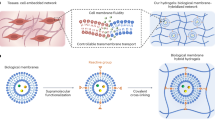
Abstract
Antibiotic contamination has emerged as a global challenge, increasing antibiotic resistance and threatening human health and ecosystems. Bioremediation using microorganism offers sustainable methods to degrade such pharmaceutical contaminants. However, these microorganisms exhibit reduced activity under high-stress conditions, and are difficult to recycle and potentially leak into environment as microbial pollutions. Here we report bioprinted retrievable microalgae hydrogel networks (MHNs) by confining living microalgae in double-network hydrogels, which achieves enhanced antibiotic degradation (>99.3%) and recyclable ability. Particularly, coating MHN with tannic acid (MHN@TA) generates a semipermeable membrane to prevent the leakage of microalgae (<0.7% for 7 days), ensuring the containment of potential microbial biohazards. The biohybrid system protects the biological activity of microalgae, enabling antibiotic degradation up to 400 mg L−1. Free-standing MHN@TA fencing systems are also manufactured to demonstrate their practical applications. This study provides insights of microalgae-material interactions in bioremediation and offers design rationales for biohybrid systems.
Similar content being viewed by others
Introduction
The emergence of antibiotics has brought great convenience to the treatment of bacterial infections in humans and animals1,2. However, bacteria can develop or acquire resistance to antibiotics, leading to approximately 1.2 million human deaths annually3. Notably, untreated antibiotics in natural waters can exacerbate the crisis of antimicrobial resistance4,5,6. Consequently, antimicrobial resistance is identified by the World Health Organization (WHO) as one of the top public health threats7,8. While antibiotic removal methods, including advanced oxidation9,10, adsorption methods11,12, and ultrafiltration methods13,14 showed good efficiency, they are energy-intensive or cost-prohibitive, and may generate hazardous solid wastes. In contrast, bioremediation harnesses microorganisms to degrade antibiotics, offering an environmentally friendly technology for addressing antibiotic contamination15,16. Particularly, photosynthetic algae are a low-carbon candidate that simultaneously enables the bioremediation of wastewater and sequestration of carbon sources17,18,19. Traditional microalgae bioremediation requires the construction of huge microalgae-containing ponds, demanding considerable land area and water resources20,21,22. Notably, these suspended microorganisms are difficult to recycle, and commonly exhibit reduced activity under high-stress conditions23,24. Moreover, the leakage of microalgae, particularly the engineered species, into natural water bodies may affect the distribution of indigenous microbiota (e.g., causing algal blooms). Developing microalgae bioremediation with optimal efficiency and recyclability is therefore imperative.
Engineered living materials, by embedding cells into a matrix, provide an isolated environment to protect cells from environmental stresses while allowing the material exchange to ensure cell growth25,26,27. Many efforts have used natural substances and synthetic polymers to encapsulate cells in the form of hydrogel systems to create biohybrid materials inheriting the functionalities of the organisms for biomedical and environmental applications28,29,30,31,32,33. For example, alginate hydrogels encapsulating engineered strains of Synechococcus elongatus (S. elongatus) were able to decontaminate chemical pollutants by regulating the production of a laccase enzyme for environmental bioremediation34. Chitosan-modified biochar was synthesized for the immobilization of Chlorella pyrenoidosa, which not only achieved efficient removal of sulfamethoxazole, but also improved algal lipid production and microalgae harvesting efficiency35. Nevertheless, these engineered systems suffer weak mechanical properties and potential leaking of the microbes, making it difficult for water treatment. 3D printed edible hydrogel-based culture system was recently developed as a prototype of regenerative food production36. Such 3D printed constructs not only allow the confined growth of microalgae but also offer a good distribution of gas, light, and nutrients throughout the constructs. We, therefore, expect that this strategy can achieve an emerging paradigm of a microalgae-driven degradation system for antibiotics.
