeuT tumor growth in mice (Figs. 1C, D and S3). The PPARγ1 K154/155Q mutant reduced the growth-enhancing function of PPARγ (N ≥ 12, WT vs. K154/155Q, P = 0.00011) (Figs. 1C, D and S3). The PPARγ1 K77R tumors grew larger than PPARγ1 WT (N ≥ 12, K77R vs. WT, P = 0.0099) (Figs. 1C, D and S3). To determine whether the tumor-enhancing function of PPARγ1 was oncogene-specific, we examined the function of the PPARγ1 acetylation site in Ha-Ras oncogene-transformed breast cancer cells (MCF10A-Ha-Ras). PPARγ1 WT enhanced MCF10A-Ha-Ras tumor growth, which was reduced by 75% by the PPARγ1 K154/155Q mutant (Figs. 1E, F and S4). These results indicate that the PPARγ1 acetylation site K154/155 plays an important role in mammary tumor growth induced by either Ha-Ras or oncogenic ErbB2 in vivo.
The PPARγ1 K154/155 acetylation site governs signaling pathways of lipogenesis, cell growth, stem cell function, autophagy and apoptosis in vivo
To determine the molecular mechanisms by which the PPARγ1 K154/155 acetylation site participates in breast adenocarcinoma xenograft growth, the tumors were subjected to Affymetrix microarray and David pathway analysis using KEGG and GO terms. A comparison of the gene expression from the PPARγ1WT and the PPARγ1 K154/155 acetylation defective mutant identified GO pathways that were enriched for the terms including “PPAR signaling” “FOXO signaling” and “Pathways in cancer” and multiple terms related to stem cell pathways (Fig. S5A). KEGG terms included “cell proliferation,” “cell growth,” “apoptotic process,” “DNA damage response” and “autophagy” (Fig. S5B).
The expression of genes related to “breast cancer stem cell like signature” [56] and “IL6 JAK STAT3” signaling were also enriched (Fig. S4C–E). Genes governing stem cell function induced by PPARγ1 WT vs. PPARγ1 K154/155Q included ALDH1 gene family members (ALDHA3, 5A1, 6A1, 1B1, 3B1), KLF4, osteopontin [57] and EphrinA4. Compared with PPARγ1 K154/155Q the PPARγ1WT induced osteopontin 2.8-fold (n = 3, SEM, P = 0.008). Target genes within modules promoting apoptosis induced by PPARγ1 K154/155Q vs. PPARγ1 WT included PRMT1 [58], EP300 [59, 60], PYCARD a bipartite protein that promotes apoptosis [61], and TP63 [62] (Fig. S4D). Gene expression related to “cancer stem cell” were differentially regulated by PPARγ1 WT vs. PPARγ1 K154/155Q (Fig. S4E).
GSEA display evidenced the enrichment of the PPARγ signaling related pathway including “peroxisome”, “fatty acid metabolism”, “cholesterol homeostasis”, “stem cell like signature” and “adipogenesis” in PPARγ1 WT xenograft tumor compared to the PPARγ1 K154/155 acetylation defective mutant (Fig. 2A) and enrichment of oncogenic and cell-cycle pathways (Fig. S6, Table S1).
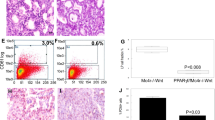
A Gene set enrichment analysis (GSEA) of ErbB2 breast adenocarcinoma xenografts expressing PPARγ1 WT or PPARγ1 K154/155Q showed the enrichment of pathways associated with peroxisomes, fatty acid metabolism, cholesterol homeostasis, adipogenesis, breast cancer stem cell like signature and IL6-JAK-Stat3 signaling. Expression was increased by PPARγ1WT compared with the PPARγ1 K154/155 acetylation defective mutant. B Schematic representation of transgenic mice expressing the ROSA26-CreERT2 mice, which encode tamoxifen inducible Cre recombinase, Pparγ1FLOX/FLOX mice and ROSA26mTmG reporter mice. C Mice were treated with a pulse of tamoxifen for 5 days to induce Cre expression. 25 weeks later mammary epithelial cells were prepared from ROSA26-CreERT2/Pparγ1FLOX/FLOX/ROSA26mTmG or ROSA26-CreERT2/Pparγ1WT/WT/ROSA26mTmG mice treated with tamoxifen. D Number of mammospheres formed (N = 3 of each genotype, Pparγ1+/+, 20.3 ± 1.5 and for Pparγ1−/−, 6.8 ± 0.7. p = 0.0011). E Representative examples of mammospheres formed from isogenic breast cell lines (MCF10A-NeuT-vector, MCF10A-NeuT-PPARγ1 and MCF10A-NeuT-PPARγ1 K154/155Q). F The number of mammospheres and G the size of mammospheres is shown as mean ± SEM for N = 3. H Representative fluorescent activated cell sorting for the markers of mammary stem cells (CD24-CD44+) with (I). mean data shown as ±SEM for N = 3.
The role of PPARγ in breast cancer stem cell function remained to be characterized in vivo. To define the role of Pparγ1 in mammary epithelial stem cell function we conducted mammosphere analysis of epithelial cells derived from transgenic mice in which the Pparγ1 locus was under control of inducible Cre recombinase expression. The ROSA26-Cre-ERT2 mice, which encode tamoxifen inducible Cre recombinase, were intercrossed with Pparγ1FLOX/FLOX mice and subsequently were intercrossed with ROSA26mT/mG reporter mice (Fig. 2B). To control for any independent effect of tamoxifen all mice were treated with a pulse of tamoxifen for 5 days to induce Cre expression. Twenty-five weeks later mammary epithelial cells were prepared from ROSA26-CreERT2Pparγ1FLOX/FLOXROSA26mTmG vs. ROSA26-CreERT2Pparγ1WT/WTROSA26mTmG mice (Fig. 2C). Pparγ1 deficient mammary epithelial cells showed a ~3-fold reduction in the number of mammospheres formed (Fig. 2D, N = 3 of each genotype. Pparγ1+/+, 20.3 ± 1.5 and for Pparγ1−/−, 6.8 ± 0.7. p = 0.0011), although the size of mammosphere was not significantly altered (Fig. S7A, B), indicating a role for Pparγ1 in the initiation and formation of mammospheres.
In the multigenic mice Cre expression mediates deletion of Pparγ1 and the conversion of RFP to GFP cells (Fig. S8A, B). If Pparγ1 were to promote mammary epithelial cellular regeneration and or survival it would be anticipated that the proportion of RFP+ cells would have a growth advantage over GFP+ cells in the mammary gland after the induction of Cre expression to delete the Pparγ1 gene. We therefore analyzed the mammary epithelial cell populations in multigenic mice for GFP, RFP and CK8 (Fig. S8C). The percentage of GFP vs. RFP positive CK8+ epithelial cells was determined in the mammary gland of the transgenic mice 25 weeks after the induction of Cre expression.
In the control mice (Pparγ1WT/WT) the proportion of RFP+ cells was 17.9 ± 5% and the proportion of GFP+ cells was 82.1 ± 5% (the ratio of GFP/RFP cells was 4.59) (Fig. S8D). In contrast, in the Pparγ1fl/fl mice (Pparγ1 deficient CK8+ epithelial cells), the proportion of RFP+ cells was 49.1 ± 6%, the proportion of GFP+ cells was 50.9 ± 6% (the ratio of GFP/RFP cells was 1.04). Thus, there was a 4.4-fold relative reduction in the relative proportion of GFP+ cells in the Pparγ1 deficient CK8+ epithelial cells. These findings are consistent with endogenous Ppar γ1 providing a replicative/survival advantage in CK8+ mammary epithelial cells in vivo.
To determine the functional significance of the PPARγ1 K154/155 acetylation site, equal numbers of mammary cells from the MCF10A-NeuT-PPARγ1 WT vs. MCF10A-NeuT-PPARγ1 K154/155Q mutant were compared. The mammosphere number and size were reduced in the MCF10A-NeuT-PPARγ1 K154/155Q (Fig. 2E–G). The mammosphere number and size were also induced by PPARγ1 WT in the MCF10A-Ras and MCF10A-Src lines (Fig. S9A–G). PPARγ1 K154/155Q reduced the size of mammospheres compared with PPARγ1 WT in MCF10A-Ras and MCF10A-Src lines (Fig. S9C, F). Fluorescent activated cell sorting for CD24-CD44+ mammary stem cell markers [38, 63] showed the abundance was enriched in the MCF10A-NeuT-PPARγ1 WT vs. MCF10A-NeuT PPARγ1 K154/155Q (Fig. 2H, I).
