Abstract
Apical and basal dendrites of pyramidal neurons receive anatomically and functionally distinct inputs, implying compartment-level functional diversity during behavior. To test this, we imaged in vivo calcium signals from soma, apical dendrites, and basal dendrites in mouse hippocampal CA3 pyramidal neurons during head-fixed navigation. To capture compartment-specific population dynamics, we developed computational tools to automatically segment dendrites and extract accurate fluorescence traces from densely labeled neurons. We validated the method on sparsely labeled preparations and synthetic data, predicting an optimal labeling density for high experimental throughput and analytical accuracy. Our method detected rapid, local dendritic activity. Dendrites showed robust spatial tuning, similar to soma but with higher activity rates. Across days, apical dendrites remained more stable and outperformed in decoding of the animal’s position. Thus, population-level apical and basal dendritic differences may reflect distinct compartment-specific input-output functions and computations in CA3. These tools will facilitate future studies mapping sub-cellular activity and their relation to behavior.
Similar content being viewed by others
Introduction
Neurons receive inputs arranged in a stratified manner, in which synapses from specific afferent brain areas arrive onto different apical and basal dendritic compartments. Beyond differences in the inputs they receive, apical and basal dendrites may differ in their molecular composition, structural and biophysical properties, and function1. Dendritic activity in the form of dendritic spikes2,3,4,5,6,7,8 and other ionic conductances9,10 can tremendously enhance the computational capacity of individual neurons11,12,13,14,15,16,17,18. However, these have primarily been studied using single neuron recordings in brain slices or with calcium imaging19,20 in sparsely labeled in vivo preparations21,22,23,24,25. While such single-neuron approaches allow faithful tracking of the activity of individual dendrites, they limit experimental throughput, are often confined to only one dendritic compartment (apical or basal), and provide limited insight into the population dynamics of large numbers of dendrites. In vivo two-photon calcium imaging from densely labeled preparations allows simultaneous recording of large populations of neurons, which enables population vector decoding of behavior26,27, and can yield insights into network properties and circuit-level function. Though calcium imaging provides subcellular resolution, thus far studies have not fully utilized the ability to track large populations of sub-cellular processes like dendrites over time.
A major limiting factor in understanding dendritic function in vivo is technical, particularly when recording from dense populations. Manually identifying regions of interest (ROIs) belonging to individual neurons or dendrites can be tedious and time-consuming. While several software suites have been developed to automate this process when recording from fields of view composed of cell bodies28,29,30,31, dendrites’ diverse shapes make morphology-based methods of dendritic ROI detection less reliable. A proper estimate of the time-varying fluorescence values from each ROI is also complicated by the highly overlapping nature of dendrites and the smaller number of pixels per ROI. Human screening of detected calcium transients can address these problems, but the amount of manual labor and time needed makes manual checking of dense datasets infeasible. Overcoming such difficulties will tremendously expand the field’s ability to investigate sub-compartment level population activity and behaviorally relevant functional properties.
To address the segmentation and demixing problems inherent to processing densely labeled dendritic populations, we developed an automated detection algorithm flexible enough to identify dendritic and somatic ROIs in dense datasets. The algorithm identifies initial ROI estimates using minimal morphological assumptions and refines them using constrained non-negative matrix factorization (CNMF)28,32. It then screens putative calcium transients with an additional fitness measure to eliminate spurious activity from undetected ROIs. To demonstrate the efficacy and utility of this approach, we applied it to several sets of data recorded from area CA3 of the mouse hippocampus8, an area containing place cells important for spatial navigation. Using this preparation, we characterized the spatial tuning properties of soma, apical dendrites, and basal dendrites during a simple navigation task. We find that apical and basal dendrites show higher calcium event rates compared to soma with faster kinetics. While all compartments show similar spatial selectivity within a single day, apical dendrites have higher stability across days in a familiar environment compared to basal dendrites. This results in CA3 apical dendrites having a better population decoding accuracy of position across days, indicating a functional divergence between sub-cellular compartments of hippocampal pyramidal neurons for long-term spatial representation.
Results
Dense dendritic fields of view are highly overlapping
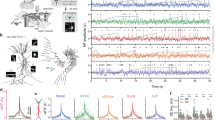
To generate dendritic imaging data sets for developing and testing our algorithm, we expressed GCaMP6f or 7b in hippocampal area CA3a/b in mice using a range of virus titers to control the labeling density (Fig. 1, Supplementary Fig. 1, Supplementary Table 1, see Methods)8,19,21,33,34. At low titers, single neurons in a given field of view (FOV) were labeled (Fig. 1d). To quantify the degree of overlap, human experts manually labeled these datasets, identifying groups of contiguous pixels with coordinated activity (Fig. 1e, see Methods). At higher titers, neurons and dendrites were labeled so densely that most ROIs overlapped with at least one other ROI, and most pixels were covered by more than one ROI. Despite the highly overlapping nature of dendrites in the more densely labeled samples, manual segmentation was possible because the neurons were sparsely active in time, such that only a few neurons were active on any given frame (Fig. 1e, Supplementary Movie 1), reducing the dense segmentation problem into a series of sparse segmentation problems. In the eight mice with the highest titer virus injected, a mean of 47% of the field of view was covered with ROIs (Fig. 1f). Manual labeling of these datasets took approximately 50 hours per field of view. Any given ROI at least partially overlapped with a mean of 5 other ROIs, and a mean of 22% of pixels belonged to a single ROI, demonstrating a high degree of spatial overlap.
a Representative confocal image of coronal section of mouse hippocampus, showing site of viral injection and cranial window implant for imaging in area CA3 (1 of 15 replicates). GCaMP+ CA3 pyramidal neurons are labeled in green. b Experimental schematic of a mouse headfixed on a textured treadmill belt. c Left, schematic of cranial window imaging setup. After aspirating the above cortex, a glass coverslip is secured above area CA3, allowing simultaneous imaging of cell bodies and dendrites. Right, maximum intensity projections of two sample fields of view (FOVs) in area CA3 (2 of 18 FOVs). d Top, maximum intensity projection image of a sample sparse FOV. Bottom, manually labeled regions of interest (ROIs) overlaid on the maximum projection image (1 of 18 FOVs). e Top-left, maximum intensity projection image of a densely labeled FOV, with many overlapping dendrites. Other panels illustrate individual frames in the imaging sequence in which dendritic and somatic ROIs can be clearly identified and labeled (1 of 18 FOVs). f Quantification of labeling density. As the number of identified ROIs (left) or percentage of pixels covered (middle) increased, the Mean Overlap increased. Right, as the Coverage increased, the percentage of pixels belonging only to one ROI decreased. See Supplementary Fig. 1 for a list of all FOVs recorded in the study.
Dendritic NMF algorithm enables efficient ROI extraction
The effort required to manually curate densely labeled neuronal datasets is massive, and the large amount of spatial overlap complicates efforts to accurately estimate activity. Hence, we sought a method to identify ROIs in an automated and unbiased way. CNMF28,32 has previously been used to identify ROIs and activities from partially overlapping neurons, so we used it as a starting point for dendritic segmentation. The goal of non-negative matrix factorization (NMF) is to identify ROIs and corresponding fluorescence traces that best approximate the original image stack. The objective function is the following:
Here, Y represents the raw calcium imaging sequence, A represents the spatial footprints (ROIs), and C represents the fluorescence traces for each ROI. Background fluorescence from out-of-plane neuropil is accounted for by additional columns and rows in A and C, respectively. The η and β terms are penalties to encourage sparsity in the fluorescence traces and ROIs, respectively. This equation is solved by iteratively solving for A and C, fixing one while optimizing the other (see Methods). This process can identify large neural populations but has not been systematically applied to densely-labeled dendritic preparations.
We developed a pipeline of unique initialization and refinement steps to be amenable to detecting soma and dendrites, which we call dendritic-NMF, or d-NMF (Fig. 2). The algorithm consists of four broad stages or steps: Initialization, Refinement, Merging, and Screening.
a Pictorial representation of the workflow. Image sequences may be filtered and/or downsampled to reduce memory requirements and improve signal-to-noise ratio. The downsampled image sequence is then split into patches. Within each patch, ROI cores are detected and their spatial footprint and temporal traces are iteratively estimated using sparse constrained NMF. Once all patches have been processed, overlapping ROIs from neighboring patches are tested to see if they can be merged. Finally, ROIs are screened for realistic activity (see panel c). b Algorithmic sketch of the ROI detection (left) and refinement (right). See Methods for further details. c Top, illustration of accepting ROIs by skewness. Left, a sample portion of a FOV containing a true ROI (orange) and false ROI (brown). Scale bar: 15 μm. Example is 1 of 11 FOVs the following analysis was performed on. Right, the activity trace for the true ROI, with a skewness value of 6.24, and the activity trace for the false ROI, with a lower skewness value of 0.58. Bottom left, receiver operative characteristic (ROC) curves for individual FOVs (light gray) overlaid with the mean ROC (thick black), parametrized on the skewness cutoff of included ROIs. The mean False Positive Rate versus True Positive Rate are plotted for four example skewness cutoff values. Bottom right, classifier performance plotted against skewness cutoff. On average, a skewness cutoff of 3.8, [3.2, 3.9] resulted in optimal performance across all tested fields of view. n = 11 FOVs.
The Initialization stage is an important step for these types of matrix factorization problems, as NMF is a non-convex problem with many local minima35. ROI initialization was performed by identifying contiguous regions of pixels active above a threshold (see Methods, Supplementary Table 2a). Such guided initialization techniques can improve performance and avoid the need to specify the number of components to be identified, but can also be influenced by poor signal-to-noise ratio (SNR) data. To address SNR issues, one can denoise data by temporal smoothing, spatial smoothing, temporal downsampling, or any combination of the three for the initialization step, tailored to the dataset and scientific question at hand. While this is not a requirement of the Initialization stage, d-NMF allows users to choose these options at their discretion. In the results presented here, we chose to temporally downsample the data from 30 Hz to 1.5 Hz for the initialization step. d-NMF operates on the image sequence in patches to reduce the memory and time requirements of the algorithm to enable processing on standard desktop computers (Fig. 2a, see Methods). Processing the image in patches had the added benefit of incorporating multiple background sources for each image patch, to better capture regional differences in neuropil signal.
The Refinement step can be performed on the original raw data or downsampled and/or otherwise filtered data. Spatial footprints and temporal traces were estimated by iterating through NMF until convergence (Fig. 2b, Supplementary Table 2b) with no explicit constraint on component size. In the final refinement step, ROIs were thresholded and split into connected components (see Methods). These final morphological operations ensured that all identified ROIs were relatively compact and easily interpretable as neuronal structures. The resulting ROIs also had the property that their activity was homogenous within an ROI. Any dendritic branch or portion of a large ROI that had independent activity was identified as a separate ROI. (Supplementary Fig. 2).
In the Merging step, we combine ROIs (Supplementary Table 2c) that are likely to correspond to the same functional unit. Long dendritic branches may span multiple image patches (Fig. 2a) or may be represented by multiple ROIs due to local variations in activity. The algorithm offers users the ability to set a threshold correlation with which to merge ROIs. A high threshold can result in a fine parcellation of dendritic structure, preserving local differences in activity, while a low threshold will merge ROIs with broadly the same activity. We observed that a general rule on the lower bound of the merging threshold is to identify when ROIs not belonging to the same neuron become linked together. This is indicated by a sudden drop in minimum within-group correlations (Supplementary Fig. 3, see Methods). Even if users decide not to merge ROIs, correlation-based linkage information can aid in scientific interpretations at later stages.
