Abstract
Background
Combining high-dose stereotactic body radiation therapy (SBRT) with FOLFIRINOX (FFX) is promising as neoadjuvant strategy for pancreatic ductal adenocarcinoma (PDAC). This study provides an in-depth histo-molecular characterisation of resected PDAC samples from patients treated with FFX ± SBRT.
Methods
Residual tumour tissues from 56 non-metastatic PDAC patients were analysed: seventeen underwent upfront surgery, seventeen received neoadjuvant FFX alone and twenty-two FFX followed by radiotherapy (sixteen SBRT, six radiochemotherapy [RT-CT]). Samples were assessed using RNAseq and immunohistochemistry/fluorescence, including multiplex.
Results
Addition of SBRT to FFX favourably remodelled PDAC, influencing stromal, immune, metabolic and molecular features. Unlike RT-CT, SBRT counteracted several detrimental effects induced by FFX alone. Notably, FFX + SBRT enriched tumours with ‘Classical’ and ‘Inactive stroma’ signatures—linked to better prognosis - while reducing ‘Basal-like’ cell enrichment. SBRT promoted COL1A1-driven stromal remodelling while globally preserving T-lymphocyte infiltration, including cytotoxic T cells, which maintained close proximity to tumour cells despite increased desmoplasia. Key transcriptional alterations induced by SBRT were identified, offering targets for future combination therapies.
Conclusions
Highlighting a more favourable stromal and molecular profile after integration of high-dose SBRT to FFX, this study supports the development, rationale and validation in prospective trials of using this treatment combination in non-metastatic PDAC.

Similar content being viewed by others
Background
Pancreatic ductal adenocarcinoma (PDAC) remains one the deadliest tumours, with a 5-year survival rate under 12% [1]. Despite the introduction of more active multi-agent chemotherapy like FOLFIRINOX [FFX], therapeutic progress has been limited [2,3,4]. Immune checkpoint inhibitors (ICIs), cancer vaccines and targeted therapy have shown little to no positive clinical impact, largely due to the PDAC’s complex and resistant tumour microenvironment (TME) [5, 6]. Therefore, understanding how current therapies used in daily-practice modulate the PDAC’s TME is essential.
Neoadjuvant therapy is increasingly used in non-metastatic PDAC patients, although the exact sequence to use remains to be determined [7]. The FFX regimen is currently preferred by many centres due to the results of several trials showing a significant superiority in survival compared to gemcitabine, as well as a safe and active profile in neoadjuvant phase II/III trials [2, 8,9,10,11]. Combining FFX with (nearly-) ablative stereotactic body radiation therapy (SBRT) in the neoadjuvant setting may offer advantages over conventional radiochemotherapy (RT-CT). These include the capacity to deliver a much higher biologically effective dose (BED) to the tumour which is associated with better prognosis, as well as shorter full-dose chemotherapy interruptions [7, 12,13,14]. Several studies reported promising results and an increasing number of ongoing randomised clinical trials are exploring this question, including ours (STEREOPAC trial – NCT05083247) [7, 14,15,16,17,18]. However, if radiation therapy is able modulate the TME, the immunological and molecular effects of high-dose SBRT (>35 Gy in 5 fractions) in PDAC remains poorly known. A better understanding of these modulations may pave the way for the development of molecularly oriented combination trials with immune and/or targeted therapies as well as stratified treatment strategies, which are urgently needed. In the last decade, identification of molecular subtypes has gained a lot of interest in PDAC and it is now clearer that these molecular signatures have the potential to improve patients’ selection, to predict treatment response, therefore leading to the development of individualised treatments [19,20,21,22,23,24,25,26]. While the relationship between molecular subtypes and chemotherapy is progressively explored, little is known regarding RT, especially (nearly) ablative SBRT combined with FFX.
In this study, we aimed to characterise, for the first time in PDAC, the molecular subtypes, transcriptomic profiles and immuno-modulations followed by FFX alone or combined with isotoxic high-dose SBRT (iHD-SBRT). We hypothesised that iHD-SBRT can induce durable alterations in the molecular and transcriptional landscape of PDAC, thereby elucidating key cellular components and pathways involved and enhancing our understanding of its therapeutic contribution and complementarity with FFX.
Results
Patients characteristics and outcomes
A total of 124 patients with non-metastatic PDAC who underwent surgical resection between 2011 and 2020 were retrospectively assessed for eligibility. Seventy-one patients were initially included, but fifteen were excluded due to failing RNA sequencing (RNAseq) quality control. Ultimately, RNAseq data from 56 patients were considered for this study. The main cohort consisted of: 1/Seventeen patients in the non-neoadjuvant (No_NAT) group; 2/Seventeen patients in the FFX group and 3/Sixteen patients in the FFX + SBRT group. An additional cohort of six patients treated with FFX + RT-CT was included to further investigate the effects of RT. The methodology workflow of the study is described in the CONSORT-like clinico-molecular diagram in Fig. 1. In the FFX + SBRT group, patients underwent an oncological resection at a median time of 44 days (range: 31–70 days) after iHD-SBRT. This group displayed significantly better tumour regression scores compared to the FFX group and included a higher proportion of locally advanced tumours. No significant difference in median overall survival (OS) or disease-free survival (DFS) were observed between groups. However, we noted that the 1-year DFS was significantly improved in the FFX + SBRT cohort (FFX + SBRT vs FFX vs No_NAT: 87.5 vs 70.6 vs 41.2%, respectively, p = 0.017) (Fig. S1). The main clinico-pathological characteristics are summarised in Table 1 and Tables S1, S2.
Detailed description of the selection process of the patients and samples cohort. PDAC pancreatic ductal adenocarcinoma, IPMN intraductal papillary mucinous neoplasm, RT radiotherapy, FFX FOLFIRINOX, RT-CT conventional radiochemotherapy, SBRT stereotactic body radiotherapy, QC quality control, RNAseq RNA sequencing, No_NAT no neoadjuvant treatment group.
iHD-SBRT following induction chemotherapy with FFX is able to reverse several of the major unfavourable transcriptomic alterations induced by FFX in PDAC
To investigate the biological functions of the identified differentially expressed genes (DEGs) between treatment groups, we performed the gene ontology (GO) functional annotation describing genes and their associations according to three ontology categories (molecular function, cellular component and biological process) (Fig. 2 and Figs. S2–S3) [27]. Compared to the No_NAT group, the FFX group demonstrated a significant enrichment in pathways related to mitotic cell cycle arrest, extracellular matrix (ECM), transcriptional activity (including histone demethylation), regulation of glucose transport, as well as for regulation of angiogenesis, the vascular endothelial growth factor (VEGF) signalling pathway and epithelial to mesenchymal transition (EMT). In contrast, the addition of iHD-SBRT to FFX resulted in significant negative enrichment in glucose transport, angiogenesis-related items, as well as ECM assembly and EMT. Furthermore, the FFX + SBRT group showed significant positive enrichment scores notably related to mitochondrial activity, glutathione biosynthetic process and apoptotic cell clearance, while a reduced level of items was detected related to cell adhesion, cell migration (including for fibroblasts), ECM organisation and cellular response to TGFβ stimulus. To complement these findings, we analysed an additional cohort of six patients treated with FFX + RT-CT. However, most of the changes observed with SBRT were not replicated, except for a similar negative enrichment in glucose transport compared to the FFX group. (Fig. S4)
Selected Gene Set Enrichement Analysis (GSEA) of Gene Ontology (GO) terms based on differential gene expression comparisons between the three main groups (No_NAT, FFX and FXX + SBRT). GO terms are grouped according to biological function (e.g. cell cycle, metabolism). Enrichment scores reflect the relative up- or downregulation of gene sets in each comparison, highlighting functional pathways associated with treatment-specific transcriptional changes. FFX FOLFIRINOX, No_NAT no neoadjuvant treatment group, SBRT stereotactic body radiotherapy, ECM extracellular matrix.
