Abstract
Cell surface glycans form various “glycan patterns” consisting of different types of glycan molecules, thus enabling strong and selective cell-to-cell recognition. We previously conjugated different N-glycans to human serum albumin to construct glycoalbumins mimicking natural glycan patterns that could selectively recognize target cells or control excretion pathways in mice. Here, we develop an innovative glycoalbumin capable of undergoing transformation and remodeling of its glycan pattern in vivo, which induces its translocation from the initial target to a second one. Replacing α(2,3)-sialylated N-glycans on glycoalbumin with galactosylated glycans induces the translocation of the glycoalbumin from blood or tumors to the intestine in mice. Such “in vivo glycan pattern remodeling” strategy can be used as a drug delivery system to promote excretion of a drug or medical radionuclide from the tumor after treatment, thereby preventing prolonged exposure leading to adverse effects. Alternatively, this study provides a potential strategy for using a single glycoalbumin for the simultaneous treatment of multiple diseases in a patient.
Similar content being viewed by others
Introduction
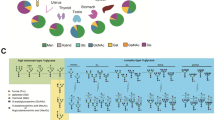
Glycoproteins, glycolipids, and proteoglycans on the cell surface play important roles in mediating cell-to-cell recognition, which is particularly relevant for immune systems1,2,3,4. Because the interaction between a monovalent glycan and the corresponding lectin is relatively weak (at millimolar KD level), the formation of glycan clusters on the cell surface enables high-avidity interactions through a multivalency effect (Fig. 1a)5,6,7,8. Cluster formation by different types of natural glycan molecules results in distinct “glycan patterns” (Fig. 1b). These glycan patterns facilitate selective cell recognition, i.e., glycan pattern recognition, which plays important roles in several biological processes9,10,11,12.
a Individual lectin–glycan interactions are generally weak and reversible (at mM KD levels). However, in the presence of glycocluster structures, they can exhibit strong cell interactions. In the presence of matching glycan patterns and lectin expression, glycoclusters can exhibit strong and selective cell interactions. b The glycocluster with various glycans would be selective for cell A (exactly matching glycan pattern) over cell B (partially matching glycan pattern) and cell C (mismatching glycan pattern). Glycans and lectins of the same color interact with each other. BIO-template: cell, antibody, and protein. c Artificial glycoalbumins were prepared by conjugating N-glycans to human serum albumins. As indicated in (I), α(2,3)-sialylated glycoalbumin targeted SW620 colon cancer cells, α(2,3)-/α(2,6)-sialylated glycoalbumin targeted HeLa cervical cancer cells, and α(2,3)-sialylated/hybrid glycoalbumin targeted A549 lung cancer cells. In the in vivo biodistribution in mice (II), α(2,3)-sialylated glycoalbumin showed preferential urinary excretion, whereas the galactosylated glycoalbumin was translocated to the intestinal excretion.
In previous work from our group, different asparagine-linked glycans (N-glycans) were conjugated to human serum albumin to mimic the glycan pattern on albumin (Fig. 1c)13,14,15,16,17. The RIKEN click reaction, which was developed by our group18,19,20,21,22, was used to introduce various large N-glycans onto albumin to synthesize glycoalbumins with different glycan patterns, including heterogeneous glycoalbumins with two different types of N-glycans. The data indicated that small changes in glycan patterns on albumin significantly alter cancer-targeting selectivity (Fig. 1c-i). Moreover, the in vivo kinetics and dissection analysis showed that the dynamics of these glycoalbumins are largely dependent on glycan pattern recognition in mice14,15. As shown in Fig. 1c-ii, compared with unmodified albumin, which is distributed over the whole body, the introduction of 10 molecules of α(2,3)-sialylated N-glycan on albumin improves blood stabilization and extends blood circulatory residence23,24, and the glycoalbumin is gradually excreted via the urinary bladder. By contrast, galactosylated glycoalbumin preferentially translocates to the intestinal excretion; the galactose-terminated N-glycan interacts with the asialoglycoprotein receptor (ASGPR) of hepatocytes25,26,27, and is then taken up by liver and eventually excreted through intestine. The results suggest that cell-targeting selectivity and in vivo biodistributions can be precisely controlled by glycan pattern recognition, which serves to elicit specific biological responses. These findings led us to hypothesize that transforming and remodeling the glycan pattern of a glycoalbumin in vivo may allow translocation of the glycoalbumin from one target to another one (Fig. 2a).
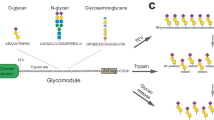
a Schematic showing the proposed strategy based on remodeling the glycan patterns of glycoalbumin-1 bound to target 1 in vivo, thereby weakening the interactions with target 1, and resulting in the translocation of the remodeled glycan pattern of glycoalbumin-2 to the second target. b Schematic of in vivo remodeling of the α(2,3)-sialylated glycan pattern to a galactosylated pattern on albumin by using a bioorthogonal chemical reaction, leading to the translocation of the glycoalbumin from the blood to the intestine. c Bioorthogonal click-to-release chemistry was used to remodel the glycan pattern on albumin in mice.
In this work, we design a method to translocate glycosylated albumin from blood to intestine by remodeling the α(2,3)-sialylated glycan pattern into a galactosylated pattern on albumin via a bioorthogonal chemical reaction (Fig. 2b). Alternatively, the α(2,3)-sialylated glycoalbumin shows a strong interaction with human colon cancer SW620 cells16,28,29,30. Therefore, we examine whether α(2,3)-sialylated glycoalbumin bound to SW620 tumors could be translocated to the intestine using the same in vivo remodeling strategy.
Results and discussion
Design and synthesis of a translocating glycoalbumin in mice
In order to efficiently achieve the remodeling of the glycan pattern recognition in vivo, we focused on the click-to-release reaction reported by Robillard et al.31,32,33,34. In this reaction, the inverse electron-demand Diels–Alder reaction (IEDDA) between trans-cyclooctene (TCO) and tetrazine (Tz) is followed by the release of payload molecules from the TCO group35,36,37. The use of the click-to-release method in cancer therapy for releasing anticancer drugs as the payload has been investigated in mouse experiments and in clinical trials38,39,40,41,42,43, and it was therefore explored in this study. On the basis of the click-to-release mechanism, a TCO derivative was bound to albumin and linked to α(2,3)-sialylated N-glycan through a self-immolative carbamate group (Fig. 2c). Following the IEDDA with a galactosylated N-glycan bearing Tz, the glycan pattern on albumin was remodeled to galactosylated albumin through the release of the α(2,3)-sialylated glycan from the TCO moiety. This reaction performed in vivo should result in the translocation of the glycoalbumin from the bloodstream or SW620 tumors to the liver, which results in the intestinal excretion (Fig. 2b).
