Abstract
Plasmodium ovale curtisi (Poc) and Plasmodium ovale wallikeri (Pow) are relapsing malaria parasites endemic to Africa and Asia that were previously thought to represent a single species. Amid increasing detection of ovale malaria in sub-Saharan Africa, we present a population genomic study of both species across the continent. We conducted whole-genome sequencing of 25 isolates from Central and East Africa and analyzed them alongside 20 previously published African genomes. Isolates are predominantly monoclonal (43/45), with their genetic similarity aligning with geography. Pow shows lower average nucleotide diversity (1.8×10−4) across the genome compared to Poc (3.0×10−4) (p < 0.0001). Signatures of selective sweeps involving the dihydrofolate reductase gene have been found in both species, as are signs of balancing selection at the merozoite surface protein 1 gene. Differences in the nucleotide diversity of Poc and Pow may reflect unique demographic history, even as similar selective forces facilitate their resilience to malaria control interventions.
Similar content being viewed by others
Introduction
Parasites in the genus Plasmodium were responsible for an estimated 249 million cases of malaria and 608,000 deaths in 20221. Ninety-four percent of these cases occurred in the World Health Organization Africa Region, where control efforts have primarily focused on the predominant species, P. falciparum (Pf)2. Yet these case counts likely underrepresent the burden of non-falciparum species, which may be rising in prevalence even where control efforts have successfully reduced P. falciparum transmission3,4,5. Over the last few decades, genomic studies of P. falciparum have enabled monitoring of drug resistance markers6, facilitated the identification of promising vaccine candidates7, uncovered the structure of parasite populations8, and identified evolutionary forces shaping their demography9,10. Much less is known about non-falciparum species, especially their comparative evolutionary history and susceptibility to malaria control interventions focused on P. falciparum.
Plasmodium ovale was first identified as a separate malaria species in 1922 based on the appearance of oval-shaped erythrocytes that contained non-ring parasite forms11. Hallmarks of this parasite species are its restriction to younger red cells and, therefore, the propensity to cause low-density infections, as well as relapses from liver hypnozoites, similar to P. vivax and P. cynomologi. The species often causes coinfection alongside P. falciparum which, along with its low parasite densities, makes it challenging to differentiate morphologically on peripheral blood smears12. The advent of polymerase chain reaction (PCR)-based diagnostics has improved the detection of ovale infections, but initial PCR surveys across Africa and Asia based on the small subunit rRNA gene revealed two apparent groups of P. ovale parasites, termed classic and variant13. The discovery of perfect sequence segregation of six genomic markers, and more recently, 12 mitochondrial loci, between classic and variant P. ovale isolates collected across Africa and Asia has led to the conclusion that this dimorphism actually represents a true species divide in the P. ovale clade14,15. The nomenclature of these species is currently evolving but will be referred to as P. ovale curtisi (Poc, formerly classic) and P. ovale wallikeri (Pow, formerly variant) herein16,17,18.
P. ovale curtisi and P. ovale wallikeri have since been confirmed to circulate within the same human populations throughout Africa and Asia19,20, with both detected by PCR at higher rates than previously appreciated5,12,21,22. Limited investigation of the genetic diversity and population genetics of the two P. ovale species have hinted at low diversity and/or small effective population size, as few unique haplotypes have been identified at antigenic gene targets like apical membrane antigen 1 (ama1) and merozoite surface protein 1 (msp1)20,23. There is some indication that drugs used to treat P. falciparum are also shaping P. ovale parasite populations; signs of a selective sweep involving a mutant dhfr allele (implicated in pyrimethamine resistance) have been detected in both Poc and Pow24,25. Until now, the low density of most P. ovale isolates combined with the lack of an in vitro culture system has hindered whole-genome sequencing of these parasites11. However, with the development of strategies for parasite DNA enrichment, as well as the construction of the first reference genomes in 2017, genome-wide analyses are now possible15,26,27,28.
In this work, we employ hybrid capture or leukodepletion to enrich P. ovale spp. DNA and perform whole-genome sequencing (WGS) of 25 clinical isolates collected from studies conducted across Ethiopia, the Democratic Republic of the Congo, Tanzania, and Cameroon. Combined with 20 additional public whole genomes from 11 countries spanning East, Central, and West Africa, we seek to better understand the comparative biology of P. ovale curtisi, P. ovale wallikeri, and co-endemic P. falciparum by examining their complexity of infection, population structure, nucleotide diversity, and genomic signatures of selection.
Results
High-quality genomic coverage of African P. ovale isolates
Parasite samples from 25 P. ovale-infected individuals collected at ten sites spanning Ethiopia, the Democratic Republic of the Congo (DRC), Tanzania, and Cameroon were selected for whole-genome sequencing (Table 1)29,30,31,32,33,34. These included 13 P. ovale curtisi and 12 P. ovale wallikeri isolates that were selected from six studies based on robust amplification of the po18S rRNA gene (Ct < 36) and predominance of one ovale species within each isolate. The majority of the samples (n = 21) underwent a custom-designed hybrid capture with RNA baits to preferentially isolate ovale DNA extracted from dried blood spots for sequencing, while four additional whole blood samples were leukodepleted (LDB) at the time of collection by CF11 filtration and directly sequenced without enrichment35. Finally, genomic data of 20 P. ovale isolates sequenced as part of four previously published studies were retrieved from the European Nucleotide Archive and the Sequence Read Archive15,28,36,37. These isolates either underwent selective whole-genome amplification (sWGA) or leukodepletion for parasite DNA enrichment prior to sequencing. Further data on all parasite isolates are found in Supplemental Table 1.
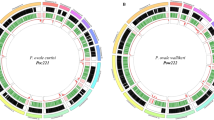
Whole-genome sequencing achieved high genome coverage, with an overall average of 86 and 87% tenfold coverage across the core genome for the 21 P. ovale curtisi and 24 P. ovale wallikeri isolates, respectively (Fig. 1). Coverage and mapping proportion were highest when aligned to the P. ovale reference genome determined by the Poc/Pow species-specific qPCR assay, corroborating initial species assignment. Compared to sWGA and LDB samples, the hybrid capture method used to enrich parasite DNA in the majority of samples yielded more complete coverage across all chromosomes except for chromosome 10 (Fig. 1). The hybrid capture was originally designed for P. ovale wallikeri, with additional P. ovale curtisi baits then selected to cover areas that differ between the two ovale genome assemblies (PowCR01 and PocGH01)28. Due to Pow chromosome 10 being incomplete in the PowCR01 reference genome (only 470 kb), this approach did not provide coverage for the full Poc chromosome 10 (1300 kb). This led to substantially lower coverage for chromosome 10 across all Poc hybrid capture isolates (40–60% 10x coverage vs. >85% for all other Poc chromosomes); thus, chromosome 10 was excluded from all genome-wide analysis in Poc isolates to limit error.
The proportion of the corresponding P. ovale reference genome covered by ≥10 reads by DNA enrichment technique (A) and species (B). The proportion of reads mapped to that reference genome by DNA enrichment technique (C) and species (D). For Poc, chromosome 10 was excluded due to incomplete coverage by hybrid capture baits. Five samples from Higgins et al. were not incorporated into this study due to having <30% 10x coverage of the corresponding genome. HC hybrid capture, LDB leukodepleted blood, sWGA selective whole-genome amplification. Source data are provided as a Source Data file.
As expected, hybrid capture led to preferential sequencing of P. ovale DNA among samples that were co-infected with P. falciparum (Pf); Pf-positive isolates that underwent hybrid capture yielded only 2–11% 10x coverage of the Pf genome compared to >90% 10x coverage of the Pf genome among leukodepleted blood samples. For genomic analysis, insertions/deletions, multiallelic sites, low-quality variants, and variants within tandem repeats and expanded gene families were excluded (see Methods), yielding final biallelic single nucleotide polymorphism (SNP) call sets of 73,015 SNPs for P. ovale curtisi and 45,669 for P. ovale wallikeri.