Further analysis using GO and Molecular signatures database (MSigDB) canonical pathways confirmed significant enrichment in mitochondrial activity, glutathione metabolism and ribosomal pathways in the FFX + SBRT vs No_NAT comparison. Additionally, applying single-nucleus signatures from Hwang et al., we observed increased ribosomal biogenesis whereas decreased TNF/NF-kB signalling following iHD-SBRT (Fig. 3) [25].
a, b Normalised Enrichment Score (NES) from enrichment analysis of PDAC subtypes RNA signatures, showing significantly higher NES for the ‘Classical’ subtypes and lower NES for ‘Basal’ associated subtypes in the FFX + SBRT vs No_NAT group (a) and the FFX + SBRT vs FFX (b) through the main transcriptomic PDAC classifications. c, d, e NES from GSEA using Hwang et al. single nucleus RNA-seq signatures. Differential gene expression comparisons between FFX vs No_NAT group (c), FFX + SBRT vs No_NAT group (d) and FFX + SBRT vs FFX group (e), reveal significant enrichment of the Mesenchymal subtype in FFX samples, while the Basaloid subtype - associated with favourable prognosis—is enriched in the FFX + SBRT group. f Application of the pancreatic adenocarcinoma molecular gradient (PAMG), a continuous gradient of PDAC pre-existing classifications, across the main treatment cohorts (No_NAT, FFX and FFX + SBRT) shows a significantly higher PAMG score in the FFX + SBRT group compared to FFX (p = 0.049), indicating a favourable shift along the continuous molecular gradient of PDAC subtypes. No_NAT no neoadjuvant treatment group, FFX FOLFIRINOX, SBRT stereotactic body radiotherapy.
Addition of iHD-SBRT to FFX is associated with transcriptomic signatures and PAMG score linked to better prognosis
The molecular subtype signatures from the main studies in the field (Puleo et al.; Moffitt et al.; Bailey et al.; Hwang et al. [20,21,22, 25]) were analysed in this cohort to assess the influence of modern neoadjuvant treatments, including high-dose SBRT (Fig. 3). Compared to both the No_NAT and FFX groups, the FFX + SBRT group showed a significant enrichment in the more favourable ‘Classical subtype’ signatures (Fig. 3a, b, in red). Furthermore, the addition of iHD-SBRT was associated with a reduced level of ‘activated stroma’ and ‘Basal-like subtype’ signatures across all major molecular classifications, which are associated with poorer prognosis (Fig. 3a, b, in blue).
To gain deeper insight into the evolution of molecular subtypes following neoadjuvant treatments, we applied the recently published single-nucleus signatures from Hwang et al. to our cohort [25]. The FFX group, compared to No_NAT, was enriched with the ‘Mesenchymal’ signature - a subtype of ‘Basal-like’ cells - and several stromal signatures associated with highly active stroma, all of which are linked to worse clinical outcomes (Fig. 3c and Fig. S5a). The neural-like progenitor and neudroendocrine –like programmes identified by Hwang et al. as significantly higher post RT-CT were not significantly enriched in our cohort (Supplementary Data 1–2) [25]. Interestingly, when compared to both the No_NAT and FFX cohorts, the FFX + SBRT group was notably significantly associated with the ‘Basaloid’ signature, a particular subtype of ‘Basal-like’ cells that is associated with better clinical outcomes (Fig. 3d, e) [25].
Finally, we applied the pancreatic adenocarcinoma molecular gradient (PAMG) - a continuous gradient of PDAC pre-existing classifications—which revealed a significant favourable shift in samples treated with FFX + SBRT toward a higher PAMG score compared to FFX alone. These findings confirm that the FFX + SBRT group is significantly enriched in ‘Classical’ subtype gene signatures, associated with better cell differentiation, as well as improved clinical outcomes (Fig. 3f) [28].
For additional insight into the effects of different RT techniques, we analysed a cohort of six patients treated with FFX + RT-CT. Similar to FFX + SBRT, the FFX + RT-CT group appeared significantly enriched in Classical and normalised stromal signatures compared to the No_NAT and FFX groups but failed to show a significant decrease in Basal-like associated signatures, as observed in the FFX + SBRT group. In direct comparison with SBRT, the FFX + RT-CT group displayed a positive enrichment in normal stroma signatures and less ‘Basaloid’ signature (Fig. S5).
The combination of FFX and SBRT modulates the metabolic state of PDAC towards an enrichment of the cholesterogenic metabolic profile
Given that FFX alone and FFX + SBRT appear to induce opposite enrichment scores regarding several transcriptional items related to metabolism, such as mitochondrial activity (including oxidative phosphorylation) and glucose import (Fig. 2 and Figs. S2–3), we performed a deeper characterisation of the metabolic state in our cohort using the metabolic gene signatures identified by Karasinska et al. [29]. Across the main treatment groups, glycolytic genes were significantly associated with ‘Basal-like’ genes expression, while cholesterogenic genes were linked to the ‘Classical’ subtype (Fig. S6). Notably, compared to FFX alone, the FFX + SBRT group showed a significant positive enrichment score for cholesterol biosynthesis, a metabolic profile correlated with favourable clinical outcomes (Fig. S6) [29].
The combination of FFX and SBRT combination induces distinct transcriptomic modulations in cancer-associated fibroblasts (CAFs) signatures compared to FFX alone
As both FFX and FFX + SBRT treatments were found to significantly impact stromal transcriptomic signatures and ECM organisation, xCell analyses were performed. These revealed a significantly higher stroma score (Fig. 4a) and increased CAFs population (Fig. 4b) following neoadjuvant FFX compared to No_NAT. To further characterise CAF-specific changes, several bulk and single-cell based CAF classifications were assessed using GSEA to evaluate the distinct modulations induced by FFX alone and FFX + SBRT (Fig. 4c–h).
Cell type enrichment analysis using xCell showing a significantly higher stroma score (p = 0.034) (a) and CAFs population (p = 0.039) (b) in FFX vs No_NAT group. c, d, e Normalised Enrichment Score (NES) after GSEA of Hwang et al. gene sets obtained with single nucleus RNA-seq: differential expression comparison between FFX vs No_NAT group (c), FFX + SBRT vs No_NAT group (d) and FFX + SBRT vs FFX group (d). NES after GSEA of state of the art CAFs gene sets: differential expression comparison between FFX vs No_NAT group (f), FFX + SBRT vs No_NAT group (g) and FFX + SBRT vs FFX group (h). i, j Gene set variation analysis (GSVA) was applied as a single sample classifier of different CAF subtypes defined in Elyada et al. to classify all the samples according to their enrichment in high and low iCAF and myCAF groups. Kaplan–Meier survival analyses were performed on high and low CAF populations. High-iCAF samples showed a significantly better overall survival (OS) compared to Low-iCAF (p = 0.0038) (i) while no statistical difference was found for myCAFs (j). k Locoregional disease free survival (LR-DFS) in the three groups stratified per high and low-iCAF samples. A significantly better LR-DFS was observed in high-iCAF in the FFX + SBRT cohort (p = 0.038) while a non-significant tendency has been observed for the No_NAT and FFX groups. No_NAT no neoadjuvant treatment group, FFX FOLFIRINOX, SBRT stereotactic body radiotherapy.
Both neoadjuvant cohorts showed an enrichment in ‘Immunomodulatory’ CAFs (from Hwang et al. [25]) and inflammatory CAFs (iCAFs) signatures compared to No_NAT. Interestingly, FFX alone, in comparison to the No_NAT and FFX + SBRT groups, was significantly enriched in myofibroblastic CAFs (myCAFs) signatures, which are associated with poorer prognosis. (Fig. 4c, f). Furthermore, patients treated with FFX + SBRT exhibited reduced levels of pancreatic stellate cells (PSCs) signatures as well as myCAFs compared to both No_NAT and FFX groups, along with an enrichment in ‘Normal Fibroblasts’ signatures relative to No_NAT. (Fig. 4d, e, g, h).
