Abstract
The lysosome serves as an essential nutrient-sensing hub within the cell, where the mechanistic target of rapamycin complex 1 (mTORC1) is activated. Lysosomal cholesterol signaling (LYCHOS), a lysosome membrane protein, has been identified as a cholesterol sensor that couples cholesterol concentration to mTORC1 activation. However, the molecular basis is unknown. Here, we determine the cryo-electron microscopy (cryo-EM) structure of human LYCHOS at a resolution of 3.1 Å, revealing a cholesterol-like density at the interface between the permease and G-protein coupled receptor (GPCR) domains. Advanced 3D classification reveals two distinct states of LYCHOS. Comparative structural analysis between these two states demonstrated a cholesterol-related movement of GPCR domain relative to permease domain, providing structural insights into how LYCHOS senses lysosomal cholesterol levels. Additionally, we identify indoxyl sulfate (IS) as a binding ligand to the permease domain, confirmed by the LYCHOS-IS complex structure. Overall, our study provides a foundation and indicates additional directions for further investigation of the essential role of LYCHOS in the mTORC1 signaling pathway.
Similar content being viewed by others
Introduction
Cholesterol is an essential building block for mammalian membranes and plays essential roles in embryonic development and other physiological processes. However, its aberrant accumulation is associated with several diseases, including atherosclerosis, obesity, neurodegenerative diseases, and cancer, underscoring the importance of maintaining cholesterol homeostasis1,2. Cellular cholesterol is tightly controlled by a feedback loop of sterol regulatory element binding protein pathway, which is activated under low cholesterol levels in the endoplasmic reticulum membrane3.
As a fundamental element of the cell membrane, cholesterol is essential for rapid cell proliferation. To match the requirement, cholesterol biosynthesis, and update are stimulated by the activated mechanistic target of rapamycin complex 1 (mTORC1) in actively proliferating cells4,5,6. mTORC1, a critical regulator of cellular biosynthetic pathways, functions as a vital nutrient sensor. Various nutrients, including glucose, amino acids, nucleotides, and lipids, have been reported to activate mTORC1 pathway7,8,9.
Cholesterol also acts as an activator of mTORC110,11,12. The endocytosed and recycled cholesterol is released in lysosomes and egressed by the cooperative work of two disease-related proteins, Niemann Pick disease type C1 (NPC1) and NPC213,14. Given that the lysosome is also the activation site for mTORC1, it has been proposed that lysosomal cholesterol may activate mTORC1 directly. Recently, the lysosomal cholesterol signaling (LYCHOS) protein was identified as a cholesterol sensor that couples lysosomal cholesterol concentrations to mTORC1 activation11.
LYCHOS, a lysosomal membrane protein, comprises 17 transmembrane segments (TMs) with an amino(N)-terminal permease domain (TMs 1-10) and a carboxyl(C)-terminal G-protein coupled receptor (GPCR) domain (TMs 11-17)11. It is suggested that LYCHOS harbors a cholesterol-binding site within the permease domain. At high cholesterol level, LYCHOS interacts with the GATOR1 complex via the LYCHOS effector domain (LED, a loop between TM15-16), releasing GATOR1 inhibition of mTORC111. Despite these advancements, it remains unknown how cholesterol induces the interaction between LED and GATOR1. It is also a mystery whether the permease domain can function as a transporter.
Here, we determine the cryo-electron microscopy (cryo-EM) structure of LYCHOS at a resolution of 3.1 Å, observing cholesterol-like density at the interface between the permease and GPCR domains. Advanced 3D classification demonstrates a cholesterol-related movement of GPCR domain relative to permease domain, providing insights into how LYCHOS senses lysosomal cholesterol levels. Furthermore, we discover that the permease domain can bind indoxyl sulfates (IS), a gut-derived uremic toxin.
Results
Structural determination of human LYCHOS
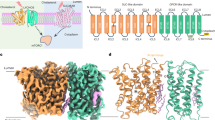
The recombinant expression and purification of human LYCHOS are detailed in the “Methods”. Briefly, LYCHOS was purified homogeneously in the presence of lauryl maltose neopentyl glycol (LMNG) and cholesteryl hemisuccinate (CHS) (Fig. 1a). The peak fractions were then concentrated for cryo-EM sample preparation, resulting in homogenous particles and promising 2D averages (Supplementary Fig. 1a). We therefore collected a large dataset for structural determination. Initially, we applied C1 symmetry to process this dataset, resulting in a 3D reconstruction at 3.3 Å resolution with 335,559 particles. Then, C2 symmetry was applied to further improve the overall resolution to 3.1 Å (Fig. 1b; Supplementary Figs. 1 and 2; Supplementary Table 1).
a Representative size exclusion chromatography (SEC) profile of LYCHOS solubilized in 0.005% LMNG and 0.0005% CHS. The indicated fractions were subjected to SDS-PAGE analysis, resolving multiple bands with different glycosylation states after Coomassie blue staining. Peak fractions were concentrated for cryo-EM sample preparation. The experiments were independently repeated more than three times with similar results. b Two perpendicular views of the final reconstructed LYCHOS. LYCHOS is a dimer with two protomers, MonoA and MonoB. c Two perpendicular views are presented to show the LYCHOS structures. Top: MonoA and MonoB are rainbow- and gray-colored, respectively. Bottom: MonoA is domain-colored with marine, light blue, and wheat for scaffold, transporter, and GPCR domains, respectively, and MonoB is gray-colored. The same color code is applied to all figures unless otherwise indicated.
