Abstract
Variations of individual lifespans within genetically identical populations in homogenous environments are remarkable, with the cause largely unknown. Here, we show the expression dynamic of the Caenorhabditis elegans fatty acid elongase ELO-6 during aging predicts individual longevity in isogenic populations. elo-6 expression is reduced with age. ELO-6 expression level exhibits obvious variation between individuals in mid-aged worms and is positively correlated with lifespan and health span. Interventions that prolong longevity enhance ELO-6 expression stability during aging, indicating ELO-6 is also a populational lifespan predictor. Differentially expressed genes between short-lived and long-lived isogenic worms regulate lifespan and are enriched for PQM-1 binding sites. pqm-1 in young to mid-aged adults causes individual ELO-6 expression heterogeneity and restricts health span and life span. Thus, our study identifies ELO-6 as a predictor of individual and populational lifespan and reveals the role of pqm-1 in causing individual health span variation in the mid-aged C. elegans.
Similar content being viewed by others
Introduction
In laboratory animals and in humans, 60–90% of the variation in lifespan is independent of genotype1. Genetically identical invertebrates in homogenous environments also experience similar variability in lifespan, indicating neither genetic nor environmental factors fully account for variability in individual longevity1. Thus far, a single reporter gene that functions as a biomarker of individual aging, which will facilitate the investigation of stochastic factors causing individual lifespan variation, remains rare. Previous studies in Caenorhabditis elegans have found that hsp-16.2 induction under heat shock, mitoflash frequency, nucleolus size, miRNA expression, and ROS level in the L2 developmental stage predict individual longevity, indicating C. elegans is a powerful model organism to investigate individual lifespan variation2,3,4,5,6.
Transcriptome analysis in multiple organisms, including yeast, nematodes, fruit flies, mice, and humans, found that significant changes in gene expression occur during aging7,8,9. These age-regulated genes generally participate in a wide variety of biological processes that are associated with age-dependent physiological changes. In mice, most age-regulated gene expression alterations can either be completely or partially delayed by caloric restriction, a well-known intervention that retards aging10. The extension of lifespan achieved by pharmacological intervention is accompanied by an enhancement of the overall transcriptional stability during aging11. These results suggest that gene expression regulation plays an important and causal role in aging. Studies demonstrate that transcription profiles can serve as a cellular aging clock for measuring ages12,13,14, indicating that gene transcriptional changes can be a potential tool to identify biomarkers of aging.
PQM-1 is a C2H2 Zn finger transcription factor that binds to genes involved in lipid metabolism15. PQM-1 binds to the DAF-16-associated element (DAE) and is an important mediator of the insulin signaling pathway. It is essential for the long-lived daf-2 mutant phenotypes of somatic aging, stress response, and reproductive aging16,17. PQM-1 is also essential for the lifespan extension of puf-8 mutant animals18. Despite its role in the lifespan extension of C. elegans mutant worms, pqm-1 mutants themselves have a wild-type lifespan, and overexpressing PQM-1 protein shortens lifespan16. Under stress conditions, PQM-1 limits survival from hypoxia and mediates the tradeoff between immunity and longevity19,20.
In this study, we sought to identify lifespan predictors through transcriptome longitudinal analysis in C. elegans somatic cells during aging. We identified genes that continuously increased or decreased expression along with age and further selected candidates by the correlation coefficient between gene expression abundance and age, the gene expression abundance in young adults, gene body histone H3.3 deposition, and histone modification patterns. We chose genes that showed a consistently reduced expression with age as candidates to be a lifespan predictor. ELO-6, a fatty acid elongase, had continuously decreased expression with age. From adult day 5, ELO-6 showed a variable expression level between individual animals, and the expression level predicted individual lifespan and health span. In addition, interventions prolonging lifespan enhance ELO-6 expression stability with age, indicating ELO-6 is a predictor of individual lifespan and populational lifespan. Through transcriptome analysis of the short-lived and long-lived animals distinguished by ELO-6 expression on adult day 5, we found that pqm-1 regulates ELO-6 expression homogeneity in young adults and impacts health span. In addition, decreasing the expression of genes highly expressed in short-lived individuals, including genes targeted by PQM-1, extends lifespan. Therefore, our study identified a single gene reporter for individual and populational lifespan prediction and suggested the function of pqm-1 in individual lifespan and health span regulation.
Results
Screen for lifespan predictors through transcriptome analysis
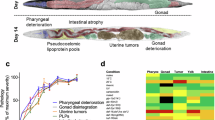
Gene expression profiles during aging are effective for age calculation12,13,14. To identify genes that have the potential for lifespan prediction, we generated whole worm gene expression profiles of C. elegans somatic cells on adult day 2 (D2A), D4A, D6A, D8A, D10A, and D12A from germline-less glp-1(e2141) growing at 25 °C by mRNA-seq. We used temperature-sensitive germline-less mutant glp-1(e2141) to avoid transcriptional signals from germlines and embryos. We cultured glp-1(e2141) at 25 °C from embryo to end without temperature shifting. Under this condition, glp-1(e2141) is germline-less, and the lifespan of glp-1(e2141) is slightly longer than that of wild-type N2 (∼10% life span extension)21. We performed a longitudinal analysis. The MDS plot shows that the transcription profiles from young to old are arranged mainly from left to right on dimension 1, which represents 51% of the gene expression difference (Supplementary Fig. 1a). When analyzing the gene expression change at the single gene level, most gene expression fluctuated during aging (Supplementary Data 4). We used 20% of FPKM (fragments per kilobase of transcript per million fragments mapped) as the cutoff for gene expression change between adjacent time points. We found 123 genes consistently increased expression, and 44 genes consistently decreased expression during aging (Supplementary Data 4). GO term analysis showed that the 44 genes that decreased expression with age are enriched for genes involved in innate immune response, structural constituent of cuticle, and oxidoreductase activity (Supplementary Data 5), implicating physiological deterioration during aging. We aim to identify genes for lifespan prediction and further develop downstream screening assays. Therefore, we sought to identify lifespan predictors from the 44 genes that decreased expression with age. To select preferred candidates from the 44 genes, we first calculated the correlation coefficient between gene expression abundance and age (Supplementary Data 4). Among the 44 genes, the expression level of elo-6 is mostly inversely correlated with age, with a correlation coefficient of −0.98. In addition, elo-6 is highly expressed on D2A compared with the other genes that have a similar inverse correlation coefficient between expression abundance and age (Supplementary Data 4). Consistent with its active expression in young worms, elo-6 is with HIS-72, one of the histone H3 variant H3.3 in C. elegans, deposited on the gene body region in somatic cells (Supplementary Fig. 1c)22. H3.3 is related to active gene expression23 and is essential for the lifespan extension of long-lived mutants24. Therefore, the expression of elo-6 may be related to lifespan regulators. In addition, elo-6 was found to possess an adult-specific histone modification pattern, with adult-specific H3K4me3 instead of H3K36me3 on its gene body region (Supplementary Fig. 1c)22,25. Therefore, we first examined whether elo-6 can be a lifespan predictor.
We used the strain MTP27 with the endogenous ELO-6 tagged by GFP to examine the expression change of ELO-6 with age. This strain was generated by CRISPR/Cas9-based genome editing, adding a GFP::3xFLAG tag to the N terminus of ELO-6 and introducing a substitution (S2A). MTP27 develops normally and has a lifespan similar to N2 (Supplementary Fig. 1d). GFP::ELO-6 is mainly located in the intestine, as previously reported (Fig. 1)26. We observed that GFP::ELO-6 shows reduced expression during aging in glp-1(e2141) at 25 °C (Fig. 1a, b) and in wild-type N2 background at 20 °C (Fig. 1c, d). On D9A in N2 at 20 °C and D7A in glp-1(e2141) at 25 °C, the GFP signal can barely be observed under a fluorescence stereo microscope, which is consistent with our mRNA-seq result from glp-1(e2141) that the mRNA abundance of elo-6 on D10A is 10% of that on D2A (Fig. 1, Supplementary Data 4). With gon-2(q388ts) mutant and the wild-type N2, the single-cell aging datasets identified gradually reduced elo-6 expression in intestinal cells27,28. In addition, ELO-6 protein level has been identified as reduced from D1A to D5A and D5A to D10A29. Thus, ELO-6 expression is an ideal candidate to monitor aging.
Fluorescent images of endogenous GFP::3xFLAG::ELO-6(S2A) in {glp-1(e2141); elo-6[kmg14(GFP::3xFLAG::elo-6(S2A))]} from D2A to D9A at 25°C (a) and quantification of the GFP level in intestine of each worm (b). Worms developed and aged at 25°C from egg-laying. Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. The numbers of analyzed worms are shown beneath the bars. Scale bar, 200 μm. c, d Fluorescent images of endogenous GFP::3xFLAG::ELO-6(S2A) in elo-6[kmg14(GFP::3xFLAG::elo-6(S2A))] with wild-type N2 background from D2A to D9A at 20 °C (c) and quantification of the GFP level in intestine of each worm (d). Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. The numbers of analyzed worms are shown beneath the bars. Images (a, c) were all taken with identical parameters. The yellow signal in fluorescent images is autofluorescence. Source data are provided as a Source Data file.
