Abstract
Glandular epithelia, including mammary gland (MG) and prostate, are composed of luminal and basal cells. During embryonic development, glandular epithelia arise from multipotent stem cells (SCs) that are replaced after birth by unipotent basal and unipotent luminal SCs. Different conditions, such as basal cell transplantation, luminal cell ablation, and oncogene expression can reinduce adult basal SC (BaSCs) multipotency in different glandular epithelia. The mechanisms regulating the reactivation of multipotency are incompletely understood. Here, we have found that Collagen I expression is commonly upregulated in BaSCs across the different multipotent conditions. Increasing collagen concentration or stiffness of the extracellular matrix (ECM) promotes BaSC multipotency in MG and prostate organoids. Single cell RNA-seq of MG organoids in stiff conditions have uncovered the importance of β1 integrin/FAK/AP-1 axis in the regulation of BaSC multipotency. Altogether our study uncovers the key role of Collagen signaling and ECM stiffness in the regulation of multipotency in glandular epithelia.
Similar content being viewed by others
Introduction
Glandular epithelia, including the mammary and the prostate gland, are composed of basal cells (BCs) and luminal cells (LCs)1,2. These two cell lineages originate from multipotent progenitors that became lineage restricted unipotent basal and luminal stem cells (SCs) in the adult mouse3,4,5,6,7. The mammary gland (MG) epithelium originates from embryonic multipotent progenitors (EMPs), that become lineage-restricted at the late stage of embryonic development around E17.5 in mice4,6,8. while the prostate BaSCs become lineage-restricted just before pubertal development3,9. In both epithelia, BaSCs can activate multipotency upon different conditions10,11,12,13.
Transplantation of BaSCs together with the appropriate microenvironmental cues (mammary mesenchyme or urogenital mesenchyme) can give rise to both BCs and LCs and reform a new mammary gland or prostate gland4,12,13,14,15,16,17. When MG BaSC are transplanted together with LCs, they maintain their unipotent fate. However, when BaSC is transplanted alone without LCs, they reactivate their multipotent differentiation potential4. Oncogenic hits (PI3KCA oncogene expression in the mammary gland and PTEN deletion) in the BaSC can induce multipotency in adult mice7,10,16,18. Finally, disrupting the basal to luminal cell communications by performing LC ablation in the adult mice can reactivate BaSC multipotency11. The mechanisms regulating the activation of BaSC multipotency in these different glandular epithelia remain poorly characterized. Also, it remains unclear whether a common mechanism controlling BaSC multipotency exists across different glandular epithelia and different experimental conditions.
Collagens are structural proteins characterized by the ability to organize into fibrillar structures. In addition to their key role in regulating mechanical proprieties of a tissue, collagens bind to different receptors and activate downstream signaling pathways. Collagen 1 (Col1) is the most abundant collagen among the different collagen members19,20,21. The mammary fat pad contains high level of Col1, which has been shown to play a key role in orienting the mammary epithelium during pubertal development pubertal22,23. In the prostate stroma, Col1 is also very abundant and it has been reported to associate with prostatic intraepithelial neoplasia and prostate cancer metastasis24,25,26. Although Col1 is important in branching morphogenesis and tumorigenesis, the role of Col1 in controlling BaSC multipotency in different glandular epithelia remains unknown.
Here, we have defined the genes that are commonly upregulated during the activation of BaSC multipotency in the prostate and the MG across different conditions associated with multipotency. We identified Col1 as being associated with multipotency in these different glandular epithelia across the different conditions associated with multipotency. We demonstrated that increasing collagen concentration or ECM stiffness promotes MG and prostate BaSC multipotency. Using scRNA-seq, we unraveled the signaling pathways and lineage trajectories activated by Collagen signaling and stiffness and uncovered that β1 integrin/FAK/AP-1 axis regulates BaSC multipotency in glandular epithelia in response to Col1 signaling and ECM stiffness.
Results
Collagen expression is associated with BaSC multipotency in the MG and the prostate
To assess whether a common mechanism is controlling BaSC multipotency in MG and prostate, we determined the genes that are upregulated in MG BaSC when they are multipotent. We have previously established the 246 gene signatures of MG SC (embryonic multipotent progenitors, EMPs) when they are multipotent during their initial stage of embryonic development (E15)6 (Fig. 1a, Supplementary Fig. 1a and Supplementary Table 1). To identify the genes associated with multipotency during BaSC transplantation, we transplanted BaSC alone or together with LCs in the mammary fat pad, recovered them 3 days following transplantation, FACS isolated, and performed transcriptional profiling of these cells in these two conditions. By comparing the transcriptional profile of BaSC in these two conditions, we found 1713 genes that were associated with multipotency during BC transplantation alone (Fig. 1a, Supplementary Fig. 1b, and Supplementary Table 1). We and others have previously shown that the expression of an oncogenic mutant of PIK3CA, the most frequently mutated oncogene in human breast cancers10,18 activates multipotency in both BaSC and LC in adult mice. We have previously established the multipotent signature of BaSCs and LCs upon oncogenic PIK3CA expression10, that are composed of 399 genes (Fig. 1a, Supplementary Fig. 1c, and Supplementary Table 1). We have also previously established the BaSC multipotency signature upon LC ablation of the MG in adult mice11 (Fig. 1a, Supplementary Fig. 1d, and Supplementary Table 1).
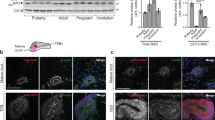
a Venn diagram illustrating the genes upregulated in the 5 multipotent conditions as indicated in MG and the prostate. The intersection shows that Col1a1, Col1a2, and Col3a1 are commonly upregulated. Immunohistochemistry of COL1A1 in control (CTL) and K5CreER/PIK3CA mice 7 months after TAM administration (b) and CTL (no dox) and K5CreER/Rosa-tdTomato/K8rtTA/TetO-DTA mice 1 week after Dox administration (c). n = 3 mice. Confocal imaging of immunostaining of MG organoid from K5CreER/td-Tomato mice in Matrigel and Col1 4 mg/ml gel (d) and in PEG 2.5% + RGD and PEG 7% + RGD conditions 1 week after TAM administration (h). tdTomato (TOM) in red, K8 in green, Hoechst in blue. Arrows indicate multipotent cells (TOM + K8+). Scale bars, 10 μm. Quantification of TOM + K8+ cells on total K8+cells at different Col1 gel (e) and PEG gel (i). n = 4 experiments (Matrigel and Col1 4 mg/ml), n = 3 experiments (Col1 2 mg/ml and 8 mg/ml), n = 5 experiments (PEG gel). Representative images of immunostaining of the prostate organoid derived from K5CreER-YFP mice in Matrigel and Col1 4 mg/ml gel (f) and in PEG 2% + RGD and PEG 9% + RGD gels (j) 5 days after TAM administration using anti-GFP (green) and anti-K8 (Red) antibodies. Hoechst in blue. Arrows indicate multipotent cells (YFP + K8+). Scale bar, 10 μm. Quantification of K8 + YFP+ cells on total K8+cells in different Col1 gel (g) and PEG gel (k). n = 4 experiments. l Representative FACS plot of K5 and K8 expression of MG organoids derived from K5CreER/td-Tomato mice at different conditions 1 week after TAM administration. m FACS quantification of TOM+ LCs on total LCs in MG organoids. n = 7 experiments. p-values are derived from two-sided unpaired t-test. n FACS quantification of YFP+ LCs on total LCs and YFP+ BCs on total BCs in MG organoids derived from K8CreER-YFP mice at different conditions 1 week after TAM administration. n = 5 experiments. Graphs are mean ± s.e.m. For (e, g, i, and k), p-values are derived from one-way ANOVA followed by Tukey’s test. Source data are provided as a Source Data file.
In the prostate, we previously demonstrated that during the early stage of prostate postnatal development, the BaSC at the tips of the ductal tree is multipotent, whereas the BaSC along the main ducts was already unipotent9. To define the multipotent gene signature of prostate BaSC, we microdissected the distal tip regions (tip—100 μm) from the main ducts of the developing prostate at postnatal day 10–12 under the stereoscope, isolated these two populations of BaSC by FACS, transcriptionally profile the unipotent and multipotent BaSC and found that 174 genes were associated with multipotency in prostate BaSC during postnatal development27 (Fig. 1a, Supplementary Fig. 1e and Supplementary Table 1).
The overlap between all the multipotent signatures revealed a common upregulation of Col1a1, Col1a2, Col3a1 from BaSC of the MG and prostate across these different conditions (transplantation, oncogenic mutation, LC ablation, and development) (Fig. 1a, Supplementary Fig. 1a–e and Supplementary Table 1). Immunohistochemistry staining confirmed that Col1a1 was expressed more abundantly within the MG stroma following the induction of an oncogenic PIK3CA mutant in BCs and after LC ablation in adult mice (Fig. 1b, c).
Collagen 1 and ECM stiffness promote BaSC multipotency in MG and the prostate
To assess whether Col1 in addition to be expressed at higher level in BaSC in multipotent conditions is directly promoting multipotency, we embedded MG and prostate organoids derived from K5CreER/Rosa-tdTomato or K5CreER/Rosa-YFP mice in collagen I gel at varying concentrations (2 mg/ml, 4 mg/ml and 8 mg/ml). The organoids were growing well within the collagen I gel. TAM was added to the culture media after embedding the organoid either with Matrigel or with collagen I to lineage trace BCs (TOM+ or YFP+). Following TAM administration, TOM expression were mainly found in BC (TOM + K8−) and very few LCs (TOM + K8+) were TOM+ in MG organoid embedded in Matrigel (Fig. 1d, e). In contrast, there was an increase in the proportion of TOM+ in LCs in organoid embedded in collagen I in a concentration-dependent manner, showing that high level of collagen promotes the differentiation of BCs into LCs by promoting BaSC multipotency in the MG and prostate organoids (Fig. 1d–g).
As Matrigel contained approximately 60% laminin and 30% collagen IV, we assessed the effect of Matrigel concentration on MG BC multipotency. To this end, organoids were embedded in 50%, 70%, and 100% Matrigel, treated with TAM for 48 h, and then cultured for an additional 5 days after TAM and the proportion of BC and LC Tomato labeled cells were quantified by FACS. Our results show a higher proportion of TOM+ LCs in 100% Matrigel compared to 50% Matrigel, showing that increasing the concentration of Matrigel promotes BC multipotency (Supplementary Fig. 2 and Supplementary Fig. 3a–c).
To assess the impact of other ECM components on BC multipotency, we embedded organoids in 70% Matrigel supplemented with either additional laminin or fibronectin. Our results showed that increasing laminin concentrations minimally enhanced BC multipotency, while fibronectin did not change BC multipotency at all (Supplementary Fig. 3d–f).
Collagen can bind to different classes of receptors, including the integrin family (integrin α1β1, α2β1, α10β1, α11β1), the discoidin domain receptors (DDRs), glycoprotein VI (GPVI), and the leukocyte-associated immunoglobulin-like receptor-1 (LAIR-1)28. Particularly, Col1 binding to the integrin α2β1 activates a signaling cascade which involves the focal adhesion kinase (FAK) activation and the related downstream pathway to promote different cellular responses, including adhesion, survival, and migration29,30,31,32. In addition, increased Col1 concentration increases ECM stiffness that can signal to the cells and activate the mechanotransduction signaling pathways33,34.