In an additional cohort of patients treated with FFX + RT-CT, similar reduction in myCAF signatures were observed. However, unlike FFX + SBRT, this group did not show enrichment in iCAF signatures. Compared to FFX + SBRT, the FFX + RT-CT group also displayed increased enrichment in PSCs signature (Fig. S5).
iCAFs but not myCAFs are significantly associated with better clinical outcome in No_NAT and FFX + SBRT cohorts
iCAF and myCAF transcriptomics signatures from Elyada et al. were independently evaluated using the single sample Gene Set Variation Analysis (GSVA) classifier to stratify the three main treatment groups in high and low enrichment groups for each CAF subtypes [30]. Interestingly, across the entire cohort, iCAF-high samples were associated with significantly better OS compared to the iCAF-low samples (p = 0.0038) (Fig. 4i). This observation was validated in an external No_NAT cohort (Puleo et al. [22]; n = 309), where iCAF enrichment was similarly associated with significantly better relapse free survival (p = 0.041; Fig. S7). In contrast, no significant differences in DFS or OS were observed between high and low myCAF groups in either our cohort or the Puleo et al. cohort (Fig. 4j and Fig. S7). In the FFX + SBRT cohort specifically, iCAF-high samples also displayed a significantly improved loco-regional disease-free survival (LR-DFS) compared to iCAF-low samples (p = 0.038; Fig. 4k). These results highlight the potential of iCAF signatures as a prognostic/predictive biomarker in PDAC.
Neoadjuvant treatments increase desmoplasia without significantly affecting tumour-infiltrating lymphocytes (TILs) except for the T helper population
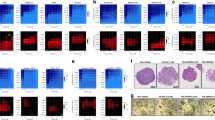
To further characterise the stromal features of treated PDAC, the percentage of the tumoral area occupied by collagen was quantified through immunohistochemistry (IHC) across the entire cohort. Consistent with our previous findings, a significant increase in Collagen1A1 (COL1A1) deposition – a marker indicative of the pan-fibroblast population - was observed in both neoadjuvant groups compared to the No_NAT group (68.4 vs 78.6 vs 83.27% for No_NAT vs FFX vs FFX + SBRT, respectively, p < 0.001) (Fig. 5a). Additionally, a non-significant trend towards a lower expression of αSMA, a marker associated with myCAFs, was noticed in the FFX + SBRT group compared to FFX (Fig. 5b). Despite the increased collagen deposition following neoadjuvant treatments, no significant changes were observed in the expression levels of CD3 TILs or cytotoxic CD8+ cells, including after SBRT (Fig. 5c, d). Among T-cell populations, only the CD4+ T helper subset was significantly decreased in the FFX + SBRT group compared to both FFX and No_NAT groups (Fig. 5e). The CD20 + B-cell population was decreased following NAT, with a significant difference observed between the FFX + SBRT and No_NAT groups (Fig. 5f). Histopathological review by specialised GI pathologists (LV and PDM), revealed frequent signs of tumour cells injury - such as cell swelling and pyknotic nuclei - following NAT (Fig. 6). Scarce tertiary lymphoid structures (TLS) were identified within the tumoral area on consecutive H&E and CD3/CD20 dual-stained slides, with no significant differences between the three main groups (Fig. 5l). Importantly, immune infiltration, including TILs, did not appear to be sequestered within the collagen-rich stroma after FFX + SBRT. Instead, TILs remained in close proximity to residual tumoral glands and were observed directly infiltrating these glands (Fig. 6 and Fig. S8). To validate these observations, we quantified immune infiltration within 15 µm of the tumoral glands using multiplex immunofluorescence (mIF). The analysis revealed no significant differences in the proximity of CD3+CD+, CD3+CD8+, except for CD3+CD4+ T cells, which showed a trend toward reduced infiltration near tumour cells after FFX alone compared to the No_NAT group (p = 0.049). (Fig. S8)
IHC analysis of stromal markers showing increased COL1 deposition after neoadjuvant treatments (a) while αSMA expression remained unchanged (b). c–f No significant differences in IHC expression were observed among the three groups for CD3+ (c), CD8+ (d). However, expression of CD4+ (e) and CD20+ cells (f) was significantly decreased in the FFX + SBRT group compared to No_NAT. No significant differences in IHC expression was observed for CD68+ (g) and CCR2 cells (h) across the three groups. i No significant differences in IHC expression was observed for FOXP3+ cells across the three groups. PD-L1 IHC expression significantly increased but remained scarce in the FFX + SBRT group (j) while PD-1 IHC expression was significantly decreased (k). No significant differences in IHC expression was observed for CD68+ (j) and CCR2 cells (k) across the three groups. l The median number of intra-tumoral tertiary lymphoid structures (TLS) did not differ significantly between groups. A representative mIHC image illustrates the rare presence of TLS adjacent to tumour glands in a No_NAT sample. m Immune cell type enrichment analysis using xCell deconvolution revealed no difference in CD4 + Th1 T cells (m(a)), but a significantly lower enrichment of CD4 + Th2 T cells (p = 0.029) (m(b)) and myeloid dendritic cells (p = 0.032) (m(e)) in the FFX + SBRT group compared to FFX. A significant enrichment of M1 tumour-associated macrophages (TAMs) (p = 0.0045) (m(c)) and M2-TAMs (p = 0.024) (m(d)) was also observed in FFX + SBRT vs FFX group. No_NAT no neoadjuvant treatment group, FFX FOLFIRINOX, SBRT stereotactic body radiotherapy, NS non-significant, IHC immunohistochemistry, mIHC multiplex immunohistochemistry, TLS tertiary lymphoid structure.
Representative images of: a Global immune infiltration in No_NAT group with high density of tumoral glands; b Global immune infiltration in FFX + SBRT group with less density of tumoral glands; c Global distribution of tumour infiltrating lymphocytes (TILs) in No_NAT group; d TILs in FFX + SBRT group are not sequestrated within the collagenous area; e TILs in No-NAT group close to the tumoral glands; f TILs in FFX + SBRT group are mainly located close and in direct with the tumoral glands; CD4+, CD8+ and CD20+ was observed within the tumoral glands. Cell swelling and pyknotic nucleus of the tumoral cells can be observed in FFX + SBRT treated PDAC; g Tumour associated macrophages (TAMs) and CCR2+ cells populations in No_NAT group; h TAMs are frequently observed within the lumen of tumoral glands in FFX + SBRT group and CCR2+ cells expression is maintained. No_NAT no neoadjuvant treatment group, FFX FOLFIRINOX, SBRT stereotactic body radiotherapy.
Immunosuppressive cells remain present after FFX ± SBRT neoadjuvant treatments
To further investigate immunosuppressive populations within the TME, IHC staining was performed for pan-macrophage marker CD68, CCR2, FOXP3 and PD-1/PD-L1 across the entire PDAC cohort. Following FFX + /-SBRT, no significant differences were observed for CD68+ and CCR2 + cells (Fig. 5g, h). However, FOXP3 expression was significantly increased in both the FFX and FFX + SBRT groups compared to the No_NAT group (Fig. 5i). In the FFX + SBRT group, CD68+ cells were frequently visualised within the lumens of residual tumour glands (Fig. 6). Quantification using mIF revealed no significant differences in the infiltration of FOXP3+ and CD68+ cells near tumour glands across the three main groups. (Fig. S8) PD-L1 and PD-1 expression was scarce on our whole cohort. PD-L1 expression was significantly increased in the FFX + SBRT group compared to both No_NAT and FFX groups, although its expression on lymphocytes-like cells remained globally low and weak with a majority of negative samples (Fig. 5j). Conversely, PD-1 expression was significantly reduced and nearly absent in the FFX + SBRT group compared to No_NAT and FFX groups (Fig. 5k). For complementary information regarding RT, an additional cohort of 6 patients treated with FFX + RT-CT was analysed through IHC. No significant differences in general stromal nor immune expression was observed when compared to FFX + RT-CT group except for a reduced expression of FOXP3+ cells and an increased expression of PD-L1 (Fig. S9).
xCell deconvolution analysis of the immune TME shows decreased CD4 Th2 population and increased macrophages polarity after FFX + SBRT
Given the significant decrease in the CD4+ population observed in the FFX + SBRT group by IHC, we further investigated the presence of transcriptomic signatures corresponding to various T helper cell sub-populations using xCell deconvolution analysis. This analysis revealed a significant reduction in the CD4 Th2 population in the FFX + SBRT group compared to the FFX group (Fig. 5m(a, b)). Consistent with the IHC findings, xCell analysis of the global macrophage population showed no differences among the groups (data not shown). However, a significant increase in both M1, and particularly, M2-macrophage subpopulations was observed in the FFX + SBRT samples compared to FFX alone (Fig. 5m(c, d)). In contrast, myeloid dendritic cells (MDCs) were significantly decreased after FFX + SBRT compared to FFX alone (Fig. 5m(e)), while no significant differences were observed for the neutrophil population (data not shown).