In this work, we report the bioprinted, retrievable, and robust microalgae hydrogel networks (MHNs) for efficient antibiotic wastewater treatment with enhanced antibiotic degradation, self-regeneration property, and recyclable ability. Specifically, the living microalgae are encapsuled in 3D double-network hydrogels (DNHs) using biocompatible poly(ethylene glycol) diacrylate (PEGDA) and sodium alginate (SA), and such hydrogels are further coated with tannic acid (TA) to generate a semipermeable membrane to prevent the leakage of microalgae (Fig. 1a, b), ensuring the containment of potential microbial biohazards. Compared with the conventional suspended microalgae (SM), the MHN@TA system achieves good antibiotic degradation (>99.3% at 100 mg L−1), and maintains the biological activity of microalgae, enabling improved microalgal proliferation and antibiotic tolerance up to 400 mg L−1 (Fig. 1c, d), as further confirmed by gene expression analysis. The diffusion behavior of antibiotics in MHN@TA is also investigated by diffusion dynamics simulation. The retrievable MHN@TA fencing system demonstrates great potential for scalable manufacturing processes and practical application. This study not only offers valuable insights into microorganism–material interactions, but also holds significance for building a sustainable water purification system.
Results
Construction of microalgae hydrogel networks (MHNs)
We first formulated the bioink suitable for wastewater treatment. SA and PEGDA were selected as the matrix materials for the robust DNHs due to their rheological properties and biocompatibility. At the content of 6% SA, the hybrid bioink containing Chlorella zofingiensis showed suitable shear-thinning behaviors for extrusion-based 3D printing, which can be crosslinked by Ca2+ and UV forming stable networks with good shape fidelity37,38 (Fig. 2a and Supplementary Figs. 1−4). TA was used to construct a conformal semipermeable coating (Fig. 2b), which allows small molecule exchange while confines microalgae to prevent leakage. The MHN exhibited improved strength than the pure PEGDA and SA hydrogels, with a tensile strength of 134.3 kPa and a stress strength of 610.0 kPa (Fig. 2c and Supplementary Figs. 5, 6), mainly attributed to the interpenetrating networks that improve the energy dissipation. The MHN@TA hydrogel showed further enhanced mechanical performance (Fig. 2c and Supplementary Figs. 5, 6), owing to the hydrogen bonding interactions between PEGDA and TA. Cyclic tensile tests of MHN@TA demonstrated the good mechanical stability of the obtained polymeric networks (Fig. 2d). In addition, the addition of microalgae did not significantly affect the mechanical properties of the hydrogels (Supplementary Fig. 7).
a Variation of viscosity with shear rate. b Photos of MHN and MHN@TA with different structures. c Tensile tests of PEGDA, SA, MHN, and MHN@TA. All hydrogels contained microalgae and they were prepared in the same protocol. d Cyclic tensile stress–strain performance of the MHN@TA. e Photos and fluorescence images depicting the distribution of live and dead CZ cells in MHN@TA on days 0 and 14. Red fluorescence from Chl indicates live cells, and green fluorescence from SYTOX indicates dead cells. f Yield of Chl a and Chl b before and after TA coating. Each experimental group consisted of three biological replicates. The statistical analysis was evaluated by a two-tailed Student’s t-test without multiple comparisons. Differences were considered statistically significant at p < 0.05. g Chl content from day 0 to day 14 in the MHN@TA system. Each experimental group consisted of three biological replicates. The statistical analysis was evaluated by one-way analysis of variance ANOVA with Tukey’s honest significant difference (HSD). Adjustment for multiple comparisons was also performed. *p < 0.05, **p < 0.01, ***p < 0.001. h Microscopy images of MHN@TA on day 0 and day 14. i SEM image of the microalgae presented in the MHN@TA on day 14. j The surface and cross-sectional SEM images and mapping of DNH@TA@Ag. k Fluorescence intensity across a typical MHN@TA filament before and after soaking in water for 24 h. l Schematic of the escape of microalgae in MHN and MHN@TA. m Comparison of microalgae leakage in the supernatant of MHN, MHN@TA, and MHN@TA3 over 7 days. Data were presented as the mean ± s.d. (n = 3). Source data are provided as a Source Data file.
