Abstract
Neurons coordinate inter-tissue protein homeostasis to systemically manage cytotoxic stress. In response to neuronal mitochondrial stress, specific neuronal signals coordinate the systemic mitochondrial unfolded protein response (UPRmt) to promote organismal survival. Yet, whether chemical neurotransmitters are sufficient to control the UPRmt in physiological conditions is not well understood. Here, we show that gamma-aminobutyric acid (GABA) inhibits, and acetylcholine (ACh) promotes the UPRmt in the Caenorhabditis elegans intestine. GABA controls the UPRmt by regulating extra-synaptic ACh release through metabotropic GABAB receptors GBB-1/2. We find that elevated ACh levels in animals that are GABA-deficient or lack ACh-degradative enzymes induce the UPRmt through ACR-11, an intestinal nicotinic α7 receptor. This neuro-intestinal circuit is critical for non-autonomously regulating organismal survival of oxidative stress. These findings establish chemical neurotransmission as a crucial regulatory layer for nervous system control of systemic protein homeostasis and stress responses.
Similar content being viewed by others
Introduction
Maintaining systemic mitochondrial function is crucial for organismal health and survival. Mitochondrial stress responses are essential molecular mechanisms that maintain metabolic homeostasis. Local stress responses can be communicated to distal tissues to systemically combat challenges, and thereby increase the chance of organismal survival. The nervous system is critical for coordinating stress responses across multiple tissues. Previous studies have revealed multiple neuronal signals that coordinate the systemic mitochondrial unfolded protein response (UPRmt) when the nervous system is exposed to mitochondrial stress encountered in disease states1,2,3,4,5,6,7. In unstressed conditions, optimizing mitochondrial function in response to metabolic and physiological fluctuations is a continual process. However, whether the nervous system tonically regulates the UPRmt in unstressed, physiological conditions is unclear. In this study, we aimed to better understand how the nervous system maintains systemic mitochondrial function and health.
Results
GABA Signalling Inhibits the Intestinal UPRmt
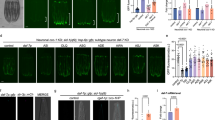
Neurotransmission is a core communication mechanism of the nervous system. To identify neurotransmitters that mediate systemic mitochondrial health, we screened neurotransmitter synthesis and transport mutants for changes to intestinal expression of the hsp-6p::gfp reporter, a well-established readout for the UPRmt 8,9,10,11,12. We screened loss-of-function mutations affecting dopamine (cat-2), octopamine and tyramine (tdc-1), serotonin (tph-1), glutamate (eat-4), gamma-aminobutyric acid (GABA) (unc-25) and acetylcholine (ACh) (cha-1) signalling (Fig. 1a, b)13,14,15,16,17,18. We found that loss of UNC-25/GAD, a glutamic acid decarboxylase required for GABA synthesis (Fig. 1c), induced intestinal hsp-6p::gfp expression (Fig. 1a, b). UNC-25 is expressed in 26 GABAergic neurons: 19 D-type motor neurons, 4 RME motor neurons, and 3 other neurons (AVL, DVB and RIS) (Fig. 1d)17. Resupplying unc-25 cDNA to all GABA expressing neurons using the unc-25 promoter rescued intestinal hsp-6p::gfp expression levels in unc-25(−) animals (Fig. 1e), confirming a role for UNC-25 in non-autonomous regulation of the intestinal UPRmt. Furthermore, induced hsp-6p::gfp expression in unc-25(−) mutant animals was rescued by unc-25 cDNA expression under the D-type motor neuron unc-30 promoter, but not under the truncated unc-47 promoter that drives expression in the RMEs, AVL, RIS and DVB neurons (Fig. 1e)19.
a, b Quantification (a) and DIC/fluorescent micrographs (b) of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type and mutant animals affecting neurotransmitter synthesis and transport. c Schematic of GABA synthesis (UNC-25/GAD), transport (UNC-47/VGAT), and release. d Schematic of GABA expressing neurons in C. elegans. e Quantification of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, unc-47(n2409), unc-25(e156), unc-25(e156); unc-25p::unc-25 DNA, unc-25(e156); unc-30p::unc-25 DNA and unc-25(e156); unc-47ptruncated::unc-25 DNA animals. n = 30 animals. The hash symbol (#) refers to independent transgenic rescue lines. P values assessed by one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Error bars indicate SEM. Scale bar, 250 μm. Source data are provided as a Source Data file.
Animals lacking UNC-25 also showed increased intestinal expression of another mitochondrial stress marker; hsp-60p::gfp (Supplementary Fig. 1a, b). Similarly, sod-3p::gfp expression—a DAF-16/FOXO transcription factor regulated gene with roles in combatting oxidative stress in mitochondria—was also induced in unc-25(−) animals (Supplementary Fig. 1c, d), consistent with previously published data20,21. We found that UNC-25 loss did not affect hsp-16.2p::gfp, a reporter for cytosolic heat stress, suggesting that disrupted GABA signalling does not induce a general stress response (Supplementary Fig. 1e, f). UPRmt induction is regulated by ATFS-1, a mitochondrial unfolded protein response transcription factor, and the chromatin remodelling proteins DVE-1 and UBL-510,22. Using RNA-mediated interference (RNAi), we silenced atfs-1, ubl-5 and dve-1 in unc-25(−) animals and found that intestinal hsp-6p::gfp induction was suppressed (Supplementary Fig. 1g, h). Therefore, GABA signalling requires canonical transcriptional regulators to activate the intestinal UPRmt.
To corroborate the role of GABA signalling in controlling the intestinal UPRmt, we analysed other components of the GABA signalling pathway (Fig. 1c, e and Supplementary Fig. 1i–k). The vesicular GABA transporter UNC-47/VGAT is required for packaging GABA into pre-synaptic vesicles for synaptic release23,24. We found that loss of unc-47 induces hsp-6p::gfp expression (Fig. 1e and Supplementary Fig. 1i). The UNC-30/Pitx2 transcription factor is a terminal selector of D-type motor neuron identity, where it directly promotes unc-25 and unc-47 expression (Supplementary Fig. 1j)25,26. We found that unc-30 loss also induced hsp-6p::gfp expression and that expressing unc-30 DNA under the D-type motor neuron specific unc-30 promoter rescued this phenotype (Supplementary Fig. 1k). In contrast, expressing unc-30 in the intestine using the ges-1 promoter did not affect hsp-6p::gfp expression (Supplementary Fig. 1k). Taken together, these data reveal that GABA signalling specifically from the D-type motor neurons represses the UPRmt in the C. elegans intestine.