Discussion
Following the failure of several clinical trials involving conventional RT-CT and low-dose SBRT, the combination of modern multi-agent chemotherapy - particularly FFX - and (nearly) ablative SBRT, has shown promising oncological outcomes in non-metastatic PDAC. This strategy is currently under investigation in several ongoing prospective randomised trials, including ours [7, 12,13,14,15,16,17,18, 31,32,33,34]. In our cohort, despite its limited size, the 1y-DFS was significantly higher in the FFX + SBRT group compared to FFX alone and No_NAT (87.5 vs 70.6 vs 41.2%, respectively; p = 0.017). Notably, the FFX + SBRT group included a higher proportion of LA patients with larger tumours at diagnosis, yet still showed favourable median DFS and OS. These findings underscore the potential of combining FFX with high-dose SBRT to improve PDAC outcomes. Nonetheless, further well-designed trials incorporating targeted therapies and stratified approaches are urgently needed to improve the dismal patients’ prognosis [5]. For this purpose, we investigated for the first time the histo-molecular modulations induced by FFX alone and FFX followed by iHD-SBRT.
We identified distinct gene expression patterns and key-pathways alterations that clearly distinguish two transcriptional profiles following neoadjuvant FFX alone versus FFX followed by iHD-SBRT. Notably, unlike RT-CT, high-dose SBRT demonstrated the ability to counteract and even reverse several detrimental transcriptional modulations induced by FFX. While FFX alone was associated with increased expression of unfavourable processes—including EMT, angiogenesis, histone demethylation and intracellular transport of glucose—the addition of iHD-SBRT reversed these effects. Moreover, the metabolic landscape differed markedly: FFX + SBRT was associated with enhanced mitochondrial activity and a shift toward a more favourable cholesterogenic profile, in contrast to the glycolytic profile observed with FFX alone [29]. These findings provide further rationale for combining high-dose SBRT with FFX and may help explain the promising oncological outcomes observed with this therapeutic strategy.
Over the past decade, transcriptomic-driven subtyping of PDAC has been performed by several groups, including ours, using various classification systems [19,20,21,22,23,24,25]. In fine, two main molecular subtypes have been consistently identified, namely the ‘Classical’ and ‘Basal-like’ (also referred to as squamous or quasi-mesenchymal) subtypes [19,20,21,22,23,24,25]. The ‘Basal-like’ subtype is associated with poorer prognosis, less differentiated tumours and EMT features, whereas the ‘Classical’ subtype is linked to better survival and well-differentiated tumours [19,20,21,22,23,24,25]. Although data are still scarce and require further validation in PDAC, emerging evidence suggests that therapeutic responses may differ by molecular subtype [24, 26, 28, 29]. Specifically, FFX appears to yield better DFS in ‘Classical’ tumours, while gemcitabine-based regimens may be more effective in ‘Basal-like’ cases [19, 23, 26, 35,36,37]. In our study, we observed significant enrichment of ‘Basal-like’ and active stroma signatures following FFX alone, consistent with previous literature - particularly Porter et al., who reported a shift from ‘Classical’ to ‘Basal-like’ states in PDAC cell lines after FFX [37, 38]. To our knowledge, this is the first study to investigate potential reprogramming of molecular expression following high-dose SBRT (>35 Gy in 5 fractions) in PDAC. Interestingly, adding iHD-SBRT to FFX resulted in a significant shift toward the ‘Classical’ subtype, associated with better prognosis. This shift was consistently observed across multiple molecular signatures and confirmed by the PAMG (pancreatic adenocarcinoma molecular gradient) score [28]. To date, the only published study exploring molecular subtypes in RT-treated patients is the recent single-nucleus RNAseq analysis by Hwang et al. which included 43 PDAC patients - 18 untreated and 25 treated with various neoadjuvant regimens, including RT-CT + FFX ± losartan (n = 19) and two patients treated with FFX + low-dose SBRT (33 Gy in 5 fractions) + losartan ± nivolumab [25]. Although not statistically significant, the authors reported lower expression of the Squamoid programme (analogous to the ‘Basal-like’ subtype), in the RT-CT group compared to No_NAT group, supporting our findings. These data suggest that high-dose SBRT may more effectively target the ‘Basal-like’ subpopulation (selection) and/or reprogramme the FFX-induced ‘Basal-like’ cells into a more ‘Classical-like’ phenotype (reprogramming). This molecular plasticity may be mediated by TGFβ signalling, as suggested by our transcriptomic data. TGFβ has been implicated as a key regulator of plasticity between ‘Basal’ and ‘Classical’ states in PDAC mouse models, with TGFβ blockade promoting a ‘Classical’ phenotype and enhancing chemosensitivity [39].
One of the main transcriptomic modulations observed after neoadjuvant treatments involved stromal remodelling. After iHD-SBRT, compared to both NT and FFX groups, we observed a clear shift toward a more normalised stromal profile associated with better prognosis, prompting further investigation into key-stromal components. Notably, ECM deposition—particularly collagen I—was significantly increased after neoadjuvant treatments, as demonstrated by RNAseq and IHC analyses, in line with previous studies [40, 41]. While desmoplasia was historically considered a driver of tumour progression—due to its role in elevating interstitial fluid pressure, impeding immune infiltration and drug delivery—recent evidence suggests that an expanded stromal compartment may correlate with improved survival and tumour restraint, depending on its cells of origin [40,41,42,43,44,45,46]. In untreated PDAC, the complex and heterogeneous CAF population accounts for ≈90% of the origin of desmoplasia. However, the effects of RT on CAFs remain poorly understood [45, 46]. Despite a significant increase in COL1A1 expression, the myCAF population - most associated with ECM deposition and poor prognosis - was not increased post iHD-SBRT, as confirmed by RNAseq and IHC [47,48,49]. These data suggest either enhanced activity of existing myCAFs and/or increased collagen production by other cell types. Furthermore, the iCAF subpopulation increased following FFX ± iHD-SBRT, aligning with recent findings from Zhou et al., who reported a similar rise in iCAFs in chemotherapy-treated samples (n = 14; FFX and/or gemcitabine/nab-paclitaxel and one case with RT-CT) [50]. High iCAF expression has been associated with improved prognosis in other No_NAT PDAC cohorts, a result we validated in two independent No_NAT cohorts [47, 51, 52]. We also demonstrated a significant association between iCAF-high status and improved LR-DFS after FFX + SBRT, supporting its potential as a prognostic and/or predictive biomarker in PDAC. Given that different neoadjuvant treatments distinctly modulate CAF populations, the efficacy of therapies targeting CAFs may vary depending on treatment context. These findings underscore the need for further studies exploring CAF-targeted strategies in combination with specific neoadjuvant regimens.