To assess whether rising Col1 concentration promotes BaSC multipotency by increasing ECM stiffness, we performed lineage tracing of BCs in MG organoids and prostate organoids embedded in polyethylene glycol gels (PEGs) at different concentrations presenting increasing stiffness. Unlike the Matrigel, PEGs are inert gels in which different components of the ECM can be added and the degree of stiffness can be modulated by changing the concentration of the polymers35,36,37. We embedded MG organoids and prostate organoids in PEG gels at different elastic modulus, including Matrigel (0.1 kPa stiffness), soft gel (2.5% PEG, 0.3 kPa stiffness), medium gel (3.5–4% PEG, 1–1.3 kPa stiffness) and stiff gel (7–9% PEG, stiffness 3.3–4 kPa stiffness), in the presence or absence of Arg-Gly-Asp (RGD) peptides, which are the minimal sequence motif on collagen bound by integrin38. These stiffness values approximate the mechanical properties of normal mammary gland stroma (0.1 kPa) and tumor (4 kPa)34, as well as collagen I gel at varying concentrations (2 mg/ml, 0.5 kPa; 4 mg/ml, 0.9 kPa; 8 mg/ml, 3.3 kPa, Advanced Biomatrix). We observed a dose-dependent enrichment of K8+ fluorescent labeled LCs arising from BaSC in the presence of increased gel stiffness in both MG and prostate (Fig. 1h–k). In addition, we found that the presence of RGD peptide was necessary for the activation of multipotency (Fig. 1h–k and Supplementary Fig. 4a–d). We used intracellular flow cytometry analysis of K5 and K8 expression to quantify MG BC multipotency mediated by increasing collagen concentration and stiffness using an orthogonal method. FACS quantification showed that similarly to what was found using immunostaining of K8 on organoid sections, high collagen concentration, and stiffness increased the proportion of TOM+ LCs (K5low K8high) coming from MG BaSCs (Fig. 1l, m), further supporting the notion that collagen I and ECM stiffness promote BaSC multipotency.
To assess whether collagen I and ECM stiffness promote LC multipotency, we embedded MG organoids derived from K8CreER/Rosa-YFP mice in four conditions: Mat, Soft (2.5% PEG + RGD), Col1 (4 mg/ml), and Stiff (7% PEG + RGD). One week following TAM administration, we observed that 15–20% LCs expressed YFP+, with only a small proportion YFP+ BCs (1–2%). Higher levels of collagen I and ECM stiffness did not lead to a further increase in YFP+ BCs (Fig. 1n and Supplementary Fig. 4e, f), suggesting that in contrast to BaSC, collagen I and ECM stiffness do not induce LC multipotency.
Altogether, these data demonstrate that Col1 and ECM stiffness promote the activation of BC multipotency in the MG and in the prostate.
Collagen 1 and ECM stiffness promote hybrid states in MG organoids
To understand the mechanisms by which collagen and stiffness promote multipotency, we performed 10x single-cell RNA sequencing (scRNA-seq) to profile MG organoids embedded in four different conditions promoting or not multipotency: MATR (Matrigel, stiffness 0.1 kPa), SOFT (2.5% PEG + RGD, stiffness 0.3 kPa), COL1 (Col1 gel, stiffness 0.9 kPa), and STIFF (7% PEG + RGD, stiffness 3.3 kPa). Following demultiplexing of the samples tagged with different antibodies and filtering out doublets/negative droplets, mitochondrial genes, and non-epithelial cells in the MG samples, we identified a total of 1390 epithelial cells in the MATR, 2511 cells in the SOFT, 718 cells in the COL1, and 2620 cells in the STIFF conditions.
Dimensionality reduction analysis using Uniform Manifold Approximation and Projection (UMAP) and Principal Component Analysis (PCA) in the various conditions showed the presence of three main clusters. Through the expression of well-known markers for BCs (Krt5), pan-LCs (Krt8), LC ER+ (Foxa1), and LC ER- (Elf5) we identified that the three main clusters were corresponding to BCs, LCs ER+, and LCs ER- found in MG in vivo6,11,39,40 (Supplementary Fig. 5a–d). In addition to these three main clusters, there were additional clusters of cells expressing both BC and LC genes, particularly in COL1 and STIFF conditions, forming non-homogeneous clusters in the UMAP plot that could represent hybrid cells between BC and LC previously identified in previous single-cell analysis studies of the MG in conditions associated with multipotency40,41,42, including embryonic MG and MG following LC ablation6,11 (Supplementary Fig. 5a–d).
To study the differences and similarities between the four samples, and to identify the changes in cell states following embedding MG organoids in different ECM gels with increased concentration of collagen and stiffness, we have integrated the four samples using a set of anchor genes calculated by Seurat integration pipeline43. After integration, we had a total of 1383 cells for MATR, 2508 cells for SOFT, 716 cells for COL1, and 2619 for STIFF. The UMAP dimensionality reduction of the integrated sample showed the presence of all expected cell types, including BC, LC ER+, and LC ER− clusters, as well as an increased enrichment of hybrid cells (HY ER+/ER−, HY BC/ER−, and a proliferative HY BC/ER−) specifically in the STIFF and COL1 conditions (Fig. 2a, b, Supplementary Fig. 5e). After data integration and plotting UMAPs split by sample (Fig. 2b), the cell type distribution mirrored what was observed in the individual datasets (Supplementary Fig. 5a–d). These observations suggest that the integration did not lead to an over-correction of the data and the disappearance of the clusters found before data integration. Additionally, we identified a new cluster called BC primed to LC differentiation (BC primed LC), which expressed BC, LC ER+, and LC ER− markers that are highly enriched in COL1 and STIFF conditions (Fig. 2a, b, Supplementary Fig. 5e).
UMAP dimensionality reduction plots with different colors representing unsupervised clustering: Integrated data using Seurat (a) and the plot of integrated data split by different conditions as indicated (b). HY stands for hybrid and Pr stands for proliferating cells. Integration of data did not bring an over-correction of the data or disappearance of certain population. Quantitative assessment of LC and BC marker gene expression in MATR (c), SOFT (d), COL1 (e), and STIFF (f) condition: Scatterplot with the x-axis representing the adjusted proportion of BC-specific marker genes and the y-axis representing the adjusted proportion of LC-specific markers. The proportion of cells which express more than 65% (0.65) of BC and LC markers is indicated as red square. g Quantification of cells in each data considered as hybrid status, indicated as red square on 2 (c–f). Slingshot pseudotime trajectory analysis: UMAP dimensionality reduction plots of integrated data for the trajectory and quantification of pseudotime as violin plot composed of trajectory 1: BC – BC Primed – LC ER+ (h), trajectory 2: BC – BC Primed – HY BC/ER- – LC ER- (i) and trajectory 3: BC – BC Primed – HY BC/ER- – LC ER- – HY ER+/ER- – LC ER+ (j). BC primed indicates BC primed LC differentiation cluster. Triangle indicates start of trajectory and square indicates termination of trajectory. Heatmaps illustrate the expression levels of genes that are differentially expressed along the trajectories in the integrated dataset, encompassing marker genes for BC, LC ER+, and LC ER-: trajectory 1 (k), trajectory 2 (l), trajectory 3 (m). Source data are provided as a Source Data file.
To determine the proportion of the cells presenting a hybrid state, we quantified in the different conditions the fraction of cells expressing high proportion of LC and BC genes (more than 65% of the genes). In the MATR and the SOFT conditions, about 1% of cells present this hybrid signature whereas in COL1 and STIFF, this proportion increased to 4% and 3% of cells respectively (Fig. 2c–g).
To determine the differentiation path taken by the BCs when they differentiate into LC, we perform a lineage trajectory analysis on the integrated dataset using slingshot44 and set the BC cluster as the starting point of the trajectory. We excluded the BC/LC ER- Pr cluster from the analysis because the genes associated with proliferation disrupt the lineage trajectories. Slingshot analysis suggested the existence of three trajectories that start from the BCs. One trajectory indicated the direction from BC toward LC ER+, passing through the BC presenting the LC priming (Fig. 2h), another trajectory from BC to LC ER− passing through the hybrid BC/ER- (Fig. 2i) and finally, a last trajectory originating from BC going toward LC ER− and then to the LC ER+ (passing through the hybrid ER+/ER−) (Fig. 2j). During the differentiation from BC to LC, there was a downregulation of BC markers following the trajectories, including Krt5, Krt14, Trp63 and Acta2. Expression levels of LC ER+ (Foxa1, Areg, Prlr, Ly6a) and LC ER- (Elf5, Ehf, Cd14) marker genes were gradually increased following the different trajectories (Fig. 2k–m).
To further define the genes associated with hybrid state, we performed analysis of the differentially expressed genes (DEG) comparing the BC cluster and the hybrid BC/ER- in Collagen and STIFF conditions. We found that 490 genes and 621 genes were significantly upregulated in HY BC/ER- under COL1 and STIFF conditions (Supplementary Fig. 6a, b). Among these differentially regulated genes, 379 genes were commonly upregulated in HY BC/ER- including many classical LC genes such as Epcam, Krt8, Krt18, Cldn3, 4 and 7, CD24a, or Elf5 (Supplementary Fig. 6a, b). This analysis also revealed that 283 genes were commonly downregulated in HY BC/ER- including canonical BC marker genes such Krt5, Krt14, Trp63, genes of the basal membrane (Col4a1, Col4a2, Col17a1, Itga6, Lama3, Lamb3), and myoepithelial markers (Acta2, Myh11, Myl9, Mylk) (Supplementary Fig. 6a, b). Additionally, three collagen genes (Col1a1, Col1a2, Col3a1) that are commonly upregulated across the different multipotent conditions (Fig. 1a) were exclusively expressed in BC primed LC differentiation during COL1 and STIFF conditions (Supplementary Fig. 6c, d).
To determine whether the hybrid state induced by high collagen and stiffness resembles to the hybrid state found in the other conditions associated with multipotency, we compared the hybrid gene signatures from EMP6 and LC ablation11 (Fig. 1a, Supplementary Table 1) with the marker genes from the clusters BC-primed LC differentiation and HY BC/ER- state found in high collagen and stiffness (Supplementary Fig. 6e, f). Three genes (Col1a1, Col1a2, Col3a1) were also shared between BC primed LC differentiation and the EMP and LC ablation signatures. Five genes (Col8a1, Emp3, Sostdc1, Timp1, Tnc) overlapped with the LC ablation condition, while 6 genes (Col18a1, Vim, Serpinh1, Bcl11b, Lgals7, Ppic) overlapped with the EMP state, mainly linked to ECM composition. For the HY BC/ER- population, two genes (Fn1, Fscn1) were common across all conditions. Compared to the EMP hybrid state, 10 genes, including those related to cytoskeleton and ECM (Col18a1, Serpinh1, Vim) and tubulins (Tuba1a, Tubb2b, Tubb5), were shared. The LC ablation comparison showed an overlap of 29 genes, including those involved in cell proliferation (Cdk6, Cdkn2b, Nupr1, Nme1), ECM composition (Col8a1, Tnc, Thbs2, Spon1), and cytoskeleton regulation (Clu, Krt7). These findings suggest that the collagen-induced hybrid state is more similar to the hybrid state found in adult mice following LC ablation.
ATAC-seq and RNA-seq performed during MG development uncover luminal genes for which the chromatin regions surrounding the promoter are also accessible in basal cells, suggesting that some luminal genes are primed in BCs45. We then assess whether these primed genes correlate with the hybrid and primed clusters induced by stiffness. Our data show that while primed genes were expressed in the BC primed to LC differentiation population, most showed higher expression in the HY ER+/ER− and LC ER+ populations (Supplementary Fig. 6g).