An atomic model was built based on the 3.1 Å reconstruction. In each protomer, 577 residues (36–545 and 652–718) could be resolved, with 421 side chains assigned and an N-acetylglucosamine molecule linked to Asn309 (Supplementary Fig. 2b). Each protomer has 17 transmembrane helices (TMs), divided into an N-terminal permease domain with 10 TMs and a C-terminal GPCR domain with 7 TMs, consistent with previous predictions11. The two protomers are dimerized by the permease domain (Fig. 1c; Supplementary Figs. 1e and 2b; Supplementary Table 1).
Within the permease domain, TMs 1/2/6/7 constitute a scaffold domain to mediate dimerization (Fig. 1c and Supplementary Fig. 1e). The left TMs (TMs 3/4/5/8/9/10) display a similar fold to the E. coli Na+/H+ antiporter (NhaA). The NhaA fold is also observed in the mammalian sodium/proton exchanger (NHEs)15,16, sodium/taurocholate cotransporters (NTCPs)17,18,19,20, and plant auxin transporters (PINs)21,22,23 (Supplementary Fig. 3a). Despite a similar fold, the membrane topology of our structure resembles that in auxin transporters, which is inverted to those in NHE9 and NTCP22 (Supplementary Figs. 1e and 3b). Even so, these structural observations suggest this domain may function as a transporter potentially, we therefore designated it as the transporter domain to facilitate description hereafter (Fig. 1c and Supplementary Fig. 1e).
The GPCR domain exhibits a canonical GPCR fold with seven TMs (Supplementary Fig. 1e). A structural similarity search within the PDB library using FoldSeek24 indicated that LYCHOS-GPCR exhibits high similarities to the GPCRs in B1 and B2 subfamilies (Supplementary Table 2). Sequence alignment revealed that the LYCHOS-GPCR domain possesses the characteristic “GWGXP” motif found in the GPCR-B family25 (Supplementary Fig. 3c). Comparative analysis with representative GPCR-B1/B2 proteins demonstrated that LYCHOS-GPCR aligns closer to structures in the inactive state (Supplementary Fig. 3d). In classical GPCRs, activation involves a significant outward movement of the cytosolic end of TM6 to facilitate G protein binding26. However, potential spatial conflicts may occur between TM8 and TM16 if the LYCHOS-GPCR were to adopt an activation conformation similar to that observed in the ADGRG2 protein, a representative B2 GPCR (Supplementary Fig. 3e).
In the B subfamily of GPCR, the extracellular loop 2 (ECL2, GPCR nomenclature) plays a crucial role in coordinating ligands in the orthosteric ligand-binding pocket27,28,29,30. However, in LYCHOS-GPCR, ECL2 appears to occupy some space in the orthosteric pocket (Supplementary Fig. 3f). The loop is fixed by a π-π stack between Phe515 and Phe447, along with some polar interactions to TM11/17 (Supplementary Fig. 3g). Together, these structural observations suggest that LYCHOS-GPCR may function without engaging G proteins. However, due to the low sequence similarity between LYCHOS-GPCR and adhesion GPCRs, we can not completely rule out the possibility of G protein involvement. Our structure provides a foundation for further exploration of this question in the future.
Lipid densities sandwiched by permease and GPCR domains
As a cholesterol sensor, LYCHOS was suggested to bind cholesterol via its permease domain, while the activation of the mTORC1 was directly related to the LED loop in the GPCR domain11. We therefore paid specific attention to the spatial relationship between permease and GPCR domains. These two domains are directly contacted with an interface formed by TM1/10 and TM11/17 (Supplementary Fig. 4a). At the interface, two residues (Phe43 and Tyr57) have been reported to be essential for LYCHOS to mediate cholesterol-dependent mTORC1 activation11, indicating functional importance of this region (Supplementary Fig. 4b).
We then checked the EM map at the interface in detail, resulting in the observation of several lipid-like densities on the cytosolic leaflet (Fig. 2a). According to the lipid densities, two phospholipids, each with two acyl chains, were easily assigned. These lipids would be derived from HEK 293 cells, as no lipids were added during the protein purification and cryo-sample preparation. These two lipids were designated as Lipid1 and Lipid2 (Fig. 2b and Supplementary Fig. 2b). The polar heads of Lipid1 and Lipid2 are coordinated with polar interactions with TM1-Tyr57/Arg61 and TM17-Asp714/Lys715 (Fig. 2c, d). The nonpolar tails of these lipids are embedded into the interface between permease and GPCR domains, serving as a molecular glue that facilitates the cytosolic interaction between these domains (Supplementary Fig. 4c). Multiple hydrophobic residues from both domains contribute to the stabilization of these nonpolar tails (Supplementary Fig. 4d, e).