Following iHD-SBRT, T-lymphocyte infiltration - including cytotoxic CD8+ T cells - was largely preserved, with immune cells maintaining proximity to, and even direct contact with tumour cells despite increased desmoplasia. This aligns with findings from Mills et al., who assessed CD4/CD8 infiltration within dense collagen regions in nine patients treated with low-dose SBRT (25 Gy in 5 fractions). The authors reported fewer T-cells in these areas in treated samples compared to No_NAT samples, suggesting that T-cell sequestration is not promoted post-SBRT [40]. Another study using spatial analysis identified several differences in immune cell marker expression after neoadjuvant treatment - including 12 patients treated with RT—across distinct tumour regions [53]. In our study, we observed an expected increase in immunosuppressive populations after FFX + SBRT, particularly FOXP3+ Treg cells and M2-macrophages. However, this was accompanied by a decrease in myeloid dendritic cells (MDCs), pancreatic stellate cells (PSCs) and CD4 + Th2 cells. PD-1 and PD-L1 expression was scarce in our whole cohort and, particularly after iHD-SBRT, with almost no PD-1 expression and rare PD-L1 positivity. These findings suggest that the PD-1/PD-L1 axis is not a dominant immunosuppressive mechanism in this context. Consequently, our data do not support the use of anti- PD-1/PD-L1 therapies in PDAC, including in combination with FFX or FFX + SBRT. This aligns with broader clinical data, where trials combining PD-1/PD-L1 inhibitors with chemotherapy and/or RT have failed to demonstrate meaningful benefit in PDAC [5, 6].
Despite certain limitations - including the absence of matched pre- and post-treatment specimens, varied resection statuses, inherent differences in time to surgery based on NAT type received and a relatively small sample size - our study demonstrates for the first time that high-dose SBRT induces durable and profound remodelling of PDAC at the stromal, metabolic and molecular levels. The key alterations identified following FFX + SBRT are summarised in Fig. S10, highlighting the ability of iHD-SBRT to reverse several unfavourable transcriptional enrichment and activations induced by chemotherapy. These findings support the complementarity of SBRT with FFX and suggest potential synergies with immune and targeted therapies. This work provides a comprehensive molecular and histological characterisation of human PDAC following high-dose SBRT, offering valuable insights to guide the future combination strategies. Prospective validation will be conducted in the ongoing randomised phase II STEREOPAC trial, enroling 256 patients with borderline resectable tumours treated with neoadjuvant FFX ± iHD-SBRT [16]. Further research is warranted to elucidate the precise mechanisms underlying the reprogramming and alterations induced by high-dose SBRT. This should include studies using preclinical models and matched human pre/post-treatment specimens to better understand the biological impact of this promising combined therapeutic approach.
Materials and methods
Patients
This study analysed formalin fixed paraffin-embedded (FFPE) residual tissue from 56 PDAC tumours resected between 2011 and 2020 at Erasme and Pitié Salpêtrière hospitals. Eligible patients were ≥18 years old, had complete clinicopathological data, no evidence of metastasis before surgery and received either no neoadjuvant therapy (No_NAT group), an induction with FFX alone (FFX group), or FFX followed by conventional RT-CT (FFX + RT-CT group) or iHD-SBRT (FFX + SBRT group). The main clinical exclusion criteria included other neoadjuvant regimens (including a shift to another type of neoadjuvant chemotherapy), other tumour histology (including PDAC associated with intraductal papillary mucinous neoplasm [IPMN]), insufficient residual tumour tissue (including College of American Pathologists [CAP] tumour regression score 0 [= complete response] to 1 [=near complete response with single cells or small rare groups of cancer cells]) and death within 30 days post-surgery.
Data collection
An aggregated standardised retrospective database was created for patients resected in Erasme and Pitié Salpêtrière hospitals. The variables included: sex, age, CA19.9 levels, clinical disease stage, tumour site, preoperative treatments received, surgery type, TNM classification, histological grade, lymphovascular and perineural invasion and relevant outcomes parameters.
Neoadjuvant treatment
Patients received a median of 6 cycles of FFX, consisting of an intravenous infusion of oxaliplatin (85 mg/m2, 2 h) then leucovorin (400 mg/m2, 2 h) concomitantly with a 90-min intravenous infusion of irinotecan (165–180 mg/m2) followed by a 46 h continuous infusion of fluorouracil (2000–2400mg/m2), administered biweekly.
For six patients, FFX was followed by conventional RT-CT at the dose prescription of 50–54 Gy in 30 fractions (5 days/week) with concomitant administration of capecitabin (800–1250 mg/m2 orally twice daily).
Sixteen patients received FFX followed by iHD-SBRT as previously described in details in [14, 54], according to the strategy implemented in our hospital since January 2018. A surgical exploration was performed in case of no progression 4–7 weeks after iHD-SBRT. Briefly, the SBRT treatment was designed to individually maximise the dose prescribed to the tumour and vessels interfaces (Dmax(0.5cc) < 53 Gy in 5 fractions) with an isotoxic dose prescription. The organs at risk dose constraints to follow were planning organ at risk volumes (PRVs) stomach, duodenum, colon and small bowel, Dmax (0.5cc) < 35 Gy, V30Gy<2cc; PRV spinal cord, V20Gy<1cc and for kidneys, Dmean < 10 Gy and V12Gy < 25% [14, 54].
Sample processing and RNA isolation
New 4 µm FFPE tissue section where H&E stained to identify tumour areas by specialised gastrointestinal pathologists (LV, PDM, NH). Five consecutive unstained 6–8 µm slides were cut under RNAse free conditions. Tumoral area were manually scraped and RNA extracted with the ALLPrep FFPE tissue kit© following the manufacturer’s instructions for semi-automated RNA extraction via Qiacube instrument (Qiagen, Venlo, The Netherlands). RNA quantity and quality were assessed via an Agilent 2100 bioanalyzer using the RNA 6000 Pico LabChip kit (Agilent, Diegem, Belgium). RNA samples with DV200 > 30% were selected and 100 ng of RNA was used for the NGS library preparation using the QuantSeq Library Prep Kit for Illumina (Lexogen) as per manufacturer recommendations’. The libraries were sequenced on NovaSeq using NovaSeq 6000 S2 Reagent Kit with 100 bp single reads.
RNA-sequencing data analysis
Sequencing quality was assessed using FASTQ [55]. Transcript abundance was quantified from raw RNA-seq files using Kallisto v0.50.0 pseudo-alignment method with 100-bootstrap value and a transcriptome index constructed from the human reference transcriptome GRCH38 from Ensembl [56]. Gene-level quantification of estimated counts was performed using the R-package tximport v1.26.1. (data available here: https://doi.org/10.5281/zenodo.10939866) [57]. Poorly covered genes (read count <10 in over half of the samples) were excluded. Differential gene expression (DGE) analysis between treatment groups was performed using the R-packages edgeR v3.40.2 and limma v3.54.2 [58, 59]. Genes with p < 0.05 were represented in heatmaps using the Complex Heatmap v2.14.0 package (Fig. S11) [60]. The PAMG classifier was applied to determine the chemosensitivity and tumour agressivity [28].
Functional analysis
Gene Set Enrichment Analysis (GSEA) was conducted using the fgsea R package v1.24.0. on pre-ranked gene lists with significance set at padj < 0.05 [61]. Gene signatures of PDAC and cancer associated fibroblast (CAF) subtypes were collected from the CancerRNASig package. Molecular Signature Database (MSigDB), Ontology and Canonical pathways gene sets were obtained by the msigdb package v1.6.0. Gene Set Variation Analysis (GSVA) was applied as a single sample classifier of different CAF subtypes [62]. Immune cell fractions were estimated using the xCell algorithm and statistical analysis between treatments of the immune populations was obtained by the package ggpubr v0.6.0 [63, 64].
Immunohistochemistry (IHC)
FFPE full-face tissue sections (4 µm) from 56 tumours were stained for immune and stromal markers (CD3/CD20, CD4/CD8, PD-1/PD-L1, CD68, CCR2, FOXP3, COL1A1 and αSMA). All antibodies and their dilution are listed in the Table S3. Chromogenic IHC was performed on a Ventana Benchmark XT automated staining instrument with the ultraVIEW DAB and ultraVIEW Red Detection Kit (Ventana Medical Systems). All antibodies were initially tested on positive and negative control tissues and staining patterns were validated by pathologists (LV and PDM). Slides were scanned at 40x with a Nanozoomer 2.0-RS Digital slide scanner (Hamamatsu) then delineation of the tumour area was performed by CB followed by experimented specialised pathologists (LV and PDM) verification. Quantification was performed with the Visiopharm© software.
