Abstract
Programmable RNA editing is harnessed for modifying mRNA. Besides mRNA, miRNA also regulates numerous biological activities, but current RNA editors have yet to be exploited for miRNA manipulation. To engineer primary miRNA (pri-miRNA), the miRNA precursor, we present a customizable editor REPRESS (RNA Editing of Pri-miRNA for Efficient Suppression of miRNA) and characterize critical parameters. The optimized REPRESS is distinct from other mRNA editing tools in design rationale, hence enabling editing of pri-miRNAs that are not editable by other RNA editing systems. We edit various pri-miRNAs in different cells including adipose-derived stem cells (ASCs), hence attenuating mature miRNA levels without disturbing host gene expression. We further develop an improved REPRESS (iREPRESS) that enhances and prolongs pri-miR-21 editing for at least 10 days, with minimal perturbation of transcriptome and miRNAome. iREPRESS reprograms ASCs differentiation, promotes in vitro cartilage formation and augments calvarial bone regeneration in rats, thus implicating its potentials for engineering miRNA and applications such as stem cell reprogramming and tissue regeneration.
Similar content being viewed by others
Introduction
MicroRNA (miRNA) is ~22-nt RNA involved in post-transcriptional gene silencing1. miRNA is typically encoded in the introns of the host gene and its biogenesis starts from transcription of an intronic long primary miRNA (pri-miRNA) along with its exonic host gene. Pri-miRNA folds up into an imperfect hairpin containing a double-stranded (ds) stem-loop region2. Nascent pri-miRNA is cleaved near the junction (called basal junction) of single-stranded RNA (ssRNA) and dsRNA hairpin by the Microprocessor (composed of Drosha/DGCR8) into a precursor miRNA called pre-miRNA3. Once processed, pre-miRNA is exported out of the nucleus and undergoes further maturation to form the mature miRNA.
miRNA dysregulation leads to cancers and diseases4, making it an attractive candidate for therapeutic intervention. Although miRNA can be regulated using overexpressed adenosine deaminase acting on RNA (ADAR)5, synthetic oligonucleotides6, miRNA sponges7 or CRISPR editing8, these methods have their respective drawbacks. Overexpressed ADAR induces serious off-target effects5. Oligonucleotides such as anti-miRs necessitate extensive chemical modifications6 and are associated with considerable off-target effects9. Using a miRNA sponge to knockdown a miRNA or a miRNA seed family could affect all relevant genes of its target, leading to undesirable effects10. CRISPR editing induces double strand break (DSB), which might trigger large-scale genomic deletions and chromosomal translocations11,12. These limitations entail the development of an alternative method for miRNA regulation.
Programmable RNA editing enables transient gene regulation without permanent genome change and promises a safer option than DNA editing13. ADAR2 naturally deaminates adenine (A) to inosine (I) in dsRNA and preferentially deaminates a target A mismatched with a cytosine (C)5. For programmable RNA editing, deaminase domain of ADAR2 (ADAR2DD) has been fused to deactivated Cas13 (dCas13) protein, which is guided by a single CRISPR RNA (crRNA) comprising a spacer sequence that base pairs with the RNA substrate14. The first system leveraging this strategy, REPAIR, comprises a fusion protein of dCas13b derived from Prevotella sp. P5-125 (dPspCas13b) and hyperactive ADAR2DD. Coupled with a site-specific crRNA, REPAIR confers precise A- > I conversion14. Subsequently, LEAPER employs engineered ADAR-recruiting RNAs (arRNAs) to recruit endogenous ADARs for A- > I conversion15. RESTORE exploits synthetic antisense oligonucleotides to recruit endogenous ADARs16. Recent studies also exploit short, chemically modified oligonucleotides or circular RNA to recruit endogenous ADAR17,18. Other programmable RNA-editing tools such as REPAIRx19, CURE20, REWIRE21, RESCUE22 and CIRTS23 are also developed. Site-specific RNA editing can also be achieved by fusing ADARs with an RNA-binding peptide or oligonucleotide24. Most of these tools edit mRNA by designing the guide RNA or oligonucleotide complementary to mRNA except a single mismatch (usually a cytosine) opposite to the adenine intended for deamination. They are harnessed to reverse pathogenic mutations on mRNA and restore native protein expression in cells and disease models. However, these methods are neither used to edit pri-miRNA, nor have they been exploited for tissue regeneration.
Because pri-miRNA processing is critical for miRNA biogenesis and current programmable RNA editing systems have yet to be harnessed to edit pri-miRNA, this study aims to create a programmable system for RNA Editing of PRi-miRNA for Efficient SuppreSsion of miRNA (REPRESS). The optimized REPRESS enables editing of various pri-miRNAs and attenuates their mature miRNAs levels in different cells including adipose-derived stem cells (ASCs). We further generate the improved REPRESS (iREPRESS) that prolongs and enhances pri-miRNA editing with minimal perturbation of transcriptome and microRNAome. iREPRESS editing of pri-miR-21 reprograms ASCs differentiation, ameliorates in vitro cartilage formation and in vivo bone regeneration, thus paving an avenue for miRNA regulation, stem cell engineering and regenerative medicine.
Results
Development of pri-miRNA base editor (REPRESS)
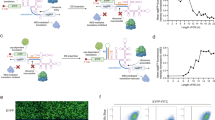
To edit pri-miRNA in a programmable fashion, we created REPRESS by fusing dCas13 and human ADAR2 deaminase domain (ADAR2DD) with a linker, in combination with a crRNA to guide the editor to the pri-miRNA (Fig. 1A). Unlike other ADAR-based RNA editing methods that introduce an A-C mismatch at the crRNA:mRNA duplex14,15,16, we designed the crRNA spacer sequence perfectly complementary to ssRNA sequences near the basal junction of pre-miRNA hairpin, such that ADAR2DD deaminates adjacent pre-miRNA hairpin (Fig. 1A).
A Mode of action of REPRESS. crRNA targeting to the ssRNA motif of pri-miRNA brings the dCas13-ADAR2DD to the vicinity of basal junction, thus catalyzing specific A- > I conversion within the pre-miRNA hairpin. B The REPRESS expression cassette. dCas13 (dPspCas13b or dRfxCas13d) was fused with ADAR2DD E488Q via various linkers. The fusion protein was flanked by two bipartite nuclear localization signals (bpNLS) to facilitate nuclear import. crRNA expression cassette was driven by U6 promoter with an architecture of spacer-direct repeat (DR) for dPspCas13b (pB-crRNA) or DR-spacer-DR for dRfxCas13d (pD-crRNA). C Schematic illustration of a segment of pri-miR-10a and the secondary structure of pre-miR-10 hairpin. The numbering of bases was defined according to the sequences retrieved from miRBase database. Translucent red box indicates the intended edited adenosine. D REPRESS variants encoding dPspCas13b (B-REPRESS 1–6) or dRfxCas13d (D-REPRESS 1–6) and various linkers. Corresponding editing efficiencies are also shown. E, F Editing efficiency using different spacer lengths (L). G Illustration of relative distance (d) between targeting position and the 5′ (‒) or 3′ (+) basal junctions. H–P Editing efficiencies at different targeting positions for various pri-miRNAs. Plasmids encoding REPRESS and crRNA were co-delivered into cells with a ratio of 1:3 (w:w). Total RNA was harvested after 1 day for reverse transcription and PCR amplification. PCR amplicons (300–500 bp for different pri-miRNA, 444 bp for pri-miR10a; see Supplementary Fig. 1 for how PCR amplicons were synthesized) encompassing the pre-miRNA and flanking sequences were Sanger sequenced. A- > I editing efficiencies were calculated by EditR. Statistical analyses were carried out with one-way (F, H, J, L, N) or two-way (P) ANOVA followed by Tukey multiple comparison test. Data represent means ± SD of three independent culture experiments. Source data are provided as a Source data file.
For proof-of concept, we first generated a series of REPRESS plasmids by fusing dPspCas13b or dRfxCas13d with the hyperactive ADAR2DD E488Q mutant (designated as ADAR2DD thereafter) using different linkers (Fig. 1B). Because we envisioned that REPRESS-mediated editing of pri-miRNA affects miRNA expression, we first tested our design on miR-10a (Fig. 1C) which is expressed abundantly in HEK293T cells25 and upregulated in a wide variety of cancers26. The fusion gene was flanked with bipartite nuclear localization signals (bpNLS) to ensure editing of nuclear pri-miRNA. The crRNA plasmids associated with dPspCas13b (pB-crRNA) or dRfxCas13d (pD-crRNA) were constructed to guide REPRESS with a 22-nt spacer to 5 nt upstream of pre-miR-10a basal junction. One day after plasmid co-transfection, total RNA was extracted for RT-PCR amplification of cDNA (444 bp) using primers flanking the pre-miRNA hairpin. The A- > I editing events were determined by Sanger sequencing of the cDNA (Supplementary Fig. 1).
