Abstract
Cytomegalovirus (CMV) infection poses risks to newborns, necessitating effective therapies. Given that the damage includes both viral infection of brain cells and immune system-related damage, here we investigate the involvement of cellular prion protein (PrP), which plays vital roles in neuroprotection and immune regulation. Using a murine model, we show the role of PrP in tempering neonatal T cell immunity during CMV infection. PrP-null mice exhibit enhanced viral control through elevated virus-specific CD8 T cell responses, leading to reduced viral titers and pathology. We further unravel the molecular mechanisms by showing CMV-induced upregulation followed by release of PrP via the metalloproteinase ADAM10, impairing CD8 T cell response specifically in neonates. Additionally, we confirm PrP downregulation in human CMV (HCMV)-infected fibroblasts, underscoring the broader relevance of our observations beyond the murine model. Furthermore, our study highlights how PrP, under the stress of viral pathogenesis, reveals its impact on neonatal immune modulation.
Similar content being viewed by others
Introduction
With no effective vaccine available1, human cytomegalovirus (HCMV) continues to infect approximately 83% of the global population, with prevalence ranging from 60% to 90% in Europe2. While primary infection in adults often goes unnoticed, HCMV can invade the fetal brain, resulting in permanent neurological impairments such as mental retardation, vision, and hearing loss3. In this regard, HCMV remains the leading cause of congenital infections, affecting approximately 7 out of every 1000 children, occurring during pregnancy or in the first few weeks after birth4,5. Regrettably, available antiviral drug options6 are limited due to potential risks to the developing child, and recent longitudinal cohort studies have revealed that treatments, including ganciclovir and valganciclovir, do not provide enduring benefits7,8. To study congenital CMV disease, we have successfully utilized a perinatal infection model in experimental mice, as HCMV is strictly species-specific9,10. While mouse CMV (MCMV) cannot pass the placenta, the brains of newborn mice are developmentally equivalent to 2nd trimester human fetuses11. Thus, in the model employed here, we infect newborn mice on day 1 after birth to mimic congenital infection in humans. In this setting, MCMV reaches the brain and causes numerous developmental sequelae reminiscent of those observed in congenitally infected human newborns12,13,14. Most of the damage is believed to be caused by an overreactive immune response, which, while essential for sufficient viral control, can also lead to tissue damage due to excessive inflammation15. Accordingly, it has been shown that anti-inflammatory drugs can reduce abnormalities and alleviate brain damage in infected newborn mice13,16. However, the precise underlying factors and contributions that determine the occurrence and severity of damage in this context remain elusive17.
Extensively studied due to its devastating role in incurable and transmissible neurodegenerative prion diseases18,19, the cellular prion protein (PrP) is a membrane-anchored protein for which recent insights have revealed to serve dual functions in immune privilege: modulating inflammation to prevent immunopathology and directly protecting cells against inflammation-induced damage (reviewed in ref. 20). Notably, PrP is highly expressed in immunologically privileged sites, such as the brain, and its phenotypic effects become evident in the presence of stress and inflammation21,22,23,24, including viral challenges25,26,27,28,29,30. Studies involving PrP knockout mice (KO) have reported several phenotypes31, with some related to immune cell functions (reviewed in refs. 32,33). However, certain functions attributed to PrP based on the analysis of specific KO mouse lines could not be replicated in others31,34,35. This discrepancy is primarily due to the influence of Prnp-flanking genes in different KO strains. Nevertheless, PrP has been implicated in hematopoietic cell development36 and binding to/interacting with multiple extracellular and transmembrane ligands37,38. It is expressed on the surface of immune cells where it affects their function32,39,40,41,42,43,44, cytokine responses23,24,45 and immunological synapses43,46,47,48,49,50. Additionally, PrP can bind to immune cells, suggesting its interaction with immune receptors51,52,53. With its involvement in various aspects of inflammation and the lack of comparable studies investigating the immunobiology of PrP during the developmental phase of the immune system, particularly in the context of early viral infection, understanding the impact of PrP on congenital HCMV infection requires comprehensive investigation.
To investigate the impact of PrP on the immune system and its potential for immune silencing, we utilized the relatively novel PrnpZH3/ZH3 mouse strain, a unique and strictly co-isogenic model with C57BL/6J mice, which possesses a specific knockout of the PrP-encoding gene (Prnp) without affecting other genes that could potentially influence immune responses35. A fraction of PrP molecules is constitutively released (shed) from cells by the metalloprotease ADAM1054,55,56. Our findings identified soluble PrP as a negative regulator of CD8 T-cell response upon MCMV infection, with PrP KO neonates exhibiting increased effector functions of virus-specific CD8 T cells, leading to improved viral control in various organs, including the brain, without significant tissue damage or inflammation. Additionally, we provide a detailed description of the molecular mechanism by which CMV infection causes increased ADAM10-mediated release of PrP from infected cells, a previously unexplored aspect. Interestingly, binding of soluble PrP to CD8 T lymphocytes and the attenuated control of CMV were specific to newborn mice, as no similar observations were documented in adult mice. This study unveils the immune-regulatory function of PrP in congenital CMV infection, shedding light on the molecular and immunological mechanisms underlying viral burden in the developing brain during acute infection.
Results
PrP KO neonates exhibit lower MCMV titers attributed to improved control of acute infection by CD8 T cells
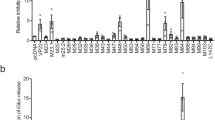
In a well-established model aimed at gaining a deeper understanding of brain damage during congenital CMV infection, newborn C57BL/6J mice (WT) are intraperitoneally infected with MCMV9,10. To assess the impact of PrP on infection consequences and immune response, we conducted a comparative analysis between WT and PrnpZH3/ZH3 mice, which are strictly co-isogenic C57BL/6J PrP KO mice that lack only a small portion of the Prnp gene, while other flanking genes potentially affecting immune responses remain unaltered35. Given that MCMV is detectable in the brain between 7 and 17 days post infection (dpi), with the most significant neurodevelopmental changes occurring between 8 and 10 dpi14, we examined virus titers at 8 and 14 dpi (Fig. 1A). Notably, a statistically significant difference in viral titer within the brain was observed on day 14.
A Viral titers in the brain of congenitally infected C57BL/6J and PrP KO mice at 8 and 14 days post-infection (dpi); dpi 8 (n = 5); dpi 14 (n = 6). B Viral titers in spleens (n = 6; both groups); livers (C57BL/6J n = 7; PrP KO n = 3), and salivary glands (n = 5; both groups) at 14 dpi. C Brain viral titers in MCMV-infected animals with and without CD8 T cell depletion at 14 dpi (C57BL/6J n = 6; PrP KO n = 6; C57BL/6J αCD8 n = 8; PrP KO αCD8 n = 8). Production of IFNγ and TNFα in CD8 T cells from spleens (D; C57BL/6J n = 8, PrP KO n = 7) and livers (E; C57BL/6J n = 8, PrP KO n = 7) of congenitally infected animals at 14 dpi determined by flow cytometry staining after a 4 h incubation in the presence of Brefeldin A. F Representative dot blots (second lowest signal in the group) showing liver MCMV-M45 tetramer staining, quantified in Fig. 1G. G Frequency of MCMV-M45 tetramer-positive CD8 T cells in the liver of congenitally infected animals at 14 dpi; (C57BL/6J n = 5; PrP KO n = 4); H Frequency (%) and total number (#) of MCMV-M45 (n = 6; both groups) and MCMV-M57 (C57BL/6 J n = 5; PrP KO n = 8) tetramer-positive CD8 T cells in the spleen of congenitally infected animals at 14 dpi. I Immunohistochemical staining of CD8 T cells (CD8 antigen) and MCMV infected nuclei (IE1 antigen) in the brain of congenitally infected PrP KO mice at 14 dpi. Nuclei were visualized with DAPI. Images were acquired using a Leica TCS SP8 confocal microscope and edited with LAS X software. Scale bar represents 50 μm (left panel) and 25 μm (right panel). Biological replicates: A (7), B ( ≥ 2), C–E (2), F, G (2), H (2–4) and I (1). Titer values for individual mice are shown, with median values indicated by horizontal bars. DL detection limit. Statistical analysis was performed with GraphPad Prism using the two-tailed Mann–Whitney (U) test. Data on graphs are shown as mean ± SEM with p-values indicated. Source data are provided as a Source Data file.
Next, we focused on day 14 pi, when the difference in viral titers in the brains of PrP KO and WT neonates was statistically significant. The analysis of other organs also revealed significantly lower MCMV titers in the PrP KO pups on day 14 (Fig. 1B). Again, no significant difference observed on day 8 (Fig. S1A). We concluded that the immunological effects of PrP absence in the context of congenital MCMV infection are not limited to the brain. In contrast, the amounts of virus in adult WT and PrP KO mice, where the virus reaches organs other than the brain, did not show significant differences, leading us to conclude that the effect is specific for neonates (Fig. S1B).
We have shown previously that, in mice infected upon birth, virus peaks in the brain between days 10 and 14 and is undetectable by day 21, congruent with the recruitment of CD8 T cells to the brain14. Next, we depleted CD8 T cells to determine whether these cells could account for the observed difference. Depletion of CD8 T cells resulted in an increase in virus titer in the brains of PrP KO mice to a level similar to that measured in WT animals (Fig. 1C). These findings supported the hypothesis of an improved control of MCMV by CD8 T lymphocytes in the absence of PrP. We observed the same in the spleen and liver, indicating that CD8 T cells are indeed the major cause of differences observed between PrP KO and WT mice. In contrast, the differences in the salivary glands were abolished only partially, in accordance with the dependence on CD4 T cells for virus control in the salivary glands57 (Fig. S1C). We therefore compared CD8 T cell effector functions and observed a stronger capacity of CD8 T cells from PrP KO mice to produce the antiviral cytokines IFNγ and TNFα (Fig. 1D, E).
