Abstract
Natural evolution has resulted in reduced cold tolerance in cultivated tomato (Solanum lycopersicum). Herein, we perform a combined analysis of ATAC-Seq and RNA-Seq in cold-sensitive cultivated tomato and cold-tolerant wild tomato (S. habrochaites). We identify that WRKY34 has the most significant association with differential chromatin accessibility and expression patterns under cold stress. We find that a 60 bp InDel in the WRKY34 promoter causes differences in its transcription and cold tolerance among 376 tomato accessions. This 60 bp fragment contains a GATA cis-regulatory element that binds to SWIBs and GATA29, which synergistically suppress WRKY34 expression under cold stress. Moreover, WRKY34 interferes with the CBF cold response pathway through regulating transcription and protein levels. Our findings emphasize the importance of polymorphisms in cis-regulatory regions and their effects on chromatin structure and gene expression during crop evolution.
Similar content being viewed by others
Introduction
The divergence of gene function, primarily driven by mutation, gene duplication, and gene loss, is fundamental to evolutionary processes1,2. Such divergence in gene function may be caused by mutations in coding regions that alter protein function. For instance, a 45 bp deletion in the ZmRR1 coding region prevents its phosphorylation by ZmMPK8, inhibiting its degradation via the 26S proteasome pathway and thereby enhancing maize cold tolerance3. Alternatively, divergence in gene function may also result from mutations in cis-regulatory regions, which interact intricately to shape expression patterns across different tissues during development4. For example, the CRISPR/Cas9 cis-regulatory allelic series of tomato SlWOX9 reveals that different pleiotropic functions can be mapped to specific cis-regulatory regions5. Evolutionary innovations in transcriptional regulation often result from changes in cis-regulatory regions, where abundant sequences directly binding to transcription factors offer significant mutational potential to alter gene expression and phenotypes6. For instance, a crucial variation in the W-box motif within the SlWRKY33 promoter suppresses its self-transcriptional activity in response to cold stress, thus contributing to the cold sensitivity observed in cultivated tomatoes compared to cold-tolerant wild tomatoes7. Nevertheless, our comprehension of the alterations and roles of cis-regulatory regions during crop evolution, as well as their regulatory mechanisms, remains limited.
In recent years, our understanding of transcriptional regulation has broadened to include the level of chromatin structure8. Chromatin remodeling, which involves the dynamic modification of chromatin structure, plays a crucial role in controlling the accessibility of transcriptional machinery to DNA9. In the medical field, chromatin remodeling has been extensively studied for its impact on cell differentiation, organ development, and its involvement in diseases such as cancer10,11. For instance, the chromatin remodeler CHD6 promotes colorectal cancer development by regulating TMEM65-mediated mitochondrial dynamics12. In botany, chromatin remodeling is increasingly recognized as pivotal in how plants respond to environmental cues. Several studies have indicated that environmental stress can alter chromatin structure, thereby influencing transcriptional regulation13,14. For example, heat stress triggers genome-wide chromatin accessibility changes in tomato, with HSFA1 binding promoting the formation of promoter-enhancer contacts to drive the expression of heat stress-responsive genes15. Similarly, cold stress enhances chromatin accessibility and leads to bivalent histone modifications of active genes in potato16. Moreover, a lamin-like protein OsNMCP1 in rice modifies chromatin accessibility by interacting with a chromatin remodeler OsSWI3C, thereby regulating numerous genes involved in root growth and drought response17. Hence, changes in chromatin structure may serve as the initial step in initiating transcriptional stress responses. Nevertheless, the mechanisms and extent to which environmental stress induces chromatin dynamics remain largely unknown. Additionally, the causal relationship between chromatin dynamics and transcriptional responses under environmental stress requires further elucidation.
The SWIB/MDM2 domain superfamily of proteins comprises a group of proteins characterized by the presence of the SWIB (SWI/SNF complex BCL7/BCL7A interacting domain) and/or MDM2 (Mouse Double Minute 2 homolog) domains18,19. These proteins are evolutionarily conserved and exist in various eukaryotes20,21. Numerous studies have highlighted the critical role of SWIB/MDM2 domain proteins in diverse cellular processes, particularly in chromatin remodeling and gene expression regulation20. For example, the SWP73 protein, a member of the SWIB/MDM2 domain superfamily, is essential for supporting yeast growth at elevated temperatures. Additionally, it plays a pivotal role in repressing seedling growth by modulating chromatin accessibility of genes regulating hypocotyl cell size in Arabidopsis22,23. Furthermore, a study underscores the importance of SWIB-4, a SWIB domain protein in spinach chloroplasts, which not only structures the nucleoid core but also binds DNA via its histone H1 motif, thus playing a crucial role in the compaction and regulation of chloroplast DNA24. Nevertheless, the functions of these proteins in plants, especially their ability for direct DNA binding in the nucleus, remain largely unexplored.
Cold stress poses significant threats to crops, leading to reduced growth, impaired development, and lower yields. Plants have evolved various pathways to withstand cold stress, including the CBF-COR pathway and hormonal pathways25. In the CBF-COR pathway, ICE (Inducer of CBF Expression) proteins act as upstream regulators, activating C-repeat binding factors (CBFs), which in turn induce the expression of cold-responsive (COR) genes to enhance cold tolerance26. Different subspecies within a species often exhibit distinct cold tolerances due to evolutionary adaptations to their specific environments. For example, a study employing a combination of genetic mapping and gene expression analysis revealed that temperate japonica rice varieties have evolved lower expression of HAN1 gene due to an increase in MYB cis-elements within its promoter during domestication. This adaptation enhances chilling tolerance mediated by jasmonic acid (JA), aiding in the adaptation to a temperate climate27. Similarly, using a metabolite genome-wide association study (mGWAS), variations in the ZmICE1 promoter were identified to affect its interaction with ZmMYB39, thereby influencing cold tolerance in maize through the regulation of metabolic reprogramming and COR gene expression28. Therefore, it is of great importance to utilize multi-omics analysis to identify key genetic loci for cold tolerance in crops. Different wild tomatoes have different levels of cold tolerance, and Solanum habrochaites is considered to be one of the most cold tolerant wild tomatoes29.
In this study, through the combined analysis of transposase-accessible chromatin sequencing (ATAC-Seq) and transcriptome sequencing (RNA-Seq), we observe that expression of WRKY34 remains largely unchanged following cold treatment in cold-sensitive cultivated tomato S. lycopersicum. Conversely, in cold-tolerant wild tomato S. habrochaites, exposure to cold leads to transcriptional suppression of WRKY34, accompanied by chromatin opening. Importantly, we identify a 60 bp InDel in the WRKY34 promoter that affects its binding to SWIBs and transcription factor GATA29, thereby influencing its chromatin accessibility and expression level under cold stress. Additionally, we demonstrate that SWIBs and GATA29 interact with each other to cooperatively suppress the expression of WRKY34. Furthermore, WRKY34 interferes with the CBF-COR cold response pathway through interaction with CBF1 or direct transcriptional inhibition, thereby negatively regulating cold tolerance. Our study elucidates that polymorphisms in cis-regulatory regions leading to differences in chromatin structure and gene expression during crop evolution, providing insights into the natural regulatory mechanism of cold tolerance in tomato.
Results
A potential role of WRKY34 in tomato response to cold stress
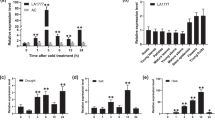
Previous research has shown substantial difference in cold tolerance between cultivated tomato and various wild tomato species7. To delve deeper into the distinctions in the potential regulatory mechanisms of cold tolerance between wild and cultivated tomatoes, we exposed the cold-sensitive cultivated tomato (Solanum lycopersicum) Ailsa Craig (AC) and the cold-tolerant wild tomato (S. habrochaites) LA1777 to cold stress for 6 h, followed by ATAC-Seq and RNA-Seq analyses, respectively. ATAC-Seq data analysis revealed that genome-wide chromatin accessibility decreased rather than increased in both wild and cultivated tomatoes after cold treatment (Fig. 1a and Supplementary Fig. 1a). High Spearman correlation coefficients between biological replicates indicate the reliability of ATAC-Seq results (Supplementary Fig. 1b). Genes near peaks exhibiting decreased chromatin accessibility (closing chromatin regions) under cold stress in both cultivated tomato AC and wild tomato LA1777 were predominantly enriched in pathways related to growth and development, such as photosynthesis, starch and sucrose metabolism and amino acid metabolism (Supplementary Fig. 1c). However, under cold stress conditions, the enrichment ratio of chromatin accessibility peaks in wild tomato LA1777 was significantly higher than that in cultivated tomato AC (Supplementary Fig. 1d). Furthermore, genes associated with differential chromatin accessibility peaks (DCAPs) between the two accessions were primarily enriched in gene ontology (GO) terms such as DNA binding, organic cyclic compound binding and heterocyclic compound binding (Supplementary Fig. 1e). Analysis of ATAC-Seq data revealed 7082 genes associated with DCAPs between the two accessions (AC and LA1777) under cold stress conditions (Fig. 1b and Supplementary Data 1), along with 337 genes showing increased chromatin accessibility peaks (opening chromatin regions) in wild tomato LA1777 post cold treatment (Fig. 1b and Supplementary Data 2). Analysis of RNA-Seq data revealed 3434 differentially expressed genes (DEGs) in wild tomato LA1777 under cold stress, whereas 25557 genes in cultivated tomato AC exhibited no significant difference under cold stress (Fig. 1b and Supplementary Data 3, 4).
a Heatmaps of ATAC-Seq read density in all samples (two replicates per sample) within a 3 kb window centered on transcriptional start sites (TSS). Detected accessible regions are showed. Each row represents one peak. The color represents the intensity of chromatin accessibility. b Venn diagram illustrating cold response candidate genes identified by combined ATAC-Seq and RNA-Seq analysis. Categories include: ATAC-Seq (7082) LA1777_4vsAC_4_Diff, indicating 7082 differential chromatin accessibility peaks (DCAPs)-associated genes between the two accessions (cultivated tomato AC and wild tomato LA1777) under cold stress conditions identified by ATAC-Seq data; ATAC-Seq (337) LA1777_4vsLA1777_25_Up, indicating 337 increased chromatin accessibility peaks (opening chromatin regions) related genes in wild tomato LA1777 after cold stress identified by ATAC-Seq; RNA-Seq (25557) AC_Insig, indicating 25557 genes with no significant change in expression in cultivated tomato AC after cold stress identified by RNA-Seq data; RNA-Seq (3434) LA1777_Sig, indicating 3434 differentially expressed genes (DEGs) in wild tomato LA1777 under cold stress identified by RNA-Seq data. c Expression levels of SlWRKY34 in S. lycopersicum (Ailsa Craig, AC/SlW34) and ShWRKY34 in S. habrochaites (LA1777/ShW34) plants under cold stress. Total RNA was isolated from leaf samples at the indicated times. d WRKY34 protein accumulation in AC and LA1777 under cold stress. The Actin protein was used as a loading control. The experiments were repeated three times with similar results (c, d). Values are expressed as means ± SD, n = 3 (*P < 0.05 and ***P < 0.001; two-tailed Student’s t-test). AC_25, cultivated tomato AC under normal temperature (25 °C); AC_4, cultivated tomato AC under cold stress (4 °C); LA1777_25, wild tomato LA1777 under normal temperature (25 °C); LA1777_4, wild tomato LA1777 under cold stress (4 °C). Source data are provided as a Source Data file.
To further identify candidate genes responsible for cold tolerance in tomato, we integrated the ATAC-Seq and RNA-Seq data as described above and generated a Venn diagram (Fig. 1b). The intersection of the Venn diagram revealed that the expression levels of 18 genes remained unchanged in the cold-sensitive cultivated tomato AC after cold treatment, but showed differential expression in the cold-tolerant wild tomato LA1777 after cold treatment. Interestingly, chromatin accessibilities of these 18 genes were significantly different between the two accessions (AC and LA1777) under cold stress, with chromatin opening observed in the cold-tolerant wild tomato LA1777 after cold stress (Fig. 1b and Supplementary Data 5). The expression of these 18 genes was validated through RT-qPCR analysis, confirming the reliability of the RNA-Seq data (Supplementary Fig. 2a). Additionally, RT-qPCR validation was performed on three genes from the RNA-Seq data that exhibited significant expression changes after cold treatment in AC but no difference in LA1777, as well as three genes that showed no difference in expression after cold treatment in both AC and LA1777 (Supplementary Fig. 2b, c). The expression patterns of these genes validated by RT-qPCR were consistent with those observed in the RNA-Seq data. Subsequently, we analyzed the 18 genes that displayed differential expression patterns and chromatin accessibilities between wild and cultivated tomatoes after cold stress. These genes, including MAPKKK86, transcription factor WRKY34, phosphatidylinositol transfer protein SFH5, and chaperone protein dnaJ, among others, play diverse roles in plant responses to cold stress, encompassing signal transduction, gene transcription, DNA repair, metabolism regulation, protein processing, and cell structure maintenance (Supplementary Data 5). The differential accessibility regions of these 18 genes were primarily located in distal intergenic regions, with four of these genes exhibiting significantly induced expression levels in LA1777 after cold treatment, while the expression of fourteen genes was significantly down-regulated following cold treatment in LA1777 (Supplementary Fig. 3 and Supplementary Data 5).