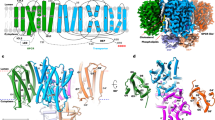
a A layer of lipid densities is observed at the interface between permease and GPCR domains. The lipid densities, contoured at 5σ, are colored light pink. b Structural modeling of two phospholipids and one cholesterol into the lipid densities. The map is contoured at 5σ. c Two perpendicular views to show the positions of lipid molecules at the interface. d A cytosolic view to show the coordination of the head groups of Lipid1 and Lipid2 at the interface. e The details of cholesterol coordination. f Biochemical verification of critical residues in cholesterol coordination. HEK-293F cells were transiently transfected with strep-tagged LYCHOS and Flag-tagged GATOR components, including DEPDC5 (Flag-DEPDC5), NPRL2 (Flag-NPRL2), and NPRL3 (Flag-NPRL3), followed by strep immunoprecipitation and immunoblotting for the indicated proteins. NPRL2 and NPRL3 were immunoblotted to represent GATOR1 complex in the assay. Multiple bands were observed for LYCHOS because of the glycosylation. The experiments were conducted independently at least three times with similar results. The lipid molecules in (d) and (e) are shown as sticks and balls and colored light pink. Two phospholipids are indicated with Lipid1 and Lipid2, and cholesterol is indicated as CHO. The potential charge interactions are indicated by dotted red lines.
According to our structural observation, Lipid1 and Lipid2 are essential for stabilizing interaction at the cytosolic side between permease and GPCR domains. Alanine substitutions at Arg61 and Phe352, critical residues involved in lipids coordination, diminished the ability of LYCHOS to recruit the GATOR1 complex (Supplementary Fig. 4f), indicating the significant role of these lipids in LYCHOS functionality.
The cholesterol-binding site
After modeling Lipid1 and Lipid2, residual densities resemble a cholesterol-like molecule, which may belong to endogenous cholesterol or exogenous CHS, a cholesterol analog added during protein purification. To describe the interaction between cholesterol-like molecule and this site, we modeled a cholesterol molecule here (Fig. 2b and Supplementary Fig. 2b). Supporting our modeling, similar cholesterol-like densities were observed at the same site in a recently published study31.
The modeled cholesterol is primarily stabilized by hydrophobic residues within the transporter domain, including Ile268 on the linker between TM1 and TM8, Phe276/Leu279/Ile280/Ile283/Leu287/Leu288 on TM8, and Ile349/Val353/Ile357 on TM10. In addition, one hydrophobic tail of Lipid1 may also contribute to the coordination of cholesterol (Fig. 2e). When mutations were introduced to disrupt the cholesterol-binding site, the LYCHOS variants exhibited reduced abilities to bind with the GATOR1 complex (Fig. 2f), suggesting an essential role of this site in LYCHOS function.
Cholesterol-related conformational changes
The observed lipids (Lipid1, Lipid2, and cholesterol) play an important role in stabilizing the interaction at the cytosolic leaflet between permease and GPCR domains of LYCHOS. In the absence of these lipids, GPCR domain may alter its position relative to permease domain. The GPCR domain was much less resolved compared to permease domain in the final EM maps (Supplementary Fig. 1d), highly suggesting the flexibility between GPCR domain and permease domain. To reveal this movement, we applied 3D variability analysis on the selected good particles (Supplementary Figs. 5a and 9), revealing a significant motion of GPCR domain relative to the permease domain (Supplementary video 1). Then, the first and last frames of 3D variability analysis were chosen as references for further 3D classification, resulting in two 3D reconstructions at 3.3 Å (tight-state, LYCHOS-T) and 3.5 Å (loose-state, LYCHOS-L) with C1 symmetry, respectively (Supplementary Fig. 5). Based on these reconstructions, two new models were generated.
The LYCHOS-T structure was almost identical to the original C2 structure (LYCHOS-C2) (Supplementary Fig. 6a). However, obviously conformational changes were observed in one protomer (MonoA) when comparing LYCHOS-T with LYCHOS-L (Fig. 3a and Supplementary Fig. 6a). Intriguingly, cholesterol-like densities were present in this protomer of LYCHOS-T but absent in LYCHOS-L (Fig. 3a). Therefore, these structures may represent the conformational states of LYCHOS in presence and absence of cholesterol, allowing further analysis on the cholesterol-sensing mechanism.
a Cholesterol and Lipid1 densities are absent in the EM map of LYCHOS-L. Top: Overlay of EM maps between LYCHOS-T (tight) and LYCHOS-L (loose). Middle & Bottom: Two perpendicular views to show the lipid densities at the interface for LYCHOS-T (Middle) and LYCHOS-L (Bottom). b Two perpendicular views showing the conformational changes between LYCHOS-T and LYCHOS-L. c The interaction between cholesterol and Lipid1 molecules. One hydrophobic tail of Lipid1 is sandwiched between cholesterol and TM17 of GPCR domain. d Biochemical verification of critical residues on TM17 in coordinating Lipid1. The experiments were conducted independently at least three times with similar results.
Along with the disappearance of cholesterol, the density of Lipid1 also disappeared in LYCHOS-L (Fig. 3a and Supplementary Fig. 6b). Lipid1 would be essential for the interaction between the GPCR and permease domains at the cytosolic leaflet. Consistent with this hypothesis, the cytosolic half of GPCR domain moves away from the permease domain in LYCHOS-L compared to LYCHOS-T (Fig. 3b).
