Abstract
Lysosome/vacuole-mediated intracellular degradation pathways, collectively known as autophagy, play crucial roles in the maintenance and regulation of various cellular functions. However, little is known about the relationship between different modes of autophagy. In the budding yeast Saccharomyces cerevisiae, nitrogen starvation triggers both macronucleophagy and micronucleophagy, in which nuclear components are degraded via macroautophagy and microautophagy, respectively. We previously revealed that Atg39-mediated macronucleophagy is important for cell survival under nitrogen starvation; however, the underlying mechanism remains unknown. Here, we reveal that defective Atg39-mediated macronucleophagy leads to the hyperactivation of micronucleophagy, resulting in the excessive transport of various nuclear components into the vacuole. Micronucleophagy occurs at the nucleus–vacuole junction (NVJ). We show that nuclear membrane proteins localized to the NVJ, including Nvj1, which is responsible for micronucleophagy, are degraded via macronucleophagy. Therefore, defective Atg39-mediated macronucleophagy results in the accumulation of Nvj1, which contributes to micronucleophagy enhancement. Blocking micronucleophagy almost completely suppresses cell death caused by the absence of Atg39, whereas enhanced micronucleophagy correlates with death in Atg39-mutant cells under nitrogen starvation. These results suggest that macronucleophagy modulates micronucleophagy in order to prevent the excess removal of nuclear components, thereby maintaining nuclear and cellular homeostasis during nitrogen starvation.
Similar content being viewed by others
Introduction
Autophagy is a collective term for the pathways that mediate degradation of intracellular components in lysosomes or vacuoles and includes macroautophagy1,2, microautophagy3,4, chaperone-mediated autophagy5, and DNautophagy/RNautophagy6. These pathways differ in terms of the mechanisms by which degradation targets are transported into lysosomes/vacuoles. Macroautophagy sequesters its targets within a double-membrane vesicle called the autophagosome, which subsequently fuses with the lysosome/vacuole, where the contents are degraded. In microautophagy, the lysosomal/endosomal/vacuolar membrane invaginates and pinches off to take up cytoplasmic material into the lumen. Both macroautophagy and microautophagy can selectively degrade specific cytoplasmic constituents, including organelles such as lipid droplets, the endoplasmic reticulum, and the nucleus7,8,9. These diverse autophagy pathways play important roles in the maintenance and regulation of various cellular functions, and their dysregulation is thought to lead to human diseases3,5,10. It is also known that some of these pathways are active under the same conditions and even mediate degradation of the same targets. However, the interplay between different autophagy pathways is poorly understood.
Recent studies have revealed that components of the nucleus, including nucleus-derived vesicles (NDVs), nuclear pore complexes, lamins, a sirtuin, and even DNA, are selectively degraded via macroautophagy and microautophagy in fungi and mammals, which are known as macronucleophagy and micronucleophagy (also referred to as piecemeal microautophagy of the nucleus), respectively11,12,13,14,15,16. In the budding yeast Saccharomyces cerevisiae, nitrogen deprivation stimulates both macronucleophagy and micronucleophagy11,17,18. In macronucleophagy, the outer nuclear membrane protein Atg39 recruits core Atg proteins and initiates autophagosome formation in the vicinity of the nucleus and also deforms the nuclear envelope, thereby generating an NDV11,19,20. Atg39 also binds to the autophagosomal membrane protein Atg8 in order to load the NDV into the autophagosome. Micronucleophagy in S. cerevisiae takes place at a membrane contact called the nucleus–vacuole junction (NVJ), which is established through the interaction between the outer nuclear membrane protein Nvj1 and the vacuolar membrane protein Vac821. The nuclear envelope forms a bleb at the NVJ in association with the invagination of the vacuolar membrane; upon fission of these membranes, an NDV surrounded by the vacuolar membrane is released into the vacuolar lumen.
We previously reported that cells lacking Atg39 exhibit abnormal nuclear morphology and die earlier than wild-type cells under nitrogen starvation, demonstrating the physiological significance of Atg39-mediated macronucleophagy. However, how this autophagy pathway supports cell viability was not known. In the present study, we reveal that modulation of micronucleophagy is a critical role for macronucleophagy that determines the survival or death of S. cerevisiae cells during nitrogen starvation.
Results
Micronucleophagy is abnormally enhanced in Atg39-mediated macronucleophagy-deficient cells under nitrogen starvation
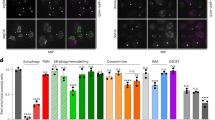
More than half of all macronucleophagy-defective cells (atg39Δ cells) die after nitrogen starvation for 10 days, whereas most wild-type cells survive11 (Supplementary Fig. 1a, b). At an earlier stage of nitrogen starvation (3 days), most atg39Δ cells are still alive but the nucleus exhibits a flattened morphology, which was visualized by mCherry-fused Pus1 (nucleoplasm protein) and mNeonGreen-fused Sec63 (endoplasmic reticulum/nuclear envelope protein) in the present study11 (Supplementary Fig. 1c). We noticed that Sec63-mNeonGreen-positive vesicles containing Pus1-mCherry appeared separately from the nucleus in these cells. These vesicles disappeared when wild-type Atg39, but not a macronucleophagy-defective mutant of Atg39 (atg39W8A V11A 52-59A or atg39AIM 11BR mut)11, was expressed from a plasmid (Supplementary Fig. 1d), suggesting that these NDVs appear in association with defects in macronucleophagy. When mTurquoise2 was fused to Nvj1 in the wild-type cells, NVJs were observed between the nucleus and vacuoles visualized by Pus1-mCherry and mNeonGreen-fused Vph1 (vacuolar membrane protein), respectively (Fig. 1a and Supplementary Fig. 1e). We found that NVJs expanded between flattened nuclei and vacuoles in atg39Δ cells under nitrogen starvation and that NDVs were associated with these NVJs and surrounded by Nvj1-mTurquoise2 signals, suggesting that these NDVs were formed via micronucleophagy. Quantification analysis revealed that these micronucleophagic vesicles significantly increased in both number and size in atg39Δ cells compared with wild-type cells (Fig. 1b, c and Supplementary Figs. 1f and 2a, b). Although these NDVs occasionally moved around within the vacuole, most remained attached to the NVJ (Fig. 1a, Supplementary Fig. 1c). However, fluorescence loss in photobleaching (FLIP) assay revealed that the NVJ-attached vesicles were also separated from the nucleus (Supplementary Fig. 2c, d). Moreover, in addition to Pus1, other nuclear proteins such as Tal1 (nucleoplasmic transaldolase), Hta2 (histone H2A), Nop56 (small nucleolar ribonucleoprotein component), Rpa49 (RNA polymerase I subunit), and Fob1 (rDNA replication fork barrier site-binding protein) were found within micronucleophagic vesicles formed in atg39Δ cells, although the extent and frequency differed among the proteins (Fig. 1d, e and Supplementary Fig. 1g). Taken together, these results suggested that micronucleophagy is abnormally enhanced in macronucleophagy-deficient cells, resulting in the excessive transport of various nuclear components into the vacuole under nitrogen starvation.
