Abstract
Cyclic nucleotide-gated ion channels (CNGCs) in plants mediate Ca2+ influx in response to environmental changes. Among numerous plant CNGCs, Medicago truncatula CNGC15a/b/c (MtCNGC15) is localized to the nuclear envelope. The opening and closing cycle of MtCNGC15 is tightly associated with the Ca2+ oscillation in symbiosis. However, the molecular mechanism underlying MtCNGC15 activity regulation remains unclear. In this study, we present the structures of MtCNGC15 in its apo form and in the presence of CaM. The apo MtCNGC15b exhibits a flexible cytoplasmic domain (CPD), whereas binding of the MtCaM inhibits Ca2+ currents and stabilizes the highly dynamic CPD. Furthermore, the activity of MtCNGC15b seems to be independent of cGMP. The hypothetical binding pocket for cGMP is occupied by an arginine residue. These findings elucidate the structural basis for the activity regulation of nuclear localized MtCNGC15.
Similar content being viewed by others
Introduction
In plant cells, intracellular Ca2+ levels fluctuate rapidly in response to environmental stresses. The basal concentration of intracellular Ca2+ is approximately 100 nM, whereas the extracellular or nuclear envelop Ca2+ can reach to the millimolar level1. Activation of calcium channels or transporters is an prerequisite event to change the cellular Ca2+ concentration when plants sense different stimulus2. For instance, the binding of Nod factors to receptor kinases localized on the root cell membrane will trigger downstream Ca2+ changes for nodulation and symbiosis. A conserved sustained and rhythmic changes in nuclear Ca2+ concentration, termed as Ca2+ oscillations, plays a central role in regulating the transcription of genes required for nodulation and symbiosis3,4. Depiction of the key components in governing such process led to the identification of nuclear membrane specifically localized Ca2+ channels CNGC15a/b/c in Medicago truncatula5. The MtCNGC15 was reported to be activated after the sensing of Nod factors and negatively regulated by camodulin (CaM) to fullfil a cycle of Ca2+ release and retrieve between the endoplasmic reticulum/nuclear envelop lumen and the nucleoplasm6. In addition, other Ca2+ transporters, such as the Ca2+ ATPase MCA8, further facilitate the Ca2+ recycling1,2.
The MtCNGC15a/b/c belongs to the superfamily of CNG ion channels, which are widely distributed from prokaryotic to eukaryotic species7. These channels mediate the permeation of various cations, including Ca2+, Na+ and K+. In animals, CNG ion channels assemble either as homo-tetramers such as TAX-4 in Caenorhabditis elegans, CNGA1 in human, or hetero-tetramers composed of different subunits in cone and rod cells8,9. In Arabidopsis, 20 homologs of CNG ion channels (CNGC1-20) participate in distinct signaling pathways10. Among the CNGCs identified in M. truncatula, CNGC15a/b/c is specifically localized to nuclear membrane. However, some remaining questions restrict the rational regulation of the Ca2+ oscillations. First, how the structure of MtCNGC15 changes upon CaM binding needs to be elucidated. Second, whether MtCNGC15 can be gated by cyclic nucleotide monophosphate (cNMP) is not examined before. Although MtCNGC15 has a probable cNMP binding domain (CNBD) and the Arabidopsis CNGCs have been reported to be affected by cNMP11,12, structural evidence remains lacking.
Here, we report the structures of MtCNGC15b in the absence or presence of CaM. Binding of CaM may stabilize the conformation of MtCNGC15b cytoplasmic domain to modulate the channel open and close cycle. Different from the several reported animal CNG channels, we prove that MtCNGC15b is not gated by cNMP. Instead, the Arg553 of MtCNGC15b occupies the corresponding cNMP pocket in animal homologs. These findings provide structural and functional insight into MtCNGCs activity regulation in symbiosis.
Results
Architecture of MtCNGC15b
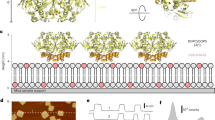
To explicate the structure of MtCNGC15, the expression level and cryo-EM performance of three isoforms MtCNGC15a/b/c were individually examined using insect cells and mammalian cells. In spite of high sequence similarities, the MtCNGC15b exhibits relatively higher expression level and better protein purity in mammalian expression system. Since MtCNGC15b is localized to the nuclear membrane, we constructed the hemagglutinin (HA) signal peptide (MKTIIALSYIFCLVFA) to the N-terminus of MtCNGC15b to aid their localization to the cell membrane13. In the two-electrode voltage clamp assay, MtCNGC15b demonstrated robust Ca²+ currents (Fig. 1a). In the following cryo-EM analysis, 2D classification indicated that the cytoplasmic domains (CPDs) of MtCNGC15b are highly flexible (Supplementary Fig. S1a; Fig. 1b).
a TEVC recording from Xenopus oocytes expressing MtCNGC15b. In Ca2+-containing bath solution buffer, 500 μM DIDS was added to inhibit the activity of the Ca2+-activated chloride channel. b Density map of MtCNGC15b with the clear transmembrane domain and the flexible cytoplasmic domain. c Architecture of the transmembrane region of MtCNGC15b channel. d Pore radius of MtCNGC15b calculated by MOLE39.