The Screening step provides a final stage to include or exclude ROIs. The raw output of d-NMF contains any set of contiguous pixels that had simultaneously high activity at least once, which can result in false positives. In many brain regions, including the hippocampus, excitatory neural activity is characterized by long periods of quiescence punctuated by short periods of activity, resulting in positive skewness values. Hence, we used the skewness of the extracted calcium traces for each ROI (Fig. 2c), to classify valid and invalid ROIs, trained on manually-labeled data (see Methods). Skewness was a very effective separator of valid and invalid ROIs (Fig. 2c), yielding comparable outcomes as signal-to-noise ratio (Supplementary Fig. 4). The optimal skewness value was relatively close across the population of fields of view, at 3.8, [3.2, 3.9]. Note that this cutoff value may vary across brain regions, cell types, or fluorescent indicators. This process yields interpretable ROIs (Supplementary Fig. 2) of varying shapes without random initialization or needing to specify the number of components to be detected beforehand. A comparison of the algorithm design choices in various methods is summarized in Supplementary Table 3.
To further validate the accuracy of d-NMF, we constructed synthetic dense datasets using one of the sparsely labeled datasets (Fig. 3). This sparse dataset had known ROI boundaries and temporal traces, serving as a ground truth. By copying, rotating, and shifting the original movie, we constructed synthetic dense datasets with up to 200 neurons with a high degree of overlap between dendrites (see Methods). We found that as we added more duplicates, the quality of signals decreased, due to the addition of more background from other ROIs and additional neuropil (Fig. 3c). This had the result that adding additional neurons beyond a certain point no longer yielded additional detectable ROIs or even decreased the amount of ROIs with a high-quality signal (Fig. 3d,e). Nevertheless, for ROIs with a signal quality above 2 standard deviations, d-NMF maintained an F1 score (Supplementary Fig. 5a, see Methods) of 0.8 out of 1, outperforming the output of suite2p, another open-source analysis package29 (Fig. 3f). This serves as evidence that our method is sound and accurate at detecting ROIs from dense fields of view.
a Maximum projection images of movies generated by copying, rotating, and shifting the movie in the first column. b ROIs derived from d-NMF for the corresponding movie in (a). c Frame of maximum activation for a sample ROI in the corresponding movies in (a). The signal quality of this ROI is noted in the titles, and decreases as more neurons are added to the synthetic data. d, The mean signal quality of ROIs decreased as more neuron replicates were added to the synthetic data. Quality is defined as the mean z-scored value of all pixels at the ROI’s most active frame. e The area of ROIs with signal quality above a certain threshold (here 1.5z, 2z, and 3z) initially increased linearly but slowed down or decreased at higher densities. f Left, restricted to ROIs with a signal quality >2z, true positive rate for d-NMF (green) and suite2p (s2p, purple) as a function of the number of neurons in the synthetic dataset. Right, F1 score for the same data. The d-NMF method maintained high performance even for very dense datasets.
d-NMF matches manual labeling and exceeds existing methods
To demonstrate the accuracy and utility of d-NMF in detecting ROIs, we evaluated its performance with human-labeled ROIs used as ground truth. At a wide range of imaging densities, d-NMF closely matched manually-labeled ROIs (Fig. 4). Segmentation similarity between algorithmic ROIs and manual ROIs was measured both based on a morphology and overall pixel-by-pixel coverage (Supplementary Fig. 5, see Methods).
a 3 sample fields of view illustrating segmentation results on sparse, medium, and dense data. Color coded FOVs indicate ROIs identified via manual segmentation (blue), d-NMF (green), CaImAn with default parameters (here labeled CNMF, gray), and suite2p (magenta). Examples are 3 of 11 FOVs the analysis in this figure was performed on. b The F1roi score of identified ROIs (compared to manual labeling) for d-NMF (green, 0.45, [0.42, 0.49]) was significantly higher (p = 9.8x10−4) than that of CNMF (gray, 0.32, [0.27, 0.38]), and significantly higher (p = 0.002) than suite2p (s2p, purple: 0.42, [0.35, 0.45]). The F1 score of suite2p was also higher than CNMF (p = 0.014). c The True Positive Rate (TPR roi) of identified ROIs (compared to manual labeling) for d-NMF (0.45, [0.31, 0.48]) was significantly higher (p = 9.8x10−4) than that of CNMF (0.21, [0.18, 0.29]), and significantly higher (p = 9.8x10−4) than suite2p (0.31, [0.23, 0.35]), which was also higher than CNMF (p = 0.0068). d d-NMF had significantly higher overall coverage than CNMF, but not suite2p (d-NMF Coverage: 0.27, [0.07, 0.55]; CNMF Coverage: 0.14, [0.09, 0.24]; suite2p Coverage: 0.28, [0.09, 0.48]; d-NMF vs CNMF: p = 0.0029; d-NMF vs suite2p: p = 0.52; CNMF vs suite2p: p = 0.002). n = 11 FOVs, two-sided Wilcoxon signed-rank test for all. Values reported as median and 95% confidence interval of the median.
We compared performance against CNMF, implemented with the CaImAn software package28, as well as suite2p29, (see Methods). The median F1 score using d-NMF was 0.45, exceeding that of CNMF (0.32) and suite2p (0.42) (Fig. 4b, Left). Similarly, the True Positive Rate of d-NMF (0.45) exceeded CNMF (0.21) and suite2p (0.31) (Fig. 4c). The percentage of the FOV covered by ROIs (coverage) using d-NMF (27%) was also higher than CNMF (14%), but not suite2p (28%) (Fig. 4d, Right). In particular, CaImAn did not identify ROIs in particularly dense regions of the FOV. While suite2p performed comparably with d-NMF in sparse preparations (Fig. 4a, Left), dense preparations were more completely labeled using d-NMF (Fig. 4a, Right). d-NMF performed well on both sparse and dense datasets with a single set of parameters, whereas other methods may need careful parameter tuning depending on the density of the preparation. The success of d-NMF compared to other methods is not due to systematic detection of a higher number of smaller ROIs; ROI size was largely comparable across methods, with several instances of ROIs identified spanning well over 100 μm (Supplementary Fig. 6). Changes in parameter settings for CNMF and suite2p led to small improvements in performance (Supplementary Fig. 7, see Methods), so we compare to the highest performing version of each in Fig. 4. d-NMF also matched or outperformed other methods when applied to other types of independently imaged data, including dendrites from motor cortex in macaque monkeys36, and entorhinal cortex axons in mouse CA1 (Supplementary Fig. 8).
Of great interest to the dendritic field is investigating localized activity, generally represented by calcium transients with rapid kinetics that remain confined to a small dendritic segment. We validated that d-NMF was able to detect localized activity from two previously published datasets, using calcium imaging in mouse hippocampal area CA121 and voltage imaging in the visual cortex37, respectively. We also identified local activity in a putative dendritic spine in our own data (Supplementary Figs. 9, 10). To explore the bounds of activity in which d-NMF could detect localized activity, we prepared synthetic datasets by injecting calcium transient waveforms of different amplitude, duration, and number into localized dendritic regions of interest with known baseline activity. This analysis demonstrated that by analyzing non-downsampled, 30 Hz data, localized transients as brief as 100 ms in width33 were sufficient to identify independent ROIs, provided the transients were of sufficient amplitude to rise above the noise level (Supplementary Fig. 11, Supplementary Table 4).
Higher sampling rates are generally required to detect rapid events. However, a higher sampling rate comes at the cost of noisier data, which may adversely impact ROI detection and refinement. Indeed, in our hippocampal data, performing the Refinement step on 30 Hz data resulted in slightly lower coverage, though the overall performance against manually-labeled data was relatively similar (Supplementary Fig. 12) to temporally downsampled data. Additionally, processing raw data increases both the size of files in memory and computation time. Depending on the expected kinetics and spatial extents of activity as well as the computing power available, users may choose to downsample or filter in time, space, or both. Because we are investigating dendritic activity, which may be rapid, all following analyses are done on data extracted from non-downsampled, 30 Hz files.
A fitness trace to refine activity estimates and eliminate cross-contamination
Densely labeled datasets present added difficulties in estimating the fluorescence activity of each ROI due to overlapping sources. Given a complete set of ROIs, NMF accurately de-mixes these signals and properly assigns activity to each ROI. However, despite the best efforts of manual labeling or automated labeling, some processes may go unlabeled. This will lead to contamination of signals from identified ROIs by unidentified ones. Such contamination can affect estimates of activity rates, tuning properties, stability, or correlation with related ROIs. While simply setting a high threshold for transient detection may reliably screen out false transients, this runs the risk of excluding low-amplitude calcium transients, which may represent different firing modes such as localized events versus back-propagating action potentials (bAPs), or single spikes versus bursts.
To both evaluate the degree of contamination and to provide an additional tool to correct this, we developed the concept of a Fitness Trace for each ROI (Fig. 5a), defined as the frame-by-frame correlation between the spatial footprint of that ROI with the fluorescence activity from the video (see Methods). The intuition behind this trace is that true activity in a given ROI occurs when all pixels show elevated activity, not just some. Significant calcium transients were detected by thresholding the Fitness trace along with the original calcium trace (Fig. 5b). Overall classifier performance was evaluated by computing the Jaccard index (the size of the intersection divided by the size of the union)38 between detected events and manually-labeled true events (see Methods). Across 9 different FOVs there was a relatively consistent optimal parameter range, at a ΔF/F threshold of 3.9z and a Fitness threshold of 0.2.
a Top, raw fluorescence trace (z-scored) of the ROI shown below. Example large amplitude true events, small amplitude true events, and false events are marked in green circles, magenta circles, and gray crosses, respectively. Middle, the Fitness Trace computed from the above fluorescence trace. Bottom, frames of large amplitude true events (green circles), false events (gray crosses), and small amplitude true events (magenta circles). b Left, classification performance, as measured by the Jaccard Index, plotted as a function of threshold on ΔF/F and Fitness Trace (see Methods). The global peak is indicated with a black circle. Right, the transient rate plotted as a function of the same thresholds. c Left, classifier performance for d-NMF with Fitness Trace (DFit) compared to a simpler detection method (2z, see Methods). DFit: 0.63, [0.54, 0.68]; 2z: 0.31, [0.24, 0.38]; DFit vs 2z: p = 3.9×10−3. Right, the detected event rate for the same 2 methods compared to manually classified events, illustrating the importance of correct classification. Manual: 0.64, [0.42, 0.77] Events/minute, DFit: 0.63, [0.51, 0.76] Events/minute; 2z: 1.94, [1.57, 2.13] Events/minute; n = 9 FOVs for all. Manual vs DFit: p = 0.36; Manual vs 2z: p = 3.9×10−3; DFit vs 2z: p = 3.9×10−3; n = 9 FOVs, two-sided Wilcoxon signed-rank test for all. Values reported as median and 95% confidence interval of the median. d Left, Schematic designating apical dendrites, soma, and basal dendrites. The half-width of detected calcium transients spanned 2 orders of magnitude in apical dendrites, basal dendrites, and soma, with a bias towards wider transients in the soma. Lines represent histograms of events averaged over sessions. Open circles indicate the median value for individual sessions. Filled circles indicate the median value across sessions. Apical: 590, [460, 770] ms; Soma: 1400, [860, 2400] ms; Basal: 590, [400, 870] ms. Apical vs Soma: p = 7.8×10−3; Apical vs Basal: p = 0.95; Soma vs Basal: p = 7.8×10−3. n = 8 sessions, two-sided Wilcoxon signed-rank test for all.
An implicit requirement of the Fitness Trace is that the entire ROI must be active for a calcium transient to be detected, which could potentially discard localized dendritic activity and bias estimates. If ROIs are too broadly defined, such that they span multiple branches containing independent activity, this would lead to a bias to exclude branch-independent transients. However, the d-NMF algorithm identifies ROIs precisely as groups of pixels with synchronous activity. Dendritic branches that show independent activity are automatically split off from other branches and analyzed as a separate ROI (Supplementary Fig. 2). In this way, accurate ROI identification assists in accurate transient detection.