Low complexity of infection
Complexity of infection (COI), or the number of unique parasite clones present in a given isolate, was estimated 1000 times using THEREALMcCOIL for all 21 P. ovale curtisi and 24 ovale wallikeri isolates, as well as 2077 geographically matched P. falciparum isolates downloaded from the publicly available MalariaGEN Pf6 dataset (Fig. 2)38. Twenty out of 21 Poc isolates (95%) and 23 out of 24 Pow isolates (96%) were estimated to be monoclonal; the remaining isolate in each P. ovale species was found to comprise two parasite clones. By comparison, roughly half (1165/2077; 56%) of P. falciparum samples were monoclonal. COI differed significantly (p = 0.005) among the three Plasmodium species. In pairwise comparisons, both Poc and Pow had significantly lower COI compared to P. falciparum (p = 0.001 and p = 0.004, respectively).
Median estimated complexity of infection (COI) among 21 Poc isolates, 24 Pow isolates, and 2077 P. falciparum isolates geographically matched to the P. ovale samples by country of origin. The distributions of COI differed significantly among the three species (p < 0.0001) by a Kruskal–Wallis test, with Pow and Poc showing significantly lower COI than P. falciparum (p = 0.0004 and 0.012, respectively) in Dunn’s multiple comparisons tests. The average read depth of coverage for Poc, Pow, and Pf isolates were 44.1, 95.7, and 147.5, respectively. Source data are provided as a Source Data file.
The two multiclonal P. ovale infections were both hybrid capture-enriched, high coverage (94 and 98% tenfold coverage in Poc and Pow, respectively) and came from high-transmission areas of the DRC39; each had a COI of 2. In order to determine whether the two clones in these samples were distinct lineages or meiotic siblings, we analyzed the distribution of heterozygous SNPs across the genome. We hypothesized that meiotic siblings would only have heterozygous SNPs in specific regions, reflecting recombination within the mosquito midgut40. In both samples, after filtering to high-confidence SNPs based on population-wide allele frequency, we saw an even distribution of heterozygous SNPs across the genome, suggestive of two distinct parasite lineages in the same host rather than meiotic siblings (Supplemental Fig. 1).
Lower nucleotide diversity in P. ovale wallikeri compared to P. ovale curtisi
Among a collection of 3339 sets of one-to-one orthologous genes between the Poc, Pow, and P. falciparum genomes, we identified 2008 sets that achieved high-quality sequencing coverage and had no overlap with masked genomic regions in any of the three species. The average species-specific nucleotide diversity (π) among these orthologues in the 20 monoclonal Poc, 23 monoclonal Pow, and 19 geographically matched monoclonal Pf samples were 2.9 × 10−4, 1.8 × 10−4, and 2.6 × 10−4, respectively. These were significantly different between species (p < 0.0001, F = 98, df = 2), with orthologues in P. ovale curtisi more diverse than in P. ovale wallikeri and P. falciparum (p values <0.0001 and 0.002, respectively), and Pow orthologues less diverse than in Pf (p < 0.0001) (Fig. 3A). To mitigate bias by geographic coverage and orthology with the P. falciparum genome, we repeated this analysis using 2,911 Poc-Pow orthologues among a group of geographically matched monoclonal Poc and Pow samples (n = 11 each, Supplemental Table 3), revealing average nucleotide diversities of 2.5 × 10−4 and 1.8 × 10−4, respectively (Fig. 3B). Nucleotide diversity was still significantly lower in Pow orthologues compared to Poc (p < 0.0001).
A Nucleotide diversity (π) per gene among 2008 sets of orthologous genes in monoclonal Poc, Pow, and Pf samples. Boxes denote the 25th, median, and 75th percentiles; whiskers are drawn at the 1st and 99th percentiles. π of 0 was coded as 1 × 10−5 to plot on a logarithmic scale. Nucleotide diversity was significantly different between orthologues of all three species by two-sided Tukey’s multiple comparisons tests, with Poc orthologues showing higher diversity than orthologues of Pow and P. falciparum, and Pow orthologues also showing lower diversity than those in P. falciparum (p values <0.001, =0.002, and <0.0001, respectively). B Nucleotide diversity (π) per gene among 2911 sets of orthologous genes in geographically matched monoclonal Poc and Pow samples. Boxes denote the 25th, median, and 75th percentiles; whiskers are drawn at the 1st and 99th percentiles. π of 0 was coded as 1 × 10−5 to plot on a logarithmic scale. Nucleotide diversity was significantly lower among Pow orthologues compared to Poc using a two-sided Wilcoxon’s matched-pair signed rank test (p < 0.0001). C SNP density in different functional regions of the genome among all Pow, Poc, and P. falciparum isolates. SNP single nucleotide polymorphism, kb kilobase. Source data are provided as a Source Data file.
This high nucleotide diversity in P. ovale curtisi was consistent with an investigation of the total number and density of genome-wide SNPs. Variant calling and filtering resulted in almost twice the number of SNPs for P. ovale curtisi (73,015) as for P. ovale wallikeri (45,669), despite a slightly smaller number of Poc isolates (21 vs. 24). This corresponded to a higher density of SNPs across the Poc genome (4.0 SNPs per kilobase[kb] in Poc vs. 2.4 SNPs/kb in Pow). However, a smaller proportion of SNPs in the Poc genome were nonsynonymous mutations, as the ratio of nonsynonymous-to-synonymous (dN/dS) mutations was 1.5 and 2.5 in Poc and Pow, respectively. Among the aforementioned geographically matched P. ovale isolates, dN/dS within 2911 orthologous genes was 1.4 for Poc and 2.5 for Pow, consistent with the broader estimates. SNP densities in both the Poc and Pow genomes were lowest in protein-coding sequences (3.9 and 1.9 SNPs/kb, respectively), and higher in introns (4.4 and 2.8 SNPs/kb) (Fig. 3C). Intergenic regions in Poc showed relatively lower SNP density similar to protein-coding sequences (4.0 SNPs/kb), but these same regions in Pow had relatively high SNP density similar to introns (2.8 SNPs/kb).
Parasite genomic similarity recapitulates geographic relationships
Genome-wide principal component (PC) analysis of the monoclonal samples of each ovale species revealed the spatial arrangement of related parasites along PC1 and PC2 that aligns with their location of origin (Fig. 4). These components accounted for 15.0 and 17.5% of the genetic differentiation in Poc and Pow, respectively. While cluster analysis by ADMIXTURE found the best fit when modeling each isolate as a separate cluster, except for one pair of isolates per species originating from the capital of Kinshasa in the DRC (in Poc) and the Amhara region in Ethiopia (in Pow), geographic alignment was evident in the PCA. For Pow, PC1 and PC2 appear to reflect an East-West axis and North-South axis, respectively, with samples from Ethiopia and South Sudan in the west divided from others by PC1. In the PCA for Poc, Ethiopian parasites were also organized separately from other samples, as did isolates from Kinshasa in the DRC. The remaining Poc samples show some division between East, Central, and West Africa, though the alignment with geography is less consistent than in Pow. In both P. ovale spp., PC3 and PC4 further separated samples from various countries (Supplemental Fig. 2).
Principal component analysis showing the first two principal components among 20 monoclonal Poc isolates (A) and 23 monoclonal Pow isolates (B) using 4116 and 3189 biallelic SNPs, respectively. Samples are colored by region of country of origin; in the map, parasites from travelers are assigned to the capital city (C). In PC2 of Pow, SNPs within both the ts-dhfr and mrp1 gene were among the top 0.5% of contributors. Source data are provided as a Source Data file.