While direct interactions between cholesterol and GPCR domain were not observed in these structures, cholesterol may stabilize Lipid1, which in turn supports the interaction between GPCR and permease domains at the cytosolic leaflet. Specifically, cholesterol interacts with one tail of Lipid1 on one side. On the opposite side, this tail is stabilized by Phe695/Ile706/Ile710 on the GPCR domain (Fig. 3c). Mutations F695A, I706A, and I710N diminished interaction between LYCHOS and GATOR1 complex (Fig. 3d), indicating the essential role of the cooperation between cholesterol and Lipid1 in LYCHOS function.
GATOR1 complex was proposed to interact with the LED loop between TM15 and TM16 in the GPCR domain under the cholesterol-loaded condition. Unfortunately, the LED loop was not resolved in our EM maps. When superimposing GPCR domains between LYCHOS-T and LYCHOS-L, most parts were almost identical with an r.m.s.d value at about 0.5 Å (Supplementary Fig. 6c).
Permease domain binds indoxyl sulfate
The permease domain exhibits a typical NhaA fold with two discontinuous helices (TM4 and TM9) crossover at the center of the transmembrane (Supplementary Figs. 1e and 7a). This signature feature was shared by the transporters, including NHEs15,16, NTCPs17,18,19,20, and PINs21,22,23, suggesting the permease domain may function as a transporter. It was reported that the pocket near the crossover site was the substrate binding site in other NhaA fold proteins15,16,17,18,19,20,21,22,23. In our LYCHOS structures, this pocket opens to the cytosol with some extra densities observed at a low threshold (5σ) (Supplementary Fig. 7a), indicating its ability to coordinate some ligands.
Lysosomes are known to degrade many kinds of biomolecules to building bricks, like nucleotides, amino acids, lipids, and monosaccharides, which are then exported for cellular utilization. Among these potential substrates, we initially screened the binding affinities of LYCHOS to amino acids with the surface plasmon resonance (SPR) assay. Among all screened amino acids, tryptophan induces the most positive response on the LYCHOS preloaded chips (Supplementary Fig. 7b). In spite of the high SPR response, tryptophan exhibited a relatively low binding affinity of 0.9 mM to LYCHOS (Supplementary Fig. 7c). We then measured the binding affinities between LYCHOS and tryptophan derivatives, including IS, indole-3-acetic acid (IAA), kynurenine (KYA), indole-3-carbaldehyde (I3A) and indole (Supplementary Fig. 7d). To our surprise, LYCHOS evidently binds to IS with a dissociation constant (Kd) at about 150 uM after quantitative measurement. The Kd value is higher than that of IAA, which was recently suggested as a LYCHOS-binding ligand31 (Supplementary Fig. 7d).
To elucidate the binding site for IS on LYCHOS, we incubated LYCHOS with 5 mM IS prior to cryo-EM sample preparation, which yielded cryo-EM map at 3.4 Å resolution with C2 symmetry (Supplementary Figs. 8 and 9). This map reveals distinct extra densities at high threshold (10σ) in the pocket of the permease domain (Fig. 4a). An IS molecule can be placed into this density perfectly (Fig. 4b), resulting in the LYCHOS-IS complex structure, which confirms the direct interaction between IS and the permease domain.
a Map comparison to show the stronger densities in the potential substrate-binding pocket in the map of LYCHOS-IS complex. The extra densities in the pocket are absent in original LYCHOS structure but are distinctly visible in the LYCHOS-IS complex when contoured at high threshold (10σ). b Structural modeling of IS molecule into the extra densities. The densities, contoured at 10σ, are shown as red mesh. c The coordination of IS molecule. Left and Middle: Two perpendicular views showing the relative position of IS in the permease domains of LYCHOS. Right: A zoomed-in view showing the coordination of IS in details. The molecule IS is shown as sticks and balls and colored magenta. The potential charge interactions are indicated by dotted red lines.
Similar to other NhaA-fold transporters, IS binds at the crossover site between discontinuous TM4 and TM9 helices (Fig. 4c). The molecule is positioned within the pocket formed by TM2/7 from the scaffold domain and TM4b/9b from the transporter domain. In detail, the sulfate moiety is primarily coordinated by polar interactions with TM4b-Phe147/Asn145 and TM9b-Pro327. Alanine substitution at these residues reduced the binding affinity between LYCHOS and IS obviously (Supplementary Fig. 7e). The indoxyl ring is stabilized by hydrophobic interactions with several residues on TM2 and TM7 and forms a hydrogen bond with Ser78. Mutations at representative residues, such as S78A, A256F, and Y259A, also resulted in decreased binding affinities (Supplementary Fig. 7e).
Upon IS binding, minor structure rearrangements are observed on the permease domain when compared to the previous C2 structure (LYCHOS-C2) (Supplementary Fig. 8c). The GPCR domain also moves away from the permease domain when IS binding (Supplementary Fig. 8d), but its position is closer to that in LYCHOS-T compared to LYCHOS-L (Supplementary Fig. 8e). In consistent with this observation, the cholesterol-like and Lipid1 densities were also observed at the interface between GPCR and permease domains in the LYCHOS-IS complex (Supplementary Fig. 8f).