We compared detection accuracy and the rate of detected transients from our optimized Fitness method to those of other event detection methods (Fig. 5c). A widely used approach is to set a threshold of 2 standard deviations above the mean on the raw ΔF/F traces, with no de-mixing (with an additional false positive rejection criterion, see Methods). We call this method 2z. We call our method using optimal values for the ΔF/F threshold and Fitness threshold DFit. Event classification performance was significantly higher using DFit (0.63) compared to 2z (0.31). Other detection methods incorporating different elements of these two approaches achieved intermediate results (Supplementary Fig. 13, Supplementary Table 5, 6). The use of the Fitness Trace also was comparable to or outperformed other related measures39 (Supplementary Fig. 13e-h, Supplementary Table 5, 6). More striking, the estimated calcium transient rate using 2z was overestimated by more than a factor of 3, while DFit closely matched the manually identified transient rate (Fig. 5c). This demonstrates that accurate event detection and screening is a vital step in processing data from densely labeled FOVs. Detected calcium transients spanned a wide range of durations, with half-widths from 100 ms up to 10 seconds. Transients in apical and basal dendrites were more rapid than those in the soma (an average of 500 ms in apical and basal dendrites, compared to an average of 850 ms in soma), with transients as fast as 100 ms in dendrites, which may be missed or severely reduced in amplitude due to averaging when imaged at slower frame rates.
Spatial coding properties of dendrites in CA3
To demonstrate the utility of d-NMF, we analyzed the activity of detected ROIs in relation to behavior as mice performed a random foraging task on a textured treadmill belt (Fig. 6, see Methods). Somata of pyramidal cells in area CA3 demonstrate place coding, where they have reliable elevated activity levels in restricted regions of space. Using d-NMF, across 8 mice we identified 709 total somatic ROIs, 3139 apical dendritic ROIs, and 3999 basal dendritic ROIs. This allowed us to examine and compare the activity and spatial tuning properties of populations of apical and basal dendrites.
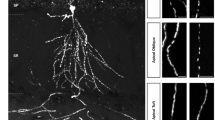
a Left, Cell compartment schematic. Right, sample FOV (1 of 8 used in this figure analysis) shows apical dendrites (magenta), soma (green), and basal dendrites (blue). b Expanded views of sample apical dendritic, somatic, and basal dendritic ROIs (Scale bars: 10 µm) with activity across time in corresponding colors. Position of the mouse is plotted in black. c, Top, middle, for the sample ROIs, ΔF/F is plotted against lap number and position. The trial-averaged tuning curve is overlaid for each ROI. Bottom, all statistically significant tuning curves for apical, somatic, and basal ROIs across all mice, demonstrating a relatively uniform tiling of the environment in each population. d Sample FOV color-coded according to the position of peak activity for each ROI. e Top, the percentage of ROIs that were tuned were similar across all 3 ROI types: Bottom, for ROIs with statistically significant tuning, the width of place fields was not significantly different between ROI types. f Left, apical and basal dendrites had a significantly higher mean transient rate than the soma, but were not different from each other. Right, apical, but not basal, dendrites showed increased transient rates as a function of distance from cell body layer. g Left, apical and basal dendrites had significantly higher transient widths compared to soma, but were not different from each other. Right, apical dendrites showed a decrease in transient width with increasing distance from the cell body layer (apical, p = 3.3x10−9, two-way ANOVA) but not basal dendrites (basal, p = 0.48, two-way ANOVA). h Left, spatial information content was lower in apical dendrites. Right, apical dendrites showed decreasing information content with increasing distance to the cell body layer (p = 3.3x10−5, two-way ANOVA). Basal dendrites did not show such a relationship (p = 0.55, two-way ANOVA). Bar graphs indicate median and 95% confidence interval of the median. n = 8 sessions for all comparisons. See Supplementary Table 7 for summary statistics.
While spatial tuning in CA3 PN somata has been well described through electrophysiology approaches40,41,42 and more recently with Ca2+ imaging43,44, we know little about how reliably CA3 dendrites encode place and their spatial tuning properties. Using our automated approaches, we identified spatially tuned CA3 dendrites (place dendrites) both in the apical and basal compartments (Fig. 6a-e). Dendritic and somatic compartments had similar spatial tuning properties in terms of the proportion of tuned ROIs (16-20%) and place field width ( ~ 25 cm) (Fig. 6e, Supplementary Table 7). Despite these similarities, dendrites showed a number of differences from soma. Apical and basal dendrites had higher activity rates than soma (0.7 events/minute (apical) and 0.6 events/minute (basal) compared to 0.5 events/minute in the soma, Fig. 6f, Supplementary Table 7), with faster kinetics (Fig. 6g). Spatial information content was lower in dendrites (1.9 bits/event) compared to soma (2.1 bits/event), possibly related to the differences in activity rates (Fig. 6h). Apical dendrites, but not basal, exhibited increasing event rates, thinner transients, lower spatial selectivity, and a higher number of place fields as a function of distance from the cell body layer (Fig. 6f-h, Supplementary Table 7, see Methods).
Basic spatial tuning characteristics were not systematically related to the ROI size or the number of overlapping ROIs (Supplementary Fig. 14, Supplementary Table 8). Additionally, these results were not biased by over splitting of dendrites into multiple compartments, as all comparisons were similar when we merged ROIs with highly correlated activity (see Methods, Supplementary Fig. 15, Supplementary Table 9).
To provide support that these findings were not influenced by the dense recording preparation or our analytical approaches, we analyzed a subset of the data with neurons labeled sparsely with GCaMP, manually identifying ROIs and calcium transients (Supplementary Fig. 16). We found corroborating evidence of higher activity rate in the apical and basal dendrites compared to soma as well as more rapid calcium kinetics at farther distance from the soma. The similar findings in dense and sparse data validate that we can accurately detect calcium transients in the dense data.
Apical dendrites are more stable across days and provide better place decoding
An advantage of optical recording techniques is the ability to track the same ROIs across long timespans. Thus, we quantified the short-term (within-day) and long-term (across-day) stability of apical dendrites, soma, and basal dendrites in a familiar environment (Fig. 7, Supplementary Table 10, see Methods). Within-day stability was not significantly different across compartments when quantified using Tuning Curve (TC) correlation (which quantifies stability per ROI), but apical dendrites showed higher within-day stability than basal dendrites when quantified using Population Vector (PV) correlation (which quantifies stability of the overall spatial representation, see Methods). More strikingly, apical dendrites were more stable across days compared to basal dendrites using TC or PV correlation. We were not able to extend these analyses to the sparse data due to the limitation of the small amount of tuned neurons in that dataset. The differences in across-day stability were not due to fields of view distorting across days (Supplementary Fig. 17), or differences in ROI size or overlap (Supplementary Fig. 18).
a Experimental timeline schematic. An environment was designated as Familiar after mice ran on that treadmill belt for 3-5 days. ROIs were tracked over following two days, Familiar Day 1 (yellow) and Familiar Day 2 (orange). b Example apical, somatic, and basal ROIs demonstrating different short- and long-term stability. Each column represents a different ROI. Raw activity and rate map for the first half of Familiar Day 1 (top row), second half of Familiar Day 1 (middle row), Familiar Day 2 (bottom row). Tuning curve correlation was compared between First and Second Half of Familiar Day 1 Session (Within Day) and Familiar Day 1 and 2 (Across Day). c Rate maps for all apical, soma, and basal ROIs in a single FOV on Familiar Day 1 (left) and Day 2 (right). ROIs are sorted according to the position of the peak of their tuning curve in Familiar Day 1, and that order is maintained for Familiar Day 2. d Distributions of across-day tuning curve correlations for data in (C) showing apical dendrites had higher correlation than soma or basal dendrites. e Population vector overlap matrices for data in (C), titles stating mean value across the diagonal. f Left, within-day tuning curve correlations were not different across compartment type. Right, in contrast, higher across-day TC correlations in apical dendrites compared to soma and basal dendrites (Apical vs Basal, p = 0.031). g Population vector correlations, apical dendrites significantly more stable than basal dendrites within-day (Left, Apical vs Basal, p = 0.023) and across-day (Right, Apical vs Basal, p = 0.031). Bar graphs indicate median and its 95% confidence interval values. n = 8 sessions for all comparisons in (f) and (g). See Supplementary Table 10 for summary statistics.
Because apical dendrites showed better across-day stability compared to basal dendrites, we predicted that apical dendrites would also outperform basal dendrites in population vector decoding. Thus, we constructed population vector decoders using only apical dendrites, soma, or basal dendrites, and tested their ability to decode position within a session (Fig. 8a) or across sessions (Fig. 8b). There was no significant difference between decoding accuracy within-day. However, apical dendrites had significantly lower decoding error compared to basal dendrites across days (Fig. 8c). Population vector decoding accuracy is highly dependent on the number of sources used to train the model, so we repeated the analysis using random subsets of data. Consistently, we found no differences across groups for within-day decoding (Fig. 8d), but a significant interaction effect of compartment group and number of ROIs on across-day decoding, with apical dendrites and soma showing significantly better decoding accuracy compared to basal dendrites (Fig. 8e).
a Left, sorted rate maps of all ROIs from a single FOV in the first half and second half of Familiar Day 1. The maps from Half 1 were used to train a decoder to decode the position in Half 2. Right, example decoded position for the same FOV using only apical, somatic, or basal ROIs. b Same as in (a) except data from the entire Familiar Day 1 was used to train a model to decode Familiar Day 2. Note that the error is generally larger compared to within-day decoding, and that apical dendrites show the lowest decoding error. c Left, within-day decoding error was not significantly different across compartment types (Apical: 22, [11, 30] cm; Soma: 25, [23, 41] cm; Basal: 27, [15, 43] cm; Apical vs Soma, p = 0.078; Apical vs Basal, p = 0.55; Soma vs Basal, p = 0.64, n = 8 sessions, two-sided Wilcoxon signed-rank test for all). Right, the across-day decoding error was lower for apical dendrites (35, [23, 43] cm) compared to basal dendrites (46, [36, 47] cm), with no significant difference between apical dendrites and soma (43, [28, 44] cm) (Apical vs Soma, p = 0.11; Apical vs Basal, p = 0.023; Soma vs Basal, p = 0.25; n = 8 sessions, two-sided Wilcoxon signed-rank test for all). d When controlling for the number of ROIs used to decode position, soma had significantly better decoding than basal dendrites (Apical vs Soma, p = 0.07; Apical vs Basal, p = 0.57; Soma vs Basal, p = 0.013; two-way ANOVA, see Methods). e Both apical dendrites and soma had better decoding than basal dendrites across days when controlling for the number of ROIs used (Apical vs Soma, p = 0.10; Apical vs Basal, p = 0.0499; Soma vs Basal, p = 3.9×10−4; two-way ANOVA, see Methods). Values reported as median and 95% confidence interval of the median.
Discussion
In summary, we developed a versatile algorithm and toolkit that facilitates analysis of densely labeled fields of view containing both apical and basal dendrites and cell bodies (Fig. 1), enabling large-throughput in vivo investigation of dendritic activity. The algorithm utilizes the well-validated mathematical framework of CNMF while providing automated initialization flexible enough to identify neural processes of varied shape and size in sparsely- and densely-labeled preparations (Figs. 2, 3, 4). The addition of the Fitness Trace post-hoc helps screen out false calcium transients to more accurately estimate dendritic event rates and tuning properties (Fig. 5). Using these tools, we demonstrate spatial tuning in apical and basal dendrites of pyramidal neurons in CA3 (Fig. 6). While the proportion of tuned ROIs and within-day tuning properties were largely comparable across all compartments, basal and apical dendrites showed higher event rates than soma. Dendritic event widths were noticeably thinner than soma, on a population (dense data) as well as on an individual neuron level (sparse data). By comparing longer-term population dynamics across days, we demonstrate that apical dendrites are more stable than basal dendrites (Fig. 7). This leads to better accuracy when constructing a population vector decoder to predict the animal’s position (Fig. 8). These population-level analyses would not be possible with sparsely labeled preparations, and we were only able to take advantage of our densely labeled preparations using the analytical tools we have developed.