Examination of the top 0.5% of variants by contribution to each of the first 4 principal components revealed that SNPs within genes encoding multidrug resistance protein 1 (mdr1) and dihydrofolate reductase-thymidylate synthase (dhfr-ts), two putative antimalarial resistance genes, were major contributors to the North-South axis in Pow PC2. Among all 24 Pow samples, three previously documented haplotypes in Pow dhfr-ts, a key gene in folate metabolism that is implicated in pyrimethamine resistance41,42, appear to drive this geographic differentiation, with the Phe57Leu + Ser58Arg haplotype existing in 45% of our Central African clones and 36% of our East African clones but none of the sequenced West African clones (Supplemental Fig. 3A)25. This haplotype is associated with resistance to pyrimethamine when expressed in E. coli. Though it did not drive differentiation in the PCA, Poc dhfr-ts haplotypes similarly showed the presence of a putative drug resistance haplotype (Ala15Ser + Ser58Arg) in the Central and East African clones but not in West Africa, though our sample size for West Africa was generally smaller for both species (three and six isolates in Pow and Poc, respectively) (Supplemental Fig. 3B).
Signatures of selection contain putative drug resistance loci, proteins involved in sexual stage differentiation, and antigenic targets
We calculated nSL and Tajima’s D across the genomes to identify loci under directional and balancing selection, respectively. The nSL statistic is considered robust to the currently unknown recombination rates across the genomes of P. ovale species43. Genetic markers of interest within 10 kb of the top 0.5% absolute normalized nSL values that may be influenced by selective sweeps are listed in Tables 2, 3. Evidence of a selective sweep involving the putative bifunctional dihydrofolate reductase–thymidylate synthase (dhfr-ts) gene41,42 was found in both P. ovale species (Fig. 5). Examination of extended haplotype homozygosity (EHH) at the selected variants show a large selective sweep in Pow spanning roughly 40 kb as well as close proximity of the dhfr-ts gene to the focal variant (Fig. 6A, B). In Poc, the positioning of the dhfr-ts gene lies at the edge of a smaller sweep (Fig. 6C, D). However, another putative marker of drug resistance, multidrug resistance-associated protein 2 (mrp2), was found in close proximity to one of the highest absolute normalized nSL values in Poc and lies near the center of a 40 kb sweep region on Poc chromosome 14 (Fig. 6E, F).
Absolute nSL of 19,205 variants among monoclonal isolates across the Poc genome (A) and 15,744 variants among monoclonal isolates across the Pow genome (B). Individual loci are depicted using alternating shapes between chromosomes for legibility. The dotted line and red color denote the top 0.5% of loci. Putative genetic markers of note within 10,000 bases of these loci are labeled, including ubiquitin transferase (ut), cysteine-rich secretory protein (crisp), cysteine repeat modular protein 2 (crmp2), early transcribed membrane protein (etramp), GPI-anchored micronemal antigen (gama), dihydrofolate reductase-thymidylate synthase (dhfr-ts), apical membrane antigen (ama1), merozoite surface protein 7-like protein (msp7), male development protein 1 (md1), AP2 domain transcription factor G (ap2-g), multidrug resistance-associated protein 2 (mrp2), dynein heavy chain (dhc), AP2 domain transcription factors (ap2), merozoite surface protein 5 (msp5), cysteine repeat modular protein 1 (crmp1), dynein light chain (dlc), 6-cysteine protein (6-cys), sporozoite protein essential for cell traversal 1 (spect1), Plasmodium interspersed repeat protein (pir), and early transcribed membrane protein (etramp). Source data are provided as a Source Data file.
Extended haplotype homozygosity (EHH) and haplotype bifurcation among monoclonal isolates at selected variants near the Pow dhfr-ts gene (A, B), the Poc dhfr-ts gene (C, D), and the Poc mrp2 gene (E, F). EHH and haplotype bifurcation show selective sweeps spanning ~30, ~40, and ~30 kb, respectively, with lineage breakdown occurring first among the unselected allele haplotypes (blue) and then in the selected allele haplotypes (red) as the distance from the focal variant increases.
Top absolute nSL hits were also found near ap2 transcription factor genes that regulate apicomplexan life cycle transitions, including sexual differentiation into gametocytes (ap2-g), and genes involved in sex-specific development of gametes, such as those coding male development protein 1 (md1) and cysteine-rich secretory protein (crisp)44,45,46. In Pow, four top nSL hits were found around genes encoding the dynein heavy and light chains, cytoskeleton components highly expressed in male gametes for motility and fusion with female gametes in the mosquito blood meal47. Top nSL hits were also found near cysteine repeat modular proteins 2 and 1 (crmp2/1) in Poc and Pow, respectively, proteins which may be involved in targeting sporozoites to the salivary glands in the mosquito prior to transmission48,49.
Finally, genes encoding putative antigenic targets at the host-parasite interface, including merozoite surface protein 7 (msp7), merozoite surface protein 5 (msp5), early transcribed membrane protein (etramp), apical membrane antigen 1 (ama1), GPI-anchored micronemal antigen (gama), and 6-cysteine protein B9 (6-cys) may be under directional selection in both P. ovale species50,51. An orthologue of sporozoite protein essential for cell traversal 1 (spect-1), a protein necessary for liver cell invasion that has been investigated as a potential vaccine target52, was also among the top hits in Pow.
Overall, Tajima’s D in both species exhibited a negative skew across the genome with an average value of −1.06 for Poc and −0.78 for Pow. This may suggest population expansion following a bottleneck or weak directional selection (Fig. 7). Among the loci with positive values in the top 0.5% of absolute Tajima’s D hits, the antigenic marker merozoite surface protein 1 (MSP1) was identified as a probable target of balancing or diversifying selection in both P. ovale species (Tables 2, 3).
Tajima’s D in 399,355 and 297,286 300 bp windows in genes across the Poc (A) and Pow (B) genomes among monoclonal isolates. Individual windows are depicted using alternating shapes between chromosomes for legibility. The dotted line and red color denote the top 0.5% of loci. For both species, the top locus with a positive Tajima’s D was located inside the merozoite surface protein 1 (msp1) gene. Source data are provided as a Source Data file.
Discussion
We present a comprehensive population genomic study of both P. ovale species within sub-Saharan Africa. Our study comprises 21 Poc and 24 Pow isolates selected from 11 studies, including both febrile and asymptomatic cases. Genome-wide analysis reveals differences in nucleotide diversity between P. ovale species, but similarity in their low complexity of infection, geographic relatedness, and signatures of selection. Our analysis was performed using genomic enrichment methods specifically designed to enable robust coverage and analysis with the 2017 reference genomes of P. ovale curtisi (from Ghana) and P. ovale wallikeri (from Cameroon), which were the available references at the time of the study28.
Compared to the “classic” P. ovale curtisi species, we observed significantly lower nucleotide diversity across orthologous genes among geographically matched P. ovale wallikeri isolates (2.5 × 10−4 for Poc and 1.8 × 10−4 for Pow, respectively). Our estimate for Poc is concordant with the genome-wide diversity calculated among six Central African Poc isolates15 as well as that derived from RNA expression data among four parasite samples from Mali53. However, our Pow estimate was substantially lower than the genome-wide estimate reported by Higgins et al. (3.4 × 10−4), despite our inclusion of their samples alongside additional Central and East African isolates. Our lower estimate may reflect the exclusion of higher-diversity intergenic regions, though we also found lower genome-wide and intergenic SNP density in Pow compared to Poc. Relatively low nucleotide diversity in Pow may indicate reduced effective population size, increased inbreeding, or a population bottleneck in the time since Poc and Pow diverged between 1.3 and 20.3 million years ago14,28. More recent population growth in both species is also suggested by the predominantly negative distribution of Tajima’s D values across their protein-coding genes, a finding that can indicate population expansion following a bottleneck54. The high ratio of nonsynonymous-to-synonymous substitutions among these protein-coding genes (2.5 for Pow, 1.5 for Poc) is similar to that seen in P. falciparum and P. vivax55,56. This finding may represent diversifying selection on proteins across either P. ovale genome, enabling maintenance of nonsynonymous substitutions, or inflation of dN/dS ratios observed among Plasmodium parasites due to the impact of the malaria life cycle on allele frequencies57. Further analysis of subpopulations of each parasite species could help to elucidate the factors driving the observed difference in genomic diversity, such as by determining whether Pow isolates from Asia have similarly low nucleotide diversity or if this finding is specific to Africa.