Discussion
LYCHOS was suggested to couple cholesterol concentrations to mTORC1 activation. Our structural investigations of human LYCHOS reveal several lipid densities, including a cholesterol-like density at the interface between the permease and GPCR domains. Further structural analysis suggests a cholesterol-related conformational change between GPCR and permease domains, offering structural insights into the cholesterol-sensing mechanism. More importantly, our structural and biochemical investigations demonstrate that the permease domain has the capacity to bind IS.
LYCHOS is recognized as a lysosomal cholesterol sensor11. In our structure, a cholesterol-like molecule is primarily coordinated by the permease domain at a site closer to the GPCR domain. The site is distinct from the previously proposed cholesterol-sensing site near TM111, which, in our structure, is occupied by Lipid1 and Lipid2 (Fig. 2d). Despite this, our biochemical investigations underscore the critical role of the newly identified site in enabling LYCHOS to activate mTORC1. In cooperation with Lipid1, cholesterol bound at this site may facilitate robust interactions between permease and GPCR domains at the cytosolic leaflet, which may make LED possess a conformation to interact with GATOR1 complex. This model provides a plausible explanation for the cholesterol-sensing mechanism of LYCHOS.
The Lipid1, sandwiched by permease and GPCR domains, disappears along with cholesterol in LYCHOS-L, indicating its essential role in cholesterol sensing. In the EM map, we observed that Lipid1 has two hydrophobic tails. We modeled a PC molecule into the density, but we cannot rule out other possibilities, like phosphatidylethanolamine (PE), phosphatidylserine (PS), and phosphatidic acid (PA). It was also reported that phospholipids can also activate mTORC1 signaling pathway, including PA, PS, and lysophosphatidic acid (LPA)32. Further identification of Lipid1 molecule is crucial for a more comprehensive understanding of LYCHOS’s functions.
The permease domain exhibits a typical NhaA fold, shared by several transporters. We identified that IS can bind to the potential substrate-binding pocket within the permease domain. Despite this binding, it remains unclear whether LYCHOS can actively transport IS. As a well-known gut-derived uremic toxin, IS plays a significant role in the progression of chronic kidney disease (CKD) and contributes to the development of cardiovascular diseases in CKD patients33. Mechanistically, IS is thought to activate the proliferation of vascular smooth muscle cells via the mTOR signaling pathway34,35. However, the exact mechanism by which IS activates mTOR pathway remains to be elucidated.
Using IAA, an indoxyl derivate, as a surrogate ligand, it was demonstrated that the ligand-induced motion of the permease domain may disrupt a latch between Phe352 and Phe705, which are suggested to be essential for LYCHOS activation31. In our study, IS, another indoxyl derivate, could also disrupt the latch between Phe352 and Phe705 by binding to a similar site in permease domain (Supplementary Fig. 8g, h). Therefore, IS may regulate LYCHOS’s ability to activate mTORC1 pathway through direct interaction. However, it cannot be ruled out that LYCHOS might regulate the accessibility of IS via potential transport activity. All these speculations will require cellular and biochemical validations in the future.
Recently, Baily-Jones et al. also reported the cryo-EM structures of LYCHOS for both the WT and F352A/W678R variants31. In their WT structure, a cholesterol-like density was also observed at a site similar to that in our structure. However, in their F352A/W678R structure, they discovered a new site near the CRAC motif on TM1, aligning with the previously proposed cholesterol-sensing site by mass spectrum analysis11. The F352A mutation may disrupt the latch between Phe352 and Phe705, potentially causing the relocation of cholesterol from our identified site to this new site. Therefore, this model might explain why our resolved cholesterol-binding site is distinct from the proposed cholesterol-sensing site near TM111. Nonetheless, further experiments are required to validate these observations and minimize potential biases introduced by the mutations.
Despite these questions, our structural and functional investigations provide insights into the molecular basis by which LYCHOS couples high cholesterol levels with mTORC1 activation and identify the ability of permease domain to bind IS. These results provide a foundation and indicate directions for further investigation of the essential roles of LYCHOS in the mTORC1 signaling pathway.
Methods
Protein expression and purification
The cDNA of human LYCHOS (NCBI reference sequence NM_001033045.4) was cloned and inserted into the pCAG vector with C-terminal FLAG and 10×His tags. All the mutants were generated with a standard two-step PCR-based strategy using primers listed in the Supplementary Data 1. FreeStyleTM 293-F suspension cells (Thermo Fisher Scientific) were cultured in SMM 293-TII (SinoBiological) at 37 °C and supplied with 5% CO2. When the cell density reached 2.0 × 106 cells per mL, the cells were transiently transfected with the plasmids and polyethylenimines (Yeasen Biotechnology (Shanghai)). Approximately 1 mg of expression plasmids was premixed with 3 mg of polyethylenimines in 50 ml of fresh medium and incubated for 15–30 min before transfection. The 50 ml mixture was then added to 1 L of cell culture and incubated for 15–30 min.