The transmembrane domains (TM) and CPD of the MtCNGC15b show contrasting features in the density map. The well-resolved EM density of TMs enabled the unambiguous assignment of residues 58-385 from MtCNGC15b (Fig. 1c; Supplementary Fig. S1b and Table S1). The TMs of MtCNGC15b channel is ~76 Å in height and ~105 Å in width when viewed perpendicular to the membrane neighboring subunits (Fig. 1b). Similar to other CNG channels, the transmembrane domains of MtCNGC15b can be divided into the voltage sensing domain (VSD, S1–S4), the pore domain (PD, S5–S6), and the selectivity filter (SF) (Fig. 1c, d). The ion conduction pathway of MtCNGC15b consists of the selectivity filter (SF) in pore loops and the inner gate formed by S6 (Fig. 1d).
The selectivity filter of MtCNGC15b for recognition of Ca2+
The SF of MtCNGC15b comprises residues Ser344-Gln347 (Fig. 2). The side chains of Gln347 restrict the permeation path, differing from a glutamic acid in reported animal CNG ion channels like TAX-4 and human CNGA1 (Fig. 2a). Sequence alignment with three other CNGCs from Arabidopsis, Oryza sativa, and Zea mays reveals the conservation of Gln residue in plant CNG channels (Fig. 2b; Supplementary Fig. S2). Substitution of Gln347 to Glu abolished the Ca2+ currents, suggesting the indispensable role of Gln347 in Ca2+ recognition (Fig. 2c). By contrast, both TAX-4 and human CNGA1 have relatively broad range of ion selectivity8,9. To investigate the ion conductivity of MtCNGC15b and the Q347E variant towards Na+ and K+, we detected the reversal potentials and calculated the ion permeability ratios (Supplementary Table S2)14. Both MtCNGC15b and Q347E are localized on the cell membrane and exhibit low conductivity of K+ (Supplementary Fig. S3 and Table S2). Based on the ion permeability ratios, MtCNGC15b WT exhibits nearly equal permeability to calcium and sodium ions (PCa/PNa: ~1.03) but demonstrates higher selectivity for Ca2+ and Na+ over K+ (PK/PCa: ~0.183, PNa/PK: ~9.44). By contrast, the PCa/PNa reduced to ~0.164 for the Q347E mutation, indicating a qualitative change of Ca2+ selectivity. Although the mutation of Q347E exhibits low ion conductivity towards all the three ions, its permeation ration reflects higher Na+ selectivity over Ca2+ and K+ (Supplementary Table S2).
a Comparison of S6 and SF of MtCNGC15b to TAX-4 (PDB: 6WEJ) and human CNGA1 (PDB: 7LFT)8,9. b Sequence alignment of the selectivity filter between representative plant and animal CNG channels. The Gln residue displays high conservation in plant CNGCs. c Substitution of Gln347 to Glu abrogates the MtCNGC15b-mediated Ca2+ currents.
Different from animal CNG channels, three pairs of disulfide bonds — Cys99/Cys281, Cys262/Cys302, and Cys267/Cys274 on MtCNGC15b is mapped to the nuclear envelop lumen side (Fig. 3a, b). Notably, the Cys99/Cys281 pair forms a bridge between the VSD and the PD of MtCNGC15b. Such structural element is conserved among plant CNG channels but not TAX-4 or human CNGA1 (Fig. 3c). Given that the lumenal environment is likely oxidative, these disulfide bonds may play a physiological role in regulating the coupling between the VSD and PD. Mutation of Cys99 and Cys281 to Ser drastically reduced Ca2+ currents without altering the localization of MtCNGC15b in the plasma membrane, while mutations in the other two cysteine pairs also led to compromised activity (Fig. 3d; Supplementary Fig. S3).
a Structural differences between MtCNGC15b with human CNGA1 or C. elegans TAX-4. b Three pairs of disulfide bonds are located at the nuclear envelop lumen side. The Cys99-Cys281 mediated the interaction between VSD and PD. c Sequence and structural comparison of the extracellular cysteine pairs in representative plant CNGCs and animal CNG channels. The Cys99-Cys281 mediated-disulfide bond is specifically observed in plant CNGCs. d Disruption of the identified three disulfide bonds decreases the Ca2+ currents. Particularly, mutations of the Cys99-Cys281 pair nearly abolish the channel activity. Current–voltage relationship of MtCNGC15b and its variants currents recorded from Xenopus oocytes. Command voltages were applied in 10-mV steps between 0 mV and –160 mV. Data are presented as means ± SEM.