Multiplex immunohistochemistry (mIHC)
FFPE tissue sections (4 µm) were processed manually for mIHC using Opal reagents (Akoya Biosciences) for illustration purposes in four representative samples (No_NAT and FFX + SBRT groups). Briefly, slides were heated at 37 °C overnight before deparaffinization. Heat-induced antigen retrieval was achieved in Antigen Retrieval (AR) 9 buffer (Akoya Biosciences) using a microwave (Panasonic with Inverter technology). Slides were labelled for CD20, CD4, CD8, CD68, CCR2, pan-cytokeratin and DAPI according to the manufacturer’s instructions (Opal 6-Plex Manual Detection Kit - for Whole Slide Imaging, NEL861001KT, Akoya Biosciences) (Table S3). Slides were mounted with Prolong Diamond Antifade Mountant (Life Technologies Europe BV) and scanned with PhenoImager HT (Akoya Biosciences). Tonsil tissue was used as positive control. Region of interests (ROIs) were selected in PhenoChart Whole Slide Viewer by an experimented gastrointestinal pathologist (LV). ROIs were unmixed using the synthetic spectral library and the tissue autofluorescence extracted from an unstained PDAC was removed in inForm Tissue Analysis Software (V.2.6.0, Akoya Biosciences).
Multiplex immunoFluorescence (mIF)
FFPE tissue sections (4 µm) were stained using the OmniVUE 8plex kit (Ultivue, Cambridge, MA, USA) according to the manufacturer protocol in twelve samples representing 4 patients per the main treatment groups (No_NAT, FFX and FFX + SBRT groups). Slides were scanned using the Zeiss AXIOSCAN Z.1 system, with Z-stack imaging (7–11 µm) employed to capture tissue depth, followed by Extended Depth of Field (EDF) computation. The resulting images were compressed using JpgXR and saved in TIFF format. Image sharpening was carried out in Zen Black software, while signal quantification of immune phenotypes (CD3⁺CD4⁺, CD3⁺CD8⁺, CD68⁺, FOXP3⁺) located within 15 µm of PanCK⁺ tumour cells in annotated tumour regions was performed using the HALO’s™ multiplex-IF module (Indica Labs).
Statistical analysis
Statistical analyses were conducted using Stata 14 and R. Data normality was assessed using histograms, boxplots and quantile–quantile plots, and the equality of variances was checked using the Levene’s test.
Categorical data were presented as percentages and numbers, while continuous data were described using median and P25–P75, and due to asymmetric distribution, analysed with nonparametric tests such as the Kruskal-Wallis rank test for group differences. Chi² tests were employed for categorical data. Bonferroni corrections were applied following multiple comparisons between the different groups.
Survival was analysed using the survival v3.5–3 and survminer v0.4.9 packages. Log-rank test was used to calculate the differences in Kaplan-Meier curves and p-values < 0.05 were considered as statistically significant. Multivariate Cox proportional hazard regression models were applied for survival with a 95% confidence interval. OS was defined as the time in months from diagnosis to death due to cancer recurrence. DFS was defined as the time from diagnosis to the first documentation of recurrent disease. Loco-regional DFS (LR-DFS) was defined as the time from diagnosis to the first documentation of loco-regional recurrence.
Non-parametric Wilcoxon test in R v4.2.3 and RStudio v2023.3.0.386 environments was used for RNAseq data analysis, assessing significant differences in treatments in PAMG, Puleo components projections, and xCell immune deconvolution outputs with p < 0.05 considered statistically significant.
For mIH, GraphPad Prism8.0 software was used. For multi-region analysis, we performed 2-way ANOVA for repeated measures. Figure legends show the number of independent experiments (N), and results as medians. Significance was accepted when p < 0,05.
Ethics approval and consent to participate
This study was approved by the Institutional Review Board of Erasme University Hospital and Pitié Salpêtrière hospital under the approval numbers P2018/392—A2020-115 and 2014/58NICB respectively. This study was performed in accordance with the Declaration of Helsinki and informed consent.
Data availability
Complete RNAseq data availability data available is available online: https://doi.org/10.5281/zenodo.10939866. For the rest, all data are available in the main text or the supplementary materials.
References
Dalmartello M, La Vecchia C, Bertuccio P, Boffetta P, Levi F, Negri E, et al. European cancer mortality predictions for the year 2022 with focus on ovarian cancer. Ann Oncol. 2022;33:330–9.
Conroy T, Hammel P, Hebbar M, Ben Abdelghani M, Wei AC, Raoul J-L, et al. FOLFIRINOX or gemcitabine as adjuvant therapy for pancreatic cancer. N Engl J Med. 2018;379:2395–406.
Tempero MA, Pelzer U, O’Reilly EM, Winter J, Oh D-Y, Li C-P, et al. Adjuvant nab-paclitaxel plus gemcitabine in resected pancreatic ductal adenocarcinoma: results from a randomized, open-label, phase III trial. J Clin Oncol. 2023;41:2007–19.
Wainberg ZA, Melisi D, Macarulla T, Cid RP, Chandana SR, De La Fouchardière C, et al. NALIRIFOX versus nab-paclitaxel and gemcitabine in treatment-naïve patients with metastatic pancreatic ductal adenocarcinoma (NAPOLI-3): a randomised, open-label, phase 3 trial. Lancet. 2023;402:1272–81.
Benkhaled S, Peters C, Jullian N, Arsenijevic T, Navez J, Van Gestel D, et al. Combination, modulation and interplay of modern radiotherapy with the tumor microenvironment and targeted therapies in pancreatic cancer: which candidates to boost radiotherapy? Cancers. 2023;15:768.
Hilmi M, Delaye M, Muzzolini M, Nicolle R, Cros J, Hammel P, et al. The immunological landscape in pancreatic ductal adenocarcinoma and overcoming resistance to immunotherapy. Lancet Gastroenterol Hepatol. 2023;8:1129–42.
Bouchart C, Navez J, Closset J, Hendlisz A, Van Gestel D, Moretti L, et al. Novel strategies using modern radiotherapy to improve pancreatic cancer outcomes: toward a new standard? Ther Adv Med Oncol. 2020;12:1758835920936093.
Conroy T, Desseigne F, Ychou M, Bouché O, Guimbaud R, Bécouarn Y, et al. FOLFIRINOX versus gemcitabine for metastatic pancreatic cancer. N Engl J Med. 2011;364:1817–25.
Janssen QP, Buettner S, Suker M, Beumer BR, Addeo P, Bachellier P, et al. Neoadjuvant FOLFIRINOX in patients with borderline resectable pancreatic cancer: a systematic review and patient-level metaanalysis. J Natl Cancer Inst. 2019;111:782–94.
Koerkamp BG, Janssen QP, van Dam JL, Bonsing BA, Bos H, van Hillegersberg R, et al. LBA83 Neoadjuvant chemotherapy with FOLFIRINOX versus neoadjuvant gemcitabine-based chemoradiotherapy for borderline resectable and resectable pancreatic cancer (PREOPANC-2): a multicenter randomized controlled trial. Ann Oncol. 2023;34:S1323.
Ghaneh P, Palmer D, Cicconi S, Jackson R, Halloran CM, Rawcliffe C, et al. Immediate surgery compared with short-course neoadjuvant gemcitabine plus capecitabine, FOLFIRINOX, or chemoradiotherapy in patients with borderline resectable pancreatic cancer (ESPAC-5): a four-arm, multicenter, randomized, phase 2 trial. Lancet Gastroenterol Hepatol. 2023;8:157–68.
Mahadevan A, Moningi S, Grimm J, Li XA, Forster KM, Palta M, et al. Maximizing tumor control and limiting complications with stereotactic body radiation therapy for pancreatic cancer. Int J Radiat Oncol Biol Phys. 2021;110:206–16.