When we used dPspCas13b and the GS linker (B-REPRESS 1, Fig. 1D), in the entire amplicon we detected very minor A- > I peaks at position 21 (A21), A93, A97 and A101 in the pre-miR-10a region. These peaks were also observed in the mock-transfected cells (Mock group, Supplementary Fig. 1) and considered background. Yet we detected minor A- > I conversion at A76 in the pre-miRNA region (Fig. 1C, D). By increasing the lengths of GS-based linker to 6-aa (GSGGGS), 9-aa (3×GGS) and 12-aa (3×GGGS), we detected elevating A- > I conversion efficiency only at A76 among 28 adenines in the pre-miR-10a hairpin (Fig. 1D and Supplementary Fig. 1). The 16-aa flexible XTEN linker (B-REPRESS 5) further improved A76 conversion efficiency to ≈12%. However, the 5-aa rigid linker (EAAAK) barely converted A76.
When we replaced dPspCas13b with dRfxCas13d (D-REPRESS 1–6, Fig. 1D), the D-REPRESS editors conferred A76 conversion in a similar trend, but with generally higher efficiency when using the same linker (Fig. 1D and Supplementary Fig. 2). Regardless of dCas13 type and linkers, only A76 was converted (Supplementary Fig. 1, 2). Among these editors, D-REPRESS 5 that exploited XTEN linker to fuse dRfxCas13d with ADAR2DD conferred the highest A76 editing efficiency (≈15%). Therefore, we abbreviated D-REPRESS 5 as REPRESS for ensuing characterization.
dRfxCas13d crRNA consists of a stem-loop and a spacer sequence for base pairing the target RNA. Because transfection of ≈70 nt synthetic oligonucleotide to base pair mRNA can recruit endogenous ADAR for mRNA editing15, we were inspired to vary the crRNA spacer length (L, Fig. 1E). We found that increasing the spacer length from 22 to 70 nt all enabled A76 conversion, but with descending efficiencies (Fig. 1E, F), indicating that 22 nt is the optimal spacer length. We next tested whether changing the crRNA targeting position would alter the A- > I conversion preference. We fabricated two crRNA spacers, one complementary to pre-miR-10a with an A-C mismatch at A76, the other with the highest predicted dRfxCas13d targeting score (Supplementary Fig. 3). Both designs, however, did not achieve A- > I conversion. Other RNA editing systems such as REPAIR, LEAPER and RESTORE that introduced an A-C mismatch targeting A76 (Supplementary Fig. 3) or targeted the ssRNA adjacent to the 5’ basal junction of pre-miR-10a (Supplementary Fig. 4) also failed to achieve A- > I conversion.
We further designed crRNAs to target bases with different distances between the crRNA binding site and the basal junction (d, Fig. 1G). We uncovered that d plays crucial roles to dictate the conversion rate although still only A76 was converted. crRNA targeting the basal junction (d = 0) conferred the poorest A76 conversion while increasing the distance to 5 nt (d = −5) yielded the highest A76 conversion rate (≈12%, Fig. 1H).
We next applied REPRESS to target other miRNAs abundant in HEK293FT. We chose miR-140 and miR-378a because miR-140 upregulation is observed in many disorders (e.g. osteoporosis27 and Alzheimer’s Disease28) while miR-378a upregulation is linked with tumor progression29. REPRESS enabled A- > I conversion of pre-miR-140 at A38 (Supplementary Fig. 5) with the highest editing efficiency at d = +5 or −10 (Fig. 1I, J). Conversely, REPRESS converted A11 of pre-miR-378a with the highest efficiency reaching ≈18% at d = −5 (Fig. 1K, L, Supplementary Fig. 5). REPRESS was also designed to target miR-21 and miR-214 as they trigger adipogenesis and block osteogenesis in adipose-derived stem cells (ASCs), respectively30,31. Likewise, we detected A29 conversion for pre-miR-21 (Supplementary Fig. 5) with the highest efficiency reaching ≈17% at d = -5 (Fig. 1M, N). By contrast, none of the REPAIR, LEAPER and RESTORE systems achieved A29 conversion, regardless of whether the guide RNAs targeted the pri-miR-21 hairpin or ssRNA sequences adjacent to the basal junction (Supplementary Figs. 6, 7). For pri-miR-214 in ASCs, we detected conversion at two adenines (A12 and A16), with the highest efficiency approaching 19% at d = −10 (Fig. 1O, P). These two adenines are in the bulge, which can be deaminated by ADAR32.
To attest whether REPRESS activity is specific, we also transfected cells with REPRESS using non-targeting (NT) crRNA. We found no off-target editing in the pre-miR-10a, pre-miR-140, pre-miR-378a regions in HEK293FT cells and pre-miR-21 and pre-miR-214 regions in ASCs (Supplementary Fig. 8). We also cross-checked the sequence in the pre-miR-214 hairpin in ASCs edited at pri-miR-21 and found no significant A- > I conversion at the hairpin region. Similarly, neither pre-miR-140 nor pre-miR-378a were adventitiously edited in HEK293FT transfected with pre-miR-10a-targeting REPRESS (Supplementary Fig. 8). These data underscored the crRNA-dependent specificity of REPRESS.
Editing of pri-miRNAs by REPRESS suppressed mature miRNAs levels
We next evaluated whether REPRESS-directed pri-miRNA editing inhibited miRNA levels, using TaqMan qPCR primers specific to mature miRNA. As a control, we constructed a deactivated REPRESS (dREPRESS) by fusing dRfxCas13d with deactivated ADAR2DD (Fig. 2A). Using the optimal crRNA targeting design (Fig. 1), dREPRESS marginally reduced the levels of several miRNAs in different cells (Fig. 2B–G), presumably because dREPRESS binding to the pri-miRNA partly disturbed the Microprocessor access to the pre-miRNA hairpin33,34 and hindered miRNA maturation. REPRESS-mediated A76 conversion at pre-miR-10a (3p strand) further inhibited miR-10a-3p level for 56% (Fig. 2B). A38 conversion at pre-miR-140 (5p strand) and A11 conversion at pre-miR-378a (5p strand) significantly knocked down miR-140-5p and miR-378a-5p levels for 25% and 79%, respectively (Fig. 2C, D). We also targeted the same pri-miR-10a region in A549 lung cancer cells and achieved ≈60% inhibition of miR-10a-3p level (Fig. 2E). The editing inhibited A549 cell proliferation, migration and invasion (Supplementary Fig. 9). In ASCs, A29 and A12/A16 conversion at pre-miR-21 (5p strand) and pre-miR-214 (5p strand) suppressed miR-21-5p and miR-214-5p levels for 72% and 58%, respectively (Fig. 2F, G).
A dREPRESS expression cassette harboring the fusion of dRfxCas13d and deactivated ADAR2DD with E488Q and E396A mutations. B–G Relative mature miRNAs levels. Plasmids encoding optimized REPRESS or dREPRESS were co-delivered with crRNA expressing plasmids into cells with a ratio of 1:3 (w:w). miRNAs were collected at 1 day for TaqMan assay using primers specific to mature miRNAs. Statistical analyses were carried out with one-way ANOVA followed by Tukey multiple comparison test. Data represent means±SD of three independent culture experiments. Source data are provided as a Source data file.
Pri-miRNAs are usually expressed from the introns together with their host genes in the exons. Editing of pri-miR-10a (in HEK293FT) and pri-miR-21 (in ASCs) did not disturb the levels of their host genes HOXB335 and VMP136, respectively (Supplementary Fig. 10). This might arise from site-specific A- > I editing in the intronic pre-miRNA hairpin, hence enabling selective inhibition of Microprocessor processing and miRNA maturation while leaving the exonic host gene expression unaffected37.