a, d Fluorescence microscopy images of yeast cells after nitrogen starvation for 3 days. Scale bars, 5 µm (a); 2 µm (d). Arrowheads, micronucleophagic vesicles (a) or micronucleophagic vesicles containing mTurquoise2-tagged nuclear proteins (d). b, e The results shown in (a, d) were quantified, and the numbers of Nvj1-positive micronucleophagic vesicles per cell (a) or Nvj1-positive micronucleophagic vesicles containing mTurquoise2-tagged nuclear proteins (d) are shown as the mean ± s.d. (n = 3 independent experiments) in (b), (e), respectively. c The size of micronucleophagic vesicles was measured as depicted in Supplementary Fig. 2a, b. Quantification results are shown as radius r ± s.d. of vesicles observed in cells after nitrogen starvation for 3 days from three independent experiments. **P < 0.01; ***P < 0.001 (unpaired two-tailed Student’s t test (b, e), Mann-Whitney U test (c), source data and the exact P values are provided as a Source Data file).
Micronucleophagy activity correlates with death in Atg39-mediated macronucleophagy-deficient cells
Next, we examined whether enhanced micronucleophagy is related to cell death in atg39Δ cells. We found that knockout of NVJ1 completely blocked micronucleophagy in atg39Δ cells and recovered the morphology of the nucleus to that of wild-type cells (Fig. 2a and Supplementary Fig. 3a, b). Indeed, NVJ1 knockout almost completely suppressed cell death in atg39Δ cells under nitrogen starvation (Fig. 2b), suggesting that the enhancement of micronucleophagy is a major cause of cell death in macronucleophagy-defective mutant cells.
a Fluorescence microscopy images of cells after nitrogen starvation for 3 days. Arrowheads, micronucleophagic vesicles. Scale bar, 2 μm. b Cells were incubated in nitrogen-starvation medium for 6 or 10 days. Dead cells stained with phloxine B were counted; the percentages are shown as the mean ± s.d. (n = 3 independent experiments). c Functional regions of Nvj1. INM, inner nuclear membrane; TM, transmembrane domain. d Schematic illustration of reconstituted NVJs. e–g Yeast cells with reconstituted NVJs were starved for a nitrogen source and examined by fluorescence microscopy. Black arrowheads, nucleus-vacuole junctions; white arrowheads, micronucleophagic vesicles. Scale bars, 5 μm. h Yeast cells were incubated in nitrogen-starvation medium and examined by fluorescence microscopy to observe Nvj1-positive micronucleophagic vesicles. The numbers of those vesicles per cell are shown graphically as the mean ± s.d. (n = 3 independent experiments). i Viability of the indicated yeast strains after nitrogen starvation for 10 days was examined by phloxine B staining. The percentages of dead cells are shown as the mean ± s.d. (n = 3 independent experiments). NS not significant; **P < 0.01; ***P < 0.001 (unpaired two-tailed Student’s t test, source data and the exact P values are provided as a Source Data file).
