Abstract
Background
Biliary atresia (BA) is a severe neonatal disease characterized by obstruction of the biliary system and hepatic fibrosis. Epithelial-mesenchymal transition (EMT) occurs during the development of liver fibrosis in BA. However, the molecular mechanism that promotes elevated MMP7 expression and mediates the EMT process remains unknown. SULT2B1 has been shown to be involved in the intrahepatocellular EMT process, but its role in BA remains unknown.
Methods
In this study, we found that SULT2B1 was overexpressed in BA livers. The increased expression of SULT2B1 exhibited a high association with the severity of fibrosis and poor prognosis in BA, and it was significantly correlated with the MMP7-mediated EMT in cholangiocytes. Further study showed that the abnormal high expression of SULT2B1 was located in cholangiocytes in BA, which was consistent with the pattern of MMP7. In vitro, upon the treatment of TGF-β1, SULT2B1 was overexpressed and promoted the EMT process via the Wnt/β-catenin/MMP7 pathway in a human intrahepatic bile duct epithelial cell line. Silencing of SULT2B1 expression blocked Wnt/β-catenin/MMP7-mediated cholangiocyte EMT.
Results
SULT2B1 was overexpressed in BA livers and correlated with fibrosis severity and poor prognosis. Its expression was associated with MMP7-mediated EMT in cholangiocytes, with overexpression localized similarly to MMP7. TGF-β1 induced SULT2B1 overexpression, activating the Wnt/β-catenin/MMP7 pathway to promote EMT, while SULT2B1 silencing inhibited this process.
Conclusion
Inhibition of the SULT2B1/Wnt/β-catenin/MMP7 axis may represent a potential therapeutic strategy for BA.
Impact
-
EMT of cholangiocytes in BA is an important process in the development of biliary obstruction and liver fibrosis.
-
SULT2B1 was a key molecule involved in regulating cholangiocyte EMT and liver fibrosis processes in BA.
-
SULT2B1 induced by TGF-β1 up-regulated MMP7 and promoted EMT in Cholangiocytes.
-
Blockage of the SULT2B1/Wnt/β-catenin/MMP7 axis may be a new therapeutic strategy to relieve BA liver fibrosis.

Similar content being viewed by others
Introduction
Biliary atresia (BA) is a serious neonatal disorder of the hepatobiliary system in which extrahepatic biliary tract fibrosis occludes, leading to obstruction of extrahepatic and intrahepatic bile ducts, progressive liver fibrosis, and liver failure.1 The prevailing treatment strategy for BA is still Kasai portoenterostomy (KPE).2 However, even after successful KPE, the majority of children progress to end-stage cirrhosis and require liver transplantation for long-term survival.3 In a recent study, the inducer of epithelial-mesenchymal transition (EMT), transforming growth factor-β1 (TGF-β1), is strongly stained in the cholangiocytes of BA, which is correlated with liver fibrosis.4 Furthermore, EMT in BA is associated with bile ductular proliferation, obliteration, and liver fibrogenesis.5 However, the mechanisms that promote cholangiocyte EMT during the disease process in BA remain largely unknown.
Matrix metalloproteinases (MMPs) are a class of zinc-dependent endoproteases that degrade the extracellular matrix (ECM) by dissolving various proteins.6 Cellular MMPs are involved in a variety of processes, including cell proliferation and migration, and may also play a role in cell death, angiogenesis, tissue regeneration, and immunological response.1 However, these processes that occur during normal development and organogenesis may lead to pathological states when unbalanced.1 MMP7, an important member of MMPs, is mainly located in the endothelium, intima, and uterus, etc., that degrades ECM components and cleaves cell surface molecules to produce soluble forms and has remodeling effects following tissue damage.6 Previous study has found that serum MMP7 level is significantly elevated in BA compared to other non-BA jaundice and normal controls,7 and the abnormally elevated MMP7 was mainly derived from intrahepatic and extrahepatic cholangiocyte epithelial cells and was associated with the progression of liver fibrosis in BA.8 However, the molecular mechanisms regulating the aberrant expression of MMP7 in BA remain unclear.
Sulfotransferase family 2b member 1 (SULT2B1), previously characterized as a cholesterol and hydroxysteroid sulfotransferase, is the key enzyme that synthesizes of cholesterol into cholesterol sulfates.9 In addition to being involved in metabolism, former studies have shown that SULT2B1 promotes angiogenesis in human gastric cancer and drives the proliferation and invasion of colorectal cancer cells,10 and SULT2B1 may be involved in the EMT process in primary mouse hepatocytes.11 However, whether SULT2B1 is involved in the promotion of EMT in BA remains unknown, and its specific molecular mechanisms require further investigation.