The observed predominance of monoclonal isolates among both P. ovale species is consistent with low within-sample haplotypic diversity seen in previous investigations of African P. ovale isolates by genome-wide RNA sequencing and amplicon sequencing53,58. The low complexity of P. ovale infections may result from efficient clonal transmission32 and/or lower transmission overall, limiting vector uptake of multiple parasite clones from either the same or different infected individuals. This low complexity is expected to limit opportunities for genetic recombination within mosquito vectors, though multiple-clone infections were identified in Kinshasa, a region with overall higher malaria endemicity and transmission intensity39.
Genomic signatures of selection within both P. ovale species highlighted the importance of antimalarials, host-vector life cycle transitions, and human immunity as evolutionary pressures impacting parasite survival. P. ovale infections are frequently subclinical and go untreated59, but likely still face substantial drug exposure from widely used antimalarials prescribed for P. falciparum22. Additionally, malaria prophylaxis using sulfadoxine-pyrimethamine, such as intermittent preventive therapy in pregnancy (IPTp) and seasonal malaria chemoprevention for infants and schoolchildren (SMC), may be applying drug pressure on P. ovale parasite populations1. Selective sweeps in dhfr-ts, a gene implicated in resistance to pyrimethamine, have been documented in both Poc and Pow, and certain mutant alleles were found to confer pyrimethamine resistance when expressed in E. coli24,25. In our dataset, sweeps near the dhfr-ts genes were among the strongest signals of directional selection in both P. ovale species, especially in Pow, possibly representing drug pressure influencing parasite survival. We also found the putative pyrimethamine resistance Pow dhfr-ts haplotype Phe57Leu + Ser58Arg to be a major contributor to principal component 2 of Pow (representing the North-South axis); the resistant haplotype composed 36% (9/25) of our Pow haplotypes, with representation in Central and East Africa but no detection in West Africa. Another putative pyrimethamine resistance haplotype (Ala15Ser + Ser58Arg) in Poc was similarly detected in East and Central Africa, though not in West Africa25. Functional evaluation of different alleles to determine their capacity to confer drug resistance, as well as monitoring of these alleles across the parasite populations over time, will further clarify how interventions targeting P. falciparum may be simultaneously rendering P. ovale parasites harder to control.
Finally, strong signals of balancing or diversifying selection were observed in both species within their genes encoding merozoite surface protein 1 (MSP1), the orthologues of the predominant antigen on blood-stage P. falciparum parasites that has been shown to induce protective immunity in some studies60. Diversifying selection on MSP1 has been documented in P. falciparum61 as well as in focused analysis among African P. ovale infections imported to China62. Our dataset provides even stronger evidence for diversifying selection at this site, as the msp1 gene showed the single highest Tajima’s D value across all genes in both P. ovale species. Such immune responses may also play a role in modulating relapse potential63.
This study has several limitations. While it represents the largest genome-wide examination of the genomic composition of both P. ovale curtisi and P. ovale wallikeri to date, the sample size for both species is nonetheless small and limits the power to detect clustering of isolates, infer population demography, and detect selection. The geographic coverage of the isolates employed differs between the 13 countries represented, and isolates from Northern or Southern Africa were not available. Whole-genome enrichment methods also differed among isolates; 21 isolates employed hybrid capture, 15 used selective whole-genome amplification, and nine relied on leukodepletion. The two former methods may have induced amplification bias, whereas leukodepletion does not amplify P. ovale DNA and, therefore, may reduce the power to identify rare variants in those isolates. Disparate average read depth between the three species (44.1, 95.7, and 147.5 for Poc, Pow, and Pf, respectively) may also have differentially impacted our ability to detect polyclonal infections, but the low complexity of infection found in both P. ovale species should be robust given the satisfactory sequencing depth overall. The source studies also differ in whether samples were derived from asymptomatic carriers (n = 11) or febrile patients (n = 34). Sample sizes were too small to analyze these populations separately. Finally, hybrid capture baits designed using the incomplete PowCR01 reference genome led to incomplete coverage of Poc chromosome 10, which was excluded from analyses. Unfortunately, newly assembled regions of Pow chromosome 10 were not available during the analysis. The hybrid capture approach also did not enable enrichment and analysis of loci in the mitochondrial and apicoplast genomes, which were excluded from analysis. We do not expect these exclusions to systematically bias the estimation of nucleotide diversity or complexity of infection, though it does prevent us from evaluating excluded loci (including mdr1, msp3, and msp8) for genomic signatures of selection. The availability of selective whole genome amplification protocols now provides a less expensive approach to targeted DNA enrichment for P. ovale spp. that does not rely on the specific design of hybrid capture baits27.
This study provides a comparative genomic analysis of the two Plasmodium ovale species sympatrically circulating in sub-Saharan Africa and presents new evidence of selective pressures on genes related to drug response, sexual differentiation, and immune evasion. Further population genomic studies of Poc and Pow should employ a larger selection of isolates from a greater geographic range, especially including Asia, and take advantage of new reference genome assemblies to build on these insights15. Functional investigation into the genes showing signatures of selection, including via orthologue replacement in closely related Plasmodium species64,65, is also an exciting new strategy in substantiating the biological relevance of key loci, with implications for transmission prevention, treatment strategies, and vaccine development for P. ovale spp. Finally, cataloging genome-wide diversity facilitates the design of targeted genotyping methods that can efficiently characterize the epidemiology of these understudied parasite species66. Combining these approaches to better evaluate P. ovale parasite relatedness, transmission, and relapse patterns can help to improve the impact of current malaria control strategies on all human-infecting malaria species36,67,68.
Methods
Ethics
This analysis was considered non-human subjects research by the University of North Carolina. The appropriate collection of samples, authorization for use in genomic studies, and IRBs involved are summarized in the original studies outlined in Supplemental Table 1.
Sample selection
Clinical isolates in the form of dried blood spots or leukodepleted blood were drawn from six studies shown in Table 1, including studies involving both asymptomatic persons and febrile patients across four countries. Across these studies, participants were screened for the presence of P. ovale spp. infection by a real-time polymerase chain reaction (qPCR) assay targeting the po18S rRNA gene22. Among 282 isolates with a po18S Ct value under 40, a species-specific (Poc and Pow) 18S rRNA qPCR assay was employed to determine the ovale species present69. Candidates were selected from isolates with only one species detected or mixed infections in which one species predominated by ≥3 Ct (corresponding to approximately 8 times as much DNA). Samples were also screened for the presence of P. falciparum coinfection using a qPCR assay for the pfldh or pf18S rRNA gene31,39. Ultimately, samples from 25 individuals were selected for whole-genome sequencing based on higher-density P. ovale infection, lack of or lower-density P. falciparum coinfection, and balance of ovale species and geographic diversity across the sample set. Characteristics of these 25 samples, and an additional 20 samples from four previously published studies15,28,36,37, are shown in Supplemental Table 1.