The PPARγ1 K154/155 acetylation site governs de novo lipogenesis in mammary tumorigenesis in vivo
The tumor samples from MCF10A-NeuT xenograft tumor model were analyzed for lipogenesis by measuring the abundance of TFs and critical enzymes required for de novo lipogenesis, including stearoyl-CoA desaturase (SCD), fatty acid synthase (FASN) and sterol regulatory element-binding protein (SREBP1,2). Western blot analysis of individual MCF10A-NeuT tumor samples expressing PPARγ1 WT, PPARγ1 K154/155Q mutant, or the PPARγ1 K77R mutant demonstrated a reduction in SCD1, FASN, SREBP1 (precursor and active form), and SREBP2 abundance in cells expressing the PPARγ1 K154/155Q mutant tumor samples, compared to PPARγ1 WT tumor samples (Fig. 3A, B, Fig. S10). MCF10A-NeuT tumors expressing PPARγ1 WT or PPARγ1 K77R mutant, but not PPARγ1 K154/155Q mutant, demonstrated robust lipogenesis by Oil Red-O staining (Fig. 3C, D). Immunohistochemical (IHC) staining confirmed the decreased expression of SCD1 and SREBP1 in PPARγ1 K154/155Q mutant MCF10A-NeuT tumor samples both within the epithelial cells and tumor stroma (Fig. 3E–H, Fig. S11A, B). These results show that the PPARγ1 K154/155Q expressing mammary adenocarcinomas was less lipogenic than PPARγ1 WT in vivo.
A Total cellular lysates from MCF10A-NeuT PPARγ1 WT or PPARγ1 mutant tumor samples (n = 8, each group) were subjected to Western blot analysis with the indicated antibodies. GDI serves as a protein loading control. All antibodies were blotted on the same membrane except FASN, which was blotted on a separate membrane using the same cellular extracts. B Quantitative analysis of the protein abundance of lipogenic proteins. Each lipogenic protein was normalized to a loading control, GDI. Data were presented using the normalized expression ratio of PPARγ1 mutants group compared to PPARγ1 WT group. C Oil Red O staining on frozen sections from MCF10A-NeuT PPARγ1 WT or PPARγ1 mutant tumor samples. D Quantitative analysis was done by the imaging software ImageJ. The relative density per cell (n = 6 separate tumors; data are mean ± SEM). E, G IHC staining of tumor samples derived from MCF10A-NeuT PPARγ1 WT or PPARγ1 mutant tumor samples using the indicated antibodies. F, H Quantitative analysis of IHC positive staining using imaging software ImageJ. Shown as the relative density per cell (n = 6 separate tumors; data are mean ± SEM). *P < 0.05, **P < 0.01, and ***P < 0.001.
The PPARγ1 K154/K155 governs breast tumor apoptosis and autophagy in vivo
TUNEL staining, used as a marker of cell death, was increased in the PPARγ1 K154/155Q mammary adenocarcinoma (Fig. 4A (n = >7)). Increased active caspase-3 is a marker of apoptotic cell death. IHC of MCF10A-NeuT tumor samples demonstrated increased active caspase 3 in tumors expressing the PPARγ1 K154/155Q mutant (Fig. 4B, Fig. S11C). Western blot analysis showed that the anti-apoptotic protein Bcl-XL, was decreased by ~90% (N = 6) in tumors expressing the PPARγ1 K154/155Q mutant (Fig. 4C, Fig. S12A). Collectively, these data demonstrate that the PPARγ1 K154/155 acetylation site participates in cellular apoptosis.
A MCF10A-NeuT tumor samples expressing PPARγ1 WT or PPARγ1 K154/155Q (n = 6 each group) were analyzed by TUNEL staining to determine apoptotic cell death. The number of TUNEL-positive cells per view (at 400× magnification was counted (>25 views each group). Data are mean ± SEM. B IHC staining of tumor samples derived from the MCF10A-NeuT PPARγ1 WT or PPARγ1 mutants group using anti-active caspase-3 antibody (Asp175). Quantitative analysis of IHC positive staining was conducted using imaging software ImageJ. Shown is the relative density per cell (n = 6 separate tumors; data are mean ± SEM). C Tumor samples expressing PPARγ1 WT or PPARγ1 mutants were analyzed by Western blot for Bcl-XL abundance. Quantitative analysis of abundance normalized to the loading control, GDI. Data were presented using the normalized ratio of PPARγ1 mutants group compared to PPARγ1 WT group. *P < 0.05, **P < 0.01, and ***P < 0.001). D–F Quantitative analysis of proteins governing autophagy. D Beclin1, E BNIP3, F Bnip3L and LC3A-1/LC3A-II, PPARγ and vinculin. Each protein was normalized to a loading control (n = 6 each group). E IHC analyzed BNIP3 abundance in tumor samples. Quantitative analysis of IHC positive staining using imaging software ImageJ. The relative density per cell (n = 6 separate tumors; data are mean ± SEM). *P < 0.05, **P < 0.01, and ***P < 0.001. G, H Western blot analysis of mitogenic kinase signaling pathways with antibodies as indicated. I (mean ± SEM, n = 8). *P < 0.05, **P < 0.01, and ***P < 0.001. J Schematic representation of PPARγ acetylation site-dependent functions.
The autophagy marker Beclin1 (Becn1) was reduced in MCF10A-NeuT tumors expressing the PPARγ1 K154/155Q mutant (Fig. 4D, Fig. S12B). BNIP3L, a marker of mitophagy/autophagy [64], was reduced in MCF10A-NeuT tumors expressing the PPARγ1 K154/155Q mutant compared with PPARγ1 WT (Fig. 4E, F, Fig. S11D, Fig. S12C). Lipidated microtubule-associated protein light chain 3A (LC3A) II, a hallmark of autophagy, was increased in MCF10A-NeuT tumors expressing PPARγ1 WT and PPARγ1 K77R, and decreased in MCF10A-NeuT tumors expressing PPARγ1 K154/155Q mutant (Fig. 4F, Fig. S12C). To determine whether the induction of apoptosis and restraint of autophagy by the PPARγ1 K154/155Q mutant induced counter-regulatory survival pathway signaling, Western blot was conducted of the tumor tissues for ERK and AKT signaling. The PPARγ1 K154/155Q mutant was associated with the induction of β-catenin, GSK3p, and activation of ERK and AKT signaling (Fig. 4G–I, Fig. S12D). These findings are consistent with prior studies showing chronic inhibition of proliferative signaling leads to feedback hyperactivation of PI3K/Akt [65,66,67,68]. Cyclin D1 levels showed a trend toward being reduced in the PPARγ1 K154/155Q compared with PPARγ1WT (Fig. 4H, S12E). Collectively, these data demonstrate that the PPARγ1 K154/155 acetylation site governs apoptosis and autophagy (Fig. 4J).
Acetylation of PPARγ1 K154/155 determines DNA binding sequence preference in the context of chromatin
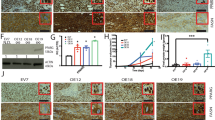
ChIP-Seq was used to define the genome-wide DNA sequence-specific binding characteristics regulated by acetylation of the PPARγ1 K154/155 residues in breast cancer cells using the identical approach as previously described [44]. ChIP and input libraries were generated from two distinct biological samples from MCF10A-NeuT cells stably expressing vector control, PPARγ1 WT or PPARγ1 K154/155Q mutant. High-confidence PPARγ1 ChIP-Seq peaks were identified in MCF10A-NeuT cells expressing PPARγ1 WT or PPARγ1 K154/155Q mutant using masc2 (FDR <5%) and consensus peaks between two replicates were compared by intersecting the peak locations. The PPARγ1 K154/155Q mutant binds to around ~11.3% of the consensus sites bound in the PPARγ WT (WT = 25,174 peaks, K154/155Q mutant = 2849). Of these, 22,882 consensus PPARγ1 peaks were present in WT only and 557 peaks present in K154,155Q mutant only (Fig. 5A). Average binding profile analysis of normalized ChIP-seq reads revealed a strong PPARγ1 ChIP-seq signal at PPARγ1 WT only peaks, while K154,155Q mutant only peaks showed binding in the K154,155Q mutant ChIP-seq and a reduced binding intensity genome-wide in the PPARγ1 WT ChIP-seq (Fig. 5B). We next compared the position of ChIP-seq peaks relative to gene transcription start site by evaluating the percentage of binding peaks upstream from the transcription start site as indicated (Fig. 5C). There was no significant difference between PPARγ1 WT and the PPARγ1 K154/155Q mutant in the distribution of binding peaks (Fig. 5C).