In situ characterization of hybrid cells
To further characterize these novel hybrid cells in situ, we performed immunostaining of basal and luminal markers following lineage tracing of BCs in MG and prostate organoids in the MATR, SOFT, COL1, and STIFF conditions. Our results revealed that both TOM+ and YFP+ cells, for the MG and the prostate organoids respectively, were restricted to K14+ basal cells under Matrigel and Soft conditions, whereas hybrid cells that co-expressed K14, K8, and TOM/YFP were found in COL1 and STIFF conditions (Fig. 3a–c and Supplementary Fig. 7a), showing that collagen and ECM stiffness promote the appearance of hybrid cells during activation BaSC multipotency. Interestingly, the appearance of hybrid cells was observed as early as two days after TAM administration in COL1 condition (Fig. 3d–f). These hybrid cells induced by Collagen and ECM stiffness fully differentiated into LCs over time as shown by the loss of K14/K5 and the gain of K8 expression at Day 7 (Figs. 3a–c and 1l, m). Taken together, these results indicate that the activation of BaSC multipotency induced by Collagen and ECM stiffness is a progressive process associated with the emergence of transient hybrid states that eventually differentiate into LCs after losing their BC identity.
a Confocal imaging of immunostaining of MG organoid from K5CreER/td-Tomato mice in Matrigel, Soft, Col1, and Stiff gel 1 week after TAM administration using anti-K14 (White) and anti-K8 (Green) antibodies. TOM in red, Hoechst in blue. Arrows indicate hybrid cells (TOM + K14 + K8+) and fully differentiated LCs (TOM + K14− K8+). Scale bars, 10 μm. FACS quantification of TOM+ hybrid (K5 + K8+) on the total cells (b) and TOM+ BCs on the total BCs (c) in MG organoids at different conditions indicated 1 week after TAM administration. n = 7 independent experiments. d Representative images of immunostaining of the organoid derived from K5CreER/td-Tomato mice in Matrigel, Col1, Soft, and Stiff gel 2 days after TAM administration. TOM in red, K8 in green, K14 in white, Hoechst in blue. Arrows indicate hybrid cells (TOM + K14 + K8+). Scale bars, 50 μm. e Representative FACS plot of K5 and K8 expression on TOM+ cells from MG organoids derived from K5CreER/td-Tomato 2 days after TAM administration. MG organoids are embedded in the different gels indicated. BC: K5-high K8-low; LC: K5-low K8-high; Hybrid: K5 + K8+. f Quantification of TOM+ LCs on total LCs, Tom+ Hybrid on total cells, Tom+ BCs on total BCs in MG organoids at different conditions indicated 2 days after TAM administration. n = 3 independent experiments. For (b, c, and f), graphs are mean ± s.e.m. p-values are derived from two-sided unpaired t-test. Source data are provided as a Source Data file.
Activation of AP-1 TFs during the promotion of BC multipotency by collagen and ECM stiffness
To understand which genes are promoting BC multipotency in different conditions, and whether common or different pathways are involved in the activation of multipotency in the stiff matrix and COL1 conditions, we used two different approaches. In the first approach, we performed DEG analysis between the BCs in Matrigel versus the other conditions (SOFT vs MATR, COL1 vs MATR, and STIFF vs MATR). Comparison of SOFT versus MATR conditions showed that only 6 genes were significantly differentially expressed, showing that these two conditions are very similar. In contrast, 26 and 40 genes were significantly different in COL1 and STIFF conditions compared to MATR (Supplementary Fig. 8a). Among those, 13 genes were commonly upregulated in the BCs in COL1 and STIFF conditions promoting BC multipotency, including AP-1-related genes such as Jun, Fos, Fosb, and Atf3 (Fig. 4a), as well as ribosomal genes like Rps29, Rsp28, and Rpl37 (Supplementary Fig. 8b). We also found upregulation of Spp1 (Osteopontin) and Dusp1 (Supplementary Fig. 8b).
a Violin plots showing the expression level of the genes which were commonly upregulated on BC cells of multipotent conditions (COL1 and STIFF) compared to unipotent conditions (MATR and SOFT). P-values were calculated using the non-parametric Wilcoxon rank sum test (two-sided). Volcano plots showing the results on the linear modeling on the BC (b), BC primed (c), and HY BC/ER- (d) for stiffness-dependent genes. BC primed indicates BC primed LC differentiation cluster. Significant genes on the linear models are marked as red (adjusted P-value < 0.01). P-value were calculated by negative binomial regression, two-sided. Violin plots showing the gene expression level of Jun (e) and Fosb (f) on BC, BC primed, HY BC/ER- cells, and HY BC/ER- proliferating on integrated data, split by different conditions. P-value is from modeling on each cell type to test the correlation between stiffness and gene expression, calculated by negative binomial regression, two-sided. g Schematic representation of the transcription factors (TFs) found as activated by SCENIC leading to BC-to-LC differentiation. Dashed arrow indicates that the lineage trajectory can go further, but not necessarily. h Violin plots showing the regulon activity of Fos, Fosb, and Atf3 on BC, which are more activated on multipotent conditions compared to the unipotent conditions. P-values were calculated using the non-parametric Wilcoxon rank sum test (two-sided) to compare AUC scores. i Representative images of immunostaining of the MG organoid embedded in Matrigel, Col1, Soft, and Stiff gel using anti-K14 (White) anti-Jun (green) antibodies, Hoechst in blue. Scale bars, 50 μm. j Quantification of nuclear Jun+ K14+ on total K14+ in MG organoids embedded in the different gels indicated (mean ± s.e.m.; n = 3 independent experiments). p-values are derived from two-sided unpaired t-test. Source data are provided as a Source Data file.
To enhance the statistical power of our analysis, we assessed using a linear model which genes are correlated with both increased stiffness and the presence of COL1. When we applied this model to the BC cluster, we identified the same set of genes previously found in the DEG analysis including the AP-1 family of TFs such as Jun, Fosb, Jund, Fos and Atf3 (Fig. 4b, Supplementary Fig. 8c). Furthermore, Jun and Fosb were also upregulated in BC primed LC and in hybrid BC/ER- cells in the stiff condition (Fig. 4c–f). These results reinforce the notion that AP-1 activation is associated with BC multipotency in both the COL1 and STIFF conditions. Additionally, ribosomal genes (Uba52, Rpl38, Rps29) which could be involved in translational regulation of stem cell function46 and transcription factors such as Klf2 and Klf6 are also showed significance on the modeling (Supplementary Fig. 8d, e).
To further strengthen the identification of the transcription factors (TFs) that mediate BaSC multipotency and BC-to-LC differentiation in response to increased collagen concentration and ECM stiffness, we used SCENIC analysis, a bioinformatics tool that identifies regulatory regions through the inference of co-expression between TFs and their target genes47 and compared TFs active in the different conditions using Fisher’s combined test. As previously found in scRNA-seq from adult MG in vivo6, we found that Trp63, Egr2, Foxp1, and Mef2c were enriched in organoid BCs. Foxa1 and Tbx3 were enriched in LC ER+, while well-known LC ER- TFs such as Elf5 and Hey1 were enriched in LC ER−. BC primed LC population showed a significant activation of the AP-1 family (Jun, Jund, and Atf3), and Klf2 as well as TFs known to promote either LC ER+ or LC ER− fate, such as Foxa1 and Creb5, respectively. Moreover, the HY BC/ER- state was enriched in regulons activated by Ets1, as well as Sox10 and Sox11, which are reported as upregulated in hybrid population of mammary epithelial cells in previous studies39,45,48,49,50 (Fig. 4g, Supplementary Fig. 8f). Also, AP-1 family of TFs (Fos, Fosb and Atf3) were predicted as activated in BC in conditions associated with multipotency (COL1 and STIFF) compared to the conditions associated with unipotency (MATR and SOFT) (Fig. 4h). Jun immunostaining in MG organoids showed that Jun was expressed in LCs, with minimal expression in BCs in Mat and Soft conditions (Fig. 4i, j), whereas in high collagen concentration and ECM stiffness promoted Jun expression in BC (Fig. 4i, j).
Altogether, these data show that AP-1 TFs and their target genes were activated during the activation of multipotency in COL1 and stiff conditions, further showing that AP-1 TFs are associated with BC multipotency in both COL1 and stiff conditions.
Integrin-β1/FAK signaling controls collagen and stiffness induced multipotency in glandular epithelia
Col1 can bind different integrin receptors and activate different intracellular signaling involved in developmental processes. Among the collagen-binding integrins, integrin β1 is the most common receptor51. To investigate the role of Col1-β1-integrin signaling in BC multipotency activation, we treated MG and prostate organoids with β1-integrin blocking antibodies under different conditions: MATR, SOFT, COL1, and STIFF. Interestingly, the addition of β1-integrin blocking antibody52 for 5–7 days after TAM administration significantly reduced the proportion of K8+ Tomato+ LC in the MG organoids from K5CreER/Rosa-tdTomato mice and K8 + YFP+ cells in the prostate organoids from K5CreER/Rosa-YFP mice compared to control IgG treated group under conditions promoting multipotency (COL1 and STIFF) (Fig. 5a, b). To further substantiate these findings, we pharmacologically inhibited α2β1 signaling by treating organoids with α2β1-inhibitor (BT 3033)53. Consistent with results of β1-integrin blocking antibody, treatment with BT 3033 for 5-7 days after TAM administration decreases the proportion of K8+ Tomato+ LCs in the MG organoids and K8 + YFP+ LCs in the prostate organoids under conditions promoting multipotency, further showing the important role of β1-integrin signaling induced by high concentration of collagen and a stiff ECM (Fig. 5c, d).
a Quantification of TOM + K8+ cells on the total K8+cells in K5CreER/td-Tomato MG organoids treated for 1 week either with IgG control antibody or with a blocking β1 antibody (HMβ1.1). MG organoids are embedded in the different gels as indicated (n = 4 independent experiments). b Quantification of YFP + K8+ cells on the total K8+cells in K5CreER/Rosa-YFP prostate organoids treated for 5 days either with IgG control antibody or with HMβ1.1 antibody. Prostate organoids are embedded in the different gels as indicated. (n = 3 independent experiments). c Quantification of TOM + K8+ cells on the total K8+cells in K5CreER/td-Tomato MG organoids treated for 1 week with control DMSO, α2β1 inhibitor (BTT 3033) at 5 μM, or FAK inhibitor (PF573228) at 10 μM. MG organoids are embedded in the different gels indicated (n = 4 independent experiments). d Quantification of YFP + K8+ cells on the total K8+cells in K5CreER/RosaYFP prostate organoids treated for 5 days with control DMSO, BTT 3033 at 2 μM, or PF573228 at 10 μM. Prostate organoids are embedded in the different gels as indicated. (n = 4 independent experiments). e Experimental design. f Representative images of YFP fluorescence of MG fat pad after DMSO and FAK inhibitor (FAKi, PF573228) treatment 8 weeks (Biweekly i.p. injection, 10 mg/kg). Scale bars, 1 mm. g Quantification of YFP+ transplantation in all transplantation. n = 16 transplantation in DMSO group, n = 18 transplantation in FAKi group. h Representative images of immunostaining of the transplanted fat pad using anti-K14 (White), anti-GFP (green), and anti-K8 (Red) antibodies. Hoechst in blue. Scale bars, 20 μm. n = 3 transplanted glands. For (a–d), graphs are mean ± s.e.m. p-values are derived from two-sided unpaired t-test. Source data are provided as a Source Data file.
Next, we assessed whether β1-integrin signaling promotes BC multipotency through its canonical downstream signaling focal adhesion kinase (FAK). To this end, we treated MG and prostate organoids with PF573228, an inhibitor of FAK54, for 5-7 days following TAM administration. Treatment with the FAK inhibitor decreased the proportion of K8+ Tomato+ LCs in the MG organoids and K8 + YFP+ LCs in the prostate organoids induced by COL1 and STIFF conditions without inducing apoptosis (Fig. 5c, d and Supplementary Fig. 9a, b), indicating that the activation of FAK is required for the promotion of BaSC multipotency downstream of β1-integrin in response to high level of Col1 or stiff ECM.
FAK inhibition in vivo prevents BaSC multipotency and MG outgrowth during BC transplantation
To assess whether FAK signaling regulates BC multipotency in vivo, we inhibited FAK signaling following the transplantation of BC into the mammary fat pad. To this end, we FACS isolated YFP+ BCs from the MG of K14Cre/YFP mice (Supplementary Fig. 10) and transplanted them into the cleared mammary fat pads of NOD-SCID mice, followed by 8 weeks of FAK inhibitor treatment (10 mg/kg, i.p., twice a week)55,56 (Fig. 5e). Interestingly, while the control mice developed well-branched mammary glands following transplantation of BCs, mice treated with FAK inhibitor showed only YFP+ cell clumps with poorly branched structures, suggesting FAK inhibition led to a decrease in MG outgrowth from BC (Fig. 5f, g). Immunostaining revealed that FAK inhibition disrupted the differentiation of BCs into LCs, resulting in smaller, less organized structures co-expressing basal and luminal markers. In contrast, the control mice showed clear segregation of BCs and LCs, forming well-developed glandular architecture (Fig. 5h). These results provide in vivo evidence that FAK signaling can regulate multipotency following BC transplantation.