Library preparation and sequencing
DNA extracted from dried blood spots using a Chelex protocol70 was sheared to 300 bp using a LE220R-plus Covaris Sonicator (Covaris, Woburn, MA). Fragment size was checked with an Agilent TapeStation 4150 (Agilent, Santa Clara, CA), and DNA concentrations were tested using a Qubit Flex fluorometer (Thermo Fisher Scientific, Waltham, MA). Isolates were then prepared for sequencing using the KAPA Hyperprep kit (Kapa Biosystems, Woburn, MA). Four Tanzanian DNA isolates extracted from blood that had been leukocyte-depleted by CF11 filtration at the time of collection35 were directly incorporated into sequencing libraries. The remaining 21 isolates derived from dried blood spots were additionally processed using a custom-designed hybrid capture protocol to enrich for ovale DNA via thousands of RNA probes specifically designed to amplify P. ovale DNA without binding to human DNA (Twist Bioscience, San Francisco, CA, USA). Hybrid-capture probes were designed first for the P. ovale wallikeri genome (PowCR01), with unique probes added for the P. ovale curtisi genome (PocGH01) at any sites that differed by more than 10% of bases28. Since the reference genome for chromosome 10 is significantly smaller for Poc compared to Pow (roughly 1300 vs 470 kb, respectively), this bait design approach led to a lack of baits covering 63% of Poc chromosome 10. Chromosome 10 was therefore excluded from the analysis of Poc isolates. Captures were performed with four samples per capture. After preliminary sequencing on a Miseq Nano flow cell (Illumina, San Diego, CA, USA), libraries were sequenced on the Novaseq 6000 S Prime (Illumina, San Diego, CA, USA) sequencing system with 150 bp paired-end chemistry. Samples from Joste et al. and Higgins et al. included in our analysis were enriched using selective whole genome amplification (sWGA), employing sets of five to ten primers designed to preferentially amplify the PowCR01 and PocGH01 genomes over human background DNA71.
Sequencing data alignment and variant calling
Data processing and analysis were performed in the bash environment using a Python-based snakemake v7.24.2 wrapper for pipeline construction, automation, and reproducibility72. Raw sequencing reads were trimmed of Illumina adapters using Trimmomatic v0.3673 before the quality of the reads was evaluated with fastQC v0.11.974. A “dual” reference genome was then produced for each P. ovale species by concatenating the reference genome of that ovale species (P. ovale curtisi: PocGH01; P. ovale wallikeri: PowCR01) to the P. falciparum strain Pf3D7 reference genome28,75. Paired reads from each isolate were aligned to their species’ corresponding dual reference genome using bwa-mem2 v2.2.176 before each alignment was sorted and given read group information using picard v2.26.1177 and deduplicated via GATK v4.4.0.078. Samtools v1.17 was then used to select for reads that aligned to the P. ovale portion of the dual reference genome rather than that of P. falciparum, thus discarding reads from contaminating P. falciparum DNA present in some isolates79. Resulting alignments of read that preferentially mapped to P. ovale were soft-clipped to reference genome edges and cleaned of unmapped reads using GATK, after which mapping proportion and coverage of the ovale reference genome were calculated by samtools and bedtools v2.3080.
Variant calling from aligned reads across each ovale species genome was also performed using the GATK best practices pipeline78. In the resulting callset, variants were masked via vcftools81 if they fell outside of the 14 chromosomes of the reference genome or were within the following expanded gene families (which were masked to reduce sequence error in repetitive genomic elements): Plasmodium interspersed repeat protein (pir), surfin-related subtelomeric protein 1 (stp1), early transcribed membrane protein (etramp), tryptophan-rich antigen (tra), Plasmodium exported protein (phist), reticulocyte binding protein (rbp), ookinete surface protein (osp), 6-cysteine protein, KELT protein, and pm-fam-a protein37. Tandem repeats across each species’ genome were identified and masked using Tandem Repeat Finder v4.09.182. GATK hard filtering was then used to remove variants with poor quality metrics using the following filter thresholds: quality by depth <3, Fisher strand bias >50, strand odds ratio >3, mapping quality >50, mapping quality rank sum <−2.5, read position rank sum <−3. Call sets were limited to biallelic single nucleotide polymorphisms (SNPs) that were present in at least 80% of individuals. SNP density across the entire genome and within specific functional regions of each genome were calculated using custom scripts (see Data availability). SNPeff v4.3 was used to annotate individual variants and determine the ratio of nonsynonymous-to-synonymous mutations28,83.
Selection of P. falciparum comparison dataset
Co-endemic P. falciparum samples were drawn from the Pf6 dataset38. Of 20,705 total P. falciparum isolates from around the globe, 2077 came from the same or nearby geographic locations as the source studies of P. ovale isolates described above. Thirty-two P. falciparum samples with over 85% base callability were randomly selected in order to have at least one co-endemic P. falciparum isolate for each P. ovale curtisi or ovale wallikeri isolate (Supplemental Table 2). Variant call sets for these P. falciparum samples were limited to the falciparum core genome75, quality filtered by Variant Quality Score Recalibration78, and restricted to sites present in at least 80% of individuals.
Complexity of infection (COI) calculation
Variant call sets were limited to variants with a minor allele frequency greater than or equal to 5%. The McCOILR R package was employed to run THEREALMcCOIL on each sample set using 1000 Markov chain Monte Carlo iterations, 100 burn-in iterations, a maximum COI of 25, a minimum number of sites for a sample to be included of 10, and a minimum number of samples for a site to be included of 10 (analysis was run on 21 and 24 P. ovale curtisi and ovale wallikeri isolates, respectively)84. This yielded 1000 estimates of the complexity of infection, or the number of unique parasite clones found in each sample, and median COI per isolate was reported. COI distributions among P. falciparum isolates were calculated in the same manner. COI distributions among the three species were compared using a Kruskal–Wallis test, with Dunn’s multiple comparisons test employed for specific pairwise comparisons85.
To investigate the distribution of heterozygous SNPs in samples that were determined to be polyclonal, the variant set was filtered to sites with >10% minor allele frequency across the whole population and a within-sample minor allele frequency >50% for at least one sample. Then, for each polyclonal sample, the remaining sites were filtered to those with a within-sample minor allele frequency greater than or equal to 5%.
Principal components analysis
After limiting to monoclonal samples (20 Poc, 23 Pow), variant call sets were filtered to remove sites with a minor allele frequency of less than 5% using vcftools v0.1.1581. Variants were processed using PLINK v1.90b6.21, first by pruning variants within windows of 50 variants between which the R2 value exceeded 0.3 (windows were shifted by steps of five variants for each pruning step)86. Then, principal components analysis was performed in PLINK, including extraction of the weights by variant. Data visualizations were performed in R using the ggplot2 v3.4.4 package87. ADMIXTURE v1.3.0 was employed to test for significant clustering among samples within each species88.
Nucleotide diversity calculation
For both ovale species and P. falciparum, the OrthoMCL database was searched for orthologous gene sets in which there was only one copy of each ortholog per genome89. Sets were removed from the total pool of orthologous genes if they were partially or totally masked by the multigene family or extrachromosomal masks within an individual species, or if any orthologs did not have ≥5x coverage of the gene at least every 10 bases in at least 60% of isolates in each species. Among the remaining ortholog sets with sufficient coverage among all species, nucleotide diversity (π) was calculated across the monoclonal samples of each species for each ortholog in the set using vcftools v0.1.1581. The P. falciparum isolate set was composed of 19 monoclonal isolates selected to ensure one geographically matched Pf sample for each P. ovale sample (Supplemental Table 2). Variant call sets in this analysis did not employ a minor allele frequency filter. Nucleotide diversity was compared between species among the remaining sets of orthologs using repeated measures ANOVA; pairwise comparisons were examined with two-tailed Tukey’s multiple comparisons test. To mitigate potential bias introduced by differing geographic coverage between the two ovale species (and by differences with the P. falciparum genome), nucleotide diversity was also calculated among a set of orthologue sets (identified among one-to-one Poc-Pow orthologues with strong coverage as detailed above) in geographically matched Poc and Pow samples (n = 11 each, Supplemental Table 3) and compared using Wilcoxon’s matched-pair signed rank test90.