Deletion of NVJ1 abrogates the NVJ, leading to the mislocalization of NVJ proteins such as Osh1, which mediates lipid transfer between the nuclear envelope and vacuolar membrane, as well as Tsc13, an enzyme that is responsible for very long chain fatty acid synthesis22,23. Therefore, the loss of NVJ functions other than micronucleophagy might suppress cell death in atg39Δ nvj1Δ cells. To eliminate this possibility, we aimed to reconstruct NJVs defective in micronucleophagy in atg39Δ cells. Nvj1 lacking the Vac8-binding region (Nvj11-206) (Fig. 2c) was reported to be defective in NVJ formation and micronucleophagy17,21. Therefore, we first engineered the chromosomal NVJ1 gene to express wild-type Nvj1 or Nvj11-206 C-terminally fused with BFP (for fluorescence observation) and a green fluorescent protein (GFP) nanobody (GNb) (Nvj1/Nvj11-206-BFP-GNb) and confirmed that Osh1 and Tsc13 localized to the NVJ together with Nvj1 and Vac8 before and after nitrogen starvation in NVJ1-BFP-GNb cells (Supplementary Fig. 3c), whereas the NVJ localization of these proteins was completely abolished in nvj11-206-BFP-GNb or nvj11-206-BFP-GNb atg39Δ cells under both conditions (Supplementary Fig. 3d, e). Subsequently, GFP was fused to the C terminus of the vacuolar membrane protein Gtr2 (EGO complex component) or Ivy1 (Ypt7 effector). The tight interaction between GNb and GFP in these fusion proteins was expected to reconstitute NVJs (Fig. 2d). Indeed, Nvj11-206-BFP-GNb formed NVJs in association with Gtr2-GFP or Ivy1-GFP (Fig. 2e and Supplementary Fig. 3f, g), and Tsc13 and Osh1 successfully localized to these NVJs (Fig. 2f, g and Supplementary Fig. 3h–k). These reconstituted NVJs differed in their ability to mediate micronucleophagy (formation of vesicles positive for Nvj11-206-BFP-GNb and Gtr2-GFP or Ivy1-GFP and containing Tal1-mCherry). Whereas Gtr2-based NVJs more strongly induced micronucleophagy compared with wild-type NVJs in atg39Δ cells, Ivy1-based NVJs caused micronucleophagy in atg39Δ cells to an extent similar to wild-type (ATG39) cells (Fig. 2e, h and Supplementary Fig. 3f). Subsequently, we examined the viability of these cells with reconstituted NVJs during nitrogen starvation. In good agreement with the extent of micronucleophagy, Gtr2-based and Ivy1-based NVJs exacerbated and suppressed cell death in atg39Δ cells, respectively (Fig. 2i and Supplementary Fig. 3l). Thus, micronucleophagy activity supported by reconstituted NVJs correlates with the rate of cell death in the absence of Atg39-mediated macronucleophagy, reinforcing our notion that enhanced micronucleophagy reduces the viability of macronucleophagy-defective cells under nitrogen starvation.
Nuclear NVJ proteins including Nvj1 are degraded via macronucleophagy
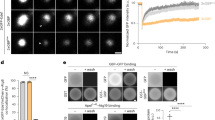
Next, we asked why micronucleophagy is enhanced in atg39Δ cells. In immunoblotting analysis, we found that Nvj1-GFP, which was expressed by fusing the GFP gene to chromosomal NVJ1, yielded GFP fragments, an indicator of vacuolar degradation of GFP-fused proteins24, in wild-type cells during nitrogen starvation (Fig. 3a). These GFP fragments were almost completely eliminated in atg39Δ cells and atg39AIM 11BR mut cells, and the level of full-length Nvj1-GFP increased accordingly (Fig. 3a–c, Supplementary Fig. 4a). We also observed that Nvj1-GFP was sequestered along with Atg39-mCherry within autophagosomes or autophagic bodies (Vac8-negative structures within the vacuole) accumulated in ypt7Δ or pep4Δ cells, which are defective in autophagosome-vacuole fusion or vacuolar degradation25,26, respectively (Fig. 3d, Supplementary Fig. 4b). The Feret’s diameter of autophagic bodies in pep4Δ cells was about half the mean diameter of micronucleophagic vesicles in wild-type cells (Fig. 1c (shown in radius) and Supplementary Fig. 4d). Collectively, these results suggest that Nvj1 is degraded by macronucleophagy, and thus defective macronucleophagy results in the accumulation of Nvj1.
a–c Vacuolar degradation of Nvj1-GFP was examined by immunoblotting, using an anti-GFP antibody (a). GFP′, GFP fragments generated by degradation of Nvj1-GFP in the vacuole. The intensities of the Nvj1-GFP and GFP′ bands in (a) were quantified; the percentages of the GFP′ value in the sum of the Nvj1-GFP and GFP′ values (Nvj1-GFP degradation) (b) and the ratio of the Nvj1-GFP values to that in wild-type cells (c) are shown as the mean ± s.d. (n = 3 independent experiments). d Fluorescence microscopy images of cells before and after nitrogen starvation for 24 h. Arrowhead, Nvj1-mNeonGreen signal contained in the Atg39-mTurquoise2-positive autophagic body in pep4Δ cells. Experiments were repeated independently three times with similar results. Scale bar, 2 μm. e, f Vacuolar degradation of Vac8-GFP, GFP-Osh1, and Tsc13-GFP under nitrogen starvation was examined and quantified as described in (a–c). The values in (f) are the mean ± s.d. (n = 3 independent experiments). g, h Yeast cells were subjected to nitrogen starvation and observed under a fluorescence microscope (g). Arrowheads, micronucleophagic vesicles. Scale bar, 2 μm. The numbers of Nvj1-positive micronucleophagic vesicles per cell are shown as the mean ± s.d. (n = 3 independent experiments) (h). *P < 0.05; **P < 0.01; ***P < 0.001 (unpaired two-tailed Student’s t test, source data and the exact P values are provided as a Source Data file).
We also examined the degradation of other NVJ proteins and found that the nuclear membrane protein Tsc13 was degraded depending on Atg39, but the vacuolar membrane proteins Vac8 and Osh1 were not (GFP-Osh1 yielded GFP fragments, which, however, were not dependent on Atg39) (Fig. 3e). Accordingly, the level of full-length Tsc13-GFP after nitrogen starvation was increased by ATG39 knockout but the levels of full-length Vac8-GFP and GFP-Osh1 were not (Fig. 3e, f). These results suggest that macronucleophagy degrades nuclear NVJ proteins such as Nvj1, and thus these proteins increase in macronucleophagy-deficient cells. Unlike Nvj1-GFP, degradation of Tsc13-GFP was not strictly dependent on Atg39 (Fig. 3e). This is likely due to the localization of Tsc13 to the endoplasmic reticulum in addition to the nuclear envelope and NVJ (Supplementary Fig. 3c).