Densely labeled dendritic fields of view present a number of challenges for existing automated detection methods, particularly the initialization step. Initialization is often based on morphology, with parameters to be set for the expected diameter of an ROI. This assumes a uniform size of circular ROIs throughout the FOV, which is not the case for dendrites. Seed pixels for initialization can be chosen from the peaks of the correlation map, constructed by computing the average correlation of a pixel with its neighbors45. However, thin dendritic segments a few pixels wide may skew towards low correlation values and be systematically missed compared to wider somatic ROIs. Furthermore, overlapping dendrites result in pixels containing activity from a combination of sources, which may obscure correlations within individual branches. Random initialization does not suffer from the drawbacks of morphology-based approaches, but there is no guarantee of complete labeling or consistent labeling across iterations. Additionally, the number of components must be specified for initialization, which may vary over the field of view. One approach is to over-specify the model and then discard unusable ROIs afterwards32, but this comes at a higher computational cost. Additionally, unusable ROIs may distort the spatial or temporal aspects of remaining, valid ROIs, so a proper estimate of the number of components is important, which we solve in a data-driven manner.
Though our approach utilizes NMF as the core extraction algorithm, there are notable differences between our approach and other published methods. Our initialization method is based on localized coactivity, and the number of components within a patch is discovered from the data, rather than prespecified. This approach works in our densely labeled datasets because soma and dendrites tended to be sparsely active in time, such that only a few ROIs are active on any given frame. This mimics the method by which manual segmentation is done, and allows the identification of ROIs that may overlap with many others (Fig. 4). We also enforce ROIs to consist of contiguous pixels, in contrast to CNMF, which imposes no spatial constraints on the ROIs. CNMF additionally constrains the temporal component by an autoregressive fit, while d-NMF has no constraint on the temporal components. This allows for the possibility of calcium transients with different dynamics to exist within a single ROI.
Different aspects of d-NMF could be used in conjunction with pre-existing methods. Other segmentation software could be initialized with the initial ROI cores or the final ROIs obtained after de-mixing. The Fitness Trace (Fig. 5) could be used to screen any activity trace regardless of the method used to obtain it46, though it would be crucial to verify that the underlying ROIs should not be further split into independent units. Ultimately, different tools may be better suited to different data sets, so it is worthwhile for investigators to be able to combine the strengths of all available approaches.
As with any data analysis pipeline, the user takes on the responsibility of curating the output of d-NMF to ensure the interpretability and robustness of their results. For instance, depending on the threshold specified for merging ROIs with similar activity, d-NMF can give a very fine or coarse parcellation of the dendritic tree (Supplementary Fig. 10). For those wanting to investigate changes in activity along the dendrite, a high merging threshold would be best, but for those wanting to capture the main global activity across an entire branch, a more permissive merging threshold would be appropriate. Furthermore, the specific optimal threshold may vary across fluorescent indicators, brain region, behavioral state, or species. Additionally, if a dendritic branch is split into multiple ROIs, these will not have independent activity, violating the assumptions of some statistical tests. This can be addressed by calculating summary statistics across ROIs for an entire FOV, or to merge or subsample ROIs with similar activity and verifying that scientific results still hold. We encourage such control analyses to be standard practice when performing population imaging.
Large population recordings allow the use of fewer animals to record from a given number of neurons, saving on time and resources. For characterizing single dendrite properties, this is particularly advantageous if a relatively small percentage of units show tuning to a given stimulus or behavioral feature. This is the case in CA3, where only ~20% of dendrites or soma show spatial selectivity (Fig. 6). Methods of inducing selectivity through artificial means47,48 can somewhat circumvent this, but the effects of these manipulations may be limited or constrained49. Larger population data not only add more power to the analysis but also provide insights into functional heterogeneity and dynamics50,51,52. Simultaneous longitudinal recording of multiple dendrites in the same animal provides within-subject control on the effect of behavior on dendritic activity, and large enough population recording enables analysis of population-level stability (Fig. 7) and decoding of behavior (Fig. 8)26,27,53. It is important to note, however, that increased density does not come for free. When we constructed our synthetic datasets by duplicating and shifting a field of view with a single neuron, the overall signal quality decreased as more neurons were added. This indicates that, for a given expected morphology and distribution of cells, a sweet spot of labeling density may exist which maximizes the number of usable neurons while minimizing the number of animals needed.
Our results demonstrate that both apical and basal dendrites show spatial selectivity on par with nearby soma (Fig. 6), and the apical dendritic signal is more stable than the basal dendritic signal (Fig. 7). Long-term recordings of large populations of neurons have revealed changing neural representations over time, a phenomenon termed representational drift54,55. This has been documented in the hippocampus56 as well as several other brain areas57,58,59,60,61,62. Previous studies have shown that population responses of areas CA1 and CA3 of the hippocampus are particularly unstable across several days44,56, though other regions of the hippocampal circuit are more stable, including the dentate gyrus44 and entorhinal cortex63. Different levels of stability in apical and basal dendrites (Figs. 7, 8) suggest that representational drift may not be homogenous across compartments of individual neurons. This may be related to the segregated inputs received and the varying functionality of CA3 as performing pattern completion or pattern separation64,65,66,67,68 by biasing propagation of inputs from apical or basal dendrites. Different compartments are likely subject to differential gating by target selective GABAergic circuits or gradient distributions of channel conductances. A similar mechanism may be at work in the neocortex, where anatomy69,70,71 and theory72,73,74 suggest that bottom-up input arrives onto the basal dendrites while top-down input arrives on to the apical dendrites, providing a spatial segregation matching the functional segregation. Apical dendrites may also be more stable due to integrating signals from several different inputs. The conjunctive activity of entorhinal cortex, dentate gyrus, and recurrent collaterals may act to trigger supra-linear dendritic spikes7,18, triggering long-term synaptic plasticity16,18,47,75,76 which could contribute to increased stability21. Differences in task demands also affect the representational stability of soma in CA153, so the stability of dendritic compartments under different behavioral tasks should be explored in the future.
It is also important to consider the contribution of forward-propagating versus back-propagating signals in defining dendritic activity. Proximal dendrites that are electrotonically close to the soma are likely to be dominated by back-propagated activity. However, the higher transient rates in apical and basal dendrites indicate a mixture of localized and back-propagated activity. Distal dendrites may enjoy a larger degree of independence because back-propagating action potentials often decay with distance77. Localized spikes likely do exist in hippocampal CA3 and CA1, supported by previous slice studies8,18,46,78. Using calcium signals as an indicator of across-compartment correlation should be done with caution though, as calcium signals may reflect large bursts of activity more robustly than single action potentials19. Future studies using voltage-sensitive indicators or more sensitive calcium indicators in large populations should provide higher temporal resolution into these questions. Due to these considerations, d-NMF does not identify different event types but rather provides the time-varying concentration of calcium in a given ROI, de-mixed from the activity of overlapping ROIs. This is a critical starting point before the identification of different event types can be done. We have demonstrated using voltage imaging and synthetic data that d-NMF can detect localized, thin transients if they are present. Knowledge of the activity of neighboring, connected ROIs is necessary to identify activity as local or more widespread.
Many valuable insights about dendritic function have been gleaned without explicitly delineating local dendritic events from back-propagating events, across different brain areas such as motor cortex22,25,79, visual cortex1,23,80,81, hippocampal CA121,33,82, and lateral amygdala83. Definitively resolving back-propagating action potentials from local dendritic activity requires deliberate experimental design involving technically challenging approaches37,84. For accurately dissociating the source of dendritic activity (forward vs. back-propagating) one needs to establish ground truth with simultaneous dendritic and somatic electrophysiology and imaging coupled with well-defined stimulation protocols85,86. Given the slower kinetics and lower single spike resolution of Ca2+ indicators, resolving bAPs requires further tool development such as in vivo optimized high signal-to-noise ratio voltage imaging sensors87. The analysis tools presented here will synergize with future developments in these experimental areas to enable principled investigation into dendritic signaling.
This study examined the activity of ensembles of apical dendrites, soma, and basal dendrites separately. In our dense datasets, we already merge together ROIs of neighboring dendrites and soma which have substantial spatial overlap and highly correlated activity. Such ROIs obtained from merging the soma and dendrites indicate a high degree of coupling between the soma and its proximal dendritic branches, as would be expected from the diffusion of calcium over short distances. But the possibility of dendrites exhibiting semi-independent activity6,22,25,84 from their cell bodies poses a challenge in linking ROIs based on correlations alone. Additional structural markers would likely help in linking connected dendrites and soma. Furthermore, many distal dendrites are connected to soma that are not in the focal plane. This could be resolved with multi-planar imaging in future studies. A more in-depth investigation of input-output transformations at a sub-cellular level would require simultaneous measurement of connected dendrites and soma, as we provided in our sparely labeled datasets (Supplementary Fig. 16). By comparing the tuning properties of the apical dendrite, soma, and basal dendrite of the same neuron within densely labeled fields of view, one could learn about branch-specific6,22,25,84 versus branch prevalent activity21,23,81 and soma-dendrite coupling dynamics while sampling large populations. Such information has so far been limited to very sparse preparations, which may overlook cellular heterogeneity within regions50,51,52.
The methods presented here were primarily tested on the calcium activity of dendrites and soma measured with GCaMP6f in hippocampal area CA3. But the approaches are broadly applicable to any cell or cellular process exhibiting dynamic fluorescence activity, including axons18,88,89 and astrocytes90,91 recorded with a range of different genetically encoded calcium indicators92 or voltage indicators93,94. These different indicators have varying signal-to-noise ratios and kinetics, so a reasonable parameter search may be necessary to obtain the optimal performance of our tools. The rapid kinetics of voltage indicators with respect to the speed of propagation within dendrites may also result in the identification of finer-grained ROIs, as an assumption of our mathematical model is that the signal in a given ROI is uniform. Hippocampal area CA3 provides the geometrical advantage of imaging large dendritic trees in a single focal plane, but our approach would easily extend to brain areas such as retrosplenial cortex with similar affordances, or those accessible with microprism approaches95. Our method can also be utilized in more traditional tuft dendrite or basal dendrite imaging, and could facilitate analysis of sparsely labeled preparations as well. Clever intersectional genetic approaches should be able to provide access to axonal input, dendritic processing, and somatic output in a single preparation, and our tools will facilitate the rigorous analysis of such datasets.
Methods
Animals
All experiments were conducted in accordance with the National Institute of Health guidelines and with the approval of the New York University Grossman School of Medicine Institutional Animal Care and Use Committee (IACUC). Imaging experiments used mice on a C57BL/6 J background, from both sexes, 15–25 weeks old, housed on a reversed light cycle.
Cranial window surgery
Surgery procedures were similar as described previously8. Mice were anesthetized using isoflurane (1.5–2.5%), a 0.5 mm hole was drilled in the skull above dorsal area CA3 of hippocampus (1.6 mm lateral, and 1.4 mm caudal of Bregma) and AAV virus was injected. For densely expressing preparations, AAV1.CamKII. GCaMP6f.WPRE.SV40 (Penn Vector Core, titer: 2.76x1013 GC/ml, 23 nL per site) was injected at two sites (1.5 mm ML, 1.3 mm AP; 1.7 mm ML, 1.5 mm AP relative to Bregma; 1.8 and 2 mm Z depths below the dural surface, covering areas CA3a/b). For sparser expression, we used a mixture of AAV1.CamKII-Cre (diluted with ACSF) and AAV1.Syn.Flex.GCaMP6f.WPRE.SV40 or AAV1.Syn.Flex.jGCaMP7b.WPRE at 23 nL per site, at different ratios8 (see Supplementary Fig. 1 for specific titers). After injection, a 3 mm glass coverslip cranial window attached to a 1.7 mm deep stainless steel cannula was implanted by performing a craniotomy centered around the injection site and aspiration of the overlying cortex and external capsule. The window was sealed to the skull using Vetbond, and a custom-designed 3D-printed plastic head post was cemented over the skull.