The growth of microalgae in the MHN and MHN@TA system was investigated. After the fabrication of MHN@TA, negligible dead cells were observed in the hydrogel (Fig. 2e and Supplementary Fig. 8), and the chlorophyll (Chl) of microalgae had no significant change (Fig. 2f), which confirmed that the bioprinting process is mild (Supplementary Figs. 9, 10). Moreover, the cell numbers and contents of Chl yield (e.g., Chl a and Chl b) exhibited a notable increase in MHN and MHN@TA systems after two-week growth, indicating the biocompatibility of MHN system and TA coating (Fig. 2g and Supplementary Figs. 11, 12). Microscopy and scanning electron microscopy (SEM) images further demonstrated the presence of microalgae within the hydrogel matrix (Fig. 2h, i). Importantly, the morphology of microalgae was intact, suggesting that the MHN@TA system did not impact the structure of cells (Supplementary Fig. 13).
We next validated the formation of semipermeable TA coatings on DNHs. In the FTIR spectrum of DNH@TA (Supplementary Fig. 14), stretching vibrations of the phenolic groups in TA shifted from 3416 to 3431 cm−1 39 and the carbonyl bonds (C=O) in TA shifted from 1718 to 1741 cm−1 40, indicating the formation of hydrogen bonds between DNH and TA41. To confirm the formation of TA coatings on the outer layer of the hydrogel rather than inside the hydrogel, we utilized the reducing property of TA to reduce silver ions42. The reduced silver particles were distributed on the surface of DNH@TA (Fig. 2j and Supplementary Fig. 15) and no silver particles were observed in the interior of DNH@TA in cross-sectional element mapping, confirming the presence of TA coating on the surface of the hydrogel (Fig. 2j and Supplementary Figs. 16, 17).
The leakage of microalgae from MHN and MHN@TA was investigated in a comparison study. After soaking in water for 24 h, the intensity and distribution of red fluorescence (indicating the living microalgae) in MHN@TA was similar to that before soaking, confirming the negligible leakage of microalgae (Fig. 2k and Supplementary Fig. 18). Even after prolonged incubation of 7 days, MHN showed a microalgae leakage of 16.1%, whereas MHN@TA exhibited a marginal leakage of 1.3% (Fig. 2l, m). Compared to single-network hydrogels, double-network hydrogels can not only prevent the leakage of microalgae but also have improved mechanical properties (Supplementary Fig. 19). Besides, we can modulate the TA coating (i.e., increasing the thickness of the TA coating) to enhance containment performance, achieving as low as 0.7% for MHN@TA3). Microalgae leaking into the solution was not even detected by microscopy (Supplementary Fig. 20). Collectively, the MHN@TA system can effectively restrict the potential leaking of microbial hazards into their surroundings during its application, which is important to gene-engineered species.
Antibiotic degradation performance
We next studied the antibiotic degradation performance of the MHN@TA system by selecting Chlorella zofingiensis as the experimental microalgae species (Supplementary Fig. 21). Using tetracycline (TC) as a model system, SM, MHN, and MHN@TA systems exhibited good degradation performance for low TC concentration (i.e., 100 mg L−1) with efficiencies of 84.5 ± 1.9%, 97.7 ± 2.7%, and 96.3 ± 2.3% for 24 h, respectively (Fig. 3a), and the degradation efficiency of MHN@TA further increased to 99.3 ± 0.5% for 72 h (Fig. 3b and Supplementary Figs. 22, 23). The degradation kinetic of MHN@TA for TC followed the pseudo-first-order kinetics with a corresponding biodegradation rate constant k1 of 0.1575 h−1 (Supplementary Fig. 24). Notably, while the SM system significantly lost the degradation ability at relatively high TC concentrations (e.g., 300 mg L−1), MHN@TA largely maintained the excellent degradation efficiency even at 400 mg L−1, which could be attributed to the protecting effects of DNH matrix. Moreover, the TA coating did not adversely affect the degradation efficiency of the system. We further investigated the effects of conditions such as pH and light intensity on the degradation performance of microalgae (Supplementary Fig. 25). The removal performance of microalgae at pH 6–10 was superior to that at pH 2–4, which could be attributed to low bioactivity of microalgae at the acidic conditions43. Meanwhile, the degradation efficiencies had no significant difference under various light intensities, which indicated that the system was suitable for most natural light conditions.