Chromatin accessibility of genes, particularly in the promoter regions, can directly impact gene transcriptional activity30. Previous studies have shown that many crucial regulatory elements, such as binding sites for most transcription factors, are typically situated within 3 kb of gene promoters31,32. Consequently, we directed our attention to genes whose differential chromatin accessibility regions were located within the 3 kb promoter region. We identified only one gene, WRKY34, that not only exhibited a differential accessibility region within 3 kb of the promoter between two accessions (AC and LA1777) under cold stress, but also corresponded to the chromatin opening region in LA1777 after cold treatment within 3 kb of the promoter (Supplementary Fig. 3 and Supplementary Data 5). The genome browser view of ATAC-Seq results for WRKY34 illustrated that the chromatin surrounding WRKY34 was predominantly closed in both AC and LA1777 plants under normal conditions. However, while chromatin remained closed in AC plants after cold stress, the chromatin within 2-3 kb upstream of the WRKY34 promoter noticeably opened in LA1777 plants after cold stress (Supplementary Fig. 3). Furthermore, the transcripts of WRKY34 exhibited minimal change in cold-sensitive cultivated tomato AC, but were significantly down-regulated in cold-tolerant wild tomato LA1777 under cold stress (Fig. 1c and Supplementary Data 5). Additionally, consistent with the transcript levels of WRKY34, we observed that WRKY34 protein accumulation in cultivated tomato AC leaves remained largely unaffected by cold stress, whereas the abundance of WRKY34 protein in wild tomato LA1777 leaves gradually declined with increasing duration of cold treatment (Fig. 1d). These results suggest that transcription factor WRKY34 may play a potential role in regulating tomato cold tolerance.
WRKY34 negatively regulates cold tolerance of tomato
Both cultivated tomato SlWRKY34 and wild tomato ShWRKY34 are situated at the end of chromosome 5 in their respective genomes. To investigate the function of WRKY34 alleles in response to cold stress in wild and cultivated tomatoes, we introduced S. habrochaites introgression line LA3942, which carries ShWRKY34 instead of SlWRKY34, along with its recurrent parent S. lycopersicum LA4024 and donor parent S. habrochaites LA1777 (Supplementary Fig. 4). We silenced SlWRKY34 (TRV-SlW34), ShWRKY34 (TRV-ShW34) in LA4024, LA3942 and LA1777 plants using virus-induced gene silencing (VIGS) technique, respectively. Silencing efficiency exceeded 65%, resulting in significantly reduced WRKY34 expression levels in silenced seedlings compared to non-silenced seedlings (TRV) (Fig. 2a). After cold stress, ShWRKY34 expression in TRV control and ShWRKY34-silenced seedlings (TRV-ShW34) of LA3942 and LA1777 was decreased significantly, while SlWRKY34 expression in TRV and SlWRKY34-silenced seedlings (TRV-SlW34) of LA4024 showed no significant difference compared to their respective control plants (Fig. 2a). Under normal conditions, no discernible phenotype change was observed in seedlings after WRKY34 silencing (Fig. 2b). Notably, TRV seedlings of LA3942 and LA1777, containing the ShWRKY34 gene, exhibited greater tolerance to cold stress than TRV seedlings of LA4024, which contains the SlWRKY34 gene. This was evidenced by lower relative electrolyte leakage (REL), higher maximum photochemical efficiency of photosystem II (Fv/Fm) and higher survival rate under cold stress (Fig. 2b–d and Supplementary Fig. 5a). Silencing of ShWRKY34 in LA3942 and LA1777 still maintained strong cold tolerance in tomato seedlings. Importantly, silencing of SlWRKY34 in LA4024 significantly increased cold tolerance, resulting in lower REL, higher Fv/Fm and higher survival rate compared with its TRV under cold stress (Fig. 2b–d and Supplementary Fig. 5a).
a Expression levels of WRKY34 in introgression line LA3942, its background material LA4024, donor parent LA1777 and their WRKY34-silenced seedlings under cold stress. Total RNA was isolated from leaf samples after 6 h of cold stress. Data are presented as means ± SD of three biological replicates. The relative expression levels of WRKY34 in TRV of LA4024, LA3942 and LA1777 under normal conditions were set to “1”. b Phenotypes of LA3942, LA4024, LA1777 and their WRKY34-silenced seedlings at 7 d after cold treatment. Eight plants of each genotype and treatment were tested with similar results. Bar: 10 cm. Relative electrolyte leakage (REL, c) and maximum photochemical efficiency of photosystem II (Fv/Fm, d) in LA4024, LA3942, LA1777 and their WRKY34-silenced seedlings at 7 d after cold treatment. The false color code depicted at the bottom of the images ranges from 0 (black) to 1 (purple). Bar: 2 cm. Data are presented as means of four biological replicates ± SD (c) or means of eight leaflets from independent plants (d). e Expression levels of CBFs and CORs in LA4024, LA3942, LA1777 and their WRKY34-silenced seedlings under cold stress. Total RNA was isolated from leaf samples after 6 h of cold stress. The relative expression levels of CBFs and CORs in TRV of LA4024, LA3942 and LA1777 under normal conditions were set to “1”. At least twice experiments were repeated independently with similar results. Data are presented as means ± SD of three biological replicates. Different letters indicate significant differences (P < 0.05) according to one-way ANOVA with Duncan’s multiple range test. TRV, non-silenced seedlings; TRV-SlW34, SlWRKY34-silenced seedlings; TRV-ShW34, ShWRKY34-silenced seedlings. Source data are provided as a Source Data file.
Furthermore, we assessed the expression levels of cold responsive genes CBFs and CORs in the aforementioned tomato plant lines. As shown in Fig. 2e, under cold stress, cold-induced up-regulation of CBFs and CORs in TRV seedlings of LA3942 and LA1777 was significantly higher than that in TRV seedlings of LA4024. Silencing ShWRKY34 in LA3942 and LA1777 maintained similar expression levels of these cold responsive genes after cold treatment compared to their respective TRV control plants. Importantly, silencing of SlWRKY34 in LA4024 significantly increased the expression of these cold responsive genes after cold treatment compared to its TRV control plants (Fig. 2e). These results indicate that SlWRKY34 does not respond to cold stress and negatively regulates cold tolerance in cultivated tomato, while the expression levels of ShWRKY34 are decreased in both wild tomato and the ShWRKY34 introgression line with strong cold tolerance.
Cold-suppressed WRKY34 expression is associated with a 60 bp InDel in its promoter region
To further elucidate the role of WRKY34 in tomato cold tolerance, we compared protein sequences of WRKY34 from six different tomato species (S. lycopersicum; S. lycopersicum var. cerasiforme; S. pimpinellifolium; S. chilense; S. pennellii; S. habrochaites) through amino acid alignments. The comparison revealed that WRKY34 proteins across different tomato species were similar and conserved. For example, there are only ten amino acid differences among the six different tomato species, with merely four amino acid distinctions between cultivated tomato S. lycopersicum and wild tomato S. habrochaites (Supplementary Fig. 6a). We subsequently constructed CaMV 35S promoter-driven SlWRKY34 and ShWRKY34 overexpressing lines (OE), as well as slwrky34 mutants through CRISPR/Cas9-mediated techniques in LA4024 background. Under normal conditions, the wrky34 mutants exhibited smaller fruits and fewer seeds per fruit compared to wild-type (WT) (Supplementary Fig. 7). Moreover, tissue-specific expression analysis revealed that tomato WRKY34 is prominently expressed in roots, followed by flowers and buds, with lower expression levels observed in leaves, and the lowest expression in fruits (Supplementary Fig. 8a). Consistently, WRKY34 protein accumulation was highest in roots and lowest in fruits (Supplementary Fig. 8b). Interestingly, both overexpression of SlWRKY34 and ShWRKY34 compromised seedlings cold tolerance, evidenced by higher REL, lower Fv/Fm and lower survival rate compared to WT (Supplementary Figs. 5b and 6b–d). Conversely, slwrky34 mutants exhibited extreme tolerance to cold stress, displaying lower REL, higher Fv/Fm and higher survival rate than WT seedlings (Supplementary Figs. 5b and 6b–d). Additionally, overexpression of WRKY34 significantly suppressed the expression of cold responsive genes CBFs and CORs under cold stress, whereas the knockout of WRKY34 promoted the expression of these genes under cold stress (Supplementary Fig. 6e). Hence, both SlWRKY34 and ShWRKY34 negatively regulate tomato cold tolerance.
Next, we concluded that differences in expression patterns of WRKY34s under cold stress, rather than their protein function, dictated the disparity in cold tolerance between wild and cultivated tomatoes. To explore the reasons behind the differential expression patterns and chromatin accessibilities of WRKY34 between wild and cultivated tomatoes under cold stress, we amplified and sequenced the 3000 bp length promoter of SlWRKY34 and the 3019 bp length promoter of ShWRKY34 (Supplementary Fig. 9). Evidently, compared to the SlWRKY34 promoter of cultivated tomato, we identified a 60 bp insertion at −2315 bp upstream of the ShWRKY34 translation initiation site (ATG) in wild tomato, precisely situated in the opening chromatin regions of the ShWRKY34 promoter under cold stress in wild tomato LA1777 (Fig. 3a and Supplementary Fig. 9). Two cis-elements, W-box and GATA-box, were identified in the 60 bp InDel by PLANTCARE software (Supplementary Fig. 9). To verify whether the 60 bp InDel of the WRKY34 promoter is accountable for the variation in its expression levels under cold stress, we performed a dual-luciferase (LUC) transcriptional activation assay in tobacco. Tobacco leaves were transformed with constructs containing the LUC reporter gene driven by SlWRKY34 and ShWRKY34 promoters. Under cold stress, the SlWRKY34 promoter (pSlW34) did not obviously affect LUC activity, whereas the ShWRKY34 promoter (pShW34) significantly inhibited the expression of LUC reporter gene. Insertion of the 60 bp InDel at position −2350 in the context of the SlWRKY34 promoter (pSlW34+60bp) led to a significant decrease in LUC activity after cold stress (Fig. 3b, c). To determine whether two cis-elements are involved in cold-suppressed WRKY34 expression, we mutated W-box and GATA-box in the 60 bp InDel of the ShWRKY34 promoter (pShW34mW-box and pShW34mGATA-box) and fused them to LUC reporter constructs, respectively (Fig. 3b). The results revealed that pShW34mGATA-box restored cold-suppressed LUC activity, whereas pShW34mW-box still exhibited significant suppression of LUC activity after cold stress (Fig. 3c), suggesting that the cis-element GATA-box within the 60 bp InDel of the ShWRKY34 promoter plays a crucial role in suppressing ShWRKY34 expression under cold stress. Furthermore, we constructed a pSlW34+30bp/LUC fusion vector, wherein only a 30 bp InDel containing the GATA-box was inserted at position −2350 in the context of the SlWRKY34 promoter, and measured LUC activity. Interestingly, compared with pShW34, pSlW34+60bp and pShW34mW-box, the LUC activity of pSlW34+30bp was only partially reduced under cold stress, suggesting that both the cis-element GATA-box and the entire 60 bp InDel fragment are important for the suppression of WRKY34 transcription after cold stress (Fig. 3c).
a WRKY34 gene structure in cultivated and wild tomatoes. The gray box represents 5’ UTR, the green box represents the coding sequence, the orange box represents the promoter and the black line in the promoter represents the 60 bp InDel region. b Schematic diagram of the reporter constructs with the full-length promoters of WRKY34 or different mutated WRKY34 promoters fused to the LUC reporter gene in the vector pGreenII 0800-LUC, while the internal control REN reporter gene was driven by the CaMV 35S promoter. pSlW34, full-length promoter of SlWRKY34; pShW34, full-length promoter of ShWRKY34; pSlW34+60bp, SlWRKY34 full-length promoter inserts a 60 bp InDel from ShWRKY34 promoter; pShW34mW-box, ShWRKY34 full-length promoter mutates W-box in the 60 bp InDel; pShW34mGATA-box, ShWRKY34 full-length promoter mutates GATA-box in the 60 bp InDel; pSlW34+30bp, SlWRKY34 full-length promoter inserts a 30 bp InDel from ShWRKY34 promoter. c LUC relative activities driven by different promoters were determined in transient transgenic tobacco plants 6 h after cold stress. The LUC relative activity driven by pSlW34 in transient expressed tobacco leaves under normal conditions was set to “1”. Data are presented as means ± SD of six biological replicates. The experiment was repeated three times with similar results. Different letters indicate significant differences (P < 0.05) according to one-way ANOVA with Duncan’s multiple range test. d The 60 bp InDel of 181 cultivated tomatoes, 74 cherry tomatoes, 58 currant tomatoes and 63 wild tomatoes. e The 60 bp InDel of 63 different wild tomatoes. f The foldchange of WRKY34 expression levels under cold stress in different tomato varieties. The horizontal line in the middle indicates mean. The experiment used three biological replicates in one experiment and was repeated twice with similar results. Source data are provided as a Source Data file.