For purification of LYCHOS, 2 L cells were collected 48 h after transfection and resuspended in 50 mL buffer containing 25 mM Tris pH 8.0, 150 mM NaCl, and protease inhibitor cocktails (Amresco). Then, the resuspended cells were solubilized with 1% (w/v) LMNG (Anatrace) at 4 °C overnight. After centrifugation at 20,000 × g for 1 h, the supernatant was collected and applied to 2 mL anti-FLAG M2 resin (Sigma). The resin was rinsed with buffer A containing 25 mM Tris pH 8.0, 150 mM NaCl, 0.005% LMNG, and 0.0005% (w/v) CHS (BLUEPUS) and eluted with buffer A plus 0.2 mg/ml FLAG peptide. Then, the eluate was loaded onto 2 mL NTA resin (Qiagen). After rinsing with buffer A, the protein was eluted with buffer A plus 250 mM imidazole. The eluate was concentrated and further purified by SEC (Superose 6 Increase 10/300 GL, Cytiva) in buffer A. The peak fractions were collected and concentrated to ~11 mg/ml for cryo-sample preparation. All the mutants were purified using the same protocol described above for the SPR assay.
Immunoprecipitation
Strep-tagged LYCHOS variants were co-expressed with Flag-DEPDC5, Flag-NPRL2 and Flag-NPRL3 in HEK293F cells. At 24 h post-transfection, cells were harvested and lysed using buffer A (30 mM Tris pH 7.5, 120 mM NaCl, 1 mM EDTA, 1 mM DTT and protease inhibitor cocktails) supplemented with 1% (w/v) LMNG and 0.1% (w/v) CHS. The cell lysate was then centrifuged at 20,000 × g for 60 min, and the supernatant was subjected to affinity purification with Strep-Tactin XT 4Flow resin (IBA-Lifesciences). After rinsing six times with buffer A plus 0.005% (w/v) LMNG and 0.0005% (w/v) CHS, the resins were denatured by adding 100 μl of sample buffer (50 mM Tris, pH 6.8, 2% SDS, 10% glycerol, 100 mM DTT, 0.1% bromophenol blue), heating to 95 °C for 5 min, resolved by SDS–PAGE and analyzed by immunoblotting with antibodies listed in the Supplementary Data 2.
Surface plasmon resonance (SPR)
SPR experiments were performed on a T200 instrument (GE Healthcare). Purified WT LYCHOS proteins and mutants were immobilized on CM7 sensor chips using amine coupling chemistry according to the manufacturer’s instructions (Cytiva). For the screen assay, 20 amino acids were dissolved in buffer A at 0.4 mM and flowed over the chip surface in buffer A at 30 μL/min. For quantitatively measuring the binding affinities, the ligands with indicated concentrations in buffer A was flowed over the chip surface in buffer A at 30 μL/min. The data were analyzed using GraphPad Prism 8.0.2.
Cryo-EM sample preparation and data collection
The cryo grids were prepared using Thermo Fisher Vitrobot Mark IV. Quantifoil R1.2/1.3 Cu grids were glow-discharged with air in a PDC-32G-2 plasma cleaner (Harrick) with mid-force for 85 s. Aliquots of 3.5 μl of purified LYCHOS, concentrated to ~11 mg/ml, were applied to the glow-discharged grids. After blotting with filter paper for 3.5 s (100% humidity and 8 °C), the grids were plunged into liquid ethane cooled with liquid nitrogen. 5 mM IS were incubated with the protein for 2 h in the tube before preparing the cryo-EM samples for the LYCHOS-IS complex.
The grid was loaded into a Titan Krios (FEI) electron microscope operating at 300 kV equipped with a BioQuantum energy filter and a K3 direct electron detector (Gatan). Images were automatically collected with EPU in the super-resolution mode. Defocus values varied from −1.5 to −2.0 μm. Image stacks were acquired with an exposure time of 3.8 s and fractionated into 32 frames with a total dose of 50 e− Å−2. The stacks were motion corrected with MotionCor236 and binned twofold, resulting in a pixel size of 1.07 Å/pixel, meanwhile dose weighting was performed37. The defocus values were estimated with CTFFIND438.
Cryo-EM data processing
For LYCHOS, a total of 4228 micrographs were collected. From these, a total of 1,828,601 particles were selected and extracted after Blob Picker in cryoSPARC39. After multiple rounds of 2D classification, 89,093 good particles were selected and subjected to an Ab-Initio Reconstruction with 3 classes, resulting in one good class. This good class was served as an initial model for further data processing. At the same time, the good 2D averages were used as templates to select more particles from micrographs using the Template Picker, resulting in 3,566,715 particles. After Heterogenous Refinements and an Ab-initio Reconstruction, 308,067 good particles were selected to yield a 4.5 Å reconstruction. These particles were then used as seeds for two rounds of seed-facilitated classification40, resulting in 689,660 good particles. These selected particles underwent Reference-based motion correction and 3D classification, producing 184,453 good particles. After another round of seed-facilitated classification, 335,559 good particles were selected to generate a 3D EM map at 3.1 Å resolution after Non-uniform Refinement with C2 symmetry.
The 335,559 selected good particles also produced a 3.3 Å reconstruction with C1 symmetry. The output was subject to 3D variability analysis with three components. Initial results were processed as a simple mode for preview. The first and last frames were chosen as references for Class 1 and Class 2, and the 335,559 good particles were applied into a Heterogeneous Refinement with these references, resulting in 130,318 particles for Class 1 and 103,110 particles for Class 2. After Non-uniform Refinement, the EM maps for Class 1 (LYCHOS-T) and Class 2 (LYCHOS-L) were determined at 3.3 Å and 3.5 Å, respectively.