Integrin-β1/FAK/AP-1 axis controls collagen and stiffness-induced multipotency in glandular epithelia
Next, we assessed whether the AP-1 transcriptional factors are promoting collagen and ECM stiffness-induced multipotency as suggested by our bioinformatic analyses (Fig. 4a–h, Supplementary Fig. 8c). Inhibition of β1-integrin and FAK signaling impaired collagen- and ECM stiffness-induced AP-1 activation, suggesting that AP-1 is activated by high collagen concentration and ECM stiffness downstream of β1-integrin and FAK signaling (Fig. 6a). To functionally investigate the role of AP-1 signaling in the activation of BC multipotency, we assessed whether T-522457, a pharmacological inhibitor of AP-1 reduced BaSC multipotency in response to high concentration of Col1 and stiff ECM in the MG and prostate organoids. Interestingly, AP-1 inhibitor treatment for 5-7 days following TAM administration decreased the proportion of K8+ Tomato+ LCs in the MG organoids as shown by immunostaining on organoid sections and FACS analysis and K8 + YFP+ LCs in the prostate organoids induced by COL1 and STIFF conditions and did not increase apoptosis (Fig. 6b–d, Supplementary Fig. 9a, b). Given that AP-1 plays a critical role in regulating cell proliferation58, and our data showed that increased collagen and ECM stiffness promote cell proliferation (Supplementary Fig. 9c, d), we investigated whether proliferation regulates BC multipotency. To this end, we inhibited cell proliferation in MG organoids embedded in different ECM by using RO-3306, a CDK inhibitor11. We found that decreasing proliferation reduced collagen- and ECM-stiffness-induced BC multipotency (Fig. 6e and Supplementary Fig. 9c, d).
a MG organoids treated for 1 week with control DMSO, α2β1 inhibitor (BTT 3033) at 5 μM, or FAK inhibitor (PF573228) at 10 μM. Relative mRNA expression levels of Jun and Fos were determined by quantitative RT-PCR. MG organoids are embedded in the different gels as indicated. n = 4 independent experiments. b Quantification of TOM + K8+ cells on total K8+ cells in K5CreER/td-Tomato MG organoids treated for 1 week with DMSO control and AP1 inhibitor (T-5224) at 10 μM. n = 3 independent experiments. c Quantification of YFP + K8+ cells on total K8+cells in K5CreER-YFP prostate organoids treated for 5 days with DMSO control and T-5224 at 10 μM. n = 3 independent experiments. d FACS quantification of TOM+ LCs on total LCs in K5CreER/td-Tomato MG organoids treated for 1 week with DMSO control and T-5224 at 10 μM. n = 5 independent experiments. e Quantification of TOM + K8+ cells on total K8+ cells in K5CreER/td-Tomato MG organoids treated for 1 week with DMSO and CDK inhibitor (RO-3306) at 10 μM. n = 3 independent experiments. f Experimental design. g Representative images of immunostaining of Ade-K5Cre infected MG organoid derived from Rosa-YFP and Junbfl/fl/Juncfl/fl /mTmG mice using anti-Jun (white) and anti-GFP (green). Hoechst in blue. Arrows indicate nuclear Jun+ YFP+ cells. Scale bars, 20 μm. h Quantification of nuclear Jun+ YFP+ cells on total YFP+ cells in Ade-K5Cre infected MG organoids from Rosa-YFP and Junbfl/fl/Juncfl/fl /mTmG mouse. n = 3 independent experiments. i Representative images of immunostaining of Ade-K5Cre infected MG organoid derived from Rosa-YFP and Junbfl/fl/Juncfl/fl /mTmG mice using anti-K8 (White) and anti-GFP (green). Hoechst in blue. Arrows indicate multipotent cells (YFP + K8+). Scale bars, 50 μm. j Quantification of YFP + K8+ cells on total K8+ cells in Ade-K5Cre infected MG organoids from Rosa-YFP and Junbfl/fl/Juncfl/fl /mTmG mice. n = 3 independent experiments. Graphs are mean ± s.e.m. Organoids are embedded in the different gels as indicated. For (a, h, and j), p-values are derived from one-way ANOVA followed by Tukey’s test. For (b–e), p-values are derived from two-sided unpaired t-test. Source data are provided as a Source Data file.
To further substantiate our findings, we performed genetic deletion of Junb/Junc and assessed the importance of these TFs in regulating BC multipotency mediated by Col1 and ECM stiffness (Fig. 6f). To this end, we generated MG organoids from Junbfl/fl/Juncfl/fl /mTmG mouse model. To delete Junb/Junc and lineage-traced BCs, we infected organoids with adenoviruses expressing Cre under the control of K5 promoter59 and embedded them in different ECM substrates. One week following Jun deletion, immunostaining showed that Jun could not be observed in BCs, and the proportion of K8 + YFP+ LCs compared to control organoids infected with CRE adenovirus was reduced (Fig. 6g-j), further demonstrating the importance of AP-1 TFs in regulating BaSC multipotency mediated by Col1 and ECM stiffness.
Taken together, these results show that collagen and ECM stiffness-induced multipotency in MG and the prostate is mediated by integrin β1/FAK/AP-1 axis.
Discussion
In this study, we uncovered a common mechanism regulating BaSC multipotency across multiple conditions associated with acquisition of multipotent features by both MG and prostate epithelial SCs.
The comparison of the gene signature of the different conditions in which BC are multipotent (embryonic development, transplantation of BC alone, PIK3CA activation, and LC ablation) has shown the common upregulation of Col1a1, Col1a2, Col3a1 in multipotent BCs, suggesting that COL1 and the mechanical properties of the matrix surrounding the SC could control the activation of BC multipotency. Different studies show that SC fate depends on the mechanical and biochemical properties of the ECM29,32,60,61,62. The role of the ECM in controlling MG and prostate lineage differentiation was demonstrated through transplantation experiments. Transplantation of non-mammary cells from various tissues including testicular cells, embryonic SCs, sweat gland SCs, or neural SCs shows that these cells can adopt a mammary epithelial cell fate upon transplantation into the mammary fat pad63,64,65,66. Similarly, urogenital mesenchyme promotes the differentiation of prostate, bladder, and urethral epithelium to differentiate into prostate epithelial cells67,68. The mechanisms by which mammary fat pad or the urogenital sinus mesenchyme promotes BaSC multipotency are unknown.
Collagens are among the most abundant proteins present in the ECM. In addition to playing a structural role, collagens also signal to epithelial cells through different receptors, such as integrins28,69,70. Integrins are functionally important for the morphogenesis and homeostasis of different epithelia69,70,71. Collagen-integrin binding activates downstream signaling that involves the focal adhesion kinase (FAK), acting as a link between the ECM, the actomyosin cytoskeleton, and the nucleus62,72,73,74. Co-transplantation of human mammary SCs with irradiated mouse fibroblasts in collagen gels under the kidney capsule recapitulates the results obtained with the transplantation of SCs in the mammary mesenchyme, suggesting that the collagen can control SC fate75. Experiments aiming at identifying the role of matrix rigidity in regulating MG growth showed that isolated mammary epithelial cells seeded in mixed collagen/basal membrane matrix at different collagen concentrations were forming bigger colonies in gels with higher collagen concentration. Importantly, the colony-forming activity, as well as the higher expression of integrin-β1 and filamentous actin (F-actin), was depending on the different degrees of stiffness associated with higher collagen content, suggesting that increased collagen in 3D structures is promoting the expression of integrin-β1 and F-actin and positively correlates with the colony-forming efficiency of MG cells34.
The first demonstration that ECM stiffness can control SC fate came from the work of Engler and colleagues that shows that mesenchymal stem cells are sensitive to matrix elasticity and differentiate into different lineages according to matrix stiffness. Soft matrix promotes neuronal differentiation, stiffer matrix promotes muscle fate, and further rigid matrix promotes osteogenic differentiation60. Moreover, high stiffness matrix promotes intestinal SC expansion, while differentiation requires a soft substrate76. Using MG and prostate organoids, we show that increased collagen 1 concentration and organoid embedding into PEG matrixes with high stiffness promote BC multipotency supporting the notion that collagen 1 signaling and/or matrix stiffness induces BC multipotency. In our culture system, we found that the addition of RGD peptide, which corresponds to smallest peptide presented on ECM that binds to integrin, induces their clustering, and activates their downstream signaling in the PEG gels. Importantly, this is necessary for BC differentiation into LC, showing that integrin signaling in BaSC is required for the activation of multipotency in stiff condition34,77.
The importance of ECM-stem cell communication in controlling SCs is suggested by the high expression of integrins in various SCs12,13. In the MG, deletion of integrin-β1 specifically in the BCs using Cre-dependent deletion, showed the reduced capacity to give rise to a new MG in secondary transplantation, supporting an important role of integrin-β1 in the maintenance of MG SCs78. Blocking antibodies against integrin-β1 and a specific inhibitor of integrin-α1β1 show that both treatments significantly decrease the proportion of LC differentiation in response to collagen signaling and ECM stiffness, suggesting that BC multipotency is mediated by the activation of integrin-dependent signaling. We show that FAK inhibition prevents the activation of BC multipotency consistent with a role of FAK to relay collagen/integrin-β1 signaling to the nucleus.
scRNA-seq of MG organoids embedded in the Matrigel and soft gel conditions shows that MG organoids in vitro present the same epithelial cell types and cell states identified in MG in vivo6,39,40. However, the LC ER- pre-committed milk-producing cells40 were not found in these organoids, possibly due to the lack of proper hormonal stimulation and stromal communications in the organoid system. Our data show that collagen signaling and stiffness promote LC commitment within the BC and the presence of a more advanced hybrid BC/LC cell population. The analysis of the cells expressing a high proportion of LC/BC markers showed that in Matrigel and soft matrix, only 1% of the cells present high proportion of hybrid BC/LC genes, whereas in collagen 1 and stiff matrix, 4% and 3% of the cells expressed high levels of BC and LC markers. Previous studies with ATAC-seq and scATAC-seq have shown that during MG development some BCs present chromatin open regions within a subset of LC genes, consistent with their priming toward LC differentiation45,79. Similarly, in this study, we identified a population of BCs primed towards the LC fate upon stimulation by a high collagen 1 content and increased stiffness, suggesting that mechanical properties could have a role in chromatin accessibility in epithelial SCs. These primed luminal genes were also expressed in the BC-primed LC differentiation population, as well as in HY ER+/ER− and LC ER+ populations upon increased collagen and stiffness.
Our analysis comparing the gene signatures of hybrid states across different biological conditions associated with multipotency shows that these different hybrid states present common and distinct features. Although some genes related to collagen, ECM, and cell proliferation are shared between EMP6, LC ablation11, and stiffness-induced hybrid populations (BC primed LC differentiation and HY BC/ER−), the overlap between the different conditions is very small. This suggests that while some components may be conserved, the hybrid state induced by mechanical cues is mediated by a different transcriptional program. Interestingly, scRNA-seq of the organoids embedded in the stiff matrix showed the presence of a subpopulation of BC expressing higher levels of Col1a1, Col1a2, and Col3a1 that may promote a feedforward loop increasing collagen production/ECM stiffening to sustain BC multipotency. Lineage differentiation analysis using Slingshot shows that BCs multipotency can be achieved through multiple differentiation paths to become either LC ER+ or LC ER-, as previously shown following LC ablation11. All these trajectories pass through the BC primed LC state, suggesting that this intermediate state is key for the initial activation of the multipotent differentiation program.
We identified commonly upregulated genes in the collagen 1 and stiff matrix conditions that contain a high proportion of AP-1-related TFs (Jun, Fos, Fosb, and Atf3) in both conditions associated with BC multipotency. scATAC-seq of embryonic MG cells also suggested that AP-1 TFs contribute to lineage differentiation during MG embryonic development79. SCENIC analysis also confirms AP-1 regulated genes are upregulated during the activation of BC multipotency, further supporting the role of AP-1 TFs and their related genes in promoting cell fate induced by Col signaling and stiffness. The AP-1 TFs play a wide range of cellular processes, such as development, cell proliferation, differentiation, and tissue regeneration58,80. AP-1 inhibitor T-5224 and genetic deletion of Junb and Junc in BCs significantly reduces multipotency activation in both collagen 1 and stiff ECM embedded organoids, demonstrating the key role of AP-1 activation to promote the multipotent transcriptional program in response to Collagen signaling and high ECM stiffness.