MtCaM negatively regulates MtCNGC15b activity by modulating CPD conformation
MtCaM has been reported to negatively regulate the activity of MtCNGC15b (Fig. 4a), but how MtCaM binding affects the CPD of MtCNGC15b remains to be elucidated. To unravel the probable molecular changes, we tried to co-express MtCaM and MtCNGC15b for cryo-EM structural study. However, the MtCaM was not stably co-purified with MtCNGC15b. We then fused the MtCaM to the C-terminus of MtCNGC15b through a short linker. The fused complex exhibited good solution behavior and dispersed evenly on cryo-EM samples. Although the cryo-EM densities of MtCaM itself were invisible, we can observe the density of the MtCNGC15b CPD (Fig. 4b; Supplementary Fig. S3). Consistent with the previous findings on the inhibitory effect on channel activity15, MtCNGC15b exhibits closed inner gate (Fig. 4b, right panel). To evaluate the impact of MtCaM, we have employed AlphaFold 316 to predict the potential binding site of MtCaM and analyzed the residues at the interaction interface (Supplementary Fig. S5a). To validate these probable binding site and its regulatory effects, we introduced mutations on MtCNGC15b and recorded the currents mediated by these variants (Supplementary Fig. S5b). Compared to wild-type MtCNGC15b, point mutations compromised the inhibitory effect of MtCaM, leading to weak recovery of currents.
a TEVC recording from Xenopus oocytes expressing MtCNGC15b and MtCaM. In Ca2+-containing bath solution buffer, 500 μM DIDS was added to inhibit the activity of the Ca2+-activated chloride channel. Command voltages were applied in 10-mV steps between 0 mV and –160 mV. The I–V curves were obtained from the peak currents at command voltages. Data are presented as means ± SEM. b The CPD of MtCNGC15b can be stabilized by CaM. The inner gate region of MtCNGC15b is regulated upon involvement of CaM. Pore radius is calculated by MOLE39.
The putative cNMP binding pocket of MtCNGC15b is occupied by an arginine
The CNG channels are widely reported to be gated by cGMP or cAMP in animals. Previous studies on plant CNGCs suggest that cyclic nucleotides (cNMPs) influence CNGC activity, which seems plausible given that plant CNGCs possess a predicted cyclic nucleotide-binding domain (CNBD) in their structures17. Based on these assumptions, we sought to validate whether cGMP binds to MtCNGC15b. First, we assessed the binding affinity of cGMP/cAMP to MtCNGC15b through surface plasmon resonance. The results show that MtCNGC15b has negligible binding capacity for cGMP/cAMP (Supplementary Fig. S6). Second, cGMP treatment has no impact on Ca2+ currents (Fig. 5a). To further validate such observations, we supplemented 5 mM cGMP to MtCNGC15b when preparing cryo-EM sample as described12. However, addition of cGMP reveals no structural changes. We observed flexible CPDs after 2D classification and even identical conformation of the transmembrane domains (Fig. 5b). Taken together, cNMP is not the ligand of MtCNGC15b.
a TEVC recording from Xenopus oocytes expressing MtCNGC15b and add 5 mM 3′5′ cGMP. In Ca2+-containing bath solution buffer, 500 μM DIDS was added to inhibit the activity of the Ca2+-activated chloride channel. The I–V curves were obtained from the peak currents at command voltages. The currents had no significant difference in the absence or presence of cGMP (inset panel). Current amplitudes at –140 mV from multiple recordings. Data are presented as means ± SEM, n = 3 biologically independent oocytes. b Structrual superimposition of MtCNGC15b in the presence of 3′5′ cGMP with the apo state. Addition of 3′5′ cGMP results in an identical density map with highly flexible CPD.
We then superimposed the MtCNGC15b CNBD with human CNGA1 bound with cGMP. In human CNGA1, the cGMP phosphor group is nestled into a positively charged pocket and coordinated by the side chain of Arg561. This arginine is absence and aligned to a serine residue in MtCNGC15b. Moreover, the putative cGMP binding pocket in MtCNGC15b is negatively charged (Fig. 6a). Instead of accommodating cGMP, the side chain of Arg553 in MtCNGC15b occupies the hypothetical pocket. The basic nature of Arg553 is compatible with the negatively charged pocket in MtCNGC15b (Fig. 6a, b). Sequence alignment reveals the cNMP-binding arginine is conserved in representative animal cNMP-gated channels, whereas in plant CNGCs, an arginine residues occupies this position (Fig. 6b; Supplementary Fig. S7). Substituting Arg553 by several kinds of residues decreased the Ca2+ influx in varing degrees, while replacement of the arginine residue with alanine nearly abolished Ca2+ currents. The results showed that the R553L and R553W mutations did not significantly reduce ion channel activity, suggesting that changes in amino acid polarity have a limited impact. Instead, side chain size appears to play a more critical role in regulating channel function (Fig. 6c). Such structural element reminiscent of that the activity regulation by intrinsic ligand in CNBD-like domains of a zebrafish channel, zKCNH. In zKCNH, a C-terminal short β-strand occupied the presumable cNMP binding pocket18.