To further explore whether the 60 bp InDel has been influenced and selected by evolution and domestication, we analyzed the variation of the 60 bp InDel in 181 cultivated tomatoes (S. lycopersicum), 74 cherry tomatoes (S. lycopersicum var. cerasiforme), 58 currant tomatoes (S. pimpinellifolium) and 63 wild tomatoes including 3 S. cheesmaniae, 2 S. galapagense, 8 S. arcanum, 3 S. chmielewskii, 9 S. neorickii, 3 S. huaylasense, 4 S. corneliomulleri, 4 S. peruvianum, 12 S. chilense, 10 S. habrochaites, 4 S. pennellii and 1 S. sitiens accessions (Supplementary Data 6). Surprisingly, we observed no 60 bp insertion in WRKY34 promoters across all cultivated tomatoes, cherry tomatoes, currant tomatoes, and two wild tomato species, S. cheesmaniae and S. galapagense accessions. However, the 60 bp insertions were prevalent in 91.4% (53 out of 58) of other wild tomatoes, including all S. chmielewskii, S. neorickii, S. corneliomulleri, S. peruvianum, S. habrochaites, S. pennellii, and S. sitiens accessions, as well as 87.5% (7 out of 8) S. arcanum, 75% (9 out of 12) S. chilense, and 66.7% (2 out of 3) S. huaylasense accessions (Fig. 3d, e and Supplementary Data 6). To verify the relationship between the 60 bp InDel and WRKY34 expression in response to cold stress, we selected a subset of cultivated tomatoes, cherry tomatoes, currant tomatoes and all wild tomatoes to measure the expression levels of WRKY34 at 6 h after cold treatment. The results showed that the WRKY34 variants harboring the 60 bp deletion exhibited no significant alteration in expression after cold stress. In contrast, the expression of WRKY34 variants with the 60 bp insertion demonstrated a notable decrease after cold stress (Fig. 3f and Supplementary Fig. 10). These results suggest that the 60 bp InDel is significantly associated with cold-suppressed WRKY34 expression in all wild and cultivated tomatoes, and the 60 bp insertion in the WRKY34 promoter is prevalent in wild tomatoes, yet it has progressively vanished during the extended evolutionary transition to currant tomatoes.
SWIBs and GATA29 directly bind to the 60 bp InDel fragment of the WRKY34 promoter
To explore how the 60 bp InDel fragment represses WRKY34 expression, we employed a yeast one-hybrid (Y1H) screen using the 60 bp InDel region as bait DNA. Prey proteins from a tomato complementary DNA library, fused with the yeast GAL4 transcription activation domain (GAL4 AD), were screened. Out of 297 putative DNA-binding proteins identified, we chose a GATA family transcription factor, SlGATA29 capable of binding to the GATA-box, and two SWIB/MDM2 domain proteins, SlSWIBa and SlSWIBb, known to influence chromatin opening (Supplementary Data 7), for further analysis.
Using full-sequence constructs of these genes, we performed gene-specific Y1H assays to determine their specific binding to the aforementioned 60 bp InDel. Yeast cells containing the 60 bp bait vector and either the pGADT7-SlGATA29 vector or pGADT7-SlSWIBa/b vectors grew on the SD-Leu media with 150 ng ml−1 aureobasidin A (AbA) (SD-Leu150). Conversely, transformants lacking SlGATA29 or SlSWIBa/b failed to grow on this media (Fig. 4a). To identify the core DNA binding sites of SWIB/MDM2 domain proteins SlSWIBa/b, we created six different 60 bp mutation probes with various mutation sites (mu1-mu6) and performed electrophoretic mobility shift assay (EMSA) and microscale thermophoresis (MST). The results revealed mu4 (TGATAA) as the common core DNA binding site of SlSWIBa and SlSWIBb, consistent with GATA-box; hence, we named it as SWIB-mu4 (Supplementary Fig. 11). Transformants with mutated GATA-box or SWIB-mu4 (mutGATA-box or mutSWIB-mu4) bait vectors did not grow on SD-Leu150 media (Fig. 4a), indicating that SlGATA29 and SlSWIBa/b could bind to the GATA-box and SWIB-mu4 of the 60 bp InDel in yeast, respectively. EMSA results further confirmed that SlGATA29 and SlSWIBa/b bound to the GATA-box and SWIB-mu4 in the 60 bp InDel, respectively (Fig. 4b). However, SlSWIBa/b could not bind to a 30 bp probe with a complete SWIB-mu4, indicating that the binding of SlSWIBa/b to the 60 bp InDel requires not only the SWIB-mu4 but also the full-length 60 bp DNA fragment (Fig. 4b). Previous studies have primarily considered SWIB/MDM2 domain proteins as regulators of chromatin accessibility through their influence on nucleosomes33. To date, there has been no report on the direct binding of SWIB/MDM2 proteins to chromatin DNA. To further identify the key amino acid sites in SWIBs for binding to the 60 bp InDel, we made an accurate prediction of protein-nucleic acid complexes using RoseTTAFoldNA34. Predicted results suggested that three conserved amino acids in two SlSWIB homologous proteins, including Arg6 in both SlSWIBa and SlSWIBb, Leu46 in SlSWIBa and Leu44 in SlSWIBb, and Lys88 in SlSWIBa and Lys86 in SlSWIBb, might be involved in binding to the 60 bp InDel through hydrogen bonds (Fig. 4c). To verify the key binding roles of these amino acids, we separately mutated these three amino acids and also mutated all three amino acids together, then tested their binding with the 60 bp InDel. Both MST and Y1H results showed that mutating any single amino acid still allowed SlSWIBa/b to bind to the 60 bp InDel, but when all three amino acids were mutated to alanine, they no longer bound to the 60 bp InDel in vitro (Fig. 4d, e). These results further indicate that SWIBs have the ability to directly bind to DNA, and we infer that they would form a helical protein to wrap around DNA, with SWIB-mu4 being the key recognition or binding site.
a Yeast one-hybrid assay (Y1H) testing the binding of SlGATA29 and SlSWIBa/b to pAbAi-60bp in the SD-Leu medium with or without 150 ng ml-1 AbA. Empty vector containing the AD serves as a negative control. b EMSA results showing SlGATA29 and SlSWIBa/b directly bind to the 60 bp InDel. Probe sequences are shown (top). muGATA-box and muSWIB-mu4, mutated probes in which the TGATAA motif was changed to AAAAAA. c Complex structures of SlSWIBa with the 60 bp DNA fragment (upper panel) and SlSWIBb with the 60 bp DNA fragment (lower panel). Blue and red indicate the SWIB/MDM2 domain and the predicted binding site, respectively. SWIB-mu4 is highlighted in red. d MST assays of the binding of SlSWIBa/b and their three amino acid mutations to the 60 bp InDel. Kd, dissociation constant. e Y1H testing the binding of SlSWIBa/b and their three amino acid mutations to pAbAi-60bp in the SD-Leu medium with or without 150 ng ml−1 AbA. f Transient dual-luciferase (dual-LUC) assay in tobacco leaves. Empty vector (EV) was included as control. Data are presented as means ± SD of six biological replicates. ChIP-qPCR assays showing the binding of SlGATA29 (g) and SlSWIBb (h) to the 60 bp InDel in vivo. Data are presented as means ± SD of three biological replicates. i Expression levels of WRKY34 in LA4024, LA3942 and SlGATA29 overexpressing (SlGATA29-OE) seedlings and SlSWIBb overexpressing (SlSWIBb-OE) seedlings in the background of LA4024 or LA3942 under cold stress. Total RNA was isolated from leaf samples after 6 h of cold stress. Data are presented as means ± SD of three biological replicates. The relative expression levels of WRKY34 in WT of LA4024 and LA3942 under normal conditions were set to “1”. Experiments in (a, b, d, e) were repeated twice, and in (f–i), three times, all yielding similar results. Different letters above bars indicate significant differences (P < 0.05) determined using one-way ANOVA with Duncan’s multiple range test. Source data are provided as a Source Data file.
To investigate the effect of SlGATA29 and SlSWIBa/b on downstream WRKY34 gene expression, we performed dual-LUC assays and found that SlGATA29 suppressed LUC activity when WRKY34 promoters contained the GATA-box (pShW34, pSlW34+60bp, pShW34mW-box and pSlW34+30bp), but had little effect on WRKY34 promoters without the GATA-box (pSlW34 and pShW34mGATA-box). Interestingly, when the reporter gene was co-transfected with SlSWIBa or SlSWIBb gene, the LUC activity of all WRKY34 promoters did not change, suggesting that SlSWIBs do not directly regulate WRKY34 expression (Fig. 4f). To verify the binding of SlGATA29 and SlSWIBs to the 60 bp InDel in vivo, we overexpressed SlGATA29 and SlSWIBb in cultivated tomato LA4024 and introgression line LA3942, respectively. ChIP-qPCR analysis showed that SlGATA29 and SlSWIBb could not bind to the SlWRKY34 promoter of LA4024 with or without cold treatment, due to the absence of the 60 bp InDel. SlGATA29 could not bind to the ShWRKY34 promoter of LA3942 under normal conditions, but could bind to its promoter under cold stress (Fig. 4g). At normal temperature, SlSWIBb could directly bind to the ShWRKY34 promoter of LA3942, and the binding was further increased after cold stress (Fig. 4h). Moreover, overexpression of SlGATA29 or SlSWIBb in LA3942 background suppressed the expression of ShWRKY34 under cold stress, while the expression of SlWRKY34 in LA4024 background did not respond to cold stress (Fig. 4i). Additionally, we compared the protein sequences of GATA29 and SWIBs between cultivated tomato S. lycopersicum and wild tomato S. habrochaites. The comparison results showed only one amino acid difference between SlGATA29 and ShGATA29 (Supplementary Fig. 12a). Compared with two homologous SlSWIBa and SlSWIBb, only one ShSWIB was identified in S. habrochaites LA1777. The protein sequence homology of SlSWIBa and SlSWIBb was 68.6%, and the protein sequence homology of ShSWIB and SlSWIBb was 99.2% with only one amino acid difference (Supplementary Fig. 12b). Y1H and EMSA results also demonstrated that ShGATA29 and ShSWIB directly bind the 60 bp InDel of WRKY34 promoter by specifically interacting with GATA-box and SWIB-mu4, respectively. Additionally, the binding of ShSWIB to the 60 bp InDel also requires a full-length 60 bp DNA (Supplementary Fig. 12c, d). Moreover, dual-LUC assays indicated that ShGATA29 suppressed LUC activity when WRKY34 promoters contained the GATA-box (pShW34, pSlW34+60bp, pShW34mW-box and pSlW34+30bp), but had little effect on WRKY34 promoters lacking the GATA-box (pSlW34 and pShW34mGATA-box). However, the LUC activity derived from all WRKY34 promoters did not exhibit LUC suppression when the reporter was co-transfected with ShSWIB (Supplementary Fig. 12e). These findings suggest that both the transcription factor GATA29 and the SWIB/MDM2 domain protein SWIBs, found in both wild and cultivated tomatoes, can bind to the 60 bp InDel fragment of the WRKY34 promoter.
SWIB and GATA29 synergistically suppress WRKY34 expression through the 60 bp InDel fragment
Chromatin remodeling factors are typically recruited to target genes via specific transcription factors, thereby synergistically regulating the expression of target genes35. Interestingly, using yeast two-hybrid (Y2H) assays, we found that SlGATA29 interacted with both SlSWIBa and SlSWIBb in yeast (Fig. 5a). To further validate this interaction, we performed a glutathione S-transferase (GST) pull-down assay. The results revealed that SlGATA29-GST successfully pulled down SlSWIBa-His or SlSWIBb-His, while the negative control GST failed to do so (Fig. 5b). Similarly, a bimolecular fluorescence complementation (BiFC) assay confirmed the interaction of SlGATA29 with SlSWIBa and SlSWIBb in the nucleus (Fig. 5c). Furthermore, the interaction between SlGATA29 and SlSWIBs was verified by co-immunoprecipitation (Co-IP) assays in tobacco leaves through co-expressing SlGATA29-HA and SlSWIBa-GFP or SlGATA29-HA and SlSWIBb-GFP (Fig. 5d). These results indicate that SlGATA29 interacts with both SlSWIBa and SlSWIBb in vitro and in vivo.
a Yeast two-hybrid assay (Y2H) testing the interactions of SlGATA29 and SlSWIBa/b in yeast. b Pull-down assays show that GST-tagged SlGATA29 physically interacts with His-tagged SlSWIBa or His-tagged SlSWIBb. The green triangle indicates the target band of SlSWIBa-His or SlSWIBb-His pulled down by SlGATA29-GST. The combination of GST and SlSWIBa-His or SlSWIBb-His was used as a negative control. c Bimolecular fluorescence complementation assay (BiFC) showing the interactions between SlGATA29 and SlSWIBa/b in tobacco. Bar: 50 μm. d Co-IP showing the interactions between SlGATA29 and SlSWIBa/b. The green triangle indicates the SlGATA29-HA target band immunoprecipitated with SlSWIBa-GFP or SlSWIBb-GFP. The combination of GFP and SlGATA29-HA was used as a negative control. e Transient dual-luciferase (dual-LUC) assay in tobacco leaves. Empty vector (EV) was included as control. Data are presented as means ± SD of six biological replicates. f ChIP-qPCR assays show that silencing SlSWIBab affects the binding of SlGATA29 to the 60 bp InDel in vivo. Data are presented as means ± SD of three biological replicates. g Expression levels of WRKY34 in SlGATA29 overexpressing lines of LA4024 or LA3942 background and their SlSWIBab-silenced seedlings under cold stress. Total RNA was isolated from leaf samples after 6 h of cold stress. Data are presented as means ± SD of three biological replicates. The relative expression levels of WRKY34 in TRV of SlGATA29 overexpressing lines of LA4024 or LA3942 background under normal conditions were set to “1”. In (a–d), experiments were repeated twice with similar results. In (e–g), experiments were repeated three times with similar results. Different letters above bars indicate significant differences (P < 0.05) determined using one-way ANOVA with Duncan’s multiple range test. TRV, non-silenced seedlings; TRV-SlSWIBab, SlSWIBa and SlSWIBb co-silenced seedlings; SlGATA29-OE/LA4024, SlGATA29 overexpressing lines in LA4024 background; SlGATA29-OE/LA3942, SlGATA29 overexpressing lines in LA3942 background. Source data are provided as a Source Data file.