a Removal efficiency of different systems at different TC concentrations for 24 h. Each experimental group consisted of three biological replicates. b Removal efficiency of TC by MHN@TA system at different time points. Each experimental group consisted of three biological replicates. c TC removal performance under different conditions (TC = 100 mg L−1). Each experimental group consisted of three biological replicates. d Reuse performance of the MHN@TA system for TC. Each experimental group consisted of three biological replicates. e Volcano plot of distribution trends for DEGs in TC treatment and control groups. f GO enrichment for DEGs. g KEGG enrichment for DEGs. h Mass chromatography of TC intermediate products during the degradation process. i Possible degradation pathways of TC in microalgae. j Toxicity of TC and TC degradation products to S. aureus. Each experimental group consisted of three biological replicates. k Removal efficiency of MHN@TA system for different concentrations of AMX. Each experimental group consisted of three biological replicates. Data were presented as the mean ± s.d. (n = 3). Source data are provided as a Source Data file.
The pathways of TC degradation in this system were studied (Fig. 3c). The degradation efficiency of TC in water was negligible (1.6 ± 1.5%) under light or dark conditions for 24 h, indicating the weak intrinsic photodegradation and hydrolysis of TC in water44. The DNH@TA hydrogel without microalgae showed an approximately 7.7 ± 1.1% decrease in TC, while MHN@TA exhibited a degradation efficiency of 96.3 ± 2.3%. These results conclude that microalgae are the main driver to degrade TC. Furthermore, such MHN@TA can be facilely recycled from the sewage for several consecutive degradation of antibiotics (Fig. 3d). In contrast to conventional adsorbents for antibiotics45,46, MHN@TA system can achieve efficient degradation of antibiotics, and undergo self-regeneration without complicate post-treatment.
For the molecular mechanism underlying this biodegradation, the expression level and biological functions of genes involved in antibiotic metabolic pathways were evaluated by transcriptomic analysis (RNA-seq). There were 1981 differentially expressed genes (DEGs, 502 up and 1479 down) in the TC treatment group compared with the control group (Fig. 3e). Based on the Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment annotations, a certain number of DEGs were enriched in the pathways involving the expression of enzymes of antibiotic degradation (Fig. 3f, g). Cytochrome P450 enzymes are generally considered the key enzyme for drug detoxification (cleavage of the aromatic rings of compounds), which involves hydrolysis, oxidation, or reduction reactions47. During the process of degradation, relevant encoding genes (Cz10g28270, Cz12g27180, and Cz13g16110) were detected with a high expression in microalgae (Supplementary Data 1). In addition, dehydrogenase as an oxidoreductase acting on the CH-CH linkage is important in the biotransformation of antibiotics, and aldo/keto reductases and decarboxylases are important in biodegradation of aromatic compounds48,49. Genes encoding dehydrogenases (Cz11g10100, Cz04g30040, and Cz13g03290), encoding aldo/keto reductases (Cz02g09230, Cz13g06240, and Cz13g06250), and encoding decarboxylases (UNPLg00178 and UNPLg00179) were upregulated (Supplementary Data 1), which contributed to the reduction and demethylation of the broken benzene ring.
The by-products from the metabolic TC degradation are consistent with the expression of the genes, suggesting that the microalgae-driven TC degradation is mainly related to the loss of functional groups and ring-opening reactions (Fig. 3h, i). Specifically, the TC removes the N-methyl group by enzymatic degradation (m/z = 430.9). Subsequently, the ring structure is cleaved to gain products (m/z = 274.1, 200.1). The ring structures are eventually decomposed into small molecules (m/z = 182.9), which are mineralized to produce CO2, H2O, and NH4+50,51. The total inorganic carbon (TIC) significantly increased in the supernatant of MHN@TA systems after degradation, suggesting the mineralization of TC (Supplementary Fig. 26). The biotoxicity of resultant products were evaluated. The growth of Staphylococcus aureus (S. aureus) was not inhibited after incubation with the MHN@TA-treated antibiotic-containing specimen (Fig. 3j and Supplementary Fig. 27), indicating the MHN@TA system could effectively denature the antibacterial functions of TC. Importantly, such MHN@TA also had excellent degradation efficiency for other type of antibiotic species i.e., amoxicillin (AMX), penicillin G (PG), and ciprofloxacin (CIP), implying that the MHN@TA system is broadly applicable for antibiotic degradation in wastewater treatment (Fig. 3k and Supplementary Figs. 28, 29).