a Structural alignment between the CNBD of MtCNGC15b and the cGMP-bound human CNGA1. The corresponding region of the cGMP binding site in MtCNGC15b is negatively charged in comparison with human CNGA1. b The basic side chain of Arg553 is compatible with the negatively charged pocket of MtCNGC15b composed by Glu533 and Glu534. The Arg553 is conserved in Medicago but not representative animal CNG channels. c Mutagenesis of Arg553 to different amino acids decreases the Ca2+ currents. Current-voltage relationship of MtCNGC15b and its variants currents recorded from Xenopus oocytes. Command voltages were applied in 10-mV steps between 0 mV and –160 mV. Data are presented as means ± SEM. d Sequence and structure clustering results of 726 sequences that share over 80% similarity with CNG channels from C. elegans, H. sapiens, and M. truncatula. The representative predicted structures are aligned to MtCNGC15b with the entries number listed.
To investigate potential evolutionary differences associated with the presence or absence of this arginine residue, we employed the ESM2-150M model to generate high-dimensional embeddings for 726 sequences derived from BLAST results in the UniProt database, using C. elegans, H. sapiens, and M. truncatula CNG channels as templates for sequences with high similarity. Among them, we utilized 505 available structures from the AlphaFold database to calculate pairwise structural similarities19. Both sequence and structure clustering results suggest that M. truncatula has followed a distinct evolutionary trajectory, potentially as an adaptation to environmental stress (Fig. 6d). As the potential intrinsic ligand observed in MtCNGC15b is conserved across all aligned plants CNGCs except for Arabidopsis thaliana AtCNGC2 and AtCNGC4, we postulate that the plants CNGCs are evolved differently from animal ones.
Comparative analysis of CNBD structures reveals consistent residues in the local environment of R553 in M. truncatula, characterized by one arginine and two glutamates at equivalent positions of R553, E533 and E534. In contrast, while the overall conformations of the CNBD domains in C. elegans and H. sapiens resemble those in M. truncatula, the local interactions between arginine and glutamates are not preserved. This discrepancy may suggest that, in an immobile state, plants need to develop mechanisms for rapid responses to environmental stress. One such mechanism could involve the “ready-to-open” of certain ion channels through occupying the “presumably ligand pocket” by this arginine but not cNMP.
MtCaM binding may regulate the local environment near the arginine
To gain insights into whether involvement of MtCaM has influence on the regulation of this arginine residue, we conducted molecular dynamics (MD) simulation of the CPD of MtCNGC15b in the absence or presence of MtCaM. MtCaM binding significantly stabilizes the cytosolic domain of MtCNGC15b, as evidenced by a lower RMSD and confirmed by the structural observation (Fig. 7a). Upon MtCaM binding, the local environment around Glu533 changes compared to its state without MtCaM. At pH 7.0, Glu533 becomes protonated in the presence of MtCaM, whereas it remains negatively charged without MtCaM (Fig. 7b). This protonation reduces the interaction between Glu533, Glu534, and Arg553 (Fig. 7c). The solvent-accessible surface area (SASA) of Arg553 and Glu533 further suggests that CaM binding disrupts their interaction, making these residues more solvent-exposed (Fig. 7d). Notably, Ser509 persistently interacts with R553 regardless the presence of CaM (Fig. 7c–e). To further validate these computational data, we performed electrophysiological experiments on E533Q and S509A. Within expectation, both mutations severely compromised the Ca2+ currents, indicating their important role in regulating the channel activity (Supplementary Fig. S8).
a RMSD of MtCNGC15b after least square fit to backbone throughout the last 500 ns of three repeated simulations. b Calculated titration curve of E533 in MtCNGC15b with or without CaM binding. Predicted pKa’s are indicated. c Three residues with the highest hydrogen bonding occupancy with R553 in each chain throughout the last 500 ns of three repeated simulations in the absence or presence of CaM. d Distribution of solvent accessible surface area of R553 with or without MtCaM binding to MtCNGC15b throughout the last 500 ns of three repeated simulations. e Local environment of Arg553.
Discussion
During the symbiosis, MtCNGC15b is considered as a Ca2+ channel for Ca2+ oscillation and regulated by camodulin. The current observation that MtCNGC15b has an intrinsic-ligand-like arginine residue indicates that MtCNGC15b channels have evolved to function without cNMP regulation. The presence of Arg553 and other unique residues reflects a natural result of their evolutionary trajectory, rather than a targeted adaptation to substitute for cNMP regulation. Such hypothesis might be true in other plants CNGCs as well because there is no adenylate cyclase homologs identified in plants20. Our structural and functional evaluations on MtCNGC15b are in agreement with the very overlapping findings on Arabidopsis CNGC1/521, including the Ca2+ selectivity determination, the extracellular/luminal side disulfide bonds and the putative intrinsic ligand.