To investigate the effect of the interaction between SlGATA29 and SlSWIBs on downstream WRKY34 gene expression, we performed transient dual-LUC assays with different types of WRKY34 promoters. LUC activity derived from WRKY34 promoters containing the GATA-box (pShW34, pSlW34+60bp, pShW34mW-box and pSlW34+30bp) decreased significantly when co-transfected with SlGATA29, and this reduction was further augmented after co-transfection with SlSWIBa or SlSWIBb, except for pSlW34+30bp. Conversely, LUC activity derived from WRKY34 promoters lacking the GATA-box (pSlW34 and pShW34mGATA-box) showed no change when co-transfected with either SlGATA29 alone or with both SlGATA29 and SlSWIBs (Fig. 5e).
To further examine the synergistic effects of SlGATA29 and SlSWIBs, we performed EMSA in vitro. As shown in Supplementary Fig. 13, while SlGATA29 could specifically bind to 60 bp probe containing the GATA-box, the addition of purified SlSWIBa or SlSWIBb proteins did not significantly affect its binding ability. Therefore, we speculated that the synergistic suppression effect of SlGATA29 and SlSWIBa/b on WRKY34 expression might be linked to chromatin accessibility, and such changes in chromatin accessibility require a eukaryotic environment. As shown in Fig. 5f, in LA4024 where the SlWRKY34 promoter lacks the 60 bp InDel, there was no binding of SlGATA29 to the SlWRKY34 promoter in both TRV and SlSWIBa and SlSWIBb co-silenced seedlings (TRV-SlSWIBab) of SlGATA29 overexpressing lines. Conversely, in LA3942 background, where ShWRKY34 promoter contains the 60 bp InDel fragment, significant binding of SlGATA29 to the ShWRKY34 promoter was observed in TRV of SlGATA29 overexpressing lines. However, when SlSWIBa and SlSWIBb were co-silenced, the binding of SlGATA29 to the ShWRKY34 promoter containing the 60 bp InDel fragment was significantly reduced (Fig. 5f). Correspondingly, the expression of SlWRKY34 in LA4024 background remained unchanged, whereas the expression of ShWRKY34 in SlGATA29 overexpressing lines of LA3942 background was significantly decreased under cold stress. Silencing SlSWIBab in SlGATA29 overexpressing lines of LA3942 background significantly alleviated the decrease in WRKY34 expression under cold stress (Fig. 5g). These results suggest that SlSWIBs can enhance the suppression effect of SlGATA29 on WRKY34 expression, and a complete 60 bp InDel in the WRKY34 promoter is indispensable for achieving the synergistic suppression effect of SlGATA29 and SlSWIBs.
Given the high protein sequence similarity between SlGATA29 and ShGATA29, as well as between SlSWIBs and ShSWIB, Y2H also detected the interaction between ShGATA29 and ShSWIB in yeast (Supplementary Fig. 14a). We further verified the interaction of ShGATA29 and ShSWIB by performing pull-down assays in vitro (Supplementary Fig. 14b). BiFC and Co-IP results also confirmed the interaction between ShGATA29 and ShSWIB in vivo (Supplementary Fig. 14c, d). Similarly, we performed dual-LUC assays to investigate the synergistic suppression effect of ShGATA29 and ShSWIB (Supplementary Fig. 14e). The results showed that ShSWIB and ShGATA29 exhibit similar functions with SlSWIBs and SlGATA29 in synergistic suppression of WRKY34 expression.
Impact of SWlB and GATA29 on chromatin accessibility and cold tolerance via the 60 bp InDel
To further elucidate the effects of SWIBs on chromatin accessibility associated with the 60 bp InDel fragment of WRKY34 promoter, we constructed SlSWIBb overexpressing lines driven by the CaMV 35S promoter (SlSWIBb-OE) and slswibab double mutants mediated by CRISPR/Cas9 in the background of LA4024 or LA3942. In an independent ATAC-qPCR assay, the region around the 60 bp InDel fragment of the WRKY34 promoter exhibited an increase in chromatin accessibility following cold treatment in LA3942 background (Fig. 6a). In contrast, the chromatin accessibility for this region did not change in response to cold stress in LA4024 background. Furthermore, in the slswibab double mutants of LA3942 background, the chromatin accessibility of this region was also lost in response to cold stress (Fig. 6a). Interestingly, overexpression of SlSWIBb in LA3942 background significantly increased chromatin accessibility of this region, and this region was more accessible under cold stress in LA3942 SlSWIBb-OE lines (Fig. 6a). RT-qPCR analysis revealed that WRKY34 did not respond to cold stress in the background of LA4024, consistent with the results of chromatin accessibility. Meanwhile, WRKY34 expression in LA3942 seedlings was significantly decreased after cold treatment, while WRKY34 expression in slswibab double mutants of LA3942 background remained unchanged regardless of cold treatment (Fig. 6b). Overexpression of SlSWIBb in LA3942 background did not affect the expression of WRKY34 under normal conditions. However, the expression of WRKY34 in SlSWIBb-OE lines of LA3942 background was significantly suppressed and lower than that in WT LA3942 seedlings after cold stress (Fig. 6b). Consistent with the expression of WRKY34, knockout of SlSWIBa and SlSWIBb or overexpression of SlSWIBb in LA4024 background had no effect on the cold tolerance of LA4024, as indicated by similar REL, Fv/Fm and survival rate (Fig. 6c–e and Supplementary Fig. 5c). However, slswibab double mutants of LA3942 exhibited compromised cold tolerance compared to WT, with higher REL, lower Fv/Fm and lower survival rate. In contrast, overexpression of SlSWIBb enhanced the cold tolerance of LA3942 resulting in lower REL, higher Fv/Fm and higher survival rate (Fig. 6c–e and Supplementary Fig. 5c).
a ATAC-qPCR results from testing chromatin accessibility in the 60 bp InDel region of the WRKY34 promoter under cold stress in slswibab double mutants and SlSWIBb overexpressing (SlSWIBb-OE) seedlings of LA4024 or LA3942 background. Data are presented as means ± SD of three biological replicates. b Expression levels of WRKY34 in slswibab double mutants and SlSWIBb-OE seedlings of LA4024 or LA3942 background under cold stress. Total RNA was isolated from leaf samples after 6 h of cold stress. Data are presented as means ± SD of three biological replicates. The relative expression levels of WRKY34 in WT of LA4024 and LA3942 under normal conditions were set to “1”. c Phenotypes of slswibab double mutants and SlSWIBb-OE seedlings in LA4024 or LA3942 background at 7 d after cold treatment. Eight plants of each genotype and treatment were tested with similar results. Bar: 10 cm. Relative electrolyte leakage (REL, d) and maximum photochemical efficiency of photosystem II (Fv/Fm, e) in slswibab double mutants and SlSWIBb-OE seedlings of LA4024 or LA3942 background at 7 d after cold treatment. The false color code depicted at the bottom of the images ranges from 0 (black) to 1 (purple). Bar: 2 cm. Data are presented as means of four biological replicates ± SD (d) or means of eight leaflets from independent plants (e). f CRISPR/Cas9-induced deletions in the 60 bp InDel region of the WRKY34 promoter of LA3942. Blue-marked area represents PAM, green-marked area indicates SgRNA, yellow-marked area represents the 60 bp InDel, and red-marked area represents the SWIB-mu4/GATA-box. g ATAC-qPCR results from testing chromatin accessibility in the 60 bp InDel region of the WRKY34 promoter in response to cold stress in different types of 60 bp InDel deletion mutants. Data are presented as means ± SD of three biological replicates. All experiments were repeated twice with similar results. Different letters above bars indicate significant differences (P < 0.05) determined using one-way ANOVA with Duncan’s multiple range test. Source data are provided as a Source Data file.
We also constructed SlGATA29 overexpressing lines driven by the CaMV 35 S promoter (SlGATA29-OE) and slgata29 mutants mediated by CRISPR/Cas9 in the background of LA4024 or LA3942. As shown in Supplementary Fig. 15a, the expression of WRKY34 showed no response to cold stress in LA4024 background. In contrast, the expression of WRKY34 in LA3942 background was significantly suppressed under cold stress, while cold-suppressed WRKY34 expression was compromised when SlGATA29 was knocked out in LA3942 background (Supplementary Fig. 15a). Conversely, overexpression of SlGATA29 further suppressed WRKY34 expression in LA3942 background under cold stress (Supplementary Fig. 15a). Consistent with the expression of WRKY34, SlGATA29 knockout or overexpression in LA4024 background had no effect on the cold tolerance of LA4024, as indicated by similar REL, Fv/Fm and survival rate (Supplementary Figs. 5d and 15b–d). However, slgata29 mutants of LA3942 exhibited compromised cold tolerance compared to WT, as indicated by higher REL, lower Fv/Fm and lower survival rate. In contrast, overexpression of SlGATA29 enhanced cold tolerance of LA3942, resulting in lower REL, higher Fv/Fm and higher survival rate (Supplementary Figs. 5d and 15b–d).
To explore the critical role of the 60 bp InDel in altering chromatin accessibility, we used the CRISPR/Cas9 to delete the 60 bp in the ShWRKY34 promoter of LA3942. We obtained two lines (named Cri-60bp-1 and Cri-60bp-2) with different deletions within the 60 bp InDel region (Fig. 6f). Specifically, Cri-60bp-1 exhibits a deletion of only 6 bp at the 3’ end of the 60 bp InDel, whereas Cri-60bp-2 has a more extensive deletion of 35 bp, which disrupts the crucial SWIB-mu4/GATA-box (Fig. 6f). As expected, ATAC-qPCR assays showed that compared with WT, the chromatin accessibility of the 60 bp InDel region was significantly weakened in Cri-60bp-2 under cold stress, while the chromatin accessibility of this region in Cri-60bp-1 was almost unchanged (Fig. 6g). Moreover, the repression effect of WRKY34 expression under cold stress was abolished in Cri-60bp-2, but not in Cri-60bp-1 (Supplementary Fig. 16a). Consistently, Cri-60bp-2 plants were more sensitive to cold stress than WT, exhibiting a significant increase in REL and a decrease in Fv/Fm under cold stress (Supplementary Fig. 16b–d). These observations confirm that the 60 bp InDel, regulated by SWIB and GATA29, is a major causal variation underlying the differential expression and chromatin accessibility of WRKY34 under cold stress.
WRKY34 disrupts the CBF-COR cold response pathway at both transcript and protein levels
WRKY34 overexpression or mutation can influence the expression of CBFs and CORs in LA4024 after cold stress (Supplementary Fig. 6e). To further analyze whether WRKY34 affects the CBF-COR cold response pathway, we detected the protein interaction between WRKY34 and CBFs or CORs. Interestingly, SlWRKY34 can interact with SlCBF1 but not SlCBF2/3 or the other three SlCORs with its C terminal domain (Fig. 7a). Moreover, the C terminal domain of ShWRKY34 also interacted with ShCBF1 or SlCBF1, but not with ShCBF2/3, ShCORs, SlCBF2/3 and SlCORs (Supplementary Fig. 17). Using GST-pull down assays, we demonstrated that SlWRKY34-GST pulled down SlCBF1-His, while the negative control GST failed to do so (Fig. 7b). BiFC results showed that the full-length SlWRKY34 and SlCBF1 proteins interacted in the nucleus (Fig. 7c). Co-IP results revealed that SlWRKY34-HA associated with SlCBF1-GFP, but not with free GFP (Fig. 7d). These results confirm the interaction between SlWRKY34 and SlCBF1.
a Yeast two-hybrid assay (Y2H) testing the interactions of SlWRKY34 with SlCBF1 in yeast. SlW34-C, C terminal of SlWRKY34 protein. b Pull-down assays show that GST-tagged SlWRKY34 physically interacts with His-tagged SlCBF1. The green triangle indicates the target band of SlCBF1-His pulled down by SlW34-GST. The combination of GST and SlCBF1-His was used as a negative control. c Bimolecular fluorescence complementation assay (BiFC) showing the interactions between SlWRKY34 and SlCBF1 in tobacco. Bar: 50 μm. d Co-IP showing the interactions between SlWRKY34 and SlCBF1. The green triangle indicates the SlW34-HA target band immunoprecipitated with SlCBF1-GFP. The combination of GFP and SlW34-HA was used as a negative control. e EMSA show that SlWRKY34 interferes with the binding of SlCBF1 to DRE elements on downstream SlCBF1 and SlCOR47 promoters. Unlabeled wild type probes were used as cold probes. mu, mutated probes in which the DRE elements (5ʹ-G/ACCGAC-3ʹ) were replaced with 5ʹ-AAAAAA-3ʹ. f Transient dual-luciferase (dual-LUC) assay in tobacco leaves. Empty vector (EV) was included as control. Data are presented as means ± SD of six biological replicates. g Phenotypes of slwrky34 mutant in LA4024 background (slw34/LA4024) and its SlCBF1-silenced seedlings at 7 d after cold treatment. Eight plants of each genotype and treatment were tested with similar results. Bar: 10 cm. Relative electrolyte leakage (REL, h) and maximum photochemical efficiency of photosystem II (Fv/Fm, i) in slwrky34 mutant of LA4024 background (slw34/LA4024) and its SlCBF1-silenced seedlings at 7 d after cold treatment. The false color code depicted at the bottom of the images ranges from 0 (black) to 1 (purple). Bar: 2 cm. Data are presented as means of four biological replicates ± SD (h) or means of eight leaflets from independent plants (i). Experiments in (a–e) were repeated twice, and in (f–i), three times, all yielding similar results. Different letters above bars indicate significant differences (P < 0.05) determined using one-way ANOVA with Duncan’s multiple range test. TRV, non-silenced seedlings; TRV-SlCBF1, SlCBF1-silenced seedlings. Source data are provided as a Source Data file.