In MCMV infection, CD8 T cells respond to several epitopes, developing virus-specific conventional and inflationary CD8 T cells. The presence of conventional T cells specific for the viral M45 or M57 epitopes indicates the early acute phase of infection, typically occurring between days 10 and 2014. Based on the titers observed at 14 dpi, we specifically examined this time-point and observed a significantly higher frequency and number of M45 and M57 tetramer-positive CD8 T cells in PrP KO neonates (Fig. 1F–H). Conversely, no difference was observed in adult mice regarding MCMV-M45 tetramer-positive and MCMV-M57 tetramer-positive CD8 T cells, which correlates with the absence of variation in viral titers (Fig. S1D). The scarcity of CD8 T cells infiltrating WT newborn mice brains during the early days post-infection when infected cells are still detectable by immunohistology (IH) prompted others to use IH for either CD8 T or MCMV detection on a single slide9,12,14,58,59. In PrP KO mouse brains, we show CD8 T cell infiltration among infected cells (Fig. 1I left) and a CD8 T cell colocalization with an MCMV-infected cell (Fig. 1I right). However, due to the limited occurrence of such events, we refrain from making quantified comparisons. As another indicator of the enhanced functionality of CD8 T cells derived from MCMV-infected PrP KO mice, we assessed the cytotoxic potential of CD8 T cells by stimulating splenocytes with MCMV-specific peptides. Flow cytometry analysis further confirmed our conclusions, as it revealed an increased expression of CD107a and an enhanced secretion of granzyme B by the PrP KO cells (Fig. S1E).
Collectively, our data indicate the importance of PrP in modulating the CD8 T cell response and influencing viral control during MCMV infection in neonatal mice.
MCMV-infected PrP KO neonates display a distinct CD8 T cell transcriptome, increased proliferative potential and heightened cytokine production
To gain a deeper understanding of the differences between CD8 T cell populations in PrP KO and WT mice, we employed single-cell RNA sequencing (scRNA-Seq) (Fig. 2A–D).
A–D scRNAseq analysis of CD8 T cells harvested from spleen tissue of MCMV-infected and naїve PrP KO and C57BL/6J mice. To obtain a purified population, CD8 T cells were FACS-sorted as FVD-CD45+CD3+CD19-CD8+. A UMAP visualization of CD8 T cell scRNAseq data with highlighted Seurat clusters. Clusters 0, 1, and 2 are marked on the UMAP. B Violin plot displaying cluster-specific gene expression. Clusters 0, 1, and 2 are indicated, with solid and dotted lines representing the most important genes. C UMAP visualization of CD8 T cell scRNAseq data with the sample names highlighted. D Bar plot showing the distribution of clusters in the MCMV-infected groups. Clusters 1 and 2 are labeled on the barplot. E Frequency of KLRG1 and CD62L (Sell) positive CD8 T cells in the spleen and liver of congenitally infected C57BL/6J and PrP KO mice at 14 dpi by flow cytometry. C57BL/6J mice (n = 8); PrP KO mice (n = 7); representative of 2 biological replicates. Statistical analysis was performed with GraphPad Prism using the two-tailed Mann–Whitney (U) test. Data on graphs are shown as mean ± SEM with p-values indicated. F Production of IFNγ in Maxi CD8 T cells upon antigen presentation by MCMV-infected BMDCs derived from the bone marrow of one female PrP KO mouse or one C57BL/6J mouse. Each symbol represents the average of a technical duplicate (adults) or a technical triplicate (young). The x-axis denotes the ratio of dendritic cells (DCs) to CD8 T cells. Source data are provided as a Source Data file.
In uninfected mice, both genotypes were characterized by the presence of cluster 0, representing naїve cells expressing the markers Sell+ (CD62L+) and IL7r+ (CD127+)60 (Fig. 2B, C). In MCMV-infected PrP KO mice cluster 1 emerged as the dominant group, represented by the highest number of cells, while cluster 2 prevailed in MCMV-infected WT mice (Fig. 2C, D). Notably, cluster 2 (WT) exhibited the expected profile associated with the effector and contraction phase (CD62L-IL7r-KLRG1 + CX3CR1+), indicative of short-lived effector cells61, while cluster 1 (PrP KO) displayed an intriguing combination of markers (CD62L-IL7r-KLRG1-CX3CR1-), deviating from the usual response to MCMV infection60. Furthermore, our flow cytometry analysis confirmed the observed differences in marker expression between the characteristic PrP KO and WT clusters at the protein level (Fig. 2E). Finally, it is worth highlighting the marker KLF2, which is the only one present in the dominant clusters of naïve and WT infected mice (clusters 0 and 2), but not in PrP KO (cluster 1) (Fig. 2B). Considering that this transcription factor is downregulated in effector T cells as a response determined by the affinity of T cell antigen receptor (TCR)62, this could speak in favor of a higher efficiency of CD8 T lymphocytes from PrP KO mice to clear MCMV.
The functional role of PrP in antigen-driven interactions with T cells has been observed in various experimental systems (reviewed in ref. 33). To assess the impact of PrP on the initial antigen presentation, we employed Maxi transgenic mice, which have TCR transgenic CD8 T cells specific for the immunodominant MCMV epitope M38316-32363. Bone marrow was collected from C57BL/6J or PrP KO mice to obtain bone marrow-derived dendritic cells (BMDCs). In vitro comparison of the priming efficiency of infected BMDCs from C57BL/6J and PrP KO mice did not show any notable differences (Fig. 2F), indicating that the observed differences are T cell-intrinsic and not dependent on the priming capacity of antigen-presenting cells. We tested differences in adult (6 weeks) and young Maxi mice (20 days) but did not observe differences in priming via PrP + or - BMDCs in any case. In conclusion, CD8 T lymphocytes in WT and PrP KO mice exhibit distinct transcriptome profiles, and PrP KO mice show improved control of MCMV infection.
Clearance of PrP from the surface of infected cells is observed in MCMV and HCMV infections across all analyzed cell types
Although generally considered optimal for studying antigen presentation, BMDCs exhibited relatively low levels of PrP on their surface. During the examination of the dendritic cell line DC2.4 to identify more pronounced membrane expression of PrP, we discovered a striking influence of CMV on surface PrP levels (Fig. 3A). Here, we show that MCMV caused disappearance of PrP from the surface of infected cells. To determine if this regulation is a widespread phenomenon, we initially confirmed it in BALB/c mouse embryonic fibroblasts (MEF) (Fig. 3B). Subsequently, the same downregulation of PrP was observed in MCMV-infected C57BL/6J MEF cells, immortalized NIH/Swiss embryo fibroblasts NIH 3T3, endothelial SVEC4-10 cells, and murine neuroblastoma Neuro-2a (N2a) cells (Fig. 3C). Furthermore, the downregulation of surface PrP was also evident in human foreskin fibroblasts (HFF), primary cells highly susceptible to HCMV infection (Fig. 3D). Building upon these observations, we then turned our attention to unraveling the molecular mechanisms underlying the CMV-induced modulation of PrP levels in infected cells.
Representative histograms showing PrP levels in mock or MCMV-infected DC2.4 cells (A) or mouse embryonic fibroblasts (MEF) (B) at 24 h post-infection (p.i.) with a 3 PFU/cell infection-dose. C Surface PrP levels in various murine cells at 24 h p.i., with the median fluorescence intensity (MFI) of the isotype control subtracted from the MFI of the PrP signal. D Surface PrP levels 48 h post HCMV infection in human foreskin fibroblast (HFF) cells. E PrP gene (Prnp) transcription in mock or MCMV-infected DC2.4 cells at 3 and 18 h p.i. (left) and RT-qPCR of mock or MCMV-infected N2a cells at 3, 12, and 30 h p.i. F Total PrP kinetics during MCMV infection, determined by intracellular flow cytometry and immunoblot at 6 and 24 h p.i. G Surface PrP kinetics during MCMV infection, determined by surface flow cytometry at 2, 6, and 24 h p.i. In mouse cell line experiments for flow cytometry, PrP staining was conducted using the D18 antibody. PrP staining on the surface of HFF was carried out utilizing the HuPrP.02 antibody. Immunoblot staining was performed with the MoPrP.03 clone, and actin was used as a loading control. A–G present typical data illustrating the key observations from the respective biological replicates, with data being repeated as follows: A (2 repetitions); B–D (over 10 times for MEFs, 1–5 times for other cells); E (1 repetition; technical replicates are shown; transcriptome 3; RT-qPCR average of two); F, G (2–10 repetitions depending on the technique and time point).
PrP is known to be stress-inducible, and our next-generation RNA sequencing dataset64 fittingly indicated PrP induction upon MCMV infection (Fig. 3E, left). The RT-qPCR data indicated a robust transcriptional upregulation in the early time points (between 3 and 12 h) and sustained induction compared to uninfected cells (between 12 and 30 h post-infection) (Fig. 3E, right). Using flow cytometry and immunoblotting, we indeed observed an increase in total PrP levels at 6 h post-infection (hpi). However, as the infection progressed to 24 hpi, infected cells exhibited significantly reduced PrP levels compared to uninfected controls (Figs. 3F, S2). To unravel the dynamics of PrP at the plasma membrane of infected cells, we conducted surface staining analysis by flow cytometry. Notably, a decline in surface PrP expression was already noticeable at 6 hpi, reaching its lowest level at 24 hpi (Fig. 3G). These results indicate that surface PrP is lost from the plasma membrane under the influence of infection, despite induction at both transcript and protein levels. Hence, our subsequent focus was to uncover the fate of PrP in MCMV infected cells.
In addition to investigating the dynamics of PrP during MCMV infection, at this point, it was crucial to explore the potential role of PrP in the initial infection of cells (as it could have a significant impact on our in vivo data), potentially leading to weaker infection and lower virus titers in tissues lacking PrP (Figs. 1, 2). To thoroughly explore this aspect, we conducted analyses using MEFs derived from PrP KO mice and N2a cells lacking PrP. Encouragingly, our findings confirmed that MCMV is capable of infecting cells independently of the presence of PrP (Fig. S3).
CMV-induced removal of PrP leads extends beyond the cell surface, leading to the progressive loss of cell-associated PrP during the course of infection
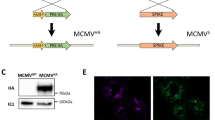
To first ensure the reliability of our observations regarding the disappearance of PrP in MCMV-infected cells (Fig. 3), we conducted additional experiments using multiple PrP-directed antibodies, in addition to our in-house-generated antibody to mouse PrP (moPrP.03, Fig. 3F). This comprehensive approach allowed us to strengthen the validity of our findings and to confirm the stark drop of PrP levels in infected cells (Fig. 4A). Furthermore, consistent reductions in PrP amounts were observed in lysates of different cell types upon infection (Fig. 4B) indicating that this phenomenon is not limited to a single cell type. Confocal microscopy analysis further supported our observations, revealing a significant decrease in the abundance of PrP in N2a cells following MCMV infection (Fig. 4C). For HCMV-infected cells, confocal microscopy with human PrP and HCMV markers IE1 or UL56.07 illustrates a clear reduction in PrP expression, confirmed by immunoblot in HFF cells (Figs. 4D and S4). Collectively, these findings show the substantial reduction of PrP in infected cells, prompting further inquiries into the fate and underlying mechanisms governing its removal.