Immunohistochemistry
To verify our imaging was in area CA3 and not neighboring CA2, we tested for localization of GCaMP6f and the CA2 marker PCP496 (Supplementary Fig. 1b) using immunohistochemistry of 50 µm PFA fixed horizontal slices in TBS. GCaMP-labeled neurons and were stained using a chicken polyclonal anti-GFP primary antibody (1:1000; Invitrogen) with a donkey anti-chicken Alexa Fluor 488 dye-conjugated IgG antibody (1:1000; Invitrogen). PCP4-positive neurons were stained using a rabbit polyclonal anti-PCP4 primary antibody (1:200; Invitrogen) with a goat polyclonal anti-rabbit Alexa Fluor 633 dye-conjugated IgG antibody (1:400; Invitrogen).
Two-photon imaging
In vivo imaging was performed on head-fixed running mice as described previously8,53,97,98 using a dual resonant galvanometric laser scanning two-photon microscope (Ultima, Bruker). The microscope was coupled to a tunable Ti:Sapphire laser (MaiTai eHP DeepSee, Spectraphysics) at 80 MHz pulse repetition rates and <70 fs pulse width with dispersion compensation, to excite GCaMP and tdTomato fluorophores at 920 nm. Images were acquired at scan speeds of 30 fps, using 512x512 frame size (1.085 mm/pixel resolution) with a resonant scanning galvanometer system mounted on a movable objective Ultima microscope, with an orbital nosepiece coupled to a 16X, 0.8NA, 3 mm water immersion objective (Nikon). The fluorescence signal was detected using high-sensitivity GaAsP photomultiplier tubes (model 7422PA-40 PMTs, Hamamatsu). Recording sessions were ~10 minutes long, yielding datasets of ~10 GB.
Head-fixed spatial navigation
Mice were trained to perform a random foraging task by running head-fixed on a 2-meter linear treadmill belt18,53 to lick for randomly located but uniformly distributed sugar-water (5% sucrose) rewards along the track. The position of the mouse, and lap onset were measured using an optical rotary encoder (S5-720, US Digital) and reading RFID tags and reader (ID-20LA, SparkFun Electronics). Behavioral programs were controlled with an Arduino Mega 2560 microcontroller. Behavior data was acquired at a sampling rate of 10 kHz synchronized to the two-photon imaging. Mice ran head-fixed on the same treadmill belt for 3 days to become acclimated to the textures and cues. The following days (Days 4-7) were designated as familiar days for the purposes of stability analysis. We always compared activity across consecutive familiar days for each animal, either days 4&5, 5&6, or 6&7, taking data recording and template matching artifacts into account.
Statistics
Unless otherwise stated, all data are reported as the median (M) and its 95% confidence interval [Lower, L and Upper, U bounds], calculated through a resampling procedure previously described6. All statistical comparisons were done using a two-sided Wilcoxon signed-rank (for paired data) or rank-sum (for unpaired data) unless otherwise specified (See section Subsampled Decoder).
All analyses were performed using custom-written codes in MATLAB R2021a (MathWorks).
d-NMF
Initialization
Pre-processsing
Image stacks were motion-corrected for XY-motion using NoRMCorre99. For temporal downsampling, each downsampled frame was computed as the mean of the surrounding frames, rounded, and stored as unsigned 16-bit integers.
Division into Patches
Image stacks were divided into 81 patches of 64x64 pixel size, with an 8-pixel overlap.
ROI Core Detection
Within each patch, the image sequence was first de-trended and transformed into ΔF/F values by subtracting and dividing by the minimum value in a rolling 30-second window. The activity threshold was set to 3 times the median ΔF/F for each pixel and optionally passed through a temporal median filter. Connected components of active pixels were then detected in the resulting 3-dimensional (X-Y-Time) image stack. Components with membership ≥ 30 pixels were projected into 2 (X-Y) dimensions. See Supplementary Table 2a.
We merged components based on the Jaccard index (size of the intersection divided by the size of the union) between components, using a 0.5 merge threshold. This step merges ROIs that are active in distinct time spans and are detected multiple times in the previous step. Pixel values of each component are then normalized to sum to 1. Two extra ROIs were included, one composed of all pixels not belonging to any other ROI and the other composed of all pixels in the area being considered, to model background neuropil fluorescence.
Refinement
Iteration and Activity Extraction
The following steps are the standard fitting procedure for constrained non-negative matrix factorization28,32 (See Supplementary Table 2b) and are performed on the original image sequence patch, without de-trending or ΔF/F transformation. An image patch with t time steps and d x d pixel size is stored as a matrix Y \(\in {R}^{d\times d\times t}\). This is reshaped into the matrix Y* \(\in {R}^{{d}^{2}\times t}\). The objective function is the following:
where ‖∙‖F denotes the Frobenius norm. For k components, A \(\in {R}^{{d}^{2}\times k}\) represents the spatial footprints (ROIs), and C \(\in {R}^{k\times t}\) represents the time-varying activity for each ROI. η and β are regularization terms to enforce sparsity of the temporal components (C) and spatial components (A), respectively.
The algorithm then iterates over solving for C and A while enforcing non-negativity. The optimal solutions for C and A are as follows:
Where I represents the identify matrix, and 1 represents a matrix with each element equal to 1.
At each iteration n, C and A are then updated from their current values with a learning rate of α (here, 0.5):
After convergence, the resulting ROIs are passed through a 2-dimensional median filter of size 3x3, and the top 10% of pixel values are kept. Resulting ROIs are split into contiguous components, and final time courses are calculated by updating C from the final ROIs with a learning rate of α = 1. Only ROIs ≥ 30 pixels are included.
Merging
Combining Results from Patches
After all image patches are processed, the resulting ROIs are evaluated to be merged. The temporal traces are detrended by subtracting the minimal value in a rolling 30 second window. In data presented here, ROIs were merged if their temporal components had a correlation coefficient above 0.8, and they shared at least 1 pixel in membership. The spatial and temporal components of merged ROIs are then updated according to the algorithm in Supplementary Table 2c.
Merging Threshold Analysis
We present a method to estimate a lower bound of a useful merging threshold in Supplementary Fig. 3. This lower bound allows users to avoid linking ROIs from different neurons. Above this lower bound, the choice of merging threshold is up to the user, depending on the scientific question. For a given FOV, all pairwise correlations between the detrended activity traces of ROIs were computed. Then, for a range of threshold values from 1 to 0 (in steps of 0.01), we merged ROIs with correlations above that value. To estimate how well the clustering worked, we identified the cluster with the largest number of member ROIs. We defined the minimum correlation as the lowest correlation value between ROIs in that cluster. This approach is similar to complete-linkage hierarchical clustering100, with distance inversely proportional to correlation. We observed that all FOVs had a critical value below which the minimum correlation substantially decreased. This corresponded to ROIs from different neurons becoming linked. We estimate this as the lower bound on an interpretable merging threshold for that FOV.
Screening
Skewness Classification
The d-NMF output contains a mixture of valid and invalid ROIs. To automatically classify ROIs as valid or invalid, we computed the skewness of the extracted temporal traces after de-trending with a rolling 30-second window. For ground truth, human labelers manually screened all extracted ROIs and labeled them as ‘valid’ or ‘invalid’ based on morphology. Using skewness as a threshold value, we quantified the performance of the classifier using two metrics: the area under the ROC curve plotting the True Positive Rate versus the False Positive Rate, and the Jaccard Index, computed as TP/(TP + FP + FN), where TP is the count of True Positives, FP is the count of False Positives, and FN is the count of False Negatives (Fig. 2c).
Signal-to-noise ratio
We also tested the efficacy of the signal-to-noise ratio as a classifier threshold (Supplementary Fig. 4). For a de-trended calcium trace, the signal-to-noise ratio was defined as SNR = P/M, where P is the 99.9th percentile of the trace, and M is the median absolute deviation of the trace (median( | x – median(x)|)).
ROI Comparisons
Manual Labeling
Human experts manually labeled ROIs using FIJI software101 on 20X temporally downsampled image stacks to generate ground truth labeling data. Image stacks were inspected frame-by-frame for neural segments showing synchronous activity. Dendritic branches were split up into multiple ROIs at branch points. Additionally, labelers were instructed to break up long dendritic branches into smaller ROIs about the length of the soma (approximately 33 µm).
CaImAn Parameters
The python implementation of CaImAn28 was run on the motion-corrected, downsampled image stacks using default parameters using the sparse_nmf initialization procedure. Patch sizes and overlaps were chosen to be 64 x 64 and 8 pixels to match those used by d-NMF. Only those ROIs labeled as valid were used for analyses (Fig. 4). K = 15 or 30 components per patch (number of clusters parameter) were chosen to be estimated, giving results for CNMF-15 and CNMF-30, respectively, in Supplementary Fig. 7. We compared against the higher-performing CNMF-30 in Fig. 4.
suite2p Parameters
Suite2p was run on the motion-corrected, downsampled image stacks using pre-configured parameters for dendrites and axons. All labeled ROIs were used for analysis (Fig. 4). Three settings of suite2p were run, with different values for the threshold parameter: 1, 0.75, and 0.5, giving results for s2p-1, s2p-0.75, and s2p-0.5, respectively, in Supplementary Fig. 7. We compared against the higher-performing s2p-0.75 in Fig. 4.
F1 score for model comparison
The output of d-NMF, CNMF, and suite2p were compared to manually labeled data in a pixel-wise fashion. The metric used was the F1 score, calculated as
This measure is a combination of recall (True Positive Rate, TP/(TP + FN)) and precision (Positive Predictive Value, TP/(TP + FP)), as human-labeled datasets may have incomplete labeling, which would yield poor estimates of false positives. The True Positive Rate and F1 score computed pixel-wise are denoted TPRpx and F1px, respectively.
The pixel-wise metric defined above has advantages in its flexibility. It is agnostic to how a neuron may be parceled up, and quantifies completeness of labeling and accuracy of the placement of ROIs. It is most interpretable for sparsely labeled datasets, but has drawbacks when applied to densely labeled datasets. Namely, a single ROI covering all pixels of all neurons in the field of view will achieve the same score as a set of individual ROIs for each dendritic branch. Hence, we defined a second metric based entirely on ROI morphology.
Algorithmically defined ROIs can often span multiple dendritic branches or encompass soma and proximal branches (Supplementary Fig. 6). To define ROIs in a systematic manner, we performed a morphological split operation on all ROIs (Supplementary Fig. 5), drawing boundaries at branch points and separating putative somatic regions, defined by connected groups of pixels with a radius of at least 10 pixels (11 μm). Resulting ROIs with a size less than 30 pixels were discarded.
Manually-drawn ROIs that sub-segmented a dendritic branch were first merged together and then put through the same morphological split operation. This ensured that manually drawn ROIs followed the same well-defined guidelines across labelers. For each ROI in set A, we found the ROI in set B that had the highest spatial correspondence, as computed by the Jaccard index (size of the intersection divided by size of the union). We then define a pixel in an ROI as “covered” if it intersects with the matching ROI.
If A is the manually-drawn ground truth and B is a test set, define:
True Positive (TP) as the number of pixels that are covered in ROIs in set A
False Negative (FN) as the number of pixels that are not covered in ROIs in set A
False Positive (FP) as the number of pixels that are not covered in ROIs in set B
The True Positive Rate (TPR) and F1 score are then computed accordingly. These measures computed ROI-wise are denoted TPRroi and F1roi, respectively.