For LYCHOS-IS complex, a total of 4373 micrographs were collected, from which 4,079,772 particles were selected and extracted. After several rounds of Heterogenous Refinements and an Ab-initio Reconstruction, 278,960 good particles were selected to yield a 4.6 Å reconstruction. Then, these good particles were used as the seed for one round of seed-facilitated classification, yeilding 1,015,981 good particles. The selected particles were applied into multiple Heterogenous Refinements and several runs of Ab-initio Reconstructions, resulting in 295,023 good particles. These particles yielded a 3.4 Å reconstruction after Non-uniform Refinement with C2 symmetry.
Resolutions were estimated with the gold-standard Fourier shell correlation 0.143 criterion41 with high-resolution noise substitution42. The local resolution maps were calculated using “Local Resolution Estimation” in cryoSPARC39.
Statistics and reproducibility
No statistical methods were used to predetermine the sample size. The experiments were not randomized, and the investigators were not blinded to allocation during experiments and outcome assessment. Each experiment was conducted independently at least three times with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The data that support this study are available from the corresponding authors upon request. The cryo-EM maps have been deposited in the Electron Microscopy Data Bank (EMDB) under accession codes EMD-37761 (LYCHOS-C2), EMD-61127 (LYCHOS-T), EMD-61128 (LYCHOS-L), and EMD-61129 (LYCHOS-IS). The atomic coordinates have been deposited in the Protein Data Bank (PDB) under accession codes 8WR3 (LYCHOS-C2), 9J3X (LYCHOS-T), 9J3Z (LYCHOS-L), and 9J40 (LYCHOS-IS). Published protein coordinates used in this study: 6Z3Y, 7PQG, 7Y9T, 6LN2, 6X18, 7WUQ, 7SF8, 6M1H, 8U5Q, 8U56. Source data are provided with this paper.
References
Duan, Y. et al. Regulation of cholesterol homeostasis in health and diseases: from mechanisms to targeted therapeutics. Signal Transduct. Target Ther. 7, 265 (2022).
Luo, J., Yang, H. & Song, B. L. Mechanisms and regulation of cholesterol homeostasis. Nat. Rev. Mol. Cell Biol. 21, 225–245 (2020).
Brown, M. S., Radhakrishnan, A. & Goldstein, J. L. Retrospective on cholesterol homeostasis: the central role of scap. Annu. Rev. Biochem. 87, 783–807 (2018).
Lu, X. Y. et al. Feeding induces cholesterol biosynthesis via the mTORC1-USP20-HMGCR axis. Nature 588, 479–484 (2020).
Duvel, K. et al. Activation of a metabolic gene regulatory network downstream of mTOR complex 1. Mol. Cell 39, 171–183 (2010).
Yecies, J. L. et al. Akt stimulates hepatic SREBP1c and lipogenesis through parallel mTORC1-dependent and independent pathways. Cell Metab. 14, 21–32 (2011).
Goul, C., Peruzzo, R. & Zoncu, R. The molecular basis of nutrient sensing and signalling by mTORC1 in metabolism regulation and disease. Nat. Rev. Mol. Cell Biol. 24, 857–875 (2023).
Emmanuel, N. et al. Purine nucleotide availability regulates mTORC1 activity through the Rheb GTPase. Cell Rep. 19, 2665–2680 (2017).
Condon, K. J. & Sabatini, D. M. Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 132, https://doi.org/10.1242/jcs.222570 (2019).
Castellano, B. M. et al. Lysosomal cholesterol activates mTORC1 via an SLC38A9-Niemann-Pick C1 signaling complex. Science 355, 1306–1311 (2017).
Shin, H. R. et al. Lysosomal GPCR-like protein LYCHOS signals cholesterol sufficiency to mTORC1. Science 377, 1290–1298 (2022).
Davis, O. B. et al. NPC1-mTORC1 signaling couples cholesterol sensing to organelle homeostasis and is a targetable pathway in Niemann-Pick type C. Dev. Cell 56, 260–276.e267 (2021).
Qian, H. et al. Structural basis of low-pH-dependent lysosomal cholesterol egress by NPC1 and NPC2. Cell 182, 98–111.e118 (2020).
Kwon, H. J. et al. Structure of N-terminal domain of NPC1 reveals distinct subdomains for binding and transfer of cholesterol. Cell 137, 1213–1224 (2009).
Dong, Y. et al. Structure and mechanism of the human NHE1-CHP1 complex. Nat. Commun. 12, 3474 (2021).
Winklemann, I. et al. Structure and elevator mechanism of the mammalian sodium/proton exchanger NHE9. EMBO J. 39, e105908 (2020).
Asami, J. et al. Structure of the bile acid transporter and HBV receptor NTCP. Nature 606, 1021–1026 (2022).
Goutam, K., Ielasi, F. S., Pardon, E., Steyaert, J. & Reyes, N. Structural basis of sodium-dependent bile salt uptake into the liver. Nature 606, 1015–1020 (2022).
Liu, H. T. et al. Structure of human NTCP reveals the basis of recognition and sodium-driven transport of bile salts into the liver. Cell Res. 32, 773–776 (2022).
Park, J. H. et al. Structural insights into the HBV receptor and bile acid transporter NTCP. Nature 606, 1027–1031 (2022).