Improved REPRESS (iREPRESS) for prolonged pri-miRNA editing and safety profiling
We reasoned that prolonged pri-miRNA editing may extend future therapeutic efficacy. Therefore, we packaged REPRESS into a hybrid Cre/loxP-based baculovirus (BV) system that transduces ASCs at >95% efficiency and prolongs transgene expression38. The hybrid vector comprises two BV: one expressing Cre recombinase and the other harboring the transgene flanked by loxP sites39. Co-transduction of ASCs with the two BV enables intracellular loxP-flanking transgene re-circularization and prolongs transgene expression39.
We generated BV vectors Bac-REPRESS encoding REPRESS and Bac-cr21 expressing cr21 to target pri-miR-21 at d = −5 as in Fig. 1F (Supplementary Fig. 11A). Both REPRESS and cr21 cassettes were flanked by two in cis loxP sequences. We co-transduced ASCs with Bac-REPRESS and Bac-cr21 (REPRESS group) and detected 12% A29 conversion at 3 days post-transduction (dpt), which rapidly decreased with time (Fig. 3A). When we co-transduced ASCs with Bac-REPRESS/Bac-cr21 and a third Bac-Cre (Supplementary Fig. 11A) that expressed Cre (iREPRESS group), A29 conversion rate increased to ≈23% at 3 dpt and remained 16% at 7 dpt and 5% at 10 dpt, indicating that iREPRESS enhances and prolongs A29 conversion (Fig. 3A). Concurrently, iREPRESS more effectively knocked down miR-21-5p levels for at least 10 days (Fig. 3B), without compromising ASCs viability, cell metabolism and proliferation (Supplementary Fig. 11B–D).
A A29 conversion rate as measured by deep sequencing. B Relative mature miR-21−5p level measured by TaqMan assay. ASCs were Mock-transduced (Mock group), co-transduced with Bac-REPRESS/Bac-cr21 (REPRESS group) or co-transduced with Bac-REPRESS/Bac-cr21/Bac-Cre (iREPRESS group). Bac-REPRESS expressed the entire REPRESS cassette flanked by two loxP sequences. Bac-cr21 expressed the crRNA targeting pri-miR-21 at d = −5 near the 5′ basal junction of the pre-miR-21 hairpin. Bac-Cre expresses Cre recombinase to excise and circularize the loxP-flanked REPRESS for sustained expression (see Supplementary Fig. 11 for schematic illustration). C, D Transcriptome-wide analysis of ASCs co-transduced with iREPRESS using cr21 (C) or non-targeting (NT) crRNA (D), with the Mock group as the reference. RNA-seq data are presented as log2(fold change) vs. log(mean expression). E, F Global miRNA analysis of ASCs co-transduced with iREPRESS using cr21 (E) or NT crRNA (F). Statistical significance was determined by two-sided Wald test followed by correction using Storey’s method to generate q values. Change with FALSE q value was cut off and volcano plot is presented in ‒log10(p value) vs. log2(fold change). Red and blue dots (C–F) represent significantly upregulated and downregulated genes in sequencing data. G, H Transcriptome-wide A-to-I off-target analysis of ASCs co-transduced with iREPRESS using cr21 (G) or NT crRNA (H). I, J miRNAome A-to-I off-target analysis of ASCs co-transduced with iREPRESS using cr21 (I) or NT crRNA (J). Orange dot represents on-target editing. Total RNA or miRNA was harvested after 3 days for RNA or small-RNA seq experiments. Statistical analyses were carried out with two-way ANOVA (A, B) followed by Tukey multiple comparison test. Data represent means±SD of three independent culture experiments. Source data are provided as a Source data file.
To evaluate the safety profile, we next performed transcriptome-wide RNA-seq to assess off-target effects of iREPRESS in rat ASCs at 3 dpt because iREPRESS editing of pri-miR-21 was most active at 3 dpt (Fig. 3A, B). Compared with the Mock group, iREPRESS triggered minimal transcriptome perturbations (p < 0.0001) with only 8 upregulated and 5 downregulated genes (Fig. 3C and Supplementary Data 1). When we replaced cr21 with a non-targeting crRNA (NT crRNA), only 9 genes were perturbed (Fig. 3D and Supplementary Data 2). Small RNA sequencing showed that iREPRESS specifically inhibited miR-21-5p levels with only one off-target miR-505-5p upregulation (Fig. 3E and Supplementary Data 3). Replacing cr21 with NT crRNA abolished miR-21-5p downregulation, with only one off-target downregulation of miR-425-3p (Fig. 3F and Supplementary Data 4). We also examined transcriptome-wide A- > I editing and revealed that iREPRESS with cr21 resulted in 15 off-target editing (Fig. 3G). iREPRESS with NT crRNA led to comparable editing pattern with 15 off-targets (Fig. 3H), possibly due to the hyperactivity of ADAR2DD14. These off-target edits occurred at coding sequence (silent and missense), 3′ UTR and antisense sequence of a gene (Supplementary Fig. 12).
We further performed global analysis of A- > I editing in mature miRNA. The miRNAome-wide analysis showed that iREPRESS with cr21 led to one off-target A15 editing of mature miR-411, but we also detected a trace of on-target A29 editing in miR-21 (Fig. 3I). The A29 editing in mature miR-21 seems counterintuitive because A- > I editing should abrogate miR-21 maturation. This might arise from partial processing of edited pri-miR-21, yielding an edited form of mature miR-21. When substituting cr21 with NT crRNA, off-target A- > I editing was detected in only two miRNAs (Fig. 3J). These data confirmed that iREPRESS prolonged pri-miRNA editing with minimal perturbation of transcriptome and miRNAome.
Switching ASCs differentiation by iREPRESS-prolonged pri-miR-21 editing
ASCs can differentiate towards adipogenic, chondrogenic and osteogenic lineages, making them useful for cartilage40 and bone41 regeneration. However, ASCs differentiate favorably towards adipogenic lineage, partly because endogenous miR-21 promotes adipogenesis30 and attenuates chondrogenesis by targeting GDF-5 during the chondrogenic differentiation42. Therefore, we next exploited iREPRESS to knockdown miR-21 and reprogram ASCs differentiation.
Along each differentiation lineage, the adipogenic marker genes C/ebpα and Ppar-γ, as well as chondrogenic markers Acan and Col2a1 are upregulated at different time points. After ASCs were mock-transduced (Mock group), co-transduced with Bac-REPRESS/Bac-cr21 (REPRESS group) or with Bac-REPRESS/Bac-cr21/Bac-Cre (iREPRESS group), miR-21 suppression by iREPRESS enabled more significant inhibition of C/ebpα and Ppar-γ both in mRNA and protein levels than REPRESS (Fig. 4A, B, Supplementary Figs. 13, 14). Both iREPRESS and REPRESS also elevated Acan and Col2a1 mRNA and protein levels (Fig. 4C, D, Supplementary Figs. 13, 14). Oil Red O staining for oil droplet formation (for adipogenesis), Alcian blue staining for glycosaminoglycan (GAG, for chondrogenesis), Alizarin Red staining for mineralization (for osteogenesis) and ensuing quantitative analyses (Fig. 4E–J) confirmed that iREPRESS suppressed adipogenesis and triggered chondrogenesis more potently than the REPRESS and Mock groups but did not apparently affect osteogenesis.
Experiments and experimental groups are identical to those shown in Fig. 3. A, B Relative expression of adipogenic marker genes C/ebpα (A) and Ppar-γ (B). C, D Relative expression of chondrogenic marker genes Acan (C) and Col2a1 (D). E–G Representative images of Safranin O, Alcian Blue and Alizarin Red staining (n = 3). H–J Spectrophotometric analysis for oil droplet formation (adipogenesis, H), GAG (for chondrogenesis, I) and mineralization (for osteogenesis, J). Scale bar = 100 μm. Statistical analyses were carried out with one-way (H–J) and two-way ANOVA (A–D) followed by Tukey multiple comparison test. Quantitative data represent means±SD of three independent culture experiments. Source data are provided as a Source data file.
iREPRESS-mediated pri-miR-21 editing in ASCs stimulated in vitro cartilage growth
Hyaline cartilage formation starts from stem cell condensation, chondrogenic differentiation and deposition of extracellular matrix (ECM) molecules such as collagen II (Col II) and GAG. We transduced ASCs as in Fig. 4, separately seeded the Mock and iREPRESS group cells into porous scaffolds (n = 3 for each group) and cultured the ASCs/scaffold constructs. At 7 and 14 dpt, the iREPRESS group constructs exhibited whitish and glassy appearance that resembled the ECM of hyaline cartilage (Fig. 5A). H&E staining revealed the onset of condensation at 7 dpt and accumulation of more ECM in the iREPRESS group than the Mock group at 14 dpt (Fig. 5B). Alcian Blue and Col II immunostaining and quantitative analyses showed that iREPRESS deposited more uniform GAG and Col II than the Mock group at both 7 and 14 dpt (Fig. 5C–F).