We next sought to determine how GABA influences mitochondria in distal tissues. Several GABA related processes regulate life and health-span, which are concepts closely related to mitochondrial health27,28. In C. elegans, GABA signalling transmits longevity signals through DAF-16/FOXO20. An intermediate component in this pathway is the PLCβ homologue, EGL-829. We found that loss of egl-8 did not induce intestinal hsp-6p::gfp expression (Supplementary Fig. 2a, c), suggesting that GABA signalling regulates the intestinal UPRmt and longevity through different mechanisms. We wondered if GABA was acting as a metabolite to influence mitochondrial health through the GABA shunt, where GABA is degraded to succinic semialdehyde (SSA) by GABA-transaminase (GTA-1) and then joins the TCA cycle30. However, loss of the C. elegans GABA transaminase, GTA-1, did not influence intestinal hsp-6p::gfp expression (Supplementary Fig. 2b, c).
Regulation of acetylcholine release by GABAB receptors controls the intestinal UPRmt
We speculated that GABA signalling mediates the UPRmt from the D-type motor neurons by signalling to downstream GABA receptors. We therefore screened available GABA receptor mutants for hsp-6p::gfp induction, focusing on those receptors previously implicated in life and health-span21. We found that loss of the inhibitory UNC-49 and excitatory EXP-1 ionotropic GABAA receptors did not influence the intestinal UPRmt (Fig. 2a–c)31,32. However, loss of both components of the metabotropic GABAB receptor—GBB-1 and GBB-2—induced intestinal hsp-6p::gfp expression (Fig. 2a–c). Intestinal hsp-6p::gfp expression in the gbb-2(−); gbb-1(−) compound mutant was not significantly different to either single mutant, indicating that both components of the GABAB receptor are required in the same pathway to control the UPRmt (Fig. 2a–c). Likewise, intestinal hsp-6p::gfp expression in the unc-25(−); gbb-2(−); gbb-1(−) triple mutant was not additive compared the unc-25(−) single mutant, showing that GABA synthesis and the metabotropic GABA receptors act within the same genetic pathway to control the systemic UPRmt (Fig. 2d). Mutant strains containing the unc-25(e156) mutation, however, exhibited slightly stronger hsp-6p::gfp expression than gbb-1(−) and gbb-2(−) mutants, suggesting that GABA signalling may also have roles outside of metabotropic signalling in controlling systemic UPRmt activation. Canonically, GBB-1 and GBB-2 act in concert to reduce neuronal excitability, however, previous studies found that GBB-1 can act independently to influence longevity through DAF-16/FOXO20. This is supported by our data, where sod-3p::gfp expression—a readout for DAF-16 activity—is regulated by GBB-1, and not GBB-2 (Supplementary Fig. 3). These data support a role for GABA in UPRmt activation independent to its role in longevity. Single cell sequencing data shows that gbb-1 and gbb-2 are expressed primarily in neurons33. Therefore, we resupplied gbb-1 cDNA under the pan-neuronal rgef-1 promoter in gbb-1(−) animals and found that this rescued the increased intestinal hsp-6p::gfp levels (Fig. 2e), confirming that the GABAB receptor complex acts in neurons to influence intestinal UPRmt activation.
a Schematic of ionotropic GABAA (UNC-49 (inhibitory) and EXP-1 (excitatory)) and metabotropic (GBB-1 and GBB-2) GABAB receptors in C. elegans. b, c Quantification (b) and DIC/fluorescent micrographs (c) of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, gbb-1(tm1406), gbb-2(tm1165), gbb-2(tm1165); gbb-1(tm1406), unc-49(e407) and exp-1(ok1131) animals. d Quantification of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, gbb-1(tm1406), gbb-2(tm1165), gbb-2(tm1165); gbb-1(tm1406), unc-25(e156), unc-25(e156); gbb-1(tm1406), unc-25(e156); gbb-2(tm1165), and unc-25(e156); gbb-2(tm1165); gbb-1(tm1406) animals. e Quantification of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, gbb-1(tm1406), gbb-1(tm1406); rgef-1p::gbb-1 cDNA and gbb-1(tm1406); acr-2p(s)::gbb-1 cDNA animals. n = 30 animals. P values assessed by one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Error bars indicate SEM. Scale bar, 250 µm. Source data are provided as a Source Data file.
Within the C. elegans ventral nerve cord, D-type GABAergic motor neurons synapse with body wall muscle and adjacent cholinergic neurons, which are the only motor neurons that express gbb-1 and gbb-2 (Fig. 3a)34,35. To determine whether the metabotropic receptor complex acts in cholinergic motor neurons to mediate the intestinal UPRmt, we resupplied gbb-1 cDNA specifically to cholinergic ventral nerve cord motor neurons using a 1882bp acr-2(s) promoter36. We found that gbb-1 expression in these cholinergic motor neurons rescued the increased intestinal hsp-6p::gfp levels of gbb-1(−) animals (Fig. 2e), confirming that the GABAB receptor complex acts in cholinergic motor neurons to influence intestinal UPRmt activation. As GABA is generally an inhibitory neurotransmitter, and ACh is the primary excitatory neurotransmitter, these neurons work in a negative feedback loop to regulate each other and the muscles they innervate37. When the inhibitory GABA signal is lost, cholinergic neurons are overactive, leading to increased ACh release35. We therefore speculated that GABA regulates the intestinal UPRmt through downstream ACh signalling. Our initial screening data showed that loss of the choline acetyltransferase CHA-1/ChAT, which is required for ACh production, significantly reduced intestinal hsp-6p::gfp expression (Fig. 1a). Likewise, loss of the ACh vesicular transporter UNC-17/VAChT also reduced intestinal hsp-6p::gfp expression (Fig. 3b, c). Importantly, loss of unc-17 rescued the increased hsp-6::gfp observed in unc-25(−) and gbb-1(−) animals (Fig. 3b, c and Supplementary Fig. 4). These data support the concept that ACh levels can influence the systemic UPRmt, and that increased ACh release in animals lacking GABA signalling would increase the intestinal UPRmt. To test this hypothesis, we examined animals lacking two acetylcholinesterases, ACE-1/2, which have an approximately two-fold increase in systemic ACh38. We found that, as with loss of GABA signalling, increased ACh in ace-2(−); ace-1(−) animals induced intestinal hsp-6p::gfp expression (Fig. 3d, e).
