Abstract
Circulating tumor DNA (ctDNA) is an emerging biomarker for the treatment of early breast cancer (EBC). We sought to evaluate a highly sensitive tumor-informed ctDNA assay in a real-world cohort of patients receiving neoadjuvant therapy (NAT) to assess clinical validity and explore prognostic outcomes. ctDNA is detected in 77.2% (88/114) of participants at baseline, with 18/88 (20.5%) having a baseline estimated variant allele frequency (eVAF) of <0.01%. Persistent detection of ctDNA, measured midway through NAT (mid-NAT), is associated with disease recurrence in all participants, reaching statistical significance in those with HER2-negative disease. Stratified analyses demonstrate that ctDNA detected mid-NAT enhances the prognostic accuracy of the residual cancer burden (RCB) score for disease recurrence. Postoperative or follow-up detection of ctDNA demonstrates a 100% positive predictive value for disease recurrence, with a median lead time of 374 days (range: 13-1010 days). These data suggest that assays with high analytical sensitivity may improve baseline ctDNA detection in patients with EBC. The ability to replicate the prognostic association of ctDNA dynamics in a real-world cohort supports further investigation. Prospective trials incorporating ctDNA testing are warranted to assess and develop the clinical utility of ctDNA-guided treatment strategies.
Similar content being viewed by others
Introduction
In jurisdictions with access to routine screening programs, most breast cancers are diagnosed in early stages when confined to the breast with or without involvement of the locoregional lymph nodes (early breast cancer, EBC). Many patients diagnosed with EBC receive neoadjuvant systemic therapy (NAT), usually in the form of multi-agent chemotherapy with or without targeted therapy, delivered in a risk-adapted strategy based on routine clinical and pathologic factors1. The pre-operative delivery of chemotherapy permits assessment of tumor response through radiographic and pathologic assessment2,3,4. Pathologic complete response (pCR) following such therapy is associated with a favorable prognosis, while increasing levels of residual disease as measured by scales such as the residual cancer burden (RCB) are associated with elevated risk of recurrence5. While residual disease assessed pathologically informs the delivery of subsequent adjuvant systemic therapy, there are no standard biomarkers used for early identification of cancers with sub-optimal or poor response to therapy, which could allow for treatment escalation or switch strategies. Furthermore, while ctDNA detected in follow-up is associated with recurrence in patients treated with curative intent, individual assay characteristics must be defined to develop clinical utility.
Liquid biopsy permits the identification, quantification, and characterization of tumor derived DNA fragments in the peripheral blood (circulating tumor DNA, ctDNA). Changes in ctDNA levels (ctDNA dynamics) are associated with treatment response in many cancers; identification of ctDNA after the delivery of curative intent therapy is associated with disease recurrence6,7,8,9,10,11,12. Various ctDNA assays exists, with differing technical approaches and levels of sensitivity. Defining the clinical validity and prognostic capabilities of relevant assays is important to understand their potential clinical utility, future positioning in clinical trials, and routine patient care. To address this, we sought to evaluate the characteristics of a high sensitivity (limit of detection, LoD 95%: 0.001%) tumor-informed assay. We evaluated a large real-world cohort of patients with EBC (all receptor subtypes) receiving standard of care NAT. Herein, we present data assessing assay sensitivity at baseline, evaluation of ctDNA dynamics on NAT, exploring their relationship to clinical outcomes, and assess serial monitoring of ctDNA to anticipate disease recurrence in the adjuvant setting.
Results
Cohort identification and assay metrics
142 participants enrolled from October 2016 to January 2021 were identified for analysis (Fig. 1). Eleven participants were excluded due to a lack of sufficient baseline tumor tissue for genomic DNA (gDNA) extraction and whole-exome sequencing (WES, assay requirement). Assays were successfully generated in 93% (122/131) of participants (Fig. 1). Three participants, for whom panels were designed, were identified to have developed metastatic disease early in their course of treatment (prior to surgery) and were excluded from the primary analysis. 119 participants had evaluable data and were included for analysis in the primary cohort (Table 1).
The median WES coverage for the 119 participants included was 246x (Supplementary Table 1; Range: 21x–711x). From this, personalized panels were designed for each participant with a median of 48 variants per panel (range: 22-53; Supplementary Table 1). Of these identified variants, a median of 39 (range: 8–49; Supplementary Table 1) passed panel quality control metrics, including exclusion of variants detected in the buffy coat (Supplementary Table 2). These final panels were used to evaluate the presence of ctDNA within extracted cell free DNA (cfDNA) from each plasma timepoint (Supplementary Table 3).
Cohort characteristics
Of the 119 participants included in the primary analysis, 118 were female, 64.4% were premenopausal (Table 1 and Supplementary Fig. 2A). Median age at diagnosis was 49 years (Table 1; Range: 24-79). Of the 119 participants, 46 (38.7%) were HER2-positive (HER2+), 41 (34.4%) estrogen receptor positive/HER2-negative (ER+), and 32 (26.9%) triple negative (TNBC). 103/119 participants (86.6%) received neoadjuvant sequential anthracycline- and taxane-based chemotherapy, the majority receiving dose-dense doxorubicin+cyclophosphamide, followed by dose dense paclitaxel with or without anti-HER2 antibody therapy (Table 1). Median clinical follow-up from diagnosis was 4.7 years (range: 0.6-7.7 years). As of the date of data cutoff (methods), 19 recurrences (1 local, 18 distant) had occurred (11 ER+, 7 TNBC, 1 HER2+) (Fig. 1).
Clinical characteristics and baseline ctDNA detection
In total, 681 individual plasma timepoints (median: 6 per participant, range: 1–12) were evaluated for the presence of ctDNA. ctDNA was detected in 24% (163/681) of all time points with 37% (61/163) having an eVAF less than 0.01% and 5% (9/163) less than 0.001% (Fig. 2A).