In addition, the CNBD domain exists widely in plant membrane proteins, exemplified by K+ channel AKT1 and Na+ exchanger SOS122,23. These ion channel or transporters are regulated by phosphorylation, with most posttranslational modification sites located on the cytoplasmic domains. No ligands have been identified to bind the CNBD of AKT1 and SOS1. Similar to MtCNGC15b, AtCNGC1/5 and zebrafish KCNH, both AKT1 and SOS1 may exhibit an intrinsic-ligand-like mode of regulation (Supplementary Fig. S9). Such hypothesis is further supported by recent studies on the intrinsic ligand-like residues of Arabidopsis potassium channel GORK21,24,25.
Based on the structural observation and electrophysiological analysis, MtCaM may close the channel through stabilizing the CPD of MtCNGC15b and modulating the local conformation of residues near the arginine. These findings support the previously reported modulatory effect of the engineered mutant MtCaMR91A. This mutation exhibits considerable flexibility, which may regulate the on-off rate of MtCaM and thereby enhance the frequency of calcium oscillations6.
In conclusion, we report a distinct regulatory mode for the nuclear the nuclear membrane-localized MtCNGC15b. We propose a regulation mode for MtCNGC15b involving an intrinsic-ligand-like residue that occupies the pocket for cNMP. Such mechanism may sensitilize the channel in a “ready-to-open” state for the rapid response and Ca2+ oscillation in symbiosis. In other plant CNGCs, calmodulin binding has been shown to negatively regulate channel activity as well. Such negative feedback mode could desensitilize the channel to avoid hyper-responses. Our results lay the molecular basis for the structural-functional relationship of this channel and pave the way for further mechanistic studies on the functional regulation of plant CNGC channels.
Materials and methods
Purification of M. truncatula CNGC15b and CNGC15b-CaM
Full-length coding sequences of M. truncatula CNGC15b (Uniprot ID: G7JND3) were synthesized and subcloned into a pCAG vector with a C-terminal Flag-TwinStrep tag between KpnI and XhoI restriction sites, respectively. The recombinant CNGC15b was expressed using mammalian cell (HEK293F) expression system. Briefly, a mixture of 1.5 mg CNGC15b encoding plasmids and 4 mg polyethylenimine (Polysciences) was added into 1 L HEK293F cells at a density of ~2.5 × 106 cells mL–1. After transfection for 60 h under 37 °C with 5% CO2, cells were collected and homogenized in lysis buffer containing 25 mM HEPES pH 7.4, 150 mM NaCl supplemented with protease inhibitor cocktails consisting of 1 mM phenylmethylsulfonyl fluoride (PMSF), 2.6 μg mL−1 aprotinin, 2 μg mL−1 pepstatin and 4 μg mL−1 leupeptin.
Then cell membranes were solubilized with 1% (w/v) decyl maltose neopentyl glycol (DMNG, Anatrace) and 0.1% (w/v) cholesteryl hemisuccinate tris salt (CHS, Anatrace) at 4 °C for 2 h with gently agitation. After centrifugation at 45,000 rpm for 30 min, the supernatant was applied to anti-Flag M2 affinity gel (Sigma) through gravity column (Bio-rad). The affinity gel was rinsed with wash buffer containing 25 mM HEPES pH 7.4, 150 mM NaCl, 0.02% (w/v) GDN supplemented with protease inhibitor cocktails consisting of 1.3 μg mL−1 aprotinin, 1 μg mL−1 pepstatin and 2 μg mL−1 leupeptin for three times. Protein was eluted with wash buffer plus 0.2 mg mL−1 flag peptide (DYKDDDDK, synthesized by GL Biochem). Then the eluent was loaded onto Strep-Tactin (Smart-lifesciences) affinity column. After being rinsed with wash buffer containing 25 mM Tris pH 8.0, 150 mM NaCl, 0.02% (w/v) GDN, target protein was eluted with wash buffer plus 2.5 mM desthiobiotin. Then eluted protein was immediately concentrated with a 100 kDa cut-off concentrator (Millipore) and further purified by size-exclusion chromatography (SEC) using Superose 6 Increase 10/300 GL column (GE Healthcare) with SEC buffer containing 25 mM Tris pH 8.0, 150 mM NaCl, 0.006% (w/v) GDN. Peak fractions were pooled together and concentrated to ~10 mg mL–1 for cryo-EM sample preparation. The MtCNGC15b that fused MtCaM2 (Medtr5g088320) by a -GGS-x6 linker was purified following the same protocol.
Cryo-EM sample preparation and data acquisition
Quantifoil 300 mesh R1.2/1.3 Au grids were glow discharged. Four microliters aliquots were applied to grids with Vitrobot Mark IV (Thermo Fisher Scientific) at 8 °C and 100% humidity. After blotted for 3 s, grids were flash-frozen in liquid ethane and transferred to liquid nitrogen for storage. The samples were screened on 200 kV Tecnai Arctica microscope, and grids with high quality were used for data acquisition. High-resolution data was collected on 300 kV Titan Krios microscope equipped with K3 Summit electron detector (Gatan) and a GIF Quantum energy filter (slit width 20 eV, Gatan) at super-resolution counting mode. A series of defocus values from –1.3 to –1.8 μm was used during data collection. Each image was dose-fractionated to 32 frames with a total electron dose of about 50 e−Å−2 and a total exposure time of 2.56 s. EPU software (Thermo Fisher) was used for data collection.