A Gross appearance of engineered cartilages. Representative images of (B) H&E, (C) Alcian Blue and (D) Col II staining (n = 3). Black arrow heads represent the positive stains. E, F Semiquantitative analysis of GAG and Col II. ASCs were mock- (Mock group) or iREPRESS-transduced (iREPRESS group) in 15-cm dishes. At 1 dpt, the cells were seeded into porous gelatin scaffolds (diameter = 6 mm; thickness = 1 mm; 5 × 106 cells/scaffold; n = 3 for each group). The ASCs/gelatin constructs continued to be cultured in chondroinductive medium and assayed at 7 or 14 dpt. Statistical analyses were carried out with two-way ANOVA followed by Tukey multiple comparison test. Quantitative data represent means±SD of three independent culture experiments. Source data are provided as a Source data file.
iREPRESS-mediated pri-miR-21 editing stimulated calvarial bone regeneration in vivo
Healing of large calvarial bone defects remains challenging for orthopedic surgeons. We previously showed that implantation of chondroinductive ASCs ameliorates calvarial bone healing by switching the repair pathway43,44. To evaluate the feasibility of pri-miRNA-21 editing to promote calvarial bone regeneration, we harvested the Mock and iREPRESS constructs at 7 or 14 dpt (as in Fig. 5) and implanted them into critical-size calvarial defects in rats (n = 4–5 for Mock and n = 6 for iREPRESS at both 7 and 14 dpt).
Micro CT (µCT) imaging and quantitative analysis revealed that the Mock constructs elicited slow and poor bone regeneration, regardless of harvesting at 7 or 14 dpt (Fig. 6), which underscores the difficulty to repair large calvarial defects using ASCs. The iREPRESS group nonetheless induced formation of bone islands as early as week 4 (W4) and progressive formation of new bone (Fig. 6A–C). Quantitative data confirmed that the iREPRESS constructs elicits significantly faster and superior bone regeneration, with the nascent bone area, volume and density at W12 approaching 44–48%, 22–24%, and 32–33% of the original defect, respectively (Fig. 6D–F). The in vitro culture time at which the constructs were collected (7 or 14 dpt) did not significantly influence the regeneration. Furthermore, histological and immune staining of the regenerated bone specimens showed evidently more bone formation, bone-specific ECM accumulation and less fibrous tissues in the iREPRESS group than the Mock group (Supplementary Fig. 15).
A–C Representative 3D and hot-and-cold projections of the transverse and sagittal views of the calvarial bone at 4 (A), 8 (B) and 12 (C) weeks post-implantation. Orange arrow heads represent new bone formation. D–F Regenerated bone area (mm2), volume (mm3) and density (HU, Hounsfield Unit) of Mock (n = 4 rats, 7 dpt; n = 5 rats, 14 dpt) and iREPRESS (n = 6 rats) implanted animals. ASCs from the Mock and iREPRESS groups (as in Fig. 5) were seeded into gelatin scaffolds, cultured and harvested at 7 or 14 dpt. The constructs were implanted into the critical-sized (diameter = 6 mm) calvarial defects of SD rats. Bone regeneration was evaluated by μCT imaging. Percentage values were calculated by normalization to the bone area (28 mm2), volume (28 mm3) and density (4600 HU) of the original defects. Statistical analyses were carried out with two-way ANOVA followed by Tukey multiple comparison test. Quantitative data are represented as means±SD. Source data are provided as a Source data file.
Discussion
Current programable RNA editing tools are used for mRNA editing but have yet to be exploited to edit pri-miRNA. Most of these tools use a guide RNA carrying an A-C mismatch at the pre-determined A in mRNA to direct ADAR-mediated A- > I conversion. Here we report a programmable pri-miRNA base editor REPRESS. REPRESS distinguishes itself in that the crRNA spacer is completely complementary to the ssRNA near the basal junction of pre-miRNA hairpin, thus re-directing the dRfxCas13d-ADAR2DD effector to deaminate adjacent pre-miRNA hairpin duplex (Fig. 1A). REPRESS is also distinct from other RNA editing tools in that REPRESS is designed to translocate to the nucleus where pri-miRNA is located, whereas other systems are introduced into the cytoplasm to edit mRNA.
We identified that REPRESS editing depends on dCas13 protein type, linker types and spacer lengths (Fig. 1D–F). dRfxCas13d generally confers higher editing efficiency than dPspCas13b used in the REPAIR system14, probably because Cas13d has a favorable accessibility toward RNA hairpin45,46. We also showed that longer linkers improve editing efficiency (Fig. 1D), presumably because the increased flexibility and accessibility enable ADAR2DD to scan more adenosines in the editing window for A- > I conversion. This observation is consistent with the role of linker characteristics in facilitating optimal enzyme-substrate interaction47.
Importantly, we identified the optimal crRNA targeting position in the window of 5 to 10 bases away from the pre-miRNA basal junction (Fig. 1G–P). These findings allow us to develop the optimal REPRESS and edit various pri-miRNAs in different cells. The editing efficiencies vary with pri-miRNAs, presumably because ADAR editing site preference and efficiency hinge on different pri-miRNAs5. REPRESS preferentially edits A within the mature miRNAs, a phenomenon observed previously5,32. Intriguingly, endogenous ADAR edits multiple adenines in pri-miRNAs5,32 but REPRESS edits only one or two adenines. The discrepancy may arise because endogenous ADAR binds the dsRNA substrate and deaminates A depending on its own structure and interaction with dsRNA. In contrast, REPRESS is guided by crRNA to ssRNA motif adjacent to the basal junction of pre-miRNA hairpin, which may constrain the accessibility of ADAR2DD active site to many A in the pre-miRNA hairpin. Further structural studies are required to decipher the rule of how REPRESS selects specific A for conversion. Nonetheless, the optimized REPRESS can edit different pri-miRNAs that are not editable by other RNA editing systems such as REPAIR, LEAPER and RESTORE (Supplementary Figs. 3–7), which may be ascribed to the distinctive ability of REPRESS to engage ADAR2DD with adjacent pre-miRNA hairpin whereas other RNA editing systems guide ADAR2DD to the immediate spacer:target duplex rather than the pre-miRNA hairpin. These data underscore the stringent requirement for REPRESS design and indicate the superiority of REPRESS for pri-miRNA editing.
Despite editing at only one or two A, REPRESS-mediated editing replaces A-U Watson–Crick pair with mismatched I·U wobble pair, which causes substantial changes in the stem structure and hinders pri-miRNA cleavage by Drosha, hence inducing pri-miRNA degradation by a ribonuclease specific to inosine-containing dsRNAs5,32. In accord, REPRESS-mediated editing knocks down different mature miRNAs in cell lines, cancer cells and stem cells (Fig. 2). The iREPRESS mediated by the hybrid BV vector prolongs and enhances pri-miRNA-21 editing and reduces mature miR-21 levels for 10 days, which is not achievable by previous ADARDD-associated RNA editing strategies. Furthermore, prior ADARDD-associated RNA editing systems usually suffer from evident off-target effects19, yet iREPRESS induces minimal off-target effects and perturbations on the transcriptome and microRNAome (Fig. 3), probably because iREPRESS is transported into nucleus, and nuclear localization of ADAR reduces off-target effects48,49. Critically, miR-21 knockdown by iREPRESS reprograms the ASCs differentiation from adipogenic to chondrogenic pathway, promotes in vitro cartilage formation (Fig. 5) and stimulates calvarial bone healing (Fig. 6 and Supplementary Fig. 14).