In this study, we found that SULT2B1 was a critical molecule involved in the regulation of cholangiocyte EMT in BA. The increased expression of SULT2B1 exhibited a high association with the severity of fibrosis and poor prognosis in BA, and it was significantly correlated with the MMP7-mediated EMT in cholangiocytes. In vitro, we found that TGF-β1 increased the expression of SULT2B1 in cholangiocytes and drove EMT via the Wnt/β-catenin/MMP7 axis. Cholangiocyte EMT is significantly attenuated by silencing SULT2B1. Therefore, the present study suggests that the SULT2B1/Wnt/β-catenin/MMP7 axis plays an important role in BA cholangiocyte EMT and that blockage of SULT2B1 may be a new therapeutic strategy for relieving BA liver fibrosis.
Methods
Human liver specimens
A total of 31 liver specimens were retrieved from BA patients undergoing Kasai surgery. Twenty normal adjacent non-tumor liver tissues taken from hepatoblastoma patients were used as healthy controls (HC) and were free of fibrosis, cholestasis, or signs of active inflammation as confirmed by histopathological examination. All patients’ guardians provided written informed consent. Patients’ general information and BA-related clinical indicators are collected preoperatively and postoperatively during the follow-up period. The patients’ characteristics preoperatively are presented in Supplementary Table 1. Children with serum total bilirubin levels at 6 months after Kasai’s surgery were followed up, and serum total bilirubin levels <1.5 mg/dL were defined as clearance of jaundice. Liver stiffness measurement (LSM) was performed preoperatively using transient elastography by trained specialist sonographers, following standardized protocols. This study was approved by the Medical Ethics Committee of the Beijing Children’s Hospital (2019-k-386).
RNA extraction, sequencing, and analysis
Total RNAs were extracted from livers and the human intrahepatic biliary epithelial cell line. The RNA libraries were constructed by the poly(A) protocol. Fragmentation was performed using the Fragmentation Buffer. The two strands of cDNA were synthesized using a random hexamer primer and M-MuLV Reverse Transcriptase (RNase H Minus) and DNA polymerase I and RNase H, respectively. The Illumina NovaSeq 6000 system was employed to sequence the cDNA libraries with paired-end 150 bp reads. The paired-end reads were aligned to the human reference genome by STAR 2.7.9a12 with Ensembl gene annotation (GRCh38/hg38 assembly). The mapped reads were then processed by Stringtie v2.1.4 to quantify the gene expression against the Ensembl transcript annotation.13 The count-based gene expression data was normalized and processed by the R DESeq2 package.14
Cell culture
The in vitro studies were performed in a human intrahepatic biliary epithelial cell line (HIBEpiC, Bluefcell, BFN608006125, Shanghai, China).15 HIBEpiC was maintained in Dulbecco’s modified Eagle medium (DMEM, Gibco, 11090099, Shanghai, China) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin at 37 °C in a humidified incubator with 5% CO2. 10 ng/ml TGF-β1 (Novoprotein, CA59, Shanghai, China) was used to stimulate HIBEpiC cells in order to establish a mesenchymal transformation model.
siRNA transfection
siNC and siRNA targeting human SULT2B1 (siSULT2B1) were obtained from AuGCT (Beijing, China). More specifically, siSULT2B1 sequence was 5′-GATCGAGATCATCTGCTTA-3′.16 Transient transfection of siRNA (40 nmol/L) was used by employing Lipofectamine RNAi MAX (Invitrogen, Carlsbad, CA) as recommended by the manufacturer. Control cells were treated with 40 nmol/L RNA interference negative control duplexes (siNC). Cells were used to fulfill further experiments after 48 h.
Western blotting
Primary antibodies used in this study were as follows: anti-SULT2B1 (Santa Cruz, sc-166423), MMP7 (Affinity Biosciences, AF0218), CK19 (Abcam, ab15463), N-Cadherin (CST, 13116T), Vimentin (Proteintech, 60330-1-Ig), Snail (Abcam, ab85931), Twist1 (CST, 69366), ZEB1 (CST, 3396), β-catenin (Abmart, M24002F), LRP6 (Abmart, T58345F). Signals were detected using the Odyssey Imaging System (LI-COR Biosciences, Lincoln, NE) or Fusion Solo Imaging system (VILBER) and analyzed with ImageJ software. Results were normalized relative to GAPDH (CST, 2118S) or β-actin (CST, 8457T) expression.
qRT-PCR
Total RNA was extracted from the HIBEpiC cell line using Trizol. qRT-PCR was performed using SYBR Green on an ABI 7500 system (Life Technologies). Primers used for qRT-PCR were as follows: SULT2B1 sense, 5′-GCTTGTGGGACACCTATGAAG-3′, and anti-sense, 5′-TCTCGATCATCCAGGTCGTGC-3′; and 18S rRNA, sense, 5′-GTAACCCGTTGAACCCCATT-3′, and anti-sense, 5′-CCATCCAATCGGTAGTAGCG-3′.