A, B Immunoblots of PrP in mock or MCMV-infected lysates at 24 h post-infection. Different antibodies were used on MEF lysates (A) or different cell types were used (B). MCMV-M57 and -m04 were used as markers of successful infection. C N2a cells, mock- or MCMV-infected, stained for PrP (D18 antibody used). Nuclei visualized with DAPI. D Left: Mock-infected HFF cells showed membrane PrP expression (clone huPrP.02), while in HCMV-infected cells, nuclei were stained with anti-IE1 HCMV (clone E13; Argene), with minimal PrP signals. Both cases utilized the huPrP.02 clone and anti-IE1 HCMV (clone E13; Argene), both of IgG1 isotype. Right: Independent analysis showed reduced PrP levels exclusively in HCMV-infected cells, confirmed by UL56.07 antibody recognizing UL56 protein in the nucleus (arrows). Distinct secondary antibodies were employed (anti-IgG1 for huPrP.02 and anti-IgG2b for UL56.07). Nuclei visualized with DAPI. E Immunoblot of mock or MCMV-infected MEF cells grown in the presence of lactacystin (Lac) or leupeptin (Leu) at 24 h p.i. F Immunoblot of PrP and PVR in mock- or MCMV-infected MEF cells at 24 h p.i. G Immunoblot of PrP in lysates of mock- or MCMV-infected cells at 24 h p.i., prepared with a higher concentration of sodium dodecyl sulfate (SDS). H Immunoblot of PrP in mock or MCMV-infected MEF cells at 22 h p.i., grown in the presence of Brefeldin A, which was added 4 hpi and then replenished at 18 hpi. Images (C, D) were acquired using a Leica TCS SP8 confocal microscope and edited with LAS X software. Scale bars: 50 μm (C), 10 μm (D); (A–H) present typical data illustrating the key observations from the respective experimental conditions, with data being repeated as follows: A (2 repetitions); B (over 10 repetitions per MoPrP.03/MEF); C (3 repetitions); D (2 repetitions); E (3 repetitions); F–H (1 repetition).
We first examined whether virus-induced loss of PrP occurs through classical cellular protein degradation pathways. However, neither a proteasome inhibitor (lactacystin) nor the lysosomal degradation inhibitor (leupeptin) showed any significant rescue of PrP levels in infected cells (Fig. 4E). To confirm that the loss of PrP in virus-infected cells is not a non-specific phenomenon, we performed a poliovirus receptor (PVR) protein immunoblot in parallel, which showed no loss of PVR (Fig. 4F). Furthermore, to address the possibility that PrP could be hidden/remained in cell lysate pellets, we utilized a buffer with increased SDS concentration, which confirmed the overall ‘loss’ of cellular PrP (Fig. 4G). The only cell pathway inhibitor that prevented the loss of PrP caused by MCMV was brefeldin A, which we used as a positive control (Fig. 4H). Brefeldin A, known to inhibit the sorting of newly synthesized proteins, retained newly synthesized PrP within the cell and impaired surface exposure. These findings suggest that mature PrP is not degraded through conventional pathways, leading us to hypothesize that it might be removed by a direct release from the plasma membrane.
PrP is cleaved (shed) from the cell surface of MCMV-infected cells by the protease ADAM10
Next, we sought to investigate the dynamics of PrP removal from the cell surface in greater detail. We conducted a comparative analysis between the rate of PrP removal from the cell surface (Fig. 5A left) and the rate of removal of PrP protein labeled with antibodies shortly after the onset of infection (Fig. 5A right). In the first case (left), we monitored the classic downregulation of PrP on the surface, reflecting its steady state as observed previously. In the second example (right), however, our focus shifted away from tracking the newly synthesized PrP reaching the surface. Instead, we aimed to understand the fate of PrP already present on the plasma membrane. As anticipated, we observed a notably accelerated removal of PrP already present on the plasma membrane in infected cells (Fig. 5A right). This was further confirmed through confocal microscopy, where the absence of PrP-antibody complexes was observed in infected cells (Fig. 5B). While it is known that PrP-directed antibodies can modulate the internalization of PrP protein65, these findings emphasized the need to investigate the underlying mechanisms by which the virus eliminates PrP located on the plasma membrane itself.
Introductory scheme to Fig. 5A, B: (left) classic detection with anti-PrP antibody after the indicated duration of infection. PrP expression is induced, and newly synthesized PrP emerges at the cell surface. All surface PrP found at the chosen time point of infection is detected. (right) PrP labeled with an anti-PrP antibody at 0 h of infection. At certain hours post infection, only previously labeled PrP (PrP-Ab complex) is detected using a secondary antibody. A Flow cytometry analysis of surface PrP in MEF cells labeled with the D18 antibody prior to a 2-h, 6-h, or 24-h infection. The data is presented as the percentage of surface PrP relative to the 2 h post-infection time-point (100%). B Confocal microscopy of N2a cells labeled with D18 antibody prior to infection, captured 24 hpi. C–E N2a and MEF cells mock- or MCMV-infected for 24 h, treated with or without GI254023X (GI), an ADAM10 inhibitor. C N2A cells stained with D18 anti-PrP antibody, anti-IE1 MCMV infection marker, and DAPI for nuclei visualization. D N2a cells stained with POM1 anti-PrP antibody, and DAPI. E MEF cells stained with D18 anti-PrP antibody, and DAPI. F MEF and MEF ADAM10 KO cells mock- or MCMV-infected. 16 hpi cells were stained with D18 anti-PrP antibody, anti-IE1 infection marker, and DAPI. G Opti-MEM cell medium was harvested from MCMV-infected MEF cells at the indicated hpi and analyzed using the Mouse Prion Protein (PRNP) ELISA kit (Abbexa). H MEF cells were mock- or MCMV-infected and treated with or without GI in Opti-MEM cell medium. Medium was harvested at 20 hpi and subjected to NaDOC/TCA precipitation. Immunoblot performed using anti-shed-PrP antibody (sPrPG227). Biological replicates: A (2 repetitions for parallel staining); B (3); C (2); D, E, F (1 repetition each); G, H (2 repetitions each). B–E Leica TCS SP8 confocal microscope, LAS X software. Scale bars: 25 μm (B, D, E), 50 μm (F); NaDOC - Sodium deoxycholate. TCA - Trichloroacetic acid. Scheme created in BioRender.com.
Endogenous proteolytic shedding of PrP is performed by the metalloprotease ADAM1054,55. Previous studies have shown that HCMV reduces the expression of TIMP3, an endogenous inhibitor of both ADAM10 and ADAM17, resulting in increased ADAM17-mediated cleavage of substrates66. To investigate the impact of MCMV on TIMP3 and ADAM10, we reanalyzed a previous RNA-Seq dataset comparing MCMV-infected and mock-infected MEF cells67. Only TIMP3 transcripts were significantly downregulated in infected cells (Fig. S5). This confirms that both MCMV and HCMV have the same effect on decreasing TIMP3 RNA expression, which could result in increased activity of ADAM proteases. To further validate the involvement of ADAM10-mediated shedding in PrP removal during CMV infection, we treated N2a cells with the ADAM10 inhibitor GI254023X (GI) prior to MCMV infection. Indeed, GI-treated N2a cells exhibited reduced capacity to remove PrP from the cell surface upon MCMV infection (Fig. 5C, Fig. S6A). We confirmed this finding using an alternative PrP-directed antibody (POM1 instead of D18, Fig. 5D) and by employing a different cell type (MEF instead of N2a, Fig. 5E). Additionally, analysis of ADAM-KO cell line showed that cells lacking ADAM10 exhibited increased PrP levels on infected cells (Fig. 5F, Fig. S6B). Using the mouse prion protein (PRNP) ELISA Kit (abx156832), we confirmed that the amount of PrP gradually started to increase in the supernatants of cells upon MCMV-infection (Fig. 5G). Finally, we employed a cleavage site-specific antibody that exclusively detects the shed form of PrP (sPrPG227) cleaved by ADAM1068 and observed a substantial increase in this fragment in the supernatant of MCMV-infected cells (Fig. 5H). Collectively, our findings provide strong evidence that ADAM10-mediated shedding is an important mechanism by which PrP is removed from the plasma membrane and released into the supernatant of CMV-infected cells.
Binding of soluble PrP to CD8 T lymphocytes dampens their effector function
Having revealed the initial upregulation and subsequent release of PrP by infected cells, our investigation aimed to explore the potential relevance of this phenomenon to the enhanced efficiency of PrP KO neonates in eliminating MCMV (Figs. 1, 2). One possible explanation might be that soluble PrP, present in WT neonates, binds to CD8 T lymphocytes and dampens their effector function. To test this hypothesis, we generated a recombinant PrP-Fc fusion protein and exposed it to CD8 T lymphocytes, but observed no binding to CD8 T cells (Fig. 6A, E, in naїve mice). Based on these findings, we hypothesized that CD8 T lymphocytes bind PrP protein only in the context of infection. In support of this, the analysis of PrP engaging with CD8 T cells from newborn mice at 9, 12, and 14 dpi revealed a significant increase in PrP binding compared to CD8 T cells from uninfected mice (Fig. 6A). In contrast, CD4 T lymphocytes and B lymphocytes did not show increased binding to recombinant PrP following infection, thus confirming the CD8 T cell specificity of this phenomenon (Fig. 6B, C).