In addition to elo-6, we also used an elo-1::3xflag::gfp knock-in strain generated by CRISPR/Cas9-based genome editing, tagging the endogenous ELO-1 with GFP and 3xFLAG. elo-1 has similar H3K4me3 and HIS-72 patterns with elo-6, and elo-1 is with H3K36me3 on its gene body region while elo-6 doesn’t have any H3K36me3 on its gene body region (Supplementary Fig. 1c). H3K36me3 maintains gene expression stability during aging, as we and others have previously reported25,30. We observed that ELO-1::GFP shows different expression dynamics during aging, with higher expression stability after D2A than GFP::ELO-6(S2A) (Supplementary Fig. 2a, b). This indicates the dynamics of GFP::ELO-6(S2A) do not reflect transcription/translational features for all genes but the expression of a subgroup gene with age. We have also generated integrated transgenic lines using the elo-6 promoter to express GFP. The expression of exogenous GFP by the elo-6 promoter shows consistent reduction with age from D3A to D9A, and the expression dynamic of exogenous GFP is similar to that of endogenous GFP::ELO-6(S2A) after D6A (Supplementary Fig. 2a, b).
Expression variation of ELO-6 in young adults predicts lifespan and health span in isogenic populations
We observed that as early as D4A, there are differences in GFP::ELO-6 expression levels between individual worms in glp-1(e2141) background at 25 °C (Fig. 1a) and on D5A in wild-type N2 background (Fig. 1c). We then quantified the heterogenicity of ELO-6 expression during aging using the coefficient of variation of individual worm GFP level which reflects the dispersion of individual worm GFP levels around the mean GFP level. We found the coefficient of variation of individual worm GFP levels increases with age (Supplementary Fig. 2c-e). We also observed individual variations of exogenous GFP levels on D6A (Supplementary Fig. 2f), indicating that the individual variation we observed is not caused by GFP tagging or S2A mutation of ELO-6. In addition, we found that the variation between individuals on D6A is also a characteristic that does not exist equally among genes. We found ELO-1::GFP showing a lower individual variation on D6A (Supplementary Fig. 2f).
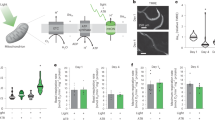
Since ELO-6 expression decreases with age, we hypothesized that the variation of ELO-6 expression represents the difference in the aging process between individuals. We separated worms according to GFP::ELO-6 levels on D5A in N2 background into 3 groups (Fig. 2a and Supplementary Fig. 3a, b) and measured their lifespan (Fig. 2b, Supplementary Fig. 3c, and Supplementary Data 10). The worms with low GFP::ELO-6 expression (GFPL) have a significantly shorter lifespan than those of medium (GFPM) and high GFP::ELO-6 (GFPH) expression groups (15.23% and 17.20% mean lifespan difference, P-value < 0.0001, P-value < 0.0001)(Fig. 2b and Supplementary Data 10), while GFPM and GFPH have a similar lifespan (P-value = 0.815) (Supplementary Data 10). In addition, we also measured the pharyngeal pumping rate and body bending rate, which represents the health span. Compared with GFPM and GFPH groups, the GFPL group shows a significantly slower bending rate and pharyngeal pumping rate (Fig. 2c, d and Supplementary Fig. 3d, e), indicating that individuals with a more rapid reduction of ELO-6 expression during aging in the GFPL group also have shorter health spans. As well, the GFPH group shows a significantly higher pharyngeal pumping rate than the GFPM group animals on D8A (Fig. 2d and Supplementary Fig. 3e), indicating its relatively slower aging process. The offspring of GFPL, GFPM, and GFPH show similar GFP expression on D5A (Supplementary Fig. 3f), confirming that the expression variation of ELO-6 on D5A is not caused by genetic factors. We also confirmed that the elo-6 mRNA level of GFPL is the lowest in the sorted groups (GFPL, GFPM, and GFPH) by RT-qPCR (Supplementary Fig. 3g) and mRNA-seq (Supplementary Data 6).
a The GFP signals in GFPL, GFPM, and GFPH group worms from elo-6[kmg14(GFP::3xFLAG::elo-6(S2A))] were photographed using a fluorescence stereo microscope with the following imaging parameters: exposure time 380 ms, gain 40, zoom 0.85x, and total video magnification 0.27x. b Survival curves of GFPL, GFPM, and GFPH. GFPL vs. GFPM, p = 2.6789E-08; GFPL vs. GFPH, p = 2.2880E-09; GFPM vs. GFPH, p = 0.8145 (log-rank test, two-sided). Age-related decline in body bending or pharyngeal pumping rate in GFPL, GFPM, and GFPH group worms. The body bending assay was performed in a 200 μL buffer droplet in a petri dish. Data are represented as mean ± SD of individual worm bends per 30 sec (c) or pumping rates (d) with individual values shown as dots. P-values are presented in the figures with p > 0.05 in purple (unpaired t-test, two-sided except otherwise indicated as one-sided; unpaired t-test assuming unequal variance or equal variance based on F-test result (source data)). The numbers of analyzed worms are shown beneath the plots. e Survival curves of GFP1, GFP2, GFP3, and GFP4 from {glp-4(bn2); elo-6[kmg14(GFP::3xFLAG::elo-6(S2A))]} at 25 °C. The corresponding fluorescent images are in Supplementary Fig. 3h. GFP1 vs. GFP2-4, p = 0.0002, 1.1292E-08, 1.3645E-10; GFP2 vs. GFP3, p = 0.0222, GFP2 vs. GFP4, p = 0.0019; GFP3 vs. GFP4, p = 0.38 (Breslow test, two-sided). f Survival curves of GFP1, GFP2, GFP3, and GFP4 from {glp-1(e2141);elo-6[kmg14(GFP::3xFLAG::elo-6(S2A))]} at 25 °C. The corresponding fluorescent images are in Supplementary Fig. 3h. GFP1 vs. GFP2-4, p = 0.1678, 0.00006, 0.0041; GFP2 vs. GFP3, p = 0.0039, GFP2 vs. GFP4, p = 0.2405; GFP3 vs. GFP4, p = 0.0167 (Breslow test, two-sided). ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗ p < 0.01, ∗p < 0.05, (ns) p > 0.05. Source data are provided as a Source Data file.
In germline-less glp-4(bn2) and glp-1(e2141) worms cultured at 25 °C, we separated worms according to GFP levels on D5A from low to high into GFP1 to GFP4 groups (Supplementary Fig. 3h). We found that worms with lower GFP::ELO-6 expression at D5A had a shorter lifespan when analyzed using the Breslow test, which is suitable for early-stage aging comparison (Fig. 2e, f and Supplementary Data 10). Thus, the expression level of ELO-6 on D5A is positively correlated with individual lifespan and health span in isogenic populations.
The lifespan curves of the germline-less mutants at 25°C show a bimodal pattern (Fig. 2e, f). There are two classes of corpses in isogenic populations identified previously: early deaths with a swollen pharynx (P deaths) and later deaths with an atrophied pharynx (p deaths)31. For glp-1(e2141), shifting to 25 °C from the egg doesn’t equally affect P deaths and p deaths, and the lifespan curve of glp-1(e2141) under this condition showed a bimodal pattern31. We found the bimodal pattern exists in the subgroups, suggesting that the cause of these two deaths (P and p) in the isogeneic population does not overlap with the ELO-6 expression variation in glp-1(e2141).
ELO-6 is a populational lifespan predictor
We further asked whether ELO-6 can predict lifespan regulated by genetic factors, in addition to the non-genetic ones that influence individual lifespan. To answer this question, we applied RNAi from hatching to reduce the expression of daf-2, utx-1, set-2, and cyc-1, which all extend lifespan32,33,34,35,36, and examined GFP::ELO-6(S2A) expression during aging. daf-2 RNAi enhanced ELO-6 expression level as early as D2A, and showed a higher GFP level than the control on D8A (Fig. 3a–d). This effect depends on daf-16. In the short-lived daf-16(mu86) mutant, the expression level of ELO-6 showed no difference between daf-2 RNAi and the control (Fig. 3b, d). In addition, ELO-6 expression in daf-16(mu86) under control treatment is reduced compared with the N2 worms from D2A and during aging (Fig. 3b, d). Previous studies have shown that daf-16 positively regulates elo-6 expression in larvae and young adults37,38. Under utx-1 RNAi treatment, ELO-6 expression level was significantly higher on D3A and D4A than the control (Fig. 3a, c). GFP::ELO-6 expression can still be well observed on D9A when the GFP expression in the control condition can only be detected (Fig. 3a). Under cyc-1 RNAi treatment, the expression level of GFP::ELO-6 is lower than that of the control in D2 adult and maintains a steady level during aging from D2A to D8A (Fig. 3a, c). Set-2 RNAi extends lifespan (Supplementary Data 10). The ELO-6 expression level is enhanced by set-2 RNAi on D4A and D5A, and the expression decreases from D6A to D8A with a significantly lower level than the control on D9A (Fig. 3a, c). Germline-less animals have prolonged longevity39. We further tested whether ELO-6 expression can also be enhanced during aging in a long-lived germline-less mutant. We observed that in long-lived glp-1(e2141) that was shifted from 25°C to 20 °C after D1A40, the ELO-6 expression was enhanced during aging (Fig. 4b, d). Therefore, ELO-6 is a predictor of lifespan regulated by genetic factors. In addition, metformin treatment, which extends lifespan41, also enhances the GFP::ELO-6 expression stability during aging (Fig. 4a, c). These results demonstrate that the expression stability of ELO-6 during aging is a general predictor of populational lifespan. These are consistent with previous findings that elo-6 expression has been reported to increase in young adults by low-copy overexpression of sir-2.142, germline ablation43, long-lived glp-1(e2141) 44, daf-2(e1370)45,46, and metformin treatment47. The single-cell aging dataset also showed that the expression of elo-6 is higher in the daf-2(lf) mutant on adult day 1 and during aging28.