Many previous studies have shown that the CBF-COR pathway is central to plant cold tolerance36. We hypothesized that SlWRKY34 interferes with the transcriptional function of SlCBF1 under cold stress by interacting with SlCBF1, thus weakening the cold tolerance of cultivated tomato. To test this hypothesis, we conducted EMSA and dual-LUC assays. As shown in Supplementary Fig. 18a, several CBF binding elements, known as Dehydration-Responsive Elements (DREs), were identified in the promoters of SlCBF1 and SlCOR47. SlCBF1 directly bound to DRE elements in SlCBF1 and SlCOR47 promoters in vitro; however, SlCBF1-bound probe signals decreased progressively with increasing concentrations of SlWRKY34 purified proteins (Fig. 7e). Additionally, SlCBF1 could transcriptionally activate the expression of itself and SlCOR47, while SlWRKY34 co-transfected with SlCBF1 significantly impaired SlCBF1-induced SlCBF1 and SlCOR47 expression (Fig. 7f). Furthermore, silencing SlCBF1 in slwrky34 mutants significantly reduced cold tolerance with higher REL and lower Fv/Fm under cold stress, compared with slwrky34 mutants (Fig. 7g–i).
Moreover, we identified many W-box elements in SlCBFs and SlCOR47 promoters (Supplementary Fig. 18a). Next, we conducted Y1H and ChIP-qPCR assays, respectively. Y1H results showed that SlWRKY34 could directly bind to the promoters of SlCBFs and SlCOR47 in yeast cells (Supplementary Fig. 18b). ChIP-qPCR analysis showed that SlWRKY34 could directly bind to the promoters of SlCBFs and SlCOR47 in vivo under cold stress (Supplementary Fig. 18c). Furthermore, dual-LUC results indicated that SlWRKY34 could transcriptionally repress the expression of SlCBFs and SlCOR47 (Supplementary Fig. 18d). Thus, WRKY34 also directly suppresses gene transcription in the CBF-COR pathway.
Discussion
The tomato likely originated in the Andes mountains of South America and adjacent tropical regions37. The diverse range of climatic and ecological conditions present across these areas has been instrumental in driving the diversification of tomatoes and their botanical relatives. Phylogenetic analyses have classified wild tomatoes into several groups: “Lycopersicon group” (S. pimpinellifolium, S. cheesmaniae, and S. galapagense), “Arcanum group” (S. arcanum, S. chmielewskii, and S. neorickii), “Eriopersicon group” (S. habrochaites, S. huaylasense, S. corneliomulleri, S. peruvianum, and S. chilense), “Neolycopersicon group” (S. pennellii); and two outgroups: Section Juglandifolia (S. juglandifolium and S. ochranthum) and Section Lycopersicoides (S. lycopersicoides and S. sitiens)38. Each group adapts to specific altitudes and average temperatures, reflecting the influence of environmental factors on their evolutionary paths38. In this study, we found a significant correlation between a 60 bp InDel in the WRKY34 promoter and WRKY34 expression under cold stress (Fig. 3). Specifically, the WRKY34 promoter in cultivated tomatoes, cherry tomatoes and “Lycopersicon group” of wild tomatoes exhibited this 60 bp deletion, while over 90% of other wild tomato groups contained the 60 bp insertion (Fig. 3d, e). Heatmap analysis revealed that the expression of WRKY34 variant with the 60 bp deletion showed no significant change post cold stress, whereas WRKY34 variant with the 60 bp insertion exhibited a marked decrease under cold stress (Supplementary Fig. 10). Notably, two wild species, S. cheesmaniae and S. galapagense, did not contain the 60 bp insertion (Fig. 3e and Supplementary Fig. 10), possibly due to their warm growing environment in low-altitude areas39,40. The S. habrochaites introgression line LA3942, which contains a single introgression fragment where ShWRKY34 replaces SlWRKY34, shows better cold tolerance than its recurrent parent S. lycopersicum LA4024 (Fig. 2). Thus, the full 60 bp InDel can be introduced into cultivated and cherry tomatoes through backcrossing and other breeding technologies to improve their cold tolerance.
Unlike changes in coding sequences, variations in cis-regulatory regions can alter gene expression in response to environmental cues and developmental processes without changing the protein they encode6. During evolution and domestication, certain variations in cis-regulatory regions can confer advantageous traits, such as enhanced yields, stress tolerance, or nutrient content. For instance, research on cotton (Gossypium hirsutum) has identified changes in cis-regulatory regions that have significantly altered gene expression during domestication, contributing to the development of desirable fiber traits41. One specific study identified a 26 bp InDel in the 5’ UTR of ZmGLK36, which modulated its expression and thus the plant’s resistance to maize rough dwarf virus (RBSDV), highlighting a key genetic adaptation for crop improvement in the face of disease challenges42. Previously, we found that a key W-box single nucleotide polymorphism (SNP) affects the self-transcriptional regulation and protein accumulation of WRKY33 under cold stress in cultivated tomato, thus contributing to the cold sensitivity of cultivated tomatoes compared with wild tomatoes7. Here, we found that the WRKY34 promoter in cold-tolerant wild tomato species contains a 60 bp insertion, which directly causes its chromatin to open and recruits the transcriptional suppressor GATA29 under cold stress, thereby diminishing WRKY34 expression and enhancing cold tolerance. Conversely, the absence of this 60 bp segment in the WRKY34 promoter of cultivated tomatoes leads to a reduced response to cold stress, contributing to the cold sensitivity observed in these domesticated tomato varieties. Both of our studies discovered SNP or InDel variations within the promoter regions of WRKY family transcription factors, leading to changes in gene expression levels and thereby affecting cold tolerance in tomatoes. Specifically, the mutation in WRKY33 resulted in the loss of a promoter cis-element during tomato evolution, while the mutation in WRKY34 involved the deletion of a fragment within the promoter. Although the functions and mechanisms of WRKY33 and WRKY34 in response to cold stress are completely different, variations in their promoters both result in decreased cold tolerance in cultivated tomato. Therefore, natural variation in the multiple genes related to cold tolerance may have occurred during the evolution of cultivated tomatoes and were preserved during domestication, resulting in an overall phenotype of cold sensitivity in cultivated tomatoes. Future research should explore how technologies such as gene editing can restore these natural variations and improve cold tolerance in cultivated tomatoes without altering other traits.
The role of SWIB/MDM2 domain-containing proteins in chromatin remodeling is increasingly recognized43,44. Typically found in SWI/SNF chromatin remodeling complexes, the SWIB/MDM2 domain is known to regulate gene transcriptional activity by altering nucleosome positioning43. This regulation can either be specific, affecting certain genes, or broadly influencing chromatin states across the genome. However, our study reveals a more direct and precise mechanism of SWIB/MDM2 domain proteins in gene regulation. We observed that under normal conditions, SWIBs were inactive, not opening chromatin or recruiting transcription factors, resulting in the low expression of WRKY34. Nevertheless, under cold stress, SWIBs not only opened chromatin and recruited the transcription factor GATA29, but also directly bound to specific sites in the WRKY34 promoter, thereby precisely inducing chromatin opening in specific regions and recruiting transcription factor GATA29 to bind to the GATA-box within the 60 bp region, further suppressing the expression of the WRKY34 gene (Figs. 4–6). Even more excitingly, through protein structure prediction and experimental validation, we have demonstrated that three evolutionarily conserved amino acids in SWIBs are involved in DNA binding, with the key DNA binding site being TGATAA. We hypothesize that proteins of this family can recognize the TGATAA motif and then form a helical tripod structure to wrap around DNA, thereby further exerting their function. Therefore, the discovery about SWIBs extends beyond the traditional understanding of their role in altering nucleosome positions through chromatin remodeling and recruiting transcription factors. It reveals that these proteins can also directly bind to specific DNA sites through three evolutionarily conserved amino acids. This direct DNA binding capacity allows SWIB/MDM2 domain proteins to be key players in activating genes in chromatin-closed regions, enhancing the precision of specific gene expression control. Moreover, it demonstrates the ability of SWIB/MDM2 domain proteins to respond to specific environmental signals and regulate gene expression by acting directly on specific DNA elements, further emphasizing their versatility and importance in gene regulation. Beyond direct impacts on gene transcription, SWIB/MDM2 domain proteins are closely related to histone modifications and epigenetic regulation45. It has been reported that SWIB domain proteins might directly recognize and bind to specific histones or their modified forms, thereby altering nucleosome stability or regulating interactions with other chromatin-associated proteins. For example, under normal conditions, SWP73A repressed NLR (NOD-like receptor) gene RPS2 through H3K9me2 modification, while this repression reduced or eliminated following pathogen infection, facilitating gene transcription and the activation of plant innate immunity46. However, the association of the SWIB/MDM2 domain proteins in our study with histone modifications warrants further investigation.
Research on WRKY34 in Arabidopsis indicates that it is specifically expressed in pollen, negatively regulates the cold tolerance of mature pollen, and may be involved in the CBF signal cascade in mature pollen47. In our study, WRKY34 also negatively impacts cold tolerance in tomato seedlings, primarily by interfering with the classic CBF-COR cold response pathway at both transcription and protein levels (Fig. 7 and Supplementary Fig. 18). WRKY34 can directly bind to W-box elements in the promoters of CBFs and CORs, transcriptionally repressing their expression. Additionally, WRKY34 interacts with CBF1, disrupting its transcriptional activation of itself and downstream CORs. Notably, while knocking out WRKY34 enhanced cold tolerance, the wrky34 mutants exhibited developmental defects, such as smaller fruits and fewer seeds per fruit than WT (Supplementary Fig. 7). Moreover, the expression and protein accumulation of WRKY34 were the highest in roots, followed by flowers and buds, with lower expression and protein accumulation in leaves and fruits (Supplementary Fig. 8). This emphasizes WRKY34’s necessity under normal conditions and suggests that complete functional loss isn’t a desirable improvement approach for cold tolerance. Gene functions are multifaceted. Although knocking out or overexpressing genes can achieve desired traits, it may also disrupt other characteristics, like growth and development. Precisely regulating gene transcription through specific promoter control is vital in crop breeding, as this approach effectively balances the enhancement of desired traits with the overall health and growth of the plant. For example, a recent study utilized a gene-editing strategy targeting the SlPIF4 binding motif in the SlCOMT2 promoter, effectively enhancing melatonin levels in tomato fruit during the ripening stage without impacting other developmental phases. In addition, this targeted approach achieved higher melatonin content and no growth defects compared to pif4 knockout mutants, demonstrating the efficacy of precise genetic modulation in crop development48. Here, we identified a 60 bp insertion in the WRKY34 promoter that diminished its expression under cold stress in Solanum species, thus boosting cold tolerance. Through multiple generations of backcrossing, we have also developed the ShWRKY34 introgression tomato line LA3942, which exhibits cold tolerance without impacting other traits. Additionally, considering that variations in cis-regulatory regions typically exert subtler phenotypic impacts and circumvent the adverse effects of coding region mutations, we advocate employing gene-editing techniques to incorporate this 60 bp sequence into the WRKY34 promoter of Solanum plants lacking this sequence.
Accessibility is generally positively correlated with expression, but examples of increased chromatin accessibility and decreased gene expression have been reported. For example, by investigating chromatin modifications and accessibility, a study suggests that although type A ARF exhibits an open chromatin configuration, it is regulated by a network of transcriptional repressors49. Therefore, the relationship between chromatin accessibility and gene expression is complex and influenced by multiple factors. Here, our results demonstrate one of these mechanisms and we thus propose a working model of WRKY34-mediated cold tolerance in wild and cultivated tomatoes (Fig. 8). Under cold stress, the presence of a 60 bp insertion in the WRKY34 promoter of wild tomato S. habrochaites leads to the binding of chromatin remodeling factor SWIBs, thereby opening chromatin in the nearby region and recruiting transcriptional repressor GATA29 to bind to the GATA-box within the 60 bp, resulting in repression of WRKY34 expression. WRKY34 interferes with CBF1-induced expression of itself and CORs by interacting with CBF1. Furthermore, WRKY34 directly binds to the promoters of downstream CBFs and CORs and represses their expression under cold stress. However, the deletion of the 60 bp DNA fragment in the WRKY34 promoter of cultivated tomatoes results in its inability to bind SWIBs under cold stress, preventing chromatin opening and recruitment of GATA29, and thus failing to suppress WRKY34 expression and contributing to the cold sensitivity of these tomatoes. Three additional points deserve to be mentioned. Firstly, ATAC-Seq data indicates that wild tomato LA1777 exhibits more chromatin opening under cold stress than cultivated tomato AC, suggesting potential functional differences in chromatin remodeling factors other than SWIBs between wild and cultivated tomatoes, which may impact cold tolerance. Secondly, in addition to the regulatory differences caused by non-coding regions, the differences in coding regions between wild and cultivated tomatoes and their potential impact on resistance traits warrant further investigation.