A–C Frequencies of PrP binding to CD8 T cells, CD4 T cells, and CD19 B cells from splenocytes of naïve and congenitally MCMV-infected C57BL/6J mice at indicated days post-infection (dpi); (CD8 T cells − 9 dpi: naive n = 4, MCMV n = 5; 12 dpi: naive n = 6, MCMV n = 8; 14 dpi: naive n = 4, MCMV n = 6; CD4 T cells − 9 dpi: naive n = 4, MCMV n = 5; 12 dpi: naive n = 5, MCMV n = 4; CD19 B cells − 9 dpi: naive n = 4, MCMV n = 5; 12 dpi: n = 5, both groups). D Frequencies of IFNγ and TNFα-positive CD8 T cells after incubation with PrP (PrP-hFc) or irrelevant protein (PVR-hFc) in the presence of cell stimulation cocktail and protein transport inhibitors. Splenocytes were harvested from congenitally MCMV-infected C57BL/6J mice at 14 dpi (n = 8; both groups). E Frequencies of PrP binding to CD8 T cells from splenocytes of naїve and congenitally MCMV-infected newborn C57BL/6J and PrP KO mice at 14 dpi (left); or naïve and MCMV-infected adult C57BL/6J and PrP KO mice at 8 dpi (right) (naïve groups: n = 3 each; newborn MCMV: n = 6; adult MCMV: n = 4). F Frequencies of IFNγ and TNFα-positive CD8 T cells after incubation with PrP or an irrelevant protein in the presence of cell stimulation cocktail and protein transport inhibitors. Splenocytes were harvested from congenitally MCMV-infected C57BL/6J mice or PrP KO mice at 14 dpi (n = 6; all groups). G Immunohistochemical staining of microglia (IBA-1) in brains of naive and congenitally MCMV infected mice (H) Thickness of the cerebellar external granular layer (EGL) in brains of uninfected and congenitally MCMV-infected C57BL/6J and PrP KO animals harvested at 9 dpi (n = 7 for MCMV PrP KO; n = 5 for other groups). Biological replicates: A–D (≥3), E (2), F (2), G (1) and H (2). Values for individual mice are shown. Statistical analysis was performed with GraphPad Prism using the two-tailed Mann–Whitney (U) test (A–F) or two-tailed unpaired t-test (H). Data on graphs are shown as mean ± SEM with p values indicated. Source data are provided as a Source Data file.
To assess the impact of PrP binding on CD8 T cells, we incubated splenocytes isolated from infected neonatal WT mice in the presence or absence of recombinant PrP and measured their ability to produce IFNγ and TNFα. According to our hypothesis, we expected that PrP binding would inhibit CD8 T lymphocytes, aligning with the stronger activation and improved virus clearance observed in PrP-deficient mice. Encouragingly, CD8 T lymphocytes isolated from MCMV-infected WT mice showed impaired cytokine production in the presence of PrP compared to controls (Fig. 6D). These findings strongly suggest that the CD8 T cell response to MCMV infection in newborn mice can be suppressed through the binding of soluble PrP. To fortify these findings, we showed that CD8 T cell inhibition is dose-dependent on PrP levels (Fig. S7). Furthermore, we demonstrated that binding of recombinant PrP to CD8 T lymphocytes occurs in cells isolated from newborn PrP KO mice infected with MCMV, effectively excluding the possibility of PrP-PrP self-association (Fig. 6E, neonates). Additionally, we confirmed that the binding of recombinant PrP to these PrP-deficient cells reduces their functional capacity (Figs. 6F and S8). This independent confirmation further supports the results presented in Fig. 6D and extends the impact of recombinant PrP on the functional capacity of both WT and PrP-deficient T cells. Consistent with the lack of differences in viral titers among strains, no binding of PrP was observed on adult CD8 T cells from MCMV-infected mice of either genotype (Fig. 6E, adults). Additionally, PrP exhibited no functional effects on non-binding T cells, including those from naїve or adult mice (Fig. S9). To further confirm the impact of PrP on the reduction of the antiviral CD8 T cell response, we adoptively transferred age-matched Maxi MCMV-specific CD8 T cells into MCMV-infected PrP KO or MCMV-infected C57BL/6J neonatal mice. IFNγ production by cells transferred to the PrP KO mice, and therefore in a milieu without PrP, was significantly higher compared to the IFNγ production by cells transferred to WT mice. Moreover, incubation of CD8 T cells with soluble PrP ex vivo suppressed IFNγ production (Fig. S10).
Having established that PrP KO mice show more effective CD8 T cell clearance of the virus, we investigated whether PrP KO mice displayed exacerbated brain pathology due to this enhanced immune response. Histological analyses monitoring typical brain development alterations caused by MCMV infection in WT mice did not reveal similar damage in the brains of PrP KO mice. Notably, in PrP KO mice, we did not observe the typical increase in the number of microglia (Fig. 6G), a contributor to brain damage in CMV infection, nor the characteristic thickening of the cerebellar external granular layer (EGL), both indicating developmental arrest in infected WT mice (Fig. 6H)10. The total number of astrocytes, serving as a control, remained similar between genotypes (Fig. S11). Altered morphogenesis of the brain in MCMV-infected mice, exemplified by a thickened EGL, is marked at earlier days post-infection, up to day 11. Our findings on CD8 T cells binding PrP already at days 9 and 12 (Fig. 6A) were further supported by monitoring the response to viremia in mice at 9 dpi, confirming once again an increased number of virus-specific CD8 T cells in PrP KO mice (Fig. S12). PrP KO mice exhibited a more favorable outcome after brain CMV infection compared to WT, showing better viral control without indications of stronger immunopathology. In conclusion, our results highlight a significant role of PrP in shaping the immune response and pathology during acute viral CMV infection.
Discussion
HCMV is the most common cause of congenital viral infection, with devastating neurological sequelae resulting from tropism for neural cells2. Despite ongoing treatments, children affected by symptomatic congenital HCMV infection still experience poor long-term outcomes3,7,8. The extent of damage tends to worsen with age, with the immune system playing a complex and often challenging role in mediating immunopathology instead of controlling the infection and limiting tissue damage7,13. Among the immune infiltrates within the brain, virus-specific CD8 T cells play a key role in virus clearance14. In our study using MCMV-infected newborn PrP KO mice, we observed significantly lower virus titers in various organs compared to WT mice, a reduction dependent on the presence and activity of CD8 T lymphocytes. Furthermore, we also report that the ex vivo binding of soluble PrP to activated CD8 T lymphocytes reduces their ability to produce IFNγ and TNFα. Previous studies have only shown PrP binding to NK cells and monocytes51,52 but not to T cells. Single-cell RNA sequencing revealed significant differences in the CD8 T cell populations between infected WT and PrP KO neonatal mice. A dominant CD62L-IL7r-KLRG1-CX3CR1-Klf2- population in PrP KO mice was nearly absent in WT pups. Finally, in vitro experiments showed that MCMV infection significantly upregulates PrP expression, followed by increased shedding of soluble PrP from infected cells via the metalloprotease ADAM10, likely facilitated by virus-mediated downregulation of the endogenous ADAM tissue inhibitor TIMP3. We also observed a similar loss of PrP from the surface of infected cells in our experiments with HCMV.
The interaction between PrP and CD8 T cells appears specific for neonatal mice, as neither this interaction nor improved MCMV control was observed in adult PrP KO mice. Previous studies have shown that neonatal CD8 T cells respond to infection differently than adult CD8 T cells. Neonatal cells exhibit enhanced proliferation, rapid recruitment during infection, a less diverse TCR repertoire, and differences in the expression of surface markers and receptors such as CD127, KLRG1, CD62L, and CD27, even when adoptively co-transferred69,70,71,72. Additionally, different expression pattern of transcription factors driving effector cell differentiation has revealed several dozen significantly different transcripts73,74. In humans, umbilical cord blood T cells express lower amounts of inhibitory receptors compared to adult peripheral blood T cells75,76. There is also a bias towards a Th2 response in human infants and neonates. Mechanisms such as suppression by TGF-β and a restricted repertoire of NK cell receptors in neonatal mice help limit inflammatory responses77,78, likely supporting colonization with commensals during the postnatal period79. In conclusion, a set of regulatory mechanisms is necessary to suppress the immune response early in life, and neonatal cells express significantly different amounts of numerous transcripts and proteins than their adult counterparts, including those in the surface proteome (reviewed in refs. 73,80). We hypothesize that PrP binding provides an additional layer of T cell regulation in neonatal mice, supporting the developing environment during in utero and postnatal exposure to environmental factors, including intracellular pathogens.
The role of PrP in viral infections has been particularly evident in influenza A infection, as demonstrated by increased pulmonary inflammation and higher viral titers in PrP KO mice27. Stimulation of PrP with antibodies induced M2 macrophage polarization, leading to reduced inflammation and viral titer in mice. Herpes simplex virus (HSV), an alpha herpesvirus belonging to the herpesvirus family, has the ability to penetrate the brain, replicate, and occasionally induce an acute inflammatory response. Notably, HSV is the sole herpesvirus that has been investigated in PrP KO mice to date, albeit with inconclusive outcomes29,81. However, all of the aforementioned studies have been performed on adult animals. By utilizing our well-established perinatal infection model, we show that the enhanced control of CMV in PrP KO mice is specifically associated with neonatal infection and was absent in adult mice. The underlying processes that are fundamentally different, the reason for the binding of PrP to T lymphocytes following MCMV infection in neonatal mice, and the potential involvement of other immune components still require further investigation.
PrP is released by ADAM10 and soluble PrP has been detected in human and mouse serum, suggesting that it might be of physiological relevance54,55,68,82. The processing of PrP is similar across different tissues, and substantial amounts of PrP are released into the medium by splenocytes and cerebellar granule cells82. In our study, we observed an increased proteolytic shedding of PrP by MCMV-infected cells, particularly during the initial hours following infection when PrP production was heightened. Notably, astrocytes were identified as a source of soluble PrP in the cerebrospinal fluid during HIV-1 CNS infection83. In the follow-up study, it was demonstrated that shedding of PrP can further increase the production of inflammatory mediators, exacerbating HIV-associated brain pathology, and it was suggested that targeting PrP shedding could be a potential therapeutic approach to mitigate the characteristic cognitive impairment observed in neuroAIDS30. Among the brain-resident cells, astrocytes represent the predominant cell type infected by HCMV during acute fetal brain infection84, making them a potential source of soluble PrP. While quantification in mice is lacking, human plasma PrP levels range from 5 to 10 ng per ml in healthy donors and exhibit a wide range in various disorders, from a few hundred nanograms in sera to several micrograms per ml in cerebrospinal fluid83,85,86,87. To contextualize our chosen PrP concentrations, we referred to a study using a concentration of 10 μg/ml for astrocytes in vitro30. Our experiments utilized 2 or 5 micrograms/sample (reported as 5) with a total volume of 200 μl, treating 1,000,000 PBMCs in each sample. Considering PrP-Fc with approximately 1 active site per fusion protein and an estimated molecular weight of around 100 kDa (compared to recombinant PrP’s 20 kDa), our calculations indicated an approximate usage of 6.25 μg/ml of PrP per 100 times more cells. Importantly, we showed a comparable effect even at 10 times lower PrP concentration (0.5 micrograms of recombinant fusion protein per sample). Acknowledging potential limitations, we recognize that our in vitro setup may not perfectly mimic physiological levels. Previous manuscripts lack discrimination between true shed PrP and overall soluble PrP. Additionally, immune cells within tissues may respond differently, facing hyperconcentration of shed PrP on one side and depletion due to cellular uptake on the other. These considerations underscore the complexity of in vivo scenarios.