Nvj1 accumulation contributes to the enhancement of micronucleophagy in Atg39-deficient cells
Next, we sought to determine whether the accumulation of Nvj1 might be a cause of micronucleophagy hyperactivation in atg39Δ cells. When the promoter of chromosomal NVJ1-GFP was replaced with that from the ADH1 gene in the wild-type cells (ADHp-NVJ1-GFP), the protein level increased about 3-fold (Fig. 3a–c), and the number of micronucleophagic vesicles per cell increased about 2-fold (Fig. 3g, h, Supplementary Fig. 4c). Thus, an increase in Nvj1 levels is suggested to be a cause of micronucleophagy enhancement in atg39Δ cells. We also showed that overexpression of Nvj1-GFP significantly promoted cell death after 10 days of starvation, even in cells expressing wild-type Atg39 (Fig. 2b). These results are consistent with the notion that micronucleophagy enhanced by Nvj1 accumulation contributes to cell death in macronucleophagy-defective cells. However, despite the level of Nvj1-GFP being higher in ADHp-NVJ1-GFP cells than in atg39Δ cells (Fig. 3a, c), both micronucleophagy enhancement and cell death were more pronounced in the latter cells (Figs. 1e, 2b, and 3h). In addition, NVJ1 overexpression did not induce NVJ expansion and abnormal nuclear morphology, which were observed in atg39Δ cells. These results suggest that NVJ1 overexpression alone is insufficient to reproduce the phenotypes observed in atg39Δ cells. It should also be noted that although NVJ1 overexpression enhanced micronucleophagy after 3 days of nitrogen starvation (Fig. 3h), cell death remained comparable to that of wild-type cells even after 6 days (Fig. 2b) (see Discussion).
atg39Δ cells die through a process different from that in atg1Δ cells under nitrogen starvation
How does enhanced micronucleophagy lead to cell death in atg39Δ cells under nitrogen starvation? As a related issue, previous studies have investigated how atg1Δ cells, in which all the macroautophagy pathways (including macronucleophagy) are disrupted, die during nitrogen starvation27,28. The results suggested that atg1Δ cells cannot supply free amino acids due to defects in non-selective cytoplasm degradation and therefore fail to support the expression of the genes required for the maintenance of mitochondrial functions, including those that encode respiratory chain components and enzymes for scavenging reactive oxygen species (ROS)27. Following the loss of mitochondrial respiration, atg1Δ cells die in media that are acidified during starvation27. We found that, as with atg1Δ cells, starvation-induced death in atg39Δ cells was suppressed by buffering the medium (Supplementary Fig. 5a). However, unlike atg1Δ cells, a decrease in ROS scavengers was not observed in atg39Δ cells (Supplementary Fig. 5b). Moreover, although a lower expression of respiratory chain proteins was observed in atg39Δ cells, the extent was much lower compared with atg1Δ cells, and importantly, this phenotype was not improved by NVJ1 knockout (Supplementary Fig. 5c). We also investigated whether micronucleophagy is enhanced in atg1Δ cells under nitrogen starvation. Since atg1Δ cells begin to die after 24 hours of starvation, we used buffered starvation medium to prevent this cell death. While enhanced micronucleophagy was observed in atg39Δ cells after 3 days in the buffered medium, no micronucleophagy induction was seen in atg1Δ cells (Supplementary Fig. 5d). The same was true for atg2Δ cells, which are also defective in all the macroautophagy pathways (Supplementary Fig. 5e). In contrast, micronucleophagy was moderately increased in atg11Δ cells, in which non-selective cytoplasm degradation via autophagy occurs normally, but macronucleophagy is partially impaired (Supplementary Fig. 5e)11. Consistent with these observations, the level of Nvj1 was lower in atg1Δ cells than in wild-type cells, probably due to decreased protein synthesis caused by amino acid shortage, while Nvj1 accumulated in atg11Δ cells (Supplementary Fig. 5f). Additionally, we showed that starvation-induced death in atg1Δ cells was not suppressed by NVJ1 knockout (Supplementary Fig. 5g). Taken together, these results suggest that micronucleophagy-related cell death in atg39Δ cells proceeds through a process distinct from mitochondrial dysfunction-triggered cell death in atg1Δ cells.
Discussion
We previously reported that Atg39-mediated macronucleophagy is important for cell survival during nitrogen starvation. However, how this macroautophagy pathway contributes to cellular homeostasis under nitrogen starvation was unknown. Meanwhile, previous studies have reported the molecular mechanisms and physiological roles of diverse modes of autophagy. However, the interplay between various autophagy pathways remains poorly understood. In the present study, we revealed that the modulation of micronucleophagy is a critical role for macronucleophagy, providing a physiological and mechanistic linkage between macroautophagy and microautophagy.
We showed that micronucleophagic vesicles increase in atg39Δ cells starved for nitrogen, suggesting that micronucleophagy is stimulated in the absence of macronucleophagy. Although retarded degradation within the vacuole could also increase micronucleophagic vesicles, vacuolar degradation activity seems unimpaired in atg39Δ cells because macroautophagy pathways other than macronucleophagy normally occurs in these cells11. However, we noticed that Nvj1-GFP hardly generated GFP fragments in atg39Δ cells (Fig. 3a, b), even though it was transported to the vacuole via enhanced micronucleophagy as observed by fluorescence microscopy (Fig. 1a, b). This was also the case for the nucleoplasm protein Tal1 fused with GFP (Fig. 1d, e, Supplementary Fig. 6a). Micronucleophagic vesicles are observed even in cells with functional vacuoles, suggesting that these vesicles, which are surrounded by the NVJ region of the vacuolar membrane, are stable (very slowly degraded) within vacuoles. Indeed, when micronucleophagy was induced by nitrogen starvation and then stopped by shifting the cells to nutrient-rich media, micronucleophagic vesicles retained over 8 hours (Supplementary Fig. 6b, c). This stability of micronucleophagic vesicles is likely to be the reason why GFP-fused nuclear proteins transported to the vacuole via micronucleophagy do not efficiently yield GFP fragments. The stable nature of micronucleophagic vesicles also suggests that vacuolar transport of nuclear components, rather than their degradation, leads to cell death in macronucleophagy-deficient cells.