The presence of the 60 bp InDel in the WRKY34 promoter of wild tomato S. habrochaites leads to the binding of chromatin remodeling factor SWIBs, thereby opening chromatin in the nearby region, and recruiting more transcriptional repressor GATA29 to bind to the GATA-box within the 60 bp, resulting in repression of WRKY34 expression. In addition, WRKY34 interferes with the transcriptional regulation of CBF1 to itself and CORs by interacting with CBF1. On the other hand, WRKY34 directly binds downstream CBFs and CORs promoters and represses their expression under cold stress. However, the deletion of the 60 bp DNA fragment in the WRKY34 promoter of cultivated tomatoes results in its inability to bind SWIBs under cold stress, preventing chromatin opening and recruitment of GATA29, and thus failing to suppress WRKY34 expression and contributing to the cold sensitivity of these tomatoes. Figure 8 Created with BioRender.com released under a Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International license.
Methods
Plant materials and growth conditions
Wild tomato (S. habrochaites accession LA1777) and cultivated tomato (S. lycopersicum cv. Ailsa Craig, AC) were used for RNA-Seq and ATAC-Seq. The S. habrochaites introgression line LA3942, containing a single introgression fragment with ShWRKY34 replacing SlWRKY34, along with its recurrent parent S. lycopersicum LA4024 and donor parent LA1777, was used for virus-induced gene silencing (VIGS) of WRKY34 genes. LA4024 and LA3942 were selected for genetic transformation.
A total of 376 tomato accessions were collected from various sources, including Tomato Genetics Resource Center (TGRC), United State Department of Agriculture (USDA), University of Florida, and European Union Solanaceae Project (EU-SOL). These accessions include 63 wild tomato accessions (3 S. cheesmaniae, 2 S. galapagense, 8 S. arcanum, 3 S. chmielewskii, 9 S. neorickii, 3 S. huaylasense, 4 S. corneliomulleri, 4 S. peruvianum, 12 S. chilense, 10 S. habrochaites, 4 S. pennellii and 1 S. sitien), 58 S. pimpinellifolium, 74 S. lycopersicum var. cerasiforme, and 181 S. lycopersicum accessions (Supplementary Data 6).
Seeds were germinated on moistened filter paper at 28 °C in the dark and subsequently sown in 72-cell plastic flats filled with a mixture of peat and vermiculite (3:1, v:v). Upon reaching the two-leaf stage, seedlings were transplanted into plastic pots (10 cm × 10 cm in height × diameter, one seedling per pot) or 32-cell plastic flats containing the same medium. The plants were cultivated in a growth room under a 12 h photoperiod, with temperature of 25/20°C (day/night), and a photosynthetic photon flux density (PPFD) of 600 μmol m−2 s−1. The relative humidity was maintained at 70%, and plants were irrigated with 1/2 strength Hoagland’s nutrient solution every 3 d.
Cold stress treatment and cold tolerance evaluation
For cold stress treatment, tomato seedlings at the five-leaf stage or tobacco plants expressing reporter vectors were transferred to a cold artificial growth chamber set at 4 °C, maintaining the same conditions as in the growth room. Each biological repeat contained eight seedlings from each tomato genotype, with three biological repeats per treatment. After 7 d of cold treatment, tomato seedlings were photographed. Then, relative electrolyte leakage (REL) was measured based on electrical conductivity and the maximum photochemical efficiency of photosystem II (Fv/Fm) was measured using an Imaging-PAM Chlorophyll Fluorometer equipped with a computer-operated PAM-control unit (IMAG-MAXI; Heinz Walz, Effeltrich, Germany), as previously described50. Survival rate assays were conducted on 20-day-old seedlings (at the three-leaf stage) grown in the growth room, which were subjected to 4 °C treatment for the specified duration before being returned to normal conditions (25 °C) for 1 week of recovery. During this process, the survival rate (percentage of green plants recovered after cold treatment) was calculated. The survival rates of the seedlings were calculated with three independent replicates for each genotype.
RNA-Seq libraries preparation and data analysis
RNA-Seq was performed as previously described7. Briefly, tomato leaves of AC and LA1777 were collected under normal conditions or after 6 h of cold stress, respectively, and used for RNA extraction. RNA-Seq library preparation and paired-end sequencing were performed on an Illumina NovaseqTM 6000 sequence platform by LC Sciences (Hangzhou, China). Approximately 4 Gb of high-quality paired-end reads were generated from each library. Clean data (clean reads) were obtained by removing reads containing adapters, poly-N sequences and low-quality reads from raw data using Trimmomatic version 0.36. These clean reads were then aligned to the tomato genome (https://solgenomics.net, SL4.0) using the Hisat2 mapping tool. Genes with FPKM’s P < 0.05 and an absolute log2-fold change ≥ 1 were considered as differentially expressed genes (DEGs).
Nuclei extraction and purification
Samples were prepared using sucrose sedimentation as previously reported51 but with slight modifications. Briefly, young leaves of AC and LA1777 were collected under normal conditions or after 6 h of cold stress, respectively, and ground to fine powder in liquid nitrogen. For each sample, 0.2 g of frozen tissue powder was homogenized in prechilled 1 ml lysis buffer (15 mM Tris-HCl pH7.5, 20 mM NaCl, 80 mM KCl, 0.5 mM spermine, 5 mM 2-ME, 0.2% TritonX-100), and the nuclear fraction was purified as described52. The nuclei pellet was resuspended in 1 ml cold lysis buffer. For ATAC-Seq and ATAC-qPCR, a nuclei aliquot (25 µl) was stained with DAPI (10 µl of 1 µg ml-1) and counted using a haemocytometer. Approximately 50,000 nuclei were used for each ATAC-Seq or ATAC-qPCR reaction.
ATAC-Seq libraries preparation and data analysis
ATAC-Seq was carried out as previously described53 with some minor modifications. Briefly, nuclei were extracted and purified from samples, and the nuclei pellet was resuspended in the Tn5 transposase reaction mix. The transposition reaction was incubated at 37 °C for 30 min. Equimolar Adapter1 and Adatper2 were added after transposition, and PCR was then performed to amplify the library. After the PCR reaction, libraries were purified with AMPure beads (Beckman, A63881) and library quality was assessed with Qubit (Thermo Fisher, Q32854). The clustering of the index-coded samples was performed on a cBot Cluster Generation System using TruSeq PE Cluster Kit v3-cBot-HS (Illumina) according to the manufacturer’s instructions. After cluster generation, the library preparations were sequenced on an Illumina platform at Novogene (Beijing, China) and 150 bp paired-end reads were generated.
Raw data was processed using fastp (version 0.20.0) to obtain clean reads, excluding adapters, poly-N, and low-quality sequences, while calculating Q20, Q30, and GC content. The reference genome and annotation were downloaded (https://solgenomics.net, SL4.0), and its index was built with BWA (version 0.7.12) for alignment of clean reads. Reads from mitochondria and chloroplast DNA, improperly paired, and PCR duplicates were excluded. Peak calling was done with MACS2 (version 2.1.0). By default, peaks with q-value threshold of 0.05 were carried out for all datasets. Peaks of different groups were merged using ‘bedtools merge’. We calculated the mean RPM of each group in the merge peak. Only peaks with an absolute log2-fold change of RPM ≥ 1 and P < 0.05 were considered as differential peaks. Genes associated with different peaks were identified using ChIPseeker. ChIPseeker was also used for gene and genomic region annotation54. GO enrichment analysis and KEGG pathway analysis were performed55. Differential peaks were identified with fold change of RPM more than 2. Genes associated with different peaks were identified using ChIPseeker. Peaks were visualized using the Integrative Genomics Viewer (version 2.12.2).
ATAC-qPCR
ATAC-qPCR was performed using the SYBR Green PCR Master Mix Kit (Takara, Shiga, Japan) on a Light Cycler 480 II detection system (Roche, Basel, Switzerland). Primers used for this analysis are shown in Supplementary Table 1. The relative accessibility and standard errors were determined using the 2−ΔΔCT method56.
VIGS
Complementary DNA (cDNA) fragments of target genes were amplified using gene-specific primers containing EcoRI and BamHI restriction sites (Supplementary Table 2). Purified PCR products were cloned into the TRV2 vector. The plasmids were then transformed into Agrobacterium tumefaciens strain GV3101. Fully expanded cotyledons of tomato seedlings were infiltrated with a mixture of A. tumefaciens strain carrying the helper vector TRV1 mixed at 1:1 with the strain carrying either TRV2 (empty vector control, TRV) or TRV2-target gene vectors57. The infiltrated plants were maintained in the growth chambers, and the silencing efficiency of the targeted genes was determined by RT-qPCR (Supplementary Fig. 19).
Constructs for genetic transformation
For overexpression constructs, the full-length coding sequences (CDS) of SlWRKY34, SlGATA29 and SlSWIBb were amplified from LA4024 cDNA and the CDS of ShWRKY34 was amplified from LA1777 cDNA using specific primers (Supplementary Data 8). For generating transgenic overexpressing lines, the SlWRKY34, ShWRKY34 and SlGATA29 CDS were inserted into a pFGC1008-3HA binary plasmid vector behind the CaMV 35S promoter. The SlSWIBb CDS was inserted into a pAC402-GFP binary plasmid vector behind the CaMV 35S promoter. All vectors were transformed into A. tumefaciens strain GV3101 for plant transformation. The resulting SlWRKY34 and ShWRKY34 overexpression plasmids were introduced into cultivated tomato LA4024, while both LA4024 and LA3942 were used as transgenic recipient materials to transform SlGATA29 and SlSWIBb overexpression plasmids. The transgenic overexpressing lines were further identified by RT-qPCR (Supplementary Fig. 20). The homozygous T2 transgenic lines were used in subsequent studies.
For the CRISPR/Cas9 constructs, single-guide (sgRNAs) or two-guide RNAs containing 20-bp targeting sequences were designed using the CRISPR-P web tool (http://cbi.hzau.edu.cn/crispr/) (Supplementary Data 8). The synthesized sequences were annealed and inserted into the BbsI site of the AtU6-sgRNA-AtUBQ-Cas9 vector as previously described58. The resulting plasmids were digested by HindIII and KpnI and then inserted into the pCAMBIA1301 binary vector digested by the same restriction enzymes. All resulting plasmids were transformed into A. tumefaciens strain GV3101 and infected into tomato cotyledons. First-generation transgenic plants were genotyped with specific primers surrounding the target sites (Supplementary Table 3). The homozygous F2 mutant lines without Cas9 were selected and used for further study. As shown in Supplementary Fig. 21a, the slwrky34-4 mutants harbored a 1 bp insertion and slwrky34-5 a 2 bp deletion in the SlWRKY34 coding region, leading to early translation termination. The protein bands of WRKY34 are barely observable in wrky34 mutants (Supplementary Fig. 21b). The slswibab double mutants in LA4024 background harbored a 4 bp deletion in the SlSWIBa coding region and a 1 bp deletion in the SlSWIBb coding region, both leading to early translation termination (Supplementary Fig. 21c). The slswibab double mutants in LA3942 background harbored a 1 bp deletion in the SlSWIBa coding region and a 4 bp deletion in the SlSWIBb coding region, both leading to early translation termination (Supplementary Fig. 21c). The slgata29 mutants in LA4024 background harbored a 2 bp deletion in the SlGATA29 coding region and the slgata29 mutants in LA3942 background harbored an 8 bp deletion in the SlGATA29 coding region, both leading to early translation termination (Supplementary Fig. 21d).
RNA isolation and RT-qPCR
Tomato leaves were collected under normal conditions or after indicated hours of cold stress. Other environmental conditions (e.g., humidity, lighting) of the growth chambers were kept consistent during sample collection. Total RNA was extracted from tomato leaves using an RNAprep Pure Plant Kit (Tiangen, DP419) following the manufacturer’s instructions. DNase I-treated extracted RNA (2 μg) was reverse-transcribed using a ReverTra Ace qPCR RT Kit (Vazyme, R223). For RT-qPCR, quantitative PCR was performed using the SYBR Green PCR Master Mix Kit (Vazyme, Q711) on a Light Cycler 480 II detection system (Roche, Basel, Switzerland). Tomato housekeeping genes Actin2 and Ubiquitin3 were used as internal references. Relative gene expression was calculated as previously described56. Primers used for RT-qPCR are listed in Supplementary Data 9.