In our long-standing research, we have investigated various immune ligands regulated by MCMV and HCMV in infected cells. Our established methodology involves identifying the specific viral regulator responsible for the observed effects. To assess the involvement of a specific MCMV gene in PrP regulation, we extensively examined several dozen viral mutants available in our repository, encompassing deletions of nearly all non-essential MCMV genes88. Despite this extensive analysis, none of the genes exhibited a specific impact on the surface expression of PrP. However, we did observe that viral mutants lacking the MCMV m154 gene displayed a relatively weaker effect on the removal of PrP from the cell. It is worth noting that, for our current study, we did not utilize these specific viral mutants due to the broader influence of m154 on a range of critical ligands involved in antiviral immunity, as previously reported in our own publications89,90. Despite the striking PrP loss from infected cells, which was confirmed to be facilitated by ADAM10-mediated shedding during infection, ADAM10 blockade could not completely prevent the reduction of PrP surface levels.
In addition to PrP, ADAM10 plays a role in the cleavage of several crucial molecules, including Notch, PD-L1, ICOS-L, TACI, EGFR/HER-, and NKG2D-ligands91,92. Therefore, blocking ADAM10 will, for instance, prevent NK and T cell activation through NKG2D. Based on our findings, simultaneous suppression of PrP shedding is expected to yield a synergistic effect on T cell activation. While targeting ADAM10 activity may appear as an attractive therapeutic strategy, considering its involvement in various cancer types, neurodegenerative diseases, and immune disorders, caution must be exercised due to potential side effects associated with its broad substrate spectrum and wide expression pattern. Given that tested ADAM10 therapies have proven ineffective, multiple factors, including PrP, must be carefully studied91,92. Notably, a recent study in Alzheimer’s disease models has shown promising beneficial effects of pharmacological ADAM10 stimulation, leading to increased shedding of PrP, a key neuronal receptor for toxic amyloid-beta peptides, from the plasma membrane93. It is worth mentioning that CMV is a herpesvirus that, once acquired, establishes lifelong infection characterized by sporadic reactivations. The influence of latent CMV infection on brain pathology is poorly understood94, but recent studies strongly suggest a connection between herpesviruses and an elevated risk of neurodegenerative diseases, including Alzheimer’s disease95.
Numerous PrP-directed antibodies and small molecules have been discovered through research focused on unraveling its devastating role in fatal and transmissible neurodegenerative prion diseases96. To further expand our understanding and explore the relevance of our findings, it is crucial to investigate whether PrP or stimulated shedding can modulate CD8 T lymphocyte responses and whether its depletion can lead to activated CD8 T lymphocytes in other relevant models. Antibodies specifically recognizing shed PrP are already available for rodent models68, and efforts are underway to characterize analogous antibodies for the human setting. Moreover, advanced studies are already exploring the specific influence of yet other PrP-directed antibodies and ligands on manipulating PrP shedding in a substrate-specific manner, without affecting the protease ADAM10 or its other substrates in the body. Notably, several PrP antibodies induce increased shedding by binding to the mature protein on the cell surface, while one antibody facilitates rapid uptake and degradation of PrP, effectively eliminating it as a substrate for shedding65. These and other means of manipulation93 should certainly be tested in virus disease models, such as ours, where the removal of PrP from cells is expected to confer benefits without evidence of causing harm, thereby opening up new possibilities for therapeutic interventions.
Methods
Mice
Mice were strictly age matched within experiments and handled in accordance with the guidelines contained in the International Guiding Principles for Biomedical Research Involving Animals. All mice were housed and bred under specific pathogen–free conditions at the Central Animal Facility of the Faculty of Medicine, University of Rijeka where they were maintained at 22 °C in a 12-h light–dark cycle, and relative humidity (40–50%). C57BL/6J mice (WT) were obtained from The Jackson Laboratory. Strictly co-isogenic C57BL/6J-Prnp−/− mice PrnpZH3/ZH3 (PrP KO)35 were a generous gift from A. Aguzzi. Maxi mice were a generous gift from A. Oxenius63. The Ethics Committee at the University of Rijeka and The National Ethics Committee for the Protection of Animals Used for Scientific Purposes (Ministry of Agriculture) approved experiments with laboratory animals (approval number HR-POK-004). Newborn mice were infected as a whole litter intraperitoneally (i.p.) with 400 PFU of MCMV prepared in volume of 50 µl on post-natal day 1. 8–10 weeks old adult male mice were infected with 2 × 105 PFU i.p. prepared in 500 µl sterile PBS. Viral titers in organs were determined by standard plaque assay. Virus titers are expressed as PFUs and correspond to the amount of infectious virus per gram tissue or per organ. CD8 T cells were depleted by i.p. injection of 50 µg of anti-CD8 (YTS 169.4, BioXcell) prepared in 50 µl of sterile PBS on days 4, 7, and 10 post infection. To isolate bone marrow to grow bone marrow-derived dendritic cells (BMDCs), 6 weeks old adult female mice were sacrificed, and bone marrow was flushed from tibia and femur. Maxi mice were sacrificed at 3 weeks of age (noted as “young”) and 6 weeks of age (noted as “adult”) for harvesting splenic cells used in the co-cultivation experiments with BMDCs. The investigated research is not influenced by sex. However, sex is consistently taken into account and clearly stated for each analysis involving adult mice. In the case of newborn mice, the analysis was not conducted based on their sex due to their extremely young age (1 day). Since it was necessary to compare mice (uninfected, infected, 2 strains) that were born on the exact same day, selecting one sex was not only deemed unnecessary but also unfeasible. Nevertheless, in an initial large titration experiment, sex was recorded for additional verification, and it was found that sex did not impact virus titer in congenitally infected PrP KO and C57BL/6J at 14 dpi.
Cells
MEF (mouse embryonic fibroblasts from BALB/c mice, C57BL/6J mice, PrP KO mice, immortalized ADAM10 KO MEF cells and their respective WT controls97), N2a (Neuro-2a; murine neuroblastoma cells) SVEC4-10 (a mouse endothelial cell line), NIH 3T3 (mouse embryo fibroblast cell line) and HFF (human foreskin fibroblasts) were cultivated in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% or 3% of fetal calf serum (FCS) and 100 U/ml of penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine10,98. DC2.4 cells (immortalized dendritic cells from C57BL/6 mouse) were cultivated in Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with 10% of FCS and 100 U/ml of penicillin, 100 μg/ml streptomycin and 2 mM L-glutamine. N2a, SVEC4-10, NIH 3T3, and DC2.4 cells are derived from the cell data bank of the Faculty of Medicine University of Rijeka. These are mouse-derived cell lines and the specific sex of the mice from which these cells are derived is generally unknown or unspecified. MEF cells were mixed sex since they were obtained by homogenization of total embryos 17 days after fertilization. BMDC cells were obtained from female mice. Opti-MEM (Reduced Serum Media) was not supplemented. To obtain bone marrow-derived dendritic cells (BMDCs), cells from the bone marrow of C57BL/6 and PrP KO mice were grown for 7 days in 10% RPMI 1640 supplemented with 20% GM-CSF (fresh medium was added every two days into the flasks, 1 ml per 106 cells) to obtain BMDCs as a floating and loosely attached fraction of derived cells99. All cells were grown in an incubator that enables conditions of 5% CO2 and 37 °C.
Viruses
Viruses were produced on BALB/c MEFs using an ultracentrifuge (Sorvall) and titrated by standard plaque assay as described10. In brief, virus stocks were titrated by serial 10-fold dilution in 48-well tissue culture plates containing MEF monolayer overlaid with methylcellulose. After incubation for 4 days, plaques were enumerated and plaque forming units (PFU) per milliliter determined. We used the BAC pSM3fr-derived MCMV (C3X), based on the MCMV Smith strain (ATCC VR-1399) as WT, Δm138-MCMV for immunofluorescence assays and Δm157-MCMV for adult mice infection. HCMV strain TB40/E was used for infection of human cells.
Cell treatment
Cells were seeded (1 × 106 cells/10 cm petri dish or 3 × 105 cells/6-well plate) and infected with the dose of 0.5–3 PFU/cell. The cells were incubated with virus (30’, 37 °C, 5% CO2), centrifuged (30’, 800 g) and further incubated. Infection of cells in suspension was carried out by resuspending cells in viral suspension (107 cells/ml of viral suspension) and incubating (60’, 37 °C). Cells were treated with Brefeldin A (5 μg/ml, eBioscience), Leupeptin (15 μg/ml, Sigma-Aldrich, L5793), Lactacystin (10 μM/ml, Sigma-Aldrich, L6785) and GI 254023X in Opti-MEM (3 μM, Merck Millipore). Infection with HCMV was performed as with MCMV, 0.5 PFU/cell for 48–90 h.
Flow cytometry
Splenic leukocytes were prepared using standard protocols including mechanical dissociation and brief erythrocytes lysis. To isolate lymphocytes from liver, the tissue was mechanically dissociated. A 40% Percoll/liver homogenate suspension was underlaid with 80% Percoll in PBS and centrifuged (25’,1050 g). Fixable Viability Dye coupled to Alexa Fluor 780 was used and Fc receptors were blocked using anti-CD16/CD32 (clone 2.4G2, produced in-house). Flow cytometric analysis included anti-mouse CD45.2 (104, AlexaFluor700 1:100 or eFluor506), CD3 (145-2C11, FITC or SuperBright600 1:100), CD8α (53-6.7, SuperBright780, 1:400), CD44 (IM7, AlexaFluor700, 1:400), CD19 (eBio1D3, PerCP-Cy5.5, 1:800), NK1.1 (PK136, PE-eFluor 610 1:100), CD4 (RM4-5, APC, 1:100), TNFα (MP6-XT22, AlexaFluor488, 1:100 or PE-eFluor610, 1:80), IFNγ (XMG1.2, PE or eFluor450, 1:100), KLRG1 (2F1, PE-eFluor610, 1:200 or APC, 1:400), CD62L (MEL-14, PE-Cy7, 1:400) from Thermo Fischer Scientific. Anti-mouse CD8α (53-6.7, BV786, 1:600) antibody was from BD Biosciences. Gating strategy for tetramer positive populations, cytokine production/cytotoxic potential, PrP binding to cells, and sorting cells for scRNA-seq is included in supplemental information as Supplemental Fig. S13. To measure cytokines, splenocytes cells were incubated in the presence of Brefeldin A (10 mg/ml, eBioscience). Flow cytometry of cell lines was performed with antibodies D18 or D13 (acquired from H.C.A; 1:150). The H-2D(b)-MCMV-M45-tetramer and H-2K(b)-MCMV-M57-tetramer were obtained through NIH Tetramer Core Facility (985HGIRNASFI993 and 816SCLEFWQRV824, respectively). Secondary antibodies included goat α-human FITC (Jackson ImmunoResearch, 1:100) and rat α-human IgG Fc PE-Cy7 (BioLegend, 1:200). For PrP detection in HFF cells, we used huPrP.02 clone (1:100) produced in-house. To assess the binding of PrP on CD8 T cells, isolated splenocytes were incubated with 5 µg of PrP-hFc fusion protein or an irrelevant fusion protein (produced in-house) for 25 min. Alternatively, cells were incubated for 4 h in the presence of Cell Stimulation Cocktail, along with protein transport inhibitors (eBioscience), added to the cell medium to measure cytokine production in the presence of PrP. Flow cytometry data were acquired using FACSAriaIIu (BD Biosciences) and analyzed using FlowJo v10 (TreeStar) software. The monoclonal antibodies and proteins generated in this study, including anti-human PrP (clone HuPrp.02), and the PrP-long-hFc (hIgG1) fusion protein, have been deposited at the Center for Proteomics, University of Rijeka, Croatia [https://products.capri.com.hr/].