We previously demonstrated that decreasing cell proliferation reduced MG BC multipotency after LC ablation11. Here, we found that increased collagen and ECM stiffness promoted cell proliferation, and decreasing proliferation with a CDK1 inhibitor reduces MG BC multipotency mediated by collagen and stiffness. This data together with our previous report shows that proliferation is a necessary step for activating MG BC multipotency.
The role of collagen in promoting BaSC multipotency has important implications for tumorigenesis. Activation of multipotency, cell fate changes, and lineage infidelity have been associated with tumor initiation, cancer progression, and resistance to therapy81,82,83,84. Increased collagen 1 expression was found in mammary epithelial cells upon oncogenic PIK3CA expression, which promotes MG epithelial cell multipotency at the earliest stage of mammary tumor initiation10. In addition, increased collagen 1 expression which correlates with stiffening of MG stroma has been associated with tumor formation and increased breast cancer invasion77,85. In MMTV-Neu model of mammary tumors, reduction of collagen cross-linking by inhibition of lysyl-oxidase (LOX) promotes the development of low-grade tumors77. Women with dense breasts that present more abundant and stiffer collagen have an increased risk of developing breast cancer86,87. Collagen abundance in human breast tumors is associated with higher mortality88 and breast tumors surrounded by aligned, linearized, and thick collagens present poorer prognosis89.
In conclusion, our study reveals that Collagen I and ECM stiffness promote BaSC multipotency in mammary gland and prostate epithelia through the activation of the β1-integrin/FAK/AP-1 signaling axis that regulates cellular plasticity in response to mechanical stress.
Methods
Mice
The generation of K5CreER and K8-CreER was previously described4. Junbfl/fl/Juncfl/fl mice were imported from Erwin F. Wagner laboratory90,91. The K14Cre, Rosa26-LSL-tdTomato, Rosa26-LSL-YFP mice, and Rosa26-LSL-mT/mG mice were purchased from the Jackson Laboratory. Mice colonies were maintained in a certified animal facility in accordance with European guidelines. The room temperature ranged from 20 to 24 °C. The relative ambient humidity at the level of mouse cages was 55 ± 10%. Each cage was provided with food, water, and two types of nesting material. A semi-natural light cycle of 12:12 was used. The experiments were approved by the corresponding ethical committee (Commission d’Ethique et du Bien Être Animal (CEBEA), Faculty of Medicine, Université Libre de Bruxelles) under protocols #671 N, #673 N, #886 N. CEBEA follows the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes. The study is compliant with all relevant ethical regulations regarding animal research. Mice were used at adult age (over 8 weeks) to isolate mammary gland organoids (females) and the prostate organoids (males).
Mammary BCs sorting, transplantation, and drug treatment
FACS sorting and Mammary fat-pad transplantation were performed as previously reported4. Briefly, Mammary glands from K14Cre/RosaYFP adult females were dissected, and lymph nodes were removed. The tissue was cut into 1 mm³ pieces and digested in HBSS containing 300 U/ml collagenase and 300 µg/ml hyaluronidase for 2 h at 37 °C with constant shaking. After digestion, EDTA was added for 10 min, followed by a 2 min treatment with trypsin-EDTA. Cells will pass the cell strainer (40 μm) and then incubate with 200 μl of 2% FBS/PBS with fluorochrome-conjugated primary antibodies for 30 min on ice, with shaking every 10 min. Primary antibodies were washed with 2% FBS/PBS, and cells were resuspended in DAPI (Invitrogen, D1306). The following primary antibodies were used: APC-conjugated anti-CD45 (1:100, clone 30-F11, eBiosciences), APC-conjugated anti-CD31 (1:100, clone 390, eBiosciences), APC-conjugated anti-CD140a (1:100, clone APA5, eBiosciences), Alexa Fluor 700-conjugated anti-CD29 Antibody (1:100, clone HMβ1-1, eBiosciences), APC-Cy7-conjugated anti-EpCAM (1:100, clone G8.8, BioLegend). Cell sorting was performed on a FACSAria sorter using the FACS DiVa software (BD Biosciences). Dead cells were excluded with DAPI; CD45+, CD31+, and CD140a+ cells were excluded (Lin+) before analysis of the YFP+ cells. Sorted BCs (DAPI− LIN− YFP+ CD29high Epcam+) were resuspended in 10 μl of a 50:50 mixture of Matrigel and FBS, then injected into the cleared mammary fat pads of 3–4 weeks NOD/SCID female mice, with each injection containing 10,000 BCs. The mice underwent an 8-week treatment with a FAK inhibitor (PF573228, 10 mg/kg, i.p., twice a week). Post-treatment, the YFP+ mammary glands were imaged using a stereo microscope (ZEISS Axio Zoom.V16), then pre-fixed in 4% PFA at room temperature for 2 h. After three 5 min washes with PBS, the samples were incubated overnight in 30% sucrose in PBS at 4 °C. The tissues were then embedded in OCT for sectioning and subsequent immunofluorescence.
Mammary gland transplantation and microarray
For the transplantation of mammary gland cells, we prepared 10,000 BC cells only as multipotent condition and mixture of 8000 LC cells and 2000 BC cells as unipotent control condition. Cells were sorted after 3 days of transplantation and 50 cells were used for RNA extraction. RNA extraction was performed using Absolutely RNA Nanoprep kit (Stratagene) according to the manufacturer’s recommendations. Total RNA was analyzed using the Mouse Genome 430 PM strip arrays. To normalize the data, the robust multi-array average expression method from the affy package92 was used. Two different biological replicates from transplanted BCs and control were analyzed. Genetic signatures were obtained by considering genes presenting a linear scale of fold change greater than 2.
MG organoids culture
For MG organoid culture, we used the protocol from Jardè, et al.93. Briefly, fat pads of 8-to-9-week-old virgin mice were dissected, and the lymph nodes removed. Tissues were briefly washed in 70% ETOH and manually chopped in 1 mm pieces. The finely minced tissue was transferred to a digestion mix consisting of serum-free Leibovitz’s L15 medium (Gibco) containing 3 mg/ml collagenase A (Sigma) and 1.5 mg/ml trypsin (Sigma). This was incubated for 1 h at 37 °C to liberate epithelial tissue fragments (‘organoids’). Isolated organoids were mixed with 50 μl of phenol red-free Matrigel (BD Biosciences) and seeded in 24-well plates. The basal culture medium contained phenol red-free DMEM/F-12 with penicillin/streptomycin, 10 mM HEPES (Invitrogen), Glutamax (Invitrogen), N2 (Invitrogen) and B27 (Invitrogen). The basal medium was supplemented with Nrg1 (100 ng/ml, R&D), Noggin (100 ng/ml, Peprotech), and R-spondin 1 (100 ng/ml, R&D). 500 μl of supplemented basal culture medium was added per well and organoids were maintained in a 37 °C humidified atmosphere under 5% CO2. After 1 week in culture, mammary organoids were released from the Matrigel by breaking the matrix with a P1000 on ice. Following 2–3 passages of washing and centrifugation at 150 × g for 5 min at 4 °C, mammary cells were resuspended in Matrigel, seeded in 24-well plates, and exposed to previously described culture conditions. To induce the expression of Tomato, mammary organoids were treated with 1 μM of 4-OH-TAM for 48 h and refreshed the new medium every 2–3 days without TAM.
Prostate organoids culture
Generation of prostate organoids was performed as previously described94. Briefly, all mouse prostate tissue including ventral prostate, dorsolateral prostate, and anterior prostate were isolated from K5CreER/RosaYFP mice. The mixed tissue was minced into small pieces in a 6 cm culture plate followed by digestion in 5 mg/ml collagenase type II (Life Technologies, cat. no. 17101-015) with 10 μM Y-27632 dihydrochloride (Abmole Bioscience, cat. no. M1817) 1–1.5 h at 37 °C on a shaking platform. We used 4 ml collagenase for one mouse. Glandular structures were then washed with Advanced DMEM/F12 (Life Technologies, cat. no. 12634-034) and centrifuged at 150 g for 5 mins at 4 °C. After that, structures were digested in 5 ml TrypLE (Life Technologies, cat. no. 12605-010) with the addition Y-27632 10 μM for 10 min at 37 °C. Trypsinized cells were washed and resuspended in growth factor reduced Matrigel (Corning, cat. no. 356231) at the dilution 2:1 (Matrigel: basal medium) and plated in the middle of 24-well tissue culture plate with 40ul drop. The tissue culture plate was flipped upside down in CO2 incubator (5% CO2, 37 °C) for 15 min to allow the Matrigel to solidify. Subsequently, organoids were cultured in 500 μL of Advanced DMEM/F12 supplemented with Glutamax (Life Technologies, cat. no. 35050-068), 10 mM HEPES (Life Technologies, cat. no. 15630-056), Penicillin/Streptomycin (Life Technologies, cat. no. 15140-122), B27 (Life Technologies, cat. no. 17504-044) and 1.25mM N-acetylcysteine (Sigma-Aldrich, cat. no. A9165) and contained following growth factors and components: 50 ng/ml EGF (PeproTech, cat. no. AF-100-15), 100 ng/ml recombinant R-spondin1 (R&D Systems, cat. no. 3474-RS-250), 100 ng/ml recombinant Noggin (Peprotech, cat. no. 250-38) and 200 nM A83-01 (Tocris Bioscience, cat. no. 2939) 1 nM dihydrotestosterone (Sigma, cat. no. A8380) with the addition of 10-μM ROCK inhibitor Y-27632 for the first week after seeding. For induction of Cre recombinase activity and BC tracing, after the organoids were fully established (5 days–1 week), organoids were added with 1 μM of 4-OH-TAM for 48 h and refresh the new medium every 2–3 days without TAM.
Collagen 1 and PEG gel preparation
Mammary gland organoids and the prostate organoids were embedded in different kinds of gels. For the pure Collagen 1 gel, we used the PhotoCol1-IRG kit from Advanced Biomatrix (catalog no 5201), which contains 100 mg of methacrylated Type I bovine collagen, 50 mL of 20 mM acetic acid and 10 mL of neutralization solution. By adding 12.5 mL of acetic acid into Col1 powder, we reconstitute the highest Col1 concentration (8 mg/ml). By serial dilution of the reconstituted Col1 gel, we obtained the other Col1 gel concentrations (4 mg/ml and 2 mg/ml). Neutralization solution was added to the gel mix according to the manufacturing protocol. Gel preparation has been performed on ice, prior to organoid embedding. Isolated organoids were resuspended in 40 μl of the Col1 gel mix and plated into 24-well plate.
The poly(ethylene-glycol)-PEG-based hydrogels, RGD peptide, and XIII factor were nicely provided by the Laboratory of Matthias Lutolf in EPFL and their preparation has been described on previous studies35,76. To embed the organoids in PEG, we resuspended the organoids in basal medium. After spinning them down, the gel mix containing PEG, 10x TRIS buffer (500 mM Tris, 1.5 M NaCl, and 100 mM CaCl2 with pH = 7.4), and RGD (1 mM) were added to the organoid pellet. Just before seeding the gel, factor XIII (10 U/ml) was added and gently mixed with the rest of the gel.
For both Col1 and PEG gels, after seeding, the plate was flipped for 2 min and flipped back before allowing gel polymerization in the incubator at 37 °C humidified atmosphere under 5% CO2 for 30 min. After polymerization, the above-described complete media with 1 μM of 4-OH-TAM for 48 h was added and refreshed with new medium without TAM every 2–3 days.
For the organoid treatment, we added to the medium described above with 2 μM or 5 μM of BTT3033, 10 μM of PF573228, 10 μM of T-5224, and 10 μM of RO-3306. BTT3033 (catalog no. 4724/10) was purchased by R&D, PF573228 (catalog no. S2012), and T-5224 (catalog no. S8966) was purchased by Selleckchem, RO-3306 (CDK inhibitor, catalog no. SML0569) was purchased by Merck. After seeding, organoids were treated with 4-OH-TAM and the inhibitor of interest for 48 h, then the 4-OH-TAM was removed, and the organoids underwent treatment for the following days. For HMB1.1 treatment isolated organoids have been previously incubated for 30 min in the incubator at 37 °C humidified atmosphere under 5% CO2. After incubation, the organoids were washed with DMEM/F12 and plated in the different gels prepared as explained above. HMB1.1 (Biolegend, 102236) and the IgG control (Biolegend, 400940) were added at a ratio of 1:50 in the base medium with 4-OH-TAM for the first 48 h, then 4-OH-TAM was removed, and the organoids underwent treatment for the following days.