Immunohistochemical analysis
Liver tissues were fixed in 4% paraformaldehyde for 24 h, dehydrated through a graded ethanol series, cleared in xylene, and embedded in paraffin. Sections (4 μm) were cut using a microtome and mounted on glass slides. Liver tissues were sectioned, dewaxed, washed, and incubated at room temperature with 0.3% H2O2 before being blocked for non-specific immunoreactivity with 3% bovine serum albumin (BSA). Furthermore, sections were incubated with the primary antibody of SULT2B1 (1:100) at 4 °C overnight. After rinsing three times with PBST, the sections were incubated with biotinylated goat anti-mouse secondary antibody (Vector Laboratories) and subsequently with streptavidin-labeled HRP (Vector Laboratories). The sections were finally visualized with 3, 3′-diaminobenzidine (DAB) substrate and counterstained with hematoxylin. Digital images were recorded by an Olympus BX53 microscope.
Immunofluorescence analysis
Liver samples were fixed in 4% paraformaldehyde and embedded in Tissue Tek OCT compound. Frozen sections of 5 μm were used for immunofluorescence. Sections were baked at 37 °C for 30 min and permeabilized for 15 min with 0.5% Triton X-100. After washing with PBS, blocking in PBS containing 2% BSA for 30 min at 37 °C. Then, the sections were incubated with primary antibodies of MMP7 and CK-19 at 37 °C for 1 h. After primary antibody incubation, sections were probed with fluorescein isothiocyanate (FITC)-conjugated secondary antibody, rabbit anti-MMP7, and Cyanine 5 (Cy5)-conjugated secondary antibody, and mouse anti-CK19 antibody for 1 h at 37 °C. Finally, the sections were stained with DAPI and observed under a confocal microscope (Leica TCS SP8, Leica Microsystems, Mannheim, Germany).
Serum MMP7 analysis
Serum samples were collected from each patient preoperatively and 1 month postoperatively, respectively, and stored promptly at −80 °C. MMP7 concentration was measured using an enzyme-linked immunosorbent assay (R&D Systems) according to the protocol.
Gene Expression Omnibus (GEO) dataset collection and preparation
RNA-Seq datasets [GSE122340] were downloaded from the GEO database, which included 171 BA liver tissue samples and 7 normal control samples. To eliminate batch effects, we selected the RNA-seq data of 121 BA samples and 7 normal control samples from the same testing batch for subsequent analysis. According to the survival, BA patients were divided into high or low survival groups.
Weighted gene co-expression network analysis (WGCNA)
The median absolute deviation (MAD) of each gene was calculated, and the top 10% of genes with the smallest MAD were removed. The goodSamplesGenes method of the “WGCNA” R package (version 1.70-3) was used to remove outlier genes and samples to build a scale-free co-expression network. Pearson’s correlation matrices and the average linkage method were both used for all pairwise genes. Then, a weighted adjacency matrix was constructed using the power function A mn = |C mn | ^β. To classify genes with similar expression profiles into gene modules, average linkage hierarchical clustering was conducted according to a topological overlap matrix (TOM)-based dissimilarity measure, with a minimum size of 30 for the gene dendrogram. We then merged modules with a distance of less than 0.25, and finally obtained 23 co-expression gene modules. Notably, genes that could not be assigned to any module were assigned to the gray module.
Gene set enrichment analysis (GSEA)
GSEA software (version 3.0) was obtained from GSEA (http://software.broadinstitute.org/gsea/index.jsp).17 The gene sets of c5.go.v7.4.symbols.gmt and c2.cp.kegg.v7.4.symbols.gmt were downloaded from the Molecular Signatures Database to assess related molecular pathways. Set the minimum gene set to 5 and the maximum gene set to 5000, with 1000 resampling. A false discovery rate (FDR) of <0.05 was considered statistically significant.
Functional enrichment analysis
The core genes of each gene module were used for gene ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) analyses. The R package “org.Hs.eg.db” (version 3.15.0) was used for gene annotations. GO and KEGG functional analyses were performed using the R package “clusterProfiler” (version 3.14.3). Differences were considered statistically significant at an adjusted p-value of less than 0.05.
Statistical analysis
The statistical analysis of the results was performed using GraphPad Prism® 7.00 software (San Diego). Results were expressed as mean ± SEM. Statistical significance was assessed by Student’s t-test or one-way ANOVA when appropriate. Correlation coefficients were calculated by Pearson’s test. p < 0.05 was considered significant. All results were verified in at least three independent experiments.