The absence of Atg39-mediated macronucleophagy strongly induces micronucleophagy (Fig. 1a, b, Supplementary Fig. 1f), and our results suggest that the mechanism of this micronucleophagy enhancement can be explained in part by an increase in Nvj1 levels (Figs. 3a, c, g, h, and 4). We demonstrated that macronucleophagy degrades nuclear NVJ proteins including Nvj1, and thus these proteins accumulate in Atg39-mediated macronucleophagy-deficient cells (Fig. 3a, c, e, f). In macronucleophagy, Atg39 forms clusters in the nuclear envelope, where NDVs are generated and sequestered within autophagosomes19. Because these Atg39 clusters are often observed at the edge of the NVJ, nuclear NVJ proteins are thought to be efficiently incorporated into NDVs and subsequently degraded via macronucleophagy. NVJ1 overexpression stimulated micronucleophagy during nitrogen starvation (Fig. 3g, h), supporting the idea that increased Nvj1 enhances micronucleophagy in macronucleophagy-deficient cells. However, NVJ1 overexpression did not fully phenocopy ATG39 deletion, including NVJ expansion and abnormal nuclear morphology. Therefore, in addition to Nvj1 accumulation, other factors such as the accumulation of other nuclear NVJ proteins as well as NVJ expansion and abnormal nuclear morphology, are thought to be involved in the stimulation of micronucleophagy in the absence of macronucleophagy (Fig. 4). These factors may also promote cell death along with micronucleophagy enhancement in atg39Δ cells. These factors might take ~10 days to reach a level that impacts the viability of NVJ1-overexpressing cells, and this may explain why significant cell death was not observed after 3 day-starvation, although micronucleophagy enhancement was already evident at that time point (Figs. 2b, 3g, h).
In wild-type cells starved for a nitrogen source, Atg39-mediated macronucleophagy is induced, leading to the degradation of nuclear NVJ proteins such as Nvj1, and micronucleophagy is maintained at a moderate level (left). If Atg39-mediated macronucleophagy is impaired, Nvj1-mediated micronucleophagy is strongly enhanced (right). This micronucleophagy enhancement can be explained in part by the accumulation of nuclear NVJ proteins (macronucleophagy substrates) and may also involve nuclear flattening and NVJ expansion caused by the absence of macronucleophagy. Enhanced micronucleophagy results in the excessive transport of various nuclear materials to the vacuole, and the removal of specific components or the disruption of normal nuclear processes is thought to lead to cell death in Atg39-mediated macronucleophagy-defective mutants under nitrogen starvation.
In a previous study, it was suggested that Atg39 is also involved in micronucleophagy, based on the observations that Atg39 clusters, along with Atg8 and Atg11, were observed around vacuole invaginations at NVJs, and that ATG39 knockout decreased Nvj1 degradation29. However, in this study, we found that Nvj1-dependent micronucleophagy is rather enhanced in the absence of Atg39, demonstrating that Atg39 is not essential for micronucleophagy. Although it remains possible that Atg39 might play an auxiliary role in micronucleophagy, the Atg39 clusters observed in the previous study may instead represent the site where nuclear NVJ components including Nvj1 are sequestered within autophagosomes during macronucleophagy.
NVJ1 knockout not only blocked micronucleophagy but also recovered the morphology of the nucleus in atg39Δ cells (Supplementary Fig. 3b), suggesting that excess micronucleophagy and/or expanded NVJs lead to a flattened nuclear morphology in atg39Δ cells. Conversely, it is also possible that flattening of the nucleus is involved in NVJ expansion in atg39Δ cells. However, the mechanisms by which NVJ expansion and nuclear flattening occur in macronucleophagy-defective cells and the relationship between these changes and micronucleophagy enhancement remain to be elucidated.
Blocking micronucleophagy by NVJ1 knockout almost completely restored cell viability in atg39Δ cells under nitrogen starvation (Fig. 2b). In contrast, overexpression of Nvj1 stimulated micronucleophagy and increased cell death (Fig. 2b). Furthermore, two different NVJs, which were constituted by artificially binding Nvj11-206 to Gtr2 or Ivy1 via the GNb-GFP interaction, increased or decreased micronucleophagy and exaggerated or suppressed cell death in atg39Δ cells, respectively (Fig. 2h, i). Collectively, these results suggest that enhanced micronucleophagy is a major cause of cell death under nitrogen starvation in the absence of macronucleophagy (Fig. 4). Notably, NVJ expansion and nuclear flattening were observed in cells with Ivy1-based NVJs, which survived nitrogen starvation comparably to wild-type cells (Fig. 2e, g, i). This observation is consistent with the idea that enhanced micronucleophagy, rather than these nucleus-related morphological changes, is responsible for starvation-induced death in macronucleophagy-defective cells. The difference in micronucleophagy activity between Gtr2- and Ivy1-based NVJs may arise from the structural/functional difference between these proteins; Ivy1 has an inverse BAR domain capable of generating membrane curvature30,31, and Gtr2, along with another GTPase, Gtr1, forms a large complex with the Ego1-Ego2-Ego3 complex, activating TORC132.
Previous studies described that the nucleolus tends to be located near the NVJ and nucleolar proteins are preferentially degraded via micronucleophagy, whereas rDNA moves away from the NVJ to escape from degradation upon micronucleophagy induction (TORC1 inactivation)33. It has been discussed that degradation of nucleolar components via micronucleophagy contributes to reducing cellular translation activity under nutrient-deprived conditions, although nvj1Δ cells do not exhibit significant defects in their viability under those conditions18. On the other hand, the preference in degradation of nuclear components via macronucleophagy remains to be investigated, but previous results suggested that nucleolar proteins are not efficiently degraded by macronucleophagy11. Therefore, nuclear proteins to be degraded seem to differ between macro- and micronucleophagy, and the balance between these two nucleophagy pathways may be important for nuclear homeostasis during nitrogen starvation. This may be the reason why enhanced micronucleophagy results in loss of cell viability rather than suppresses defects caused by the absence of macronucleophagy in atg39Δ cells.