Fluorescent images of GFP::ELO-6(S2A) during aging with daf-2, utx-1, set-2, cyc-1 RNAi treatment in wild-type N2 background strain (a) and quantification (c). L4440 is an RNAi treatment control. RNAi knockdown samples were compared with L4440 control at the same age. Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. The numbers of analyzed worms are shown beneath the bars. P-values are presented in the source data (unpaired t-test or unpaired t-test with Welch’s correction, two-sided). Fluorescent images of GFP::ELO-6(S2A) during aging with daf-2 RNAi knockdown in wild-type N2 background strain and daf-16(mu86) strain (b) and quantification (d). The RNAi knockdown sample was compared with the corresponding control sample at the same age. Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. The numbers of analyzed worms are shown beneath the bars. P-values are presented in the source data (unpaired t-test or unpaired t-test with Welch’s correction, two-sided). For all RNAi treatments in this figure, iOP50 overnight cultures were diluted 1:10 and grown till O.D. ~ 0.8 and then concentrated 5-fold before being seeded onto plates. The yellow signal in fluorescent images is autofluorescence. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗ p < 0.01, ∗p < 0.05, (ns) p > 0.05. Source data are provided as a Source Data file.
Fluorescent images of GFP::ELO-6(S2A) during aging with metformin treatment (a) and quantification (c). Metformin treatment samples were compared with the untreated controls at the same age. Data are represented as mean ± SD of individual worm GFP levels. The numbers of analyzed worms are shown beneath the bars. P-values are shown in the figures with p > 0.05 in purple (multiple unpaired t-test with Welch correction, two-sided). Fluorescent images of GFP::ELO-6(S2A) during aging in long-lived glp-1(e2141) that was shifted from 25 °C to 20 °C at D1A (b) and quantification (d). The GFP signals in glp-1(e2141) were compared with the wild-type N2 background controls at the same age in (a). Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. The numbers of analyzed worms are shown beneath the bars. P-values are shown in the figures with p > 0.05 in purple (multiple unpaired t-test with Welch correction, two-sided). The yellow signal in fluorescent images is autofluorescence. Source data are provided as a Source Data file.
Since the ELO-6 expression during aging is correlated with lifespan, we further asked whether elo-6 is a lifespan regulator. We used RNAi to reduce the expression of ELO-6 (Supplementary Fig. 4a, b). We reduced elo-6 expression by RNAi from D2A or D5A and found that elo-6 RNAi does not change lifespan (RNAi from D2A, P-value = 0.166; RNAi from D5A, P-value = 0.667) (Supplementary Fig. 4c, d and Supplementary Data 10). Supplementation of C17iso, the product of ELO-626, can slightly extend the populational lifespan when the treatment begins at D4A (3% mean lifespan extension, P-value = 0.03) (Supplementary Fig. 4e, Supplementary Data 10). Therefore, the expression of elo-6 in the adult intestine doesn’t regulate lifespan but is simply a lifespan predictor under the standard laboratory culture condition. The role of elo-6 in lifespan regulation can be further systematically investigated with an elo-6 loss-of-function mutant. We further quantified C17iso levels in GFPL, GFPM, and GFPH group worms. Interestingly, we found there’s no positive correlation between C17iso level and ELO-6 expression, with the GFPL groups having the highest C17iso levels when normalized using the total fatty acid amount (Supplementary Fig. 4f). We also measured C17iso levels during aging in wild-type N2 worms and the long-lived daf-2(e1370) mutant (Supplementary Fig. 4g). We didn’t observe decreased C17iso with age in wild-type N2 worms from D2A to D8A. C17iso levels in long-lived daf-2(e1370) were similar to those of N2 until D8A. We only observed a slight increase of C17iso in wild-type N2 worms on D12A compared to D4A and D6A (Supplementary Fig. 4g).
Since ELO-6 expression stability during aging is influenced by the insulin/IGF-1 signaling pathway, and previous studies showed that the daf-16 downstream gene sod-3 shows expression variation between individuals on D9A reflecting the pathogenic effect of OP5048, we asked whether the expression variation of ELO-6 on D5A can be caused by OP50 infection as well. We transferred worms for two generations onto OP50 plates treated with UV and found that GFP expression on D5A still shows variation between individuals (Supplementary Fig. 4h), indicating that ELO-6 expression variation on D5A is not caused by OP50 infection. For N2 background animals, worms are transferred every day from D1A. To exclude the possibility that the transfer by worm picker might cause the variation expression of ELO-6 between individuals, we also washed worms to get rid of offspring, and we still observed GFP level variation between individuals from D5A (Supplementary Fig. 4i).
Transcriptome analysis of short-lived and long-lived individuals
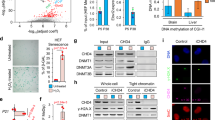
To understand the molecular mechanism related to ELO-6 prediction of individual worm lifespan, we performed mRNA-seq to identify differentially expressed genes (DEGs) between the short-lived and long-lived animals distinguished by ELO-6 expression variation on D5A. We performed mRNA-seq in germline-less mutants growing at 25 °C to avoid transcriptional signals from germlines and embryos. We used both germline-less mutant glp-1(e2141) and glp-4(bn2) backgrounds to avoid the bias caused by glp-1(e2141) or glp-4(bn2) mutation. On D5A, we separated the isogenic worms into GFP1, GFP2, GFP3, and GFP4 groups according to GFP levels from low to high, similar to the grouping for lifespan analysis, and pooled 100 worms in each group for mRNA-seq. Transcriptome analysis showed no striking gene expression differences between different GFP groups, with all correlation coefficients higher than 0.85 (Supplementary Data 3). The MDS plot of mRNA-seq data showed that the difference of transcriptomes followed similar trends as the GFP signal intensity (Fig. 5a). Comparing mRNA-seq data from adjacent GFP groups from the glp-1(e2141) background showed that GFP1 vs. GFP2 groups had 73 significantly differentially expressed genes when applying FDR < 0.1, while there were only 40 genes between GFP2 and GFP3, and 10 genes between GFP3 and GFP4 that showed expression differences (Supplementary Data 6). Since we observed that there is a large fraction of the population that dies on D6A in GFP1 groups in both the glp-1(e2141) and glp-4(bn2) background (Fig. 2e, f), the transcriptome of GFP1 might reflect the features that occur before death. Considering that the transcriptomes of GFP3 and GFP4 are highly similar, we then compared the transcriptome between GFP2 and GFP4 to identify differentially expressed genes (DEGs) between the shorter-lived and longer-lived groups. We found 65 genes increased expression and 113 genes decreased expression in the short-lived worms compared with the long-lived ones in both glp-1(e2141) and glp-4(bn2) backgrounds (Fig. 5b). Functional clustering analysis showed that xenobiotic metabolic process and structural constituent of cuticle-related genes are enriched in the decreased expression genes, and ribosome biogenesis and CUB domain related genes are enriched in increased expression genes in short-lived worms (Supplementary Data 7).
a MDS plot of mRNA-seq performed in GFP1, GFP2, GFP3, and GFP4 animals in the glp-1(e2141) or glp-4(bn2) background. b The Venn diagrams show genes that were differentially expressed between GFP2 and GFP4 worms in the glp-1(e2141) and glp-4(bn2) background. Red: genes highly expressed (increased) in GFP2 in glp-1(e2141); Lime: genes low expressed (decreased) in GFP2 in glp-1(e2141); Fuchsia: genes highly expressed in GFP2 in glp-4(bn2); Aqua: genes low expressed in GFP2 in glp-4(bn2). Gene numbers for each group are shown. c Motif analysis with genes differentially expressed between GFP2 and GFP4 worms in both the glp-1(e2141) and glp-4(bn2) background. Genomic regions surrounding TSS [-400, +100] of the 65 increased genes and 113 decreased genes in GFP2 were analyzed by Homer. Genes that are highly expressed and low expressed in GFP2 animals are both enriched in the PQM-1/ELT-3 binding motif. TSS: transcriptional start site. d The Venn diagrams show the occupancy of PQM-1/ELT-3 binding motif and PQM-1 peak in differentially expressed genes between GFP2 and GFP4. The genes showing expression differences between GFP2 and GFP in both the glp-1(e2141) and glp-4(bn2) background were subjected to this analysis. Gene numbers for each group are shown. Source data are provided as a Source Data file.