The organoids were analyzed after 1 week of treatment for MG organoids and after 5 days of treatment for prostate organoids following the initial addition of TAM.
Immunofluorescence
For collecting organoids from different gels, organoids in Matrigel were collected by cold basal medium, organoids in collagen were digested in collagenase (1 mg/ml, Collagenase A, cat. no. 11088793001) in 37 °C for 5 min followed by collected in cold PBS, organoids in PEG gel were digested in Dispase II (Sigma, cat. no. D4693) followed by collected in cold PBS. After matrix digestion, organoids were collected, fixed for 30 min in 4% PFA, and, after washing, embedded in OCT to be kept at −80 °C.
Sections of 4 µm were cut using an HM560 Microm cryostat (Mikron Instruments). Tissue sections were incubated in blocking buffer (BSA 1%, HS 5%, Triton-X 0.2% in PBS) for 1 h at RT. The different primary antibodies were incubated overnight at 4 °C. Sections were then rinsed in PBS and incubated with the corresponding secondary antibodies diluted at 1:400 in blocking buffer for 1 h at RT. The following primary antibodies were used: anti-K8 (rat, 1:1000, Troma-I, Developmental Studies Hybridoma Bank, University of Iowa), anti-GFP (Goat, 1:500, Abcam, cat. no. ab6673). Anti-K14 (chicken, 1:500, ThermoFisher Scientific cat. no. MA5-11599), Anti-c-Jun (Rabbit, 1:200, Cell signaling, cat. no. 9165S), Anti-Ki67 (Rabbit, 1:200, Abcam, cat. no. ab15580) and Anti-cleaved caspase 3 (Rabbit, 1:100, Cell Signaling, cat. no. 9664S).
The following secondary antibodies, AlexaFluor 488-conjugated anti-rat (1:400; Invitrogen, cat. no. A-11006), AlexaFluor 488-conjugated anti-goat (1:400; Invitrogen, cat. no. A-11055), AlexaFluor 488-conjugated anti-rabbit (1:400; Invitrogen, cat. no. A-21206), Rhodamine Red RRX-anti-rat (1:400, JacksonImmunoResearch, 712-296-153). Cy5-anti-chicken (1:400, JacksonImmunoResearch, 703-175-155), Cy5-anti-Rabbit (1:400, JacksonImmunoResearch, 711-175-152). Nuclei were stained with Hoechst solution (1:2000) and slides were mounted in DAKO mounting medium supplemented with 2.5% Dabco (Sigma).
Immunohistochemistry
MG tissues were fixed in 4% PFA for 2 h at RT, embedded in paraffin, sectioned at 5 μm, and mounted on glass slides according to standard laboratory protocols. The paraffin samples of oncogenic PIK3CA mutant in BCs10 and LC ablation11 in adult mice are previously described. The paraffin sections were deparaffinized and rehydrated through a series of graded ethanol. Antigen retrieval was performed by heating the slides in citrate buffer (pH 6.0) for 10 min using a steam pressure pot. Endogenous peroxidase was blocked using 3% H2O2 in methanol for 10 min at RT. Endogenous avidin and biotin were blocked using the Endogenous Blocking kit (Invitrogen, E21390) for 20 min each at RT. The sections were then incubated overnight at 4 °C with anti-COL1A1 antibody (1:200, Cell Signaling Technology, #72026S). Standard ABC-HRP kit (Vector Laboratories, PK-4001) and ImmPACT DAB (Vector Laboratories, SK-4105) were used for the detection of HRP activity according to the manufacturer’s instructions. Nuclear counterstaining was performed with hematoxylin for 3 min, after which the sections were dehydrated and mounted using SafeMount.
Microscope image acquisition and quantification
Images were acquired at RT using a Zeiss LSM780 confocal microscope fitted on an Axiovert M200 inverted microscope equipped with a C-Apochromat (40X, A. = 1.2) water immersion objective (Carl Zeiss Inc.) or using a Zeiss Axio Imager M1 (Thornwood) fluorescence microscope with a Zeiss Axiocam MR3 camera using Axiovision release 4.6 software. Brightness, contrast, and picture size were adjusted using Adobe Photoshop CS6 and ZEN Microscopy Software. The images were displayed and analyzed with the ZEN software and with Fiji (ImageJ). We manually counted TOM + /YFP+ and K8+ double-positive cells by identifying TOM/YFP+ expression within K8+ cells. Cells were considered positive if they were clearly Hoechst positive and their nuclei were intact.
Intracellular flow cytometry for organoids
Intracellular flow cytometry of BCs and LCs was performed as previously described95. Briefly, organoids were digested with 1 ml of TrypLE (Life Technologies, cat. no. 12605-010) for 10–15 min at 37 °C. After digestion, the cells were washed, filtered through a 40 μm cell strainer, and thoroughly resuspended in 200 μl of fixation/permeabilization solution (BD Cytofix/Cytoperm kit, cat. no. 554714) for 20 min on ice. The cells were then washed and stained in 1x BD Perm/Wash buffer with anti-K5-Alexa Fluor 647 (1:200, Abcam; ab193895), anti-K8-Alexa Fluor 488 (1:200, Abcam; ab192467), or anti-K8-Alexa Fluor 405 (1:200, Abcam; ab210139), followed by incubation on ice for 30 min in the dark. After the staining, the cells were washed in 1x BD Perm/Wash buffer and resuspended in FACS buffer (2% FBS in PBS) for flow cytometric analysis. We gated BCs: K5high K8low and LCs: K5low K8high.
Adenoviral infection of organoids
MG Organoids were generated from Rosa/YFP mice and Junbfl/fl/Juncfl/fl /Rosa-mTmG mice. After one week of establishment, organoids were washed and collected in complete medium containing 1 μl of high titer (8E + 10 to 2E + 10 pfu/ml) Ade-K5Cre virus (Ad5-bk5-Cre, Dr. Anton Berns, Netherlands Cancer Institute) obtained from Viral Vector Core at University of Iowa. The mixture was gently mixed and transferred to a pre-Matrigel-coated 24-well plate, then incubated at 37 °C for 12–14 h. After the infection, the organoids on Matrigel surface were collected and embedded in different ECM substrates, including Matrigel, collagen I (4 mg/ml), soft (2.5% PEG + RGD), and stiff (7% PEG + RGD) hydrogels for one week. The organoids were then harvested, fixed, and sectioned for immunofluorescence as previously described.
Quantitative RT-PCR
Total RNA from organoids was extracted using the Direct-zol RNA Microprep Kit (Zymo) following the manufacturer’s protocol. Genomic DNA was eliminated via on-column DNase digestion. cDNA synthesis was performed with the RevertAid First Strand cDNA Synthesis Kit (ThermoFisher). Quantitative real-time PCR (qRT-PCR) was conducted using SYBR Green Supermix (Applied Bioscience) on a Light Cycler 96 (Roche). Gene expression levels were normalized to housekeeping gene Gapdh for relative quantification. The following probes from Eurogentec were used: Gapdh forward: AGGTCGGTGTGAACGGATTTG, Gapdh reverse: TGTAGACCATGTAGTTGAGGTCA, Fos forward: CGGGTTTCAACGCCGACTA, Fos reverse: TTGGCACTAGAGACGGACAGA, Jun forward: CCTTCTACGACGATGCCCTC, Jun reverse: GGTTCAAGGTCATGCTCTGTTT.
Single-cell RNA sequencing
Single cells from the mammary gland organoid from Matrigel (MATR), soft (SOFT), collagen (COL1), and stiff (STIFF) conditions were dissociated and labeled with TotalSeq-A hashtag oligonucleotide (HTO) antibodies96. Matrigel and soft, collagen and stiff were pooled and loaded onto each channel of the Chromium Single Cell 3′ microfluidic chips (V2-chemistry, PN-120232, 10X Genomics) and barcoded with a 10X Chromium controller according to the manufacturer’s recommendations (10X Genomics).
RNA from the barcoded cells was subsequently reverse transcribed, followed by amplification, shearing 5′ adapter, and sample index attachment. The libraries were prepared using the Chromium Single Cell 3′ Library Kit (V3-chemistry, PN-120233, 10X Genomics) and sequenced on an Illumina Novaseq 6000 (paired-end 100 bp reads).
Single-cell transcriptomic data analysis
Sequencing reads were aligned and annotated with the mm10-2020-A reference dataset as provided by 10X Genomics and demultiplexed using CellRanger (v.5.0.0)97 with default parameters. Data was demultiplexed based on data from cell hashing, by the function HTODemux() included in Seurat R package (v.4.2.0)98 with default parameters. Further downstream analyses were carried out individually for each of the four samples (MATR, SOFT, COL1, and STIFF). After de-multiplexing, the cells annotated as either doublet or negative on HTO global classification are excluded for further QC and downstream analysis.
Quality control and downstream analysis were performed using the Seurat R package (v.4.2.0). For each sample, all the cells passed the following criteria: showed expression of more than 2000 and less than 7000 unique genes and had less than 10% UMI counts belonging to mitochondrial sequences. Read counts were normalized using the NormalizeData() function of Seurat, with parameter ‘normalization.method = “LogNormalize” and scale.factor = 10000’. A PCA for each sample was calculated using the scaled expression data of the 2000 most variable genes (identified as outliers on a mean/variability plot, implemented in the FindVariableGenes() function). UMAP calculation and Louvain graph-based clustering were performed for each sample using Seurat (default parameters) with the respective 30 PCA results as input99.
The clusters which did not express the epithelial marker (Epcam) but expressed high levels of stromal markers (Vim, Zeb2, Fscn), fibroblast (Pdgfra, Fn1, Col1a1, Fbn1), and macrophage markers (Ptprc, Cd86, Cd68, Ccr1) were excluded and dimensionality were recalculated after the exclusion of these cells. The final clustering resolution for individual datasets was set to 0.5, after testing a range from 0.3 to 0.9. This resolution best reflected the biological heterogeneity that emerged from the different cell types consisting of the mammary gland, identifying a BC (highly enriched in Krt14, Krt5, Acta2), LC ER+ (highly enriched in Esr1, Foxa1, Ly6a), LC ER- (highly enriched in Elf5, Kit, Cd14) and general LC (highly enriched in Krt8, Cdh1, Epcam) populations.
A Wilcoxon rank-sum test was used to define marker genes for each cluster using the FindAllMarkers() function. Benjamini–Hochberg FDR correction for potential cluster marker genes across all samples using the p.adjust method in R and only markers expressed in at least 25% of cells of the cluster, having an average log2-fold change of at least 0.25 were reported.
For visualization and comparison between different samples, we integrated the MATR, SOFT, COL1, and STIFF samples using the Seurat package’s standard CCA-MNN-based data integration workflow. Feature selection was performed using the FindVariableFeatures() function from Seurat with default parameters, selecting the 2000 most variable genes. Canonical correlation analysis (CCA) followed by integration anchors selection was then performed on the selected features using the FindIntegrationAnchors() function from Seurat, taking the first 30 dimensions from the CCA into account, as described on the standard workflow of Seurat. These anchors were then used to integrate the data with the IntegrateData() function. Following the annotation of clusters, clusters sharing the same cell identities were merged into a single cluster. This merging process was performed after confirming that there were no differences in terms of gene expression between these clusters with the same cell identity. The assessment of gene expression was based on both well-known marker genes and marker genes determined using the Wilcoxon rank-sum test.
On the individual and integrated datasets, to identify cell proliferation stages, the S-phase and G2/M-phase scores were computed by CellCycleScoring() function, implemented in the Seurat.
BC/LC-specific markers were determined by filtering marker genes identified in the MATR sample, retaining only marker genes with log2-fold changes higher than 0.6, adjusted p-value is less than 0.01, and that were expressed in at least 25% of their respective population. The proportion of expressed lineage-specific markers for each cell was computed by counting the number of specific LC/BC markers over the total number of LC/BC-specific markers. This proportion for differences in sensitivity is adjusted by modeling the linear relationship between the proportion of markers detected and the total number of genes detected for each cell, by MASS R Package (v.7.3-60.0.1)100.