Immunoblot
Cell lysates were prepared using RIPA buffer (NaCl 150 mM, Triton X-100 1%, 0.5% Sodium deoxycholate, 0.1% SDS, 50 mM Tris-HCl pH 8.0; cOmplete™, Mini, EDTA-free Protease Inhibitor Cocktail). 50–100 mg of protein lysate was separated by 12% SDS-PAGE electrophoresis. Incubation with primary antibodies was performed overnight at 4 °C. α-mouse IgG (1:8000) and α-rabbit IgG (1:2000) HRP-coupled secondary antibody (Jackson Immuno Research) were incubated at room temperature for 1 h. Supernatants of cells treated with GI 254023X were collected after 5–24 h of infection and precipitated with 0.1% Na-DOC and 10% trichloroacetic acid. Primary antibodies used: MoPrP.03 (1:500), IE1.01 (1:1000), m04.10 (1:1000), M55.01 (1:500), E1.01/CROMA103 (1:1000), mPVR.13 (1:1000) and M57.02 (1:1000) (produced in-house), W226, D18 (1:1000) and sPrPG227 (1:1000) (acquired from G.L. and H.C.A.). Immunoblot signals were visualized using Image-Quant LAS 4000 mini-instrument (GE Healthcare). Cleavage site-specific antibody for the detection of murine shed PrP (sPrPG227) may be provided by Dr. Altmeppen (Hamburg, Germany) upon reasonable request. The monoclonal antibodies generated in this study, including anti-MCMV (clone m04.10, M57.02, IE1.01, E1.01), anti-mouse PrP (clone MoPrp.03), have been deposited at the Center for Proteomics, University of Rijeka, Croatia [https://products.capri.com.hr/].
Immunofluorescence
MEF and N2A cells grown on coverslips were infected with 1–3 PFU/cell of Δm138-MCMV. HFF cells were infected with 0.5 PFU/cell of TB40/E HCMV for 48 h. Cells were fixed and permeabilized in ice cold methanol. Antibodies were incubated for 1 h in humid conditions. Primary antibodies: IE1.01, UL56.07, and HuPrP.02 (produced in-house, 1:100), D18 (acquired from G.L., 1:150), POM1 (Millipore, 1:100) to recognize PrP and anti HCMV IE1 (Argene, 1:100). In the antibody internalization assay, cells were incubated with primary antibodies for 20 min in PBS on ice and then washed twice before incubation. DAPI was used to visualize nuclei. The samples were stained with secondary antibodies coupled to fluorescein isothiocyanate (FITC, 1:100) or tetramethylrhodamine (TRITC, 1:100) fluorophores. Cell preparations were imaged using an inverted confocal microscope Leica DMI8 (confocal unit: TCS SP8; Leica Microsystems GmbH, Wetzlar, Germany), equipped with UV (diode 405), Ar 488, DPSS 561, and He/Ne 633 lasers, and with 4 detectors (2 × PMT and 2 × HyD). An HC PLAPO CS2 (63×/1.40 oil) oil immersion objective was used. Images (515 × 515 pixels) were acquired using LAS (Leica Application Suite) × software (Leica Application Suite × 3.5.6.21594), in sequential mode, with pixel sizes ranging from 481.47 × 481.47 nm to 120.37 × 120.37 nm. Each experiment was acquired under the same parameters, referencing control preparations. Adjustments included minimal thresholding consistently applied to all samples within the same group. No gamma correction, pseudocoloring, contrast adjustment, or similar modifications were applied. The monoclonal antibodies generated in this study, including anti-HCMV (clone UL56.07) and anti-human PrP (clone HuPrp.02), have been deposited at the Center for Proteomics, University of Rijeka, Croatia [https://products.capri.com.hr/].
Histology
For morphometric analyzes of cerebellum, serial sagittal sections (3 µm thick) were stained with cresyl violet (Thickness of the cerebellar EGL was measured at eight points along the primary cerebellar fissure). Immunohistochemical staining was performed using anti-mouse-Iba-1 antibody for microglia (FUJIFILM; Wako Chemicals) and GFAP for astrocytes. Antibody binding was visualized with peroxidase-conjugated goat anti-rabbit IgG antibody (Abcam), followed with 3,3′-diaminobenzidine (Dako). Slides were counterstained with hematoxylin. The liver staining was performed with hematoxylin and eosin. Histological slides were analyzed with BX 51 microscope (Olympus) and CellSens Dimension software were used (cellSens Standard V1.14).
qPCR
RNA was extracted from N2A cells using an RNA extraction kit (NucleoSpin RNA Plus; MN), and cDNA was generated with a reverse transcription kit (iScript Reverse Transcription Supermix for RT-qPCR; BioRad). The expression of mRNA was examined by qPCR with a 7500 Fast Real Time PCR machine 7500 software (v2.3) (ABI). TaqMan assays were used to quantify the expression of Prnp (Mm00448389_m1). Relative mRNA expression was normalized by quantification of Gapdh (Mm05724508_g1) RNA in each sample.
ELISA
MEF cells were seeded in a 6-well plate and infected as described in the Cell treatment section. Supernatants were harvested from MCMV-infected cells at 6, 12, 18, and 24 h post-infection (from independent wells). Cells were grown in Opti-MEM medium (Thermo Fisher). The Mouse Prion Protein (PRNP) ELISA Kit (Abbexa, abx156832) was used to measure the absorbance in samples, with the Opti-MEM absorbance subtracted as a blank. The absorbance was measured using a TriStar LB 941 multimode microplate reader (Berthold) with the wavelength set at 450 nm and MikroWin2010 software (v5.24).
Statistical analysis
Data are presented as mean or mean ± SEM. Statistical significance was determined by either two-tailed Mann–Whitney U test or two-tailed unpaired t-test using GraphPad Prism v8. A value of p > 0.05 was deemed not statistically significant, if significant p value is denoted in the figure. Differences between values were considered significant when *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001. In the figure legends, detailed information is provided regarding the experimental design, including the number of replicates for each experiment, the number of animals per group, and other relevant data.
Single cell RNA-sequencing
Splenic leukocytes were prepared from congenitally MCMV infected and naïve PrP KO and C57BL/6J mice 14 days post-infection using standard protocols including mechanical dissociation and brief erythrocyte lysis. Cells were counted and cryopreserved in fetal bovine serum (FBS) supplemented with 10% DMSO at −80 °C. Gently thawed samples were stained with Fixable Viability Dye (FVD) and fluorochrome-conjugated antibodies and two target populations were FACS sorted by MoFlo EQ Astrios (Beckman Coulter): FVD−CD45.2+CD3+CD19-CD8+ T cells and FVD-CD45.2+CD3-CD19-NK1.1+ NK cells.
For single-cell RNA-seq experiments, the Chromium Next GEM Single Cell 3’ v3.1 kit (10X Genomics) was used according to manufacturer instructions. Each sample was composed of 10,000 freshly sorted T and NK cells in a ratio of 9:1, respectively. Briefly, the single-cell suspension was loaded on the 10X Chromium controller to generate the Gel bead-in Emulsion (GEM). After the encapsulation, cells were lysed, and the barcoded mRNA was reverse-transcribed. The generated cDNA was amplified and used to prepare libraries for next-generation sequencing (NGS). The sequencing was performed on a Novaseq S4 (Illumina) sequencer aiming at 20k reads/cell.
Sequences and data processing
Genome mapping and count matrix generation were done following 10X Genomics Cell Ranger pipeline (v6.0.1) with the refdata-gex-mm10-2020-A reference genome. scRNA-seq datasets were processed using Seurat V3 ((https://satijalab.org/seurat/)) in R (R Core Team (2022). R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/.). Cells with more than 10% of mitochondrial gene fraction, less than 500 detected genes, and less than 1000 UMI were discarded. Based on marker genes expression and singleR annotation, cells not corresponding to either NK or T-cells were also discarded. Normalization was performed using the SCTransform Seurat function with 2500 variable features and a regression on the mitochondrial fraction. UMAP generation and clustering were performed following Seurat workflow with the first 30 PC. All 5 samples were merged using the Seurat merge function. The 2 replicates (PrP-KO-MCMV) overlapped completely, suggesting that no corrections or data integrations was required.
Graphics
BioRender was used to create the respective images, with the licenses provided as follows: Fig. 5a, b: Agreement Number: ZN272VJ45S; Graphical Abstract: Agreement Number: CG272VK2TH.
Additionally, all graphs were generated in GraphPad Prism (v8), then exported and imported into Adobe Illustrator CS5 for panel organization and font adjustment.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Single-cell RNA sequencing (scRNA-seq) data generated in this study have been deposited in the NCBI Gene Expression Omnibus (GEO) archive platform database under accession code GSE245338. The data source file, containing raw data of ex vivo experiments and uncropped Western blots, is in Excel format and available at https://doi.org/10.6084/m9.figshare.24290398.v2. Source data are provided with this paper.
Code availability
The R scripts used for the scRNA-seq analysis will be available in public repositories by the end of 2024 as part of the Institut Pasteur’s joint script deposition. Until then, the scripts are available upon request from Sébastien Mella (sebastien.mella@pasteur.fr) or the corresponding author.
References
Plotkin, S. A. et al. The status of vaccine development against the human cytomegalovirus. J. Infect. Dis. 221, S113–S122 (2020).