We further asked whether the DEGs between short-lived and long-lived isogenic animals are under a common regulatory mechanism. Motif analysis with the 65 increasingly expressed and 113 decreased expressed genes showed that both gene groups are enriched for the PQM-1/ELT-3 binding motif in the gene promoter regions (Fig. 5c), indicating common transcription factors control the differentially expressed genes. In addition to the motif analysis, we inspected the PQM-1 ChIP-seq profile from L3 stage worms15. In the 65 genes with increased expression, 26 have PQM-1 peaks in gene upstream regions, with 12 overlapping with the PQM-1/ELT-3 binding motif prediction. Of the 113 genes with decreased expression, 43 have PQM-1 peaks in gene upstream regions, and 19 coincided with PQM-1/ELT-3 binding motif prediction (Fig. 5d, Supplementary Data 6). When analyzing DEGs from the adjacent GFP group from the glp-1(e2141) background, we found that the PQM-1/ELT-3 binding motif and PQM-1 peak are enriched as well (Supplementary Fig. 5e,f, Supplementary Data 6). Therefore, the transcriptome difference between short-lived and long-lived individuals might be regulated by PQM-1/ELT-3. To verify this, we also performed mRNA-seq with GFPL, GFPM, and GFPH worms with wild-type N2 background growing at 20 °C (Supplementary Fig. 5a and Supplementary Data 6). The DEGs between GFP2 and GFP4 from germline-less mutants are partially overlapped with DEGs between GFPL and GFPH (Supplementary Fig. 5b). Motif analysis showed that for the 902 low expressed genes in GFPL, the most enriched motif TCTTATCA from Homer de novo motif analysis matches the ELT-3 binding motif (score 0.99), also matching the binding motif of ELT-2 (sore 0.97) and PQM-1 (score 0.96) (Supplementary Fig. 5c, Supplementary Data 6). 32.9 % of GFPL low-expressed genes (264 genes) are with this motif; among them, 185 genes are bound by PQM-1 peak (Supplementary Fig. 5d, Supplementary Data 6). For 1179 highly expressed genes in GFPL, the most enriched motif is CTCTTATY, matching the binding motif of ELT-2 (score 0.86), ELT-3 (score 0.81), and PQM-1 (score 0.80) (Supplementary Fig. 5c, Supplementary Data 6). 45.28% of GFPL high-expressed genes (412 genes) are with this motif; among them, 159 genes are bound by PQM-1 peak (Supplementary Fig. 5d). Altogether, 476 genes out of 902 GFPL low-expressed genes are with TCTTATCA motif and/or PQM-1 peak, and 638 genes out of 1179 GFPL high-expressed genes are with CTCTTATY and/or PQM-1 peak (Supplementary Fig. 5d). Therefore, the transcriptome difference between short-lived and long-lived individuals in both germline-less and the wild-type N2 background might be regulated by PQM-1/ELT-3.
pqm-1 promotes ELO-6 expression variation between individuals in mid-aged adults and regulates health span and life span
To investigate whether PQM-1 or ELT-3 regulates ELO-6 expression variation between individual worms on D5A and during subsequent aging, we used RNAi to knockdown pqm-1 or elt-3 starting from D2A or D3A and examined GFP::ELO-6 expression. We found that pqm-1 RNAi decreased the variation of ELO-6 expression between individuals, with fewer worms with low GFP::ELO-6 expression on D6A compared to the control (Fig. 6a–c and Supplementary Fig. 6a–c). To test whether pqm-1 regulates the aging process, we measured the worm bending frequency and pharyngeal pumping rate of the mid-aged worms upon pqm-1 RNAi treatment starting from D2A. The results show that decreased pqm-1 expression delayed the aging process, with the bending frequency on D8A being significantly higher in the pqm-1 RNAi group than in the control group (Fig. 6d, Supplementary Fig. 6d). The pharyngeal pumping rate under pqm-1 RNAi treatment is also higher than the control on D7A to D9A (Fig. 6e and Supplementary Fig. 6e). In addition, we also observed the reduction of variation in pumping rate from D6A to D7A between individuals with pqm-1 RNAi (Fig. 6e and Supplementary Fig. 6e).
Fluorescent images of GFP::ELO-6(S2A) on D6A with pqm-1 RNAi treatment (a) and quantification (b, c). *Worms without a GFP signal in some part of the intestine and with a quantified GFP level lower than 50. The yellow signal is autofluorescence. The numbers of analyzed worms are shown beneath the bars. The occurrence of worms with low GFP level (quantified GFP < 50) in L4440 control and pqm-1 RNAi knockdown (Fisher’s exact test, two-sided)(b). Coefficient of variation of individual worm GFP levels in L4440 control and pqm-1 RNAi knockdown (c). Coefficient of Variation = Standard deviation (σ) / Mean (μ). Age-related decline in body bending rate (d) or pharyngeal pumping rate (e) with pqm-1 RNAi knockdown and control. RNAi treatment started from D2A. The body bending assay was performed in 1 ml buffer in a 24-well Tissue culture-treated plate. Data are represented as mean ± SD of individual worm bends per 30 sec (d) or pumping rates (e) with individual values shown as dots. The numbers of analyzed worms are shown beneath the plots. P-values are presented in the figures with p < 0.05 in red (two-sided unpaired t-test assuming unequal variance or equal variance based on F-test result). f Differentially expressed genes caused by pqm-1 RNAi treatment and those between GFP2 and GFP4 are compared with Venn diagrams. Red: genes increased expressed (increased) upon pqm-1 RNAi treatment; Lime: genes with reduced expression (decreased) upon pqm-1 RNAi treatment; Fuchsia: genes highly expressed in GFP2; Aqua: genes low expressed in GFP2. Gene numbers for each group are shown. g Survival curves of pqm-1 RNAi treatment from D3A to D6A followed by dcr-1 RNAi treatment from D7A (log-rank test, two-sided). Quantitative data are presented in Supplementary Data 10. Source data are provided as a Source Data file.
We further examined the transcriptome change on D5A upon pqm-1 RNAi treatment starting from D2A in the germline-less mutant glp-1(e2141) growing at 25 °C. The MDS plot of the mRNA-seq data from the pqm-1 RNAi treatment along with the transcriptome data from D2A to D12A (Supplementary Fig. 1a) shows that the transcriptome upon pqm-1 RNAi is shifted towards a younger age on dimension 1, which represents 38% gene expression difference (Supplementary Fig. 1b). There were 267 genes with increased expression upon pqm-1 RNAi treatment (fold change > 0.5, FDR < 0.05) and 124 genes with reduced expression (fold change >0.5, FDR < 0.05) compared to the control. We then further compared the DEGs between the short-lived and long-lived individual animals (Fig. 5b) with the expression difference caused by pqm-1 RNAi treatment. We found that 10 out of the 65 genes that are highly expressed in short-lived worms had decreased expression upon pqm-1 RNAi treatment, and among the 113 genes that are expressed at a lower level in short-lived worms, pqm-1 RNAi treatment increased the expression of 25 genes and reduced the expression of 8 genes (Fig. 6f). Thus, pqm-1 RNAi influences the transcriptome to reduce the transcriptional difference that would occur between short-lived and long-lived individuals during aging.
We further asked whether pqm-1 regulates lifespan. We found that pqm-1 RNAi treatment from D3A by culturing worms on pqm-1 RNAi plate from D3A to the end doesn’t change lifespan, which is consistent with the previous reports16,18 (Supplementary Fig. 6f, Supplementary Data 10). Then, we controlled the timing of pqm-1 RNAi. We started pqm-1 RNAi from D3A and transferred the worms to dcr-1 RNAi plates from D7A to reduce RNAi efficiency33. We found that pqm-1 RNAi treatment limited to 4 days (D3A to D6A) extends lifespan (7.65 % mean lifespan difference, P-value = 0.014) (Fig. 6g, Supplementary Fig. 6g and Supplementary Data 10). The RNA expression levels of pqm-1 are confirmed by RT-qPCR (Supplementary Fig. 6h, i).
ceh-60 and unc-62 suppress ELO-6 expression during aging
We noticed that both the high and low-expressed genes in the short-lived isogenic worms are enriched in the PQM-1/ELT-3 binding site. We speculated that this dual function of PQM-1 in gene expression regulation can be caused by the interaction of PQM-1 with different factors, and only a fraction of PQM-1 in the cell that interacts with specific co-factors suppresses the expression of ELO-6. RNAi pqm-1 alone might not be specific and efficient in changing the expression of elo-6. We examined factors that have been reported physically or functionally interacted with PQM-1 with RNAi knockdown from D3A and found that RNAi of ceh-60 enhances ELO-6 expression level compared to the control on D6A and D7A (Fig. 7e, f). It has been reported that elo-6 expression is significantly increased in ceh-60(ok1485) mutant on D4A (log2FC = 1.20)49. UNC-62 physically interacts with CEH-6049,50 and regulates target gene expression49. We found that RNAi of unc-62 from D3A also enhances ELO-6 expression level compared to the control from D6A to D9A (Fig. 7a, b, e, f) and reduces ELO-6 expression variation between individuals on D5A, D6A, and D7A (Fig. 7e, h, g). We used the loss-of-function mutation strain pqm-1(ok485) to examine the functional interaction between pqm-1 with ceh-60 and unc-62. The GFP::ELO-6 expression shows no difference in pqm-1(ok485) compared to wild-type background on D3A before RNAi treatment (Supplementary Fig. 6j, k). We found that ELO-6 expression level increased compared to the control in pqm-1(ok485) until D9A (Fig. 7a, d), which doesn’t occur under pqm-1 RNAi treatment (Fig. 7a, b). unc-62 functions synergistically with pqm-1 to suppress ELO-6 expression on D7A with unc-62 RNAi in pqm-1(ok485) resulted in a much higher GFP level than pqm-1(ok485) and unc-62 RNAi treatment alone (Fig. 7e, f). Previous studies showed that RNAi ceh-60 or unc-62 extends lifespan49,51. Therefore, PQM-1 might repress ELO-6 expression during aging through cooperation with these life span regulators.