Data processing
For the cryo-EM analysis of MtCNGC15b, MtCNGC15b-CaM, movie stacks have been collected and processed following the tutorial of CryoSPARC26. The average resolution of MtCNGC15b or MtCNGC15b-CaM is 3.1 Å or 3.6 Å on the basis of the Fourier shell correlation (FSC) 0.143 criterion. The FSC curves were corrected for the effects of a soft mask using high-resolution noise substitution. Local resolution variations were estimated using CryoSPARC26.
Model building, refinement and validation
The models were manually adjusted based on the predicted model from AlphaFold2 database19,27. The structure was then refined in real space using PHENIX with secondary structure and geometry restraints28. The atomic model was manually improved using COOT29. The final atomic models were refined in real space using PHENIX. The final atomic model was evaluated using MolProbity30.
Two-electrode voltage clamping recording from Xenopus oocytes
The cDNAs for MtCNGC15b, MtCNGC15b variants and MtCaM2 were cloned into the pGEMHE oocyte expression vector. Since MtCNGC15b is localized to the nuclear membrane, we fused the hemagglutinin (HA) signal peptide (MKTIIALSYIFCLVFA) to the N-terminus of all MtCNGC15b and its variants to aid their localization to the cell membrane13. For making HA-MtCNGC15b-EGFP fusion, the EGFP tag was amplified by PCR and inserted in-frame to the C terminus of the MtCNGC15b into pGEMHE-HA-MtCNGC15b. Transcription and capped RNAs (cRNAs) were synthesized from 2 μg of linearized plasmid DNA template using the T7 High Yield RNA Transcription Kit and Vaccinia Capping Enzyme Kit (Vazyme), according to the manufacturer’s recommendations. The quality of the cRNA was checked by agarose gel electrophoresis. 50 ng of each cRNA was injected into each oocyte. For recording of CNGC15b + CaM2, a 50 ng-mixture of two cRNAs (25 ng for CNGC15b and 25 ng for CaM2) was injected into one oocyte. Injected oocytes were incubated in ND96 solution (96 mM NaCl, 2 mM KCl, 1 mM MgCl2, 1.8 mM CaCl2, 10 mM HEPES/NaOH, pH 7.4) supplemented with 0.1 mg/mL gentamycin and streptomycin at 18 °C for 2 days before electrophysiological recording. Oocytes were voltage-clamped using a Axoclamp 900 A amplifier (Axon Instruments) and monitored by computer through Digidata 1440 A converter (Axon CNS) and Clampex 11.2 software (Axon Instruments). The pipette solution contained 3 M KCl. The oocytes were continuously perfused during the voltage-clamp experiment. The standard bath solution contained 5 mM CaCl2, 2 mM KCl, 1 mM MgCl2 and 10 mM MES/Tris (pH 5.6). Moreover, 500 μM DIDS was added to inhibit the activity of the Ca2+-activated chloride channel. The osmolalities of all solutions were adjusted to 220 mOsmol kg–1 using mannitol. Voltage steps were applied from +40 to –200 mV in –10/–20 mV decrements during 0.8 s, from a holding potential of –60 mV. Each step begins with 0.03 s and ends with 1.4 s at the resting potential of the oocyte membrane in the tested bath solution. For recording of CNGC15b + 3′5′ cGMP, 50 nL of 5 mM 3′5′ cGMP was injected into one oocyte which has already been injected with CNGC15b cRNA 2 days ago, and then incubated in ND96 solution at 18 °C for 1 h before TEVC experiment.
Sequence-based clustering and structure-based clustering
The ESM2-150M model was utilized to generate high-dimensional representations of each original sequence. Subsequently, we employ t-SNE method to reduce these representations to two dimensions for visualization. Structural data for the sequences were obtained from AlphaFold database, yielding structures for 505 of the 726 original sequences. We calculated pairwise structural similarity using the tmscoring package, determining the TM-score between structures and defining the structural distance as (1 − TM-score). Through this approach we generated a 505 × 505-dimensional distance matrix. The t-SNE method was applied to reduce these structural representations to two dimensions for visualization.
Molecular dynamics simulation
MtCNGC15b structures with or without calmodulin were modeled with AlphaFold3. Considering the region of interest, the cytosolic domain of MtCNGC15b (T387-C620) was retained. The systems were solvated in 0.15 M NaCl solution. GROMACS31 version 2024.2 with the CHARMM36 force field32 and TIP3P water model33 were used to perform the simulations. During NVT, NPT equilibrations, position restraints on protein heavy atoms were applied with a force constant of 1000 kJ/mol nm2. To minimize the impact of the absence of the transmembrane domain, a position restraint with force constant of 1000 kJ/mol nm2 was applied to the backbone atoms of the C-linter region (T387-T428) throughout the entire simulations. Three repeats of 700 ns atomistic production simulations were then performed for each system.