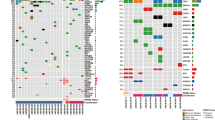
A Distribution of eVAF across baseline timepoints evaluated (n = 114 baseline samples). B Number of variants included in personalized ctDNA panels, by receptor subtype (One-way ANOVA, p = 0.0003; ER+ n = 40, TNBC n = 29, HER2+ n = 45). C Number of variants per personalized panel in those with and without ctDNA detected at baseline, by receptor subtype (t-test, p=ns; ER+/ctDNA+ n = 28, ER+/ctDNA- n = 12, TNBC/ctDNA+ n = 26, TNBC/ctDNA- n = 3, HER2+/ctDNA+ n = 34, HER2+/ctDNA- n = 11). D Baseline eVAF distribution in all participants and by receptor subtype (One-way ANOVA, p = 0.68; ER+ n = 40, TNBC n = 29, HER2+ n = 45). eVAF in baseline samples for participants by: E clinical T-size (One-way ANOVA, p = 0.082; T1 n = 13, T2 n = 61, T3 n = 32, T4 n = 8), F clinical stage (One-way ANOVA, p = 0.19; IA n = 6, IIA n = 27, IIB n = 50, IIIA n = 20, IIIB n = 7, IIIC n = 4), G clinical N-stage (One-way ANOVA, p = 0.048; N0 n = 42, N1 n = 60, N2 n = 7, N3 n = 4) H Nottingham clinical grade as assessed on diagnostic biopsies (One-way ANOVA, p = 0.039; Grade 1 or 2 n = 32, Grade 2 to 3 n = 22, Grade 3 n = 60) I Recurrence status (t-test, p = 0.024; Recurrence n = 19, No Recurrence n = 95). J Representative Oncoplot of common mutations in breast cancer-related genes, clinical receptor subtype, mutation classification, TMB (mut/Mb), and baseline detection of ctDNA. Box plots demonstrate median, minimum, and maximum values. All values are illustrated individually where horizontal lines represent the median. All p-values are 2-sided, with p < 0.05 indicating statistical significance. ER+: estrogen receptor positive/HER2-negative, TNBC triple negative breast cancer, HER2+ human epidermal growth factor receptor positive, ctDNA+ ctDNA detected, ctDNA- ctDNA not detected, NS not significant, T clinical tumor size, N clinical nodal status.
There were fewer variants identified for panels in ER+ participants (n = 23, range: 7–48) than those with TNBC (n = 37, range: 14–48) or HER2+ (n = 36, range: 13–47) disease (Fig. 2B, p = 0.0003). Despite this, tumor mutational burden (TMB, mutations/Mb; mut/Mb) did not differ by baseline ctDNA detection status (Supplementary Fig. 2B) or between ER+ or HER2+ subtypes (Supplementary Fig. 2C, D). TMB was lower in individuals with TNBC and no ctDNA detected at baseline, but this analysis is limited by the small number of cases (Supplementary Fig. 2E, p = 0.024). Within subtypes, there was no difference in the number of variants per panel in those with or without ctDNA detected at baseline by receptor subtype (Fig. 2C).
The overall rate of ctDNA detection at baseline was 77.2% (88/114) (Table 2); baseline detection differed by receptor subtype: ER+ 70.0% (28/40), TNBC 89.7% (26/29), and HER2+ 75.6% [34/45; HER2+/ER−: 13/15 (87.0%) and HER2+/ER+: 21/30 (70.0%)] (Table 2, p = 0.15). Median estimated variant allele frequency (eVAF) in baseline samples with ctDNA detected was 0.0823% (Fig. 2D; range: 2.90×10-5 to 7.5%) with 18/88 (20.5%) participants having a baseline eVAF <0.01%, which did not differ between clinical receptor subtypes (Fig. 2D, p = 0.68).
Baseline ctDNA detection was not associated with clinical tumor size (Table 2, p = 0.60), nodal involvement (Table 2, p = 0.31), or anatomic stage (Table 2, AJCC 8th Edition; p = 0.74), but was significantly associated with a higher Nottingham grade as assessed on the diagnostic biopsy (Table 2, p = 0.0498). There were non-significant associations between baseline eVAF, tumor size (T-stage; Fig. 2E, p = 0.082) and anatomic stage (AJCC 8th Edition; Supplementary Fig. 2F, p = 0.19). Clinical nodal status (N-stage; Fig. 2G, p = 0.048) and Nottingham grade as measured on the diagnostic biopsy (Fig. 2H, p = 0.039) were associated with a higher eVAF at baseline.
All but one participant (18/19, 94.7%; low grade ER+) with clinical recurrence had ctDNA detected at baseline. Baseline eVAF was higher in those who experienced clinical recurrence than those who did not (Fig. 2I; p = 0.024).
Alterations in commonly mutated breast cancer genes were evaluated using a tumor-only filtering strategy (Fig. 2J). The presence of common breast cancer mutations was not associated with ctDNA detection at baseline including TP53 (n = 52; p = 0.20), PIK3CA (n = 15; p = 1.0), and BRCA2 (n = 14; p = 1.0).
Treatment-related changes in ctDNA and clinical outcome
Detection of ctDNA at baseline was associated with a shorter recurrence-free interval (RFI) (Fig. 3A; HR: 2.89, 95% CI: 1.003–8.31, p = 0.049) albeit with high sensitivity but low predictive value (Supplementary Table 5); the 3-year recurrence-free rate was 96.2% in those without ctDNA detected and 83.4% in those with ctDNA detected at baseline. In an exploratory analysis by receptor subtype, this association was present but not statistically significant in participants with ER + EBC (Supplementary Fig. 3A; HR: 3.03, 95% CI: 0.87–10.62, p = 0.083). Analysis of TNBC and HER2+ subgroups were not performed given the high baseline sensitivity and lack of recurrence events, respectively. ctDNA clearance was observed in most participants following initiation of NAT (Supplementary Fig. 3B; pre-cycle 2 (C2), mid-NAT, preoperative, postoperative, and follow-up).
A Baseline ctDNA detection and association with RFI (HR: 2.89, 95% CI: 1.003–8.31; Log-rank (Mantel–Cox) p = 0.049). B Mid-NAT ctDNA detection and association with RFI in all participants (HR: 3.59, 95% CI: 0.99–12.87; Log-rank (Mantel–Cox) p = 0.050). C Mid-NAT ctDNA detection and association with RFI in participants with HER2-negative disease (HR: 14.43, 95% CI: 3.00–69.53; Log-rank (Mantel-Cox) p = 0.0009). HR hazard ratio, 95% CI: 95% confidence interval, NR not reached.