Zuhair, M. et al. Estimation of the worldwide seroprevalence of cytomegalovirus: a systematic review and meta-analysis. Rev. Med. Virol. 29, e2034 (2019).
Marsico, C. & Kimberlin, D. W. Congenital cytomegalovirus infection: advances and challenges in diagnosis, prevention and treatment. Ital. J. Pediatr. 43, 38 (2017).
Gantt, S., Bitnun, A., Renaud, C., Kakkar, F. & Vaudry, W. Diagnosis and management of infants with congenital cytomegalovirus infection. Paediatr. Child Health 22, 72–74 (2017).
Pass, R. F., Fowler, K. B., Boppana, S. B., Britt, W. J. & Stagno, S. Congenital cytomegalovirus infection following first trimester maternal infection: symptoms at birth and outcome. J. Clin. Virol. 35, 216–220 (2006).
Chiopris, G. et al. Congenital cytomegalovirus infection: update on diagnosis and treatment. Microorganisms 8, https://doi.org/10.3390/microorganisms8101516 (2020).
Lanzieri, T. M., Caviness, A. C., Blum, P., Demmler-Harrison, G. & Longitu, C. C. Progressive, long-term hearing loss in congenital CMV disease after ganciclovir therapy. J. Pediat. Infect. Dis. Soc. 11, 16–23 (2022).
Kimberlin, D. W. et al. Valganciclovir for symptomatic congenital cytomegalovirus disease. N Engl. J. Med. 372, 933–943 (2015).
Slavuljica, I. et al. Immunobiology of congenital cytomegalovirus infection of the central nervous system-the murine cytomegalovirus model. Cell. Mol. Immunol. 12, 180–191 (2015).
Brizic, I. et al. Mouse models for cytomegalovirus infections in newborns and adults. Curr. Protoc. 2, e537 (2022).
Workman, A. D., Charvet, C. J., Clancy, B., Darlington, R. B. & Finlay, B. L. Modeling transformations of neurodevelopmental sequences across mammalian species. J. Neurosci. 33, 7368–7383 (2013).
Koontz, T. et al. Altered development of the brain after focal herpesvirus infection of the central nervous system. J. Exp. Med. 205, 423–435 (2008).
Kvestak, D. et al. NK/ILC1 cells mediate neuroinflammation and brain pathology following congenital CMV infection. J. Exp. Med. 218, https://doi.org/10.1084/jem.20201503 (2021).
Brizic, I. et al. Brain-resident memory CD8(+) T cells induced by congenital CMV infection prevent brain pathology and virus reactivation. Eur. J. Immunol. 48, 950–964 (2018).
Bradford, R. D. et al. Murine CMV-induced hearing loss is associated with inner ear inflammation and loss of spiral ganglia neurons. PLoS Pathog. 11, e1004774 (2015).
Kosmac, K. et al. Glucocorticoid treatment of MCMV infected newborn mice attenuates CNS inflammation and limits deficits in cerebellar development. PLoS Pathog. 9, e1003200 (2013).
Pass, R. F. & Arav-Boger, R. Maternal and fetal cytomegalovirus infection: diagnosis, management, and prevention. F1000Res 7, 255 (2018).
Atkinson, C. J., Zhang, K., Munn, A. L., Wiegmans, A. & Wei, M. Q. Prion protein scrapie and the normal cellular prion protein. Prion 10, 63–82 (2016).
Legname, G. Elucidating the function of the prion protein. PLoS Pathog. 13, e1006458 (2017).
Bakkebo, M. K. et al. The cellular prion protein: a player in immunological quiescence. Front. Immunol. 6, 450 (2015).
Tsutsui, S., Hahn, J. N., Johnson, T. A., Ali, Z. & Jirik, F. R. Absence of the cellular prion protein exacerbates and prolongs neuroinflammation in experimental autoimmune encephalomyelitis. Am. J. Pathol. 173, 1029–1041 (2008).
Gourdain, P., Ballerini, C., Nicot, A. B. & Carnaud, C. Exacerbation of experimental autoimmune encephalomyelitis in prion protein (PrPc)-null mice: evidence for a critical role of the central nervous system. J. Neuroinflamm. 9, 25 (2012).
Ezpeleta, J. et al. Protective role of cellular prion protein against TNFalpha-mediated inflammation through TACE alpha-secretase. Sci. Rep. 7, 7671 (2017).
Salvesen, O. et al. LPS-induced systemic inflammation reveals an immunomodulatory role for the prion protein at the blood-brain interface. J. Neuroinflamm. 14, 106 (2017).
Nakamura, Y. et al. Transfection of prion protein gene suppresses coxsackievirus B3 replication in prion protein gene-deficient cells. J. Gen. Virol. 84, 3495–3502 (2003).
Nasu-Nishimura, Y. et al. Cellular prion protein prevents brain damage after encephalomyocarditis virus infection in mice. Arch. Virol. 153, 1007–1012 (2008).
Chida, J. et al. Prion protein signaling induces M2 macrophage polarization and protects from lethal influenza infection in mice. PLoS Pathog. 16, e1008823 (2020).
Chida, J. et al. Prion protein protects mice from lethal infection with influenza A viruses. PLoS Pathog. 14, e1007049 (2018).
Korom, M. et al. A proautophagic antiviral role for the cellular prion protein identified by infection with a herpes simplex virus 1 ICP34.5 mutant. J. Virol. 87, 5882–5894 (2013).
Megra, B. W., Eugenin, E. A. & Berman, J. W. The role of Shed PrP(c) in the Neuropathogenesis of HIV Infection. J. Immunol. 199, 224–232 (2017).
Wulf, M. A., Senatore, A. & Aguzzi, A. The biological function of the cellular prion protein: an update. BMC Biol. 15, 34 (2017).
Isaacs, J. D., Jackson, G. S. & Altmann, D. M. The role of the cellular prion protein in the immune system. Clin. Exp. Immunol. 146, 1–8 (2006).
Zhang, B. et al. Expression and functions of cellular prion proteins in immunocytes. Scand. J. Immunol. 91, e12854 (2020).
Steele, A. D., Lindquist, S. & Aguzzi, A. The prion protein knockout mouse: a phenotype under challenge. Prion 1, 83–93 (2007).
Nuvolone, M. et al. Strictly co-isogenic C57BL/6J-Prnp-/- mice: a rigorous resource for prion science. J. Exp. Med. 213, 313–327 (2016).
Zhang, C. C., Steele, A. D., Lindquist, S. & Lodish, H. F. Prion protein is expressed on long-term repopulating hematopoietic stem cells and is important for their self-renewal. Proc. Natl Acad. Sci. USA 103, 2184–2189 (2006).
Linden, R. The biological function of the prion protein: a cell surface Scaffold of signaling modules. Front. Mol. Neurosci. 10, 77 (2017).
Beland, M. & Roucou, X. The prion protein unstructured N-terminal region is a broad-spectrum molecular sensor with diverse and contrasting potential functions. J. Neurochem. 120, 853–868 (2012).
Cashman, N. R. et al. Cellular isoform of the scrapie agent protein participates in lymphocyte activation. Cell 61, 185–192 (1990).
Ingram, R. J. et al. A role of cellular prion protein in programming T-cell cytokine responses in disease. FASEB J. 23, 1672–1684 (2009).
Durig, J. et al. Differential constitutive and activation-dependent expression of prion protein in human peripheral blood leucocytes. Br. J. Haematol. 108, 488–495 (2000).
Biasini, E., Turnbaugh, J. A., Unterberger, U. & Harris, D. A. Prion protein at the crossroads of physiology and disease. Trends Neurosci. 35, 92–103 (2012).
Ballerini, C. et al. Functional implication of cellular prion protein in antigen-driven interactions between T cells and dendritic cells. J. Immunol. 176, 7254–7262 (2006).
Mariante, R. M. et al. Neuroimmunoendocrine regulation of the prion protein in neutrophils. J. Biol. Chem. 287, 35506–35515 (2012).
Wu, G. R. et al. Prion protein is required for tumor necrosis factor alpha (TNFalpha)-triggered nuclear factor kappaB (NF-kappaB) signaling and cytokine production. J. Biol. Chem. 292, 18747–18759 (2017).
Paar, C., Wurm, S., Pfarr, W., Sonnleitner, A. & Wechselberger, C. Prion protein resides in membrane microclusters of the immunological synapse during lymphocyte activation. Eur. J. Cell Biol. 86, 253–264 (2007).
Hu, W. et al. Pharmacological prion protein silencing accelerates central nervous system autoimmune disease via T cell receptor signalling. Brain 133, 375–388 (2010).
Mabbott, N. A., Brown, K. L. & Bruce, M. E. T lymphocyte activation and the cellular form of the prion protein, PrPc. Biochem. Soc. Trans. 25, 307S (1997).
Wurm, S. et al. Co-localization of CD3 and prion protein in Jurkat lymphocytes after hypothermal stimulation. FEBS Lett. 566, 121–125 (2004).
Mattei, V. et al. Prion protein is a component of the multimolecular signaling complex involved in T cell activation. FEBS Lett. 560, 14–18 (2004).
Seong, Y. J. et al. Activation of human natural killer cells by the soluble form of cellular prion protein. Biochem. Biophys. Res. Commun. 464, 512–518 (2015).
Jeon, J. W. et al. The soluble form of the cellular prion protein enhances phagocytic activity and cytokine production by human monocytes via activation of ERK and NF-kappaB. Immune Netw. 13, 148–156, (2013).
Krebs, B. et al. Prion protein induced signaling cascades in monocytes. Biochem. Biophys. Res. Commun. 340, 13–22 (2006).
Taylor, D. R. et al. Role of ADAMs in the ectodomain shedding and conformational conversion of the prion protein. J. Biol. Chem. 284, 22590–22600 (2009).
Altmeppen, H. C. et al. Lack of a-disintegrin-and-metalloproteinase ADAM10 leads to intracellular accumulation and loss of shedding of the cellular prion protein in vivo. Mol. Neurodegener. 6, 36 (2011).
Vanni, I. et al. An optimized Western blot assay provides a comprehensive assessment of the physiological endoproteolytic processing of the prion protein. J. Biol. Chem. 299, 102823 (2023).
Al-Talib, M., Dimonte, S. & Humphreys, I. R. Mucosal T-cell responses to chronic viral infections: Implications for vaccine design. Cell. Mol. Immunol. https://doi.org/10.1038/s41423-024-01140-2 (2024).
Bantug, G. R. et al. CD8+ T lymphocytes control murine cytomegalovirus replication in the central nervous system of newborn animals. J. Immunol. 181, 2111–2123 (2008).