Cytoprotective effects for defensing antibiotic threat
We next investigated the rationales for the improved degradation performance of the MHN@TA system. We hypothesize that the microenvironment provided by MHN@TA protects the cells from environmental stresses, for example, acting as a buffering zone against external antibiotic threats. Fluorescence images demonstrated high mortality of microalgae in the SM system at high TC concentrations (LC50 = 368.30 mg L−1, Supplementary Fig. 30), whereas that was negligible in the MHN and MHN@TA systems (Fig. 4a–c and Supplementary Fig. 31). The photosynthetic Chl of microalgae in the SM system significantly decreased, while there were no obvious changes in MHN and MHN@TA systems (Fig. 4d, e and Supplementary Fig. 32). The maximum potential quantum efficiency of Photosystem II (Fv/Fm) represents the level of photosynthesis in microalgae, which can be affected by external environmental stresses. After TC treatment (400 mg L−1), the value of Fv/Fm in the SM system showed a significant decrease of 48.1%, while that of the MHN@TA system showed a slight decrease of 17.8% (Fig. 4f). These results validate that the encapsulation system effectively shields microalgae from antibiotic-induced damage. Reactive oxygen species (ROS) play a crucial role in the metabolic activities of microalgae and mitigate the toxic effect of antibiotics52. When exposed to TC, ROS of microalgae in MHN and MHN@TA remained at regular levels, whereas ROS of microalgae in the SM system dropped significantly, which may be due to the malfunction of the microalgae biological system and restrict its capability to degrade antibiotics (Supplementary Figs. 33, 34).
a–c Fluorescence images depicting the distribution of live and dead CZ cells in SM (a), MHN (b), and MHN@TA (c) systems before and after incubation in 400 mg L−1 TC. Red indicates live cells and green indicates dead cells. d, e Relative Chl content of microalgae in SM (d), and MHN@TA (e) systems under different TC concentrations. Each experimental group consisted of three biological replicates. f Changes of Fv/Fm in SM, MHN, and MHN@TA systems under different TC concentrations. Each experimental group consisted of four biological replicates. g Diffusion dynamics simulation in SM and MHN@TA systems. h Photo of a free-standing and flexible MHN@TA fencing system (70 cm × 40 cm × 0.5 cm). i Photo of wastewater sampled from a contaminated pond in Sichuan, China. j Removal efficiency of MHN@TA system for simulated antibiotic-containing wastewater. Each experimental group consisted of three biological replicates. k Comparison of the degradation performance of MHN@TA with other reported methods. Data were presented as the mean ± s.d. (n = 3). Source data are provided as a Source Data file.
To further understand the protection mechanism, we performed RNA-seq on the microalgae in MHN@TA and SM system under a high-concentration antibiotic environment (400 mg L−1). The gene expression levels of the MHN@TA and SM groups verified the consistency and comparability of the sequencing data among the samples (Supplementary Fig. 35a). The volcano plot and heatmap-plot of DEGs in the SM system indicated 2909 downregulated genes and 2947 upregulated genes compared to MHN@TA (Supplementary Fig. 35b, c). Most DEGs in GO enrichment enriched in oxidoreductase activity, antioxidant activity, lyase activity, monooxygenase activity, enzyme activator activity, and nucleotide binding were downregulated, revealing that the enzyme-related metabolic activities of SM were inhibited (Supplementary Fig. 35d). KEGG enrichment analysis further revealed that the biological functions of photosynthesis and cell proliferation were suppressed (Supplementary Figs. 35e, 36)53. These results unveiled at the genetic level that the MHN@TA system can protect the biological activity of microalgae.