The association between treatment-related clearance in ctDNA and disease recurrence was evaluated; patients receiving neoadjuvant endocrine therapy (n = 2, both of whom did not have ctDNA detected at baseline) were excluded from these analyses. Most participants experienced early clearance of ctDNA. Persistent detection of ctDNA pre-C2 was not associated with an increased risk of recurrence in all participants (Supplementary Fig. 3C; HR: 1.13, 95%CI: 0.44-2.96, p = 0.80). However, persistent detection of ctDNA mid-NAT was associated with a worse RFI in all participants, with 6/14 evaluable patients experiencing disease recurrence (Fig. 3B; HR: 3.59, 95%CI: 0.99-12.87, p = 0.050). Mid-NAT ctDNA clearance was not predictive of residual disease but all participants with ctDNA detected had residual disease in their surgical specimens (data not shown, p = 0.36). While mid-NAT detection identified a high-risk population, the positive predictive value (PPV) for clinical recurrence was only 35% (Supplementary Table 5; sensitivity: 43%, specificity: 81%). In a time-dependent cox-regression using only time points collected prior to surgery, persistent ctDNA detection was significantly associated with disease recurrence (p = 0.001). The delivery of anti-HER2 therapy in the second half of NAT may be a confounding factor between those with HER2+ and negative disease. Of those with ctDNA detected mid-NAT, 47% (8/17) were HER2+, of which no participant experienced a subsequent clinical recurrence while 67% (6/9) of the HER2-negative participants experienced disease recurrence at the time of the data cutoff. In an analysis excluding HER2+ participants, the detection of ctDNA mid-NAT was associated with a worse RFI (Fig. 3C; HR: 14.43, 95% CI: 3.00–69.53, p = 0.0009) with a PPV of 67% (Supplementary Table 5; sensitivity: 46%, specificity: 91%). This association was present irrespective of ER status (Supplementary Fig. 3D, E).
Treatment-related changes in ctDNA and clinical outcome stratified by RCB
The presence of residual invasive disease at surgery is known to be associated with a higher risk of recurrence. RCB status was prognostic of RFI in this cohort (Supplementary Fig. 4A, p = 0.0014). To account for this, we evaluated the relationship between treatment-related ctDNA changes and outcome, considering the effect in patients with or without residual disease using stratified analyses.
When stratified by RCB, mid-NAT ctDNA detection was associated with a worse RFI in the entire cohort (Fig. 4A, p = 0.011) and in those with HER2-negative disease (Fig. 4B, p = 0.0016). The prevalence of mid-NAT ctDNA positivity was similar between RCB groups (mid-NAT ctDNA detected vs. not detected: RCB-1: 18% vs. 82%, RCB-2: 19% vs. 81%, and RCB-3: 23% vs. 77%). No participants with ER+ or TNBC and RCB-0 disease had ctDNA detected mid-NAT; no clinical recurrence has occurred in these patients as of the data cutoff. The association between residual disease and mid-NAT ctDNA detection was maintained in stratified subgroup analyses by receptor subtype in participants with ER+ (Supplementary Fig. 4B, p = 0.084) and TNBC (Supplementary Fig. 4C, p = 0.017).
A Mid-NAT ctDNA clearance and RFI in all participants stratified by RCB-score (Log-rank (Mantel–Cox) p = 0.011). Note: curves for RCB-0 Detected and Not Detected are superimposed. B Mid-NAT ctDNA clearance and RFI in participants with HER2-negative EBC stratified by RCB-score (Log-rank (Mantel–Cox) p = 0.0016). Note: no participants with RCB-0 disease had ctDNA detected and are thus not represented. NR not reached, RCB residual cancer burden, RFI recurrence-free interval.
Associations between ctDNA dynamics and recurrence were not explored in the HER2+ cohort given the limited number of recurrences (only one recurrence, suspected to be a new primary, discussed below).
Landmark postoperative and follow-up ctDNA detection and clinical outcome
Representative swimmer plots by receptor subtype showing the ctDNA status of each longitudinally collected plasma timepoint are provided (Fig. 5A–C). Few participants (5/119, 4.2%) had ctDNA detected at perioperative timepoints; however, residual disease was present in all individuals where this occurred. Following surgery, two participants (ER+) had ctDNA detected at their postoperative timepoint, both had subsequent clinical recurrence (Fig. 5A). Of the three participants with ctDNA detected at their preoperative timepoint (post-NAT), two had a subsequent clinical recurrence (Fig. 5A, B). No participants with HER2+ disease had ctDNA detected at their perioperative timepoints (Fig. 5C).
Participants’ clinical timeline and timeline of plasma collection for ctDNA analysis for A ER+ B TNBC and C HER2 + EBC. Participants who received adjuvant HER2-targeted therapy who were not classified as HER2+ at baseline had ASCO/CAP-defined HER2+ disease on surgical resection and received one year of trastuzumab as standard of care. RCB residual cancer burden, cN clinical nodal status at baseline, cT clinical tumor size at baseline.
Of the 19 participants with disease recurrence, 12 had evaluable postoperative and adjuvant follow-up samples. In a binary analysis, any ctDNA detection postoperatively or in follow-up was associated with disease recurrence (Fig. 6; HR: 35.7, 95%CI: 4.30-297; p < 0.0001). This remained significant when analyzed using a time-dependent Cox regression, accounting for differences in collection frequency, when including only postoperative and follow-up timepoints (p < 0.0001) or all timepoints analyzed in this study (p < 0.0001). All participants with ctDNA detected postoperatively or in follow-up experienced subsequent clinical recurrence. One participant, without ctDNA detected at baseline (ER+ , low grade) experienced a subsequent clinical recurrence ~3.5 years after completing definitive surgery while on adjuvant endocrine therapy; ctDNA was never detected for this participant in clinical follow-up (Fig. 5A). The PPV for recurrence with ctDNA detected in the post-operative or follow-up setting was 100% (Supplementary Table 5; sensitivity: 75%, specificity: 100%, negative predictive value: 96%).
Lead time assessment and clinical recurrence
A lead time from ctDNA detection to clinical recurrence was evaluable in 8/12 participants; one had ctDNA detected at recurrence. The median lead time from detection of ctDNA until clinical recurrence was 374 days (Table 3; range: 13–1010 days). Of the four participants without a positive test at their most proximal time point prior to recurrence, one had an ipsilateral HER2+ local recurrence (grade 2, pT1a) with a change in the ER-status suggestive of a new primary. The other, a patient with TNBC, had tumor-derived variants detected 72 days prior to recurrence, but detection was below the threshold of high confidence calling (eVAF: 2.71 × 10−5). This participant was diagnosed with an asymptomatic solitary sub centimeter lung nodule (confirmed by pathology). The third and fourth participants had an interval of approximately one year between their last sample collected and recurrence, limiting interpretation (Table 3).