Result
Identify the key molecules in the EMT process of BA
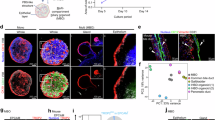
To identify the key molecules regulating the EMT process of BA, we first performed the WGCNA based on the RNA-seq data of GSE122340. The hierarchical clustering dendrogram showed the identified co-expressed gene modules (Fig. 1a), and the appropriate soft thresholding power was selected according to the network topology (Fig. 1b). The eigengene adjacency plot showed the distances between gene modules (Fig. 1c). A total of 23 co-expressed gene modules were obtained by WGCNA, of which 9 gene modules were positively associated with lower native liver survival (Fig. 1d). To screen for key molecules involved in EMT, the core genes of each module were analyzed by GO biological process and molecular function analysis. We found that the core genes of the lightsteelblue1 module were mainly enriched in signal pathways such as EMT and extracellular matrix (Fig. 1e). Subsequently, the samples were divided into high and low survival groups based on the native liver survival. The differential analysis of all genes in the lightsteelblue1 module was performed between the two groups, and 119 genes were highly expressed in the low survival group (Fig. 2a). As shown in Fig. 2a, except for 5 differentially expressed genes (DEGs) which were up-regulated in normal liver tissues, most of DEGs were obviously down-regulated in normal liver tissues compared to BA. The top 10 DEGs with the highest fold change are shown in Fig. 2b. Among them, SULT2B1 was the highest fold change DEG. Moreover, it was worth noting that MMP7 was also a significantly up-regulated DEG.
The cluster dendrogram (a), soft threshold (b), and module eigengene adjacency plot (c) of WGCNA in a dataset of 121 BA liver samples. d The relationship between native liver survival and 23 different gene modules obtained by WGCNA. e The functional analyses of hub genes in the lightsteelblue1 module enriched in EMT, cell adhesion, and extracellular matrix. BA biliary atresia, EMT epithelial-mesenchymal transition.
a The DEGs between the BA of high survival, low survival, and normal control groups in the lightsteelblue1 module of WGCNA. b The volcano plot of DEGs in the lightsteelblue1 module of WGCNA. c The results of GSEA showed that SULT2B1 was enriched in EMT and Wnt/β-catenin pathways. d The relative mRNA expression levels of SULT2B1, MMP7, KRT19, and EMT-related genes in the BA and HC groups. e The correlation heatmap between SULT2B1 and MMP7 with other genes. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). WGCNA Weighted Gene Co-expression Network Analysis modules, GSEA Gene Set Enrichment Analysis results.
Next, we performed GSEA to explore the potential function of SULT2B1 in BA livers. The results showed that the high expression of SULT2B1 was significantly involved in EMT and Wnt/β-catenin pathways (Fig. 2c). RNA-seq results showed that the expression levels of SULT2B1, MMP7, KRT19, VIM, SNAI1, TWIST1, CDH2 and ZEB1 were increased in BA livers compared with HC, especially in low survival group (Fig. 2d). Meanwhile, the correlation heatmap showed a significant co-expressed trend between SULT2B1 and MMP7 (Fig. 2e). Furthermore, SULT2B1 and MMP7 were positively correlated with EMT and fibrosis-related genes, such as VIM, SNAI1, TWIST1, ACTA2, COL1A1, and COL3A1 (Fig. 2e). In addition, the mRNA expression level of cholangiocyte marker KRT19 was also co-expressed with SULT2B1 and MMP7 (Fig. 2e). Thus, these results indicated that SULT2B1 and MMP7 may be involved in the cholangiocyte EMT in BA through public RNA-seq data.
SULT2B1 and MMP7 were upregulated in BA livers and positively correlated with the BA pathogenesis
To validate the results from the public database, we applied RNA-seq in 31 BA and 20 HC with detailed clinicopathological and prognostic features (Supplementary Table 1). The results showed that SULT2B1, MMP7, KRT19 and mesenchymal-associated genes including VIM, SNAI1 and TWIST1 were significant highly expressed in BA group (Fig. 3a). The correlation heatmap also showed that VIM, SNAI1, TWIST1 and KRT19 were co-expressed with SULT2B1 and MMP7 (Fig. 3b). Then, the correlation heatmap of clinicopathological features revealed that the up-regulation of SULT2B1 and MMP7 were significantly correlated with higher liver stiffness measurement (LSM) (Fig. 3c). Furthermore, patients with BA were divided into two groups according to the presence or absence of cytomegalovirus (CMV) infection. Among 35 BA patients, 8 were CMV-IgM positive (22.9%). SULT2B1 expression was significantly higher in CMV-positive vs. CMV-negative groups (p < 0.0001). Compared to the CMV-negative group, MMP7 (p = 0.0332) was significantly overexpressed in the CMV-positive group (Fig. 3d). Moreover, BA patients were divided into two groups according to the clearance of jaundice within 6 months postoperatively. Similarly, SULT2B1 was significantly up-regulated in the jaundice-positive group (p = 0.0448; Fig. 3d). In conclusion, these results further suggested that increased SULT2B1 and MMP7 may be involved in the EMT and liver fibrosis processes in BA.
a The relative mRNA expression levels of SULT2B1, MMP7, KRT19, and EMT-related genes in BA and HC livers. b Correlation heatmap of SULT2B1 and MMP7 with other genes. c Correlation heatmap of SULT2B1 and MMP7 with clinical indicators. d The relative mRNA expression levels of SULT2B1 and MMP7 among different subgroups of BA. (*p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001). BA biliary atresia, HC healthy controls, EMT epithelial-mesenchymal transition.