We designed the unsaturated aldehyde probe 1 containing the TCO motif (Fig. 3). On the basis of the “in vivo glycan pattern remodeling” depicted in Fig. 2b, c, the α(2,3)-sialylated glycan (to be released by the click-to-release reaction) was conjugated to the TCO moiety via a carbamate linker (Fig. 3). N-glycan structures, such as sialylated compounds, are hydrophilic, complex, and large, and conjugating other hydrophobic molecules such as the aldehyde probe 1 is challenging. We therefore explored the use of the strain-promoted azide–alkyne click reaction (SPAAC)44 for the conjugation of TCO and N-glycans. The unsaturated ester–aldehyde function of probe 1 is essential for conjugation with the lysine residues of albumin via the RIKEN click reaction, i.e., 6π-azaelectrocyclization of the unsaturated imines, which was developed earlier by our group18,19,20,21,22.
The synthesis of compound 1 is shown in Fig. 3. To efficiently introduce the anilino amino group in 745 to the TCO 346, the sterically hindered carboxylic acid of 3 was initially elongated by a glycine methyl ester linker to yield 4 at 88%. After the allylic alcohol of 4 was activated to 4-nitrophenyl carbonate, the reaction with 547 yielded the cyclooctyne/TCO 6 at 78% in two steps. Following hydrolysis of the methyl ester group of 6, compound 8 was obtained by amidation with 7 through an acyl chloride intermediate at 52% in two steps. After the allylic alcohol in 8 was oxidized by Dess–Martin periodinane at 60%, the resultant aldehyde 1 was conjugated with azide-functionalized α(2,3)-sialylated glycan 2 (Fig. 3) via the SPAAC reaction, providing the desired glycan aldehyde 9 at a quantitative yield. Alternatively, the galactosylated Tz (Gal-Tz) could be prepared from the activated ester of Tz 11 and the galactosylated glycan 10 at 52%.
Examination of “glycan pattern remodeling” by click-to-release chemistry
Glycoalbumin-I was efficiently generated using the RIKEN click reaction, which consisted of treating albumin with 15 equivalents of the glycan-aldehyde probe 9 at 37 °C for 3.5 h (Fig. 4a). Small molecules were filtered by Amicon centrifugation, and the number of conjugated glycans was evaluated by MALDI-TOF mass spectroscopy. As shown in Fig. 4b, the average molecular weight of glycoalbumin-I was 108 kDa (blue line), whereas that of unmodified albumin was 66.5 kDa (red), indicating that approximately 12 molecules of probe 9 were introduced into albumin. Because of the high reactivity of the RIKEN click reaction, approximately 80% of probe 9 reacted with the albumin surface to form the α(2,3)-sialylation.
a The RIKEN click reaction and glycan-aldehyde 9 were used to synthesize glycoalbumin-I conjugated with 12 α(2,3)-sialylated glycans. Following treatment with Gal-Tz, glycoalbumin-I was remodeled into glycoalbumin-II, which showed a glycan pattern consisting of 6 α(2,3)-sialylated glycans and 10 galactosylated glycans via click-to-release. b The molecular weight change of glycoalbumin-I was measured by MALDI-TOF-MS. c The real-time molecular weight changes of glycoalbumin-I during the glycan pattern remodeling process were monitored by MALDI-TOF-MS.
To test the “glycan pattern remodeling” on albumin (Fig. 4a), glycoalbumin-I was reacted with an excess amount (60 equivalents) of Gal-Tz at 37 °C, and the process of replacement of α(2,3)-sialylated glycans with galactosylated ones was evaluated by real-time monitoring of the molecular weight of the glycoalbumins (Fig. 4c). The molecular weight of glycoalbumin-I increased rapidly to 128 kDa for 5–10 min (Fig. 4c-ii, c-iii), indicating that approximately 10 molecules of Gal-Tz were conjugated to glycoalbumin-I via the IEDDA. The average molecular weight gradually decreased to 112 kDa over 4 h (Fig. 4c-vii) and eventually to 110 kDa after 16 h (Fig. 4c-viii), indicating that six molecules of α(2,3)-sialylated glycan were released from glycoalbumin-I. As a result, the glycan pattern of the “remodeled” glycoalbumin-II consisted of 6 molecules of α(2,3)-sialylated glycan and 10 molecules of galactosylated glycan. This in vitro outcome demonstrated that the chemistry-based “glycan pattern remodeling” of albumin was feasible.
In vitro cell interaction with glycoalbumins
ASGPR is expressed on the plasma membrane of hepatocytes (and highly expressed in liver cancer cells such as HepG2 cells), and it binds preferentially to galactosylated glycans. The main function of ASGPR is to maintain serum glycoprotein homeostasis through endocytosis of desialylated glycoproteins with terminal galactose residues25,26,27. In the next stage of this study, we used HepG2 as the target cells for investigating whether “glycan pattern remodeling” of albumin could alter cell binding selectivity. Both TAMRA-labeled glycoalbumin-I and -II were incubated with HepG2 cells at 37 °C for 3 h (Fig. 5a, c). After washing with PBS to remove nonspecifically bound glycoalbumins, cell fluorescence imaging was performed. The galactosylated pattern on glycoalbumin-II showed a higher interaction with HepG2 cells than the sialylated pattern on glycoalbumin-I (Fig. 5a, c, e). Following the same incubation conditions, in the co-treatment of TAMRA-labeled glycoalbumin-I and Gal-Tz with HepG2 cells, the TAMRA fluorescence displayed a similar intensity to that in the treatment of TAMRA-labeled glycoalbumin-II only (Fig. 5b, c, e). This result indicated that the remodeling of the α(2,3)-sialylated glycan pattern on glycoalbumin-I into the galactosylated glycan pattern by the click-to-release reaction, resulting in the increased binding to HepG2 cells. To confirm if the ASGPR involves binding to glycoalbumin-II, a competitive ligand-binding assay was conducted by incubating HepG2 cells with a mixture of TAMRA-labeled glycoalbumin-II and excess galactose (Fig. 5d). As a result, the significantly weakened interaction of glycoalbumin-II against HepG2 illustrated that the ASGPR bound with the galactosylated glycan pattern of glycoalbumin-II (Fig. 5e). Moreover, the confocal fluorescence images in Fig. 5f showed that the ASGPR-mediated endocytosis transported glycoalbumin-II into the intracellular compartments of HepG2. In addition, we previously showed that galactosylated albumin is translocated to the intestine of mice through liver uptake14. Given these data in Fig. 5, in a mouse model, the chemistry-based remodeling of the α(2,3)-sialylated glycan pattern on glycoalbumin-I leads to the uptake of the remodeled galactosylated glycoalbumin-II by ASGPR in hepatocytes, which results in the intestinal excretion.