Construction of Synthetic Data
Movie Generation
To validate the accuracy of d-NMF, we constructed a series of synthetic datasets using one of our sparse FOVs (Fig. 3). To define ground truth, we took FOV2 (Supplementary Fig. 1), manually drew ROIs, and computed the activity of those ROIs by averaging over member pixels. A template movie was constructed by shifting the movie by -100 pixels in the Y dimension and -40 pixels in the X dimension, to center the visible neuron. This template was then modified by shifting the X-dimension of the entire movie by a random amount -50 to 50 μm, shifting the Y-dimension by -50 to -50 μm, then rotating by a random angle between 0 and 360 degrees. The altered template was then shifted in time by a random amount between -250 and 250 seconds. The ground truth ROIs and activity traces were modified accordingly. ROIs that fell outside of the original 512x512 pixel frame after shifting and rotating were discarded. The frames from this new movie were then added to the original template. This process was repeated 200 times, to generate synthetic movies with 1 to 201 neurons.
ROI Signal Quality
The overlaying of movie replicates increased the noise present in each pixel, which had the potential to degrade overall signal quality of the known ROIs. To quantify this degradation, we computed the signal quality of each ROI in our synthetic datasets (Fig. 3c-e). For a given ROI, the frame of maximum activity was identified from its ground truth activity trace. From the z-scored movie, the mean value of pixels within that ROI at that frame was computed to define the ROI signal quality. This value gets smaller as more neurons are added to the synthetic data, an unavoidable consequence of adding uncorrelated variables.
Evaluating Performance
Because different methods or experimenters may segment dendrites into more fine or coarse-grained ROIs, we defined an approach to compute the similarity between ground truth ROIs and ROIs obtained through automated methods. To do this, we introduce the concept of Coverage.
Define 2 sets of ROIs, set A and B and their corresponding activity traces. For any ROIi in set A, identify all ROIs in set B that 1) Spatially overlap ROIi; and 2) Are correlated in time above a value of 0.5. This excludes contributions from unrelated crossing ROIs. Take the union of these ROIs, and quantify the pixels covered by that union. Repeat this process by covering B with A. Define classification accuracy in a similar fashion as above:
If A is the ground truth and B is a test set, define:
True Positive (TP) as the number of pixels that are covered in ROIs in set A
False Negative (FN) as the number of pixels that are not covered in ROIs in set A
False Positive (FP) as the number of pixels that are not covered in ROIs in set B
The True Positive Rate and F1 score (Fig. 3e) are then computed accordingly, denoted as TPRcov and F1cov, respectively.
Note this measure differs from the ROI-based measure above because we know the ground truth activity to use to link together ROIs and thus are not restricted to choosing a single ROI with the highest overlap.
In Fig. 3f, these values were only computed for ROIs with a minimum Signal Quality of 2.
Temporal Fidelity Testing Using Synthetic Data
To test the limits of d-NMF in detecting localized activity, we generated synthetic data in two ways. For Supplementary Fig. 11a-b, entirely synthetic data was simulated in a dendritic segment 6 pixels in width by 90 pixels long, diagonally embedded in a 64 x 64-pixel space. Background fluorescence was generated independently for each pixel by drawing from an exponential distribution with mean 7, raised to the power of 1.8, to match the statistics of noise observed in our data.
Calcium transients were simulated by a difference of exponentials of the form
Where B is chosen to normalize values such that the peak of the waveform has amplitude A. When simulated at 30 Hz, a transient characterized by the time constant τ has a full width at half max of approximately τ/10 seconds.
3 transients with τ = 24 (2.4-second half-width), A = 700 were generated across the entire synthetic dendrite to provide global activity. A variable number N of additional transients with variable values of A and τ were present in a portion of the dendrite with variable length L. The full array of parameters is listed in Supplementary Table 4.
Each parameter combination was simulated 5 times, resulting in 5200 simulations to generate Supplementary Fig. 11b.
Data for Supplementary Fig. 11d was generated in a similar fashion. A patch of dendrite from FOV 2 (Supplementary Fig. 1) was taken as background activity, and calcium transients were added to the movie only for pixels belonging to a small branch. The array of parameters is listed in Supplementary Table 4.
Each parameter combination was simulated 5 times, resulting in 3600 simulations to generate Supplementary Fig. 11d.
Spatial accuracy was computed as the maximum correlation coefficient between any detected ROI and the target ROI. Temporal accuracy was computed as the proportion of injected transients that were detected in the ROI with the highest spatial correlation with the target ROI. Total accuracy was calculated as the product of these two measures.
Fitness Trace
To generate ground truth data to evaluate the Fitness Trace (Fig. 5), 90 ROIs generated from d-NMF (30 apical dendrites, 30 soma, and 30 basal dendrites, when possible) were chosen at random from 9 FOVs. Their activity traces were re-estimated by taking a weighted average across the ROI, which was then de-trended using a rolling minimum of 30-second frames, then z-scored. Putative transients were identified as contiguous time bins where the z-scored trace exceeded 1 standard deviation. These event detection criteria were meant to be permissive, to include examples of valid and invalid transients. Transients were then manually reviewed and classified as valid or invalid using a previously published graphical user interface39.
The image stack was de-trended and transformed into ΔF/F values, then z-scored across time. For a given ROI, a rectangular bounding box was defined around all non-zero values with a padding of 3 pixels in each dimension. The Fitness Trace for a given ROI at time t was defined as the correlation coefficient between the z-scored image stack at time t and the ROI, calculated only using the pixels in that bounding box.
Transient Detection Methods
To describe the different detection methods (Fig. 5, Supplementary Fig. 13), it is useful to define two types of activity traces. The first is the activity trace arrived at through d-NMF, that is, a signal de-mixed from overlapping sources; we denote this as De-mixed ΔF/F. The second is a simple signal obtained through taking a weighted average across all of the pixels of an ROI. We denote this as Simple ΔF/F. Detection method differences are summarized in Supplementary Table 5. Unless otherwise noted, activity traces are all de-trended by subtracting the rolling minimum across 30 seconds, then z-scored.
Sparse Data Processing
In addition to expressing GCaMP, animals used for the Sparse Data analyses (Supplementary Fig. 16) expressed a Cre-dependent tdTomato. ROIs were manually drawn and the time-varying activity in the green (G) and red (R) channels was computed by a simple average and temporally smoothed by convolving with a Gaussian kernel with σ = 33 ms. The activity of a given ROI (S) was then defined as the ratio of the calcium-dependent green fluorescence versus the calcium-independent red fluorescence (G/R)86. This step was critical to correct for motion in the Z plane. Calcium transients were manually identified and verified with the original image stacks for each ROI in the Sparse Data.
Spatial Tuning Properties
Single session analyses (Fig. 6, Supplementary Figs. 11-12) were performed on data from Familiar Day 1. Behavioral data was first downsampled to match the frame rate of imaging. Only data during movement (speed > 2 cm/s) was used for all spatial analyses. To construct firing rate maps, the track was divided into 40 equally spaced bins of width 5 cm. Occupancy and transient event count were computed, circularly smoothed with a Gaussian smoothing kernel of width 5 cm, and then divided to compute the rate.
ROIs were manually classified as apical dendrites, soma, or basal dendrites based on position in the field of view. ROIs encompassing both a soma and one or more dendritic branches were classified as soma. In all other instances we treat apical dendrites, soma, and basal dendrites as independent populations, without assigning parent soma to individual dendrites. Thus, when we refer to apical dendrites or basal dendrites, we are referring to dendrites, which are some distance from their parent cell body, which is often not visible in the same field of view.
We used two measures to quantify the degree of tuning of each rate map. Maps were divided into 40 bins as described above. Spatial information content102 was computed as
Where Pi is the normalized occupancy in the ith bin, such that the sum across all Pi = 1, and Ri is the normalized value of the rate map in the ith bin, such that the sum across all Ri = 1.
Sparsity was computed as
Where Ri is the value of the rate map in the ith bin.
Each ROI served as its own control to determine the statistical significance of spatial tuning. For each ROI, the transient times were shifted between -166 and 166 seconds (500 total shifts per ROI). The information content at a shift of 0 was then normalized to the 99th percentile of information content at non-0 shift. A normalized information content value greater than 1 indicated significant tuning. Additionally, a minimum of 4 transients must have been detected for an ROI to be called significantly tuned.
Tuning Parametrization
Tuning curves were parametrized as a mixture of Von Mises functions, as described previously103. Rate maps were circularly smoothed with a sigma of 15 cm (3 bins), and the curve was fit by increasing the number of Von Mises functions until the residuals were below 25% of the maximum value of the original rate map. Place field width was defined as the width of a fitted component at 50% of the component’s amplitude (i.e. full width at half max). For ROIs with multiple place fields, their width was defined as the mean width of all place fields.
Merging ROIs to Address Oversplitting
It is likely that extended dendrites could be split into multiple ROIs, especially if a portion of the dendrite dives out of the focal plane or if it is obscured by another neuron. This could lead to a problem of oversplitting, where multiple dendrites from the same neuron with highly correlated signals contribute multiple times to statistical estimates. To investigate if this biased our results, we merged dendritic ROIs with highly correlated activity into single ROIs (Supplementary Fig. 15, Supplementary Table 9). Correlation was calculated on a smoothed binary activity vector. This avoids issues of correlation being affected by different decay time constants or contaminated activity from overlapping dendrites. Dendrites of the same type (apical or basal) were merged if their activity was correlated above a value of 0.7. When 2 or more ROIs were merged, the statistics (activity rate, information content, rate map, etc.) were computed as the median value of the statistics of the ROIs being merged.
Quantifying Overlap
We defined two ROIs as overlapping if they shared at least 8 pixels and had a correlation coefficient between their activity traces less than 0.75. This avoids inflating the overlap amount from ROIs that are part of the same dendritic tree.
Distance Dependence
Distance dependence in the sparse dataset was defined as the path distance from the center of a neuron’s soma to the center of an ROI, only traversing ROIs that belong to that neuron.
To quantify distance-dependence in the dense dataset, a line was fit to the center of all somatic ROIs to define the cell body layer. Distance to any dendritic ROI was defined as the tangential distance to the cell body layer, as not all dendritic ROIs could be directly traced back to their parent soma.
Stability
Within-day stability (Fig. 7) was assessed by splitting the data from Familiar Day 1 in half. Rate maps were re-estimated for each ROI using only the data in each session half. The tuning curve (TC) correlation was defined as the correlation coefficient between the tuning curves for each half. This measure was computed for each ROI and then averaged to obtain a single value for the session. To be included in the calculation for TC correlation, an ROI must have had a minimum of 4 calcium transients and be significantly tuned across the entire session. Population vector (PV) correlation was computed by constructing the rate estimates of all ROIs for a given position bin in the first half and taking the correlation coefficient between that vector and the corresponding vector in the second half. This yields a value for each position bin, which was then averaged to obtain a single value for a session. To be included in the calculation for PV correlation, an ROI must have had a minimum of 4 calcium transients, with no criteria for significant tuning.
To quantify across-day stability (Fig. 7, Supplementary Figs. 14-15), image stacks from Familiar Days 1 and 2 were stitched into a single image stack and motion-corrected together. d-NMF was then run on the combined image stack to obtain ROIs that spanned both sessions. Stability analysis was then performed as for within-day estimates. To be included in across-day stability analyses, an ROI must have had at least 4 detected calcium transients in both days and significantly tuned in at least one day. The activity criterion ensured that correlation estimates were not skewed by ROIs that were not visible on one of the recording days.
Structural Stability
To verify that differences in tuning stability were not due to the inability to track ROIs across days, we additionally quantified the structural stability of ROIs (Supplementary Fig. 17). For each ROI we took the average of frames at which significant calcium transients were detected for Familiar Days 1 and 2. The structural stability was defined as the correlation coefficient between these averages, using only data within the modified boundary of the ROI. The modified boundary of an ROI was defined by first thresholding the ROI above 20% of its maximum pixel intensity, then dilating the resulting mask by one pixel.