Serum MMP7 overexpressed in BA patients and derived from cholangiocytes
The above results have confirmed that the level of MMP7 was significantly elevated in BA livers and correlated with KRT19 and EMT-related molecules, therefore, we further performed cellular localization of MMP7 by immunofluorescence, and the confocal results showed that MMP7 was co-localized with the cholangiocyte marker CK19, thus confirming that the high expression of MMP7 in BA liver tissues mainly originated from cholangiocytes (Fig. 4a).
a Immunofluorescence co-staining of MMP7 (green) and CK19 (red) in BA liver tissues. Nuclei were counterstained with DAPI (blue). White arrows indicate co-localized signals (scale bar: 20 μm). b MMP7 levels in serum of BA and non-BA jaundiced patients. c Correlation heatmap of preoperative serum MMP7 with clinical indicators in BA patients. d MMP7 levels in serum of pre- and post-operative BA patients. e Correlation heatmap of postoperative serum MMP7 with clinical indicators in BA patients. (****p < 0.0001). DAPI 4′,6-Diamidino-2-Phenylindole.
Next, we collected preoperative serum from 45 BA patients and 17 non-BA jaundiced patients. As shown in Fig. 4b, compared to non-BA patients, the serum level of MMP7 was statistically up-regulated in BA patients (p < 0.0001). Moreover, serum MMP7 was significantly correlated with several clinical indicators, including gamma-glutaryl transferase (r-GGT), liver stiffness measurement (LSM), and total bile acid (TBA) (Fig. 4c). In addition, postoperative (3–6 months) serum MMP7 levels were detected in 20 BA patients, and the result showed that serum MMP7 levels remained consistently elevated after Kasai surgery, with no difference from preoperative levels (p > 0.05; Fig. 4d). Furthermore, postoperative serum MMP7 levels were also correlated with clinical indicators of liver fibrosis (Fig. 4e). In summary, these results demonstrated that significantly elevated serum MMP7 levels in BA patients may originate from cholangiocytes and were associated with the progression of liver fibrosis pre- and postoperatively in BA.
SULT2B1 was overexpressed in BA livers and localized in cholangiocytes
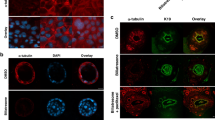
The above results showed that mRNA levels of SULT2B1 and MMP7 were significantly elevated in BA and may be associated with cholangiocyte EMT. Then, we detected the protein levels of SULT2B1, MMP7 and CK19, and the western blot results showed that they were significantly overexpressed in BA livers compared to HC (Fig. 5a). Additionally, the protein levels of several mesenchymal markers, such as TWIST1 and N-Cadherin, were slightly up-regulated in BA livers (Fig. 5a). Of particular note, β-catenin, a downstream transcription factor of Wnt signaling pathway, were elevated in BA livers (Fig. 5a). Relative protein level results showed that the expression levels of SULT2B1 (p < 0.01), MMP7 (p < 0.05), CK19 (p < 0.01), N-Cadherin (p < 0.05), and β-catenin (p < 0.05) were significant elevated in BA livers (Fig. 5b). Moreover, immunohistochemistry was used to detect the SULT2B1 expression and localization in BA liver tissue, as shown in Fig. 5c, highly expressed SULT2B1 was mainly localized in cholangiocytes, which was consistent with the expression of MMP7 in cholangiocytes.
a Western blot of SULT2B1, MMP7, CK19, and EMT-related proteins in BA and HC liver tissues. b Relative protein levels in BA and HC groups. c Localization of SULT2B1 in the immunohistochemistry of BA liver tissue and HC liver (normal liver tissue resected due to trauma in an 18-month-old girl). (*p < 0.05, ** p < 0.01). BA biliary atresia, HC healthy controls.
TGF-β1 induced the expression of SULT2B1 and EMT in cholangiocytes
TGF-β1 is considered to be an important cytokine to promote EMT.18 We next treated HIBEpiC cells with TGF-β1 at different time points. Western blot results showed that the protein expression of SULT2B1, MMP7, and mesenchymal markers, including Vimentin, SNAIL, SLUG, N-Cadherin, and ZEB1, was increased significantly over time after TGF-β1 treatment (Fig. 6a). The above results showed that the increased expression of SULT2B1 was associated with the activation of the Wnt/β-catenin pathway in BA. Thus, we examined the protein levels of β-catenin and LRP6, markers of the Wnt signaling pathway. Western blot results showed that β-catenin and LRP6 were increased after the treatment of TGF-β1 for 48 h (Fig. 6a).
a Western blot of SULT2B1, MMP7, Wnt/β-catenin signaling pathway-related proteins, and EMT-related proteins in HIBEpiC cells treated with/without TGF-β1 for different times. b Functional analysis (left panel: GO terms; right panel: KEGG) in DEGs from RNA-seq data for HIBEpiC cells treated with/without TGF-β1 for 48 h. TGF-β1 transforming growth factor beta 1.