a Schematic of the C. elegans motor circuit. Orange = inhibitory GABAergic motor neurons; blue = excitatory cholinergic motor neurons; Pink = body wall muscle. b, c Quantification (b) and DIC/fluorescent micrographs (c) of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, unc-25(e156), unc-17(e113), and unc-25(e156); unc-17(e113) animals. d, e Quantification (d) and DIC/fluorescent micrographs (e) of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type and ace-2(g72); ace-1(p1000) animals. f Quantification of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type and ace-2(g72); ace-1(p1000) animals grown on empty vector (EV), acr-6, acr-7 or acr-11 RNAi from the mother’s L4 stage. g Quantification of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type and unc-25(e156) animals grown on empty vector (EV) or acr-11 RNAi from the mother’s L4 stage. n = 30 animals. P values assessed by one-way analysis of variance (ANOVA) Tukey’s post hoc test (b, f, g) and two-way unpaired t test with Welch’s correction (d). Error bars indicate SEM. Scale bars, 250 µm. Source data are provided as a Source Data file.
The intestinal ACR-11 nicotinic α7 receptor regulates mitochondrial health
We theorized that excess ACh acts directly on intestinal cells to induce the UPRmt. Therefore, we used RNAi to knock down intestinally-expressed ACh receptors (ACR-6, ACR-7 and ACR-11) in the ace-2(−); ace-1(−) compound mutant, screening for inhibition of hsp-6p::gfp induction (Fig. 3f). This analysis revealed that only knockdown of the nicotinic α7 receptor (α7 nAChR) ACR-11, an ortholog of human CHRNA7, abrogated intestinal hsp-6p::gfp induction in ace-2(−); ace-1(−) animals (Fig. 3f). Importantly, ACR-11 knockdown also prevented hsp-6p::gfp induction in unc-25(−) animals (Fig. 3g). To corroborate our RNAi results, we used CRISPR/Cas9 to generate independent acr-11 deletions in wild type and ace-2(−); ace-1(−) mutant animals (Fig. 4a). As acr-11 and ace-2 are tightly linked on chromosome I, the acr-11 deletions were generated separately. Confirming our RNAi data, the acr-11(rp192) deletion prevented hsp-6p::gfp induction in ace-2(−); ace-1(−) animals (Fig. 4b). However, loss of acr-11 alone was not sufficient to lower hsp-6p::gfp expression, as observed in unc-17 mutant animals (Fig. 4b, c, e). We wondered whether other intestinal α7 nAChR may support optimal UPRmt activation in conditions of physiological ACh levels. We therefore used CRISPR/Cas9 to generate acr-6 and acr-7 deletions (Fig. 4a) and measured hsp-6p::gfp levels in single, double and triple mutant combinations with acr-11(rp191) animals. We found that hsp-6p::gfp was only reduced when all three intestinally expressed α7 nAChR were deleted (Fig. 4c). This implies a small but significant role for ACR-6 and ACR-7 in UPRmt activation under basal ACh conditions, with ACR-11 the main responder to elevated ACh.
a (i) Structure of the acr-11 locus. rp191 = 3038 bp deletion in wild-type animals; rp192 = 3046 bp deletion in ace-2(g72); ace-1(p1000) animals. (ii) Structure of the acr-6 locus. rp209 = 3209 bp deletion in wild-type animals; rp210 = 3158 bp deletion in acr-11(rp191) animals. (iii) Structure of the acr-7 locus. rp212 = 1556 bp deletion. Black boxes, coding regions; grey boxes, untranslated regions; red lines, CRISPR/Cas9-generated deletion alleles. b Quantification of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, acr-11(rp191), ace-2(g72); ace-1(p1000), and ace-2(g72) acr-11(rp192); ace-1(p1000) animals. c Quantification of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, acr-6(rp209), acr-7(rp212), acr-11(rp191), acr-6(rp209); acr-7(rp212), acr-6(rp210) acr-11(rp191), acr-11(rp191); acr-7(rp212) and acr-6(rp210) acr-11(rp191); acr-7(rp212) animals. d, e Quantification (d) and DIC/fluorescent micrographs (e) of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, ace-2(g72); ace-1(p1000), acr-11(rp191), ace-2(g72) acr-11(rp192); ace-1(p1000) and ace-2(g72) acr-11(rp192); ace-1(p1000); ges-1p::acr-11 DNA animals. f, g Quantification (f) and DIC/fluorescent micrographs (g) of UPRmt reporter (hsp-6p::gfp) expression in L4 larvae of wild-type, unc-17(e113), ges-1p::acr-11 DNA and unc-17(e113); ges-1p::acr-11 cDNA animals. n = 30 animals. P values assessed by one-way analysis of variance (ANOVA) with Tukey’s post hoc test. Error bars indicate SEM. Scale bars, 250 µm. Source data are provided as a Source Data file.
To examine the expression pattern of acr-11, we generated transgenic animals expressing green fluorescent protein (GFP) under control of the 2024 bp acr-11 promoter and detected expression in the intestine and pharynx, and not in neurons or body wall muscle (Supplementary Fig. 5). To determine whether ACR-11 is required in the intestine, we resupplied acr-11 cDNA in the intestine (ges-1 promoter) to the ace-2(−) acr-11(−); ace-1(−) triple mutant, which restored the hsp-6p::gfp expression levels to that of the ace-2(−); ace-1(−) double mutant (Fig. 4d, e). Furthermore, we found that acr-11 overexpression in the intestine greatly induced hsp-6p::gfp expression in wild-type animals, and that this induction is rescued by preventing ACh release using an unc-17 mutation (Fig. 4f, g). These data reveal that ACh acts through ACR-11 in the intestine to regulate the UPRmt.