Our results suggest that the process of cell death in atg39Δ cells is distinct from that in atg1Δ cells. In atg1Δ cells, the inability to degrade cytoplasmic proteins results in the depletion of free amino acids under nitrogen starvation, leading to mitochondrial respiration deficiency and subsequent cell death in a medium acidified during starvation27. We showed that unlike atg39Δ cells, atg1Δ cells did not exhibit enhanced micronucleophagy, and cell death was not suppressed by NVJ1 knockout, suggesting that micronucleophagy does not contribute to cell death in atg1Δ cells. We also showed that the level of Nvj1 was rather reduced in atg1Δ cells, likely due to amino acid depletion caused by defects in non-selective autophagic degradation of cytoplasmic proteins (Supplementary Fig. 5f), and this may at least partly explain why micronucleophagy is not enhanced in atg1Δ cells even though macronucleophagy is abolished in these cells. In contrast, our data suggest that micronucleophagy enhancement is responsible for cell death in atg39Δ cells. Although unlike atg1Δ cells, the respiratory function of mitochondria was not suggested to be severely impaired in atg39Δ cells, cell death in these cells was also suppressed by medium buffering (Supplementary Fig. 5a). In atg39Δ cells, excessive transport of nuclear components to the vacuole via enhanced micronucleophagy likely leads to cell death. Vacuolar transport of specific nuclear components may be lethal in acidified nitrogen-starvation media. Alternatively, excessive removal of nuclear components may have a more global impact on gene expression, rendering yeast cells vulnerable to that harsh environment. Future research will elucidate how macronucleophagy maintains cellular homeostasis during nitrogen starvation through the regulation of micronucleophagy.
Methods
Yeast strains, media and plasmids
The yeast strains used in this study were all derived from SEY621034 and are listed in Table S1. Knockout and tagging of chromosomal genes were performed based on a standard method, using PCR-amplified DNA cassettes35. Yeast cells were cultured at 30 °C in YPD medium [1% yeast extract (Becton, Dickinson, and Company), 2% peptone (Becton, Dickinson, and Company), and 2% glucose (Wako)], or SD + CA medium [0.17% yeast nitrogen base without amino acids and ammonium sulfate (Becton, Dickinson, and Company), 0.5% ammonium sulfate (Nacalai Tesque), 0.5% casamino acid (Becton, Dickinson, and Company), and 2% glucose] with appropriate supplements [0.002% adenine sulfate (Wako), 0.008% tryptophan (Wako), and 0.002% uracil (Wako)]. To induce macronucleophagy, cells were grown to mid-log phase (OD600 = ~1.5) in SD + CA medium and transferred to SD-N medium [0.17% yeast nitrogen base without amino acids and ammonium sulfate and 2% glucose].
The plasmids used in this study were listed in Table S2. To construct the pRS426-atg39W8A V11A 52-59A−6HA, residues 52–59 of Atg39 were replaced with alanine using pRS426-atg39W8A V11A−6HA, a plasmid generated in our previous work11, by site-directed mutagenesis with Quik Change kit (Agilent Technologies).
Immunoblotting analysis
Harvested cells were treated with 20% trichloroacetic acid (Nacalai Tesque) on ice for 15 min, centrifuged at 15,000 × g for 5 min, and washed with cold acetone. Cells were then pelleted by centrifugation at 15,000 × g for 5 min, and dried at room temperature for 10 min. The cell pellets were dissolved in (the value of OD600 units×50) µL of sodium dodecyl sulfate (SDS) sample buffer [100 mM Tris-HCl (pH 7.5), 2% SDS, 20 mM dithiothreitol (Nacalai Tesque), and 10% glycerol], and incubated at 65 °C for 10 min. Urea sample buffer [37.5 mM MOPS-NaOH (pH 6.8), 2% SDS, 100 mM DTT, and 4 M urea] was used to prepare samples for immunoblotting analysis of Atg39. Cells were then disrupted using a FastPrep-24 (MP Biomedicals) and 0.5-mm zirconia beads. Proteins in these samples were separated by SDS–polyacrylamide gel electrophoresis, and transferred to polyvinylidene difluoride membranes (Millipore) using Transblot Turbo Blotting System (Bio-Rad). The membranes were incubated with primary antibodies against GFP (Clontech, 632381; 1:2000 dilution) or HA (Roche, 11867431001; 1:1000 dilution), followed by secondary antibodies: anti-mouse IgG conjugated with horseradish peroxidase (Jackson ImmunoResearch, 315-035-003) or anti-rat IgG conjugated with horseradish peroxidase (Jackson ImmunoResearch, 112-035-003). Immunoblots were visualized using an ImageQuant LAS 4000 CCD imager (GE Healthcare) or the FUSION FX/EvolutionCapt Version 18.0.12.0 system (Vilber, Eberhardzell, Germany).
Fluorescence microscopy
A Delta Vision Elite microscope system (GE Healthcare) equipped with a scientific CMOS camera (pco.edge 5.5; PCO AG) and either a ×60 (PLAPON, NA/1.42; Olympus) or ×100 objective lens (UPlanSApo, NA/1.40; Olympus) was used. For imaging, 25 z-stack images (Fig. 2b, i; Supplementary Figs. 1b, 3l, and 5a, d) or 15 z-stack images (Figs. 1a, d, 2a, h and 3d, g; Supplementary Figs. 1c–g, 2b, 3a, b, 4b, c, 5d, e, and 6b) at 0.2 μm intervals were acquired and pre-processed using SoftWoRx 7.0.0 software (GE Healthcare).