For participants with recurrences and serial collections in the adjuvant setting, eVAF values increased over time, with the highest values observed closer to clinical recurrence (Fig. 7A–C). Clearance in ctDNA occurred in participants after initiation of both adjuvant capecitabine (ER-low on biopsy, TNBC on resection specimen; Fig. 7A) and endocrine therapy (ER+ tumors; Fig. 7B, C). One participant (Fig. 7D) had early clearance of ctDNA during NAT, but subsequent preoperative detection, with no delays from the completion of NAT to definitive surgery. The postoperative time point was negative but was later followed by detection of ctDNA at metastatic recurrence (Table 3).
Representative time course plots illustrating changes in eVAF over time with respect to initiation or changes in therapy for representative patients of interest: A a participant with a diagnosis of ER-low EBC on biopsy, TNBC on resection (B) a participant with ER + EBC with a routine endocrine therapy switch (C) a participant with ER + EBC on adjuvant AI and (D) a participant with TNBC and rapid ctDNA clearance on NAT followed by pre-operative detection. C cycle, FU follow-up pre-op: preoperative time point, post-op postoperative time point, ET endocrine therapy.
Discussion
Neoadjuvant therapy, typically chemotherapy, is standard for patients with high-risk EBC, defined by aggressive disease biology (e.g., TNBC, HER2+) or a combination of disease burden and biological features (ER+/HER2−). While the response to NAT, measured by the extent of residual disease at surgery, helps guide risk stratification and informs adjuvant systemic therapy, these estimates are imprecise at the individual level. Sensitive and specific ctDNA assays offer the potential to develop more individualized treatment approaches. The largest studies of tumor-informed assays in the neoadjuvant setting have mainly been conducted in clinical trials with non-standard treatments13,14. The real-world cohort described here represents a clinically relevant population with several key distinctions. Most participants received standard anthracycline-taxane combinations (+/- anti-HER2 therapy) without investigational agents, with anthracycline given before taxane, aligning with real-world practice at the time (prior to the introduction of chemo-immunotherapy as standard for TNBC)15. Additionally, most assays were successfully generated using limited material from archival diagnostic cores, or in rare cases, residual disease from surgical specimens not designated for research at treatment initiation. Despite tissue limitations, only 9 out of 131 patients (7%) were excluded for technical reasons, with adequate sequencing depth achieved from the WES.
Prospective collection of plasma and clinical outcomes enabled retrospective evaluation of this assay across the neoadjuvant and adjuvant time course, including comprehensive assessment of dynamic changes in the pre-operative setting and their relationship with RCB status. Reflecting the clinical risk of the enrolled population, recurrences were observed in 19 out of the 119 participants (16.0%) over a median follow-up of 4.7 years. This follow-up period allowed for the characterization of both early changes and clinical outcomes, as well as evaluation of ctDNA detection in the adjuvant setting.
Baseline ctDNA detection was consistent with previous reports that the rate of detection with tumor-informed WES-based assays varies with breast cancer subtype; lower in ER+ tumors and highest in TNBC, thought to reflect cell turnover and disease biology14. Of note, our cohort of ER+ participants was not restricted to a high genomic risk population (as defined by MammaPrint), unlike the requirement in iSPY213,14. This may offer a more accurate representation of the baseline ctDNA detection rate in ER + EBC using this assay, as only 20-35% of ER+ breast cancers are typically classified as high genomic risk by MammaPrint. In most jurisdictions, patients are routinely selected for NAT based on clinicopathologic factors rather than genomic risk16,17.
The detection of ctDNA at baseline may be achievable in patients with a lower tumor fraction in cfDNA using more sensitive research assays. The relationship between ctDNA detection and clinical outcomes should be considered in the context of an assay’s analytical properties. This is illustrated in this cohort, where fewer variants were selected for ctDNA panels in ER+ participants compared to other receptor subtypes (by the locked assay workflow), despite no difference in TMB by receptor subtype (Supplementary Fig. 2B–E). Interestingly, those with TNBC and undetectable ctDNA at baseline had a lower TMB, suggesting a possible biological relationship (Supplementary Fig. 2E). Among those without baseline ctDNA detection, no significant differences were observed in the number of variants selected per panel so it is unlikely that ctDNA was missed due to variant selection. Instead, ctDNA may have been present at very low levels or not actively shed (Fig. 2C). Across all timepoints in the cohort, ctDNA was detected at an eVAF of less than 0.01% in 37% (61/163) of positive samples, underscoring the need for highly sensitive ctDNA assays to capture prognostic and/or predictive dynamics in real-world cohorts with standard clinical risk. The premature termination of the Phase III ZEST trial, which evaluated ctDNA in the adjuvant setting using an assay with lower technical sensitivity (LoD95%: 0.01%), suggests that earlier generation assays may not be optimal for clinical intervention strategies18,19,20,21. New ctDNA assays that evaluate hundreds or thousands of tumor-specific variants may further improve assay sensitivity in this setting22. Evaluation of detection rates and other parameters of such assays will be important to assess potential clinical advantages in the development of ctDNA-based strategies.
An interesting observation in this cohort is that nearly all participants who experienced recurrence had ctDNA detected at baseline, with only one exception. The exception was a participant with low-grade ER + EBC who received neoadjuvant endocrine therapy; the lack of ctDNA detection may be due to the low cell turnover and minimal ctDNA shed from this tumor. ctDNA detection using an assay with this level of sensitivity may reflect intrinsic disease biology not captured by routine clinical variables, with ctDNA shedding acting as an additional surrogate measure correlating with individual clinical risk. The ability to detect ctDNA in nearly all participants who eventually recurred highlights the importance of high technical sensitivity in ctDNA assays for monitoring those at risk of recurrence. However, the association between ctDNA detection at baseline using assays with a higher technical sensitivity is unknown. These findings warrant further investigation in a more homogenous patient cohort.