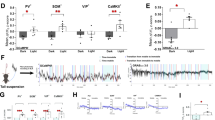
Calcium plays key roles in mitochondrial signalling, function and health, including in stress response activation39,40. As ACR-11 is an α7 nAChR, which are highly permeable to calcium ions41, we investigated intracellular calcium storage using an intestine-specific fluorescence resonance energy transfer (FRET)-based calcium indicator42,43 in animals with perturbed GABA and ACh signalling (Fig. 5a-b). We found that both unc-25(−) and ace-2(−); ace-1(−) animals had increased intracellular calcium levels (Fig. 5a, b). Furthermore, animals lacking ACR-11, either alone or when combined with the ace-2(−); ace-1(−) compound mutant, had wild-type calcium levels (Fig. 5a, b). These data mirror hsp-6p::gfp reporter levels in these mutants, supporting our hypothesis that ACR-11 responds to ACh release by increasing intracellular calcium levels in the intestine, which likely activates the UPRmt.
a, b Quantification (a) and micrographs (b) of calcium imaging by FRET microscopy of the two anterior intestinal cells of wild-type, unc-25(e156), ace-2(g72); ace-1(p1000), acr-11(rp191) and ace-2(g72) acr-11(rp192); ace-1(p1000) animals expressing nhx-2p::D3cpv, a ratiometric indicator. n = 38, 36, 36, 35, 36 animals (left to right). Scale bar, 50 µm. Note that induced expression of the ratiometric reporter in animals with excess ACh (unc-25 and ace2; ace-1 mutants) in intestinal cells Int1-3 was consistent in all animals and not due to mosaicism. c Survival analysis of wild-type, unc-25(e156), gbb-1(tm1406) and gbb-1(tm1406); rgef-1p::gbb-1 cDNA animals exposed to 200 mM paraquat from the L4 larval stage. n = 67, 74, 70, 67 animals (top to bottom). d Survival analysis of wild-type, ace-2(g72); ace-1(p1000), acr-11(rp191) and ace-2(g72) acr-11(rp192); ace-1(p1000) animals exposed to 200 mM paraquat from the L4 larval stage. n = 73, 72, 65, 67 animals (top to bottom). e Survival analysis of wild-type, unc-25(e156), unc-17(e113) and unc-25(e156); unc-17(e113) animals exposed to 200 mM paraquat from the L4 larval stage. n = 73, 74, 72, 71 animals (top to bottom). f Survival analysis of wild-type, unc-17(e113), ges-1p::acr-11 cDNA and unc-17(e113); ges-1p::acr-11 cDNA animals exposed to 200 mM paraquat from the L4 larval stage. n = 74, 74, 64, 75 animals (top to bottom). g–i Quantification of (g) autophagosome puncta (AP), (h) autolysosome puncta (AL), and micrographs (i) of GFP and mCherry fluorescence in the two anterior intestinal cells of wild-type and acr-11(rp191) L4 larvae expressing LGG-1p::mCherry::GFP::LGG-1. White arrows = green puncta/autophagosomes, yellow arrows = red puncta/autolysosomes. n = 62 (wild type) and 59 (acr-11(rp191) animals). Scale bar, 5 µm. P values assessed by one-way analysis of variance (ANOVA) with Tukey’s post hoc test (a), two-way analysis of variance (ANOVA) with Tukey’s post hoc test (c–f), and two-way unpaired t test with Welch’s correction (g, h). Error bars indicate SEM. Source data are provided as a Source Data file.
UPRmt activation can have a positive or negative effect on mitochondrial health44. An overactive stress response in the absence of stressors may prime mitochondria to manage subsequent stress exposure. Conversely, UPRmt induction due to poor mitochondrial health may cause sensitivity to mitochondrial stressors. To assess whether perturbed GABA/ACh signalling influences mitochondrial stress resistance, we examined acute paraquat sensitivity. Paraquat exposure induces mitochondrial stress by disrupting complex I of the electron transport chain and increases superoxide levels45. We found that loss of GABA biosynthesis (unc-25 mutant) or metabotropic GABA receptors (gbb-1 and gbb-2 mutants) were sensitive to paraquat (Fig. 5c and Supplementary Fig. 6a). Paraquat sensitivity was rescued in the gbb-1(tm1406) mutant by resupplying gbb-1 cDNA in neurons (Fig. 5c). Further, we found that ace-2(−); ace-1(−) animals were sensitive to paraquat exposure (Fig. 5d). Thus, two conditions where ACh signalling is amplified cause sensitivity to a mitochondrial stressor. In contrast, acr-11(−) single and ace-2(−) acr-11(−); ace-1(−) triple mutant animals exhibited increased paraquat resistance (Fig. 5d). Neither acr-6(−) nor acr-7(−) caused changes to paraquat survival (Supplementary Fig. 6b), highlighting the specific importance of ACR-11. Furthermore, reduced ACh signalling in unc-17(−) animals conferred paraquat resistance, and was sufficient to rescue paraquat sensitivity induced by loss of unc-25 (Fig. 5e). We found that intestinal acr-11 cDNA overexpression severely impeded paraquat survival (Fig. 5f), mirroring the dramatic increase in hsp-6p::gfp expression observed when acr-11 is overexpressed in the intestine. This sensitivity to paraquat was rescued by reducing ACh signalling via loss of unc-17 (Fig. 5f). Therefore, increased UPRmt activation via increased ACh signalling to ACR-11 is associated with mitochondrial damage and paraquat sensitivity, whilst decreased ACh signalling to ACR-11 is associated with increased mitochondrial fitness, leading to increased paraquat survival.