Identification of signatures of selection
Variant call sets were limited to monoclonal isolates and sites which had no missingness in any samples. After employing the default minor allele frequency filter of >5%, selscan v1.2.0 was used to calculate nSL (a metric of directional selection) for all remaining variants in each species and the norm function used to normalize values in allele frequency bins91. nSL was chosen as it is more robust to the currently unknown recombination rates across either P. ovale genome than metrics like iHS and because it can evaluate selection within a single population of organisms. Loci with an nSL in the most extreme 0.5% were investigated in PlasmoDB for proximity to genes of interest within 10 kb89. Extended haplotype homozygosity (EHH) was calculated at select sites using rehh92. Tajima’s D (a metric of possible directional and balancing selection) was calculated in 300 base pair sliding windows shifted by 10 bp using all non-missing variants across all known genes in each genome using vcf-kit v0.2.993,94. Variants with positive Tajima’s D values in the top 0.5% of absolute values were investigated in PlasmoDB.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All new sequence data are available at NCBI SRA: BioProject ID PRJNA1092086. Public data used include the contents of the following projects/studies: European Nucleotide Archive Study accession number PRJEB51041, Run accession numbers: ERR10738334, ERR10738339, ERR10738341, and ERR10738346. SRA Study accession number PRJEB13344 https://www.ncbi.nlm.nih.gov/bioproject/PRJEB13344], Run accession numbers: ERR1739852, and ERR1739853. SRA Study accession number PRJEB12679, Run accession numbers: ERR1428159, ERR1254542, and ERR1254543. SRA Study accession number PRJNA1015456, Run accession numbers: SRR26037552, SRR26037551, SRR26037550, SRR26037549, SRR26037548, SRR26037546, SRR26037545, SRR26037544, SRR26037543, SRR26037542, and SRR26037541. Additional Run accession numbers: ERR404145. ERR404154. ERR377533. ERR404191. ERR404207. ERR1045266. ERR1045267. ERR676479. ERR1106575. ERR1106579. ERR1106586. ERR1106587. ERR1106590. ERR449901. ERR449903. ERR405238. ERR405244. ERR666939. ERR562889. ERR636018 [https://www.ncbi.nlm.nih.gov/sra/?term=ERR562889]. ERR912913 [https://www.ncbi.nlm.nih.gov/sra/?term=ERR636018]. ERR1514567. ERR1045287. ERR1172616. ERR1172593. ERR1172615. ERR1172608. ERR059405. ERR045598. ERR666937. ERR580480. ERR701763 Source data are provided with this paper.
Code availability
All code for processing and analysis of samples is available at: Carey-Ewend, K., Population genomics of Plasmodium ovale species in sub-Saharan Africa, https://github.com/bailey-lab/Po_popgen_snakemake/tree/main/final, https://doi.org/10.5281/zenodo.1402678695.
References
World Health Organization. World Malaria Report 2023 (World Health Organization, 2023).
Lover, A. A., Baird, J. K., Gosling, R. & Price, R. N. Malaria elimination: time to target all species. Am. J. Trop. Med. Hyg. 99, 17–23 (2018).
Betson, M., Clifford, S. & Stanton, M. Emergence of nonfalciparum Plasmodium infection despite regular artemisinin combination therapy in an 18-month longitudinal study of Ugandan children and their mothers. J. Infect. Dis. 217, 1099–1109 (2018).
Yman, V. et al. Persistent transmission of Plasmodium malariae and Plasmodium ovale species in an area of declining Plasmodium falciparum transmission in eastern Tanzania. PLoS Negl. Trop. Dis. 13, e0007414 (2019).
Akala, H. M. et al. Plasmodium interspecies interactions during a period of increasing prevalence of Plasmodium ovale in symptomatic individuals seeking treatment: an observational study. Lancet Microbe 2, e141–e150 (2021).
Amato, R. et al. Genetic markers associated with dihydroartemisinin–piperaquine failure in Plasmodium falciparum malaria in Cambodia: a genotype–phenotype association study. Lancet Infect. Dis. 17, 164–173 (2017).
Bailey, J. A. et al. Microarray analyses reveal strain-specific antibody responses to Plasmodium falciparum apical membrane antigen 1 variants following natural infection and vaccination. Sci. Rep. 10, 3952 (2020).
Amambua-Ngwa, A. et al. Major subpopulations of Plasmodium falciparum in sub-Saharan Africa. Science 365, 813–816 (2019).
Volkman, S. K. et al. A genome-wide map of diversity in Plasmodium falciparum. Nat. Genet. 39, 113–119 (2007).
Ghansah, A. et al. Monitoring parasite diversity for malaria elimination in sub-Saharan Africa. Science 345, 1297–1298 (2014).
Collins, W. E. & Jeffery, G. M. Plasmodium ovale: parasite and disease. Clin. Microbiol. Rev. 18, 570–581 (2005).
Mueller, I., Zimmerman, P. A. & Reeder, J. C. Plasmodium malariae and Plasmodium ovale-the ‘bashful’malaria parasites. Trends Parasitol. 23, 278–283 (2007).
Calderaro, A. et al. Genetic polymorphisms influence Plasmodium ovale PCR detection accuracy. J. Clin. Microbiol. 45, 1624–1627 (2007).
Sutherland, C. J. et al. Two nonrecombining sympatric forms of the human malaria parasite Plasmodium ovale occur globally. J. Infect. Dis. 201, 1544–1550 (2010).
Higgins, M. et al. New reference genomes to distinguish the sympatric malaria parasites, Plasmodium ovale curtisi and Plasmodium ovale wallikeri. Sci. Rep. 14, 3843 (2024).
Šlapeta, J., Sutherland, C. J. & Fuehrer, H.-P. Calling them names: variants of Plasmodium ovale. Trends Parasitol. https://doi.org/10.1016/j.pt.2023.12.010 (2023)
Snounou, G., Sharp, P. M. & Culleton, R. Appropriate naming of the two Plasmodium ovale species. Trends Parasitol. 40, 207–208 (2024).
Snounou, G., Sharp, P. M. & Culleton, R. The two parasite species formerly known as Plasmodium ovale. Trends Parasitol. 40, 21–27 (2024).
Oguike, M. C. et al. Plasmodium ovale curtisi and Plasmodium ovale wallikeri circulate simultaneously in African communities. Int. J. Parasitol. 41, 677–683 (2011).
Fuehrer, H.-P. et al. Plasmodium ovale in Bangladesh: genetic diversity and the first known evidence of the sympatric distribution of Plasmodium ovale curtisi and Plasmodium ovale wallikeri in southern Asia. Int. J. Parasitol. 42, 693–699 (2012).
Fuehrer, H.-P., Campino, S. & Sutherland, C. J. The primate malaria parasites Plasmodium malariae, Plasmodium brasilianum and Plasmodium ovale spp.: genomic insights into distribution, dispersal and host transitions. Malaria J. 21 https://doi.org/10.1186/s12936-022-04151-4 (2022).
Sendor, R. et al. Similar prevalence of Plasmodium falciparum and non–P. falciparum malaria infections among schoolchildren, Tanzania. Emerg. Infect. Dis. J. 29, 1143 (2023).
Lei, Y. et al. Low genetic diversity and strong immunogenicity within the apical membrane antigen-1 of Plasmodium ovale spp. imported from africa to china. Acta Trop. 210, 105591 (2020).