Gene regulatory network analysis was performed using pySCENIC47 with default parameters and the run was done on python 3.7.10101. To correct for stochastic variation, the SCENIC pipeline was run 10 times for each dataset and the average AUC (Area Under the Curve) scores were calculated for further analysis. Differentially activated regulons for each cluster were determined by performing a Wilcoxon rank-sum test on the regulon AUC values of cells in the various clusters, p-values were FDR-adjusted using the Benjamini–Hochberg method, and regulons with an adjusted p-value less than 0.01 were considered as differentially activated. On the integrated data, to define the regulons activated on certain clusters and certain samples, Fisher’s combined test was performed by MADAM R package (v.1.2.2)102 and adjusted p-value less than 0.01 were counted as differentially activated on one cluster than the others.
Pseudotime ordering of cells was calculated using slingshot (v.2.0.0)44. To prevent the distortion of pseudotime due to cell proliferation, we excluded clusters composed of proliferating cells from this analysis. We checked the robustness of the resulting trajectories by performing the analysis on PCA and UMAP reductions, these different reductions and permutations did not affect the described trajectories.
Differentially expressed genes (DEG) between different cell populations in individual samples and comparison between different samples on same cluster were computed as two-sided Wilcoxon ranked sum tests using the FindMarkers() function in Seurat and p-values were FDR adjusted for the total number of comparisons. For the running DEG analysis, ‘min.pct = 0.25, logfc.threshold = 0.25, only.pos=T’ were given threshold. Also, to calculate the set of DEGs specifically regulated on certain sample compared to the others on the same cluster, the comparison between two samples was taken for Fisher’s combined test and the genes with an adjusted p-value less than 0.01 were considered as differentially regulated on one specific sample.
To define the cell marker genes of BC primed LC differentiation and HY BC/ER− for comparison to the hybrid state found in the other conditions associated with multipotency, we applied FindAllMarkers() function of Seurat package, ‘min.pct = 0.25, logfc.threshold = 0.35, only.pos=T’ were given threshold. The genes with adjusted P-value higher than 0.01 are excluded from the marker gene lists.
To calculate the genes whose gene expression profiles were either increasing or decreasing by stiffness, linear regression using NEBULA103 (version 1.2.0), which allows for the use of negative binomial generalized linear mixed effects modeling, were applied to the integrated data. For calculating the effect of stiffness, we fitted a model of the form: ~ stiffness (MATR = 0.1kpa, SOFT = 0.3kpa, COL1 = 0.9kpa, STIFF = 3.3kpa) + Collagen (1 for COL1, 0 for the rest). Since this model did not have term considering cell type information, the population for the linear regression were selected with subset() function of Seurat package by giving the cell type on ‘idents’ parameter.
Statistics and reproducibility
All experiments presented in the manuscript were repeated in at least three independent experiments or biological replicates. All statistical analyses were based on these replicates and were conducted using GraphPad Prism v.8.00, R (v.4.2.0), and Microsoft Excel. For pairwise comparisons between two groups, two-sided unpaired t-tests were performed. For comparisons involving more than two groups, one-way ANOVA followed by Tukey’s test were used. The number of mice, experiments, cells, and corresponding p-values are provided in the figures and figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are available within the article and its supplementary files. Single-cell RNAseq data generated in this study have been deposited in the Gene Expression Omnibus (GEO) database under accession code GSE243335. RNA-seq data generated in this study have been deposited in the GEO database under accession code GSE243336 (prostate duct BC). Microarray data generated in this study have been deposited in the GEO database under accession code GSE243337. Previously published RNA-seq data that were re-analyzed here are available under accession codes GSE222786 (prostate tip BCs) and GSE127975 (LC ablated). Previously published microarray data that was re-analyzed here are available under accession code GSE69290 (Gene upregulated on PIK3CA). Correspondence and requests for materials should be addressed to C.B. Source data are provided with this paper.
References
Toivanen, R. & Shen, M. M. Prostate organogenesis: tissue induction, hormonal regulation and cell type specification. Development 144, 1382–1398 (2017).
Lloyd-Lewis, B., Harris, O. B., Watson, C. J. & Davis, F. M. Mammary stem cells: premise, properties, and perspectives. Trends Cell Biol. 27, 556–567 (2017).
Ousset, M. et al. Multipotent and unipotent progenitors contribute to prostate postnatal development. Nat. Cell Biol. 14, 1131–1138 (2012).
Van Keymeulen, A. et al. Distinct stem cells contribute to mammary gland development and maintenance. Nature 479, 189–193 (2011).
Wuidart, A. et al. Quantitative lineage tracing strategies to resolve multipotency in tissue-specific stem cells. Genes Dev. 30, 1261–1277 (2016).
Wuidart, A. et al. Early lineage segregation of multipotent embryonic mammary gland progenitors. Nat. Cell Biol. 20, 666–676 (2018).
Choi, N., Zhang, B., Zhang, L., Ittmann, M. & Xin, L. Adult murine prostate basal and luminal cells are self-sustained lineages that can both serve as targets for prostate cancer initiation. Cancer Cell 21, 253–265 (2012).
Lilja, A. M. et al. Clonal analysis of Notch1-expressing cells reveals the existence of unipotent stem cells that retain long-term plasticity in the embryonic mammary gland. Nat. Cell Biol. 20, 677–687 (2018).
Tika, E., Ousset, M., Dannau, A. & Blanpain, C. Spatiotemporal regulation of multipotency during prostate development. Development 146, https://doi.org/10.1242/dev.180224 (2019).
Van Keymeulen, A. et al. Reactivation of multipotency by oncogenic PIK3CA induces breast tumour heterogeneity. Nature 525, 119–123 (2015).
Centonze, A. et al. Heterotypic cell-cell communication regulates glandular stem cell multipotency. Nature 584, 608–613 (2020).
Shackleton, M. et al. Generation of a functional mammary gland from a single stem cell. Nature 439, 84–88 (2006).
Stingl, J. et al. Purification and unique properties of mammary epithelial stem cells. Nature 439, 993–997 (2006).
Deome, K. B., Faulkin, L. J. Jr., Bern, H. A. & Blair, P. B. Development of mammary tumors from hyperplastic alveolar nodules transplanted into gland-free mammary fat pads of female C3H mice. Cancer Res. 19, 515–520 (1959).
Lawson, D. A., Xin, L., Lukacs, R. U., Cheng, D. & Witte, O. N. Isolation and functional characterization of murine prostate stem cells. Proc. Natl Acad. Sci. USA 104, 181–186 (2007).
Wang, Z. A. et al. Lineage analysis of basal epithelial cells reveals their unexpected plasticity and supports a cell-of-origin model for prostate cancer heterogeneity. Nat. Cell Biol. 15, 274–283 (2013).
Goldstein, A. S., Huang, J., Guo, C., Garraway, I. P. & Witte, O. N. Identification of a cell of origin for human prostate cancer. Science 329, 568–571 (2010).
Koren, S. et al. PIK3CA(H1047R) induces multipotency and multi-lineage mammary tumours. Nature 525, 114–118 (2015).
Devos, H., Zoidakis, J., Roubelakis, M. G., Latosinska, A. & Vlahou, A. Reviewing the regulators of COL1A1. Int. J. Mol. Sci. 24, https://doi.org/10.3390/ijms241210004 (2023).
Henriksen, K. & Karsdal, M. A. Biochemistry of Collagens, Laminins and Elastin: Structure, Function and Biomarkers. 3rd edn, 1–11 (Elsevier Inc., 2016).
Bächinger, H. P., Mizuno, K., Vranka, J. A. & Boudko, S. P. In Comprehensive Natural Products II (eds Hung-Wen Liu & Lew Mander) 469–530 (Elsevier, 2010).
Nerger, B. A. et al. Local accumulation of extracellular matrix regulates global morphogenetic patterning in the developing mammary gland. Curr. Biol. 31, 1903–1917.e1906 (2021).
Brownfield, D. G. et al. Patterned collagen fibers orient branching mammary epithelium through distinct signaling modules. Curr. Biol. 23, 703–709 (2013).
Docheva, D., Padula, D., Schieker, M. & Clausen-Schaumann, H. Effect of collagen I and fibronectin on the adhesion, elasticity and cytoskeletal organization of prostate cancer cells. Biochem. Biophys. Res. Commun. 402, 361–366 (2010).
Hall, C. L., Dai, J., van Golen, K. L., Keller, E. T. & Long, M. W. Type I collagen receptor (alpha 2 beta 1) signaling promotes the growth of human prostate cancer cells within the bone. Cancer Res 66, 8648–8654 (2006).
Luthold, C., Hallal, T., Labbé, D. P. & Bordeleau, F. The extracellular matrix stiffening: a trigger of prostate cancer progression and castration resistance? Cancers. 14, https://doi.org/10.3390/cancers14122887 (2022).
Giafaglione, J. M. et al. Prostate lineage-specific metabolism governs luminal differentiation and response to antiandrogen treatment. Nat. Cell Biol. 25, 1821–1832 (2023).
Leitinger, B. Transmembrane collagen receptors. Annu Rev. Cell Dev. Biol. 27, 265–290 (2011).
Geiger, B., Spatz, J. P. & Bershadsky, A. D. Environmental sensing through focal adhesions. Nat. Rev. Mol. Cell Biol. 10, 21–33 (2009).
Mitra, S. K., Hanson, D. A. & Schlaepfer, D. D. Focal adhesion kinase: in command and control of cell motility. Nat. Rev. Mol. Cell Biol. 6, 56–68 (2005).
Zhao, X. & Guan, J. L. Focal adhesion kinase and its signaling pathways in cell migration and angiogenesis. Adv. Drug Deliv. Rev. 63, 610–615 (2011).
Weng, S., Shao, Y., Chen, W. & Fu, J. Mechanosensitive subcellular rheostasis drives emergent single-cell mechanical homeostasis. Nat. Mater. 15, 961–967 (2016).
Dupont, S. et al. Role of YAP/TAZ in mechanotransduction. Nature 474, 179–183 (2011).
Paszek, M. J. et al. Tensional homeostasis and the malignant phenotype. Cancer Cell 8, 241–254 (2005).
Gjorevski, N. & Lutolf, M. P. Synthesis and characterization of well-defined hydrogel matrices and their application to intestinal stem cell and organoid culture. Nat. Protoc. 12, 2263–2274 (2017).
Lutolf, M. P., Gilbert, P. M. & Blau, H. M. Designing materials to direct stem-cell fate. Nature 462, 433–441 (2009).
Rezakhani, S., Gjorevski, N. & Lutolf, M. P. Extracellular matrix requirements for gastrointestinal organoid cultures. Biomaterials 276, 121020 (2021).
Arnold, M. et al. Activation of integrin function by nanopatterned adhesive interfaces. Chemphyschem 5, 383–388 (2004).
Giraddi, R. R. et al. Single-cell transcriptomes distinguish stem cell state changes and lineage specification programs in early mammary gland development. Cell Rep. 24, 1653–1666.e1657 (2018).
Bach, K. et al. Differentiation dynamics of mammary epithelial cells revealed by single-cell RNA sequencing. Nat. Commun. 8, 2128 (2017).
Li, C. M. et al. Aging-associated alterations in mammary epithelia and stroma revealed by single-cell RNA sequencing. Cell Rep. 33, 108566 (2020).
Pal, B. et al. Construction of developmental lineage relationships in the mouse mammary gland by single-cell RNA profiling. Nat. Commun. 8, 1627 (2017).
Butler, A., Hoffman, P., Smibert, P., Papalexi, E. & Satija, R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018).
Street, K. et al. Slingshot: cell lineage and pseudotime inference for single-cell transcriptomics. BMC Genomics 19, 477 (2018).
Dravis, C. et al. Epigenetic and transcriptomic profiling of mammary gland development and tumor models disclose regulators of cell state plasticity. Cancer Cell 34, 466–482.e466 (2018).
Saba, J. A., Liakath-Ali, K., Green, R. & Watt, F. M. Translational control of stem cell function. Nat. Rev. Mol. Cell Biol. 22, 671–690 (2021).