Another indicator of mitochondrial health is mitochondrial morphology. In general, more mitochondrial fusion indicates increased OXPHOS functionality, whereas more mitochondrial fragmentation indicates increased autophagy and/or biogenesis46. Based on our paraquat data, we hypothesized that unc-25(−) and ace-2(−); ace-1(−) animals would have more fragmented mitochondria, and that acr-11(−) single and ace-2(−) acr-11(−); ace-1(−) triple mutants would have increased mitochondrial fusion. Using a mitochondrial-targeted GFP reporter expressed in the intestine (ges-1p::gfpmt)47, we found that unc-25(−) and the ace-2(−); ace-1(−) animals had increased mitochondrial fragmentation in the intestine compared to wild type animals, as predicted (Supplementary Fig. 7a). This phenotype was intestine-specific, as we did not detect mitochondrial morphology changes in body wall muscle using a ubiquitously expressed reporter (cox-4p::gfpmt) in unc-25(−) animals (Supplementary Fig. 7c, d)48. Unexpectedly, the acr-11(−) single mutant also displayed a fragmented mitochondria phenotype (Supplementary Fig. 7a). Further, the ace-2(−) acr-11(−); ace-1(−) triple mutant exhibited more mitochondrial fragmentation than the acr-11(−) single and ace-2(−); ace-1(−) double mutants (Supplementary Fig. 7a). These data imply that, in the context of mitochondrial morphology, ACh signalling and the ACR-11 receptor act in distinct pathways.
We wondered how similar mitochondrial fragmentation phenotypes observed in unc-25(−), ace-2(−); ace-1(−) and acr-11(−) animals could lead to opposing mitochondrial fitness in terms of paraquat resistance. We posited that, in a pathway separate from GABA/ACh signalling, ACR-11 may repress autophagy and thus mitochondrial turnover, perhaps as an energy conservation mechanism. This means that ACR-11 loss would induce fragmentation as mitochondria are excessively cleared from the system. Using an mCherry::gfp::lgg-1 reporter to measure autophagy49, we indeed found that the number of intestinal autophagosomes and autolysosomes in acr-11(−) L4 larvae is higher compared to control animals (Fig. 5g–i). This suggests that animals lacking ACR-11 exhibit elevated mitochondrial turn-over in unstressed conditions that conveys a survival advantage when exposed to oxidative stress.
Discussion
In summary, we discovered a neuro-intestinal circuit that regulates the UPRmt and organismal survival. By screening animals lacking specific neurotransmitters, we found that balanced GABAergic and cholinergic signalling is essential for non-cell-autonomous UPRmt regulation. We found that GABA released from ventral nerve cord motor neurons controls the UPRmt through two metabotropic GABAB receptor subunits that are expressed on, and inhibit, cholinergic motor neurons. Appropriate control of ACh levels is critical for UPRmt regulation, as animals lacking the ACh-degradative enzymes exhibit a two-fold induction of the UPRmt. Further, we discovered that UPRmt induction by increased systemic ACh is dependent on ACR-11, an α7 nAChR receptor acting in the intestine. Other intestinally expressed α7 nAChR receptors—ACR-6 and ACR-7—also play a minor role, in that all three receptors are required for mediating basal intestinal UPRmt levels when systemic ACh is within a normal range. In addition to regulating the UPRmt, elevated ACh induces mitochondrial fragmentation and reduces survival of oxidative stress. Interestingly, we found that the ACR-11 α7 nAChR receptor can act independently to ACh, with animals lacking ACR-11 exhibiting more intestinal mitochondrial fragmentation and autophagy irrespective of ACh levels. These distinctions likely provide animals lacking ACR-11 a survival advantage in oxidative stress conditions. In vertebrates, nicotinic α7 receptors can act in the liver (some functions of which are performed by the C. elegans intestine) to promote cell survival50, suggesting that mechanisms we describe here are phylogenetically conserved.
Methods
C. elegans culture
All C. elegans strains were grown at 20 °C on NGM agar seeded with Escherichia coli OP50, unless otherwise stated. All mutant strains were backcrossed to N2 at least three times and maintained well fed for a minimum of two generations prior to analysis. Strains used in this study are listed in Supplementary Table 1.
Fluorescence microscopy and quantification
Worms were anesthetised in 0.1 ng/ml levamisole on a 5% agarose pad. Images for analysis were taken using a Zeiss AXIO Imager M2 upright fluorescence microscope and ZEN 2.0 software at 20× objective, unless otherwise specified. ImageJ software was used to determine whole intestinal fluorescence by isolating the intestine area and measuring integrated density (IntDensity). The corrected cellular fluorescence was calculated as: CTCF = Intestinal Integrated Density—(area * background MeanGrey)
RNA-mediated interference (RNAi) by feeding
HT115 bacteria containing control (L4440 plasmid) or experimental plasmids were cultured overnight in LB containing 50 μg/ml ampicillin and seeded uniformly over plates containing IPTG (Isopropyl β- d-1-thiogalactopyranoside). Plates were dried at 37 °C and brought to room temperature before worms were applied. For atfs-1 and ubl-5 RNAi experiments, five L4 hermaphrodites were picked to RNAi plates and the subsequent generation was scored at L4. For dve-1 RNAi experiments, five L4 hermaphrodites were picked to NGM and allowed to grow for five days. Animals were then washed from the NGM plates using M9 and bleached for 4 min using a 1:1 ratio of bleach (White King Premium) and 5 M NaOH to extract eggs. Extracted eggs were washed three times in M9 and filtered through a 40 μm mesh before being applied to RNAi plates. Animals were grown to L4 and imaged as described above.
C. elegans expression constructs and transgenic strain generation
Reporter gene constructs were generated by PCR amplifying DNA elements and cloning into Fire vectors. Constructs were injected into young adult hermaphrodites using standard methods.
acr-11p::gfp reporter construct
A 2024 bp sequence upstream of the acr-11 start codon was amplified from C. elegans genomic DNA with forward (GAAATGAAATAAGCTTGCGAAGAGAGCGAGGAGG) and reverse (CCAATCCCGGGGATCCTTCAAAAAAATGTGGCTAAG) primers from Sigma-Aldrich incorporating HindIII-BamHI restriction sites. The HindIII-BamHI digested promoter fragment was ligated into HindIII-BamHI digested pPD95.75 vector. The resultant acr-11p::gfp plasmid was injected at 50 ng/µl, with 50 ng/µl of ttx-3p::mCherry vector and 120 ng/µl bacterial DNA.