Image results of HepG2 cells incubated with a α(2,3)-sialylated glycoalbumin-I (1 μM), b a mixture of α(2,3)-sialylated glycoalbumin-I (1 μM) and Gal-Tz (100 μM), c galactosylated glycoalbumin-II (1 μM), or d a mixture of galactosylated glycoalbumin-II (1 μM) and galactose (50 mM) at 37 °C for 3 h. The shown images of (a–d) were obtained at ×40 magnification via Hoechst fluorescence (cell nuclei staining), TAMRA fluorescence (glycoalbumins), brightfield, and the combined overlay. e Comparison of the TAMRA fluorescence intensity per cell of the images in a-d. Data are presented as the mean ± SD. Each data point indicated an independent experiment (n = 3). Significance p values were determined by two-way ANOVA. **p = 0.0038 (glycoalbumin-I with Gal-Tz vs. glycoalbumin-II); ****p < 0.0001 (glycoalbuin-I vs. glycoalbumin-I with Gal-Tz and glycoalbuin-I vs. glycoalbumin-II). f Confocal microscopy fluorescence images of HepG2 cells incubated with galactosylated glycoalbumin-II (1 μM) at 37 °C for 3 h. The shown images obtained at ×60 magnification via Hoechst fluorescence (cell nuclei staining), TAMRA fluorescence (glycoalbumins), and the combined Overlay. Three experiment was done in three biological replicates (n = 3). The scale bar corresponds to 50 μm.
Because α(2,3)-sialylated glycoalbumin binds to human colon cancer SW620 cells16,28,29,30, we will explore using the same in vivo glycan remodeling strategy for weakening the interaction of glycoalbumin-I against SW620 tumors to examine whether this eventually results in its movement to the intestine. Before moving onto animal experiments, the interactions of glycoalbumin-I and -II against SW620 cells were investigated. The experiment result of SW620 cells incubated with TAMRA-labeled albumin as a control (Fig. 6a), TAMRA-labeled glycoalbumin-I and -II (Fig. 6b, d) clearly showed that the α(2,3)-sialylated pattern on glycoalbumin-I displayed a stronger interaction with SW620 cells than the galactosylated glycan pattern on glycoalbumin-II (Fig. 6e). In comparison with TAMRA-labeled glycoalbumin-I (Fig. 6b), the interaction of TAMRA-labeled glycoalbumin-I against SW620 cells was significantly reduced by co-treating Gal-Tz as the result of the click-to-release reaction (Fig. 6c, e). The confocal fluorescence images in Fig. 6f showed that glycoalbumin-I mainly bound to SW620 cells on the cell surface, indicating that the remodeling of the α(2,3)-sialylated glycan pattern to the galactosylated glycan pattern on glycoalbumin-I was possible on the cell surface. These outcomes in Fig. 6 illustrated that translocation of glycoalbumin-I from the SW620 tumors to the intestine by using the glycan pattern remodeling will be feasible.
Image results of SW620 cells incubated with a albumin (5 μM), b α(2,3)-sialylated glycoalbumin-I (5 μM), c a mixture of α(2,3)-sialylated glycoalbumin-I (5 μM) and Gal-Tz (300 μM), or d galactosylated glycoalbumin-II (5 μM) at 37 °C for 30 min. The shown images of a–d were obtained at ×40 magnification via Hoechst fluorescence (cell nuclei staining), TAMRA fluorescence (glycoalbumins), brightfield, and the combined overlay. e Comparison of the TAMRA fluorescence intensity per cell of the images in a–d. Data are presented as the mean ± SD. Each data point indicated an independent experiment (n = 3). Significance p values were determined by two-way ANOVA. *p = 0.0268 (lactalbumin-I vs. lactalbumin-I with Gal-Tz); **p = 0.0075 (lactalbumin-I with Gal-Tz vs. lactalbumin-II); ***p = 0.0004 (lactalbumin-I vs. lactalbumin-II). f Confocal microscopy images of SW620 cells incubated with α(2,3)-sialylated lactalbumin-I (5 μM) at 37 °C for 30 min. The shown images obtained at ×60 magnification via Hoechst fluorescence (cell nuclei staining), TAMRA fluorescence (glycoalbumins), and the combined Overlay. Three experiment was done in three biological replicates (n = 3). The scale bar corresponds to 50 μm.
Glycoalbumin translocation through glycan pattern remodeling in mice
On the basis of in vitro data in Fig. 5, “in vivo glycan pattern remodeling” triggered by the click-to-release reaction was examined in animal studies (Fig. 7). Glycoalbumin-I and -II were labeled with the near-infrared fluorophore, HiLyte Fluor 750®, to analyze the in vivo kinetics and organ-specific translocation by molecular imaging. As shown in Fig. 7a, three groups of mice received the indicated glycoalbumins (glycoalbumin-I, co-injection of glycoalbumin-I with Gal-Tz, and glycoalbumin-II) via intravenous injection. α(2,3)-Sialylated glycoalbumin-I or galactosylated glycoalbumin-II was used as the control. The “in vivo glycan pattern remodeling” experimental group received glycoalbumin-I followed by Gal-Tz. After 3 h, the mice were dissected, and the fluorescence intensity (ROI) of the whole abdominal cavity and the collected intestine, blood, and urine was measured.
a In vivo fluorescence imaging of glycoalbumin-I, co-injection of glycoalbumin-I and Gal-Tz, and glycoalbumin-II: Translocation of glycoalbumin-I from the bloodstream to the intestine induced by treatment with Gal-Tz for glycan pattern remodeling in mice. The indicated compounds were injected intravenously into 10-week-old BALB/cAJcl-nu/nu mice (n = 5). Control groups received glycoalbumin-I (1.5 nmol in 200 μL saline) or glycoalbumin-II (1.5 nmol in 200 μL saline) alone. The “in vivo glycan pattern remodeling” experimental group received glycoalbumin-I (1.5 nmol in 100 μL saline) followed by Gal-Tz (500 nmol in 100 μL saline, 333 equivalents relative to glycoalbumin-I) after 10 min. The mice were then anesthetized with isoflurane and placed in a fluorescence imager. Abdominal images were acquired at 1-h intervals. After 3 h of observation, the mice were sacrificed and b fluorescence images of the whole abdominal cavity and the intestine, blood, and urine were obtained. Fluorescence intensity from the dissected intestine (c), collected urine (d), and blood (e) after 3 h is shown. Each data point indicated an independent experiment (n = 5). Fluorescence intensity was calculated within an arbitrarily defined region of interest (ROI). Data are presented as the mean ± SD. Significance p values were determined by two-way ANOVA. c **p = 0.0015 (glycoalbumin-I vs. glycoalbumin-I with Gal-Tz); ***p = 0.0004 (glycoalbumin-I vs. glycoalbumin-II). d *p = 0.0277 (glycoalbumin-I vs. glycoalbumin-I with Gal-Tz); *p = 0.0354 (glycoalbumin-I vs. glycoalbumin-II).
Mice treated with α(2,3)-sialylated glycoalbumin-I showed weak fluorescence signals in the intestinal tract (Fig. 7b-i), whereas mice treated with glycoalbumin-II (Fig. 7b-iii), which corresponded to the galactosylated glycan pattern, showed obvious fluorescence activity in the intestine. In the group co-injected with glycoalbumin-I and Gal-Tz (Fig. 7b-ii), fluorescence activity in the intestine was similar to that in the control glycoalbumin-II group. These results indicate that the click-to-release reaction caused remodeling of the α(2,3)-sialylated glycan pattern on glycoalbumin-I into the galactosylated glycan pattern in mice, resulting in intestinal translocation. This was confirmed by quantification of the fluorescence intensity in the intestine, urine, and blood and comparison of the groups receiving glycoalbumin-I, co-injection of glycoalbumin-I with Gal-Tz, and glycoalbumin-II (Fig. 7b, c–e). As shown in Fig. 7c, fluorescence intensity in the intestine was 3-fold higher in the groups receiving single administration of glycoalbumin-II and co-injection of glycoalbumin-I with Gal-Tz than in the group injected with glycoalbumin-I. Consistent with the longer circulatory residence in blood of α(2,3)-sialylated glycoalbumin, i.e., glycoalbumin-I, and its eventual excretion via the urinary bladder, the group injected with glycoalbumin-I showed notably higher fluorescence activity in the urine (Fig. 7b-i, d) and blood (Fig. 7b-i) than the other groups, in which intestinal excretion (Supplementary Fig. 52) was induced.