A ChIP-Seq analysis was performed on samples derived from MCF10A-NeuT transduced with either PPARγ1 WT or PPARγ1 K154/155Q mutant, and peak calling defined by MACS2 software is shown in the Venn diagram. B Quantitative analysis of PPARγ1 ChIP-Seq signal. Normalized read density is shown in blue for PPARγ WT and in green for PPARγ 154/155Q. Scatterplots of the maximum stack height (tag counts) at each PPARγ1 peak, color-coded based on whether each peak was called alone in PPARγ1 WT or in PPARγ1 K154/155Q or common to both PPARγ1 WT and PPARγ1 K154/155Q. C Distribution of PPARγ1 WT or PPARγ1 K154/155Q peaks relative to the TSS. The region upstream from the TSS was divided as indicated. D Comparison of the enrichment of PPARγ1 WT alone or PPARγ1 K154/155Q for each canonical or non-canonical PPARγ1 motif. E Comparison of the percentage of peaks within PPARγ1 WT alone or PPARγ1 K154/155Q for each PPARγ1 motif. F The -fold enrichment (observed/expected) for PPARγ1 WT or PPARγ1 K154/155Q at CEBP sites defined in MCF7 cells. G–I Integrated genome browser visualization of tag density profiles for ChIP-Seq PPARγ1 WT and K154/155Q. Selected genes are: LSM12, NTNG1 and ABACB.
The differences in the number of peaks, reflecting chromatin occupancy, may relate to differences in motif recognition and transcription factor (TF) binding determined by the PPARγ acetylation site. The ChIP-seq peak regions were scanned using position weight matrices generated from TF binding site motifs collected in the HOMER database. TFs associated with non-canonical PPARγ regulation, including AP-1, Atf3, BATF, Fosl2, JunB, Jun-AP1 and Fra1/2, were significantly associated with PPARγ1 binding in both PPARγ1 WT and PPARγ1 K154/155Q mutant (Fig. 5D). Canonical PPARE and RXR motifs were only significantly enriched at PPARγ1 WT only peaks as compared to matched background regions, while PPARγ1 K154/155Q mutant only peaks were specifically enriched at C/EBP motifs as compared to matched background regions (Fig. 5D).
To study the specific binding motifs enriched in PPARγ1 in an acetylation-dependent manner, the TF motifs were individually analyzed in the peak regions of PPARγ1 WT only and PPARγ1 K154/155Q mutant only. Of the consensus PPARγ1 WT only binding peaks, about 32% of the peaks matched the PPARE (NR) or RXR (NR) motif (32.23% and 32.59%, respectively) (Fig. 5E). In contrast, only 14% of the PPARγ1 K154/155Q mutant only binding peaks matched the PPARE (NR) or RXR (NR) motif (12.99% and 15.78%, respectively) (Fig. 5E). We next compared the proportion of PPARγ1 peaks that were located at AP-1 and C/EBP motifs between PPARγ1 WT and K154/155Q mutant. For the non-canonical AP-1 motif, there was no significant difference between PPARγ1 WT alone (39.78%) and the PPARγ1 K154/155Q mutant alone (32.25%). Interestingly, PPARγ K154/155Q mutant alone binding peaks (32.48%) more frequently overlapped the C/EBP motif than PPARγ1 WT alone (21.88%) (Fig. 5E). To further confirm the putative change in CEBPB binding at these K154/155Q mutant only peaks, we quantified the enrichment of CEBPB binding (ENCODE MCF7; ENCAN087WJW) at both WT only and K154/155Q mutant only peaks. CEBPB binding sites were significantly more enriched at K154/155Q mutant only peaks as compared to WT only peaks (Fig. 5F). These studies suggest that PPARγ1 K154/155 contributes to the binding specificity of the canonical (PPARE and RXR/155 mutant were preferentially enriched for CEBPB binding as illustrated in Fig. 5G (Fig. 5G–I).
To establish whether the ChIP seq for PPARγ conducted on cell lines overexpressing PPARγ in our studies faithfully represented PPARγ binding sites, we first integrated PPARγ ChIP-seq data from ReMap2022 [69] and compared the location of the PPARγ peaks identified in our study to these described in other cell types (HUVEC, ACS and HT29). We observed a significant overlap between called peaks in each of the experiments (Fig. S13A) with ~75% of peaks identified in our study being present in at least one other PPARγ ChIP-seq experiment. Thus, in our study PPARγ is binding mostly to previously established, functional PPARγ binding sites. Secondly, we assessed the frequency of PPARγ ChIP-Seq site binding to the frequency of endogenous PPARγ site binding in prior studies. We performed analyses of publicly available PPARγ ChIP-seq in different tissues and cell lines from ReMap2022 and compared the total number of peaks detected in each of the experiments to our study (Fig. S13B). The number of consensus peak binding sites in our studies was similar to the number of consensus peak binding sites identified by others, suggesting that the PPARγ expression system used in our study did not induce random PPARγ binding throughout the chromatin. Thirdly, we determined the concordance of PPARγ peaks between the two individual PPARγ ChIP experiments. This analysis showed over 65% overlap between detected PPARγ peaks in the two replicates (Fig. S13C), very high correlation between the ChIP-seq signal at identified peaks (Pearson’s R = 0.9223) (Fig. S13D) and clear concordant ChIP-seq signal at peaks in both replicates (Fig. S13E). Fourthly, we assessed whether the distribution of PPARγ binding site locations within the genome was similar to that identified for endogenous PPARγ. We compared the RefSeq distribution of PPARγ peaks from our study (WT Rep1 and WT Rep2) to that of previously published datasets (HUVEC, ACS and HT29 from ReMap2022). These analyses demonstrated a similar distribution of peaks detected in our study to that described in previous studies (Fig. S13F).
The PPARγ1 K154/155 determines CBP recruitment and H3K9 acetylation
To assess whether the alteration in DNA binding, dependent upon the PPARγ1 K154/155 residues, may determine the regulation of cellular functions observed in the breast tumor xenografts in vivo, we interrogated the depth of coverage of ChIP-Seq reads in promoters of the genes involved in lipid biogenesis and autophagy. ChIP-Seq analysis showed reduced binding of PPARγ1 K154/155Q compared with PPARγ1 at the regulatory regions of genes involved in lipid biogenesis (FABP4, SCD, SREBF2 (encodes SREBP2 protein), and ADIPOQ) (Fig. 6A). In addition, regulatory regions of genes that promote autophagy (BNIP3L, ATG10) also showed enhanced binding to PPARγ1 WT (Fig. 6B). To validate the results of the ChIP-Seq, PPARγ1 enrichment was studied by ChIP-qPCR at the PPARγ1 summit peak regions of FABP4, SCD, SREBF2, AdipoQ, ATG10 and Bnip3L genes (Fig. 6C, D). The sites located 2 kb upstream from the binding region of PPARγ1 were selected for the AdipoQ and Bnip3L genes as negative controls (Fig. S14). PPARγ1 binding was enhanced in MCF10A-NeuT cells expressing PPARγ1 WT compared with vector control or the PPARγ1 K154/155 mutant at the canonical PPAREs. We determined the intersection of genes that preferentially bound and were regulated by PPARγ vs. PPARγ1 K154/155 in the breast tumor xenografts (Fig. S15). A representative gene example IL6 preferentially bound PPARγ1 vs PPARγ1 K154/155 in ChIP-Seq and was induced 2-to 3-fold by PPARγ1 (Fig. S16).
A, B Integrated genome browser visualization of tag density profiles for ChIP-Seq PPARγ1 WT and K154/155Q. Selected genes are: FABP4, SCD, SREBF2, ADIPOQ, BNIP3L and ATG10. C, D ChIP-qPCR analysis of PPARγ1 enrichment for PPARγ1 WT and PPARγ1 K154/155Q at the four genomic regions highlighted in (A, B). Primers were designed against the peak interval sequence. Data were mean ± SEM (n = 3). E ChIP-qPCR analysis of H3K9Ac enrichment at the same genomic regions shown in (C, D). F, G ChIP-qPCR analysis of CBP enrichment at the same genomic regions shown in (E).
Histone acetylation, including acetylated lysine 9 of histone H3 (H3K9Ac), is highly enriched at active promoter regions [70,71,72] and was therefore used to assess the transcriptional functionality of genome-wide PPARγ binding in adipocytes and macrophages [23, 73]. The occupancy of PPARγ in chromatin influences the recruitment of other TFs and local histone chromatin modifications in a cell-type-specific manner [73]. Consistent with PPARγ1 enrichment, H3K9Ac binding was also increased at PPARγ1 binding locations in PPARγ1 WT compared with vector control or the PPARγ1 K154/155Q mutant (Fig. 6E). The PPARγ1 coactivator CBP was recruited with PPARγ1 to the FABP, AdipoQ, ATG10, and BNIP3L promoter regions (Fig. 6F, G) while a reduced recruitment of CBP was detected to these PPAR binding elements with PPARγ1 K154/155Q mutant.