Analysis of ctDNA dynamics was undertaken in the entire cohort and separately in those with HER2-negative disease given the standard approach to the delivery of anti-HER2 antibodies with taxanes in the second half of NAT. This approach acknowledged the potential of targeted therapy to influence response and ctDNA dynamics in participants with HER2+ disease. Interestingly, HER2+ participants had a higher rate of mid-NAT ctDNA positivity and none experienced clinical recurrence. Outcomes overall for the HER2+ participants were excellent, as is observed with modern therapy. The prognostic value of mid-NAT ctDNA detection and clinical recurrence was improved when excluding HER2+ participants; those with HER2-negative disease, who had ctDNA detected midway through NAT were more likely to have disease recurrence than participants where ctDNA was not detected. While none exhibited clinically progressive disease during NAT, persistent ctDNA detection may serve as an early marker of treatment-resistant disease and tumor persistence, which could contribute to a failure to eradicate micro-metastases and eventual recurrence. Nearly all participants with HER2-negative disease with mid-NAT detection had subsequent clinical recurrence (67%, 6/9 participants), in contrast to those without ctDNA detected mid-NAT (14.5%, 8/55 participants). These data suggest that future analyses should consider both subtype and the inclusion of targeted therapy when designing prospective analyses to evaluate clinical utility. While not identifying all patients who subsequently recurred, the ability to identify those at the highest risk of recurrence may allow more targeted treatment intensification. Adjuvant treatment escalation in those with residual disease is common treatment strategy, but despite this, only a small minority of treated patients benefit while all are exposed to treatments associated with toxicities23,24. Incorporation and validation of on-treatment ctDNA surveillance into trial design in breast cancer may allow for better patient selection, improved outcomes, and reduced toxicity, as seen in other solid tumor sites25.
Stratified analyses explored the relationship between pCR (RCB-0), residual disease (RCB-1,2,3), recurrence, and whether ctDNA detection added to standard risk stratification. While non-pCR status is correlated with RFI, it offers a probabilistic estimate rather than precise identification of individual risk. Given the limited events in the study, stratified analyses were performed as sufficient power was lacking for a multivariable analysis. Nonetheless, mid-NAT ctDNA detection identified the highest-risk participants with residual disease, suggesting an additive effect beyond pathologic outcomes. In HER2-negative EBC, no participants with RCB-0 disease had mid-NAT ctDNA detected, and none experienced clinical recurrence. However, mid-NAT ctDNA detection was not sensitive enough to identify those with subsequent residual disease at surgery. Many participants without detectable ctDNA at mid-NAT had residual disease, but all participants with mid-NAT ctDNA detection went on to have RCB-2/3 disease.
Adjuvant therapy is delivered in a preventive fashion, precluding determination of its efficacy in individual patients. In contrast, perioperative and adjuvant ctDNA detection was strongly linked to imminent recurrence risk, with a median lead time of 374 days. Operational changes during the COVID-19 pandemic, such as variable sample collection, may have affected detection lead time. Nonetheless, a 100% PPV, 75% sensitivity, and 100% specificity of ctDNA detection was observed, emphasizing the assay’s specificity in predicting recurrence. Among the three participants who recurred without detectable ctDNA, two had technical factors (e.g., no collection within ~1-year, likely new primary tumor) that limited interpretation, while the third had a small volume recurrence with low levels of tumor-derived variants detected (below the assay’s threshold of positivity). Despite these issues, the assay’s 100% specificity is an important factor when ctDNA surveillance is contemplated to guide salvage escalation therapies. However, most participants who recurred did not have detectable ctDNA immediately postoperatively, but rather during follow-up, suggesting that a single landmark timepoint may be insufficient to guide adjuvant therapy. Careful evaluation of assay performance and suitability for the intended purpose is necessary in designing prospective trials, balancing intervention risks with assay properties.
Our study has several limitations. Recruitment and sample collection occurred during the COVID-19 pandemic, which impacted the timing of collections, particularly in perioperative and follow-up phases, due to efforts to reduce hospital visits. Consequently, some research samples were missed when clinical testing was performed outside the primary center. In addition, although the ctDNA results are derived from a locked assay which includes the filtering of germline variants, our mutation calls for describing the tumor somatic landscape are limited by the lack of matched germline (Fig. 2J). This may have led to a higher-than-expected frequency of some alterations, despite the use of tumor-only filtering strategies. Additionally, while the study evaluated a large cohort, subtype-specific analyses were limited by the small sample sizes, particularly the low number of recurrences in the HER2+ group, reflecting the good prognosis with modern therapy. This limits the interpretation of ctDNA dynamics and clinical outcomes, as well as lead time assessments. Further studies focusing on high-risk HER2+ populations are needed. The retrospective, single-center nature of the cohort is another limitation, as NAT selection and clinical follow-up may vary and affect generalizability. Although most patients receiving NAT were approached for enrollment, not all participated. Finally, expert pathology review for selecting pre-treatment biopsy tissue may not be feasible in all settings, potentially limiting the use of tumor-informed assays where suitable tissue is unavailable for genomic DNA extraction.
In conclusion, a high sensitivity, tumor-informed, ctDNA assay successfully detected ctDNA at baseline in a substantial proportion of participants prior to the initiation of NAT in a real-world cohort of patients EBC. Undetectable ctDNA at baseline was associated with a favorable outcome. Further, ctDNA detection on treatment was associated with clinical outcomes, particularly in HER2-negative EBC. Adjuvant ctDNA surveillance with this assay had a 100% PPV for clinical recurrence, with potentially clinically meaningful lead times, favoring those with ER+ disease. Prospective trials incorporating ctDNA assessment with this assay are warranted to evaluate the clinical utility of ctDNA-guided treatment strategies.
Methods
Patient recruitment
Patients diagnosed with EBC of any receptor subtype (triple negative, TNBC; Human epidermal growth factor receptor-positive, HER2+; and estrogen receptor positive/HER2-negative, ER+) receiving standard of care NAT at the Princess Margaret Cancer Centre were enrolled prospectively from October 2016 onwards for serial blood collection and banking (Supplementary Fig. 1A). Formal inclusion in the Liquid Biopsy Evaluation and Repository Development at Princess Margaret (LIBERATE) cohort (NCT03702309) began in August 2017 with the last participant included in this analysis enrolled in January 2021. Initiation of NAT and choice of therapy was at the discretion of the treating physician in discussion with their patient. This study was approved by the University Health Network Research Ethics Board (REB#: 17-5962 and #23-5446) and all participants provided informed consent prior to study enrolment and participation. ctDNA results were not returned to patients given the retrospective nature of these analyses. Standard clinical follow-up was delivered by treating physicians. The date of clinical recurrence was the date of imaging confirmed metastatic disease or local recurrence.