With the promising cell experiment data in Fig. 6, we finally explored the translocation of glycoalbumin-I, which was initially targeted to SW620 cancer–xenografted mice, to the intestinal tract using a similar strategy. As shown in Fig. 8a, the three groups of mice received the indicated compounds (glycoalbumin-I, co-injection of glycoalbumin-I followed by Gal-Tz after 10 min, and glycoalbumin-II) via intratumoral administration. The time-dependent confocal fluorescence images (Supplementary Fig. 37) showed that glycoalbumin-I bound with the cell surface of SW620 within 30 min and was slowly taken up intracellularly after 3 h. Therefore, a 10-min interval was set up between the co-injection of glycoalbumin-I and Gal-Tz before SW620 tumor completely took up glycoalbumin-I intratumorally. After 5 h, the mice were dissected, and the fluorescence intensity of the collected intestine, urine, and blood was measured (Fig. 8b–e).
a In vivo fluorescence imaging of glycoalbumin-I, co-injection of glycoalbumin-I and Gal-Tz, and glycoalbumin-II: translocation of glycoalbumin-I from the SW620 tumor to the intestine induced by Gal-Tz treatment for glycan pattern remodeling in mice. The indicated compounds were injected into 10-week-old BALB/cAJcl-nu/nu mice bearing SW620 tumors via intratumoral injection (n = 5). The control groups received glycoalbumin-I (0.5 nmol in 20 μL saline) or glycoalbumin-II (0.5 nmol in 20 μL saline) alone. The “in vivo glycan pattern remodeling” experimental group was injected with glycoalbumin-I (0.5 nmol in 10 μL saline) followed by Gal-Tz (200 nmol in 10 μL saline, 400 equivalents relative to glycoalbumin-I) after 10 min. The mice were then anesthetized with isoflurane and placed in a fluorescence imager. Abdominal images were acquired at 1-h intervals. After 5 h of observation, the mice were sacrificed and b fluorescence images of the whole abdominal cavity and the collected intestine, blood, and urine were obtained. Fluorescence intensity in the dissected intestine (c), collected urine (d), and blood (e) after 5 h is shown. Each data point indicated an independent experiment (n = 5). Fluorescence was calculated within an arbitrarily defined region of interest (ROI). Data are presented as the mean ± SD. Significance p values were determined by two-way ANOVA. c *p = 0.0387 (glycoalbumin-I vs. glycoalbumin-I with Gal-Tz); *p = 0.0116 (glycoalbumin-I vs. glycoalbumin-II). d *p = 0.0347 (glycoalbumin-I vs. glycoalbumin-I with Gal-Tz); *p = 0.0293 (glycoalbumin-I vs. glycoalbumin-II).
Because the α(2,3)-sialylated glycan pattern shows high affinity for SW620 tumors, direct intratumoral injection of glycoalbumin-I into the cancer region caused notable tumor accumulation (Fig. 8a and Supplementary Fig. 45), and intestinal translocation was not observed (Fig. 8b-i). By contrast, the group co-injected with glycoalbumin-I and Gal-Tz showed fluorescence activity in the intestine, similar to the group receiving glycoalbumin-II (Fig. 8b-ii,-iii and c). These groups showed lower fluorescence intensity in urine (Fig. 8d) and blood (Fig. 8e). These results indicate that in vivo remodeling of the glycan pattern in sialylated glycoalbumin-I weakened the interaction with SW620 tumors, which promoted movement into blood vessels, trapping by hepatocytes, and secretion via the intestine.
Thus, the results of mouse experiments shown in Figs. 7 and 8 demonstrate that the chemistry-based glycan remodeling from the α(2,3)-sialylated glycan pattern (which binds to tumors and/or increases stability in blood and leads to urinary excretion) to the galactosylated glycan pattern (which causes a shift from urinary to rapid intestinal excretion) was successfully induced in mice, resulting in the translocation of the glycosylated proteins.
In summary, we developed a unique glycoalbumin capable of undergoing transformation and remodeling of its glycan pattern in vivo through the bioorthogonal click-to-release reaction. A single glycoalbumin was successfully translocated from the initial target to a second target. Thus, replacing α(2,3)-sialylated N-glycans on glycoalbumin with galactosylated glycans induced the translocation of glycoalbumin from the blood or tumor region to the intestine in mice. Through testing in vitro and in vivo experiments, a limitation of the “in vivo glycan pattern remodeling” strategy was noted. The efficiency of remodeling the glycan pattern on albumin was not so prompt since the glycan release process took over 4 h. This limitation may result in the inefficient translocation of proteins of interest to the target organs in vivo. Conceivable approaches to overcome this limitation could be to modify the structure of glycan aldehyde probe and the glycan-tetrazine to increase the reactivity of the click-to-release reaction on albumin.
Such “in vivo glycan pattern remodeling” strategy could be used as an innovative drug delivery system to promote excretion of drugs and/or radionuclide medicines from the tumor after treatment, thereby preventing unnecessarily prolonged exposure, which may lead to adverse effects. Alternatively, this study provides a possible strategy for the use of a single glycoalbumin for the simultaneous treatment of multiple diseases in a patient through combination treatment with albumin-based biocompatible artificial metalloenzymes (ArMs)48,49,50,51,52,53,54.
Methods
General information
Reagents and buffer components were purchased from Sigma-Aldrich, Fisher Scientific, TCI, Wako Chemicals without further purification. All N-glycans were supplied from Glytech, Inc. Human serum albumin, lyophilized powder (Product no. A1653-5G), was purchased from Sigma-Aldrich. HiLyteTM Fluor 750 succinimidyl ester (Product no. AS-81266) was purchased from Anaspec. Inc. 5-(and-6)-Carboxytetramethylrhodamine succinimidyl ester (5(6)-TAMRA, SE) (Product no. ABD-371-100) was purchased from AAT Bioquest, Inc. Compound 11 (Product no. CP-6117-100MG) was purchased from Conju-Probe, LLC. DMEM medium (Product no. 044-29765) and Leivovitz’s L-15 medium (Product no. 128-06075) were purchased from Wako Chemicals. Fetal Bovine Serum (Mexico origin, Product no.173012) was purchased from Sigma-Aldrich. 1% penicillin-streptomycin (Product no. 15140-122) was purchased from Gibco. Amicon Ultra Centrifugal Filters (10 kDa) were purchased from Merck Millipore. All experiments dealing with air and moisture-sensitive compounds were conducted under an atmosphere of argon. Anhydrous solvents were used in this study, which were acquired from Kanto Chemical Co., Inc., including dichloromethane, dimethylformamide, dimethyl sulfoxide, and acetonitrile. Thin layer chromatography (TLC) analyses (F-254) were performed with 60Å silica gel from Merck. MALDI-TOF-MS data in Fig. 4b, c were plotted on software Microsoft Excel (Version 16.87) according to the raw data. Graphs in Figs. 5e, 6e, 7c–e, 8c–e and kinetic data in Supplementary Fig. 10 were plotted on a software Graphpad prism (Version 8.0.2) according to the raw data.