Su, N. et al. Structures and mechanisms of the Arabidopsis auxin transporter PIN3. Nature 609, 616–621 (2022).
Ung, K. L. et al. Structures and mechanism of the plant PIN-FORMED auxin transporter. Nature 609, 605–610 (2022).
Yang, Z. S. et al. Structural insights into auxin recognition and efflux by Arabidopsis PIN1. Nature 609, 611–615 (2022).
van Kempen, M. et al. Fast and accurate protein structure search with Foldseek. Nat. Biotechnol. 42, https://doi.org/10.1038/s41587-023-01773-0 (2024).
Hollenstein, K. et al. Structure of class B GPCR corticotropin-releasing factor receptor 1. Nature 499, 438–443 (2013).
Rasmussen, S. G. et al. Crystal structure of the beta2 adrenergic receptor-Gs protein complex. Nature 477, 549–555 (2011).
Barros-Alvarez, X. et al. The tethered peptide activation mechanism of adhesion GPCRs. Nature 604, 757–762 (2022).
Xiao, P. et al. Tethered peptide activation mechanism of the adhesion GPCRs ADGRG2 and ADGRG4. Nature 604, 771–778 (2022).
Wang, J. et al. Cryo-EM structures of PAC1 receptor reveal ligand binding mechanism. Cell Res. 30, 436–445 (2020).
Zhang, X. et al. Differential GLP-1R binding and activation by peptide and non-peptide agonists. Mol. Cell 80, 485–500.e7 (2020).
Bayly-Jones, C. et al. LYCHOS is a human hybrid of a plant-like PIN transporter and a GPCR. Nature, https://doi.org/10.1038/s41586-024-08012-9 (2024).
Joy, J. M. et al. Phosphatidic acid enhances mTOR signaling and resistance exercise induced hypertrophy. Nutr. Metab. 11, 29 (2014).
Hung, S. C., Kuo, K. L., Wu, C. C. & Tarng, D. C. Indoxyl sulfate: a novel cardiovascular risk factor in chronic kidney disease. J. Am. Heart Assoc. 6, https://doi.org/10.1161/JAHA.116.005022 (2017).
Lin, C. Y. et al. Enhanced expression of glucose transporter-1 in vascular smooth muscle cells via the Akt/tuberous sclerosis complex subunit 2 (TSC2)/mammalian target of rapamycin (mTOR)/ribosomal S6 protein kinase (S6K) pathway in experimental renal failure. J. Vasc. Surg. 57, 475–485 (2013).
Nakano, T. et al. Indoxyl sulfate contributes to mTORC1-induced renal fibrosis via the OAT/NADPH oxidase/ROS pathway. Toxins 13, https://doi.org/10.3390/toxins13120909 (2021).
Zheng, S. Q. et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods 14, 331–332 (2017).
Grant, T. & Grigorieff, N. Measuring the optimal exposure for single particle cryo-EM using a 2.6 A reconstruction of rotavirus VP6. Elife 4, e06980 (2015).
Rohou, A. & Grigorieff, N. CTFFIND4: fast and accurate defocus estimation from electron micrographs. J. Struct. Biol. 192, 216–221 (2015).
Punjani, A., Rubinstein, J. L., Fleet, D. J. & Brubaker, M. A. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods 14, 290–296 (2017).
Wang, N. et al. Structural basis of human monocarboxylate transporter 1 inhibition by anti-cancer drug candidates. Cell 184, 370–383.e313 (2021).
Rosenthal, P. B. & Henderson, R. Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J. Mol. Biol. 333, 721–745 (2003).
Chen, S. et al. High-resolution noise substitution to measure overfitting and validate resolution in 3D structure determination by single particle electron cryomicroscopy. Ultramicroscopy 135, 24–35 (2013).
Acknowledgements
We thank the Cryo-EM Center of the University of Science and Technology of China (USTC) for the EM facility support. We are grateful to Dr. Yong-Xiang Gao and all the other staff members at the Cryo-EM Center for their technical support on cryo-EM data collection. This work was supported by the National Natural Science Foundation of China (32271241 to H.Q.), the Fundamental Research Funds for the Central Universities (WK9100000031 to H.Q.), “the Talent Fund Project of Biomedical Sciences and Health Laboratory of Anhui Province, University of Science and Technology of China” (BJ9100000003 to H.Q.), the start-up funding from the University of Science and Technology of China (KY9100000034 and KJ2070000082 to H.Q.), and “USTC Research Funds of the Double First-Class Initiative” (YD9100002058 to Z.W.).
Author information
Authors and Affiliations
Contributions
H.Q. conceived the project and designed the experiments. Y.H., Y.Y., and Z.W. performed cloning and protein purification. Z.W. prepared cryo-EM samples and collected data. Z.W. and H.Q. determined the structures. Z.W. did the SPR assays. J.H. and Z.W. performed the pull-down assay between LYCHOS and GATOR1. All authors contributed to the data analysis and preparation of the manuscript. H.Q., Z.W., and Y.H. prepared the figures, and H.Q. wrote the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewers for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Wang, Z., He, J., Yang, Y. et al. Structural basis for cholesterol sensing of LYCHOS and its interaction with indoxyl sulfate. Nat Commun 16, 2815 (2025). https://doi.org/10.1038/s41467-025-58087-9
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58087-9