Clinical specimens
Formalin fixed, paraffin embedded (FFPE) tumor tissue obtained from a standard diagnostic biopsy and/or surgical resection with residual disease were identified to fulfill requirements for assay generation. Pathology review was performed to assess overall tumor cellularity and ensure adequacy for genomic DNA (gDNA) extraction, targeting 20-80% tumor content. Either 10 µm slides (10) or 1 mm2 cores (2-3) were used for nucleic acid extraction. Institutional requirements prevented diagnostic tissue from being used to exhaustion. All archival tissue was sent to NeoGenomics Laboratories, Inc. (Durham, North Carolina) where DNA was extracted and whole-exome sequencing (WES) was performed as previously described26,27. Serial blood samples (3× Streck BCT tubes) were collected at baseline, on treatment, perioperatively, and during follow-up associated with routine standard of care clinical visits. Following surgery, blood samples were collected approximately every 6-12 months at the discretion of the treating physician. Collected blood samples were drawn, double-spun, separated into plasma and buffy coat, aliquoted, and stored at −80 °C until analysis. Approximately 4 mL of plasma for each timepoint and 1.5 mL of baseline buffy coat was used for cell-free DNA (cfDNA) and control genomic DNA extraction, respectively.
Baseline time points were defined as those collected at any point prior to initiation of NAT. Preoperative (pre-op) timepoints were defined as those collected after the completion of chemotherapy and prior to surgery. Postoperative (post-op) timepoints were defined as those occurring up to 60 days after the participant’s definitive surgery for their breast cancer. Adjuvant or follow-up (FU) timepoints were those collected beyond 60 days from a participants’ primary surgery with or without the initiation of adjuvant therapy (Supplementary Fig. 1A).
Clinical variables
Baseline clinical stage was evaluated via retrospective analysis of the medical record and is reported as per the AJCC 8th edition prognostic staging of breast cancer. Tumor size (T-stage) and nodal involvement (N-stage) were assessed through retrospective evaluation of diagnostic MRI, mammogram, and/or ultrasound and pathologic assessment (axillary fine-needle aspirate). Estrogen receptor and HER2 status were evaluated on each participants’ clinical diagnostic biopsy using ASCO/CAP guidelines as reported by participating institutional pathologists28,29. Participant characteristics including age at diagnosis, sex, systemic therapy (including neoadjuvant and adjuvant chemotherapy, targeted therapy and/or endocrine therapy), surgeries, pathology (including residual cancer burden score, RCB), and clinical recurrence were derived from manual review of the electronic health record. The residual cancer burden (RCB) score was calculated as previously described30. Recurrence outcomes were last updated on July 26th, 2024. Patients without clinical recurrence were censored at their last follow-up date.
Tumor whole-exome sequencing analysis
Deduplicated and filtered BAM files were provided by NeoGenomics. Tumor whole-exome sequencing was performed independently from the RaDaR® ctDNA workflow. To ensure quality, base quality score recalibration (BQSR) was done using GATK, and the resulting BAM files were sorted and indexed using Samtools31,32,33. Variants were identified using Mutect2 in GATK and compared to a human reference genome (Homo sapiens assembly 38), with filtering for germline variants and sequencing errors. These variants were further filtered and annotated using in-house tools with criteria such as depth >100, variant allele frequency (VAF) > 5%, and specific mutation types (e.g., missense or nonsense mutations). Known driver genes from the COSMIC Cancer Gene Census were prioritized, and visualizations were created using maftools and RColorBrewer in R34,35. Additional details are outlined in the Supplemental Methods.
ctDNA sequencing analysis
ctDNA analysis followed the RaDaR assay protocol as previously outlined26,36. NeoGenomics was blinded to clinical data and outcomes during sample processing. FFPE tumor samples underwent WES using extracted tumor DNA, which was prepared with the KAPA Hyper Prep kit and enriched for exome regions using the IDT xGen Exome Research panel26,37. Sequencing was performed on the Illumina HiSeq4000 platform. The tumor-only WES data were processed through a custom workflow, involving alignment to the human genome (hg38), duplicate marking, and variant calling. Germline variants were filtered using public SNP databases, and somatic variants identified in the tumor tissue were prioritized via a proprietary algorithm. This algorithm generated a personalized primer panel targeting up to 48 somatic variants. To validate these variants, targeted sequencing was performed on both tumor DNA and baseline buffy coat samples to remove CHIP and sequencing artifacts. Plasma cfDNA was analyzed using the personalized panels, with the buffy coat sample serving to filter out germline variants, CHIP-associated variants, and act as a positive control for amplification. The full RaDaR assay workflow, including these steps, has been previously described in detail and is included in the Supplemental Methods26,36,37.
Statistical analysis
Predefined analyses included assessment of baseline sensitivity, evaluation of ctDNA detection through each participant’s clinical timeline, and exploratory assessment of prognostic association with ctDNA detection and clinical outcomes in the entire cohort and in those with HER2+ and HER2-negative disease. The recurrence-free interval (RFI) was defined as the time from pathologic diagnosis of invasive breast cancer to localized or metastatic recurrence of invasive breast malignancy or the development of a new primary invasive breast cancer. Participants who did not have a recurrence by the time of last follow-up were censored at their last follow-up date. Pre-cycle 2 (C2) was defined as a specimen collected prior to the second cycle of NAT. Mid-NAT was defined as a specimen collected prior to the midway point of their predefined neoadjuvant treatment [ie. docetaxel and cyclophosphamide, pre-cycle (C3); anthracycline and taxane, pre-cycle (C5)]. Those receiving neoadjuvant endocrine therapy only contributed to the baseline assay sensitivity and recurrence lead time analysis but were excluded from on-treatment ctDNA dynamic analyses. Participants without a baseline sample were excluded from analyses unless ctDNA was detected in a subsequent test in the NAT setting. The time to RFI outcome was calculated in years from the date of diagnosis (core biopsy date) to the date of the last follow-up along with their recurrence status on this date. RFI rates were calculated using the Kaplan-Meier product-limit method. In addition to landmark analyses, ctDNA detection was assessed at different time points during the follow-up period. Accordingly, ctDNA positivity was considered as a time-dependent variable based on the time interval of subsequent measures until last follow-up and used a proportional hazards model to assess the impact on RFI. Lead time was calculated (in days) as the interval between the first sample with ctDNA detected in the post-operative or adjuvant setting and clinical recurrence. Statistical analyses were performed using PRISM GraphPad (v9.0) and an open-source statistical software R version 4.1.0 (R Core Team (2021), R Foundation for Statistical Computing, Vienna, Austria). Chi-squared methods were used to calculate statistical significance from contingency tables. Hazard ratios are calculated using the Mantel-Haenszel method. All p-values are 2-sided, with p < 0.05 indicating statistical significance. Graphs were generated using R-studio and PRISM GraphPad (v9.0). Oncoplots and swimmer plots were created using R-studio.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The whole exome data generated in this study have been deposited in the European Genome-phenome Archive (EGA) under controlled access to ensure that data use is not for profit or commercial purposes. Access can be obtained by submitting a data access request via the EGA portal [https://ega-archive.org/access/request-data/how-to-request-data/] under the dataset ID: EGAD50000001133. Datasets can be accessed directly via: https://ega-archive.org/datasets/EGAD50000001133 and the study can be found at: https://ega-archive.org/studies/EGAS50000000771. The code for selecting variants to include in the RaDaR assay panels are not available due to data privacy laws and copyright restriction. The summary of the variants selected in personalized RaDaR assays and the results of each timepoint can be found in the Supplementary Tables. Given the single institution and geographically restricted nature of the cohort, in concordance with the REB approved clinical data sharing guidelines at the host institution, additional de-identified participant data are available for academic purposes on request from the corresponding authors, Dr. David W. Cescon (dave.cescon@uhn.ca) or Dr. Mitchell J. Elliott (mitchell.elliott@uhn.ca). Requests will be reviewed within 60 days by the host institution and any additional information needed will be discussed with the requesters. Source data are provided with this paper.