Population Vector Decoding
To test the impact of increased across-day stability in apical dendrites, we performed population vector decoding in a manner similar to that described previously53 (Fig. 8). A separate decoder was constructed for each FOV for each of apical dendrites, soma, and basal dendrites. Template tuning curves for each ROI were constructed as above. For within-day decoding, data from the first half of Familiar Day 1 was used to define the template. For across-day decoding, all data from Familiar Day 1 was used to define the template. As above for defining Population Vector correlations, only ROIs that were active in both days and significantly tuned in at least one day were included for decoding.
Time-varying rate vectors for each ROI were constructed using data from either the second half of Familiar Day 1 (within-day decoding) or all data from Familiar Day 2 (across-day decoding), using 33 ms bins smoothed with a Gaussian smoothing kernel with σ = 667 ms. For each time point in the decoded portion of the data, the decoded position was the position corresponding to the highest correlation with the template matrix. Any time points with zero activity across all relevant ROIs had an undefined correlation with any position. The most recently decoded position was copied in to those undefined frames.
Decoding error was defined as the median absolute error between decoded position and actual position, defined circularly so the beginning of the track (position 0 cm) and the end of the track (position 200 cm) were 0 cm away.
Subsampled decoder
The performance of a population vector decoder is highly dependent on the number of units used to construct it53. To control for variation in the number of ROIs across compartments, we constructed additional population vector decoders using subsets of eligible ROIs (Fig. 8d,e). For each session and compartment type (apical, soma, basal), we randomly selected (without replacement) 5, 15, 25, 35, 45, 55, or 65 ROIs if available to decode position and record decoding error. This process was repeated 100 times and the median value across the 100 iterations was used as the decoding error of the subsampled classifier.
The statistical significance of decoding for subsampled decoders was assessed using a two-way ANOVA with the number of ROIs as a continuous predictor, the ROI type (Apical, Soma, or Basal) as a categorical predictor, and the decoding error as the dependent variable. The p-values reported in Fig. 8 are the p-values for an interaction effect between the number of ROIs and the ROI type, performed in a pairwise manner (Apical vs Soma, Apical vs Basal, Soma vs Basal).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Downsampled (1.5 Hz) imaging data and intermediate data analysis files generated in this study have been deposited in the Zenodo database: https://doi.org/10.5281/zenodo.14207307 and https://doi.org/10.5281/zenodo.14207412. The raw, 30 Hz imaging data are available under restricted access due to large file sizes, and access can be obtained by request to the corresponding authors, with an expected timeframe of two weeks for response.
Code availability
Code and data needed to generate the figures in this study can be downloaded from Zenodo: https://doi.org/10.5281/zenodo.14207307 and https://doi.org/10.5281/zenodo.14207412. The most updated version of the d-NMF code is available at https://github.com/basulab-nyu/d-NMF.
References
Park, J. et al. Contribution of apical and basal dendrites to orientation encoding in mouse V1 L2/3 pyramidal neurons. Nat. Commun. 10, 1–11 (2019).
Amitai, Y., Friedman, A., Gutnick, M. J. & Connors, B. W. Regenerative activity in apical dendrites of pyramidal cells in neocortex. Cereb. Cortex 3, 26–38 (1993).
Kim, H. G. & Connors, B. W. Apical Dendrites of the Neocortex: Correlation between Sodium- and Calcium-dependent Spiking and Pyramidal Cell Morphology. J. Neurosci. 13, 5301–5311 (1993).
Larkum, M. E., Zhu, J. J. & Sakmann, B. Dendritic mechanisms underlying the coupling of the dendritic with the axonal action potential initiation zone of adult rat layer 5 pyramidal neurons. J. Physiol. 533, 447–466 (2001).
Schiller, J., Major, G., Koester, H. J. & Schiller, Y. NMDA spikes in basal dendrites of cortical pyramidal neurons. Nature 404, 285–289 (2000).
Moore, J. J. et al. Dynamics of cortical dendritic membrane potential and spikes in freely behaving rats. Science 355, (2017).
Bilash, O. M., Chavlis, S., Johnson, C. D., Poirazi, P. & Basu, J. Lateral entorhinal cortex inputs modulate hippocampal dendritic excitability by recruiting a local disinhibitory microcircuit. Cell Rep 42, 111962 (2023).
Moore, J. J., Robert, V., Rashid, S. K. & Basu, J. Assessing Local and Branch-specific Activity in Dendrites. Neuroscience 489, 143–164 (2022).
Kupferman, J. V. et al. Reelin signaling specifies the molecular identity of the pyramidal neuron distal dendritic compartment. Cell 158, 1335–1347 (2014).
Tran-Van-Minh, A. et al. Contribution of sublinear and supralinear dendritic integration to neuronal computations. Front. Cell. Neurosci. 9, 1–15 (2015).
Mel, B. W. Synaptic integration in an excitable dendritic tree. J. Neurophysiol. 70, 1086–1101 (1993).
Poirazi, P., Brannon, T. & Mel, B. W. Pyramidal neuron as two-layer neural network. Neuron 37, 989–999 (2003).
Mehta, M. R. Cooperative LTP can map memory sequences on dendritic branches. Trends Neurosci 27, 69–72 (2004).
London, M. & Häusser, M. Dendritic computation. Annu. Rev. Neurosci. 28, 503–532 (2005).
Johnston, D. & Narayanan, R. Active dendrites: colorful wings of the mysterious butterflies. Trends Neurosci 31, 309–316 (2008).
Golding, N. L., Staff, N. P. & Spruston, N. Dendritic spikes as a mechanism for cooperative long-term potentiation. Nature 418, 326–331 (2002).
Gambino, F. et al. Sensory-evoked LTP driven by dendritic plateau potentials in vivo. Nature 515, 116–119 (2014).
Basu, J. et al. Gating of hippocampal activity, plasticity, and memory by entorhinal cortex long-range inhibition. Sciene (80-.). 351, aaa5694 (2016).
Chen, T. W. et al. Ultrasensitive fluorescent proteins for imaging neuronal activity. Nature 499, 295–300 (2013).
Dana, H. et al. High-performance calcium sensors for imaging activity in neuronal populations and microcompartments. Nat. Methods 16, 649–657 (2019).
Sheffield, M. E. J. & Dombeck, D. A. Calcium transient prevalence across the dendritic arbour predicts place field properties. Nature 517, 200–204 (2015).
Cichon, J. & Gan, W. B. Branch-specific dendritic Ca2+ spikes cause persistent synaptic plasticity. Nature 520, 180–185 (2015).
Francioni, V., Padamsey, Z. & Rochefort, N. L. High and asymmetric somato-dendritic coupling of V1 layer 5 neurons independent of visual stimulation and locomotion. Elife 8, 1–25 (2019).
Kerlin, A. et al. Functional clustering of dendritic activity during decision-making. Elife 8, 1–32 (2019).
Otor, Y. et al. Dynamic compartmental computations in tuft dendrites of layer 5 neurons during motor behavior. Science (80-.). 376, 267–275 (2022).
Huber, D. et al. Multiple dynamic representations in the motor cortex during sensorimotor learning. Nature 484, 473–478 (2012).
Mau, W. et al. The Same Hippocampal CA1 Population Simultaneously Codes Temporal Information over Multiple Timescales. Curr. Biol. 28, 1499–1508.e4 (2018).
Giovannucci, A. et al. CaImAn an open source tool for scalable calcium imaging data analysis. Elife 8, 1–45 (2019).
Pachitariu, M. et al. Suite2p: beyond 10,000 neurons with standard two-photon microscopy. bioRxiv 061507 https://doi.org/10.1101/061507 (2016).
Mukamel, E. A., Nimmerjahn, A. & Schnitzer, M. J. Automated Analysis of Cellular Signals from Large-Scale Calcium Imaging Data. Neuron 63, 747–760 (2009).
Kaifosh, P., Zaremba, J. D., Danielson, N. B. & Losonczy, A. SIMA: Python software for analysis of dynamic fluorescence imaging data. Front. Neuroinform. 8, 1–10 (2014).
Pnevmatikakis, E. A. et al. Simultaneous Denoising, Deconvolution, and Demixing of Calcium Imaging Data. Neuron 89, 285 (2016).
Sheffield, M. E. J., Adoff, M. D. & Dombeck, D. A. Increased Prevalence of Calcium Transients across the Dendritic Arbor during Place Field Formation. Neuron 96, 490–504.e5 (2017).
Rajasethupathy, P. et al. Projections from neocortex mediate top-down control of memory retrieval. Nature 526, 653–659 (2015).
Lee, D. D. & Seung, H. S. Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791 (1999).
Trautmann, E. M. et al. Dendritic calcium signals in rhesus macaque motor cortex drive an optical brain-computer interface. Nat. Commun. 12, (2021).
Cornejo, V. H., Ofer, N. & Yuste, R. Voltage compartmentalization in dendritic spines in vivo. Science (80-.) 375, 82–86 (2022).
Jaccard, P. The Distribution of the Flora in the Alpine Zone. New Phytol 11, 37–50 (1912).
Gauthier, J. L. et al. Detecting and correcting false transients in calcium imaging. Nat. Methods https://doi.org/10.1038/s41592-022-01422-5 (2022).
Leutgeb, S., Leutgeb, J. K., Treves, A., Moser, M. B. & Moser, E. I. Distinct ensemble codes in hippocampal areas CA3 and CA1. Science (80-.). 305, 1295–1298 (2004).
Muller, R. U., Kubie, J. L. & Ranck, J. B. Spatial firing patterns of hippocampal complex-spike cells in a fixed environment. J. Neurosci. 7, 1935–1950 (1987).
Barnes, C. A., McNaughton, B. L., Mizumori, S. J. Y., Leonard, B. W. & Lin, L.-H. Comparison of spatial and temporal characteristics of neuronal activity in sequential stages of hippocampal processing. Prog. Brain Res. 83, 287–300 (1990).
Dong, C., Madar, A. D. & Sheffield, M. E. J. Distinct place cell dynamics in CA1 and CA3 encode experience in new environments. Nat. Commun. 12, 1–13 (2021).
Hainmueller, T. & Bartos, M. Parallel emergence of stable and dynamic memory engrams in the hippocampus. Nature 558, 292–296 (2018).
Zhou, P. et al. Efficient and accurate extraction of in vivo calcium signals from microendoscopic video data. Elife 7, 1–37 (2018).
Robert, V. et al. Entorhinal cortex glutamatergic and GABAergic projections bidirectionally control discrimination and generalization of hippocampal representations. bioRxiv (2023).
Bittner, K. C. et al. Conjunctive input processing drives feature selectivity in hippocampal CA1 neurons. Nat. Neurosci. 18, 1133–1142 (2015).
Bittner, K. C., Milstein, A. D., Grienberger, C., Romani, S. & Magee, J. C. Behavioral time scale synaptic plasticity underlies CA1 place fields. Science (80-.). 357, 1033–1036 (2017).
McKenzie, S. et al. Preexisting hippocampal network dynamics constrain optogenetically induced place fields. Neuron 109, 1040–1054.e7 (2021).
Sun, Q. et al. Proximodistal Heterogeneity of Hippocampal CA3 Pyramidal Neuron Intrinsic Properties, Connectivity, and Reactivation during Memory Recall. Neuron 95, 656–672.e3 (2017).
Hunt, D. L., Linaro, D., Si, B., Romani, S. & Spruston, N. A novel pyramidal cell type promotes sharp-wave synchronization in the hippocampus. Nat. Neurosci. 21, 985–995 (2018).
Soltesz, I. & Losonczy, A. CA1 pyramidal cell diversity enabling parallel information processing in the hippocampus. Nat. Neurosci. 21, 484–493 (2018).
Zemla, R., Moore, J. J., Hopkins, M. D. & Basu, J. Task-selective place cells show behaviorally driven dynamics during learning and stability during memory recall. Cell Rep 41, 111700 (2022).