Reconstituted NVJs (Fig. 2e–g; Supplementary Fig. 3e, f and 3h–k) were observed using an inverted fluorescence microscope (IX83; Olympus) equipped with an electron-multiplying CCD camera (ImagEM C9100-13; Hamamatsu Photonics), a ×150 objective lens (UAPON 150XOTIRF, NA/1.45; Olympus), and 405-nm, 488-nm, and 588-nm lasers (Coherent). Fluorescence was filtered with a dichroic mirror (Olympus) reflecting these wavelengths and split into two channels using the DV2 multichannel imaging system (Photometrics) with a Di02-R594-25×36 dichroic mirror (Semrock). Images were acquired using MetaMorph Version 7.10.0.119 (Molecular Devices, San Jose, CA).
The FLIP experiments (Supplementary Fig. 2c, d) were conducted using a confocal laser scanning microscope (LSM 780; Zeiss), equipped with an AxioCam MRm CCD monochromatic camera (Zeiss), a ×63 objective lens (NA/1.4 M27 OIL DIC Plan-Apochromat; Zeiss), and a 32-channel Quasar detector (32 GaAsP-PMT detector array; Zeiss). During the FLIP experiments, after capturing the initial image, the selected area was photobleached with the laser power set to 100%. Images were then captured at 5-second intervals, with photobleaching performed after every 3 scans. ZEN2012 software was used for image acquisition and pre-processing.
Yeast cells were harvested by centrifugation and observed immediately. Image processing and analysis were conducted using Fiji (ImageJ)36.
Electron microscopy
Electron microscopy was conducted by Tokai-EM Inc. using rapid freezing and freeze-substitution methods. Yeast cells were sandwiched between copper disks and rapidly frozen in liquid propane at −175 °C. Subsequently, they were freeze-substituted with ethanol containing 2% glutaraldehyde and 0.5% tannic acid. The samples underwent dehydration through three 30 min ethanol washes, followed by infiltration with propylene oxide twice for 30 minutes each. The samples were then placed in a 1:1 mixture of propylene oxide and resin (Quetol-651, Nisshin EM) for 3 hours, and transferred to 100% resin overnight. Ultra-thin sections were prepared from these samples, mounted on copper grids, and stained with 2% uranyl acetate at room temperature for 15 minutes. The sections were rinsed with distilled water and subsequently stained with Lead stain solution (Sigma-Aldrich) at room temperature for 3 minutes. The stained samples were observed under a transmission electron microscope (JEM-1400Plus; JEOL Ltd.) equipped with a CCD camera (EM-14830RUBY2, JEOL Ltd.).
Cell viability assay
Yeast cells were grown to mid-log phase in SD + CA medium, transferred to SD-N medium, and incubated for the indicated time periods. Dead cells were stained with 2.5 μg/mL phloxine B for about 1 min at room temperature. The cells were harvested by centrifugation and then washed with and resuspended in distilled water for observation. The proportion of phloxine B-positive cells observed under a fluorescence microscope with an RFP filter to total cells observed under a bright field was calculated.
Statistics and reproducibility
All experiments were independently repeated at least three times with similar results. No statistical methods were used to predetermine the sample size. For quantification, Nvj1-positive vesicles and phloxine B-positive dead cells were manually counted across three independent experiments. The number of cells examined varied, with a minimum of 150 cells analyzed per experiment. The actual number of cells examined in each experiment is provided in the Source Data file. Two out of 22 cells analyzed in Supplementary Fig. 2d were excluded from the analysis, as they exhibited connectivity between the nucleus and the micronucleophagic vesicle. No other data were excluded from the analysis. Statistical analysis of microscopy data and immunoblots was conducted using Microsoft Excel and GraphPad Prism. Unpaired two-tailed Student’s t tests were applied, except for Fig. 1c, under the assumption of normal data distribution, though this was not formally tested. For Fig. 1c, the Mann-Whitney U test was applied as the data were confirmed not to be normally distributed using the Shapiro-Wilk test.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data supporting the findings of this study are available from the authors upon request. The blots and graphs (Figs. 1b, c, e, 2b, h, i, and 3a–f and Supplementary Figs. 1b, f, 2d, 3l, 4a, d, 5b–f, and 6a, c) generated in this study are provided in the Source Data file. Source data are provided with this paper.
References
Ohsumi, Y. Historical landmarks of autophagy research. Cell Res. 24, 9–23 (2014).
Yang, Z. & Klionsky, D. J. Eaten alive: a history of macroautophagy. Nat. Cell Biol. 12, 814–822 (2010).
Schuck, S. Microautophagy – distinct molecular mechanisms handle cargoes of many sizes. J. Cell Sci. 133, jcs246322 (2020).
Oku, M. & Sakai, Y. Three distinct types of microautophagy based on membrane dynamics and molecular machineries. BioEssays 40, 1800008 (2018).
Kaushik, S. & Cuervo, A. M. The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 (2018).
Hase, K. et al. Cytosolic domain of SIDT2 carries an arginine-rich motif that binds to RNA/DNA and is important for the direct transport of nucleic acids into lysosomes. Autophagy 16, 1974–1988 (2020).
Schott, M. B., Rozeveld, C. N., Weller, S. G. & McNiven, M. A. Lipophagy at a glance. J. Cell Sci. 135, jcs259402 (2022).
Mochida, K. & Nakatogawa, H. ER‐phagy: selective autophagy of the endoplasmic reticulum. EMBO Rep. 23, e55192 (2022).