ges-1p::acr-11 rescue construct
The 1386 bp acr-11 cDNA was amplified from a C. elegans cDNA library with forward (AGGACCCTTGGCTAGCATGATATTTAATCTAATTAATAG) and reverse (GATATCAATACCATGGTTAGGCAATAATATGAGG) primers from Sigma-Aldrich incorporating NheI-NcoI restriction sites. The Pges-1p-sphk-1 vector was digested with NheI-NcoI and the acr-11 cDNA was inserted using the In-Fusion HD Cloning Kit (Takara Bio) to replace the sphk-1 sequence. The resultant ges-1p::acr-11cDNA plasmid was injected at 10 ng/µl, with 50 ng/µl of ttx-3p::mCherry vector and 120 ng/µl bacterial DNA into acr-11 mutant animals for rescue, or wild-type for overexpression.
unc-25 rescue strains
The plasmids pHP03 [unc-25p::unc25::unc-25 3’ UTR], pNK06 [unc-47ptruncated::unc-25::SL2::GFP::unc-54 3’ UTR], pNK07 [unc-30p::unc-25::SL2::GFP::unc-54 3’UTR] were kindly provided by Haijun Tu. pNK06, was injected at 20 ng/µl19. pHP03 and pNK07 were injected at 1 ng/µl. All plasmids were injected with 50 ng/µl of Pttx-3::mCherry vector and 120 ng/µl bacterial DNA into unc-25(e156); hsp-6::GFP animals.
unc-30 rescue strains
pJL24 [unc-30p::unc-30::SL2::GFP::unc-54 3’ UTR], kindly provided by Haijun Tu, was injected at 1 ng/µl. RJP807 ges-1p::unc-30 cDNA was constructed by amplifying the unc-30 cDNA from pJL24 with forward (AGGACCCTTGGCTAGCATGGATGACAATACGGCCACAC) and reverse (GATATCAATACCATGGCTAAAGTGGTCCACTGTACTGAC) primers incorporating NheI-NcoI restriction sites. RJP807 was injected at 10 ng/µl. All plasmids were injected with 50 ng/µl of ttx-3p::mCherry vector and 120 ng/µl bacterial DNA into unc-30(e191); hsp-6::GFP animals.
gbb-1 motor neuron rescue
The acr-2p(s)::gbb-1 sequence was amplified by PCR from pBS77 (Pacr-2-gbb-1 cDNA) with forward (GCTCACCACCGACTGATTTTC) and reverse (TTGCGGTCATAAACTGAAACG) primers and injected at 5 ng/µl into gbb-1(tm1406); hsp-6::GFP animals with 50 ng/µl of ttx-3p::mCherry vector and 120 ng/µl bacterial DNA.
CRISPR-Cas9 genome editing
To generate acr-11 deletion mutants, adult wild-type or ace-2(p1000)I; ace-1(g72)X hermaphrodites were microinjected with Cas9 protein and two crRNAs: 5’ crRNA (CCTGTGCGACGGAAGTGTTG), 3’ crRNA (CGAATCTCCAATCCGTTTGA) from IDT. acr-11 deletions were identified by PCR and confirmed by Sanger sequencing. The acr-11(rp191) allele generated in wild-type animals is a 3038 bp deletion and the acr-11(rp192) allele generated in ace-2(g72)I; ace-1(p1000)X animals is a 3046 bp deletion. Both deletion alleles remove most of the acr-11 gene (Fig. 4a(i)). Each allele was backcrossed to wild-type males prior to analysis.
To generate acr-6 deletion mutants, adult wild-type or acr-11(rp191) mutant hermaphrodites were microinjected with Cas9 protein and two crRNAs: 5’ crRNA (GTATTGTTGAACTCACCAGA), 3’ crRNA (ATTCACGGATTGATAGCAAT) from IDT. acr-6 deletions were identified by PCR and confirmed by Sanger sequencing. The acr-6(rp209) allele generated in wild-type animals is a 3209 bp deletion and the acr-6(rp210) allele generated in acr-11(rp191) animals is a 3158 bp deletion. Both deletion alleles remove most of the acr-6 gene (Fig. 4a(ii)). Each allele was backcrossed twice to wild-type males prior to analysis.
To generate the acr-7 deletion mutant, adult wild-type hermaphrodites were microinjected with Cas9 protein and two crRNAs: 5’ crRNA (ATTCACTCCTCATAGACGAG), 3’ crRNA (TTTTCACGTATTTACTACCAT) from IDT. The acr-7 deletion was identified by PCR and confirmed by Sanger sequencing. The acr-7(rp212) allele is a 1556 bp deletion that removes the first 8 exons (Fig. 4a(iii)). The acr-7(rp212) allele was backcrossed twice to wild-type males prior to analysis.
Calcium imaging
Calcium levels were visualised using the calcium indicator d3cpv expressed in the intestine using the nhx-2 intestine-limited promoter43. Worms were anesthetised with in 0.1 ng/ml levamisole diluted in M9 and mounted to 5% agarose pads. Images were taken using Leica Stellaris5 Invert Confocal Microscope and objective lens 63×/1.40 Oil (WD 140 μm). CFP (405 excitation, 455–495 emission) and FRET (405 excitation, 515–555 emission) were collected. The FRET ratio in the first three intestinal cell pairs was calculated as (FRETint − FRETbg)/(CFPint − CFPbg).
Mitochondrial morphology analysis
Worms were anesthetised with 50 mM sodium azide diluted in M9 and mounted to 5% agarose pads. Images were taken using Leica Stellaris5 Invert Confocal Microscope and objective lens 63×/1.40 Oil (WD 140 μm). Optical slice thickness was 0.2 μm. Z stack images were blinded to genotype and mitochondrial morphology classified as fused, intermediate or fragmented6.
Acute paraquat sensitivity assays
200 mM paraquat plates were prepared51 and seeded with 50 μl 10× concentrated OP50. 25 L4 worms were transferred to each plate and survival was assessed each hour for 15 consecutive hours. Worms that left the agar were excluded from analysis.
Autophagy analysis
Intestinal autophagy was analyzed using the lgg-1p::mCherry::gfp::lgg-1 reporter49. A single image was taken in line with the nucleus of the first two intestinal cells using a Zeiss AXIO Imager M2 upright fluorescence microscope as described above. Images were blinded to genotype and green and red LGG-1 puncta were counted, with green puncta representing autophagosomes and red puncta representing autolysosomes.