Mau, W., Hasselmo, M. E. & Cai, D. J. The brain in motion: How ensemble fluidity drives memory-updating and flexibility. Elife 9, 1–24 (2020).
Qin, S. et al. Coordinated drift of receptive fields in Hebbian/anti-Hebbian network models during noisy representation learning. Nat. Neurosci 26, 339–349 (2023).
Ziv, Y. et al. Long-term dynamics of CA1 hippocampal place codes. Nat. Neurosci. 16, 264–266 (2013).
Schoonover, C. E., Ohashi, S. N., Axel, R. & Fink, A. J. P. Representational drift in primary olfactory cortex. Nature 594, 541–546 (2021).
Deitch, D., Rubin, A. & Ziv, Y. Representational drift in the mouse visual cortex. Curr. Biol. 31, 4327–4339.e6 (2021).
Marks, T. D. & Goard, M. J. Stimulus-dependent representational drift in primary visual cortex. Nat. Commun. 12, 1–16 (2021).
Rokni, U., Richardson, A. G., Bizzi, E. & Seung, H. S. Motor Learning with Unstable Neural Representations. Neuron 54, 653–666 (2007).
Driscoll, L. N., Pettit, N. L., Minderer, M., Chettih, S. N. & Harvey, C. D. Dynamic Reorganization of Neuronal Activity Patterns in Parietal Cortex. Cell 170, 986–999.e16 (2017).
Lecoq, J. et al. Visualizing mammalian brain area interactions by dual-axis two-photon calcium imaging. Nat. Neurosci. 17, 1825–1829 (2014).
Diehl, G. W., Hon, O. J., Leutgeb, S. & Leutgeb, J. K. Stability of medial entorhinal cortex representations over time. Hippocampus 29, 284–302 (2019).
Rolls, E. T. The mechanisms for pattern completion and pattern separation in the hippocampus. 7, 1–21 (2013).
Yassa, M. A. & Stark, C. E. L. Pattern separation in the hippocampus. Trends Neurosci 34, 515–525 (2011).
Guzman, S. J., Schlögl, A., Frotscher, M. & Jonas, P. Synaptic mechanisms of pattern completion in the hippocampal CA3 network. Science (80-.). 353, 1117–1123 (2016).
Leutgeb, J. K., Leutgeb, S., Moser, M. B. & Moser, E. I. Pattern separation in the dentate gyrus and CA3 of the hippocampus. Science (80-.). 315, 961–966 (2007).
Neunuebel, J. P. & Knierim, J. J. CA3 retrieves coherent representations from degraded input: Direct evidence for CA3 pattern completion and dentate gyrus pattern separation. Neuron 81, 416–427 (2014).
Callaway, E. M. Feedforward, feedback and inhibitory connections in primate visual cortex. Neural Networks 17, 625–632 (2004).
Spruston, N. Pyramidal neurons: Dendritic structure and synaptic integration. Nat. Rev. Neurosci. 9, 206–221 (2008).
Larkum, M. A cellular mechanism for cortical associations: An organizing principle for the cerebral cortex. Trends Neurosci 36, 141–151 (2013).
Sacramento, J., Bengio, Y., Costa, R. P. & Senn, W. Dendritic cortical microcircuits approximate the backpropagation algorithm. (2018).
Golkar, S., Tesileanu, T., Bahroun, Y., Sengupta, A. M. & Chklovskii, D. B. Constrained Predictive Coding as a Biologically Plausible Model of the Cortical Hierarchy. 1–24 (2022).
Siegel, M., Körding, K. P. & König, P. Integrating Top-Down and Bottom-Up Sensory Processing by Somato-Dendritic Interactions. 173, 161–173 (2000).
Losonczy, A., Makara, J. K. & Magee, J. C. Compartmentalized dendritic plasticity and input feature storage in neurons. Nature 452, 436–441 (2008).
Brandalise, F., Carta, S., Helmchen, F., Lisman, J. & Gerber, U. Dendritic NMDA spikes are necessary for timing-dependent associative LTP in CA3 pyramidal cells. Nat. Commun. 7, 1–9 (2016).
Golding, N. L., Kath, W. L. & Spruston, N. Dichotomy of action-potential backpropagation in CA1 pyramidal neuron dendrites. J. Neurophysiol. 86, 2998–3010 (2001).
Kim, S., Guzman, S. J., Hu, H. & Jonas, P. Active dendrites support efficient initiation of dendritic spikes in hippocampal CA3 pyramidal neurons. Nat. Neurosci. 15, 600–606 (2012).
Kerlin, A. et al. Functional clustering of dendritic activity during decision-making. Elife 8, 1–33 (2019).
Smith, S. L., Smith, I. T., Branco, T. & Häusser, M. Dendritic spikes enhance stimulus selectivity in cortical neurons in vivo. Nature 503, 115–120 (2013).
Beaulieu-Laroche, L., Toloza, E. H. S., Brown, N. J. & Harnett, M. T. Widespread and Highly Correlated Somato-dendritic Activity in Cortical Layer 5 Neurons. Neuron 103, 235–241.e4 (2019).
O’Hare, J. K. et al. Compartment-specific tuning of dendritic feature selectivity by intracellular Ca2+ release. Science (80-.). 375, eabm1670 (2022).
d’Aquin, S. et al. Compartmentalized dendritic plasticity during associative learning. Science (80-.). 376, eabf7052 (2022).
Jia, H., Rochefort, N. L., Chen, X. & Konnerth, A. Dendritic organization of sensory input to cortical neurons in vivo. Nature 464, 1307–1312 (2010).
Branco, T., Clark, B. A. & Häusser, M. Dendritic Discrimination of Temporal Input Sequences in Cortical Neurons. Science (80-.). 329, 1671–1675 (2010).
Tsay, D., Dudman, J. T. & Siegelbaum, S. A. HCN1 Channels Constrain Synaptically Evoked Ca2+ Spikes in Distal Dendrites of CA1 Pyramidal Neurons. Neuron 56, 1076–1089 (2007).
Popovic, M. A., Carnevale, N., Rozsa, B. & Zecevic, D. Electrical behaviour of dendritic spines as revealed by voltage imaging. Nat. Commun. 6, (2015).
Kaifosh, P., Lovett-Barron, M., Turi, G. F., Reardon, T. R. & Losonczy, A. Septo-hippocampal GABAergic signaling across multiple modalities in awake mice. Nat. Neurosci. 16, 1182–1184 (2013).
Lovett-Barron, M. et al. Dendritic Inhibition in the Hippocampus Supports Fear Learning. Science (80-.). 343, 857–864 (2014).
Srinivasan, R. et al. New Transgenic Mouse Lines for Selectively Targeting Astrocytes and Studying Calcium Signals in Astrocyte Processes In Situ and In Vivo. Neuron 92, 1181–1195 (2016).
Bindocci, E. et al. Three-dimensional Ca2+ imaging advances understanding of astrocyte biology. Science (80-.). 356, eaai8185 (2017).
Zhang, Y. et al. Fast and sensitive GCaMP calcium indicators for imaging neural populations. bioRxiv 2021.11.08.467793 https://doi.org/10.1101/2021.11.08.467793 (2021).
Piatkevich, K. D. et al. A robotic multidimensional directed evolution approach applied to fluorescent voltage reporters article. Nat. Chem. Biol. 14, 352–360 (2018).
Abdelfattah, A. S. et al. Bright and photostable chemigenetic indicators for extended in vivo voltage imaging. Science (80-.) 364, 699–704 (2019).
Andermann, M. L. et al. Chronic cellular imaging on entire cortical columns in awake mice using microprisms. Neuron 80, 900–913 (2013).
Kohara, K. et al. Cell type-specific genetic and optogenetic tools reveal hippocampal CA2 circuits. Nat. Neurosci. 17, 269–279 (2014).
Danielson, N. B. et al. Distinct Contribution of Adult-Born Hippocampal Granule Cells to Context Encoding. Neuron 90, 101–112 (2016).
Magnus, C. J. et al. Ultrapotent chemogenetics for research and potential clinical applications. Science (80-.). 364, (2019).
Pnevmatikakis, E. A. & Giovannucci, A. NoRMCorre: An online algorithm for piecewise rigid motion correction of calcium imaging data. J. Neurosci. Methods 291, 83–94 (2017).
Everitt, B., Landau, S., Leese, M. & Stahl, D. Cluster analysis. In Cluster Analysis 62–64 (Arnold, 2001). https://doi.org/10.1007/BF00154794.
Schindelin, J. et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Skaggs, W. E., McNaughton, B. L., Gothard, K. M. & Markus, E. J. An Information-Theoretic Approach to Deciphering the Hippocampal Code. Adv. neural Inf. Process. 5, 1030–1037 (1993).
Moore, J. J., Cushman, J. D., Acharya, L., Popeney, B. & Mehta, M. R. Linking hippocampal multiplexed tuning, Hebbian plasticity and navigation. Nature https://doi.org/10.1038/s41586-021-03989-z (2021).
Acknowledgements
This work was supported by funding to J.B. from the NIH NINDS BRAIN INITIATIVE grant 1R01NS109994, NINDS 1R01NS109362, NINDS 1RM1NS132981, NIH NIMH 5R01MH122391, Mathers Charitable Foundation Investigator Award, McKnight Scholar Award in Neuroscience, Klingenstein-Simons Fellowship Award in Neuroscience, Alzheimer’s Association Research Grant to Promote Diversity – New to the Field (AARG-D-NTF), Alfred P. Sloan Research Fellowship, Whitehall Research Grant, American Epilepsy Society Junior Investigator Award, Blas Frangione Young Investigator Research Grant, New York University Whitehead Fellowship for Junior Faculty in Biomedical and Biological Sciences, and Leon Levy Foundation; and funding to D.B.C. from the Flatiron Institute, Center for Computational Neuroscience. J.J.M. was supported by an HHMI extension grant to D.B.C., a T32AG052909 grant (Wisniewski, Scharfman), and the Emerald Foundation. S.K.R was supported by a NIH T32GM007308. N.C. was supported by a McKnight Endowment Fund for Neuroscience Mathew Pecot URM Award. We thank Martial Dufour for early development of the experimental preparation and Roland Zemla for his contribution in developing the hardware and software for the custom treadmill and behavioral tracking system. We are indebted to Andrea Giovannucci, Eric Thomson, Pat Gunn, and Johannes Friedrich for their expertise and support in modifying CNMF and CaImAn parameters. NYU High Performance Computing resources were used for data analysis. We thank Tanvi Butola for generously providing axonal calcium imaging data (Data reused from Butola et al., Nature Neuroscience 2025 in Press, NN-A80353B manuscript with permission from Nature Neuroscience); Rafael Yuste and Victor Cornejo for generously providing dendritic voltage imaging data (Data reused with permission from 37); and Daniel Dombeck and Mark Sheffield for generously providing dendritic calcium imaging data (Data reused with permission from 21). We thank Pauline Rahal and Buyong Kim for help with data curation. We thank Vincent Robert, Keelin O'Neil and Tanvi Butola for valuable discussions and critical review of previous versions of the manuscript.
Author information
Authors and Affiliations
Contributions
J.J.M. and J.B. conceived the project and designed experiments. S.K.R. and J.J.M. performed surgeries and recorded data. J.J.M. developed all software code and performed all analyses with guidance and supervision from J.B. and D.B.C. J.J.M. and E.B. wrote software. J.J.M., S.K.R., E.B., C.D.J., and N.C. performed manual curation of data. J.J.M. and J.B. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Moore, J.J., Rashid, S.K., Bicker, E. et al. Sub-cellular population imaging tools reveal stable apical dendrites in hippocampal area CA3. Nat Commun 16, 1119 (2025). https://doi.org/10.1038/s41467-025-56289-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56289-9
This article is cited by
-
Hippocampus shapes entorhinal cortical output through a direct feedback circuit
Nature Neuroscience (2025)