The genes that preferentially bound PPARγ1 K154/155Q compared with PPARγ1 WT were assessed for changes in gene expression in MCF10A-NeuT cell tumors. The relative levels of gene expression are shown as a heat map (Fig. S17). The direction of expression regulated by the K154/155Q mutant is consistent with a model in which these genes may contribute to the demise of cancer cells (ACSL1, ARID5B, C1QTNF1, IDO1, IQGAP2, PPFIBP2, RGS2, SERTAD4 and SLC16A4 (Fig. S17)).
A PPARγ1 K154/155 acetylation-specific lipid signature (PASS2) is associated with prognosis in basal and ErbB2 breast cancer patients
The current studies suggest a transcriptional output of PPARγ1, mediated via its K154/155 acetylation site, governs breast cancer tumor growth and may therefore potentially contribute to the prognostic significance in human breast cancer. SREBP1 and SREBP2, which were induced by PPARy via its K154/155 site, were previously shown to correlate with poor prognosis in breast cancer [74, 75]. We therefore conducted analysis of human samples. We first assessed PPARγ1 mRNA expression. Secondly, we generated a gene signature representing the intersection of PPARγ1 acetylation site specific gene expression with ChIP binding (PASS1). Thirdly, we generated a signature that represented the intersection of PPARγ1 acetylation site specific lipid metabolism with PPARγ1 acetylation site specific binding in ChIP (PASS2).
First, we used a gene expression database consisting of 2254 breast cancer patients (Fig. S18A) to catalog patients by breast cancer subtype, and PPARG gene expression. As SIRT1 is a key deacetylase of PPARγ [7, 30, 76], we also cataloged SIRT1 abundance (Fig. S18A). We compared the relative expression of PPARG and SIRT1 in breast epithelium of 55 healthy women with the 2254 breast cancer samples. These studies showed a reduction in PPARG and SIRT1, in breast cancer (Fig. S18A). The relative abundance of PPARG was increased in breast cancer patients compared to healthy women (Fig. S18B) and inversely correlated with SIRT1 (r = −0.13, p = 1.875E−009).
Secondly, we derived a PPARγ1 K154/155 acetylation site-mediated gene expression module (Fig. 7A), then identified its intersection with a PPARγ1-ChIP module (Fig. 7B), to define a PPARγ1 K154/155 acetylation site-specific gene signature. In our prior publication, we performed microarray gene expression analysis using MCF10A-NeuT cells stably expressing PPARγ1 WT, PPARγ1 K154/155Q, or control vector [7]. Genome-wide expression analysis identified 995 genes differentially expressed in PPARγ1 WT-transduced cells compared with vector control (Fig. 7A). To study the specific genes regulated by PPARγ1 K154/155 acetylation, we identified the 612 genes regulated by the PPARγ1 K154/155Q mutant compared with vector control (Fig. 7A). The intersection of the 995 genes regulated by PPARγ1 WT with the 612 genes regulated by PPARγ1 K154/155Q mutant identified 250 genes regulated by both (Fig. 7A, yellow). Thus 745 genes were regulated by PPARγ1 WT and not regulated by the PPARγ1 K154/155Q mutant, suggesting these genes were regulated in a manner that is dependent upon the PPARγ1 K154/155 acetylation site (Fig. 7A). In contrast with analysis of ChIP-Seq that included all binding sites (Fig. 5), we next analyzed the number of genes that selectively bound PPARγ1 WT vs. PPARγ1 γ1 K154/155 as defined by the limits of within 10 kb upstream of the transcriptional start site (TSS), and 10 kb downstream of the transcription end sites (TES) (Fig. 7B). This analysis identified 5162 genes that were selectively bound by PPARγ1 WT (Fig. 7B). The intersection between the microarray gene expression data of PPARγ1-acetylation sites regulated genes and the ChIP-Seq data of genes bound by PPARγ1 WT vs. PPARγ1 K154/155Q, identified 230 genes that were both bound and regulated by PPARγ1 in a PPARγ1 K154/155 acetylation-site dependent manner (Fig. 7C). The top 23 up-regulated genes (>2 fold) were used to define the PPARγ Acetylation-Specific Signature (PASS) (Fig. 7C). This set of genes was then used in a blinded analysis of breast cancer patient’s clinical datasets (Fig. 7D–F). We studied PPARγ mRNA expression using combined microarray datasets [77] encompassing 55 healthy women and 2254 breast cancer patient samples. 22 genes from the 23 genes were present within the breast cancer dataset and were used in the analyses. The PPARγ Acetylation-Specific Signature 1 (PASS1) was significantly associated with worse outcomes and shorter distant metastasis-free survival (DMFS) in all breast cancer patients (Fig. 7Dp = 0.04) and ERα-negative breast cancer patients (Fig. 7E, p = 0.007). PASS1 was further investigated in the 5 breast cancer subtypes, and the high expression of PASS was significantly associated with shorter overall-free survival (OS) in the luminal B breast cancer subtype (Fig. 7F).
A Derivation of the PPARγ acetylation site-specific gene expression signature was based on the intersection of the genes differentially regulated by PPARγ1 WT vs. the PPARγ1 K154/155Q in MCF10A-NeuT breast cancer cells and is shown by the Venn diagram. B PPARγ acetylation site-specific ChIP-Seq was derived by comparing the ChIP-Seq of the PPARγ1 WT and PPARγ1 K154/155Q mutant. Genes bound by PPARγ1 within 10 kb upstream of the transcription start site (TSS), 10 kb downstream of the TSS and within the gene were generated for PPARγ1 WT or PPARγ1 K154/155Q. Genes only bound by in a manner dependent upon the PPARγ acetylation site (5162) were applied for the pathway analysis. C The PPARγ acetylation site-specific signature (PASS) generation was derived by the intersection of the PPARγ acetylation site-specific gene expression and ChIP-Seq. The top 23 up-regulated genes (>2 fold) were selected for the clinical analysis. D–F Kaplan-Meier survival curves for distant metastasis-free survival (DMFS) and overall survival (OS) in subsets of breast cancer samples and F human breast cancer subtypes.
Thirdly, to determine the prognostic significance of lipid metabolism regulated by the PPARγ acetylation site, we conducted pathway analysis comparing gene expression of MCF10A-NeuT-PPARγ1 vs. MCF10A-NeuT-PPARγ1 K154/155Q. We identified 126 entities determined as “associated with lipid metabolism” (Fig. S18C). We then identified 513 genes from the ChIP-seq analysis “associated with lipid metabolism” pathway analysis that bound PPARγ1 WT vs. PPARγ1 K154/155Q as defined by the limits of within 10 kb upstream of the transcriptional start site (TSS), and 10 kb downstream of the TES (Fig. S18D). The intersection of these two gene sets, (the genes that were regulated in an acetylation-site specific manner in MCF10A cells, and genes bound by PPARγ1 in ChIP Seq), identified promoter regions that were selectively bound by PPARγ1 WT and that were regulated by PPARγ1 WT compared with PPARγ1 K154/155Q (Fig. S18E). The top 23 upregulated genes (>2 fold) were selected for the clinical analysis of breast cancer patients. 20 genes from the 23 genes were present within the breast cancer set. This signature, referred to as PASS2 was significantly associated with ERα-negative breast cancer patients (Fig. S18F). PASS2 was further investigated in 5 canonical breast cancer subtypes; the high expression of PASS2 was significantly associated with poor overall survival in Her2-positive subtype (Fig. S18G) and basal subtype (Fig. S18H) of breast cancer. These data indicate that PASS2 is associated with poor outcome in basal and ErbB2 positive breast cancer patients.
Discussion
The current studies determined the function of the PPARγ1 acetylation site K154/K155 in vivo. PPARγ1 acetylation is regulated by both endogenous TSA-sensitive and NAD-dependent deacetylases [7]. Acetylation of lysine 154 was identified by mass spectrometry and deacetylation of lysine 155 by SIRT1 was confirmed by in vitro deacetylation assay [7]. In vivo labeling assays revealed K154/K155 as bona fide acetylation sites. Wild-type Pparγ1 was acetylated while the K154/155 mutation significantly reduced the incorporation of [3H] acetyl-CoA [7].