Cekinovic, D. et al. Passive immunization reduces murine cytomegalovirus-induced brain pathology in newborn mice. J. Virol. 82, 12172–12180 (2008).
van den Berg, S. P. H. et al. The hallmarks of CMV-specific CD8 T-cell differentiation. Med. Microbiol. Immunol. 208, 365–373 (2019).
Herndler-Brandstetter, D. et al. KLRG1(+) effector CD8(+) T cells lose KLRG1, differentiate into all memory T cell lineages, and convey enhanced protective immunity. Immunity 48, 716–729.e718 (2018).
Preston, G. C., Feijoo-Carnero, C., Schurch, N., Cowling, V. H. & Cantrell, D. A. The impact of KLF2 modulation on the transcriptional program and function of CD8 T cells. PLoS ONE 8, e77537 (2013).
Torti, N., Walton, S. M., Brocker, T., Rulicke, T. & Oxenius, A. Non-hematopoietic cells in lymph nodes drive memory CD8 T cell inflation during murine cytomegalovirus infection. PLoS Pathog. 7, e1002313 (2011).
Lucin, P. et al. Cytomegaloviruses exploit recycling Rab proteins in the sequential establishment of the assembly compartment. Front. Cell Dev. Biol. 6, 165 (2018).
Linsenmeier, L. et al. Ligands binding to the prion protein induce its proteolytic release with therapeutic potential in neurodegenerative proteinopathies. Sci. Adv. 7, eabj1826 (2021).
Esteso, G. et al. Altered microRNA expression after infection with human cytomegalovirus leads to TIMP3 downregulation and increased shedding of metalloprotease substrates, including MICA. J. Immunol. 193, 1344–1352 (2014).
Juranic Lisnic, V. et al. Dual analysis of the murine cytomegalovirus and host cell transcriptomes reveal new aspects of the virus-host cell interface. PLoS Pathog. 9, e1003611 (2013).
Linsenmeier, L. et al. Structural and mechanistic aspects influencing the ADAM10-mediated shedding of the prion protein. Mol. Neurodegener. 13, 18 (2018).
Rudd, B. D., Venturi, V., Davenport, M. P. & Nikolich-Zugich, J. Evolution of the antigen-specific CD8+ TCR repertoire across the life span: evidence for clonal homogenization of the old TCR repertoire. J. Immunol. 186, 2056–2064 (2011).
Rudd, B. D. et al. Acute neonatal infections ‘lock-in’ a suboptimal CD8+ T cell repertoire with impaired recall responses. PLoS Pathog. 9, e1003572 (2013).
Smith, N. L. et al. Rapid proliferation and differentiation impairs the development of memory CD8+ T cells in early life. J. Immunol. 193, 177–184 (2014).
Venturi, V. et al. The Neonatal CD8+ T cell repertoire rapidly diversifies during persistent viral infection. J. Immunol. 196, 1604–1616 (2016).
Tabilas, C., Smith, N. L. & Rudd, B. D. Shaping immunity for life: Layered development of CD8(+) T cells. Immunol. Rev. 315, 108–125 (2023).
Maymi, V. I. et al. Neonatal CD8+ T cells resist exhaustion during chronic infection. J. Immunol. 212, 834–843 (2024).
Dietz, S. et al. Expression of immune checkpoint molecules on adult and neonatal T-cells. Immunol. Res. 71, 185–196 (2023).
Miller, R. E. et al. Reduced CTLA-4 protein and messenger RNA expression in umbilical cord blood T lymphocytes. Exp. Hematol. 30, 738–744 (2002).
Marcoe, J. P. et al. TGF-beta is responsible for NK cell immaturity during ontogeny and increased susceptibility to infection during mouse infancy. Nat. Immunol. 13, 843–850 (2012).
Rozmanic, C. et al. Perinatal murine cytomegalovirus infection reshapes the transcriptional profile and functionality of NK cells. Nat. Commun. 14, 6412 (2023).
Zaghouani, H., Hoeman, C. M. & Adkins, B. Neonatal immunity: faulty T-helpers and the shortcomings of dendritic cells. Trends Immunol. 30, 585–591 (2009).
Dowling, D. J. & Levy, O. Ontogeny of early life immunity. Trends Immunol. 35, 299–310 (2014).
Thackray, A. M. & Bujdoso, R. Elevated PrPC expression predisposes to increased HSV-1 pathogenicity. Antivir. Chem. Chemother. 17, 41–52 (2006).
Parizek, P. et al. Similar turnover and shedding of the cellular prion protein in primary lymphoid and neuronal cells. J. Biol. Chem. 276, 44627–44632 (2001).
Roberts, T. K. et al. PrPC, the cellular isoform of the human prion protein, is a novel biomarker of HIV-associated neurocognitive impairment and mediates neuroinflammation. Am. J. Pathol. 177, 1848–1860 (2010).
Teissier, N. et al. Cytomegalovirus-induced brain malformations in fetuses. J. Neuropathol. Exp. Neurol. 73, 143–158 (2014).
Mitsios, N. et al. Cellular prion protein is increased in the plasma and peri-infarcted brain tissue after acute stroke. J. Neurosci. Res. 85, 602–611 (2007).
Meyne, F. et al. Total prion protein levels in the cerebrospinal fluid are reduced in patients with various neurological disorders. J. Alzheimers Dis. 17, 863–873 (2009).
Volkel, D. et al. Immunochemical determination of cellular prion protein in plasma from healthy subjects and patients with sporadic CJD or other neurologic diseases. Transfusion 41, 441–448 (2001).
Chaudhry, M. Z., Messerle, M. & Cicin-Sain, L. Construction of human cytomegalovirus mutants with markerless BAC mutagenesis. Methods Mol. Biol. 2244, 133–158 (2021).
Strazic Geljic, I. et al. Cytomegalovirus protein m154 perturbs the adaptor protein-1 compartment mediating broad-spectrum immune evasion. Elife 9, https://doi.org/10.7554/eLife.50803 (2020).
Strazic Geljic, I. et al. Viral interactions with adaptor-protein complexes: a ubiquitous trait among viral species. Int J. Mol. Sci. 22, https://doi.org/10.3390/ijms22105274 (2021).
Smith, T. M. Jr., Tharakan, A. & Martin, R. K. Targeting ADAM10 in cancer and autoimmunity. Front. Immunol. 11, 499 (2020).
Wetzel, S., Seipold, L. & Saftig, P. The metalloproteinase ADAM10: a useful therapeutic target? Biochim. Biophys. Acta Mol. Cell Res. 1864, 2071–2081 (2017).
Jarosz-Griffiths, H. H. et al. Proteolytic shedding of the prion protein via activation of metallopeptidase ADAM10 reduces cellular binding and toxicity of amyloid-beta oligomers. J. Biol. Chem. 294, 7085–7097 (2019).
Krstanovic, F., Britt, W. J., Jonjic, S. & Brizic, I. Cytomegalovirus infection and inflammation in developing brain. Viruses 13, https://doi.org/10.3390/v13061078 (2021).
Cairns, D. M., Itzhaki, R. F. & Kaplan, D. L. Potential involvement of Varicella Zoster Virus in Alzheimer’s Disease via reactivation of Quiescent Herpes Simplex Virus Type 1. J Alzheimers Dis. 88, 1189–1200 (2022).
Zhu, C. & Aguzzi, A. Prion protein and prion disease at a glance. J. Cell Sci. 134, https://doi.org/10.1242/jcs.245605 (2021).
Hartmann, D. et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum. Mol. Genet. 11, 2615–2624 (2002).
Sprenger, A. et al. Comparative quantitation of proteome alterations induced by aging or immortalization in primary human fibroblasts and keratinocytes for clinical applications. Mol. Biosyst. 6, 1579–1582 (2010).
Alloatti, A., Kotsias, F., Hoffmann, E. & Amigorena, S. Evaluation of cross-presentation in bone marrow-derived dendritic cells in vitro and splenic dendritic cells ex vivo using antigen-coated beads. Bio Protoc. 6, https://doi.org/10.21769/BioProtoc.2015 (2016).
Acknowledgements
We would like to express our gratitude to S. Malić and K. Miklić for their invaluable assistance in laboratory analyses. Additionally, we extend our appreciation to T. Rudančić, E. Ražić, D. Rumora, and L. Mikša for their excellent support throughout the project. We thank Paul Saftig (Kiel, Germany) for ADAM10 KO (and respective WT) MEFs. We also thank A. Aguzzi (Zürich, Switzerland) and A. Oxenius (Zürich, Switzerland) for generously providing PrnpZH3/ZH3 and Maxi mice, respectively. This research has been supported by: Croatian Science Foundation grant IP-2020-02-6617 (T.L.R.) University of Rijeka, uniri-biomed-18-233 (T.L.R.) University of Rijeka, uniri-iskusni-biomed-23-197 (T.L.R.) University of Rijeka, uniri-biomed-18-205 (S.J.) “Strengthening the capacity of CerVirVac for research in virus immunology and vaccinology“, KK.01.1.1.01.0006, awarded to the Scientific Centre of Excellence for Virus Immunology and Vaccines and cofinanced by the European Regional Development Fund (S.J.). Croatian French program Cogito partnership Hubert Curien (T.L.R., M.H.).
Author information
Authors and Affiliations
Contributions
Conceptualization, T.L.R., S.J., and D. Karner.; Investigation, validation and formal analysis, T.L.R., D. Karner, D. Kvestak, P.K.B., M.C.B., B.L., I.B., V.J.L., M.G., J.T., A.K. M.H., E.K., V.L., S.M., G.L., and H.A.; Funding acquisition and project administration, T.L.R., S.J., and M.H.; Writing, T.L.R., S.J., D. Karner, M.H., G.L., and H.A.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Declaration of Generative AI and AI-assisted technologies in the writing process
During the preparation of some parts [Introduction, Results, Disscussion] of this manuscript, the authors used ChatGPT to improve the readability of the manuscript. After using this tool, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.
Ethics
The Ethics Committee at the University of Rijeka and The National Ethics Committee for the Protection of Animals Used for Scientific Purposes (Ministry of Agriculture) approved experiments with laboratory animals (approval number HR-POK-004).
Peer review
Peer review information
Nature Communications thanks Laura Lewis, and the other, anonymous, reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Karner, D., Kvestak, D., Kucan Brlic, P. et al. Prion protein alters viral control and enhances pathology after perinatal cytomegalovirus infection. Nat Commun 15, 7754 (2024). https://doi.org/10.1038/s41467-024-51931-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-51931-4