Chen, J. et al. Disparate selection of mutations in the dihydrofolate reductase gene (dhfr) of Plasmodium ovale curtisi and P. o. wallikeri in Africa. PLoS Negl. Trop. Dis. 16, e0010977 (2022).
Joste, V. et al. Plasmodium ovale spp dhfr mutations associated with reduced susceptibility to pyrimethamine in sub-Saharan Africa: a retrospective genetic epidemiology and functional study. Lancet Microbe 5, 669–678 (2024).
Bright, A. T. et al. Whole genome sequencing analysis of Plasmodium vivax using whole genome capture. BMC Genomics 13, 262 (2012).
Joste, V., Guillochon, E., Clain, J., Coppée, R. & Houzé, S. Development and optimization of a selective whole-genome amplification to study Plasmodium ovale Spp. Microbiol. Spectr. 10, e0072622 (2022).
Rutledge, G. G. et al. Plasmodium malariae and P. ovale genomes provide insights into malaria parasite evolution. Nature 542, 101–104 (2017).
Feleke, S. M. et al. Plasmodium falciparum is evolving to escape malaria rapid diagnostic tests in Ethiopia. Nat. Microbiol. 6, 1289–1299 (2021).
Parr, J. B. et al. Analysis of false-negative rapid diagnostic tests for symptomatic malaria in the Democratic Republic of the Congo. Sci. Rep. 11, 6495 (2021).
Mwandagalirwa, M. K. et al. Individual and household characteristics of persons with Plasmodium falciparum malaria in sites with varying endemicities in Kinshasa Province, Democratic Republic of the Congo. Malar. J. 16, 456 (2017).
Tarimo, B. B. et al. Seasonality and transmissibility of Plasmodium ovale in Bagamoyo District, Tanzania. Parasit. Vectors 15, 56 (2022).
Rogier, E. et al. Plasmodium falciparum pfhrp2 and pfhrp3 gene deletions among patients enrolled at 100 health facilities throughout Tanzania: February to July 2021. Sci Rep 14, 8158 (2024).
Ali, I. M. et al. Arboviruses as an unappreciated cause of non-malarial acute febrile illness in the Dschang Health District of western Cameroon. PLoS Negl. Trop. Dis. 16, e0010790 (2022).
Venkatesan, M. et al. Using CF11 cellulose columns to inexpensively and effectively remove human DNA from Plasmodium falciparum-infected whole blood samples. Malar. J. 11, 41 (2012).
Joste, V. et al. Genetic profiling of Plasmodium ovale wallikeri relapses with microsatellite markers and whole-genome sequencing. J. Infect. Dis. 228, 1089–1098 (2023).
Ansari, H. R. et al. Genome-scale comparison of expanded gene families in Plasmodium ovale wallikeri and Plasmodium ovale curtisi with Plasmodium malariae and with other Plasmodium species. Int. J. Parasitol. 46, 685–696 (2016).
MalariaGEN et al. An open dataset of Plasmodium falciparum genome variation in 7,000 worldwide samples. Wellcome Open Res. 6, 42 (2021).
Sendor, R. et al. Epidemiology of Plasmodium malariae and Plasmodium ovale spp. in Kinshasa Province, Democratic Republic of Congo. Nat. Commun. 14, 6618 (2023).
Bright, A. T. et al. A high resolution case study of a patient with recurrent Plasmodium vivax infections shows that relapses were caused by meiotic siblings. PLoS Negl. Trop. Dis. 8, e2882 (2014).
Sirawaraporn, W., Sathitkul, T., Sirawaraporn, R., Yuthavong, Y. & Santi, D. V. Antifolate-resistant mutants of Plasmodium falciparum dihydrofolate reductase. Proc. Natl Acad. Sci. USA 94, 1124–1129 (1997).
Hastings, M. D. et al. Dihydrofolate reductase mutations in Plasmodium vivax from Indonesia and therapeutic response to sulfadoxine plus pyrimethamine. J. Infect. Dis. 189, 744–750 (2004).
Ferrer-Admetlla, A., Liang, M., Korneliussen, T. & Nielsen, R. On detecting incomplete soft or hard selective sweeps using haplotype structure. Mol. Biol. Evol. 31, 1275–1291 (2014).
Gomes, A. R. et al. A transcriptional switch controls sex determination in Plasmodium falciparum. Nature 612, 528–533 (2022).
Russell, A. J. C. et al. Regulators of male and female sexual development are critical for the transmission of a malaria parasite. Cell Host Microbe 31, 305–319.e10 (2023).
Tadesse, F. G. et al. Gametocyte sex ratio: the key to understanding Plasmodium falciparum transmission? Trends Parasitol. 35, 226–238 (2019).
Khan, S. M. et al. Proteome analysis of separated male and female gametocytes reveals novel sex-specific Plasmodium biology. Cell 121, 675–687 (2005).
Douradinha, B. et al. Plasmodium cysteine repeat modular proteins 3 and 4 are essential for malaria parasite transmission from the mosquito to the host. Malar. J. 10, 71 (2011).
Thompson, J. et al. Plasmodium cysteine repeat modular proteins 1-4: complex proteins with roles throughout the malaria parasite life cycle. Cell. Microbiol. 9, 1466–1480 (2007).
Lyons, F. M. T., Gabriela, M., Tham, W.-H. & Dietrich, M. H. Plasmodium 6-cysteine proteins: functional diversity, transmission-blocking antibodies and structural scaffolds. Front. Cell. Infect. Microbiol. 12, 945924 (2022).
Arumugam, T. U. et al. Discovery of GAMA, a Plasmodium falciparum merozoite micronemal protein, as a novel blood-stage vaccine candidate antigen. Infect. Immun. 79, 4523–4532 (2011).
Patarroyo, M. E., Alba, M. P. & Curtidor, H. Biological and structural characteristics of the binding peptides from the sporozoite proteins essential for cell traversal (SPECT)-1 and -2. Peptides 32, 154–160 (2011).
Tebben, K. et al. Malian children infected with Plasmodium ovale and Plasmodium falciparum display very similar gene expression profiles. PLoS Negl. Trop. Dis. 17, e0010802 (2023).
Tajima, F. Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123, 585–595 (1989).
Singh, G. P. & Sharma, A. South-East Asian strains of Plasmodium falciparum display higher ratio of non-synonymous to synonymous polymorphisms compared to African strains. F1000Res. 5, 1964 (2016).
Feng, X. et al. Single-nucleotide polymorphisms and genome diversity in Plasmodium vivax. Proc. Natl Acad. Sci. USA 100, 8502–8507 (2003).
Chang, H.-H. et al. Malaria life cycle intensifies both natural selection and random genetic drift. Proc. Natl Acad. Sci. USA 110, 20129–20134 (2013).
Saralamba, N. et al. Genetic dissociation of three antigenic genes in Plasmodium ovale curtisi and Plasmodium ovale wallikeri. PLoS ONE 14, e0217795 (2019).
Faye, F. B. K. et al. Diagnostic criteria and risk factors for Plasmodium ovale malaria. J. Infect. Dis. 186, 690–695 (2002).
Thomson-Luque, R., Stabler, T. C., Fürle, K., Silva, J. C. & Daubenberger, C. Plasmodium falciparum merozoite surface protein 1 as asexual blood stage malaria vaccine candidate. Expert Rev. Vaccines 23, 160–173 (2024).
Escalante, A. A., Lal, A. A. & Ayala, F. J. Genetic polymorphism and natural selection in the malaria parasite Plasmodium falciparum. Genetics 149, 189–202 (1998).
Chu, R. et al. Limited genetic diversity of N-terminal of merozoite surface protein-1 (MSP-1) in Plasmodium ovale curtisi and P. ovale wallikeri imported from Africa to China. Parasit. Vectors 11, 596 (2018).