We deployed the acetylation defective mutant PPARγ1 K154/155Q for several reasons. Firstly, prior in vivo labeling studies demonstrated that mutation of PPARγ1 K154/155 reduced the incorporation of [3H] acetyl Co-A into PPARγ [7]. Secondly, mutation of lysyl (K) residues to either alanine (A) or glutamine (Q) in other acetylated proteins rendered the mutants functionally defective [7, 78, 79]. Substitutions of the progesterone receptor (PR) acetylation site, with either K to A or K to Q mutations, resulted in receptors with reduced phosphorylation and reduced transcriptional activation of acute response target genes [78] (reviewed in [80]). Secondly, acetylation-defective cortactin mutants (K9Q and K9R) showed defective induction of GN11 cell migration [79]. Thirdly, SIRT1, which participates in the function of the PPARγ transcriptional complex [76], and plays an essential role in PPARγ-mediated remodeling of brown adipose tissue [30], was previously shown to be defective in binding to PPARγ1 K154/155Q compared with the PPARγ1 WT receptor [7]. One limitation of the studies however is that the exogenous PPARγ was expressed in the presence of, all be it low levels, of endogenous PPARγ. These current studies are consistent with a model in which substitutions of lysyl residues with either A or Q mutations inactivated PPARγ functions.
Herein, PPARγ induced tumor growth in oncogenic ErbB2- and Ha-Ras-induced mammary tumors in immune-deficient mice. The growth of the PPARγ1 K154/155Q expressing tumors was reduced by ~90%. Mechanistic analysis showed the PPARγ1 K154/155 lysine motif serves as a molecular switch of breast tumor growth, apoptosis, autophagy, cancer stem cell expansion and lipogenesis in vivo. Mutation of the PPARγ1 K154/155 acetylation site to residues that could not be acetylated (PPARγ1 K154/155Q), abrogated the induction of breast tumor de novo lipogenesis in xenograft tumors. Analysis of the PPARγ1 K154/155Q acetylation defective mutant breast tumors linked lipogenesis to tumor growth. The loss of PPARγ-dependent lipid synthesis in the PPARγ1 K154/155Q mutant expressing tumors is predicted to reduce the supply of lipids required to support the rapid proliferation of tumor cells. The PPARγ1 K154/155Q mutant expressing breast tumors showed induction of β-catenin/GSKβ consistent with activation of Wnt signaling. GSEA confirmed the induction of Wnt signaling. The induction of β-catenin/Wnt signaling and reduced adipogenesis is consistent with studies in which Wnt activation inhibited adipogenesis [81,82,83,84,85]. The PPARγ1 WT tumors showed reduced ERK and AKT signaling compared with K154/155Q mutant tumors consistent with the reduced AKT/MAPK signaling that occurs with chronic pro-proliferative or oncogenic pathway activation [65,66,67,68].
We showed mammary epithelial cells derived by genetic deletion of Pparγ1 in the mammary gland, conveyed a reduced capacity to form mammospheres. Compared with PPARγ1 WT, the PPARγ1 K154/155 mutant tumors conveyed reduced “stem cell like” gene expression by GSEA, reduced size and number of mammospheres and reduced expression of the CD24-CD44+ mammary stem cell markers. These findings are consistent with a previous study in which GW9662, which preferentially inhibits PPARγ compared with PPARα, reduced the renewing subpopulation of breast cancer stem cells in tissue culture [86]. The population of mammary stem cells can be characterized by the CD24-CD44+ markers [38, 63]. Mutation of PPARγ1 K154/155 in the context of human breast cancer cells expressing oncogenic ErbB2, reduced the proportion of CD24-CD44+ cells. Resistance to HER2 inhibition correlated with increased CD24-CD44+ BCSCs resulted from activation of an IL6-driven inflammatory loop [87]. Our prior Ingenuity Pathway Analysis (IPA) analysis of the pathways governed by endogenous Pparγ1 in ErbB2 mammary tumors identified IFNG and IL6 as the most induced by several genes and Z score [22]. A significant correlation between PPARγ and IL6 (n = 136, Pearson 0.55, p = 6.1 × 10−13) was identified in human metastatic breast cancer (Metastatic Breast Cancer Project). Although the mechanism by which PPARγ1 augments stem cell expansion via the acetylation site remains to be further assessed, the IL-6 promoter bound PPARγ1 WT but not PPARγ1 K154/155Q and IL-6 mRNA was induced 2.6-fold in PPARγ1 WT compared with PPARγ1 K154/155Q (data not shown) suggesting a potential role for IL6 in PPARγ1 WT-mediated breast cancer stem cell expansion.
The PPARγ1 acetylation site restrained apoptosis as evidenced by increased TUNEL staining and activated caspase 3 (Asp175) in the PPARγ1 acetylation site defective mutant tumor samples. Apoptotic signaling modules, defined by gene expression and GO term for apoptosis, were contingent upon the PPARγ1 acetylation site. The PPARγ1 acetylation site-dependent anti-apoptotic function may be due, in part, to the induction of the anti-apoptotic protein Bcl-XL. Mutation at the PPARγ1 acetylation site reduced Bcl-XL abundance by 90%. Bcl-XL is overexpressed in a substantial proportion of human breast cancers, promotes breast cancer therapeutic resistance, anti-apoptosis and metastasis, and has a more potent anti-apoptotic function than Bcl-2 in breast cancer cells [88]. Breast tumor mitophagy and autophagy were evidenced by the abundance of the mitophagy-specific marker BNIP3 and the autophagy marker lipidated LC3A in both the tumors and cell lines. Enhanced binding of PPARγ1 to the regulatory regions of autophagy-related genes Bnip3L and ATG10 occurred in an acetylation site-dependent manner. In addition to affecting direct binding to TF sites, acetylation of PPARγ1 increased local H3K9 acetylation at its cognate binding sites. H3K9 acetylation, which is mediated by a dynamic interaction between acetylation (PCAF, GCN5) and deacetylation (SIRT1, HDACs), is enriched at active gene promoters correlating positively with high levels of gene expression. Our findings are consistent with studies in which NRs either induced autophagy (PPARγ [16], PPARα [89] and the androgen receptor (AR) [90]), or suppressed autophagy (FXR/NR1H4 [89] and NR1D1/Rev-Erba [91]). The PPARα-mediated induction of autophagy in the liver was associated with the induction of autophagy gene expression [89]. The PPARγ acetylation site motif is conserved between many NRs, including PPARα, AR, and Rev-Erba [33]. Therefore, it will be of interest to determine whether acetylation of other NRs contributes to their autophagy function.
The PPARγ1 K154/155 residues, which reside in close proximity to the DNA binding region, determined recruitment in chromatin. The number of sites bound by PPARγ1 WT, including all cis-elements, was comparable to the ~23,000 and ~21,000 sites identified in human adipocyte cell lines [92]. In contrast with PPARγ1 WT, which bound 25,174 sites, the PPARγ1 K154/155Q bound 2849 sites. ChIP of the PPARγ1 K154/155Q for associated TF binding sites was either not significantly altered (AP-1) or increased (C/EBP). In adipocytes, C/EBPs co-localize with PPARγ1 at most of its binding sites and cooperate in target gene transcription [23, 92], whereas in macrophages, PPARγ1 ChIP is enriched with the hematopoietic TF PU.1 [93]. The finding herein that the PPARγ1 K154/155Q mutant conveyed enhanced C/EBP site binding, and reduced growth promotion, is consistent with the understanding that the C/EBPs function as growth inhibitors in breast tissue [94].
We did not conduct studies on the role of PPARγ ligands in breast cancer cells for several reasons. Firstly, because the ligands for PPARγ in breast or breast cancer are not known [95], and whether PPARγ is ligand bound in breast cells is unknown. Our studies were designed to study the impact of the PPARγ acetylation site in the basal state without an added ligand. PPARγ conveys ligand-dependent and independent functions [96] and the acetylation function of PPARγ is ligand independent [97], as are several functions including the PPARγ mediated recruitment of P300 and RAD21 to the DNA, governing the M2-like phenotype in macrophages [98]. An expansive number of natural PPARγ ligands have been described [99,100,101,102,103,104], however their biological role in humans is not known [103].
The current studies extend our understanding of Her2-mediated mammary tumor growth via PPARγ through several novel findings. Herein, PPARγ induced mammary tumor growth in xenografted immune-deficient mice via a conserved acetylation site (K154/155) that served as a key determinant of breast tumor growth, apoptosis, mammary cancer stem cell expansion and autophagy. The acetylation site defective mutant tumors showed altered lipogenesis, stemness and autophagy (Fig. S15). Secondly, using mammary epithelial cells derived from inducible Pparγ1 deletion transgenic mice, we showed that endogenous Pparγ1 contributes to the expansion of mammary stem cells. We showed that the PPARγ acetylation site contributes to breast cancer tumor stem cell expansion using multiple different assays. Thirdly, we showed that the PPARγ1 K154/155 site governs gene expression for canonical peroxisome and lipid signaling and contributes to anti-apoptotic and stem cell signaling. We showed enhanced binding of PPARγ1 K154/155Q to C/EBP sites, similar binding to AP-1 sites, and reduced binding to canonical PPARγ binding sites. In turn, this binding preference altered binding to genes participating in autophagy, anti-apoptosis, and lipogenesis. Fourth, we show that the PPARγ acetylation site governs the recruitment of the co-activator and PPARγ acetylase CBP and changes in local chromatin acetylation, including local H3K9 acetylation. Finally, we showed the PPARγ acetylation site-dependent gene expression signature provided prognostic significance in human breast cancer. Together, these results are consistent with a model in which the PPARγ1 K154/155 residues contribute to mammary epithelial cell lipid biogenesis, stem cell expansion, autophagy, and apoptosis via changes in cis associations and thereby chromatin binding to the regulatory regions of genes that govern these functions.