Statistics and reproducibility
Statistical analysis was performed using GraphPad Prism 9 software. Statistical tests and n numbers are indicated in corresponding figure legends. Differences with a P value < 0.05 were considered significant. All experiments were performed with at least three independent biological replicates, with similar results; the investigator was blinded to the genotype. No statistical method was used to pre-determine sample size. No data were excluded from analysis. The experiments were not randomized.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data is available in the main text or supplementary materials. There are no accession codes, unique identifiers, or weblinks in our study and no restrictions on data availability. Materials will be available upon request from the Pocock laboratory. Source data are provided with this paper.
References
Durieux, J., Wolff, S. & Dillin, A. The cell-non-autonomous nature of electron transport chain-mediated longevity. Cell 144, 79–91 (2011).
Berendzen, K. M. et al. Neuroendocrine coordination of mitochondrial stress signaling and proteostasis. Cell 166, 1553–1563.e1510 (2016).
Shao, L.-W., Niu, R. & Liu, Y. Neuropeptide signals cell non-autonomous mitochondrial unfolded protein response. Cell Res. 26, 1182–1196 (2016).
Zhang, Q. et al. The mitochondrial unfolded protein response is mediated cell-non-autonomously by retromer-dependent Wnt signaling. Cell 174, 870–883.e817 (2018).
Lan, J. et al. Translational regulation of non-autonomous mitochondrial stress response promotes longevity. Cell Rep. 28, 1050–1062.e1056 (2019).
Zhang, Y. et al. Neuronal TORC1 modulates longevity via AMPK and cell nonautonomous regulation of mitochondrial dynamics in C. elegans. eLife 8, e49158 (2019).
Chen, L. T. et al. Neuronal mitochondrial dynamics coordinate systemic mitochondrial morphology and stress response to confer pathogen resistance in C. elegans. Dev. Cell 56, 1770–1785.e1712 (2021).
Yoneda, T. et al. Compartment-specific perturbation of protein handling activates genes encoding mitochondrial chaperones. J. Cell Sci. 117, 4055–4066 (2004).
Andrew, M. & Cole, M. H. UPRmt regulation and output: a stress response mediated by mitochondrial-nuclear communication. Cell Res. 28, 281–295 (2018).
Haynes, C. M., Petrova, K., Benedetti, C., Yang, Y. & Ron, D. ClpP mediates activation of a mitochondrial unfolded protein response in C. elegans. Dev. Cell 13, 467–480 (2007).
Nargund, A. M., Pellegrino, M. W., Fiorese, C. J., Baker, B. M. & Haynes, C. M. Mitochondrial import efficiency of ATFS-1 regulates mitochondrial UPR activation. Science 337, 587 (2012).
Nargund, A. M., Fiorese, C. J., Pellegrino, M. W., Deng, P. & Haynes, C. M. Mitochondrial and nuclear accumulation of the transcription factor ATFS-1 promotes OXPHOS recovery during the UPR(mt). Mol. Cell 58, 123 (2015).
Omura, D. T., Clark, D. A., Samuel, A. D. & Horvitz, H. R. Dopamine signaling is essential for precise rates of locomotion by C. elegans. PLoS ONE 7, e38649 (2012).
Alkema, M. J., Hunter-Ensor, M., Ringstad, N. & Horvitz, H. R. Tyramine functions independently of octopamine in the Caenorhabditis elegans nervous system. Neuron 46, 247–260 (2005).
Sze, J. Y., Victor, M., Loer, C., Shi, Y. & Ruvkun, G. Food and metabolic signalling defects in a Caenorhabditis elegans serotonin-synthesis mutant. Nature 403, 560–564 (2000).
Rankin, C. H. & Wicks, S. R. Mutations of the Caenorhabditis elegans brain-specific inorganic phosphate transporter eat-4 affect habituation of the tap-withdrawal response without affecting the response itself. J. Neurosci. 20, 4337–4344 (2000).
Jin, Y., Jorgensen, E., Hartwieg, E. & Horvitz, H. R. The Caenorhabditis elegans gene unc-25 encodes glutamic acid decarboxylase and is required for synaptic transmission but not synaptic development. J. Neurosci. 19, 539–548 (1999).
Rand, J. B. Genetic analysis of the cha-1-unc-17 gene complex in Caenorhabditis. Genetics 122, 73–80 (1989).
Liu, J. et al. GABAergic signaling between enteric neurons and intestinal smooth muscle promotes innate immunity and gut defense in Caenorhabditis elegans. Immunity 56, 1515–1532.e1519 (2023).
Chun, L. et al. Metabotropic GABA signalling modulates longevity in C. elegans. Nat. Commun. 6, 8828 (2015).
Yuan, F. et al. GABA receptors differentially regulate life span and health span in C. elegans through distinct downstream mechanisms. Am. J. Physiol. 317, C953–C963 (2019).
Haynes, C. M., Yang, Y., Blais, S. P., Neubert, T. A. & Ron, D. The matrix peptide exporter HAF-1 signals a mitochondrial UPR by activating the transcription factor ZC376.7 in C. elegans. Mol. Cell 37, 529–540 (2010).
The C. elegans Deletion Mutant Consortium. Large-scale screening for targeted knockouts in the Caenorhabditis elegans genome. G3 (Bethesda) 2, 1415–1425 (2012).
McIntire, S. L., Reimer, R. J., Schuske, K., Edwards, R. H. & Jorgensen, E. M. Identification and characterization of the vesicular GABA transporter. Nature 389, 870–876 (1997).
Jin, Y., Hoskins, R. & Horvitz, H. R. Control of type-D GABAergic neuron differentiation by C. elegans UNC-30 homeodomain protein. Nature 372, 780–783 (1994).
Eastman, C., Horvitz, H. R. & Jin, Y. Coordinated transcriptional regulation of the unc-25 glutamic acid decarboxylase and the unc-47 GABA vesicular transporter by the Caenorhabditis elegans UNC-30 homeodomain protein. J. Neurosci. 19, 6225–6234 (1999).
Shore, D. E., Carr, C. E. & Ruvkun, G. Induction of cytoprotective pathways is central to the extension of lifespan conferred by multiple longevity pathways. PLoS Genet. 8, e1002792 (2012).