RNA editing offers numerous opportunities for fundamental research and medical applications50. Besides mRNA, miRNAs regulate gene expression and are also accountable for numerous cancers and diseases51. Although miRNA can be regulated using oligonucleotide-based methods (e.g. anti-miRs), translation of miRNA-based therapeutics into clinical use is hampered by their off-target effects caused by either sequence similarities or overdosing to levels much higher than expected (for review see ref. 52). For instance, anti-miRNA-132 therapy, which showed promising results in a phase 1b clinical trial, leads to side effects such as severe immune response and even death (for review see ref. 53). Likewise, other methods (e.g. CRISPR editing) have respective limitations such as off-target effects and requirement to induce DSB (see Introduction). The DSB-free CRISPR-guided REPRESS specifically suppresses miRNA with minimal off-target effects and does not disturb host gene expression (Supplementary Fig. 10). REPRESS also successfully edits pri-miRNAs that are not editable using other RNA editing tools, lending REPRESS a promising tool for miRNA regulation. REPRESS editing of miR-10a inhibits cancer cell proliferation, migration and invasion (Supplementary Fig. 9), rendering it a potential tool for cancer treatment. iREPRESS further enhances and prolongs the pri-miRNA editing effects, inducing minimal off-target effects and disturbance of transcriptome and miRNAome (Fig. 3), hence enabling its use in stem cell engineering and tissue regeneration. These data collectively implicate the potentials of iREPRESS for broad applications.
One limitation of our system is the stringent preference to deaminate adenosines within the stem duplex of pre-miRNA hairpins, which may miss potentially crucial adenosines in the apical loop that influence Microprocessor processing3. Future development of structure-guided modification of ADAR2DD may enhance REPRESS versatility. Furthermore, current version of REPRESS is too large (≈6.8 kb) to fit into the commonly used adeno-associated viral (AAV) vector, which may impede its clinical applications. This delivery bottleneck may be tackled by using the split dual-AAV system54 or truncated/smaller Cas13 such as mini Cas13d55 or Cas13X.156 for AAV delivery. Another challenge for human use is the pre-existing immunity against RfxCad13d57, which may mitigate the editing efficacy of iREPRESS in humans. This obstacle may be circumvented by replacing dRfxCad13d with other dCas13 protein (e.g. dCas13X.1) capable of RNA targeting.
Methods
Ethics statement
Animal experiments were performed in compliance with the Guide for the Care and Use of Laboratory Animals (National Council of Science and Technology, Taiwan) and were approved by the Institutional Animal Care and Use Committee of National Tsing Hua University.
Construction of REPRESS plasmids and recombinant BV
The loxP-CMV enhancer-EF1α promoter element41, dPspCas13b, human ADAR2DD E488Q (referred to as ADAR2DD in this article) from pC0039 (Addgene #133849)14, and WPRE-SV40 poly A signal-loxP41 were PCR-amplified with PrimeSTAR Max DNA polymerase (Takara). The N-terminus of dPspCas13b and the C-terminus of ADAR2DD E488Q were tethered with the bipartite nuclear localization signal sequence to facilitate nuclear translocation. Amplicons were joined with NEBuilder HiFi DNA assembly master mix (NEB) to the templated REPRESS plasmid. Various peptide linkers between dPspCas13b and ADAR2DD were introduced via inverse PCR with divergent primers appended with desired peptide sequence, followed by phosphorylation with T4 Polynucleotide Kinase (NEB). The phosphorylated amplicons were self-ligated with RBC Rapid Ligation kit (RBC Bioscience) at room temperature for 30 min to generate B-REPRESS 1 to 6 plasmids. Similarly, D-REPRESS 1 to 6 plasmids were cloned following the same manner with dRfxCas13d amplification from pXR002 (Addgene #109050)45. Full-length FL hADAR2 E488Q was cloned from pmGFP-ADAR2 (Addgene # 117929). dREPRESS plasmid was constructed by replacing ADAR2DD E488Q in pD-REPRESS 5 with deactivated ADAR2DD E488Q E396A via inverse PCR and self-ligation.
Spacer sequences were selected based on two criteria: (i) the relative distance between the target site and the basal junctions of pre-miRNA hairpin and (ii) scores for Cas13d guide design using web-based algorithm (https://cas13design.nygenome.org/58). The selected sequences (Supplementary Table 1) were chemically synthesized and cloned into BbsI-digested pC0043 (encoding crRNA for dPsbCas13b) (Addgene #103862) or pXR004 (encoding precursor crRNA for dRfxCas13d) (Addgene #109054) to generate pB-crRNA or pD-crRNA plasmids. Plasmids expressing arRNA and gRNA (for LEAPER15 and RESTORE16 systems, respectively) were constructed by annealing chemically synthesized oligonucleotides and subcloning into EcoRI/BbsI-digested psgRNAa. Alternatively, pXR004 carrying the original BbsI cloning sites was used as a non-targeting (NT) crRNA.
Donor plasmids for recombinant bacmid and BV production were generated by joining two PCR amplicons: one from the entire REPRESS from D-REPRESS 5 or dREPRESS expression cassette; and the other from the donor backbone of pFastBac® Dual expression vector (Gibco). The PCR amplicon joining yielded pBac-REPRESS and pBac-dREPRESS. Similarly, expression cassette harboring the pri-miR-21-targeting (cr21) or non-targeting (NT crRNA) spacers (Supplementary Table 1) were joined with the donor backbone of pFastBac® to generate pBac-cr21 or pBac-NT crRNA.
Cell culture, transfection and electroporation
HEK293FT (Invitrogen) and A549 cells were maintained in DMEM high glucose (Gibco) supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (PS) (Gibco). Cells were passaged at ≈80% confluency and seeded to 12-well plates (1 × 105 cells/well). After overnight culture, 4 sets of plasmids were co-transfected into cells using Lipofectamine 3000 (Invitrogen) with a DNA:lipid ratio of 1:1.5: (i) REPRESS (300 ng) and crRNA (600 ng) plasmids; (ii) pC0039 (300 ng) and crRNA (600 ng) plasmids for REPAIR; (iii) 500 ng of U6-driven arRNA plasmid for LEAPER; or (iv) pmGFP-ADAR2 (250 ng) and U6-driven gRNA plasmid (750 ng) for RESTORE.
ASCs were isolated from SD rats (Biolasco, Taiwan) as described59. The cells were cultured in complete αMEM (Gibco) containing 10% FBS, 1% PS and 4 ng/mL basic fibroblast growth factor (bFGF). ASCs at passage 2 to 5 were used for subsequent experiments. ASCs were resuspended in Opti-MEM (Gibco) and mixed with 3 µg of REPRESS and 6 µg of crRNA plasmids for REPRESS; 3 µg of pC0039 and 6 µg of crRNA plasmid for REPAIR; 5 µg of U6-driven arRNA plasmid for LEAPER; or 2.5 µg of pmGFP-ADAR2 and 7.5 µg of U6-driven gRNA plasmids for RESTORE. The plasmids were electroporated into cells in a 2-mm gap cuvette with NEPA21 Electroporator (NEPAGENE) at 2 pulses of 275 V/2.5 ms with an interval of 50 ms for poring phase and 5 pulses of 20 V/50 ms with an interval of 50 ms.
Recombinant BV production and ASCs transduction
Recombinant BV were produced using the Bac-to-BacTM system (Invitrogen) as described38. Briefly, E. coli DH10Bac (Gibco) was transformed with donor plasmids (pBac-REPRESS, pBac-dREPRESS, pBac-cr21 or pBac-NT crRNA) and plated on Luria plate containing gentamycin (10 µg/mL), kanamycin (50 µg/mL), tetracycline (10 µg/mL), X-gal (200 µg/mL) and IPTG (40 µg/mL) at 37 °C overnight. White colonies were selected and further cultured in LB broth containing gentamycin, kanamycin, tetracycline of the same concentration. The recombinant bacmids were purified from the cultured cells for infecting SF9 cells to generate P0 viruses. P0 cells were used to infect SF9 cells for BV amplification. Cre-expressing baculovirus (Bac-Cre) was constructed previously59.
For transduction of ASCs, cells were seeded to 12-well plates (5 × 104 cells/well) and cultured overnight at 37 °C under 5% CO2. The cells were washed and incubated with BV suspended in surrounding medium (complete culture medium without sodium bicarbonate) at optimal multiplicity of infection (MOI) and mixed on a rocking plate (10 rpm, room temperature). After 6 h, the cells were cultured in fresh medium containing 3 mM sodium butyrate. After 18 h, the sodium butyrate-containing medium was removed, and the cells continued to be cultured in fresh or induction medium for subsequent analysis.