Rudra S, Jiang N, Rosenberg SA, Olsen JR, Roach MC, Wan L, et al. Using adaptive magnetic resonance image-guided radiation therapy for treatment of inoperable pancreatic cancer. Cancer Med. 2019;8:2123–32.
Bouchart C, Engelholm J-L, Closset J, Navez J, Loi P, Gökburun Y, et al. Isotoxic high-dose stereotactic body radiotherapy integrated in a total multimodal neoadjuvant strategy for the treatment of localized pancreatic ductal adenocarcinoma. Ther Adv Med Oncol. 2021;13:17588359211045860.
Chuong MD, Lee P, Low DA, Kim J, Mittauer KE, Bassetti MF, et al. Stereotactic MR-guided on-table adaptive radiation therapy (SMART) for borderline resectable and locally advanced pancreatic cancer: a multi-center open-label phase 2 study. Radiother Oncol. 2024;191:110064.
Bouchart C, Navez J, Borbath I, Geboes K, Vandamme T, Closset J, et al. Preoperative treatment with mFOLFIRINOX (or Gemcitabine/Nab-paclitaxel) ± isotoxic high-dose Stereotactic Body Radiation Therapy (iHD-SBRT) for borderline resectable pancreatic adenocarcinoma (the STEREOPAC trial): study protocol for a randomised comparative multicentre phase II trial. BMC Cancer. 2023;23:1–13.
Oar A, Lee M, Le H, Wilson K, Aiken C, Chantrill L, et al. ACITG MASTERPLAN: a randomised phase II study of modified FOLFIRINOX alone or in combination with stereotactic body radiotherapy for patients with high-risk and locally advanced pancreatic cancer. BMC Cancer. 2021;21:936.
Portales F, Assenat E, Samalin E, Mazard T, Adenis A, Gallet BS, et al. Sequential treatment with gemcitabine/nab-paclitaxel (GA) and FOLFIRINOX (FFX) followed by stereotactic MRI-guided adaptive radiation therapy (SMART) in patients with locally advanced pancreatic cancer (LAPC): GABRINOX-ART phase 2, multicenter trial. J Clin Oncol. 2022;40:TPS4191.
Collisson EA, Sadanandam A, Olson P, Gibb WJ, Truitt M, Gu S, et al. Subtypes of pancreatic ductal adenocarcinoma and their differing responses to therapy. Nat Med. 2011;17:500–3.
Moffitt RA, Marayati R, Flate EL, Volmar KE, Herrera Loeza SG, Hoadley KA, et al. Virtual microdissection identifies distinct tumor- and stroma-specific subtypes of pancreatic ductal adenocarcinoma. Nat Genet. 2015;47:1168–78.
Bailey P, Chang DK, Nones K, Johns AL, Patch A-M, Gingras M-C, et al. Genomic analyses identify molecular subtypes of pancreatic cancer. Nature. 2016;531:47–52.
Puleo F, Nicolle R, Blum Y, Cros J, Marisa L, Demetter P, et al. Stratification of pancreatic ductal adenocarcinomas based on tumor and microenvironment features. Gastroenterology. 2018;155:1999–2013.e3.
Martens S, Lefesvre P, Nicolle R, Biankin AV, Puleo F, Van Laethem J-L, et al. Different shades of pancreatic ductal adenocarcinoma, different paths towards precision therapeutic applications. Ann Oncol. 2019;30:1428–36.
Chan-Seng-Yue M, Kim JC, Wilson GW, Ng K, Figueroa EF, O’Kane GM, et al. Transcription phenotypes of pancreatic cancer driven by genomic events during tumor evolution. Nat Genet. 2020;52:231–40.
Hwang WL, Jagadeesh KA, Guo JA, Hoffman HI, Yadollahpour P, Reeves JW, et al. Single-nucleus and spatial transcriptome profiling of pancreatic cancer identifies multicellular dynamics associated with neoadjuvant treatment. Nat Genet. 2022;54:1178–91.
Nicolle R, Bachet J-B, Harlé A, Iovanna J, Hammel P, Rebours V, et al. Prediction of adjuvant gemcitabine sensitivity in resectable pancreatic adenocarcinoma using the GemPred RNA signature: an ancillary study of the PRODIGE-24/CCTG PA6 clinical trial. J Clin Oncol. 2023;14:JCO2202668.
Thomas PD. The Gene Ontology and the meaning of biological function. Methods Mol Biol. 2017;1446:15–24.
Nicolle R, Blum Y, Duconseil P, Vanbrugghe C, Brandone N, Poizat F, et al. Establishment of a pancreatic adenocarcinoma molecular gradient (PAMG) that predicts the clinical outcome of pancreatic cancer. EBioMedicine. 2020;57:102858.
Hammel P, Huguet F, Van Laethem J-L, Goldstein D, Glimelius B, Artru P, et al. Effect of chemoradiotherapy vs chemotherapy on survival in patients with locally advanced pancreatic cancer controlled after 4 months of gemcitabine with or without erlotinib: the LAP07 randomized clinical trial. JAMA. 2016;315:1844–53.
Fietkau R, Ghadimi M, Grützmann R, Klautke G, Krug D, Liersch T, et al. Randomized phase III trial of induction chemotherapy followed by chemoradiotherapy or chemotherapy alone for nonresectable locally advanced pancreatic cancer: first results of the CONKO-007 trial. J Clin Oncol. 2022;40:S4008. abstract S4008.
Lambert A, Bouche O, Ayav A, Locher C, Lecomte T, Gourgou S, et al. LBA62 Preoperative modified FOLFIRINOX (mFOLFIRINOX) with or without chemoradiation (CRT) in borderline resectable pancreatic cancer (BRPC): results from the randomized phase II trial PANDAS/PRODIGE 44. Ann Oncol. 2024;35:S1252. abstract LBA62.
Katz MHG, Shi Q, Meyers JP, Herman JM, Marsh R, de Willett C, et al. Efficacy of preoperative mFOLFIRINOX vs mFOLFIRINOX plus hypofractionated radiotherapy for borderline resectable adenocarcinoma of the pancreas: the A021501 phase 2 randomized clinical trial. JAMA Oncol. 2022;8:1263–70.
Karasinska JM, Topham JT, Kalloger SE, Jang GH, Denroche RE, Culibrk L, et al. Altered gene expression along the glycolysis–cholesterol synthesis axis is associated with outcome in pancreatic cancer. Clin Cancer Res. 2020;26:135–46.
Elyada E, Bolisetty M, Laise P, Flynn WF, Courtois ET, Burkhart RA, et al. Cross-species single-cell analysis of pancreatic ductal adenocarcinoma reveals antigen-presenting cancer-associated fibroblasts. Cancer Discov. 2019;9:1102–23.
Daemen A, Peterson D, Sahu N, McCord R, Du X, Liu B, et al. Metabolite profiling stratifies pancreatic ductal adenocarcinomas into subtypes with distinct sensitivities to metabolic inhibitors. Proc Natl Acad Sci USA. 2015;112:E4410–E4417.
O’Kane GM, Grünwald BT, Jang G-H, Masoomiam M, Picardo S, Grant RC, et al. GATA6 expression distinguishes classical and basal-like subtypes in advanced pancreatic cancer. Clin Cancer Res. 2020;26:4901–10.
Aung KL, Fischer SE, Denroche RE, Jang G-H, Dodd A, Creighton S, et al. Genomics-driven precision medicine for advanced pancreatic cancer: early results from the COMPASS trial. Clin Cancer Res. 2018;24:1344–54.
Porter RL, Magnus NKC, Thapar V, Morris R, Szabolcs A, Neyaz A, et al. Epithelial to mesenchymal plasticity and differential response to therapies in pancreatic ductal adenocarcinoma. Proc Natl Acad Sci USA. 2019;116:26835–45.
Qiang L, Hoffman MT, Ali LR, Castillo JI, Kageler L, Temesgen A, et al. Transforming growth factor-β blockade in pancreatic cancer enhances sensitivity to combination chemotherapy. Gastroenterology. 2023;165:874–90.