The long-range electrostatics (< 1 nm) was modeled using the Particle Mesh Ewald (PME) method34. The first step of equilibration was conducted under an NVT ensemble for 125 ps with integration time step of 1 fs, followed by an NPT equilibration phase with the same integration time step for 125 ps. Berendsen thermostat35 with a reference temperature of 301.15 K was used for temperature coupling during the entire equilibration phase. Berendsen barostat35 with a reference pressure of 1 bat and a compressibility of 4.5 × 10−5/bar was applied for pressure control during NPT equilibration. For production runs, v-rescale thermostat36 and Parrinello-Rahman barostat37 were used for temperature and pressure coupling, respectively. Covalent bonds are constrained to their equilibrium length by the LINCS algorithm38 for all simulations.
Radius of gyration (ROG) was calculated using MDAnalysis (https://docs.mdanalysis.org/2.7.0/index.html). Since the tetrameric structure was aligned with respect to the Z-axis, the ROG essentially describes the average distance between each site to the center of the protein in X–Y plane. Based on the equilibrium MD simulations of CNGC with and without calmodulin (between 200 and 700 ns), we averaged ROG over three MD trajectories, and then computed the change of average ROG (n) from without to with calmodulin to quantify the calmodulin-induced expansion/contraction at residue position n.
Data availability
The cryo-EM maps of the whole protein and the transmembrane domain of MtCNGC15b and the MtCNGC15b-CaM complex have been deposited in the Electron Microscopy Data Bank (EMDB) with the accession code EMD-62261, EMD-62262. The atomic coordinates for the corresponding model have been deposited in the Protein Data Bank (PDB) under the accession code 9KCU, 9KCV, respectively.
Materials availability
Requests for materials should be addressed to G.Y. (guanghuiyang@cau.edu.cn).
References
Capoen, W. et al. Nuclear membranes control symbiotic calcium signaling of legumes. Proc. Natl. Acad. Sci. USA 108, 14348–14353 (2011).
Tian, W., Wang, C., Gao, Q., Li, L. & Luan, S. Calcium spikes, waves and oscillations in plant development and biotic interactions. Nat. Plants 6, 750–759 (2020).
Chiasson, D. M. et al. A quantitative hypermorphic CNGC allele confers ectopic calcium flux and impairs cellular development. Elife 6, e25012 (2017).
Martin, F. M., Uroz, S. & Barker, D. G. Ancestral alliances: plant mutualistic symbioses with fungi and bacteria. Science 356, eaad4501 (2017).
Charpentier, M. et al. Nuclear-localized cyclic nucleotide–gated channels mediate symbiotic calcium oscillations. Science 352, 1102–1105 (2016).
del Cerro, P. et al. Engineered CaM2 modulates nuclear calcium oscillation and enhances legume root nodule symbiosis. Proc. Natl. Acad. Sci. USA 119, e2200099119 (2022).
Talke, I. N., Blaudez, D., Maathuis, F. J. M. & Sanders, D. CNGCs: prime targets of plant cyclic nucleotide signalling? Trends Plant Sci. 8, 286–293 (2003).
Zheng, X. et al. Mechanism of ligand activation of a eukaryotic cyclic nucleotide−gated channel. Nat. Struct. Mol. Biol. 27, 625–634 (2020).
Xue, J., Han, Y., Zeng, W., Wang, Y. & Jiang, Y. Structural mechanisms of gating and selectivity of human rod CNGA1 channel. Neuron 109, 1302–1313.e4 (2021).
Jarratt-Barnham, E., Wang, L., Ning, Y. & Davies, J. M. The complex story of plant cyclic nucleotide-gated channels. Int J. Mol. Sci. 22, 874 (2021).
Balagué, C. et al. HLM1, an essential signaling component in the hypersensitive response, is a member of the cyclic nucleotide–gated channel ion channel family. Plant Cell 15, 365–379 (2003).
Leng, Q., Mercier, R. W., Hua, B. G., Fromm, H. & Berkowitz, G. A. Electrophysiological analysis of cloned cyclic nucleotide-gated ion channels. Plant Physiol. 128, 400–410 (2002).
Guan, X. M., Kobilka, T. S. & Kobilka, B. K. Enhancement of membrane insertion and function in a type IIIb membrane protein following introduction of a cleavable signal peptide. J. Biol. Chem. 267, 21995–21998 (1992).
Kim, S. et al. Ca(2+)-regulated Ca(2+) channels with an RCK gating ring control plant symbiotic associations. Nat. Commun. 10, 3703 (2019).
Tian, W. et al. A calmodulin-gated calcium channel links pathogen patterns to plant immunity. Nature 572, 131–135 (2019).