Nuclear magnetic resonance (NMR) spectroscopy
1H- and 13C-NMR characterizations were done using a Bruker Avance III NMR (400 MHz) instrument with the solvent peaks as internal standard δH 7.26 and δC 77.0 for CDCl3, δH 5.32 and δC 53.8 for CD2Cl2. Multiplicities are reported using the following abbreviations: s = singlet, d = doublet, t = triplet, q = quartet, m = multiplet, br = broad. J = coupling constant values are expressed in Hertz. NMR spectroscopy was analyzed using MestReNova (Version 14.21).
HPLC analysis
To identify compounds from reaction mixtures, reverse-phase HPLC was used with a Shimadzu system consisting of two LC-20AP pumps, an SPD-20AV photodiode array detector. The column was an analytical 4.6 × 250 mm Cosmosil 5C18-AR-300 from Nacalai Tesque. Samples were eluted using a combination of mobile phases A (H2O with or w/o 0.1% TFA) and B (acetonitrile with or w/o 0.1% TFA).
For absorbance, the detector was set to 220 and 254 nm. Product peaks are identified by retention times and mass spectrometry (MS) analysis. HPLC data was analyzed on LabSolutions Postrun Analysis (version 5.81 SP1).
Mass spectrometry
For chemical synthesis, mass spectra were collected using Bruker Daltonics Hystar version 3.2 SR4 software on a Bruker Daltonics MicroTOF-QIII spectrometer by electron spray ionization time-of-flight (ESI-TOF-MS). The data was analyzed by Bruker Daltonics Compass Data Analysis (version 4.2). Sample preparations of synthetic compounds used MeOH as a solvent. The mass spectra were analyzed on Bruker Daltonics Compass DataAnalysis version 4.2 (Build 383.1). High weight mass characterizations (i.e., albumin and glycoalbumin) were done using matrix-assisted laser desorption ionization MS (MALDI-TOF-MS) analysis on a Shimadzu Benchtop Linear MALDI-8020 mass spectrometer. The data was analyzed by Shimadzu MALDI Solutions (Version 2.6.0). Sample preparations of albumins and glycoalbumins used sinapic acid as a matrix. MALDI-TOF spectrometry data was analyzed by Shimadzu MALDI Solutions (Version 2.6.0).
Preparation of compounds
The related compounds used in this study were prepared as described in the supplementary information.
Synthesis of glycan aldehyde 9
The synthesis was performed in the dark because of the light-labile character of aldehydes 1 and 9. To a solution of α(2,3)-disialoglycan-azide 2 (1.25 mg, 500 mol) in DMSO (150 μL) was added the solution of aldehyde 1 (0.26 mg, 300 nmol) in MeCN (300 μl). The resulting mixture was reacted for 3 h at 70 °C and monitored by ESI-TOF-MS and HPLC (solvent A: H2O + 0.1% TFA; solvent B: MeCN + 0.1% TFA; gradient: from 10% B/90% A to 100% B over 30 min). The retention time of glycan aldehyde 9 was observed at 18.0 min. After 3 h, the mixture was cooled down to room temperature, followed by the removal of MeCN under vacuum to give a 5.0 mM stock solution in DMSO without further purification.
General protocol for preparation of glycoalbumin-I and fluorophore-labeled glycoalbumin-I
The glycan aldehyde 9 solution (75 nmol, 15 μl from 5.0 mM stock solution in DMSO) was added to either of albumin (5 nmol), TAMRA-labeled albumin (5 nmol, Supplementary Fig. 19), or FL750-labeled albumin (5 nmol, Supplementary Fig. 23) in milli-Q water (135 μl). The resulting solution was mildly mixed and incubated for 3.5 h at 37 °C in the dark. To remove the unreacted 9, the solution was concentrated and washed with milli-Q water using Amicon 10 K® ultra centrifugal filters (10 kDa cut-off). The solution of glycoalbumin-I or fluorophore-labeled glycoalbumin-I was then diluted in milli-Q water to necessary volumes as required: 100 μl to obtain 50 μM. To confirm formation of glycoalbumins, MALDI-TOF-MS was done. Analysis under positive mode, as for glycoalbumin-I, an average molecular weight of 108 kDa was detected, which indicated 12.4 molecules of α(2,3)-sialylated glycan are conjugated per albumin. As for TAMRA-labeled glycoalbumin-I, an average molecular weight of 109.9 kDa was detected, which indicated 12.7 molecules of α(2,3)-sialylated glycan are conjugated per albumin (Supplementary Fig. 20). As for FL750-labeled glycoalbumin-I, an average molecular weight of 99.2 kDa was detected, which indicated 9.3 molecules of α(2,3)-sialylated glycan are conjugated per albumin (Supplementary Fig. 24).
A protocol for preparation of glycoalbumin-II by glycan remodeling
To a solution of glycoalbumin-I (4.0 nmol) in milli-Q water (216 μl) was added by the Gal-Tz (240 nmol) solution in DMSO (24 μl). The resulting mixture was mildly mixed and incubated for 16 h at 37 °C. To monitor the process of replacing α(2,3)-sialylated glycans with galactosylated ones, a portion (1/40) of the mixture solution was washed with milli-Q water using Amicon 10 K® ultra centrifugal filters (10 kDa cut-off), followed by measurement using MALDI-TOF-MS for a specific time (5, 10, and 30 min, 1.5, 3.0, 4.0 and 16.0 h). The measured data were presented in Fig. 4c. After 16 h, the solution was concentrated and washed with milli-Q water using Amicon 10 K® ultra centrifugal filters (10 kDa cut-off) to remove the unreacted Gal-Tz to afford glycoalbumin-II, which consisted of 6 molecules of α(2,3)-sialylated glycan and 10 molecules of galactosylated glycan.
General protocol for preparation of fluorophore-labeled glycoalbumin-II by glycan remodeling
To a solution of fluorophore-labeled glycoalbumin-I (4.0 nmol) in milli-Q water (216 μl) was added by the Gal-Tz (240 nmol) solution in DMSO (24 μl). The resulting mixture was mildly mixed and incubated for 16 h at 37 °C. To monitor the process of replacing α(2,3)-sialylated glycans with galactosylated ones, a portion (1/40) of the mixture solution was washed with milli-Q water using Amicon 10 K® ultra centrifugal filters (10 kDa cut-off), followed by measurement using MALDI-TOF-MS for a specific time (30 min and 16.0 h). The measured data were presented in Supplementary Figs. 21–22 and 25–26. After 16 h, the solution was concentrated and washed with milli-Q water using Amicon 10 K® ultra centrifugal filters (10 kDa cut-off) to remove the unreacted Gal-Tz to afford fluorophore-labeled glycoalbumin-II. The TAMRA-labeled glycoalbumin-II consisted of 8.1 molecules of α(2,3)-sialylated glycan and 7.6 molecules of galactosylated glycan. The FL750-labeled glycoalbumin-II consisted of 5.7 molecules of α(2,3)-sialylated glycan and 5.8 molecules of galactosylated glycan.