Fluorescent images of GFP::ELO-6(S2A) during aging with unc-62, ceh-60, and pqm-1 RNAi treatment in wild-type N2 background and unc-62 and ceh-60 RNAi treatment in pqm-1(ok485) (a) and quantification (b–d). The numbers of analyzed worms are shown beneath the bars. Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. P-values are presented in the source data (unpaired t-test or unpaired t-test with Welch’s correction, two-sided). L4440 is a control for RNAi knockdown. The quantification and comparison of GFP levels among N2 background worms with N2 L4440 treatment at the same age (b), or among pqm-1(ok485) background worms with pqm-1(ok485) L4440 treatment at the same age (c). The quantification and comparison between pqm-1(ok485) mutant worms under L4440 treatment, unc-62 RNAi, and ceh-60 RNAi with N2 L4440 treatment at the same ages (d). Fluorescent images of GFP::ELO-6(S2A) from D5A to D7A with unc-62 or ceh-60 RNAi treatment in N2 background or pqm-1(ok485) (e) and quantification of this biological replicate (f–h). f Quantification and comparison of GFP levels between N2 L4440 control worms and the rest at the same age (low panel t-test); comparison of GFP levels between pqm-1(ok485) L4440 control worms and pqm-1(ok485) with unc-62 or ceh-60 RNAi knockdowns at the same age (up panel t-test); the interaction between 2 factors (pqm-1(ok485) and ceh-60 RNAi, pqm-1(ok485) and unc-62 RNAi) (2way-ANOVA). The numbers of analyzed worms are shown beneath the bars. Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. P-values are presented in the source data (unpaired t-test or unpaired t-test with Welch’s correction, two-sided; 2way-ANOVA full model). The percentage of worms with a GFP level less than 50 was calculated for each image (g). The coefficient of variation of individual worm GFP levels was calculated for each image (h). The yellow signal in fluorescent images is autofluorescence. ∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗ p < 0.01, ∗p < 0.05, (ns) p > 0.05. Source data are provided as a Source Data file.
Reduction of the expression of genes highly expressed in short-lived individuals enhances ELO-6 expression stability and extends lifespan
We next investigated whether the DEGs between the short-lived and long-lived worms on D5A regulate GFP::ELO-6 expression stability during aging and lifespan. In this study, we focused on the genes with increased expression in the short-lived GFP2 group. We knocked down 61 of the 65 genes by RNAi treatment from D2A and examined the GFP level change with age and lifespan in the N2 background. We found that RNAi of 18 genes delayed the decrease of GFP::ELO-6 expression with age (Fig. 8, Supplementary Fig. 7, and Supplementary Data 9), with 10 out of the 18 genes possessing a PQM-1 binding motif and/or PQM-1 peaks in the gene upstream regions (Supplementary Data 9). Interestingly, among the 18 genes, smf-2 and B0238.13 reduce expression upon pqm-1 RNAi treatment (Supplementary Data 8), although there is no PQM-1 binding motif or PQM-1 binding site identified by ChIP-seq in their gene upstream region. Among the 18 genes, RNAi of ribosomal genes tag-151, E02H1.1, C16A3.6, B0511.6, and T23D8.3 extended lifespan in the N2 background (Fig. 8g, h, Supplementary Data 10). RNAi tag-151 enhanced the GFP signal on D6A (Fig. 8a, d), and RNAi of C16A3.6, E02H1.1, B0511.6, T23D8.3 delayed GFP level reduction until D8A (Fig. 8b, c, e, f). The additional 3 genes, nst-1, F53F4.11, and lpd-7, also extended the lifespan. Interestingly, all eight of these lifespan-regulating genes are related to nucleolus function (Supplementary Data 9), which is consistent with previous findings that long-lived mutants are with reduced ribosomal function and small nucleoli are correlated with extended longevity in multiple organisms4,52. Among these 8 genes, 3 have PQM-1 binding motif and/or PQM-1 peaks in the gene upstream regions (Supplementary Data 9), including T23D8.3, which extends lifespan most profoundly (Supplementary Data 10). T23D8.3 has been identified as a longevity regulator through longevity network prediction53. Therefore, ELO-6 expression and lifespan are regulated by these genes, which are highly expressed in short-lived animals. Understanding the regulatory mechanism that influences the expression of these genes will elucidate the cues that trigger the difference in the aging process between individuals.
Representative fluorescent image of GFP::ELO-6(S2A) on D6A upon RNAi treatment of tag-151 or B0238.13 from D2A (a) and its quantification (d). L4440 treatment is the control for comparison. The numbers of analyzed worms are shown beneath the bars. Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. P-values are presented in the figure (unpaired t-test or unpaired t-test with Welch’s correction, two-sided). b, c, e, f Representative images of GFP::ELO-6(S2A) on D8A with RNAi knockdown the genes highly expressed in short-lived individuals from D2A (b, c) and their quantifications (e, f). L4440 treatment is the control for comparison. The numbers of analyzed worms are shown beneath the bars. Data are represented as mean ± SD of individual worm GFP levels with individual values shown as dots. P-values are presented in the figure (unpaired t-test or unpaired t-test with Welch’s correction, two-sided). g, h Survival curves of RNAi knockdown of the indicated ribosome-related genes that are highly expressed in short-lived individuals. Quantitative data are presented in Supplementary Data 10. P-values are presented in the figures (log-rank test, two-sided). The yellow signal in fluorescent images is autofluorescence. Source data are provided as a Source Data file.
Discussion
In this study, we used longitudinal analysis of transcriptome profiles during aging in germline-less mutant glp-1(e2141) at 25 °C to identify lifespan predictor genes. Generally, the transcriptome changes remarkably with age. In this study, when analyzing the transcriptome at an interval every two days during aging, we found only a few genes that could be identified as significantly changed in expression using the general RNA-seq data analysis pipeline. When combining the data from all aging intervals to identify genes with consistent trends of expression change, no genes fulfilled the criteria. This suggests that gene expression during aging occurs with subtle changes. In this study, we used an arbitrary 20% FPKM difference as the criteria for gene expression changes between adjacent aging time points. We found 123 genes consistently increased and 44 genes consistently decreased expression with age. When inspecting the expression of the lifespan predictor candidate gene elo-6, we found that ELO-6 expression decreased consistently, as expected, indicating the arbitrary criteria is meaningful to measure trends of gene expression change during aging.
Reduced expression of elo-6 from D2A was confirmed by GFP::ELO-6 level and the single-cell aging datasets27,28. We also observed the variation of GFP::ELO-6 expression between individual worms on D5A. This difference between individuals is neither caused by genetic variations nor OP50 infection. We speculate that gene expression variation between individual animals during aging is not unique to elo-6 but should be a common feature for dynamically expressed genes with age. Other genes with expression differences between long-lived and short-lived individuals on D5A and consistently changed expression levels during aging might also be proper reporters (Supplementary Data 6). elo-6 is highly expressed in young adults with an expression level ranked 224 out of all the expressed genes on D2A, and elo-6 expression is dramatically reduced during subsequent aging. Therefore, the abundance of expression change is adequate for detecting differences between individuals.
In the wild-type N2 background and germline-less mutants, the animals with low GFP::ELO-6 are significantly short-lived. elo-6 encodes a fatty acid elongase synthesizing C17iso, and it is partially functionally redundant with elo-526. Unlike elo-5, which is essential for development, elo-6 deficiency does not cause developmental defects26. While ELO-6 expression on D5A is positively correlated with lifespan and health span, we found that RNAi of elo-6 in adult stages does not affect lifespan. A previous study showed that C17iso supplementation from D1A does not affect lifespan54. Therefore, we supplied C17iso from D4A and found that the supplement slightly extended the whole populational lifespan, which might reflect the lifespan extension of the short-lived animals.