Zhu, D. et al. NuRD mediates mitochondrial stress–induced longevity via chromatin remodeling in response to acetyl-CoA level. Sci. Adv. 6, eabb2529 (2020).
Miller, K. G., Emerson, M. D. & Rand, J. B. Goalpha and diacylglycerol kinase negatively regulate the Gqalpha pathway in C. elegans. Neuron 24, 323–333 (1999).
Yogeeswari, P., Sriram, D. & Vaigundaragavendran, J. The GABA shunt: an attractive and potential therapeutic target in the treatment of epileptic disorders. Curr. Drug Metab. 6, 127–139 (2005).
Bamber, B. A., Beg, A. A., Twyman, R. E. & Jorgensen, E. M. The Caenorhabditis elegans unc-49 locus encodes multiple subunits of a heteromultimeric GABA receptor. J. Neurosci. 19, 5348–5359 (1999).
Beg, A. A. & Jorgensen, E. M. EXP-1 is an excitatory GABA-gated cation channel. Nat. Neurosci. 6, 1145–1152 (2003).
Taylor, S. R. et al. Molecular topography of an entire nervous system. Cell 184, 4329–4347 e4323 (2021).
Cook, S. J. et al. Whole-animal connectomes of both Caenorhabditis elegans sexes. Nature 571, 63–71 (2019).
Dittman, J. S. & Kaplan, J. M. Behavioral impact of neurotransmitter-activated G-protein-coupled receptors: muscarinic and GABAB receptors regulate Caenorhabditis elegans locomotion. J. Neurosci. 28, 7104–7112 (2008).
Gao, S. et al. Excitatory motor neurons are local oscillators for backward locomotion. Elife 7, e29915 (2018).
Schultheis, C., Brauner, M., Liewald, J. F. & Gottschalk, A. Optogenetic analysis of GABAB receptor signaling in Caenorhabditis elegans motor neurons. J. Neurophysiol. 106, 817–827 (2011).
Lee, J., Kim, K. Y. & Paik, Y. K. Alteration in cellular acetylcholine influences dauer formation in Caenorhabditis elegans. BMB Rep. 47, 80–85 (2014).
Glancy, B., Willis, W. T., Chess, D. J. & Balaban, R. S. Effect of calcium on the oxidative phosphorylation cascade in skeletal muscle mitochondria. Biochemistry 52, 2793–2809 (2013).
Joiner, M.-lA. et al. CaMKII determines mitochondrial stress responses in heart. Nature 491, 269–273 (2012).
Shen, J. X. & Yakel, J. L. Nicotinic acetylcholine receptor-mediated calcium signaling in the nervous system. Acta Pharm. Sin. 30, 673–680 (2009).
Palmer, A. E. & Tsien, R. Y. Measuring calcium signaling using genetically targetable fluorescent indicators. Nat. Protoc. 1, 1057–1065 (2006).
Zhang, F. et al. Bacillus thuringiensis crystal protein Cry6Aa triggers Caenorhabditis elegans necrosis pathway mediated by aspartic protease (ASP-1). PLOS Pathog. 12, e1005389 (2016).
Sprenger, H. G. & Langer, T. The good and the bad of mitochondrial breakups. Trends Cell Biol. 29, 888–900 (2019).
Cochemé, H. M. & Murphy, M. P. Complex I is the major site of mitochondrial superoxide production by paraquat. J. Biol. Chem. 283, 1786–1798 (2008).
Adebayo, M., Singh, S., Singh, A. P. & Dasgupta, S. Mitochondrial fusion and fission: The fine-tune balance for cellular homeostasis. FASEB J. 35, e21620 (2021).
Benedetti, C., Haynes, C. M., Yang, Y., Harding, H. P. & Ron, D. Ubiquitin-like protein 5 positively regulates chaperone gene expression in the mitochondrial unfolded protein response. (Author abstract). Genetics 174, 229 (2006).
Raiders, S. A., Eastwood, M. D., Bacher, M. & Priess, J. R. Binucleate germ cells in Caenorhabditis elegans are removed by physiological apoptosis. PLOS Genet. 14, e1007417 (2018).
Chang, J. T., Kumsta, C., Hellman, A. B., Adams, L. M. & Hansen, M. Spatiotemporal regulation of autophagy during Caenorhabditis elegans aging. Elife 6, e18459 (2017).
Gergalova, G. et al. Mitochondria express alpha7 nicotinic acetylcholine receptors to regulate Ca2+ accumulation and cytochrome c release: study on isolated mitochondria. PLoS ONE 7, e31361 (2012).
Senchuk, M. M., Dues, D. J. & Van Raamsdonk, J. M. Measuring oxidative stress in Caenorhabditis elegans: paraquat and juglone sensitivity assays. Bio Protoc. 7, e2086 (2017).
Acknowledgements
We thank members of the Pocock laboratory for advice and comments on the paper. Imaging for this project was performed at Monash Microimaging. Some strains were provided by the Caenorhabditis Genetics Center (University of Minnesota), which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440) and National BioResource Project of Japan. We thank Shawn Xu, Haijun Tu and Jianfeng Liu for strains and plasmids. This work was supported by the following grants: National Health and Medical Research Council GNT1105374, GNT1137645, GNT2000766 and veski Innovation Fellowship (VIF23) to R.P.
Author information
Authors and Affiliations
Contributions
Conceptualization: R.C., R.P. Methodology: R.C., W.C., A.H., B.H., R.G., R.P. Investigation: R.C., W.C., A.H., B.H., R.G., R.P. Visualization: R.C., W.C., A.H., B.H. Funding acquisition: R.P. Project administration: R.P. Supervision: A.H., R.P. Writing—original draft: R.C. Writing—review & editing: W.C., A.H., B.H., R.G., R.P.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Cornell, R., Cao, W., Harradine, B. et al. Neuro-intestinal acetylcholine signalling regulates the mitochondrial stress response in Caenorhabditis elegans. Nat Commun 15, 6594 (2024). https://doi.org/10.1038/s41467-024-50973-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-50973-y
This article is cited by
-
Unlocking neuronal health: leveraging C. elegans for drug repurposing studies
Cell Communication and Signaling (2026)
-
Beyond the gut: decoding the gut–immune–brain axis in health and disease
Cellular & Molecular Immunology (2025)