Joyner, C. J. et al. Humoral immunity prevents clinical malaria during Plasmodium relapses without eliminating gametocytes. PLoS Pathog. 15, e1007974 (2019).
Thiam, L. G., Mangou, K., Ba, A., Mbengue, A. & Bei, A. K. Leveraging genome editing to functionally evaluate Plasmodium diversity. Trends Parasitol. 38, 558–571 (2022).
Mohring, F. et al. Cation ATPase (ATP4) orthologue replacement in the malaria parasite Plasmodium knowlesi reveals species-specific responses to ATP4-targeting drugs. MBio 13, e0117822 (2022).
Siegel, S. V. et al. Lineage-informative microhaplotypes for recurrence classification and spatio-temporal surveillance of Plasmodium vivax malaria parasites. Nat. Commun. 15, 6757 (2024).
Liu, P. et al. Increasing proportions of relapsing parasite species among imported malaria in China’s Guangxi Province from Western and Central Africa. Travel Med. Infect. Dis. 43, 102130 (2021).
Wångdahl, A. et al. Relapse of Plasmodium vivax and Plasmodium ovale malaria with and without primaquine treatment in a nonendemic area. Clin. Infect. Dis. 74, 1199–1207 (2022).
Potlapalli, V. R. et al. Real-time PCR detection of mixed Plasmodium ovale curtisi and wallikeri infections in human and mosquito hosts. PLoS Negl. Trop. Dis. 17, e0011274 (2023).
Mitchell, C. L. et al. Under the radar: epidemiology of Plasmodium ovale in the Democratic Republic of the Congo. J. Infect. Dis. 223, 1005–1014 (2021).
Clarke, E. L. et al. swga: a primer design toolkit for selective whole genome amplification. Bioinformatics 33, 2071–2077 (2017).
Mölder, F. et al. Sustainable data analysis with Snakemake. F1000Res. 10, 33 (2021).
Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120 (2014).
Wingett, S. W. & Andrews, S. FastQ Screen: a tool for multi-genome mapping and quality control. F1000Res. 7, 1338 (2018).
Miles, A. et al. Indels, structural variation, and recombination drive genomic diversity in Plasmodium falciparum. Genome Res. 26, 1288–1299 (2016).
Vasimuddin, M., Misra, S., Li, H. & Aluru, S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In 2019 IEEE International Parallel and Distributed Processing Symposium (IPDPS) 314–324 (IEEE, 2019).
Institute, B. ‘Picard Toolkit’, Broad institute, GitHub repository (2021).
Van der Auwera, G. A. & O’Connor, B. D. Genomics in the Cloud: Using Docker, GATK, and WDL in Terra (O’Reilly Media, Inc., 2020).
Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079 (2009).
Quinlan, A. R. & Hall, I. M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics 26, 841–842 (2010).
Danecek, P. et al. The variant call format and VCFtools. Bioinformatics 27, 2156–2158 (2011).
Benson, G. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res. 27, 573–580 (1999).
Cingolani, P. et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly 6, 80–92 (2012).
Chang, H.-H. et al. THE REAL McCOIL: a method for the concurrent estimation of the complexity of infection and SNP allele frequency for malaria parasites. PLoS Comput. Biol. 13, e1005348 (2017).
Dunn, O. J. Multiple comparisons among means. J. Am. Stat. Assoc. 56, 52–64 (1961).
Purcell, S. et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 81, 559–575 (2007).
Wickham, H. ggplot2: Elegant Graphics for Data Analysis (Springer Science & Business Media, 2009).
Alexander, D. H., Novembre, J. & Lange, K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 19, 1655–1664 (2009).
Amos, B. et al. VEuPathDB: the eukaryotic pathogen, vector and host bioinformatics resource center. Nucleic Acids Res. 50, D898–D911 (2022).
Conover, W. J. Practical Nonparametric Statistics 3rd edn (John Wiley & Sons, 1999).
Szpiech, Z. A. & Hernandez, R. D. selscan: an efficient multithreaded program to perform EHH-based scans for positive selection. Mol. Biol. Evol. 31, 2824–2827 (2014).
Gautier, M. & Vitalis, R. rehh: an R package to detect footprints of selection in genome-wide SNP data from haplotype structure. Bioinformatics 28, 1176–1177 (2012).
Parobek, C. M. et al. Selective sweep suggests transcriptional regulation may underlie Plasmodium vivax resilience to malaria control measures in Cambodia. Proc. Natl Acad. Sci. USA 113, E8096–E8105 (2016).
Cook, D. E. & Andersen, E. C. VCF-kit: assorted utilities for the variant call format. Bioinformatics 33, 1581–1582 (2017).
Carey-Ewend, K. Population genomics of Plasmodium ovale species in sub-Saharan Africa. Zenodo https://doi.org/10.5281/zenodo.14026786 (2024).
Acknowledgements
We thank all the participants of the parent studies for providing the biological material for the study. The following reagents were obtained through BEI Resources, NIAID, NIH: (1) P. falciparum, strain 3D7A, MRA-151, contributed by David Walliker and (2) Diagnostic plasmid containing the small subunit ribosomal RNA gene (18S) from Plasmodium ovale, MRA-180, contributed by Peter A. Zimmerman. We also thank Dr. Parul Johri for her feedback on the final manuscript and Dr. Kevin Wamae for his support in bioinformatic pipeline construction. This study was funded by the National Institute for Allergy and Infectious Diseases, National Institutes of Health (R01AI137395 and R21AI152260 to J.T.L., R21AI148579 to J.B.P. and J.T.L., R01AI65537 to J.J.J., K24AI134990 to J.J.J., R01AI129812 to A.T., R01AI132547 to J.J.J., R01TW010870 to J.J.J., T32AI070114 supporting K.C.-E.), the Fogarty Center, the Global Fund to Fight AIDS, Tuberculosis, and Malaria, and the Bill and Melinda Gates Foundation (Inv. No. 002202 to D.I., J.J.J., and J.A.B.). Under the grant conditions of the Bill and Melinda Gates Foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the Author Accepted Manuscript version that might arise from this submission.
Author information
Authors and Affiliations
Contributions
K.C.-E., J.T.L., J.J.J., and J.B.P. conceived and designed the study. S.F., F.P., K.M., O.A., D.I., I.A., B.B., B.N., A.K., and A.T. contributed to clinical sample collection. K.C.E., Z.P.H., M.M., C.H., and C.G. conducted laboratory work. K.C.-E., Z.P.H., K.N., W.H., F.A., and A.S. conducted data analysis. K.C.-E. drafted the first version of the manuscript. K.C.-E., J.T.L., J.J.J., Z.P.H., J.B., C.S., and J.B.P. edited and finalized the manuscript.
Corresponding author
Ethics declarations
Competing interests
J.B.P. reports research support from Gilead Sciences, non-financial support from Abbott Laboratories, and consulting for Zymeron Corporation, all outside the scope of this study. The remaining authors declare no competing interests. The findings and conclusions in this report are those of the author(s) and do not necessarily represent the official position of the Bill and Melinda Gates Foundation or other funders.
Peer review
Peer review information
Nature Communications thanks Sarah Auburn, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Carey-Ewend, K., Popkin-Hall, Z.R., Simkin, A. et al. Population genomics of Plasmodium ovale species in sub-Saharan Africa. Nat Commun 15, 10297 (2024). https://doi.org/10.1038/s41467-024-54667-3
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54667-3
This article is cited by
-
Immunogenicity and gene polymorphism analysis of Plasmodium ovale wallikeri glycosylphosphatidylinositol-anchored micronemal antigen (GAMA)
Malaria Journal (2025)
-
Metagenomic complexity of high, seasonal transmission of Plasmodium spp. in asymptomatic carriers in Northern Sahelian Ghana
Communications Medicine (2025)