We also found that the expression dynamics of elo-6 is a populational lifespan predictor. In this study, reducing the expression of daf-2 and utx-1, which extends lifespan through the insulin/IGF-1 signaling pathway, enhances the expression level of ELO-6 during aging. Accordingly, in the short-lived daf-16(mus86) mutant, the ELO-6 expression level was reduced on D2A and during aging. The ELO-6 expression level during aging is also enhanced in a long-lived germline-less mutant and with metformin treatment. Therefore, ELO-6 expression levels can predict longevity under diverse interventions. In the interventions extending lifespan we tested in this study, set-2 RNAi does not affect ELO-6 expression before D4A, increases ELO-6 expression on D4A and D5A, and decreases ELO-6 expression after D6A, indicating neither the active expression of elo-6 in young animals nor the expression stability during aging after D6A is dependent on set-2. Previous studies have shown that reducing H3K4me3 by set-2 and ash-2 RNAi both extend lifespan34. ash-2 RNAi leads to increased lipid droplet numbers in intestinal cells, and the lipid droplet numbers in the intestine on adult days 1 and 6 predict the remaining lifespan of isogenic worms55. We then inspected elo-6 expression with ash-2 RNAi and found that elo-6 has lower expression levels in the intestine in relatively long-lived ash-2 RNAi worms on adult day 2.5 (log2 FC = −0.14)56.
To investigate the mechanism of regulating individual lifespan, we performed mRNA-seq to compare the transcriptome of short-lived and long-lived isogenic worms distinguished by ELO-6 expression in germline-less mutants and the wild-type N2 background. Surprisingly, in germline-less mutants background, the genome transcriptomes show high similarity between different GFP groups, despite the GFP1 groups with many worms dying the next day. We could still identify genes with significantly changed expression in short-lived animals compared to long-lived ones. Xenobiotic metabolic process and structural constituent of cuticle-related genes are enriched in downregulated genes in short-lived worms, and ribosome biogenesis and CUB domain-related genes were enriched in upregulated genes in short-lived worms. The PQM-1 binding site was enriched in both downregulated and upregulated genes in both germline-less mutant background and the wild-type N2 background. In germline-less mutants background, upon pqm-1 RNAi treatment, the transcriptome of the animals shifted to a relatively younger state. The health span and life span were improved with pqm-1 RNAi in wild-type N2. This is consistent with the previous finding that overexpression of PQM-1 shortens wild-type C. elegans lifespan16. Since PQM-1 mediates the tradeoff between survival and lifespan under stress conditions, we speculate that the short-lived individuals might undergo certain stress conditions, which might be associated with malfunction of xenobiotic metabolic process, cuticle properties, or enhanced expression of ribosomal synthesis genes. From mRNA-seq analysis with GFPL, GFPM, and GFPH with wild-type N2 background, we found the enriched binding motifs in the DEGs also match ELT-2 binding motif, suggesting ELT-2 might also be involved in heterogenous gene expression between individuals. In this study, we found that ribosomal genes are enriched in the highly expressed genes in short-lived animals, and RNAi of some of them extended lifespan. This is consistent with the previous findings that small nucleoli on adult day 1 predicts longer individual lifespan4. C17iso mediates the coordination between growth and amino acids supplement under a dietary restriction condition54. We found that C17iso in short-lived individuals is relatively higher than in long-lived ones. Enhanced expression of ribosomal synthesis genes in short-lived animals might reflect enhanced ribosomal function and, therefore, cause amino acids supplement stress, leading to the downregulation of elo-6 to reduce the synthesis of C17iso. We also inspected whether elo-6 expression is correlated with developmental ROS level, which predicts individual lifespan in isogenic worms at the larval L2 stage, and it shows that elo-6 has a lower expression level in relatively long-lived more oxidized state worms at the L2 stage (log2 FC = –0.19897)5. Further analysis of whether developmental ROS can affect elo-6 expression in adults with proper control will provide information on whether these two phenotypes are related.
Our study identified elo-6 as a single gene reporter for predicting individual and population lifespan. Investigating the mechanism by which PQM-1 regulates the differentially expressed genes between short-lived and long-lived isogenic animals will help us understand the lifespan variation between individuals.
Methods
C. elegans strains and culture
C. elegans strains were cultured under standard growth conditions except otherwise indicated57. All strains were grown and maintained on NGM plates seeded with Escherichia coli OP50 except for the RNAi experiments. The strains that have been used in this study are listed in Supplementary Data 1. For the temperature-sensitive germline-less mutant glp-4(bn2) and glp-1(e2141) background strains, worms were maintained at the permissive temperature of 16°C. To prepare germline-less animals, eggs were laid at 16°C for 4~6 h, and the adults were removed from the plates; eggs were then shifted to 25 °C to hatch, develop, and age. To prepare long-lived germline-less glp-1(e2141) animals, eggs were laid at 16 °C for 4~6 h, and the adults were removed from the plates; eggs were then shifted to 25 °C to hatch and develop till D1A at 25°C, then shifted to 20 °C. Metformin treatment was performed as previously reported41. Metformin was added to agar at the concentration of 50 mM while preparing plates. For OP50 UV treatment, OP50 was seeded onto NGM plates and grew overnight at room temperature. The plates were then treated with 5 J/cm2 UV with the lids opened. Supplementation of C17iso was performed as previously reported58. C17iso (Cayman) was dissolved in 100% DMSO at 10 mM concentration and mixed with OP50 to 1 mM final concentration before spotting plates, with equivalent DMSO concentration as the control treatment. To separate offspring from adult worms by washing from D1A, worms were washed off the plates with 13 ml M9. Adult worms settled to the bottom of a 15 ml tube by gravity within 2–3 min. The 11 ml supernatant was removed, and the worms in 2 ml M9 were resuspended with 11 ml M9 for the 2nd round of wash. A total of 4–5 rounds of wash were used to remove most of the offspring. After the last round, the worms in 2 ml M9 were poured onto an OP50-seeded NGM plate and blow-dried within 10 min. The plate was then checked under a dissection scope to remove the remaining offspring if there were any. All worms were cultured without FUdR.
RNA-seq library preparation and data analysis
Worm samples were collected, washed 3 times with cold M9 to remove bacteria, frozen with liquid nitrogen, and stored at -80 °C. Total RNA was extracted from frozen worm pellet using TRI reagent (Molecular Research Center) and isopropanol precipitation. For RNA extraction from ~ 100 worms from GFP groups, after worms were mixed and incubated with TRI reagent for 10 min, the mixture was homogenized 5 x A and 10 x B with a 2 ml Kontes Dounce. Total RNAs were sent to sequencing facilities with dry ice. All RNA samples were quantified for RNA integrity. mRNA libraries were prepared and sequenced with PE150 at Annoroad for D2A to D12A aging samples or at Novogene for GFP groups and pqm-1 RNAi treatment. mRNA-seq data were analyzed as previously described22. Sequencing reads were trimmed by cutadapt (v2.9). tRNA, and rRNA reads were first filtered out using Bowtie2, and the remaining reads were further aligned to WBcel235 by TopHat2 (v2.1.1) with no novel junctions allowed59. The uniquely aligned reads with a maximum of two mismatches were kept for differential expression analysis. The relative expression levels at different ages were calculated as FPKM using CuffDiff of Cufflinks software (v 2.2.1)59. Differential gene expression was analyzed by edgeR60.
CRISPR/Cas9 alleles generation
CRISPR/Cas9-based gene editing to knock in the gfp::3xflag coding sequence to the 3’-end of elo-1 or the 5′-end of elo-6 was performed as previously described61. Repair templates in pDD282 (Addgene 66823), sgRNA in pDD162 (Addgene 47549), and a pharyngeal fluorescence selection marker pCFJ90 (Addgene 19327) were injected into wild-type N2 young adults. The editing also introduced a S2A substitution into ELO-6. The drug selection cassette was removed by heat shock. Homozygous knock-in alleles were verified by Sanger sequencing. The knock-in strains were backcrossed with wild-type N2 at least 3 times. sgRNA sequences are listed in Supplementary Data 2.
Transgenic line generation
The 1540 bp upstream region of elo-6 was amplified and cloned into C. elegans expression construct pPD95.77. The plasmid containing elo-6p::SL2::GFP::unc-54 3’UTR was injected into wild-type N2 young adults. Progenies were screened by GFP expression in the intestine. The transgenic lines with over 50% of the population with GFP expression in the intestine were used to generate integrated lines by UV. The integrated line was backcrossed with wild-type N2 8 times before being used for analysis. The primers for elo-6 promoter amplification were listed in Supplementary Data 2.
Imaging
Images of worms on a 3% agarose pad in 10 mM levamisole were taken with the Olympus BX53/DP74 by the software cellSens Standard (Olympus). Images of free-moving worms on NGM plates were taken with the Leica M205FCA/K3C by the software LAS X (Leica). Olympus BX53 was equipped with ET485/20x, 69,000 m, and 69,000bs filters (Chroma), and Leica M205FCA was equipped with ET485/10x and 69,000 m filters (Chroma) for GFP imaging and observation to distinguish autofluorescence from GFP62. Images acquired by Olympus BX53/DP74 using 30 ms exposure time with a 10x objective lens and 200 ms exposure time with a 5x objective lens were presented, except otherwise indicated. Imaging parameters for Leica M205FCA were presented in figures or figure legends.
Sorting GFP worms
Worms were synchronized by egg-laying for 4–6 h on 6 cm plates. Worms were transferred from egg-laying plates on D1A to new plates with around 100 worms per 6 cm plate and further cultured at this worm density on 6 cm plates. For N2 background animals, worms were transferred every day from D1A. For the germline-less animals cultured at 25 °C, worms were transferred once on D1A and untouched before sorting. Worms with different GFP levels were picked manually with the Leica M205FCA equipped with ET485/10x and 69,000 m filters before the end of D5A.