Mills BN, Qiu H, Drage MG, Chen C, Mathew JS, Garrett-Larsen J, et al. Modulation of the human pancreatic ductal adenocarcinoma immune microenvironment by stereotactic body radiotherapy. Clin Cancer Res. 2022;28:150–62.
Perez VM, Kearney JF, Yeh JJ. The PDAC extracellular matrix: a review of the ECM protein composition, tumor cell interaction, and therapeutic strategies. Front Oncol. 2021;11:751311.
Jiang B, Zhou L, Lu J, Wang Y, Liu C, You L, et al. Stroma-targeting therapy in pancreatic cancer: one coin with two sides? Front Oncol. 2020;10:576399.
Torphy, RJ, Wang, Z, True-Yasaki, A, Volmar, KE, Rashid, N, Yeh, B, et al. Stromal content is correlated with tissue site, contrast retention and survival in pancreatic adenocarcinoma. JCO Precis Oncol. 2018. https://doi.org/10.1200/PO.17.00121.
Jiang H, Torphy RJ, Steiger K, Hongo H, Ritchie AJ, Kriegsmann M, et al. Pancreatic ductal adenocarcinoma progression is restrained by stromal matrix. J Clin Investig. 2020;130:4704–9.
Tian C, Clauser KR, Öhlund D, Rickelt S, Huang Y, Gupta M, et al. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc Natl Acad Sci USA. 2019;116:19609–18.
Chen Y, Yang S, Tavormina J, Tampe D, Zeisberg M, Wang H, et al. Oncogenic collagen I homotrimers from cancer cells bind to α3β1 integrin and impact tumor microbiome and immunity to promote pancreatic cancer. Cancer Cell. 2022;40:818–34.
Hu B, Wu C, Mao H, Gu H, Dong H, Yan J, et al. Subpopulations of cancer-associated fibroblasts link the prognosis and metabolic features of pancreatic ductal adenocarcinoma. Ann Transl Med. 2022;10:262.
Krishnamurty AT, Shyer JA, Thai M, Gandham V, Buechler MB, Yang YA, et al. LRRC15+ myofibroblasts dictate the stromal setpoint to suppress tumour immunity. Nature. 2022;611:148–54.
Mucciolo G, Henriques JA, Jihad M, Teles SP, Manansala JS, Ashworth S, et al. EGFR-activated myofibroblasts promote metastasis of pancreatic cancer. Cancer Cell. 2023;19:S1535–6108.
Zhou DC, Jayasinghe RG, Chen S, Herndon JM, Iglesia MD, Navale P, et al. Spatially restricted drivers and transitional cell populations cooperate with the microenvironment in untreated and chemo-resistant pancreatic cancer. Nat Genet. 2022;54:1390–405.
Croft W, Pearce H, Margielewska-Davies S, Lim L, Nicol SM, Zayou F, et al. Spatial determination and prognostic impact of the fibroblast transcriptome in pancreatic ductal adenocarcinoma. ELife. 2023;12:e86125.
Dings MPG, Manoukian P, Waasdorp C, Quik JSE, Stijker M, Lodestijn SC, et al. Serum levels of iCAF-derived osteoglycin predict favorable outcome in pancreatic cancer. Int J Cancer. 2023;152:511–23.
Farren MR, Sayegh L, Ware MB, Chen H-R, Gong J, Liang Y, et al. Immunologic alterations in the pancreatic cancer microenvironment of patients treated with neoadjuvant chemotherapy and radiotherapy. JCI Insight. 2020;5:e130362.
Manderlier M, Navez J, Hein M, Engelholm J-L, Closset J, Bali MA, et al. Isotoxic high-dose stereotactic body radiotherapy (iHD-SBRT) versus conventional chemoradiotherapy for localized pancreatic cancer: a single cancer center evaluation. Cancers. 2022;14:5730.
Andrews S. FastQC: a quality control tool for high throughput sequence data. 2010. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc. Accessed 29 Mar 2024.
Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7.
Soneson C, Love MI, Robinson MD. Differential analyses for RNA-seq: transcript-level estimates improve gene-level inferences. F1000Res. 2015;4:1521 30.
McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–97.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Gu Z, Eils R, Schlesner M. Complex heatmaps reveal patterns and correlations in multidimensional genomic data. Bioinformatics. 2016;32:2847–9.
Korotkevich G, Sukhov V, Sergushichev A. Fast gene set enrichment analysis.BioRxiv, Preprint posted online 01 February 2021, https://doi.org/10.1101/060012 (2021).
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-Seq data. BMC Bioinforma. 2023;14:7.
Sturm G, Finotello F, Petitprez F, Zhang JD, Baumbach J, Fridman WH, et al. Comprehensive evaluation of transcriptome-based cell-type quantification methods for immuno-oncology. Bioinformatics. 2019;35:i436–i445.
Aran D, Hu Z, Butte AJ. xCell: digitally portraying the tissue cellular heterogeneity landscape. Genome Biol. 2017;18:220.
Acknowledgements
The authors want to thank all our collaborators, including in particular Ms Ligia Craciun and the Pathology Labs of Brussels University Hospital, Visual and Spatial Tissue Analysis (Vrije Universiteit Brussel (VUB), Brussels, Belgium; https://vsta.research.vub.be) and La Salpêtrière Hospital.
Funding
The first authors disclosed receipt of the following financial support for the research: ‘Les Amis de l’Institut Bordet/L’Association Jules Bordet’ grant numbers: [2019-31] (CB, DVG, LM) and [2021-03] (CB), ‘Fonds de la Recherche Scientifique—FNRS’ grant numbers [FC 33593] (CB) and [PDR T0011.22] (OAS), ‘Fonds Erasme’ (JN), ‘Fondation Contre le Cancer—FCC’ grant number [F/2020/1402] (VD) and [FAF-C/2018/1203] (IR and JLVL).
Author information
Authors and Affiliations
Contributions
CB: study design and conceptualisation; Clinical data collection; (m)IHC quantification, data analysis, interpretation and related figure design; mIF quantification, data analysis, interpretation and related figure design; RNAseq data analysis, interpretation and related figures design; drafted the manuscript and obtained funding for the study. OAS: RNAseq data analysis, interpretation and related figures design; statistical analysis and drafted the manuscript. JN: provided human samples; Clinical data collection and RNAseq data analysis. LV: provided human samples; (m)IHC experiments, quantification, data analysis, interpretation and related figure design. AB: (m)IHC experiments, quantification, data analysis, interpretation and related figure design. MH: (m)IHC data analysis, interpretation, statistical analysis and related figure design. KS: (m)IHC experiments, RNA isolation, RNAseq data analysis, Graphical abstract and Fig. S10 design. EQ: RNA isolation, RNAseq data analysis. VT: RNA isolation, RNAseq data analysis. SZ: Clinical data collection. LMa: Clinical data collection. ND: provided human samples. DVG: obtained funding for the study and performed editing. LMo: obtained funding for the study and performed editing. EM: mIF experiments, quantification, data analysis, interpretation and related figure design. IR: mIF experiments, quantification, data analysis, interpretation and related figure design and obtained funding for the study. VD: RNAseq data analysis, interpretation and related figures. JBB: provided human samples and clinical data collection. PDM: provided human samples and (m)IHC experiments, quantification, data analysis, interpretation. KWG: (m)IHC experiments, quantification, data analysis, interpretation and related figure design. RN: RNAseq data analysis, interpretation and related figures. TA: study design and conceptualisation; RNA isolation, RNAseq data analysis, interpretation and related figures, drafted the manuscript. JLVL: study design and conceptualisation and obtained funding for the study. All authors performed critical revisions. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Bouchart, C., Azurmendi Senar, O., Navez, J. et al. Favorable histo-molecular remodeling of pancreatic ductal adenocarcinoma after neoadjuvant FOLFIRINOX followed by high-dose stereotactic body radiotherapy. Br J Cancer (2026). https://doi.org/10.1038/s41416-025-03274-0
Received:
Revised:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41416-025-03274-0