Cell culture
HepG2 cell line (Human hepatocellular carcinoma cells) was obtained from RIKEN Cell Bank (RCB1648). SW620 cell line (human colon cancer cells) was obtained from American Type Culture Collection (ATCC) (CCL-227). Cell lines were typically incubated at 37 °C with a 5% CO2 humidified environment. HepG2 cancer cell line and SW620 were cultured in DMEM medium with supplemented with 10% FBS and 1% penicillin-streptomycin.
Cell imaging studies
To determine the degree of interactions of α2,3-sialylated glycoalbumin-I or glycan-remodeled glycoalbumin-II against HepG2 or SW620 cells, fluorescent imaging studies were performed. HepG2 or SW620 cells were plated onto 96-well clear-bottomed plates at a density of 2 × 104 cells per well and grown. After 2 days, the medium was then removed, followed by treatment with the indicated compound solutions. As for treating HepG2 cells, TAMRA-labeled glycoalbumin-I (1 μM), co-treatment of TAMRA-labeled glycoalbumin-I (1 μM) and Gal-Tz (100 μM), TAMRA-labeled glycoalbumin-II (1 μM), and co-treatment of TAMRA-labeled glycoalbumin-II (1 μM) and galactose (50 mM) in DMEM medium with 10% FBS were used. As for treating SW620 cells, TAMRA-labeled albumin (5 μM), TAMRA-labeled glycoalbumin-I (5 μM), co-treatment of TAMRA-labeled glycoalbumin-I (5 μM) and Gal-Tz (300 μM), and TAMRA-labeled glycoalbumin-II (5 μM) in DMEM medium with 10% FBS were used. HepG2 cells were incubated for 3 h at 37 °C and SW620 cells were incubated for 30 min at 37 °C. After the medium was removed, cells were washed (3×) with PBS buffer, followed by the addition of 200 μl nuclear staining agent hoechst (20,000× diluted solution by PBS buffer). Following an incubation period of 15 min at room temperature, cells were washed (2×) with PBS buffer 200 μl and fixed to the plates using a 4% paraformaldehyde, and then prepared for microscopy imaging using a Keyence BZ-X710 all-in-one fluorescence microscope with a software BZ-X Analyzer (Version 1.4.0.1). For TAMRA dye observation, fluorescence was measured at λEX = 545 nm/λEM = 605 nm, while for Hoechst-dye observation, fluorescence was measured at λEX = 360 nm/λEM = 460 nm. Cell fluorescence images were obtained at ×40 magnification. Bright field images were obtained at a 1/500 s exposure setting, Hoechst images were obtained at a 1/10 s exposure setting, TAMRA images were obtained at a 2 s exposure setting for SW620 or a 3 s exposure setting for HepG2. The obtained TAMRA images were proceeded with black balance on a software BZ-X Analyzer (Version 1.4.0.1). The obtained TAMRA and Hoechst images were analyzed on a free software ImageJ (Version 1.54). For TAMRA images, fluorescent regions were specified by Threshold, and the brightness value of the region was measured and designated as the fluorescence intensity; for Hoechst images, the number of nuclei in the image was manually counted and designated as the number of cells. In Figs. 5e and 6e, the fluorescence intensity per cell was presented. Cell confocal microscopy images were obtained at ×60 magnification using an Olympus Fluoview FV3000RS confocal laser scanning microscope. Cell confocal microscopy was analyzed on a software FV31S-SW (Version 2.4.1).
Animal experiments
All animal experiments were carried out with approval by RIKEN’s Animal Ethics Committee (W2019-2-049). The maximal tumour size/burden permitted by RIKEN’s Animal Ethics Committee is 10% of body weight, which corresponds to a tumor volume of 2000 mm3 in adult nude mice (~20 g). The maximal tumour size/burden was not exceeded in any experiment of this study. In general, mice were anesthetized with 2.5% isoflurane in oxygen at a flow rate of 2.5–3.0 L/min. Mice were housed in a facility with controlled temperature (24–26 °C), controlled humidity (30–60%), aeration, ad libitum access to food and water, and a regular 12-h light-dark cycle.
Intravenous injection of FL750-linked glycoalbumin-I and Gal-Tz into mice
10-week-old female nude mice (BALB/cAJcl-nu/nu) were randomly divided into 3 groups; FL750-linked glycoalbumin-I only (group 1, n = 5); co-injection of FL750-linked glycoalbumin-I and Gal-Tz (group 2, n = 5); FL750-linked glycoalbumin-II only (group 3, n = 5). The FL750-linked glycoalbumin-I and -II were prepared at a concentration of 1.5 nmol/100 μl stock solution in saline. The Gal-Tz was prepared at a concentration of 500 nmol/100 μl stock solution in saline. For each mouse, total injection volume was adjusted to 200 μl. By intravenous administration, each mouse in group 1 received FL750-linked glycoalbumin-I (100 μl stock solution), followed by 100 μl saline 10 min later; each mouse in group 2 received FL750-linked glycoalbumin-I (100 μl stock solution), followed by Gal-Tz (100 μl stock solution) 10 min later; each mouse in group 3 received FL750-linked glycoalbumin-II (100 μl stock solution), followed by 100 μl saline 10 min later. After treatment, fluorescence whole body imaging of mice (dorsal side) by using a Clairvivo OPT kinetics fluorescence imager (Shimadzu, Japan) with a software Clairvivo OPT measurement/viewer (Version 2.6.0.0) was immediately performed. The mice were exposed at the same shooting time and intensity. To image the mice, the excitation and emission wavelengths of fluorescence were adjusted to 785 and 849 nm, respectively and a 5-s exposure setting was used. After 3 h from treatment, fluorescence imaging with the abdomen of the mouse split open was performed. Then, mice were sacrificed, and fluorescence imaging of each organ was performed. Fluorescence was calculated within an arbitrarily defined region of interest (ROI) on a software Clairvivo OPT measurement/viewer (Version 2.6.0.0).
SW620 bearing mice xenograft models
The SW620 cancer xenograft tumors were established in 6-week-old female nude mice (BALB/cAJcl-nu/nu) by subcutaneous injection of cells (~1.1 × 106 cells in 100 μl of cold 50% Matrigel in unnourished DEME) into the right shoulder. Tumor growth was monitored while mice were housed in a facility with controlled temperature (24–26 °C), controlled humidity (30–60%), aeration, ad libitum access to food and water, and a regular 12-h light-dark cycle. When SW620 tumors reached 700–1100 mm3, the mice were ready to be used for the experiment of translocating a α(2,3)sialyled-glycoalbumin from the tumor to the intestine.