ASCs differentiation and characterization
Transduced ASC cells were differentiated in adipogenic, chondrogenic or osteogenic induction medium. Half of the media was removed and replaced by fresh induction media until analysis time. Differentiated cells were fixed in 4% buffered formaldehyde (Macron) for 15 min at room temperature and stained with Oil Red O, Alcian Blue, or Alizarin Red for adipogenesis, chondrogenesis, or osteogenesis, respectively. The dyes in the stained cells were extracted with isopropanol, guanidine hydrochloride, or cetylpyridinium chloride and read at 492, 595, or 550 nm on the plate reader (SpectraMax M2, Molecular Devices) for quantitation.
Cell metabolism and proliferation assay
ASCs cells were seeded to 96-well plates (2.5 × 103 cells/well) prior to transduction. Mock or transduced cells were cultured in fresh medium and replenished every 2 days. For metabolism assay, cells were washed with PBS and incubated in fresh medium with 0.5 mg/mL MTT (Sigma) at 37 °C for 3 h. After medium removal, cells were incubated with 100 µL solution of 4 µM HCl and 0.1% Triton X-100 in isopropyl alcohol at room temperature for 15 min on a rocking plate. The adsorption was read on SpectraMax M2 at 560 nm. Alternatively, cells were stained with BrdU for proliferation assay (Cell BioLabs). Briefly, the stained cells were fixed and subsequently stained with anti-BrdU antibody followed by HRP-conjugated secondary antibody staining. The cells were allowed to react with substrate. The reaction was stopped before reading at 450 nm on SpectraMax M2.
Adenine conversion rate by Sanger sequencing
Total RNA was extracted with Gene-Spin™ Total RNA Purification Kit (Protech) and quantified with Nanodrop One (ThermoFisher). cDNA synthesis was performed using pri-miRNA-specific primers (Supplementary Table 2) with High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems) to amplify the pri-miRNA region of interest using the following temperature cycle: RNA and the primer mixtures were heated at 65 °C/5 min and incubated at 4 °C prior to the addition of reaction mixture, followed by 37 °C/2 h and 85 °C/2 min. The cDNA was PCR-amplified with primers flanking the pre-miRNA region (Supplementary Table 2). The amplicons were purified, and Sanger sequenced (Genomics, Taiwan). The ab1. chromatogram files were used to calculated A conversion rate by EditR60.
Quantitative reverse-transcription real-time PCR (qRT-PCR)
Total RNA (500 ng) was reverse transcribed in 20 μL reaction with random hexamers using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems). Two microliter of cDNA was used for qPCR using primers specific for C/ebpα, Ppar-γ, Acan, Col2a1, HOX3B and Vmp1 (Supplementary Table 2) and qPCRBIO SyGreen Mix Lo-ROX (PCRBIOSYTEMS) on LightCycler® 96 (Roche). Primer sequences were reported previously (C/ebpα, Ppar-γ, Acan, Col2a1)41 or designed by Primer-BLAST with amplicon size set at 100–200 bp (HOX3B and Vmp1).
Alternatively, miRNA-enriched total RNA was collected using NucleoSpin miRNA Plus Mini Extraction Kit (MACHEREY NAGEL) and quantified on NanoDrop One (ThermoFisher). miRNA quantitation and calculation were performed with a TaqManTM miRNA assay (ThermoFisher) (Supplementary Table 3) and measured on LightCycler® 96 (Supplementary Information). Details of minimum information for publication of quantitative real-time PCR experiments (MIQE) is provided in Supplementary Data 5.
Amplicon library preparation and analysis
The cDNA reverse transcribed from total RNA was PCR amplified with pre-miRNA-flanking primers carrying TruSeq indexes for high throughput sequencing. PCR amplification was carried out in 25-µL reaction of PrimeSTAR Max DNA polymerase (Takara), containing 0.2 µM of forward and reverse primers. Each amplicon (5 µL) was resolved on 2% agarose gel and pooled together in equivalent amount prior to purification by GeneJET PCR Purification Kit (ThermoFisher). Library preparation was performed by Genomics, BioSci & Tech Co (Taiwan) with TruSeq® Nano DNA Library Prep (Illumina). Library quality was assessed on a Qubit 2.0 Fluorometer (ThermoFisher) and Agilent Bioanalyzer 2100 and sequenced on NovaSeq 6000 (Illumina) with 150 bp paired-end reading. Paired-end reads were trimmed by Trimmomatics61 and demultiplexed by in-house script. Demultiplexed paired-end reads were joined with PEAR v0.9.662, and unique joined sequences were counted by in-house Perl script.
RNA sequencing and differential expressed gene analysis
Total RNA was used for library preparation by TruSeq Stranded mRNA Library Prep Kit (Illumina). Briefly, mRNA was purified from 1 μg of total RNA by oligo(dT)-coupled magnetic beads and fragmented into small pieces under elevated temperature. cDNA was synthesized using reverse transcriptase and random primers, followed by double stranded cDNA generation, 3′ adenylation and adapter ligation. The library was purified with AMPure XP system (Beckman Coulter) and the quality was assessed on Qsep400 with N1 High Sensitivity Cartridge Kit (Bioptic Inc., Taiwan). The qualified library was sequenced on NovaSeq 6000 (Illumina) with 150 bp paired-end reading (Genomics, BioSci & Tech Co., Taiwan). Bases with low quality and sequences from adapters in raw data were removed using program fastp (version 0.20.0)63. The filtered reads were aligned to the reference genomes using HISAT2 (version 2.1.0). FeatureCounts (v2.0.1) in Subread package was applied for the quantification of gene abundance64. EdgeR was used to perform differentially expressed gene (DEG) analysis. The sequencing depth was ≈20 million reads per sample. DEG was deemed significant using the following thresholds: p value < 0.0001 and absolute log2(Fold-Change) value >= 1.
Transcriptome-wide A- > I editing analysis
The analysis was performed as previously described14 by Genomics, BioSci & Tech Co., Taiwan. Briefly, the data were down sampled to 5 million reads using seqtk (https://github.com/lh3/seqtk) and further trimmed and filtered with fastp63. The resulted reads were mapped to a pre-indexed rat reference genome mRatBN7.2 using RSEM65. BAM files were subject to base calling and quantification using REDitool66. To collect meaningful A-to-I editing, significant edits in the Mock groups were removed and considered as SNPs. Edits in experiment groups were deemed significant after Fisher’s exact test with a p value < 0.05 and occurring at least in 2 out of 3 replicates.
Small RNA sequencing and differential expressed gene analysis
The analysis was performed by Genomics, BioSci & Tech Co, Taiwan. miRNA-enriched total RNA (100 ng) was used for small RNA library preparation with QIAseq® miRNA Library Kit (QIAGEN) as instructed. Briefly, 3′ and 5′ adapters were directly and specifically ligated to 3′ and 5′ end of small RNA, respectively. First-strand cDNA was synthesized using QIAseq miRNA NGS RT Enzyme and Primer. cDNA was purified by QIAseq beads prior to PCR amplification with QIAseq miRNA NGS Library Buffer and HotStarTaq® DNA Polymerase. The library was purified by QMN Beads and the quality was assessed on the Qsep400. The qualified library was sequenced on NovaSeq 6000 (Illumina) with trimmed 75 bp single-end reading (Genomics, BioSci & Tech Co.). Adapter sequences were trimmed with TrimGalore! and mapped to reference genome, mature and hairpin miRNAs with Bowtie (v1.3.0) to obtain proper miRNA reads. Bam files were processed with SAMtools and the expression profile of miRNAs were calculated and normalized with EdgeR. Differentially expressed miRNAs (DEmiRNAs) were identified using DEGSeq. The sequencing depth was ≈5 million reads per sample. DEG was deemed significant using Storey’s method with the following thresholds: q value = TRUE and absolute log2(Fold-Change) value >= 167.
miRNAome-wide A- > I editing analysis
The analysis was performed as previously described68. Briefly, reads were trimmed and filtered using fastp and mapped against a pre-indexed rat reference genome mRatBN7.2 using Bowtie. BAM files were transformed by an in-house script with additional inputs from pre-miRNA hairpin (miRBase) mapped against mRatBN7.2 and from mature miRNA (miRBase). Edits were deemed significant after multiple test followed by Benjamini-Hochberg correction with a p value < 0.05 by a script running with Math-CDF package in Perl69 and occurring at least in 2 out of 3 replicates. Finally, SNPs were removed from the significant edits manually at UCSC Genome Browser.