References
Korde, L. A. et al. Neoadjuvant chemotherapy, endocrine therapy, and targeted therapy for breast cancer: ASCO guideline. J. Clin. Oncol. 39, JCO2003399 (2021).
Hylton, N. M. et al. Neoadjuvant chemotherapy for breast cancer: functional tumor volume by MR imaging predicts recurrence-free survival-results from the ACRIN 6657/CALGB 150007 I-SPY 1 TRIAL. Radiology 279, 44–55 (2016).
DeMichele, A. et al. The neoadjuvant model is still the future for drug development in breast cancer. Clin. Cancer Res. 21, 2911–2915 (2015).
Berry, D. A. & Hudis, C. A. Neoadjuvant therapy in breast cancer as a basis for drug approval. JAMA Oncol. 1, 875–876 (2015).
Cortazar, P. et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet 384, 164–172 (2014).
Siravegna, G., Marsoni, S., Siena, S. & Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 14, 531–548 (2017).
Donaldson, J. & Park, B. H. Circulating tumor DNA: measurement and clinical utility. Annu. Rev. Med. 69, 223–234 (2018).
Cescon, D. W., Bratman, S. V., Chan, S. M. & Siu, L. L. Circulating tumor DNA and liquid biopsy in oncology. Nature Cancer 1, 276–290 (2020).
McDonald, B. R. et al. Personalized circulating tumor DNA analysis to detect residual disease after neoadjuvant therapy in breast cancer. Sci. Transl. Med. 11, eaax7392 (2019).
Coombes, R. C. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
Zhou, Q. et al. Persistence of ctDNA in patients with breast cancer during neoadjuvant treatment is a significant predictor of poor tumor response. Clin. Cancer Res. 28, 697–707 (2022).
Riva, F. et al. Patient-specific circulating tumor DNA detection during neoadjuvant chemotherapy in triple-negative breast cancer. Clin. Chem. 63, 691–699 (2017).
Magbanua, M. J. M. et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. 32, 229–239 (2021).
Magbanua, M. J. M. et al. Clinical significance and biology of circulating tumor DNA in high-risk early-stage HER2-negative breast cancer receiving neoadjuvant chemotherapy. Cancer Cell 41, 1091–1102.e4 (2023).
Schmid, P. et al. Pembrolizumab for early triple-negative breast cancer. N. Engl. J. Med. 382, 810–821 (2020).
Whitworth, P. W. et al. Distinct neoadjuvant chemotherapy response and 5-year outcome in patients with estrogen receptor-positive, human epidermal growth factor receptor 2-negative breast tumors that reclassify as basal-type by the 80-gene signature. JCO Precis. Oncol. 6, e2100463 (2022).
Cardoso, F. et al. 70-Gene signature as an aid to treatment decisions in early-stage breast cancer. N. Engl. J. Med. 375, 717–729 (2016).
GSK Q1 2023 Press Release. https://www.gsk.com/media/10013/q1-2023-results-announcement.pdf (2023).
Turner, N. C. et al. Results of the c-TRAK TN trial: a clinical trial utilising ctDNA mutation tracking to detect molecular residual disease and trigger intervention in patients with moderate- and high-risk early-stage triple-negative breast cancer. Ann. Oncol. 34, 200–211 (2023).
Coakley, M. et al. Abstract PD5-03: PD5-03 comparison of a personalized sequencing assay and digital PCR for circulating tumor DNA based Molecular Residual Disease detection in early-stage triple negative breast cancer in the cTRAK-TN trial. Cancer Res. 83, PD5-03 (2023).
Turner, N. GS3-01: Circulating tumor DNA surveillance in ZEST, a randomized, phase 3, double-blind study of niraparib or placebo in patients w/ triple-negative breast cancer or HER2+ BRCA-mutated breast cancer with molecular residual disease after definitive therapy. in San Antonio Breast Cancer Symposium 2024.
Garcia-Murillas, I. et al. Ultra-sensitive ctDNA mutation tracking to identify molecular residual disease and predict relapse in patients with early breast cancer. J. Clin. Oncol. 42, 1010–1010 (2024).
Masuda, N. et al. Adjuvant capecitabine for breast cancer after preoperative chemotherapy. N. Engl. J. Med. 376, 2147–2159 (2017).
Dixon-Douglas, J. & Loi, S. Immunotherapy in early-stage triple-negative breast cancer: where are we now and where are we headed? Curr. Treat. Options Oncol. 24, 1004–1020 (2023).
Tie, J. et al. Circulating tumor DNA analysis guiding adjuvant therapy in stage II colon cancer. N. Engl. J. Med. 386, 2261–2272 (2022).
Flach, S. et al. Liquid BIOpsy for MiNimal RESidual DiSease Detection in Head and Neck Squamous Cell Carcinoma (LIONESS)-a personalised circulating tumour DNA analysis in head and neck squamous cell carcinoma. Br. J. Cancer 126, 1186–1195 (2022).
Gale, D. et al. Residual ctDNA after treatment predicts early relapse in patients with early-stage non-small cell lung cancer. Ann. Oncol. 33, 500–510 (2022).
Allison, K. H. et al. Estrogen and Progesterone Receptor Testing in Breast Cancer: ASCO/CAP Guideline Update. J. Clin. Orthod. 38, 1346–1366 (2020).