Aibar, S. et al. SCENIC: single-cell regulatory network inference and clustering. Nat. Methods 14, 1083–1086 (2017).
Zvelebil, M. et al. Embryonic mammary signature subsets are activated in Brca1-/- and basal-like breast cancers. Breast Cancer Res. 15, R25 (2013).
Dravis, C. et al. Sox10 regulates stem/progenitor and mesenchymal cell states in mammary epithelial cells. Cell Rep. 12, 2035–2048 (2015).
Chung, C. Y. et al. Single-cell chromatin analysis of mammary gland development reveals cell-state transcriptional regulators and lineage relationships. Cell Rep. 29, 495–510.e496 (2019).
Emsley, J., Knight, C. G., Farndale, R. W., Barnes, M. J. & Liddington, R. C. Structural basis of collagen recognition by integrin alpha2beta1. Cell 101, 47–56 (2000).
Freeman, S. A. et al. Lipid-gated monovalent ion fluxes regulate endocytic traffic and support immune surveillance. Science 367, 301–305 (2020).
Kanamoto, T. et al. Integrin α2β1 plays an important role in the interaction between human articular cartilage-derived chondrocytes and atelocollagen gel. Sci. Rep. 11, 1757 (2021).
Liu, Z. et al. Heterogeneous responses to mechanical force of prostate cancer cells inducing different metastasis patterns. Adv. Sci. 7, 1903583 (2020).
Zheng, Y. et al. Matrix stiffness triggers lipid metabolic cross-talk between tumor and stromal cells to mediate bevacizumab resistance in colorectal cancer liver metastases. Cancer Res. 83, 3577–3592 (2023).
Zhang, H. et al. Gain-of-function RHOA mutations promote focal adhesion kinase activation and dependency in diffuse gastric cancer. Cancer Discov. 10, 288–305 (2020).
Aikawa, Y. et al. Treatment of arthritis with a selective inhibitor of c-Fos/activator protein-1. Nat. Biotechnol. 26, 817–823 (2008).
Shaulian, E. & Karin, M. AP-1 in cell proliferation and survival. Oncogene 20, 2390–2400 (2001).
Ferone, G. et al. SOX2 is the determining oncogenic switch in promoting lung squamous cell carcinoma from different cells of origin. Cancer Cell 30, 519–532 (2016).
Engler, A. J., Sen, S., Sweeney, H. L. & Discher, D. E. Matrix elasticity directs stem cell lineage specification. Cell 126, 677–689 (2006).
Watt, F. M. & Huck, W. T. Role of the extracellular matrix in regulating stem cell fate. Nat. Rev. Mol. Cell Biol. 14, 467–473 (2013).
Vining, K. H. & Mooney, D. J. Mechanical forces direct stem cell behaviour in development and regeneration. Nat. Rev. Mol. Cell Biol. 18, 728–742 (2017).
Lu, C. P. et al. Identification of stem cell populations in sweat glands and ducts reveals roles in homeostasis and wound repair. Cell 150, 136–150 (2012).
Bruno, R. D. et al. Mammary extracellular matrix directs differentiation of testicular and embryonic stem cells to form functional mammary glands in vivo. Sci. Rep. 7, 40196 (2017).
Boulanger, C. A., Mack, D. L., Booth, B. W. & Smith, G. H. Interaction with the mammary microenvironment redirects spermatogenic cell fate in vivo. Proc. Natl Acad. Sci. USA 104, 3871–3876 (2007).
Booth, B. W. et al. The mammary microenvironment alters the differentiation repertoire of neural stem cells. Proc. Natl Acad. Sci. USA 105, 14891–14896 (2008).
Donjacour, A. A. & Cunha, G. R. Assessment of prostatic protein secretion in tissue recombinants made of urogenital sinus mesenchyme and urothelium from normal or androgen-insensitive mice. Endocrinology 132, 2342–2350 (1993).
Chung, L. W. & Cunha, G. R. Stromal-epithelial interactions: II. Regulation of prostatic growth by embryonic urogenital sinus mesenchyme. Prostate 4, 503–511 (1983).
Glukhova, M. A. & Streuli, C. H. How integrins control breast biology. Curr. Opin. Cell Biol. 25, 633–641 (2013).
Streuli, C. H. Integrins as architects of cell behavior. Mol. Biol. Cell 27, 2885–2888 (2016).
Raymond, K., Deugnier, M. A., Faraldo, M. M. & Glukhova, M. A. Adhesion within the stem cell niches. Curr. Opin. Cell Biol. 21, 623–629 (2009).
DuFort, C. C., Paszek, M. J. & Weaver, V. M. Balancing forces: architectural control of mechanotransduction. Nat. Rev. Mol. Cell Biol. 12, 308–319 (2011).
Nelson, C. M. & Bissell, M. J. Of extracellular matrix, scaffolds, and signaling: tissue architecture regulates development, homeostasis, and cancer. Annu Rev. Cell Dev. Biol. 22, 287–309 (2006).
Zeltz, C. & Gullberg, D. The integrin-collagen connection-a glue for tissue repair? J. Cell Sci. 129, 653–664 (2016).
Eirew, P. et al. A method for quantifying normal human mammary epithelial stem cells with in vivo regenerative ability. Nat. Med 14, 1384–1389 (2008).
Gjorevski, N. et al. Designer matrices for intestinal stem cell and organoid culture. Nature 539, 560–564 (2016).
Levental, K. R. et al. Matrix crosslinking forces tumor progression by enhancing integrin signaling. Cell 139, 891–906 (2009).
Taddei, I. et al. Beta1 integrin deletion from the basal compartment of the mammary epithelium affects stem cells. Nat. Cell Biol. 10, 716–722 (2008).
Pervolarakis, N. et al. Integrated single-cell transcriptomics and chromatin accessibility analysis reveals regulators of mammary epithelial cell identity. Cell Rep. 33, 108273 (2020).
Sarate, R. M. et al. Dynamic regulation of tissue fluidity controls skin repair during wound healing. Cell 187, 5298–5315.e5219 (2024).
Pérez-González, A., Bévant, K. & Blanpain, C. Cancer cell plasticity during tumor progression, metastasis and response to therapy. Nat. Cancer 4, 1063–1082 (2023).
Lin, S., Margueron, R., Charafe-Jauffret, E. & Ginestier, C. Disruption of lineage integrity as a precursor to breast tumor initiation. Trends Cell Biol. 33, 887–897 (2023).
Malinova, A., Veghini, L., Real, F. X. & Corbo, V. Cell lineage infidelity in PDAC progression and therapy resistance. Front. Cell Dev. Biol. 9, 795251 (2021).
Davies, A. et al. An androgen receptor switch underlies lineage infidelity in treatment-resistant prostate cancer. Nat. Cell Biol. 23, 1023–1034 (2021).
Provenzano, P. P. et al. Collagen reorganization at the tumor-stromal interface facilitates local invasion. BMC Med. 4, 38 (2006).
Boyd, N. F., Martin, L. J., Yaffe, M. J. & Minkin, S. Mammographic density and breast cancer risk: current understanding and future prospects. Breast Cancer Res. 13, 223 (2011).
Northey, J. J. et al. Stiff stroma increases breast cancer risk by inducing the oncogene ZNF217. J. Clin. Invest 130, 5721–5737 (2020).
Hasebe, T., Tsuda, H., Tsubono, Y., Imoto, S. & Mukai, K. Fibrotic focus in invasive ductal carcinoma of the breast: a histopathological prognostic parameter for tumor recurrence and tumor death within three years after the initial operation. Jpn J. Cancer Res. 88, 590–599 (1997).
Conklin, M. W. et al. Aligned collagen is a prognostic signature for survival in human breast carcinoma. Am. J. Pathol. 178, 1221–1232 (2011).
Behrens, A. et al. Impaired postnatal hepatocyte proliferation and liver regeneration in mice lacking c-jun in the liver. Embo j. 21, 1782–1790 (2002).
Kenner, L. et al. Mice lacking JunB are osteopenic due to cell-autonomous osteoblast and osteoclast defects. J. Cell Biol. 164, 613–623 (2004).
Gautier, L., Cope, L., Bolstad, B. M. & Irizarry, R. A. affy-analysis of Affymetrix GeneChip data at the probe level. Bioinformatics 20, 307–315 (2004).
Jardé, T. et al. Wnt and neuregulin1/ErbB signalling extends 3D culture of hormone responsive mammary organoids. Nat. Commun. 7, 13207 (2016).
Drost, J. et al. Organoid culture systems for prostate epithelial and cancer tissue. Nat. Protoc. 11, 347–358 (2016).
Crowell, P. D. et al. Expansion of luminal progenitor cells in the aging mouse and human prostate. Cell Rep. 28, 1499–1510.e1496 (2019).
Stoeckius, M. et al. Cell Hashing with barcoded antibodies enables multiplexing and doublet detection for single cell genomics. Genome Biol. 19, 224 (2018).
Zheng, G. X. et al. Massively parallel digital transcriptional profiling of single cells. Nat. Commun. 8, 14049 (2017).
Hao, Y. et al. Integrated analysis of multimodal single-cell data. Cell 184, 3573–3587.e3529 (2021).
Becht, E. et al. Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol, https://doi.org/10.1038/nbt.4314 (2018).
Venables, W. N. & Ripley, B. D. Modern Applied Statistics with S. 4th edn, (Springer New York, NY, 2002).
Rossum, G. V. & Drake, F. L. Python 3 Reference Manual. (CreateSpace, 2009).
Kugler, K. G., Mueller, L. A. & Graber, A. MADAM—an open source meta-analysis toolbox for R and Bioconductor. Source Code Biol. Med 5, 3 (2010).
He, L. et al. NEBULA is a fast negative binomial mixed model for differential or co-expression analysis of large-scale multi-subject single-cell data. Commun. Biol. 4, 629, (2021).
Acknowledgements
We thank the ULB animal facility and ULB genomic core facility (F. Libert and A. Lefort); J.M.Vanderwinden and LiMif for the help with confocal microscopy and B. Lloyd-Lewis for help with the organoids culture. This work was supported by the ERC and the FNRS. C.J. is supported by a long-term EMBO Postdoctoral Fellowships (ALTF 982-2021) and Foundation Against Cancer (2023-041). A.C. is supported by the FNRS/FRIA. Y.S. is supported by Télévie. C.B. is supported by WELBIO, FNRS, Télévie, Fond Erasme, Fondation Contre le Cancer, ULB Foundation, European Research Council, and the Foundation Baillet Latour. A.Sifrim., is supported by KULeuven (SymBioSys—C14/18/092), the Fondation Contre le Cancer (2015-143).
Author information
Authors and Affiliations
Contributions
C.J., A.C., Y.S., and C.B. designed the experiments and performed data analysis. C.J. and A.C. performed most of the biological experiments. Y.S. and A.Sifrim. performed the bioinformatic analysis. C.J. and E.T. performed experiments and data analysis on prostate glands and organoids. A.Chris., S.R., and M.L. generated PEG gel. Z.Z. and G.B. provided technical help. C.J. and C.D. performed FACS and sorting experiments. A.V.K. performed the transplantation experiments. C.B. wrote the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Andrew Goldstein, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, C., Centonze, A., Song, Y. et al. Collagen signaling and matrix stiffness regulate multipotency in glandular epithelial stem cells in mice. Nat Commun 15, 10482 (2024). https://doi.org/10.1038/s41467-024-54843-5
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-54843-5
This article is cited by
-
Mechanically tunable fiber-based hydrogel activates PIEZO1–integrin axis for enhanced bone repair
Journal of Nanobiotechnology (2025)
-
Collagen-based biomaterials: recent advances on regulating cell migration and correlated biomedical applications
Collagen and Leather (2025)
-
Podocalyxin and ciliary neurotrophic factor receptor are novel components of the surfaceome of chondrogenic cells
Cell Communication and Signaling (2025)
-
ECM formation and degradation during fibrosis, repair, and regeneration
npj Metabolic Health and Disease (2025)
-
Innate immunity and the NF-κB pathway control prostate stem cell plasticity, reprogramming and tumor initiation
Nature Cancer (2025)