Intratumoral injection of FL750-linked glycoalbumin-I and Gal-Tz into SW620 tumor-bearing mice
SW620 bearing mice were randomly divided into 3 groups; FL750-linked glycoalbumin-I only (group 1, n = 5); co-injection of FL750-linked glycoalbumin-I and Gal-Tz (group 2, n = 5); FL750-linked glycoalbumin-II only (group 3, n = 5). The FL750-linked glycoalbumin-I and -II were prepared at a concentration of 0.5 nmol/10 μl stock solution in saline. The Gal-Tz was prepared at a concentration of 200 nmol/10 μl stock solution in saline. For each mouse, total injection volume was adjusted to 20 μl. By intratumoral administration, each mouse in group 1 received FL750-linked glycoalbumin-I (10 μl stock solution), followed by 10 μl saline 10 min later; each mouse in group 2 received FL750-linked glycoalbumin-I (10 μl stock solution), followed by Gal-Tz (10 μl stock solution) 10 min later; each mouse in group 3 received FL750-linked glycoalbumin-II (10 μl stock solution), followed by 10 μl saline 10 min later. After treatment, fluorescence whole body imaging of mice (dorsal side) by using a Clairvivo OPT kinetics fluorescence imager (Shimadzu, Japan) with a software Clairvivo OPT measurement/viewer (Version 2.6.0.0) was immediately performed. The mice were exposed at the same shooting time and intensity. To image the mice, the excitation and emission wavelengths of fluorescence were adjusted to 785 and 849 nm, respectively and a 5-s exposure setting was used. After 5 h from treatment, mice were sacrificed, and fluorescence imaging of each organ was performed. Fluorescence was calculated within an arbitrarily defined ROI on a software Clairvivo OPT measurement/viewer (Version 2.6.0.0)
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data on synthetic compounds, cell-based experiments, and animals generated in this study are provided in the Supplementary Information/Source Data file and can also be obtained from the corresponding author upon request. The source data underlying Figs. 4b, c; 5, 6; 7b, c, and 8, Supplementary Figs. 6–10, and 27–53 are provided as a Source Data file. Source data are provided with this paper.
References
Sperandio, M., Gleissner, C. A. & Ley, K. Glycosylation in immune cell trafficking. Immunol. Rev. 230, 97–113 (2009).
Rudd, P. M., Elliot, T., Cresswell, P., Wilson, I. A. & Dwek, R. A. Glycosylation an the immune system. Science 291, 2370–2376 (2001).
Davicino, R. C., Eliçabe, R. J., Genaro, M. S., Di & Rabinovich, G. A. Coupling pathogen recognition to innate immunity through glycan-dependent mechanisms. Int. Immunopharmacol. 11, 1457–1463 (2011).
Rabinovich, G. A. & Toscano, M. A. Turning “sweet” on immunity: galectin-glycan interactions in immune tolerance and imflammation. Nat. Rev. Immunol. 9, 338–352 (2009).
Lis, H. & Sharon, N. Lectins: carbohydrate-specific proteins that mediate cellular recognition. Chem. Rev. 98, 637–674 (1998).
Gestwicki, J. E., Cairo, C. W., Strong, L. E., Oetjen, K. A. & Kiessling, L. L. Influencing receptor−ligand binding mechanisms with multivalent ligand architecture. J. Am. Chem. Soc. 124, 14922–14933 (2002).
Dam, T. K. & Brewer, C. F. Effects of clustered epitopes in multivalent ligand-receptor interactions. Biochemistry 47, 8470–8476 (2008).
Dam, T. K. & Brewer, C. F. Lectins as pattern recongnition molecules: the effects of epitope density in innate immunity. Glycobiology 20, 270–279 (2010).
Gabius, H.-J. Antitumor potential and other emerging natural compounds (Eds.: E. F. Fang, T. Ng), Springer Netherlands, Dordrecht, pp. 21–32 (2013).
Jayaraman, N. Multivalent ligand presentation as a central concept to study intricate carbohydrate–protein interactions. Chem. Soc. Rev. 38, 3463 (2009).
Vong, K., Yamamoto, T. & Tanaka, K. Artificial glycoproteins as a scaffold for targeted drug therapy. Small 16, 1906890 (2020).
Kim, Y., Hyun, J. Y. & Shin, I. Multivalent glycans for biological and biomedical applications. Chem. Soc. Rev. 50, 10567–10593 (2021).
Ogura, A. et al. Glycan multivalency effects toward albumin enable N-glycan-dependent tumor targeting. Bioorg. Med. Chem. Lett. 26, 2251–2254 (2016).
Ogura, A. et al. Visualizing trimming dependence of biodistribution and kinetics with homo- and heterogeneous N-glycoclusters on fluorescent albumin. Sci. Rep. 6, 21797 (2016).
Latypova, L. et al. Sequential double “clicks” toward structurally well-defined heterogeneous N-glycoclusters: the importance of cluster heterogeneity on pattern recognition in vivo. Adv. Sci. 4, 1600394 (2017).
Ogura, A. et al. A viable strategy for screening the effects of glycan heterogeneity on target organ adhesion and biodistribution in live mice. Chem. Commun. 54, 8693–8696 (2018).
Kurbangalieva, A., Zamalieva, R., Nasibullin, I., Yamada, K. & Tanaka, K. Homo- and heterogeneous glycoconjugates on the basis of n-glycans and human serum albumin: synthesis and biological evaluation. Molecules 27, 1285 (2022).
Fujiki, K. & Tanaka, K. RIKEN click reagent for protein labeling, e-ROS Encyclopedia of reagents for organic synthesis. (2019)
Tanaka, K., Fukase, K. & Katsumura, S. New strategy in synthetic biology: from enzyme inhibition and natural products synthesis to PET imaging by 6π-azaelectrocyclization. Chem. Rec. 10, 119–139 (2010).
Tanaka, K. et al. A submicrogram-scale protocol for biomolecule-based PET imaging by rapid 6pi-azaelectrocyclization: visualization of sialic acid dependent circulatory residence of glycoproteins. Angew. Chem. Int. Ed. 47, 102–105 (2008).
Tanaka, K. et al. Whole-body imaging of tumor cells by azaelectrocyclization: visualization of metastasis dependence on glycan structure. Bioorg. Med. Chem. 21, 1074–1077 (2013).
Tanaka, K. et al. Cell surface biotinylation by azaelectrocyclization: easy-handling and versatile approach for living cell labeling. Bioorg. Med. Chem. 20, 1865–1868 (2012).
Sato, M. et al. Glycoinsulins: dendritic sialyloligosaccharide-displaying insulins showing a prolonged blood-sugar-lowering activity. J. Am. Chem. Soc. 126, 14013–14022 (2004).
Imai, N. et al. Physicochemical and biological characterization of asialoerythropoietin. Eur. J. Biochem. 194, 457–462 (1990).
Morell, A. G., Irvine, R. A., Sternlieb, I., Scheinberg, I. H. & Ashwell, G. Physical and chemical studies on ceruloplasmin. V. Metabolic studies on sialic acid-free ceruloplasmin in vivo. J. Biol. Chem. 243, 155–159 (1968).