Wolff, A. C. et al. Human epidermal growth factor receptor 2 testing in breast cancer: American Society of Clinical Oncology/College of American Pathologists clinical practice guideline focused update. Arch. Pathol. Lab. Med. 142, 1364–1382 (2018).
Symmans, W. F. et al. Measurement of residual breast cancer burden to predict survival after neoadjuvant chemotherapy. J. Clin. Oncol. 25, 4414–4422 (2007).
Van der Auwera, G. A. et al. From FastQ data to high confidence variant calls: the Genome Analysis Toolkit best practices pipeline. Curr. Protoc. Bioinformatics 43, 11.10.1–11.10.33 (2013).
McKenna, A. et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 20, 1297–1303 (2010).
Danecek, P. et al. Twelve years of SAMtools and BCFtools. Gigascience 10, giab008 (2021).
Tate, J. G. et al. COSMIC: the catalogue of somatic mutations in cancer. Nucleic Acids Res. 47, D941–D947 (2019).
Mayakonda, A., Lin, D.-C., Assenov, Y., Plass, C. & Koeffler, H. P. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 28, 1747–1756 (2018).
van Dorp, J. et al. Author Correction: High- or low-dose preoperative ipilimumab plus nivolumab in stage III urothelial cancer: the phase 1B NABUCCO trial. Nat. Med. https://doi.org/10.1038/s41591-023-02500-7 (2023).
Lipsyc-Sharf, M. et al. Circulating tumor DNA (ctDNA) and late recurrence in high-risk, hormone receptor–positive, HER2-negative breast cancer (CHiRP). J. Clin. Orthod. 40, 103–103 (2022).
Acknowledgements
The authors would like to thank the participants and their families for their participation in this study. This study was supported by the BMO Financial Group Chair in Precision Genomics at the Princess Margaret Cancer Centre through the Liquid Biopsy Evaluation and Repository Development at Princess Margaret (LIBERATE) program (NCT03702309), the Canadian Cancer Society Research Institute (to D.W.C), and the Princess Margaret Foundation (DH Gales Family Foundation). M.J.E. is supported by the Hold ‘Em for Life and Canadian Association of Medical Oncology (CAMO) fellowships. S.M. is supported by a Canadian Cancer Society Research Training Award - PhD (CCS award #708002). Assay production and results were provided by NeoGenomics.
Author information
Authors and Affiliations
Contributions
M.J.E. and D.W.C. wrote the manuscript. M.J.E. and E.G.A. performed the primary analyses included in this manuscript. E.G.A. provided statistical support for the manuscript. M.J.E. and S.M. performed the genomic analyses. P.E. and H.K.B. performed pathology review for participants in this cohort. M.J.E., J.F.A., Z.V., A.D., E.A., M.B.N., N.M., P.L.B., and D.W.C. participated in study recruitment. Study support was provided by E.S., C.Y., and L.L.S. Assays and vendor support was provided by N.C., R.V., and C.P. All authors reviewed the manuscript and provided feedback.
Corresponding authors
Ethics declarations
Competing interests
M.J.E, P.E., J.F.A., S.M., E.G.A., A.D., Z.V., N.M., E.S., C.Y., and H.K.B. report no competing interests. E.A. reports receiving honoraria from Sandoz and Seagen and consulting fees from AstraZeneca and Novartis. M.B.N. reports speaker honoraria and consulting fees from Novartis and Exact Sciences, outside the scope of this submitted work. N.C., R.V., and C.P. were employees of NeoGenomics at the time of data generation and manuscript preparation. P.L.B. reports research funding (to the institution) from Amgen, AstraZeneca, Bayer, Bicara, Bristol Myers Squibb, Genentech/Roche, Gilead, GlaxoSmithKline, Lilly/Loxo, Medicenna, Merck, Nektar, Novartis, PTC Therapeutics, Sanofi, SeaGen, Servier, SignalChem Life Sciences, Takeda, and Zymeworks. He also reports uncompensated honoraria/consultancy with Zymeworks, Lilly, Seattle Genetics, Merck, Amgen, Gilead, Janssen, and Repare. L.L.S. serves on scientific advisory boards for Merck, Pfizer, AstraZeneca, Roche, GlaxoSmithKline, Voronoi, Arvinas, Navire, Relay, Daiichi Sankyo, Amgen, Marengo, Medicenna, Tubulis, LTZ Therapeutics, Pangea, and Break Through Cancer. She also reports clinical trial support (to the institution) from Novartis, Bristol Myers Squibb, Pfizer, Boehringer Ingelheim, GlaxoSmithKline, Roche/Genentech, AstraZeneca, Merck, Celgene, Astellas, Bayer, AbbVie, Amgen, Symphogen, Mirati, BioNTech, 23andMe, and EMD Serono. Additionally, her spouse holds stock ownership in Agios and has leadership roles at Treadwell Therapeutics. D.W.C. reports consultancy and advisory relationships with AstraZeneca, Daiichi Sankyo, GenomeRx, Gilead, GlaxoSmithKline, NeoGenomics, Lilly, Merck, Novartis, Pfizer, Roche and SAGA; research funding (to the institution) from AstraZeneca, Guardant Health, Grail, Gilead, GlaxoSmithKline, NeoGenomics, Knight, Merck, Pfizer, ProteinQure, and Roche.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Elliott, M.J., Echelard, P., Pipinikas, C. et al. Longitudinal evaluation of circulating tumor DNA in patients undergoing neoadjuvant therapy for early breast cancer using a tumor-informed assay. Nat Commun 16, 1837 (2025). https://doi.org/10.1038/s41467-025-56658-4
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-56658-4
This article is cited by
-
High mid-treatment tumour RNA disruption in patients with HER2-negative breast cancer is associated with improved disease-free survival after neoadjuvant chemotherapy
Breast Cancer Research (2025)
-
Personalized ctDNA detection and genomic profiling in the NeoRHEA Study
npj Breast Cancer (2025)
-
Evolutionary Overview and Future Perspectives: ESR1 Mutations, Liquid Biopsy, and Artificial Intelligence for a New Era of Personalized Medicine in ER+ Breast Cancer
Molecular Diagnosis & Therapy (2025)
-
Artificial intelligence and multi-omics integration in liquid biopsy for genitourinary cancers: a systematic scoping review
International Urology and Nephrology (2025)
-
Personalized ctDNA monitoring in metastatic HR+/HER2− breast cancer patients during endocrine and CDK4/6 inhibitor therapy
npj Breast Cancer (2025)