Data availability
All sequencing data created in this study have been uploaded to the Gene Expression Omnibus (GEO; https://www.ncbi.nlm.nih.gov/geo/) and are available under primary accession code GSE263175.
References
Chaffer CL, Weinberg RA. How does multistep tumorigenesis really proceed?. Cancer Discov. 2015;5:22–24.
Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144:646–74.
Martinez-Outschoorn UE, Peiris-Pages M, Pestell RG, Sotgia F, Lisanti MP. Cancer metabolism: a therapeutic perspective. Nat Rev Clin Oncol. 2017;14:113.
Marino N, German R, Rao X, Simpson E, Liu S, Wan J, et al. Upregulation of lipid metabolism genes in the breast prior to cancer diagnosis. NPJ Breast Cancer. 2020;6:50.
Snaebjornsson MT, Janaki-Raman S, Schulze A. Greasing the wheels of the cancer machine: the role of lipid metabolism in cancer. Cell Metab. 2020;31:62–76.
Peck B, Schulze A. Lipid desaturation - the next step in targeting lipogenesis in cancer? FEBS J. 2016;283:2767–78.
Tian L, Wang C, Hagen FK, Gormley M, Addya S, Soccio R, et al. Acetylation-defective mutant of Ppargamma is associated with decreased lipid synthesis in breast cancer cells. Oncotarget. 2014;5:7303–15.
Porstmann T, Santos CR, Griffiths B, Cully M, Wu M, Leevers S, et al. SREBP activity is regulated by mTORC1 and contributes to Akt-dependent cell growth. Cell Metab. 2008;8:224–36.
Baumann J, Sevinsky C, Conklin DS. Lipid biology of breast cancer. Biochim Biophys Acta. 2013;1831:1509–17.
Menendez JA. Fine-tuning the lipogenic/lipolytic balance to optimize the metabolic requirements of cancer cell growth: molecular mechanisms and therapeutic perspectives. Biochim Biophys Acta. 2010;1801:381–91.
Ponde N, Brandao M, El-Hachem G, Werbrouck E, Piccart M. Treatment of advanced HER2-positive breast cancer: 2018 and beyond. Cancer Treat Rev. 2018;67:10–20.
Zhang L, Huang Y, Zhuo W, Zhu Y, Zhu B, Chen Z. Identification and characterization of biomarkers and their functions for Lapatinib-resistant breast cancer. Med Oncol. 2017;34:89.
Miles PD, Barak Y, He W, Evans RM, Olefsky JM. Improved insulin-sensitivity in mice heterozygous for PPAR-gamma deficiency. J Clin Investig. 2000;105:287–92.
Tontonoz P, Hu E, Spiegelman BM. Stimulation of adipogenesis in fibroblasts by PPAR gamma 2, a lipid-activated transcription factor. Cell. 1994;79:1147–56.
Mueller E, Sarraf P, Tontonoz P, Evans RM, Martin KJ, Zhang M, et al. Terminal differentiation of human breast cancer through PPAR gamma. Molecular cell. 1998;1:465–70.
Zhou J, Zhang W, Liang B, Casimiro MC, Whitaker-Menezes D, Wang M, et al. PPARgamma activation induces autophagy in breast cancer cells. Int J Biochem Cell Biol. 2009;41:2334–42.
Suh N, Wang Y, Williams CR, Risingsong R, Gilmer T, Willson TM, et al. A new ligand for the peroxisome proliferator-activated receptor-gamma (PPAR-gamma), GW7845, inhibits rat mammary carcinogenesis. Cancer Res. 1999;59:5671–3.
Mehta RG, Williamson E, Patel MK, Koeffler HP. A ligand of peroxisome proliferator-activated receptor gamma, retinoids, and prevention of preneoplastic mammary lesions. J Natl Cancer Inst. 2000;92:418–23.
Girnun GD, Smith WM, Drori S, Sarraf P, Mueller E, Eng C, et al. APC-dependent suppression of colon carcinogenesis by PPARgamma. Proc Natl Acad Sci. 2002;99:13771–6.
Lefebvre AM, Chen I, Desreumaux P, Najib J, Fruchart JC, Geboes K, et al. Activation of the peroxisome proliferator-activated receptor gamma promotes the development of colon tumors in C57BL/6J-APCMin/+ mice. Nature Med. 1998;4:1053–7.
Yuan H, Kopelovich L, Yin Y, Lu J, Glazer RI. Drug-targeted inhibition of peroxisome proliferator-activated receptor-gamma enhances the chemopreventive effect of anti-estrogen therapy. Oncotarget. 2012;3:345–56.
Jiao X, Tian L, Zhang Z, Balcerek J, Kossenkov AV, Casimiro MC, et al. Ppargamma1 facilitates ErbB2-mammary adenocarcinoma in mice. Cancers. 2021;13:2171.
Lefterova MI, Zhang Y, Steger DJ, Schupp M, Schug J, Cristancho A, et al. PPARgamma and C/EBP factors orchestrate adipocyte biology via adjacent binding on a genome-wide scale. Genes Dev. 2008;22:2941–52.
Pascual G, Fong AL, Ogawa S, Gamliel A, Li AC, Perissi V, et al. A SUMOylation-dependent pathway mediates transrepression of inflammatory response genes by PPAR-gamma. Nature. 2005;437:759–63.
Hou Y, Moreau F, Chadee K. PPARgamma is an E3 ligase that induces the degradation of NFkappaB/p65. Nat Commun. 2012;3:1300.
Harmon GS, Lam MT, Glass CK. PPARs and lipid ligands in inflammation and metabolism. Chem Rev. 2011;111:6321–40.
Choi JH, Banks AS, Kamenecka TM, Busby SA, Chalmers MJ, Kumar N, et al. Antidiabetic actions of a non-agonist PPARgamma ligand blocking Cdk5-mediated phosphorylation. Nature. 2011;477:477–81.
Hu E, Kim JB, Sarraf P, Spiegelman BM. Inhibition of adipogenesis through MAP kinase-mediated phosphorylation of PPARgamma. Science. 1996;274:2100–3.
Khandekar MJ, Banks AS, Laznik-Bogoslavski D, White JP, Choi JH, Kazak L, et al. Noncanonical agonist PPARgamma ligands modulate the response to DNA damage and sensitize cancer cells to cytotoxic chemotherapy. Proc Natl Acad Sci USA. 2018;115:561–6.
Qiang L, Wang L, Kon N, Zhao W, Lee S, Zhang Y, et al. Brown remodeling of white adipose tissue by SirT1-dependent deacetylation of Ppargamma. Cell. 2012;150:620–32.
Ji S, Park SY, Roth J, Kim HS, Cho JW. O-GlcNAc modification of PPARgamma reduces its transcriptional activity. Biochem Biophys Res Commun. 2012;417:1158–63.
Fu M, Wang C, Reutens AT, Wang J, Angeletti RH, Siconolfi-Baez L, et al. p300 and p300/cAMP-response element-binding protein-associated factor acetylate the androgen receptor at sites governing hormone-dependent transactivation. J Biol Chem. 2000;275:20853–60.
Wang C, Fu M, Angeletti RH, Siconolfi-Baez L, Reutens AT, Albanese C, et al. Direct acetylation of the estrogen receptor alpha hinge region by p300 regulates transactivation and hormone sensitivity. J Biol Chem. 2001;276:18375–83.
Kraakman MJ, Liu Q, Postigo-Fernandez J, Ji R, Kon N, Larrea D, et al. PPARgamma deacetylation dissociates thiazolidinedione’s metabolic benefits from its adverse effects. J Clin Invest. 2018;128:2600–12.
Casimiro MC, Di Sante G, Di Rocco A, Loro E, Pupo C, Pestell TG, et al. Cyclin D1 restrains oncogene-induced autophagy by regulating the AMPK-LKB1 signaling axis. Cancer Res. 2017;77:3391–405.
Jiao X, Rizvanov AA, Cristofanilli M, Miftakhova RR, Pestell RG. Breast cancer stem cell isolation. Methods Mol Biol. 2016;1406:121–35.