Abramson, J. et al. Accurate structure prediction of biomolecular interactions with AlphaFold 3. Nature 630, 493–500 (2024).
Dietrich, P., Moeder, W. & Yoshioka, K. Plant cyclic nucleotide-gated channels: new insights on their functions and regulation. Plant Physiol. 184, 27–38 (2020).
Brelidze, T. I., Carlson, A. E., Sankaran, B. & Zagotta, W. N. Structure of the carboxy-terminal region of a KCNH channel. Nature 481, 530–533 (2012).
Jumper, J. et al. Highly accurate protein structure prediction with AlphaFold. Nature 596, 583–589 (2021).
Wong, A. et al. Plant adenylate cyclases have come full circle. Nat. Plants 9, 1389–1397 (2023).
Wang, J. et al. Cryo-EM structures of Arabidopsis CNGC1 and CNGC5 reveal molecular mechanisms underlying gating and calcium selectivity. Nat. Plants 11, 632–642 (2025).
Lu, Y. et al. Structural basis for the activity regulation of a potassium channel AKT1 from Arabidopsis. Nat. Commun. 13, 5682 (2022).
Zhang, Y. et al. Structural basis for the activity regulation of Salt Overly Sensitive 1 in Arabidopsis salt tolerance. Nat. Plants 9, 1915–1923 (2023).
Li, Q.-y. et al. Structural and mechanistic insights into symmetry conversion in plant GORK K+ channel regulation. bioRxiv https://doi.org/10.1101/2024.12.17.628833 (2024).
Zhang, X. et al. GORK K(+) channel structure and gating vital to informing stomatal engineering. Nat. Commun. 16, 1961 (2025).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Varadi, M. et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Res. 50, D439–d444 (2022).
Adams, P. D. et al. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D. Biol. Crystallogr. 58, 1948–1954 (2002).
Emsley, P. & Cowtan, K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 60, 2126–2132 (2004).
Chen, V. B. et al. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D. Biol. Crystallogr. 66, 12–21 (2010).
Abraham, M. J. et al. GROMACS: High performance molecular simulations through multi-level parallelism from laptops to supercomputers. SoftwareX 1-2, 19–25 (2015).
Best, R. B. et al. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone ϕ, ψ and side-chain χ1 and χ2 dihedral angles. J. Chem. Theory Comput. 8, 3257–3273 (2012).
Jorgensen, W. L., Chandrasekhar, J., Madura, J. D., Impey, R. W. & Klein, M. L. Comparison of simple potential functions for simulating liquid water. J. Chem. Phys. 79, 926–935 (1983).
Essmann, U. et al. A smooth particle mesh Ewald method. J. Chem. Phys. 103, 8577–8593 (1995).
Berendsen, H. J. C., Postma, J. P. M., van Gunsteren, W. F., DiNola, A. & Haak, J. R. Molecular dynamics with coupling to an external bath. J. Chem. Phys. 81, 3684–3690 (1984).
Bussi, G., Donadio, D. & Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 126, 014101 (2007).
Parrinello, M. & Rahman, A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 52, 7182–7190 (1981).
Hess, B., Bekker, H., Berendsen, H. J. C. & Fraaije, J. G. E. M. LINCS: A linear constraint solver for molecular simulations. J. Comput. Chem. 18, 1463–1472 (1997).
Pravda, L. et al. MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update). Nucleic Acids Res. 46, W368–w373 (2018).
Acknowledgements
We thank the SM10 Cryo-EM Facility at the Institute of Physics, Chinese Academy of Science and Cryo-EM Facility at Peking University for the support in cryo-EM data collection. This work was supported by the National Natural Science Foundation of China (32422038 to G.Y.); the National Key R&D Program of China (2022YFA1303400); the 2115 Talent Development Program of China Agricultural University; Chinese Universities Scientific Fund funds (2024TC032; 31051301; 00114332 to G.Y.); China Agricultural University Young Talent Program in Life Science (Grant No. 008 to G.Y.), Pinduoduo-China Agricultural University Research Fund (Grant No. PC2023B01010 to G.Y.), and the Ministry of Science and Technology of China (2022YFF1201901 to H.W.), STI2030-Major Project (2022ZD0212700 to H.W.)
Author information
Authors and Affiliations
Contributions
G.Y. initiated and supervised the project. X.X., T.S., and H.G. prepared the sample. X.X. performed the electrophysiological experiment. X.X. and J.Z. collected the EM data. J.Y. and P.F. analyzed the EM data and calculated the EM map. G.Y. built and refined the atomic model. Q.W., R.G. and H.W. performed and analyzed the sequence and structural clustering, and molecular dynamics simulation. All of the authors discussed the results. G.Y. wrote the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, X., Wang, Q., Sun, T. et al. Structural basis for the activity regulation of Medicago calcium channel CNGC15. Cell Discov 11, 63 (2025). https://doi.org/10.1038/s41421-025-00815-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41421-025-00815-y