Cell/scaffold construct preparation and characterization
ASCs were mock or co-transduced in a 15-cm dish as described previously. At 1 dpt, gelatin discs (d = 6 mm, thickness 1 mm) were cut from Spongostan sheets (Ethicon, MS0003) with a biopsy puncher (Integra Miltex), and prewetted in PBS. ASCs cells were collected and seeded to gelatin discs (5 × 106 cells/disc). The resultant ASCs/gelatin constructs were incubated at 37 °C, 5% CO2 for 4 h prior to culturing in chondroinduction medium. At 7 and 14 dpt, the constructs were collected for calvarial defect implantation. Alternatively, the constructs were fixed in 4% buffered formaldehyde (Macron) overnight at room temperature before embedding in paraffin and sliced into 3-µm thick sections. The sections were cleaned in xylene and hydrated through a series of descending alcohol. For histochemical analysis, the hydrated sections were stained with H&E and Alcian Blue/Nuclear Fast Red. Alternatively, antigen retrieval of the sections were carried out with 0.05% trypsin EDTA (Gibco) at 37 °C for 20 min followed by blocking in PBS containing 5% bovine serum albumin and 0.1% Tween 20 for 1 h before incubating with rabbit anti-collagen type II (Abcam, 1:100) primary antibody at 4 °C overnight. The sections were then incubated with HRP-conjugated goat anti-rabbit (Abcam, 1:500) secondary antibody at room temperature for 1 h and developed with SIGMAFAST™ 3,3′-Diaminobenzidine (Sigma). Sections were photographed on Eclipse TS100-F (Nikon). Positively stained areas of the images were processed by Fiji for semi-quantification.
Animal implantation
Mock (Mock group) and transduced (iREPRESS) ASCs/gelatin constructs cultured in chondroinduction medium were harvested at 7 or 14 dpt for implantation. On the day of implantation, 6-week-old female SD rats were injected intramuscularly with Zoletil® 50 (25 mg/kg body weight, Virbac Animal Health) and 2% Rompun® (0.15 mL/kg body weight, Bayer Health Care) for anesthesia, followed by cefazolin injection (160 mg/kg body weight). The calvaria were exposed by a 2-cm midline incision and the removal of the periosteal layers.
A 6-mm defect was created by punching through the exposed calvarium with a biopsy puncher (Integra Miltex) gradually with occasional saline buffer supplement to minimize damage to the neighboring bone tissue and underlying dura mater. The bone flaps were gently discarded and replaced with the ASCs/scaffold constructs followed by suturing with 4-0 absorbable stitch (PolySorb, Covidien). Post-operative procedure was performed with an administration of topical neomycin/bacitracin and intramuscular injection of ketoprofen (5 mg/kg).
Qualitative and quantitative characterization of bone healing
µCT imaging was performed on a nanoScan SPECT/CT (Mediso) at Chang Gung Memorial Hospital to assess bone regeneration of the animals at 4 (W4), 8 (W8) and 12 (W12) weeks post-implantation. 3D and hot-and-cold projections of the calvarial bones were rendered by InterViewTM FUSION (Mediso) and PMOD (PMOD Technologies). DICOM files from scanning were further processed by PMOD to generate bone area, volume and density.
Statistics and reproducibility
In vitro data and images are representative of at least three independent culture experiments. All quantitative data are expressed as mean±standard deviation (SD). The sample size of animal studies was not predetermined and experiments were not randomized. No data were excluded from the analyses. Statistical analyses were carried out with one-way or two-way ANOVA followed by Tukey multiple comparison test. A p value less than 0.05 was considered significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the results of this study are available within the paper and Supplementary Information. High-throughput sequencing data are available from the NCBI Sequence Read Archive database PRJNA1153716. Source data are provided with this paper.
Code availability
All the code used in the study for processing DNA and RNA seq data, analyzing differential gene expression, calculating A- > I editing are derived from the original articles referenced in the Methods section.
References
Filipowicz, W., Bhattacharyya, S. N. & Sonenberg, N. Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102–114 (2008).
Jin, W., Wang, J., Liu, C.-P., Wang, H.-W. & Xu, R.-M. Structural basis for pri-miRNA recognition by Drosha. Mol. Cell 78, 423–433.e425 (2020).
Rice, G. M., Shivashankar, V., Ma, E. J., Baryza, J. L. & Nutiu, R. Functional atlas of primary miRNA maturation by the microprocessor. Mol. Cell 80, 892–902.e894 (2020).
Rupaimoole, R. & Slack, F. J. MicroRNA therapeutics: towards a new era for the management of cancer and other diseases. Nat. Rev. Drug Discov. 16, 203–222 (2017).
Yang, W. et al. Modulation of microRNA processing and expression through RNA editing by ADAR deaminases. Nat. Struct. Mol. Biol. 13, 13–21 (2006).
Lima, J. F., Cerqueira, L., Figueiredo, C., Oliveira, C. & Azevedo, N. F. Anti-miRNA oligonucleotides: a comprehensive guide for design. RNA Biol. 15, 338–352 (2018).
Chen, C. L. et al. Baculovirus-mediated miRNA regulation to suppress hepatocellular carcinoma tumorigenicity and metastasis. Mol. Ther. 23, 79–88 (2015).
Chang, H. et al. CRISPR/cas9, a novel genomic tool to knock down microRNA in vitro and in vivo. Sci. Rep. 6, 22312 (2016).
Stenvang, J., Petri, A., Lindow, M., Obad, S. & Kauppinen, S. Inhibition of microRNA function by antimiR oligonucleotides. Silence 3, 1 (2012).
Tay, F. C., Lim, J. K., Zhu, H., Hin, L. C. & Wang, S. Using artificial microRNA sponges to achieve microRNA loss-of-function in cancer cells. Adv. Drug Deliv. Rev. 81, 117–127 (2015).
Adikusuma, F. et al. Large deletions induced by Cas9 cleavage. Nature 560, E8–E9 (2018).
Kosicki, M., Tomberg, K. & Bradley, A. Repair of double-strand breaks induced by CRISPR-Cas9 leads to large deletions and complex rearrangements. Nat. Biotechnol. 36, 765–771 (2018).
Sheridan, C. Shoot the messenger: RNA editing is here. Nat. Biotechnol. 41, 306–308 (2023).
Cox, D. B. T. et al. RNA editing with CRISPR-Cas13. Science 358, 1019–1027 (2017).
Qu, L. et al. Programmable RNA editing by recruiting endogenous ADAR using engineered RNAs. Nat. Biotechnol. 37, 1059–1069 (2019).
Merkle, T. et al. Precise RNA editing by recruiting endogenous ADARs with antisense oligonucleotides. Nat. Biotechnol. 37, 133–138 (2019).
Yi, Z. et al. Engineered circular ADAR-recruiting RNAs increase the efficiency and fidelity of RNA editing in vitro and in vivo. Nat. Biotechnol. 40, 946–955 (2022).
Monian, P. et al. Endogenous ADAR-mediated RNA editing in non-human primates using stereopure chemically modified oligonucleotides. Nat. Biotechnol. 40, 1093–1102 (2022).
Liu, Y. et al. REPAIRx, a specific yet highly efficient programmable A > I RNA base editor. EMBO J. 39, e104748 (2020).
Huang, X. et al. Programmable C-to-U RNA editing using the human APOBEC3A deaminase. EMBO J. 40, e108209 (2021).
Han, W. et al. Programmable RNA base editing with a single gRNA-free enzyme. Nucleic Acids Res. 50, 9580–9595 (2022).
Abudayyeh, O. O. et al. A cytosine deaminase for programmable single-base RNA editing. Science 365, 382–386 (2019).
Rauch, S. et al. Programmable RNA-guided RNA effector proteins built from human parts. Cell 178, 122–134.e112 (2019).
Vogel, P. et al. Efficient and precise editing of endogenous transcripts with SNAP-tagged ADARs. Nat. Methods 15, 535–538 (2018).
Koh, T.-C., Lee, Y.-Y., Chang, S.-Q. & Nissom, P. M. Identification and expression analysis of miRNAs during batch culture of HEK-293 cells. J. Biotechnol. 140, 149–155 (2009).
Lund, A. H. miR-10 in development and cancer. Cell Death Differ. 17, 209–214 (2010).
Yin, R. et al. miR-140-3p aggregates osteoporosis by targeting PTEN and activating PTEN/PI3K/AKT signaling pathway. Hum. Cell 33, 569–581 (2020).
Akhter, R. et al. Regulation of ADAM10 by miR-140-5p and potential relevance for Alzheimer’s disease. Neurobiol. Aging 63, 110–119 (2018).