Liu M, Casimiro MC, Wang C, Shirley LA, Jiao X, Katiyar S, et al. p21CIP1 attenuates Ras- and c-Myc-dependent breast tumor epithelial mesenchymal transition and cancer stem cell-like gene expression in vivo. Proc Natl Acad Sci USA. 2009;106:19035–9.
Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci USA. 2003;100:3983–8.
Liu D, Sun J, Zhu J, Zhou H, Zhang X, Zhang Y. Expression and clinical significance of colorectal cancer stem cell marker EpCAM(high)/CD44(+) in colorectal cancer. Oncol Lett. 2014;7:1544–8.
Velasco-Velazquez MA, Yu Z, Jiao X, Pestell RG. Cancer stem cells and the cell cycle: targeting the drive behind breast cancer. Expert Rev Anticancer Ther. 2009;9:275–9.
Wu K, Jiao X, Li Z, Katiyar S, Casimiro MC, Yang W, et al. Cell fate determination factor Dachshund reprograms breast cancer stem cell function. J Biol Chem. 2011;286:2132–42.
Jiao X, Katiyar S, Willmarth NE, Liu M, Ma X, Flomenberg N, et al. c-Jun induces mammary epithelial cellular invasion and breast cancer stem cell expansion. J Biol Chem. 2010;285:8218–26.
Murakami-Nishimagi Y, Sugimoto K, Kobayashi M, Tachibana K, Kojima M, Okano M, et al. Claudin-4-adhesion signaling drives breast cancer metabolism and progression via liver X receptor b. eta Breast Cancer Res. 2023;25:41.
Casimiro MC, Crosariol M, Loro E, Ertel A, Yu Z, Dampier W, et al. ChIP sequencing of cyclin D1 reveals a transcriptional role in chromosomal instability in mice. J Clin Investig. 2012;122:833–43.
Jiao X, Di Sante G, Casimiro MC, Tantos A, Ashton AW, Li Z, et al. A cyclin D1 intrinsically disordered domain accesses modified histone motifs to govern gene transcription. Oncogenesis. 2024;13:4.
Ewels PA, Peltzer A, Fillinger S, Patel H, Alneberg J, Wilm A, et al. The nf-core framework for community-curated bioinformatics pipelines. Nat Biotechnol. 2020;38:276–8.
Feng JX, Liu T, Qin B, Zhang Y, Liu XS. Identifying ChIP-seq enrichment using MACS. Nat Protoc. 2012;7:1728–40.
Ramirez F, Ryan DP, Gruning B, Bhardwaj V, Kilpert F, Richter AS, et al. deepTools2: a next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016;44:W160–165.
Welch RP, Lee C, Imbriano PM, Patil S, Weymouth TE, Smith RA, et al. ChIP-Enrich: gene set enrichment testing for ChIP-seq data. Nucleic Acids Res. 2014;42:e105.
Heger A, Webber C, Goodson M, Ponting CP, Lunter G. GAT: a simulation framework for testing the association of genomic intervals. Bioinformatics. 2013;29:2046–8.
Ju X, Casimiro MC, Gormley M, Meng H, Jiao X, Katiyar S, et al. Identification of a cyclin D1 network in prostate cancer that antagonizes epithelial-mesenchymal restraint. Cancer Res. 2014;74:508–19.
Chandra V, Huang P, Hamuro Y, Raghuram S, Wang Y, Burris TP, et al. Structure of the intact PPAR-gamma-RXR- nuclear receptor complex on DNA. Nature. 2008;456:350–6.
Diezko R, Suske G. Ligand binding reduces SUMOylation of the peroxisome proliferator-activated receptor gamma (PPARgamma) activation function 1 (AF1) domain. PLoS ONE. 2013;8:e66947.
Yamashita D, Yamaguchi T, Shimizu M, Nakata N, Hirose F, Osumi T. The transactivating function of peroxisome proliferator-activated receptor gamma is negatively regulated by SUMO conjugation in the amino-terminal domain. Genes Cells. 2004;9:1017–29.
Sherman MY, Meng L, Stampfer M, Gabai VL, Yaglom JA. Oncogenes induce senescence with incomplete growth arrest and suppress the DNA damage response in immortalized cells. Aging Cell. 2011;10:949–61.
Gwili N, Jones SJ, Amri WA, Carr IM, Harris S, Hogan BV, et al. Transcriptome profiles of stem-like cells from primary breast cancers allow identification of ITGA7 as a predictive marker of chemotherapy response. Br J Cancer. 2021;125:983–93.
Pietras A, Katz AM, Ekstrom EJ, Wee B, Halliday JJ, Pitter KL, et al. Osteopontin-CD44 signaling in the glioma perivascular niche enhances cancer stem cell phenotypes and promotes aggressive tumor growth. Cell Stem Cell. 2014;14:357–69.
Li M, An W, Xu L, Lin Y, Su L, Liu X. The arginine methyltransferase PRMT5 and PRMT1 distinctly regulate the degradation of anti-apoptotic protein CFLAR(L) in human lung cancer cells. J Exp Clin Cancer Res. 2019;38:64.
Kramer D, Schon M, Bayerlova M, Bleckmann A, Schon MP, Zornig M, et al. A pro-apoptotic function of iASPP by stabilizing p300 and CBP through inhibition of BRMS1 E3 ubiquitin ligase activity. Cell Death Dis. 2015;6:e1634.
Albanese C, D’Amico M, Reutens AT, Fu M, Watanabe G, Lee RJ, et al. Activation of the cyclin D1 gene by the E1A-associated protein p300 through AP-1 inhibits cellular apoptosis. J Biol Chem. 1999;274:34186–95.
Schroder K, Tschopp J. The inflammasomes. Cell. 2010;140:821–32.
Flores ER, Tsai KY, Crowley D, Sengupta S, Yang A, McKeon F, et al. p63 and p73 are required for p53-dependent apoptosis in response to DNA damage. Nature. 2002;416:560–4.
Liu S, Cong Y, Wang D, Sun Y, Deng L, Liu Y, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2014;2:78–91.
Amaravadi R, Kimmelman AC, White E. Recent insights into the function of autophagy in cancer. Genes Dev. 2016;30:1913–30.
Pratilas CA, Solit DB. Targeting the mitogen-activated protein kinase pathway: physiological feedback and drug response. Clin Cancer Res. 2010;16:3329–34.
Fremin C, Meloche S. From basic research to clinical development of MEK1/2 inhibitors for cancer therapy. J Hematol Oncol. 2010;3:8.
Solit DB, Garraway LA, Pratilas CA, Sawai A, Getz G, Basso A, et al. BRAF mutation predicts sensitivity to MEK inhibition. Nature. 2006;439:358–62.
Sos ML, Fischer S, Ullrich R, Peifer M, Heuckmann JM, Koker M, et al. Identifying genotype-dependent efficacy of single and combined PI3K- and MAPK-pathway inhibition in cancer. Proc Natl Acad Sci USA. 2009;106:18351–6.
Hammal F, de Langen P, Bergon A, Lopez F, Ballester B. ReMap 2022: a database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 2022;50:D316–D325.
Consortium EP. An integrated encyclopedia of DNA elements in the human genome. Nature. 2012;489:57–74.
Gates LA, Shi J, Rohira AD, Feng Q, Zhu B, Bedford MT, et al. Acetylation on histone H3 lysine 9 mediates a switch from transcription initiation to elongation. J Biol Chem. 2017;292:14456–72.
Gates LA, Foulds CE, O’Malley BW. Histone marks in the ‘Driver’s Seat’: functional roles in steering the transcription cycle. Trends Biochem Sci. 2017;42:977–89.
Lefterova MI, Steger DJ, Zhuo D, Qatanani M, Mullican SE, Tuteja G, et al. Cell-specific determinants of peroxisome proliferator-activated receptor gamma function in adipocytes and macrophages. Mol Cell Biol. 2010;30:2078–89.
Bao J, Zhu L, Zhu Q, Su J, Liu M, Huang W. SREBP-1 is an independent prognostic marker and promotes invasion and migration in breast cancer. Oncol Lett. 2016;12:2409–16.
Jie Z, Xie Z, Xu W, Zhao X, Jin G, Sun X, et al. SREBP-2 aggravates breast cancer associated osteolysis by promoting osteoclastogenesis and breast cancer metastasis. Biochim Biophys Acta Mol Basis Dis. 2019;1865:115–25.
Picard F, Kurtev M, Chung N, Topark-Ngarm A, Senawong T, Machado De Oliveira R, et al. Sirt1 promotes fat mobilization in white adipocytes by repressing PPAR-gamma. Nature. 2004;429:771–6.