Furthermore, RNA-seq was performed using HIBEpiC cells treated with/without TGF-β1 for 48 h, and the functional analyses also showed that biological processes such as EMT, ECM-receptor interaction, and focal adhesion were significantly activated in TGFβ1-treated HIBEpiC cells (Fig. 6b). These results showed that SULT2B1 was involved in the process of cholangiocyte EMT induced by TGF-β1.
Cholangiocyte EMT was mediated by SULT2B1/Wnt/β-catenin/MMP7 axis
To further determine whether SULT2B1 mediated the induction of cholangiocyte EMT triggered by TGF-β1, we knocked down the SULT2B1 expression in HIBEpiC cells by using specific siRNA. The qRT-PCR analysis revealed that siRNA-SULT2B1 significantly decreased the level of SULT2B1 mRNA in HIBEpiC cells with a knockdown efficiency of approximately 70% (Fig. 7a). Furthermore, siRNA-SULT2B1 significantly decreased the mRNA and protein levels of SULT2B1 upon TGF-β1 treatment (Fig. 7b). Next, western blot results showed that the transfection of siRNA-SULT2B1 significantly inhibited the protein levels of β-catenin and LRP6 (Fig. 7b). Importantly, MMP7 and further EMT-related proteins of Snail and N-Cadherin were decreased after SULT2B1 knockdown (Fig. 7b).
a The mRNA level of SULT2B1 was assessed by qRT-PCR in HIBEpiC cells. b Western blot of SULT2B1, MMP7, Wnt/β-catenin signaling pathway-related proteins, and EMT-related proteins in HIBEpiC cells with/without siSULT2B1 and TGFβ1 treatment. c Functional analysis (left panel: GO terms; right panel: KEGG) in DEGs from RNA-seq data for HIBEpiC cells treated with siRNA-SULT2B1 or siRNA-NC upon stimulation with TGF-β1 for 48 h. (**p < 0.01).
Furthermore, we performed RNA-seq on siRNA-SULT2B1 and siRNA-NC infected HIBEpiC cells upon treatment with TGF-β1. Functional analyses also showed that silencing of SUTL2B1 could significantly inhibit biological processes such as EMT, ECM-receptor interaction, and TGF-β1 signaling pathway (Fig. 7c). The above findings confirmed that SULT2B1 promoted cholangiocyte EMT process by regulating the Wnt/β-catenin/MMP7 pathway.
Discussion
BA is the leading cause of neonatal cholestasis with a complex etiology, and the main pathological changes of BA were collagen deposition and liver fibrosis.19 Cholangiocytes are highly specialized epithelial cells that line the intrahepatic and extrahepatic bile ducts. Despite accounting for only 3%–5% of the total liver cell population, cholangiocytes are critical for liver function and health.20 In BA, Kupffer cells and macrophages produce hedgehog (Hh) ligands, and Hh ligand-stimulated cholangiocytes produce a variety of cytokines, including TGF-β1, as well as chemokines that attract different inflammatory cell populations.21 In addition, the transformation of cholangiocytes into myofibroblasts through EMT was an important process leading to progressive liver fibrosis in BA.2,5 Several studies have reported that factors such as viruses or bacteria may induce the EMT of cholangiocytes,5,22 and EMT might be a survival response of cholangiocytes to resist virus-induced apoptosis, which persisted even after virus clearance.22 In the present study, we found that several mesenchymal markers, such as Snail, Twist1, and N-Cadherin, in BA livers were up-regulated and that SULT2B1/MMP7 was involved in mediating EMT in cholangiocytes.
SULT2B1, one of the most important sterol sulfates in human plasma, is present as a normal component in various human tissues,9 and it was detected to be highly expressed in the liver of fetal rats and gradually decreased after birth.23 The EMT process occurs during embryonic development, suggesting that SULT2B1 is associated with a significant proliferative capacity for EMT and liver physiological function.23 However, the role of SULT2B1 in BA has rarely been reported. In this study, we identified SULT2B1 as a key molecule involved in regulating the cholangiocyte EMT process of BA through WGCNA. GSEA analysis also suggested that highly expressed SULT2B1 was enriched in EMT. Furthermore, RNA-seq results suggested that SULT2B1 showed a clear trend of co-expression with a variety of EMT-related molecules, such as Snai1, Twist1, and VIM. Immunohistochemical staining showed that the highly expressed SULT2B1 was mainly distributed in the bile duct epithelial cells of BA. Further in vitro studies confirmed that SULT2B1 mediated the EMT process in HIBEpiC cells via the Wnt/β-catenin/MMP7 signaling pathway.