GFP quantification
GFP images for quantification were acquired by Olympus BX53/DP74 with ET485/20x, 69,000 m, and 69,000 bs filters and saved as TIF files. The exposure settings were identical across all conditions within all experiments, with multiple exposure times for each image. Blank fields were used as black balances for GFP image acquisition. Raw images were split into color channels (green, red, blue) in ImageJ. “free hand selection” was used to select the worm body part of an individual worm from the front of the intestine to the end of the intestine and saved to the Region of Interest (ROI). The total signal (Area x Mean) per selected region was quantified in the green and red channels. Signals in the green channel were subtracted by the signals in the red channel to eliminate the autofluorescence signal. The normalized signal in the green channel is presented as GFP level.
Lifespan experiments
For N2 background strains, eggs were laid and always cultured at 20 °C on 35 mm plates, transferred every 1 or 2 days from D1A to D9A. For glp-1(e2141) and glp-4(bn2) background strains, eggs from 16 °C cultured worms were laid at 16 °C onto lifespan plates, then hatched and cultured at 25 °C on 6 cm plates. For RNAi treatment, worms were transferred to freshly IPTG-induced iOP5063 or xu36364 RNAi plates at the specific age and further transferred every 1 or 2 days to freshly IPTG-induced RNAi plates until approximately D9A. Lifespan was measured by scoring worms every other day or every day. Worms that died from internal hatching, bursting in the vulval region, or sticking to the plate walls were not included in the lifespan analysis.
RNAi treatment
All RNAi clone was obtained from the Ahringher bacteria library65. Control L4440 and RNAi constructs were transformed into iOP50 or xu363. Transformed iOP50 was plated on LB plates with 50 μg/ml tetracycline, 17 μg/ml chloramphenicol, and 50 μg/ml carbenicillin. Transformed xu363 were plated on LB plates with 50 μg/ml tetracycline and 50 μg/ml carbenicillin. Transformed iOP50 and xu363 clones were further inoculated into LB with 50 μg/ml carbenicillin at 37°C overnight, concentrated 10 fold, seeded on NGM plates, and stored at 4 °C for less than 2 weeks. iOP50 or xu363 on NGM plates were induced with 4 mM IPTG for 4 h at room temperature, and worms were transferred to freshly induced RNAi plates every day or every other day to the 10-fold concentrated iOP50 or xu363 on NGM plates for RNAi treatment except otherwise indicated in the figure legends. For mRNA-seq of pqm-1 RNAi treatment, eggs were laid at 16 °C for 4~6 h onto OP50 plates, and the adults were removed from the plates; eggs were then shifted to 25°C to hatch and developed till D2A at 25°C, then transferred onto the iOP50 plates at 25°C and collected on D5A for RNA extraction.
RT-qPCR
Worm samples were collected, washed 3 times with cold M9 to remove bacteria, frozen with liquid nitrogen, and stored at –80 °C. Total RNA was extracted from frozen worm pellet using TRI reagent (Molecular Research Center) and isopropanol precipitation. After worms were mixed and incubated with TRI reagent for 10 min, the mixture was homogenized 5xA and 10xB with a 2 ml Kontes Dounce. 0.5–1 μg total RNA was reverse transcribed by HiScript III All-in-one RT SuperMix (Vazyme) to generate cDNA. qPCR was performed by using ChamQ Universal SYBR qPCR Master Mix (Vazyme) on a Light Cycler 480 II (Roche). Relative gene expression levels were calculated using the 2-ΔCt method with act-1 for normalization. Primer sequences for qPCR are listed in Supplementary Data 2.
Body bending rate
Worms were individually transferred into a 200 μl S buffer (100 mM NaCl and 50 mM potassium phosphate, pH 6.0) droplet in a petri dish, or 1 ml S buffer in a 24-well Tissue culture-treated plate to avoid worm sticking to the plastic surface, and let rest for 1 min. The number of body-bending within 30 sec in the liquid was counted single-blinded.
Pharyngeal pumping rate
Worms’ pharyngeal pumping rates were scored directly on NGM plates without transferring to new plates. Worms continuously feeding within 10 sec regardless of pumping speed were chosen for the analysis. The number of pharyngeal contractions in one minute was counted using Leica M205FCA.
Motif analysis and GO term analysis
Motif analysis of gene promoter region was performed by Homer (findMotifs.pl genelist.txt worm output directory/) using the default setting (-start -400 -end 100 -len 8,10). The motif is annotated by Homer (findMotifs.pl genelist.txt worm motif directory -find motif file > output).GO term analysis was performed using DAVID.
Statistical analysis
Statistical analyses were performed by SPSS software, Excel, or Prism 10. The survival function was estimated using the Kaplan Meier estimator (SPSS software), and statistical analysis was done using a two-sided log-rank (Mantel-Cox) test or Breslow test. We tested the normality of the data with the Shapiro-Wilk normality test. We used the F-test to examine differences in variance between groups. We compared means between 2 samples with unpaired t-test. We used an unpaired t-test assuming unequal or equal variance when the F-test showed significant differences or no significant differences in variances in Excel or an unpaired t-test with Welch correction when the F-test showed significant differences in GraphPad. We used two-way ANOVA to analyze interactions between two variables. We used Fisher’s exact test to compare two categorical variables. We used the coefficient of variation (Coefficient of Variation = Standard deviation / Mean) to measure dispersion for comparison of variation between samples. P-value < 0.05 was considered significantly different from the control. Detailed descriptions of tests performed to determine the statistical significance are included in figure legends. The significance of statistical differences is indicated as (∗∗∗∗) P-value < 0.0001, (∗∗∗) P-value < 0.001, (∗∗) P-value < 0.01, (∗) P-value < 0.05, (ns) P-value > 0.05. Experiments are performed with at least 2 independent biological replicates.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data are available within this article and its Supplementary Information. The mRNA-sequencing datasets generated in this study have been deposited in the Genome Sequence Archive in the National Genomics Data Center (NGDC) PRJCA021128. Source data are provided with this paper.
References
Finch, C. E. & Tanzi, R. E. Genetics of aging. Science 278, 407–411 (1997).
Rea, S. L., Wu, D., Cypser, J. R., Vaupel, J. W. & Johnson, T. E. A stress-sensitive reporter predicts longevity in isogenic populations of Caenorhabditis elegans. Nat. Genet. 37, 894–898 (2005).
Shen, E. Z. et al. Mitoflash frequency in early adulthood predicts lifespan in Caenorhabditis elegans. Nature 508, 128–132 (2014).
Tiku, V. et al. Small nucleoli are a cellular hallmark of longevity. Nat. Commun. 8, 16083 (2017).
Bazopoulou, D. et al. Developmental ROS individualizes organismal stress resistance and lifespan. Nature 576, 301–305 (2019).
Pincus, Z., Smith-Vikos, T. & Slack, F. J. MicroRNA predictors of longevity in Caenorhabditis elegans. PLoS Genet. 7, e1002306 (2011).
Frenk, S. & Houseley, J. Gene expression hallmarks of cellular ageing. Biogerontology 19, 547–566 (2018).
Tabula Muris, C. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature 583, 590–595 (2020).
Schaum, N. et al. Ageing hallmarks exhibit organ-specific temporal signatures. Nature 583, 596–602 (2020).
Lee, C. K., Klopp, R. G., Weindruch, R. & Prolla, T. A. Gene expression profile of aging and its retardation by caloric restriction. Science 285, 1390–1393 (1999).
Rangaraju, S. et al. Suppression of transcriptional drift extends C. elegans lifespan by postponing the onset of mortality. Elife 4, e08833 (2015).
Peters, M. J. et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun. 6, 8570 (2015).
Fleischer, J. G. et al. Predicting age from the transcriptome of human dermal fibroblasts. Genome Biol. 19, 221 (2018).
Buckley, M. T. et al. Cell-type-specific aging clocks to quantify aging and rejuvenation in neurogenic regions of the brain. Nat. Aging 3, 121–137 (2023).
Niu, W. et al. Diverse transcription factor binding features revealed by genome-wide ChIP-seq in C. elegans. Genome Res. 21, 245–254 (2011).
Tepper, R. G. et al. PQM-1 complements DAF-16 as a key transcriptional regulator of DAF-2-mediated development and longevity. Cell 154, 676–690 (2013).
Templeman, N. M. et al. Insulin signaling regulates oocyte quality maintenance with age via Cathepsin B Activity. Curr. Biol. 28, 753–760 e754 (2018).
Xu, Z. et al. Structural recognition of the mRNA 3’ UTR by PUF-8 restricts the lifespan of C. elegans. Nucleic Acids Res 49, 10082–10097 (2021).
Heimbucher, T., Hog, J., Gupta, P. & Murphy, C. T. PQM-1 controls hypoxic survival via regulation of lipid metabolism. Nat. Commun. 11, 4627 (2020).