Ertel A, Dean JL, Rui H, Liu C, Witkiewicz AK, Knudsen KE, et al. RB-pathway disruption in breast cancer: differential association with disease subtypes, disease-specific prognosis and therapeutic response. Cell cycle. 2010;9:4153–63.
Daniel AR, Gaviglio AL, Czaplicki LM, Hillard CJ, Housa D, Lange CA. The progesterone receptor hinge region regulates the kinetics of transcriptional responses through acetylation, phosphorylation, and nuclear retention. Mol Endocrinol. 2010;24:2126–38.
Zhang X, Yuan Z, Zhang Y, Yong S, Salas-Burgos A, Koomen J, et al. HDAC6 modulates cell motility by altering the acetylation level of cortactin. Mol Cell. 2007;27:197–213.
Ashton AW, Dhanjal HK, Rossner B, Mahmood H, Patel VI, Nadim M, et al. Acetylation of nuclear receptors in health and disease: an update. FEBS J. 2024;291:217–36.
Ross SE, Hemati N, Longo KA, Bennett CN, Lucas PC, Erickson RL, et al. Inhibition of adipogenesis by Wnt signaling. Science. 2000;289:950–3.
Kennell JA, MacDougald OA. Wnt signaling inhibits adipogenesis through beta-catenin-dependent and -independent mechanisms. J Biol Chem. 2005;280:24004–10.
Cantwell MT, Farrar JS, Lownik JC, Meier JA, Hyun M, Raje V, et al. STAT3 suppresses Wnt/beta-catenin signaling during the induction phase of primary Myf5+ brown adipogenesis. Cytokine. 2018;111:434–44.
Geoghegan G, Simcox J, Seldin MM, Parnell TJ, Stubben C, Just S, et al. Targeted deletion of Tcf7l2 in adipocytes promotes adipocyte hypertrophy and impaired glucose metabolism. Mol Metab. 2019;24:44–63.
Nguyen-Tu MS, Martinez-Sanchez A, Leclerc I, Rutter GA, da Silva Xavier G. Adipocyte-specific deletion of Tcf7l2 induces dysregulated lipid metabolism and impairs glucose tolerance in mice. Diabetologia. 2021;64:129–41.
Wang X, Sun Y, Wong J, Conklin DS. PPARgamma maintains ERBB2-positive breast cancer stem cells. Oncogene. 2013;32:5512–21.
Korkaya H, Kim GI, Davis A, Malik F, Henry NL, Ithimakin S, et al. Activation of an IL6 inflammatory loop mediates trastuzumab resistance in HER2+ breast cancer by expanding the cancer stem cell population. Mol Cell. 2012;47:570–84.
Oakes SR, Vaillant F, Lim E, Lee L, Breslin K, Feleppa F, et al. Sensitization of BCL-2-expressing breast tumors to chemotherapy by the BH3 mimetic ABT-737. Proc Natl Acad Sci USA. 2012;109:2766–71.
Lee JM, Wagner M, Xiao R, Kim KH, Feng D, Lazar MA, et al. Nutrient-sensing nuclear receptors coordinate autophagy. Nature. 2014;516:112–5.
Blessing AM, Rajapakshe K, Reddy Bollu L, Shi Y, White MA, Pham AH, et al. Transcriptional regulation of core autophagy and lysosomal genes by the androgen receptor promotes prostate cancer progression. Autophagy. 2017;13:506–21.
Woldt E, Sebti Y, Solt LA, Duhem C, Lancel S, Eeckhoute J, et al. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nat Med. 2013;19:1039–46.
Schmidt SF, Jorgensen M, Chen Y, Nielsen R, Sandelin A, Mandrup S. Cross species comparison of C/EBPalpha and PPARgamma profiles in mouse and human adipocytes reveals interdependent retention of binding sites. BMC Genom. 2011;12:152–68.
Dispirito JR, Fang B, Wang F, Lazar MA. Pruning of the adipocyte peroxisome proliferator-activated receptor gamma cistrome by hematopoietic master regulator PU.1. Mol Cell Biol. 2013;33:3354–64.
Lourenco AR, Coffer PJ. A tumor suppressor role for C/EBPalpha in solid tumors: more than fat and blood. Oncogene. 2017;36:5221–30.
Augimeri G, Giordano C, Gelsomino L, Plastina P, Barone I, Catalano S, et al. The role of PPARgamma ligands in breast cancer: from basic research to clinical studies. Cancers. 2020;12:2623.
Werman A, Hollenberg A, Solanes G, Bjorbaek C, Vidal-Puig AJ, Flier JS. Ligand-independent activation domain in the N terminus of peroxisome proliferator-activated receptor gamma (PPARgamma). Differential activity of PPARgamma1 and -2 isoforms and influence of insulin. J Biol Chem. 1997;272:20230–5.
Jiang X, Ye X, Guo W, Lu H, Gao Z. Inhibition of HDAC3 promotes ligand-independent PPARgamma activation by protein acetylation. J Mol Endocrinol. 2014;53:191–200.
Daniel B, Nagy G, Czimmerer Z, Horvath A, Hammers DW, Cuaranta-Monroy I, et al. The Nuclear receptor PPARgamma controls progressive macrophage polarization as a ligand-insensitive epigenomic ratchet of transcriptional memory. Immunity. 2018;49:615–26.e616.
Mansoori A, Sotoudeh G, Djalali M, Eshraghian MR, Keramatipour M, Nasli-Esfahani E, et al. Effect of DHA-rich fish oil on PPARgamma target genes related to lipid metabolism in type 2 diabetes: a randomized, double-blind, placebo-controlled clinical trial. J Clin Lipido. 2015;9:770–7.
Gionfriddo G, Plastina P, Augimeri G, Catalano S, Giordano C, Barone I, et al. Modulating tumor-associated macrophage polarization by synthetic and natural PPARgamma ligands as a potential target in breast cancer. Cells 2020;9:174.
Wang L, Waltenberger B, Pferschy-Wenzig EM, Blunder M, Liu X, Malainer C, et al. Natural product agonists of peroxisome proliferator-activated receptor gamma (PPARgamma): a review. Biochem Pharm. 2014;92:73–89.
Bonofiglio D, Giordano C, De Amicis F, Lanzino M, Ando S. Natural products as promising antitumoral agents in breast cancer: mechanisms of action and molecular targets. Mini Rev Med Chem. 2016;16:596–604.
Schopfer FJ, Lin Y, Baker PR, Cui T, Garcia-Barrio M, Zhang J, et al. Nitrolinoleic acid: an endogenous peroxisome proliferator-activated receptor gamma ligand. Proc Natl Acad Sci USA. 2005;102:2340–5.
Ipsen JH, Jorgensen K, Mouritsen OG. Density fluctuations in saturated phospholipid bilayers increase as the acyl-chain length decreases. Biophys J. 1990;58:1099–107.
Acknowledgements
RGP was supported in part by NIH R01CA132115, R21CA235139-01, and R43HL164131, and the DOD Breast Cancer Research Program Breakthrough Awards (W81XWH1810605, and W81XWH-22-BRCP). The project was implemented with the support from the National Research, Development and Innovation Fund of the Ministry of Culture and Innovation under the RGH_24 (RGH 151464) Grant Agreement with the National Research, Development and Innovation Office (RGP). BG was supported by the NVKP_16-1-2016-0037 grant of the National Research, Development and Innovation Office, Hungary. RP was supported by the FK-142285 grant of the National Research, Development and Innovation Office, Hungary. RP is a holder of the Bolyai Fellowship (BO/00174/22) from the Hungarian Academy of Sciences. There are no conflicts of interest associated with this manuscript.
Author information
Authors and Affiliations
Contributions
Designed research: RGP. Performed research: LT, XJ, CW, DL, SA, RS, ERC, BG, GDS, ZZ, PAM. Analyzed data: LT, XJ, CW, DL, AE, JA-K, SA, RS, ERC, BG, GDS, HA, NE, PAM, EMC, AVK, and SC. Wrote and/or revised the paper: RGP, XJ, CW, PT, and RP.
Corresponding author
Ethics declarations
Competing interests
The authors have declared that no conflict of interest exists. RGP discloses ownership interests in Shenandoah Pharmaceuticals, EcoGenome, StromaGenesis, LightSeed, ioROC.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Tian, L., Jiao, X., Wang, C. et al. PPARγ acetylation governs mammary adenocarcinoma tumor growth via acetylated residues that determine DNA sequence-specific binding. Oncogene 44, 3476–3492 (2025). https://doi.org/10.1038/s41388-025-03492-z
Received:
Revised:
Accepted:
Published:
Version of record:
Issue date:
DOI: https://doi.org/10.1038/s41388-025-03492-z