DNA extraction, PCR and sequencing
Genomic DNAs of different tomato varieties were extracted using the TIANamp Genomic DNA Kit (Tiangen, DP304) and stored at -80°C. The nucleotide sequences of the 60 bp InDel in WRKY34 promoters of different tomato varieties were amplified with the following general primers: F: 5’-TGATATGAAAACCATTCACAAGTTGA-3’; R: 5’- TAGGGTGGTGAAAATGAGGTACATA-3’. The amplified products were sequenced by Sanger sequencing using an ABI 3730xl instrument by Youkang Biotechnology (Hangzhou, China). The sequencing results are shown in Supplementary Data 6.
Transient expression assays in tobacco leaves
Transient expression assays in tobacco leaves were performed as previously described in ref. 7. For promoter activity assays, promoters of pSlW34, pShW34, pSlW34+60bp, pShW34mW-box, pShW34mGATA-box and pSlW34+30bp were inserted into pGreenII 0800-LUC vectors as reporter genes. Renilla luciferase (REN) gene driven by CaMV 35S promoter in pGreenII 0800-LUC was used as an internal control to quantify transformation efficiency. Then, the above constructs were transformed into A. tumefaciens strain GV3101 and infiltrated into three-week-old tobacco leaves. After inoculation for 36 h, tobacco plants were treated at 4 °C for 6 h and proteins were extracted using Dual Luciferase Reporter Assay Kit (Vazyme, DL101).
For dual-luciferase (LUC) transcription activity assays, full-length CDSs of SlGATA29, ShGATA29, SlSWIBa/b, ShSWIB, SlCBF1 and SlWRKY34 were inserted into pGreenII 0029 62-SK vectors as effectors. The empty vector SK was used as a control. Promoters of pSlW34, pShW34, pSlW34+60bp, pShW34mW-box, pShW34mGATA-box, pSlW34+30bp, pSlCBF1 and pSlCOR47 were inserted into pGreenII 0800-LUC vectors as reporter genes. Then, all the constructs were transformed into A. tumefaciens strain GV3101. The tobacco leaves were transfected with different combinations of vectors (A. tumefaciens strain carrying the pGreenII 0800-LUC vector or pGreenII 0029 62-SK vector in a 1:10 ratio) for 36 h, then collected and lysed for the detection of dual luciferase activity (Vazyme, DL101) according to the manufacturer’s recommendations.
The activities of firefly LUC and renilla luciferase REN were measured using the Glomax 96 microplate luminometer (Promega, Fitchburg, USA). The measured levels were normalized by calculating the LUC/REN ratio. Primers used for plasmid construction are listed in Supplementary Data 8.
Y1H assay
Y1H assays were performed as previously described59. The 60 bp InDel was amplified and cloned into the pAbAi vector to generate pAbAi-60bp, while the 60 bp InDel containing mutant GATA-box or SWIB-mu4 was also amplified and cloned into the pAbAi vector to generate pAbAi-mutGATA-box or pAbAi-mutSWIB-mu4. The promoters of SlCBF1/2/3 and SlCOR47 were amplified and cloned into the pAbAi vector to generate pAbAi-SlCBF1, pAbAi-SlCBF2, pAbAi-SlCBF3 and pAbAi-SlCOR47. Full-length CDSs of SlGATA29, ShGATA29, SlSWIBa, SlSWIBb, ShSWIB and SlWRKY34 were amplified and cloned into the pGADT7 vector as prey plasmids. The mutant SlSWIBa/b CDSs (SlSWIBaR6A, SlSWIBaL46A, SlSWIBaK88A, SlSWIBaallmut, SlSWIBbR6A, SlSWIBbL44A, SlSWIBbK86A, and SlSWIBballmut) were synthesized and cloned into the pGADT7 vector by Youkang Biotechnology (Hangzhou, China). The linearized pAbAi constructs were transformed into Y1HGold yeast strain as bait strains, and screened with different Aureobasidin A (AbA) concentrations to detect background AbAr expression of bait strains. Then, prey plasmids were transformed into bait strains, and the transformed yeast cells were selected on selective plates (SD-Leu) supplemented with 150 ng ml−1 or 200 ng ml−1 AbA. The empty pGADT7 was used as the negative control. Primers used for plasmid construction are listed in Supplementary Data 8.
Recombinant proteins and electrophoretic mobility shift assays (EMSA)
Full-length CDSs of SlGATA29, ShGATA29, SlSWIBa, SlSWIBb, ShSWIB, SlCBF1 and SlWRKY34 were PCR amplified and cloned into the pET-28a vector (Supplementary Data 8). All recombinant vectors were transformed into Escherichia coli strain BL21 (DE3) and expressed at 37°C until OD600 reached 0.6, and then induced by 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, SIGMA, 092M4001V) at 16 °C for 14 h. The recombinant His-fusion proteins were purified according to the instructions provided with the Novagen pET purification system. To carry out EMSAs, oligonucleotide probes (Supplementary Table 4) were biotin-labelled using the Biotin 3′-End DNA Labeling Kit (Thermo Fisher Scientific, 89818) according to the manufacturer’s protocol and annealed to double-stranded DNA. EMSAs were performed using the Light Shift Chemiluminescent EMSA kit (Thermo Fisher Scientific, 20148) according to the manufacturer’s instructions60. Briefly, purified recombinant proteins were incubated with biotin-labelled probes at 28 °C for 30 min in 20 μl binding buffer (10× binding buffer, 50% glycerol, 25 ng μl−1 poly-dI-dC, 1% NP-40). For competition assays, 10-, 20- or 100-fold non-labelled competitor DNA was added to the reaction. The reaction products were resolved on a 6% polyacrylamide gels in 0.5 × TBE at 100 V for 1-2 h on ice. Probe-protein complexes and free probes were transferred to a charged Hybond-N membrane and detected by western blotting with 1:5000 diluted anti-biotin antibodies (Abcam, ab53494).
Microscale thermophoresis (MST) assay
The mutant SlSWIBa/b CDSs (SlSWIBaR6A, SlSWIBaL46A, SlSWIBaK88A, SlSWIBaallmut, SlSWIBbR6A, SlSWIBbL44A, SlSWIBbK86A, and SlSWIBballmut) were synthesized and cloned into the pET-28a vector by Youkang Biotechnology (Hangzhou, China). All recombinant His-proteins were induced and purified as described above. The Cy5-labeled double-stranded DNA was synthesized and diluted by MST buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl and 0.05% tween-20. 16 micro reaction tubes were set up, with tube 1 containing His-tag SlSWIBa/b proteins and tubes 2 to 16 filled with MST buffer. A dilution process was initiated by transferring a sample from tube 1 to tube 2. A serial dilution was obtained by repeating 15 times and remove 10 μl from tube number 16 after mixing. 10 μl of Cy5-labeled DNA were mixed with 10 μl of purified proteins and incubated at room temperature for 10 minutes, then loaded into silica capillaries (Polymicro Technologies, TSP010150). Binding reactions were measured using a Monolith NT.115 instrument (NanoTemper Technologies) at 25 °C, 40% MST power and 20% LED power. The Kd values were calculated using the mass action equation via the NanoTemper MO. Affinity Analysis software (NanoTemper Technologies).
ChIP-qPCR
ChIP experiments were performed using the EpiQuiK Plant ChIP kit (Epigentek, 50-109-6154) as described in the manufacturer’s protocol7. Approximately 1.5 g of leaf tissue was harvested from transgenic plants and WT plants under normal conditions or after 6 h of cold stress, respectively. The harvested tissues were crosslinked in 1× PBS buffer containing 1% formaldehyde for 15 min under vacuum. Fixation was stopped by adding 1× PBS buffer containing 0.125 M glycine under vacuum for 5 min. After washing three times with cold sterilized water, the tissues were homogenized, dried and ground into powder in liquid nitrogen, followed by isolation and sonication of chromatin. Sonicated chromatin fragments were immunoprecipitated with either anti-HA antibody or anti-GFP antibody, while a goat anti-mouse IgG antibody was served as the negative control. The enriched DNA was amplified by qPCR using specific primers (Supplementary Table 5). Relative enrichment was calculated by comparing the percentage of anti-HA- or anti-GFP-immunoprecipitated DNA to the percentage of IgG-immunoprecipitated DNA.
Y2H assay
The CDSs of SlSWIBa, SlSWIBb, ShSWIB and the C-terminal fragment of SlWRKY34 and ShWRKY34 were amplified and cloned into the pGBKT7 vector. The CDSs of SlGATA29, ShGATA29, SlCBF1/2/3, ShCBF1/2/3, SlCOR47, SlCOR15a, SlCOR27, ShCOR47, ShCOR15a and ShCOR27 were amplified and cloned into the pGADT7 vector. The resulting constructs were co-transformed in various combinations into Saccharomyces cerevisiae strain AH109 according to the manufacture’s instruction (Yeastmaker™ Yeast Transformation System 2). The transfected yeast cells were grown on SD-Leu-Trp plates at 28 °C for 3 d and then transferred to selective plates (SD-Leu/-Trp/-His/-Ade) at 28 °C for 4 d. Primers used for plasmid construction are listed in Supplementary Data 8.
Pull-down assay
Full-length CDSs of SlSWIBa, SlSWIBb, ShSWIB and SlCBF1 were amplified and cloned into the pET-28a vector. Full-length CDSs of SlGATA29, ShGATA29 and SlWRKY34 were amplified and cloned into the pGEX-4T-3 vector. The recombinant vectors were transformed into E. coli BL21 (DE3). The recombinant His-fusion proteins were purified according to the instructions provided with the Novagen pET purification system. To carry out pull-down assays, GST and GST-fusion proteins were extracted with extraction buffer and kept immobilized on Glutathione Sepharose 4B beads (Cytiva, 17075601)61. Glutathione beads containing the GST and GST-fusion proteins were incubated with equal amounts of different His-fusion proteins at 4 °C for 3 h and then were washed five times with PBS buffer (containing 0.1% tween-20). The proteins were detected by immunoblotting with anti-His antibody and anti-GST antibody. Primers used for plasmid construction are listed in Supplementary Data 8.
Bimolecular fluorescence complementation (BiFC)
For BiFC assay, full-length CDSs of SlSWIBa, SlSWIBb, ShSWIB and SlWRKY34 were amplified and cloned into the binary vector p2YN. Full-length CDSs of SlGATA29, ShGATA29 and SlCBF1 were amplified and cloned into the binary vector p2YC. They were then transformed into A. tumefaciens strain GV3101. For transient expression, different combinations of A. tumefaciens carrying different constructs at OD600 = 0.8 were co-infiltrated into four-week-old Nicotiana benthamiana leaves. Nucleus-located H2B-mCherry was used as a nucleus marker. After 36 h of infiltration, the fluorescence signals of the infiltrated leaves were observed under a confocal laser scanning microscope (Zeiss LSM 780, Oberkochen, Germany) using preset settings of YFP (Ex: 488 nm, Em: 520-540 nm) and mCherry (Ex: 561 nm, Em: 610-630 nm). Primers used for plasmid construction are listed in Supplementary Data 8.
Co-immunoprecipitation (Co-IP)
Full-length CDSs of SlSWIBa, SlSWIBb, ShSWIB and SlCBF1 were amplified and cloned into GFP tag vector, while full-length CDSs of SlGATA29, ShGATA29 and SlWRKY34 were amplified and cloned into HA tag vector. They were then transformed into A. tumefaciens strain GV3101. For transient expression, different combinations of A. tumefaciens carrying different constructs at OD600 = 0.8 were co-infiltrated into four-week-old N. benthamiana leaves. After 36 h of infiltration, the proteins co-expressed in the infiltrated leaves were extracted using Co-IP buffer and analyzed by immunoblotting with anti-HA antibody and anti-GFP antibody60. Extracts of equal total proteins were incubated with anti-GFP Magnetic Beads (Chromotek) for 3 h with gentle rotation at 4°C. Beads were washed five times with the Co-IP buffer. The immunoprecipitated proteins were analyzed by immunoblotting with anti-HA antibody. Primers used for plasmid construction are listed in Supplementary Data 8.
Protein extraction and western blotting
Protein extraction and western blotting were performed as described60,62. Proteins were separated by SDS-PAGE using 10% (w:v) acrylamide gels and then transferred onto nitrocellulose membranes. WRKY34 protein was detected with anti-WRKY34 polyclonal antibody (No.230608037). Anti-WRKY34 polyclonal antibody was customized by the Laboratory Animal Center of Zhejiang University.
Statistical analysis
Data were subjected to statistical analysis of variance using the SPSS package (SPSS 19.0). Data were represented as the mean ± SD. The difference between the two groups was assessed by two tailed Student’s t-tests. Statistically significant differences among multiple groups were evaluated by one-way ANOVA followed by a Duncan’s multiple range test. Details of each statistical test are indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The RNA-Seq data generated in this study have been deposited in the NCBI Sequence Read Archive database under BioProject accession PRJNA825093. The ATAC-Seq data generated in this study have been deposited in the NCBI GEO data libraries under accession GSE254893. All cultivated tomato genes involved in this study can be found at the Sol genomics network with the following accession numbers: SlWRKY34 (Solyc05g055750 [https://solgenomics.net/locus/25311/view]), SlCBF1 (Solyc03g026280 [https://solgenomics.net/locus/4512/view]), SlCBF2 (Solyc03g124110 [https://solgenomics.net/locus/76752/view]), SlCBF3 (Solyc03g026270 [https://solgenomics.net/locus/17440/view]), SlCOR47 (Solyc04g082200 [https://solgenomics.net/locus/8351/view]), SlCOR15a (Solyc06g083920 [https://solgenomics.net/locus/28101/view]), SlCOR27 (Solyc04g078880 [https://solgenomics.net/locus/22587/view]), SlGATA29 (Solyc12g008830 [https://solgenomics.net/locus/40868/view]), SlSWIBa (Solyc08g075400 [https://solgenomics.net/locus/32384/view]), SlSWIBb (Solyc08g005590 [https://solgenomics.net/locus/30733/view]). All wild tomato genes involved in this study can be found at the NCBI or our RNA-Seq data with the following accession numbers or gene IDs: ShWRKY34 (g49463), ShCBF1 (ACB45087.1), ShCBF2 (ACB45080.1), ShCBF3 (ACB45078.1), ShCOR47 (AHB20199.1), ShCOR15a (g53092), ShCOR27 (g44172), ShGATA29 (g19325), and ShSWIB (g55). Source data are provided with this paper.