Otarigho, B. & Aballay, A. Immunity-longevity tradeoff neurally controlled by GABAergic transcription factor PITX1/UNC-30. Cell Rep. 35, 109187 (2021).
Ni, Z., Ebata, A., Alipanahiramandi, E. & Lee, S. S. Two SET domain containing genes link epigenetic changes and aging in Caenorhabditis elegans. Aging Cell 11, 315–325 (2012).
Pu, M., Wang, M., Wang, W., Velayudhan, S. S. & Lee, S. S. Unique patterns of trimethylation of histone H3 lysine 4 are prone to changes during aging in Caenorhabditis elegans somatic cells. PLoS Genet 14, e1007466 (2018).
Szenker, E., Ray-Gallet, D. & Almouzni, G. The double face of the histone variant H3.3. Cell Res 21, 421–434 (2011).
Piazzesi, A. et al. Replication-independent Histone variant H3.3 controls animal lifespan through the regulation of pro-longevity transcriptional programs. Cell Rep. 17, 987–996 (2016).
Pu, M. et al. Trimethylation of Lys36 on H3 restricts gene expression change during aging and impacts life span. Genes Dev. 29, 718–731 (2015).
Kniazeva, M., Crawford, Q. T., Seiber, M., Wang, C. Y. & Han, M. Monomethyl branched-chain fatty acids play an essential role in Caenorhabditis elegans development. PLoS Biol. 2, E257 (2004).
Roux, A. E. et al. Individual cell types in C. elegans age differently and activate distinct cell-protective responses. Cell Rep. 42, 112902 (2023).
Gao, S. M. et al. Aging atlas reveals cell-type-specific effects of pro-longevity strategies. Nat. Aging, https://doi.org/10.1038/s43587-024-00631-1 (2024).
Narayan, V. et al. Deep Proteome analysis identifies age-related processes in C. elegans. Cell Syst. 3, 144–159 (2016).
Sen, P. et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 29, 1362–1376 (2015).
Zhao, Y. et al. Two forms of death in ageing Caenorhabditis elegans. Nat. Commun. 8, 15458 (2017).
Kenyon, C., Chang, J., Gensch, E., Rudner, A. & Tabtiang, R. A C. elegans mutant that lives twice as long as wild type. Nature 366, 461–464 (1993).
Dillin, A. et al. Rates of behavior and aging specified by mitochondrial function during development. Science 298, 2398–2401 (2002).
Greer, E. L. et al. Members of the H3K4 trimethylation complex regulate lifespan in a germline-dependent manner in C. elegans. Nature 466, 383–387 (2010).
Jin, C. et al. Histone demethylase UTX-1 regulates C. elegans life span by targeting the insulin/IGF-1 signaling pathway. Cell Metab. 14, 161–172 (2011).
Maures, T. J., Greer, E. L., Hauswirth, A. G. & Brunet, A. The H3K27 demethylase UTX-1 regulates C. elegans lifespan in a germline-independent, insulin-dependent manner. Aging Cell 10, 980–990 (2011).
Wu, C. W., Deonarine, A., Przybysz, A., Strange, K. & Choe, K. P. The Skp1 Homologs SKR-1/2 Are Required for the Caenorhabditis elegans SKN-1 Antioxidant/Detoxification Response Independently of p38 MAPK. PLoS Genet 12, e1006361 (2016).
Amrit, F. R. et al. DAF-16 and TCER-1 facilitate adaptation to germline loss by restoring lipid homeostasis and repressing reproductive physiology in C. elegans. PLoS Genet 12, e1005788 (2016).
Hsin, H. & Kenyon, C. Signals from the reproductive system regulate the lifespan of C. elegans. Nature 399, 362–366 (1999).
Arantes-Oliveira, N., Apfeld, J., Dillin, A. & Kenyon, C. Regulation of life-span by germ-line stem cells in Caenorhabditis elegans. Science 295, 502–505 (2002).
Onken, B. & Driscoll, M. Metformin induces a dietary restriction-like state and the oxidative stress response to extend C. elegans Healthspan via AMPK, LKB1, and SKN-1. PLoS One 5, e8758 (2010).
Viswanathan, M., Kim, S. K., Berdichevsky, A. & Guarente, L. A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span. Dev. Cell 9, 605–615 (2005).
Sinha, A. & Rae, R. A functional genomic screen for evolutionarily conserved genes required for lifespan and immunity in germline-deficient C. elegans. PLoS One 9, e101970 (2014).
Shioda, T. et al. Neuronal MML-1/MXL-2 regulates systemic aging via glutamate transporter and cell nonautonomous autophagic and peroxidase activity. Proc. Natl Acad. Sci. USA 120, e2221553120 (2023).
Gao, A. W. et al. Identification of key pathways and metabolic fingerprints of longevity in C. elegans. Exp. Gerontol. 113, 128–140 (2018).
Mack, H. I. D. et al. Further extension of lifespan by Unc-43/CaMKII and Egl-8/PLCbeta mutations in germline-deficient caenorhabditis elegans. Cells 11, https://doi.org/10.3390/cells11223527 (2022).
Pryor, R. et al. Host-microbe-drug-nutrient screen identifies bacterial effectors of Metformin therapy. Cell 178, 1299–1312 e1229 (2019).
Sanchez-Blanco, A. & Kim, S. K. Variable pathogenicity determines individual lifespan in Caenorhabditis elegans. PLoS Genet 7, e1002047 (2011).
Dowen, R. H. CEH-60/PBX and UNC-62/MEIS coordinate a metabolic switch that supports reproduction in C. elegans. Dev. Cell 49, 235–250 e237 (2019).
Van de Walle, P. et al. CEH-60/PBX regulates vitellogenesis and cuticle permeability through intestinal interaction with UNC-62/MEIS in Caenorhabditis elegans. PLoS Biol. 17, e3000499 (2019).
Curran, S. P. & Ruvkun, G. Lifespan regulation by evolutionarily conserved genes essential for viability. PLoS Genet. 3, e56 (2007).
Hansen, M. et al. Lifespan extension by conditions that inhibit translation in Caenorhabditis elegans. Aging Cell 6, 95–110 (2007).
Tacutu, R. et al. Prediction of C. elegans longevity genes by human and worm longevity networks. PLoS One 7, e48282 (2012).
Zhu, M. et al. Monomethyl branched-chain fatty acid mediates amino acid sensing upstream of mTORC1. Dev. Cell 56, 3171 (2021).
Papsdorf, K. et al. Lipid droplets and peroxisomes are co-regulated to drive lifespan extension in response to mono-unsaturated fatty acids. Nat. Cell Biol. 25, 672–684 (2023).
Han, S. et al. Mono-unsaturated fatty acids link H3K4me3 modifiers to C. elegans lifespan. Nature 544, 185–190 (2017).
Brenner, S. The genetics of Caenorhabditis elegans. Genetics 77, 71–94 (1974).
Kniazeva, M., Euler, T. & Han, M. A branched-chain fatty acid is involved in post-embryonic growth control in parallel to the insulin receptor pathway and its biosynthesis is feedback-regulated in C. elegans. Genes Dev. 22, 2102–2110 (2008).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012).
Chen, Y., Lun, A. T. & Smyth, G. K. From reads to genes to pathways: differential expression analysis of RNA-Seq experiments using Rsubread and the edgeR quasi-likelihood pipeline. F1000Res 5, 1438 (2016).
Dickinson, D. J., Pani, A. M., Heppert, J. K., Higgins, C. D. & Goldstein, B. Streamlined genome engineering with a self-excising drug selection cassette. Genetics 200, 1035–1049 (2015).
Teuscher, A. C. & Ewald, C. Y. Overcoming autofluorescence to assess GFP expression during normal physiology and aging in Caenorhabditis elegans. Bio. Protoc. 8, https://doi.org/10.21769/BioProtoc.2940 (2018).
Neve, I. A. A. et al. Escherichia coli Metabolite Profiling Leads to the Development of an RNA Interference Strain for Caenorhabditis elegans. G3 (Bethesda 10, 189–198 (2020).
Xiao, R. et al. RNAi interrogation of dietary modulation of development, metabolism, behavior, and aging in C. elegans. Cell Rep. 11, 1123–1133 (2015).
Kamath, R. S. & Ahringer, J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods 30, 313–321 (2003).
Acknowledgements
We thank the Caenorhabditis elegans Genetics Center for worm strains, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). We thank Dr. Di Chen for CRISPR/cas-9 reagents and technical support. We thank Dr. Bin Liang for supporting C17iso quantification and lipid droplet analysis. We thank Dr. Leonard Krall for critical reading and comments. This work was supported by funding from the National Science Foundation of China (32160163) and Yunnan Fundamental Research Projects (202201BF070001-009) to M.P.
Author information
Authors and Affiliations
Contributions
M.P. conceived the study. M.P., W.K., G.G., T.D., B.C., Y.W., and Z.Z carried out the experiments. M.P. designed the experiments and interpreted the results. M.P. wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kong, W., Gu, G., Dai, T. et al. ELO-6 expression predicts longevity in isogenic populations of Caenorhabditis elegans. Nat Commun 15, 9470 (2024). https://doi.org/10.1038/s41467-024-53887-x
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-53887-x