As an important member of the MMP family, MMP7 promotes fibrosis by inducing E-cadherin protein degradation, ECM accumulation, and TGF-β1 signaling activation.24 Otherwise, several studies have confirmed that MMP7 can be used as a specific diagnostic serum marker for BA.7,8,25,26,27 A recent study in vivo also showed that silencing MMP7 can significantly reduce virus-induced bile duct injury and obstruction in RRV mice, suggesting that MMP7 was directly involved in biliary fibrosis of BA.7 In the present study, we found that MMP7 was significantly up-regulated in serum and liver of BA compared with controls, and MMP7 was also positively correlated with a variety of liver function and liver fibrosis indicators. The above results fully confirm the importance of MMP7 in BA, but the mechanism of MMP7 upregulation in BA remains unclear. The pronounced upregulation of SULT2B1 in CMV-positive BA patients suggests that viral infection may enhance cholangiocyte EMT via inflammatory pathways (e.g., NF-κB). Further studies are needed to dissect the interplay between CMV and SULT2B1. In the present study, we found that the expression levels of SULT2B1 and MMP7 were also consistent with the trend of mesenchymal markers, suggesting that SULT2B1 and MMP7 are jointly involved in the EMT process. MMP7 is a known downstream target gene of Wnt/β-catenin;28 we also found that Wnt/β-catenin was significantly activated in BA livers. In addition, silencing of SULT2B1 significantly reduced the expression of multiple Wnt/β-catenin pathway-related genes, including MMP7, in HIBEpiC cells. The above findings confirmed that SULT2B1 regulated MMP7 through the Wnt/β-catenin pathway and mediated the EMT process.
TGF-β1 is a multifunctional cytokine that is a potent inducer of EMT.1 A recent study based on sea lamprey found that aberrant expression of TGF-β in BA underlay EMT, which was associated with degeneration of the biliary system during the development of BA.29 Besides, several studies have found that TGF-β1 also played an important role in the fibrosis formation of BA.30,31 In our study, we found that TGF-β1 could significantly upregulate the expression of SULT2B1 and further induce the EMT process in HIBEpiC cells. Silencing SULT2B1 could inhibit the EMT process of cholangiocytes induced by TGF-β1. These findings suggest that SULT2B1 could act as a downstream molecule of TGF-β1 in the regulation of EMT in cholangiocytes. However, the mechanism of TGF-β1-induced SULT2B1 upregulation still needs to be further investigated in the future. And the future study will focus on establishing animal models of BA and validating the impact of cholangiocyte-specific SUL2B1 knockdown in EMT and liver fibrosis. Furthermore, MMP7 is reported to cleave and activate latent TGF-β1, which may establish a positive feedback loop in BA pathogenesis. Specifically, TGF-β1-induced SULT2B1 upregulates MMP7, and MMP7 in turn activates more TGF-β1, thereby amplifying cholangiocyte EMT and fibrosis. Future studies will explore this potential loop by combining MMP7 inhibition with SULT2B1 knockdown.
In summary, our findings showed that SULT2B1 was a key molecular involved in regulating the cholangiocyte EMT and liver fibrosis processes of BA. SULT2B1 could promote the cholangiocyte EMT via the Wnt/β-catenin/MMP7 pathway upon TGF-β1 stimulation. In the future, blocking of SULT2B1/Wnt/β-catenin/MMP7 axis may provide insight into potential targeted therapeutic interventions in BA. Although targeting the SULT2B1/Wnt/β-catenin/MMP7 axis is promising, potential off-target effects (e.g., disruption of cholesterol metabolism by SULT2B1 inhibition) require careful evaluation. Future studies should prioritize developing cholangiocyte-specific delivery systems (e.g., nanoparticle carriers) to minimize systemic toxicity.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors under reasonable request.
References
Scheau, C. et al. The role of matrix metalloproteinases in the epithelial-mesenchymal transition of hepatocellular carcinoma. Anal. Cell. Pathol. 2019, 9423907 (2019).
Wang, J.-Y. et al. Suppressing microRNA-29c promotes biliary atresia-related fibrosis by targeting DNMT3A and DNMT3B. Cell. Mol. Biol. Lett. 24, 10 (2019).
Asai, A., Miethke, A. & Bezerra, J. A. Pathogenesis of biliary atresia: defining biology to understand clinical phenotypes. Nat. Rev. Gastroenterol. Hepatol. 12, 342–352 (2015).
Li, F. B. et al. Expression of transforming growth factor-β1 and connective tissue growth factor in congenital biliary atresia and neonatal hepatitis liver tissue. Genet. Mol. Res. https://doi.org/10.4238/gmr.15017217 (2016).
Shen, W.-J., Chen, G., Wang, M. & Zheng, S. Liver fibrosis in biliary atresia. World J. Ppediatr. 15, 117–123 (2019).
Cui, N., Hu, M. & Khalil, R. A. Biochemical and biological attributes of matrix metalloproteinases. Prog. Mol. Biol. Transl. Sci. 147, 1–73 (2017).
Lertudomphonwanit, C. et al. Large-scale proteomics identifies MMP-7 as a sentinel of epithelial injury and of biliary atresia. Sci. Transl. Med. https://doi.org/10.1126/scitranslmed.aan8462 (2017).
Kerola, A. et al. Increased MMP-7 expression in biliary epithelium and serum underpins native liver fibrosis after successful portoenterostomy in biliary atresia. J. Pathol. Clin. Res. 2, 187–198 (2016).