Change history
27 September 2024
A Correction to this paper has been published: https://doi.org/10.1038/s41467-024-52843-z
References
Rodríguez-Leal, D., Lemmon, Z. H., Man, J., Bartlett, M. E. & Lippman, Z. B. Engineering quantitative trait variation for crop improvement by genome editing. Cell 171, 470–480.e8 (2017).
Panchy, N., Lehti-Shiu, M. & Shiu, S. H. Evolution of gene duplication in plants. Plant Physiol. 171, 2294–2316 (2016).
Zeng, R. et al. Natural variation in a type-A response regulator confers maize chilling tolerance. Nat. Commun. 12, 4713 (2021).
Wray, G. A. et al. The evolution of transcriptional regulation in eukaryotes. Mol. Biol. Evol. 20, 1377–1419 (2003).
Hendelman, A. et al. Conserved pleiotropy of an ancient plant homeobox gene uncovered by cis-regulatory dissection. Cell 184, 1724–1739.e16 (2021).
Wittkopp, P. J. & Kalay, G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 13, 59–69 (2012).
Guo, M. Y. et al. A single-nucleotide polymorphism in WRKY33 promoter is associated with the cold sensitivity in cultivated tomato. New Phytol. 236, 989–1005 (2022).
Dupont, S. & Wickström, S. A. Mechanical regulation of chromatin and transcription. Nat. Rev. Genet. 23, 624–643 (2022).
Ho, L. & Crabtree, G. R. Chromatin remodelling during development. Nature 463, 474–484 (2010).
Jones, P. A. & Baylin, S. B. The fundamental role of epigenetic events in cancer. Nat. Rev. Genet. 3, 415–428 (2002).
Wang, Z. X. et al. Dual ARID1A/ARID1B loss leads to rapid carcinogenesis and disruptive redistribution of BAF complexes. Nat. Cancer 1, 909–922 (2020).
Zhang, B. Y. et al. The chromatin remodeler CHD6 promotes colorectal cancer development by regulating TMEM65-mediated mitochondrial dynamics via EGF and Wnt signaling. Cell Discov. 8, 130 (2022).
Probst, A. V. & Scheid, O. M. Stress-induced structural changes in plant chromatin. Curr. Opin. Plant Biol. 27, 8–16 (2015).
Song, Z. T., Liu, J. X. & Han, J. J. Chromatin remodeling factors regulate environmental stress responses in plants. J. Integr. Plant Biol. 63, 438–450 (2021).
Huang, Y. et al. HSFA1a modulates plant heat stress responses and alters the 3D chromatin organization of enhancer-promoter interactions. Nat. Commun. 14, 469 (2023).
Zeng, Z. X. et al. Cold stress induces enhanced chromatin accessibility and bivalent histone modifications H3K4me3 and H3K27me3 of active genes in potato. Genome Biol. 20, 123 (2019).
Yang, J. et al. A lamin-like protein OsNMCP1 regulates drought resistance and root growth through chromatin accessibility modulation by interacting with a chromatin remodeler OsSWI3C in rice. New Phytol. 227, 65–83 (2020).
Wang, J. Y. et al. The conserved domain database in 2023. Nucleic Acids Res. 51, D384–D388 (2023).
Bennett-Lovsey, R., Hart, S. E., Shirai, H. & Mizuguchi, K. The SWIB and the MDM2 domains are homologous and share a common fold. Bioinformatics 18, 626–630 (2002).
Vieira, W. A. & Coetzer, T. L. Localization and interactions of Plasmodium falciparum SWIB/MDM2 homologues. Malar. J. 15, 32 (2016).
Hernández-García, J. et al. Comprehensive identification of SWI/SNF complex subunits underpins deep eukaryotic ancestry and reveals new plant components. Commun. Biol. 5, 549 (2022).
Cairns, B. R., Levinson, R. S., Yamamoto, K. R. & Kornberg, R. D. Essential role of SWP73p in the function of yeast SWI/SNF complex. Gene Dev. 10, 2131–2144 (1996).
Jégu, T. et al. The SWI/SNF protein BAF60 mediates seedling growth control by modulating DNA accessibility. Genome Biol. 18, 114 (2017).
Melonek, J., Matros, A., Trösch, M., Mock, H. P. & Krupinska, K. The core of chloroplast nucleoids contains architectural SWIB domain proteins. Plant Cell 24, 3060–3073 (2012).
Ding, Y. L., Shi, Y. T. & Yang, S. H. Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol. 222, 1690–1704 (2019).
Liu, J. Y., Shi, Y. T. & Yang, S. H. Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 60, 780–795 (2018).
Mao, D. H. et al. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl Acad. Sci. USA 116, 3494–3501 (2019).
Jiang, H. F. et al. Natural polymorphism of ZmICE1 contributes to amino acid metabolism that impacts cold tolerance in maize. Nat. Plants 8, 1176–1190 (2022).
Cao, X., Jiang, F. L., Wang, X., Zang, Y. W. & Wu, Z. Comprehensive evaluation and screening for chilling-tolerance in tomato lines at the seedling stage. Euphytica 205, 569–584 (2015).
Miyamoto, K. et al. Chromatin accessibility impacts transcriptional reprogramming in oocytes. Cell Reports 24, 304–311 (2018).
De Witte, D. et al. BLSSpeller: exhaustive comparative discovery of conserved cis-regulatory elements. Bioinformatics 31, 3758–3766 (2015).
Song, Q. et al. Prediction of condition-specific regulatory genes using machine learning. Nucleic Acids Res. 48, e62 (2020).
Han, Y., Reyes, A. A., Malik, S. & He, Y. Cryo-EM structure of SWI/SNF complex bound to a nucleosome. Nature 579, 452–455 (2020).
Baek, M. et al. Accurate prediction of protein-nucleic acid complexes using RoseTTAFoldNA. Nat. Methods 21, 117–121 (2023).
Sang, Q. et al. Mutagenesis of a quintuple mutant impaired in environmental responses reveals roles for CHROMATIN REMODELING4 in the Arabidopsis floral transition. Plant Cell 32, 1479–1500 (2020).
Shi, Y. T., Ding, Y. L. & Yang, S. H. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 23, 623–637 (2018).
Razifard, H. et al. Genomic evidence for complex domestication history of the cultivated tomato in Latin America. Mol. Biol. Evol. 37, 1118–1132 (2020).
Ramirez-Ojeda, G. et al. Edaphoclimatic descriptors of wild tomato species (Solanum Sect. Lycopersicon) and closely related species (Solanum Sect. Juglandifolia and Sect. Lycopersicoides) in South America. Front. Genet. 12, 748979 (2021).
Yu, X. F. et al. Chromosome-scale genome assemblies of wild tomato relatives Solanum habrochaites and Solanum galapagense reveal structural variants associated with stress tolerance and terpene biosynthesis. Hortic. Res. 9, uhac139 (2022).
Nuez, F., Prohens, J. & Blanca, J. M. Relationships origin, and diversity of Galápagos tomatoes: Implications for the conservation of natural populations. Am. J. Bot. 91, 86–99 (2004).
Zhang, T. Z. et al. Sequencing of allotetraploid cotton (Gossypium hirsutum L. acc. TM-1) provides a resource for fiber improvement. Nat. Biotechnol. 33, 531–537 (2015).
Xu, Z. N. et al. A transcription factor ZmGLK36 confers broad resistance to maize rough dwarf disease in cereal crops. Nat. Plants 9, 1720–1733 (2023).
Singh, A., Modak, S. B., Chaturvedi, M. M. & Purohit, J. S. SWI/SNF chromatin remodelers: structural, functional and mechanistic implications. Cell Biochem. Biophys. 81, 167–187 (2023).
He, S. et al. Structure of nucleosome-bound human BAF complex. Science 367, 875–881 (2020).
Jian, Y., Shim, W.-B. & Ma, Z. Multiple functions of SWI/SNF chromatin remodeling complex in plant-pathogen interactions. Stress. Biology 1, 18 (2021).
Huang, C. Y. et al. The chromatin-remodeling protein BAF60/SWP73A regulates the plant immune receptor NLRs. Cell Host Microbe 29, 425–434.e4 (2021).
Zou, C. S., Jiang, W. B. & Yu, D. Q. Male gametophyte-specific WRKY34 transcription factor mediates cold sensitivity of mature pollen in Arabidopsis. J. Exp. Bot. 61, 3901–3914 (2010).
Zhang, Z. X. et al. Understanding the mechanism of red light-induced melatonin biosynthesis facilitates the engineering of melatonin-enriched tomatoes. Nat. Commun. 14, 5525 (2023).
Truskina, J. et al. A network of transcriptional repressors modulates auxin responses. Nature 589, 116–119 (2021).
An, S. M. et al. Brassinosteroid signaling positively regulates abscisic acid biosynthesis in response to chilling stress in tomato. J. Integr. Plant Biol. 65, 10–24 (2023).
Lu, Z. F., Hofmeister, B. T., Vollmers, C., DuBois, R. M. & Schmitz, R. J. Combining ATAC-Seq with nuclei sorting for discovery of cis-regulatory regions in plant genomes. Nucleic Acids Res. 45, e41 (2017).
Bajic, M., Maher, K. A. & Deal, R. B. Identification of open chromatin regions in plant genomes using ATAC-Seq. Methods Mol. Biol. 1675, 183–201 (2018).
Corces, M. R. et al. An improved ATAC-Seq protocol reduces background and enables interrogation of frozen tissues. Nat. Methods 14, 959–962 (2017).
Yu, G. C., Wang, L. G. & He, Q. Y. ChIPseeker: an R/Bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015).
Bu, D. et al. KOBAS-i: intelligent prioritization and exploratory visualization of biological functions for gene enrichment analysis. Nucleic Acids Res. 49, 317–325 (2021).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(-Delta Delta C) method. Methods 25, 402–408 (2001).
Chen, X. L. et al. Role of selective autophagy receptors in tomato response to cold stress. Environ. Exp. Bot. 213, 105426 (2023).
Song, J. N. et al. SlMPK1-and SlMPK2-mediated SlBBX17 phosphorylation positively regulates CBF-dependent cold tolerance in tomato. New Phytol. 239, 1887–1902 (2023).
Xia, X. J. et al. Brassinosteroid signaling integrates multiple pathways to release apical dominance in tomato. Proc. Natl. Acad. Sci. USA 118, e2004384118 (2021).
Zou, J. P. et al. Autophagy promotes jasmonate-mediated defense against nematodes. Nat. Commun. 14, 1–16 (2023).
Lin, R. et al. CALMODULIN6 negatively regulates cold tolerance by attenuating ICE1-dependent stress responses in tomato. Plant Physiol. 193, 2105–2121 (2023).
Sang, K. Q. et al. The APETALA2a/DWARF/BRASSINAZOLE-RESISTANT 1 module contributes to carotenoid synthesis in tomato fruits. Plant J. 112, 1238–1251 (2022).
Acknowledgements
We thank Prof. Gang Lu for providing 181 cultivated tomatoes, 74 cherry tomatoes and 58 currant tomatoes seeds. We also thank Prof. Xingxing Shen and Prof. Mingfang Zhang for their meaningful suggestions on this article. This work was supported by the National Natural Science Foundation of China (32272790) and the Starry Night Science Fund of Zhejiang University Shanghai Institute for Advanced Study (SN-ZJU-SIAS-0011) to J.Z. and partially by China Agriculture Research System of MOF and MARA (CARS -23-B01) and Zhejiang Province Science and Technology Plan (2023C02001) to J.Y.
Author information
Authors and Affiliations
Contributions
M.G., J.Y. and J.Z. conceived and designed the experiments. M.G., F.Y., L.Z., L.W., Z.L. and Z.Q. performed the experiments. M.G. and J.Z. analyzed the data. V.F. provided the critical discussion. M.G. and J.Z. wrote the article. All authors reviewed and revised the article.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Cécile Raynaud, Shuhua Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Guo, M., Yang, F., Zhu, L. et al. Loss of cold tolerance is conferred by absence of the WRKY34 promoter fragment during tomato evolution. Nat Commun 15, 6667 (2024). https://doi.org/10.1038/s41467-024-51036-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51036-y
This article is cited by
-
Fine-mapping of sm6.1, a novel locus conferring resistance to gray leaf spot from Solanum habrochaites accession LA1777
Theoretical and Applied Genetics (2025)