Li, Z. & Nakatogawa, H. Degradation of nuclear components via different autophagy pathways. Trends Cell Biol. 32, 574–584 (2022).
Levine, B. & Kroemer, G. Biological functions of autophagy genes: a disease perspective. Cell 176, 11–42 (2019).
Mochida, K. et al. Receptor-mediated selective autophagy degrades the endoplasmic reticulum and the nucleus. Nature 522, 359–362 (2015).
Lee, C. W. et al. Selective autophagy degrades nuclear pore complexes. Nat. Cell Biol. 22, 159–166 (2020).
Tomioka, Y. et al. TORC1 inactivation stimulates autophagy of nucleoporin and nuclear pore complexes. J. Cell Biol. 219, e201910063 (2020).
Dou, Z. et al. Autophagy mediates degradation of nuclear lamina. Nature 527, 105–109 (2015).
Li, Y. et al. Nuclear accumulation of UBC9 contributes to SUMOylation of lamin A/C and nucleophagy in response to DNA damage. J. Exp. Clin. Cancer Res. 38, 67 (2019).
Xu, C. et al. SIRT1 is downregulated by autophagy in senescence and ageing. Nat. Cell Biol. 22, 1170–1179 (2020).
Kvam, E. & Goldfarb, D. S. Nucleus-vacuole junctions and piecemeal microautophagy of the nucleus in S. cerevisiae. Autophagy 3, 85–92 (2007).
Roberts, P. et al. Piecemeal microautophagy of nucleus in Saccharomyces cerevisiae. Mol. Biol. Cell 14, 129–141 (2003).
Mochida, K. et al. Atg39 links and deforms the outer and inner nuclear membranes in selective autophagy of the nucleus. J. Cell Biol. 221, e202103178 (2022).
Chandra, S. et al. Atg39 selectively captures inner nuclear membrane into lumenal vesicles for delivery to the autophagosome. J. Cell Biol. 220, e202103030 (2021).
Pan, X. et al. Nucleus-vacuole junctions in Saccharomyces cerevisiae are formed through the direct interaction of Vac8p with Nvj1p. Mol. Biol. Cell 11, 2445–2457 (2000).
Levine, T. P. & Munro, S. Dual targeting of Osh1p, a yeast homologue of oxysterol-binding protein, to both the Golgi and the nucleus-vacuole junction. Mol. Biol. Cell 12, 1633–1644 (2001).
Kohlwein, S. D. et al. Tsc13p is required for fatty acid elongation and localizes to a novel structure at the nuclear-vacuolar interface in Saccharomyces cerevisiae. Mol. Cell. Biol. 21, 109–125 (2001).
Torggler, R., Papinski, D. & Kraft, C. Assays to monitor autophagy in Saccharomyces cerevisiae. Cells 6, 23 (2017).
Kirisako, T. et al. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147, 435–446 (1999).
Takeshige, K., Baba, M., Tsuboi, S., Noda, T. & Ohsumi, Y. Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J. Cell Biol. 119, 301–311 (1992).
Suzuki, S. W., Onodera, J. & Ohsumi, Y. Starvation induced cell death in autophagy-defective yeast mutants is caused by mitochondria dysfunction. PLoS One 6, 17412 (2011).
Onodera, J. & Ohsumi, Y. Autophagy is required for maintenance of amino acid levels and protein synthesis under nitrogen starvation. J. Biol. Chem. 280, 31582–31586 (2005).
Otto, F. B. & Thumm, M. Mechanistic dissection of macro- and micronucleophagy. Autophagy 17, 626–639 (2021).
Itoh, Y., Kida, K., Hanawa-Suetsugu, K. & Suetsugu, S. Yeast Ivy1p is a putative I-BAR-domain protein with pH-sensitive filament forming ability in vitro. Cell Struct. Funct. 41, 1–11 (2016).
Nepal, B., Sepehri, A. & Lazaridis, T. Mechanism of negative membrane curvature generation by I-BAR domains. Structure 29, 1440–1452.e4 (2021).
Powis, K. et al. Crystal structure of the Ego1-Ego2-Ego3 complex and its role in promoting Rag GTPase-dependent TORC1 signaling. Cell Res. 25, 1043–1059 (2015).
Golam Mostofa, M. et al. CLIP and cohibin separate rDNA from nucleolar proteins destined for degradation by nucleophagy. J. Cell Biol. 217, 2675–2690 (2018).
Robinson, J. S., Klionsky, D. J., Banta, L. M. & Emr, S. D. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol. Cell. Biol. 8, 4936–4948 (1988).
Janke, C. et al. A versatile toolbox for PCR-based tagging of yeast genes: new fluorescent proteins, more markers and promoter substitution cassettes. Yeast 21, 947–962 (2004).
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Acknowledgements
We thank the members of our laboratory for materials, discussions, and technical and secretarial support as well as the Biomaterials Analysis Division of the Open Facility Center at Institute of Science Tokyo for DNA sequencing. This work was supported in part by KAKENHI Grants-in-Aid for Scientific Research (JP24H00553, JP19H05708, JP17H01430, and JP23K20044) (to H.N.) from the Ministry of Education, Culture, Sports, Science and Technology of Japan, an AMED Grant (JP21gm1410004) (to H.N.), Takeda Science Foundation (to H.N.), and a STAR Grant funded by the Tokyo Tech Fund (to H.N.).
Author information
Authors and Affiliations
Contributions
Z.L., K.M., and H.N. designed the project, Z.L. performed the experiments, and Z.L. and H.N. wrote the manuscript. All authors analyzed and discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Li, Z., Mochida, K. & Nakatogawa, H. Macronucleophagy maintains cell viability under nitrogen starvation by modulating micronucleophagy. Nat Commun 15, 10670 (2024). https://doi.org/10.1038/s41467-024-55045-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-024-55045-9