Lee, D. Y., Deng, Z., Wang, C. H. & Yang, B. B. MicroRNA-378 promotes cell survival, tumor growth, and angiogenesis by targeting SuFu and Fus-1 expression. Proc. Natl Acad. Sci. USA 104, 20350–20355 (2007).
An, X. et al. miR-17, miR-21, and miR-143 enhance adipogenic differentiation from porcine bone marrow-derived mesenchymal stem cells. DNA Cell Biol. 35, 410–416 (2016).
Li, K.-C. et al. Baculovirus-mediated miR-214 knockdown shifts osteoporotic ASCs differentiation and improves osteoporotic bone defects repair. Sci. Rep. 7, 16225 (2017).
Kawahara, Y. et al. Redirection of silencing targets by adenosine-to-inosine editing of miRNAs. Science 315, 1137–1140 (2007).
Beisel, C. L., Chen, Y. Y., Culler, S. J., Hoff, K. G. & Smolke, C. D. Design of small molecule-responsive microRNAs based on structural requirements for Drosha processing. Nucleic Acids Res. 39, 2981–2994 (2011).
Lee, D. & Shin, C. Emerging roles of DROSHA beyond primary microRNA processing. RNA Biol. 15, 186–193 (2018).
Tehler, D., Hoyland-Kroghsbo, N. M. & Lund, A. H. The miR-10 microRNA precursor family. RNA Biol. 8, 728–734 (2011).
Ribas, J. et al. A novel source for miR-21 expression through the alternative polyadenylation of VMP1 gene transcripts. Nucleic Acids Res. 40, 6821–6833 (2012).
Nishikura, K. Functions and regulation of RNA editing by ADAR deaminases. Annu. Rev. Biochem. 79, 321–349 (2010).
Sung, L.-Y. et al. Efficient gene delivery into cell lines and stem cells using baculovirus. Nat. Protoc. 9, 1882–1899 (2014).
Sung, L.-Y. et al. Enhanced and prolonged baculovirus-mediated expression by incorporating recombinase system and in cis elements: a comparative study. Nucleic Acids Res. 41, e139 (2013).
Lu, C.-H. et al. Regenerating cartilages by engineered ASCs: Prolonged TGF-β3/BMP-6 expression improved articular cartilage formation and restored zonal structure. Mol. Ther. 22, 186–195 (2014).
Truong, V. A. et al. Bi-directional gene activation and repression promote ASC differentiation and enhance bone healing in osteoporotic rats. Mol. Ther. 30, 92–104 (2022).
Zhang, Y. et al. MicroRNA-21 controls the development of osteoarthritis by targeting GDF-5 in chondrocytes. Exp. Mol. Med. 46, e79–e79 (2014).
Lin, C.-Y. et al. The use of ASCs engineered to express BMP2 or TGF-β3 within scaffold constructs to promote calvarial bone repair. Biomaterials 34, 9401–9412 (2013).
Nguyen, N. T. K. et al. CRISPR activation of long non-coding RNA DANCR promotes bone regeneration. Biomaterials 275, 120965 (2021).
Konermann, S. et al. Transcriptome engineering with RNA-targeting type VI-D CRISPR effectors. Cell 173, 665–676 (2018).
Morelli, K. H. et al. An RNA-targeting CRISPR–Cas13d system alleviates disease-related phenotypes in Huntington’s disease models. Nat. Neurosci. 26, 27–38 (2023).
Chen, X., Zaro, J. L. & Shen, W. C. Fusion protein linkers: property, design and functionality. Adv. Drug Deliv. Rev. 65, 1357–1369 (2013).
Katrekar, D. et al. In vivo RNA editing of point mutations via RNA-guided adenosine deaminases. Nat. Methods 16, 239–242 (2019).
Vallecillo-Viejo, I. C., Liscovitch-Brauer, N., Montiel-Gonzalez, M. F., Eisenberg, E. & Rosenthal, J. J. C. Abundant off-target edits from site-directed RNA editing can be reduced by nuclear localization of the editing enzyme. RNA Biol. 15, 104–114 (2018).
Booth, B. J. et al. RNA editing: expanding the potential of RNA therapeutics. Mol. Ther. 31, 1533–1549 (2023).
Kim, T. & Croce, C. M. MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies. Exp. Mol. Med. 55, 1314–1321 (2023).
Winkle, M., El-Daly, S. M., Fabbri, M. & Calin, G. A. Noncoding RNA therapeutics-challenges and potential solutions. Nat. Rev. Drug Discov. 20, 629–651 (2021).
Huang, S. et al. Advances in microRNA therapy for heart failure: Clinical trials, preclinical studies, and controversies. Cardiovasc. Drugs Ther. https://doi.org/10.1007/s10557-023-07492-7 (2023).
Levy, J. M. et al. Cytosine and adenine base editing of the brain, liver, retina, heart and skeletal muscle of mice via adeno-associated viruses. Nat. Biomed. Eng. 4, 97–110 (2020).
Zhao, F. et al. A strategy for Cas13 miniaturization based on the structure and AlphaFold. Nat. Commun. 14, 5545 (2023).
Xu, C. et al. Programmable RNA editing with compact CRISPR-Cas13 systems from uncultivated microbes. Nat. Methods 18, 499–506 (2021).
Tang, X.-Z. E., Tan, S. X., Hoon, S. & Yeo, G. W. Pre-existing adaptive immunity to the RNA-editing enzyme Cas13d in humans. Nat. Med. 28, 1372–1376 (2022).
Wessels, H. H. et al. Massively parallel Cas13 screens reveal principles for guide RNA design. Nat. Biotechnol. 38, 722–727 (2020).
Lo, S. C. et al. Enhanced critical-size calvarial bone healing by ASCs engineered with Cre/loxP-based hybrid baculovirus. Biomaterials 124, 1–11 (2017).
Kluesner, M. G. et al. EditR: A method to quantify base editing from Sanger sequencing. CRISPR J. 1, 239–250 (2018).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Zhang, J., Kobert, K., Flouri, T. & Stamatakis, A. PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620 (2014).
Chen, S., Zhou, Y., Chen, Y. & Gu, J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890 (2018).
Liao, Y., Smyth, G. K. & Shi, W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014).
Li, B. & Dewey, C. N. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinform. 12, 323 (2011).
Lo Giudice, C., Tangaro, M. A., Pesole, G. & Picardi, E. Investigating RNA editing in deep transcriptome datasets with REDItools and REDIportal. Nat. Protoc. 15, 1098–1131 (2020).
Storey, J. D. & Tibshirani, R. Statistical significance for genomewide studies. Proc. Natl Acad. Sci. USA 100, 9440–9445 (2003).
Alon, S. et al. Systematic identification of edited microRNAs in the human brain. Genome Res. 22, 1533–1540 (2012).
Baiocchi, G. Using Perl for statistics: Data processing and statistical computing. J. Statis Soft 11, 1–75 (2004).
Acknowledgements
The authors thank the Laboratory Animal Center, Chang Gung Memorial Hospital, Linkou, Taiwan for the molecular imaging and technical support. The authors also acknowledge the financial support from the National Science and Technology Council (NSTC 112-2223-E-007-002 (Y.C.H.), 112-2314-B-007-004-MY3 (Y.C.H.), 111-2223-E-007-003 (Y.C.H.), 111-2634-F-007-007 (Y.C.H.), 111-2622-E-007-003 (Y.C.H.), 109-2314-B-007-002-MY3 (Y.C.H.)), Chang Gung Memorial Hospital (CMRPG3M0782 (Y.C.H.), CMRPG3M0781 (Y.C.H.)) and National Health Research Institutes (NHRI-EX112-11014BI (Y.C.H.), NHRI-EX113-11329EI (Y.C.H.)), Taiwan.
Author information
Authors and Affiliations
Contributions
V.A.T. conceptualized the project, designed and performed experiments and wrote the paper. Yu-Han C. acquired funding and wrote the paper. T.Q.D., Y.T., J.T., C.W.C., and Yi-Hao C. performed experiments. G.S.L. wrote the paper. Y.C.H. supervised the project, acquired funding and wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Truong, V.A., Chang, YH., Dang, T.Q. et al. Programmable editing of primary MicroRNA switches stem cell differentiation and improves tissue regeneration. Nat Commun 15, 8358 (2024). https://doi.org/10.1038/s41467-024-52707-6
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-52707-6