Lee, R. T. & Lee, Y. C. Affinity labeling of the galactose/N-acetylgalactosamine-specific receptor of rat hepatocytes: preferential labeling of one of the subunits. Biochemistry 26, 6320–6329 (1987).
Lauc, G., Lee, R. T., Dumiae, J. & Lee, Y. C. Photoaffinity glycoprobes—a new tool for the identification of lectins. Glycobiology 10, 357–364 (2000).
Carlsson, S. et al. Affinity of galectin-8 and its carbohydrate recognition domains for ligands in solution and at the cell surface. Glycobiology 17, 663–676 (2007).
Ideo, H., Matsuzaka, T., Nonaka, T., Seko, A. & Yamashita, K. Galectin-8-N-domain recognition mechanism for sialylated and sulfated glycans. J. Biol. Chem. 286, 11346–11355 (2011).
Lahm, H. et al. Comprehensive galectin fingerprinting in panel of 61 huamn tumor cell lines by RT-PCR and its implications for diagnostic and therapeutic procedures. J. Cancer Res. Clin. Oncol. 127, 375–386 (2001).
Yi, W. et al. Recent advances in developing active targeting and multi-functional drug delivery systems via bioorthogonal chemistry. Signal Transduct. Target. Ther. 7, 386 (2022).
Versteegen, R. M., Rossin, R., Hoeve, W. T., Janssen, H. M. & Robillard, M. S. Click to release: instantaneous doxorubicin elimination upon tetrazine ligation. Angew. Chem. Int. Ed. 52, 14112–14116 (2013).
Rossin, R. et al. Triggered drug release from an antibody-drug conjugate using fast “click-to-release” chemistry in mice. Bioconjug. Chem. 27, 1697–1706 (2016).
Rossin, R. et al. Chemically triggered drug release from an antibody-drug conjugate leads to potent antitumour activity in mice. Nat. Commun. 9, 1484 (2018).
Wang, J., Wang, X., Fan, X. & Chen, P. R. Unleashing the power of bond cleavage chemistry in living systems. ACS Cent. Sci. 7, 929–943 (2021).
Wang, Y., Zhang, C., Wu, H. & Feng, P. Activation and delivery of tetrazine-responsive bioorthogonal prodrugs. Molecules 25, 5640 (2020).
Ji, X. et al. Click and release: bioorthogonal approaches to “on-demand” activation of prodrugs. Chem. Soc. Rev. 48, 1077–1094 (2019).
Oneto, J. M. M., Khan, I., Seebald, L. & Royzen, M. In vivo bioorthogonal chemistry enables local hydrogel and systemic pro-drug to treat soft tissue sarcoma. ACS. Cent. Sci. 2, 476–482 (2016).
Wu, K. et al. Click activated protodrugs against cancer increase the therapeutic potential of chemotherapy through local capture and activation. Chem. Sci. 12, 1259–1271 (2021).
Srinivasan, S. et al. SQ3370 activates cytotoxic drug via click chemistry at tumor and elicits sustained responses in injected & non-injected lesions. Adv. Ther. 4, 2000243 (2021).
Mcfarland, J. M. et al. Click chemistry selectively activates an auristatin protodrug with either intratumoral or systemic tumor-targeting agents. ACS Cent. Sci. 9, 1400–1408 (2023).
Vlastara, M. et al. Click-to-release: cleavable radioimmunoimaging with [89Zr]Zr-DFO-trans-cyclooctene-trastuzumab increases tumor-to-blood ratio. Theranostics 13, 4004–4015 (2023).
Li, H., Conde, J., Guerreiro, A. & Bernardes, G. J. L. Tetrazine carbon nanotubes for pretargeted in vivo “click‐to‐release” bioorthogonal tumour imaging. Angew. Chem. Int. Ed. 59, 16023–16032 (2020).
Agard, N. J., Prescher, J. A. & Bertozzi, C. R. A strain-promoted [3 + 2] azide−alkyne cycloaddition for covalent modification of biomolecules in living systems. J. Am. Chem. Soc. 126, 15046–15047 (2004).
Debets, M. F. et al. Aza-dibenzocyclooctynes for fast and efficient enzyme PEGylation via copper-free (3+2) cycloaddition. Chem. Commun. 46, 97–99 (2010).
Geus, M. A. R. D. et al. Fluorogenic bifunctional trans-cyclooctenes as efficient tools for investigating click-to-release kinetics. Chemistry 26, 9900–9904 (2020).
Nakamura, K. et al. Efficient route to RIKEN click probes for glycoconjugation. J. Carbohydr. Chem. 38, 127–138 (2019).
Tsubokura, K. et al. In vivo gold complex catalysis within live mice. Angew. Chem. Int. Ed. 56, 3579–3584 (2017).
Vong, K. et al. An artificial metalloenzyme biosensor can detect ethylene gas in fruits and Arabidopsis leaves. Nat. Commun. 10, 5746 (2019).
Vong, K. et al. Disrupting tumor onset and growth via selective cell tagging (SeCT) therapy. Sci. Adv. 7, eabg4038 (2021).
Nasibullin, I. et al. Synthetic prodrug design enables biocatalytic activation in mice to elicit tumor growth suppression. Nat. Commun. 13, 39 (2022).
Tanaka, K. & Vong, K. Unlocking the therapeutic potential of artificial metalloenzymes. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 96, 79–94 (2020).
Yamada, K., Muguruma, K. & Tanaka, K. Therapeutic in vivo synthetic chemistry using an artificial metalloenzyme with glycosylated human serum albumin. Adv. Carbohydr. Chem. Biochem. 82, 11–34 (2022).
Chang, T.-C. et al. Evaluation of acute toxicity of cancer-targeting albumin-based artificial metalloenzymes. Bioorg. Med. Chem. 73, 117005 (2022).
Acknowledgements
This work was financially supported by the AMED Grant JP15KM0908001 (K.T.), a research grant from the Astellas Foundation (K.T.), Mizutani Foundation for Glycoscience (K.T.), and JSPS KAKENHI Grant Numbers, JP21H02065 (K.T.), JP21K19042 (K.T.), and JP21K05302 (T.-C.C.). The authors thank Glytech, Inc. for supplying various N-glycans.
Author information
Authors and Affiliations
Contributions
Conceptualization: T.-C.C., A.R.P., and K.T.; methodology: K.Y., T.-C.C., and K.T.; compound synthesis: K.Y.; cell-based experiments: K.Y., A.M., and T.-C.C.; animal-based experiments: K.Y., A.N., and Y.K.; visualization: T.-C.C., A.R.P., and K.T.; funding acquisition: T.-C.C., and K.T.; project administration and supervision: K.T.; manuscript writing: K.Y., T.-C.C., and K.T. manuscript revision: K.Y., T.-C.C., and K.T.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yamada, K., Mukaimine, A., Nakamura, A. et al. Chemistry-driven translocation of glycosylated proteins in mice. Nat Commun 15, 7409 (2024). https://doi.org/10.1038/s41467-024-51342-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51342-5