Wang, Y. et al. Sult2b1 deficiency exacerbates ischemic stroke by promoting pro-inflammatory macrophage polarization in mice. Theranostics 11, 10074–10090 (2021).
Chen, W., Zhou, H., Ye, L. & Zhan, B. Overexpression of SULT2B1b promotes angiogenesis in human gastric cancer. Cell. Physiol. Biochem. 38, 1040–1054 (2016).
Yang, X. et al. SULT2B1b promotes epithelial-mesenchymal transition through activation of the β-catenin/MMP7 pathway in hepatocytes. Biochem. Biophys. Res. Commun. 510, 495–500 (2019).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Pertea, M. et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295 (2015).
Love, M. I., Huber, W. & Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014).
Zhou, T. et al. Knockdown of vimentin reduces mesenchymal phenotype of cholangiocytes in the Mdr2-/- mouse model of primary sclerosing cholangitis (PSC). EBioMedicine 48, 130–142 (2019).
Hong, W. et al. Hydroxysteroid sulfotransferase 2B1 affects gastric epithelial function and carcinogenesis induced by a carcinogenic agent. Lipids Health Dis. 18, 203 (2019).
Subramanian, A. et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA 102, 15545–15550 (2005).
Qiu, J.-L. et al. Ligustrazine attenuates liver fibrosis by targeting miR-145 mediated transforming growth factor-β/Smad signaling in an animal model of biliary atresia. J. Pharmacol. Exp. Ther. 381, 257–265 (2022).
Cielecka-Kuszyk, J. et al. The usefulness of immunohistochemical staining of bile tracts in biliary atresia. Clin. Exp. Hepatol. 7, 41–46 (2021).
Banales, J. M. et al. Cholangiocyte pathobiology. Nat. Rev. Gastroenterol. Hepatol. 16, 269–281 (2019).
Ortiz-Perez, A. et al. Innate immunity and pathogenesis of biliary atresia. Front. Immunol. 11, 329 (2020).
Harada, K. Sclerosing and obstructive cholangiopathy in biliary atresia: mechanisms and association with biliary innate immunity. Pediatr. Surg. Int. 33, 1243–1248 (2017).
Wang, Z. et al. Upregulation of hydroxysteroid sulfotransferase 2B1b promotes hepatic oval cell proliferation by modulating oxysterol-induced LXR activation in a mouse model of liver injury. Arch. Toxicol. 91, 271–287 (2017).
Ke, B., Fan, C., Yang, L. & Fang, X. Matrix metalloproteinases-7 and kidney fibrosis. Front. Physiol. 8, 21 (2017).
Yang, L. et al. Diagnostic accuracy of serum matrix metalloproteinase-7 for biliary atresia. Hepatology 68, 2069–2077 (2018).
Nomden, M. et al. Current concepts of biliary atresia and matrix metalloproteinase-7: a review of literature. Front. Med. 7, 617261 (2020).
Jiang, J. et al. Serum MMP-7 in the diagnosis of biliary atresia. Pediatrics https://doi.org/10.1542/peds.2019-0902 (2019).
Wang, C. et al. Silencing of KIF3B suppresses breast cancer progression by regulating EMT and Wnt/β-catenin signaling. Front. Oncol. 10, 597464 (2020).
Chung-Davidson, Y.-W. et al. TGF-β Signaling plays a pivotal role during developmental biliary atresia in sea lamprey (Petromyzon marinus). Hepatol. Commun. 4, 219–234 (2020).
Lee, S.-Y. et al. Identification of transforming growth factors actively transcribed during the progress of liver fibrosis in biliary atresia. J. Pediatr. Surg. 39, 702–708 (2004).
Ahmed AFKU et al. In situ expression of fibrogenic growth factors and their receptors in biliary atresia: comparison between early and late stages. J. Pathol. 192, 73–80 (2000).
Acknowledgements
We appreciate all the subjects who participated in the study.
Funding
This work was supported by the National Natural Science Foundation of China grant (82400592).
Author information
Authors and Affiliations
Contributions
H.J.S., Y.T., and W.D.D. conceptualized and supervised the study; Y.T., W.D.D., Y.S., M.W.J., and Z.J.W. managed the resources. H.J.S., Y.T., W.D.D., Z.L., and Y.S. developed the methodology. Y.T., W.D.D., Y.S., C.Y.J., F.L.B., and W.H.M. performed the investigation. Y.T. and Y.S. wrote the manuscript. H.J.S. and W.D.D. reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate
All patients’ guardians provided written informed consent. This study was approved by the Medical Ethics Committee of the Beijing Children’s Hospital (2019-k-386).
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, T., Yang, S., Mou, W. et al. SULT2B1 promotes cholangiocyte epithelial-mesenchymal transition via Wnt/β-catenin/MMP7 pathway in biliary atresia. Pediatr Res (2025). https://doi.org/10.1038/s41390-025-04304-6
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41390-025-04304-6