The mechanism of the MHN@TA system protecting microalgae was further analyzed by diffusion dynamics simulation (Fig. 4g). Antibiotic molecules diffused quickly throughout the SM system within 1.0 h, making microalgae exposed to high-concentration antibiotics abruptly and causing severe damage to microalgae. In contrast, the hydrogel matrix slowed the diffusion of antibiotics, creating a retention zone for microalgae. Specifically, it required 55.7 h for antibiotics to diffuse throughout the MHN@TA system. Therefore, by ensuring a relatively low concentration of antibiotic in close proximity to the microalgae, the MHN@TA system allows microalgae to gradually adapt to the antibiotic stresses and dynamically degrade the antibiotic molecules.
The industrial application of the MHN@TA system for removing antibiotics in sewage was evaluated by using wastewater spiked with TC as a proof of concept. We first fabricated an MHN@TA fencing system (70 cm × 40 cm × 0.5 cm) with a custom 3D printer (Fig. 4h and Fig. S37, Supporting Information), demonstrating the potential of modular large-scale manufacturing and good mechanical properties for industrial use. Water samples were collected from contaminated and natural ponds in Sichuan (30.64°N, 104.08°E) and Jiangxi (28.65°N, 115.79°E) of China (Fig. 4i, Supplementary Fig. 38, and Supplementary Table 1), and were spiked with TC to simulate the real wastewater conditions with concentrations of 2 ppm and 20 ppm54,55. Notably, the MHN@TA fencing system can effectively remove TC from the wastewater with a degradation efficiency of more than 99.1 ± 0.4% after 24 h treatment (Fig. 4j). Furthermore, microalgae can survive and proliferate in real wastewater (Supplementary Fig. 39), and the activated sludge in the wastewater had negligible effects on the degradation performance of the MHN@TA system (Supplementary Fig. 40). Together with the degradation performance of high-concentration antibiotics aforementioned, the MHN@TA can treat antibiotic in a wide range of concentrations (2–400 ppm) with high removal efficiency (>95.6%), which is an improvement over previously reported microalgae-driven or bacteria-driven bioremediation strategies56,57,58,59,60,61,62,63,64,65 (Fig. 4k and Supplementary Table 2).
Discussion
We develop a retrievable MHN@TA system that achieves efficient degradation of antibiotics in a wide concentration range with good recyclability. Moreover, the semipermeable membrane generated by TA coating prevents the leakage of microalgae, ensuring the containment of potential microbial biohazards. Compared to the conventional SM system, the MHN@TA system isolates the microalgae from harsh external environments and largely protects their biological activity even in high concentrations of antibiotics. The improved performance of MHN@TA for antibiotic degradation is revealed by gene expression analysis and computational simulations. We also demonstrate that MHN@TA can be manufactured on a large scale and exhibit industrial potential for treating antibiotics. This study not only provides a low-carbon, environmentally friendly strategy for addressing global water pollution, but also provides insights into microalgae-material interactions for designing robust biohybrid systems.
Methods
Chemicals and materials
All chemical reagents were used without further purification. Alginic acid sodium salt (viscosity 500-1000 mPa·s), calcium chloride anhydrous (CaCl2), polyethylene glycol diacrylate (PEGDA, Mw = 1000), lithium phenyl (2,4,6-trimethylbenzoyl) phosphinate (LAP, Mw = 294), tannic acid (TA, Mw = 1701), methyl alcohol, zirconium chloride (ZrCl4), tetracycline hydrochloride (macrolide antibiotic), amoxicillin (β-lactam antibiotic), Penicillin G (β-lactam antibiotic) and Ciprofloxacin (quinolone antibiotic) were purchased from Shanghai Aladdin Biochemical Technology CO., Ltd. Agar was purchased from Beijing Coolaibo Technology Co., Ltd. BG11 powder was purchased from Hope Bio-Technology Co., Ltd. High-purity water with a resistivity of 18.2 MΩ.cm was obtained from a Millipore water purification system.
Characterization
The rheological properties of hydrogels were evaluated on an Anton Paar MCR302 rotational rheometer. UV–vis absorption measurements were analyzed on a PerkinElmer LAMBDA 1050 + UV/Vis/NIR spectrophotometer. Fluorescent images were captured on an Olympus IX73 inverted microscope. Three-dimensional (z-stack) fluorescence images were captured on a Leica STELLARIS 5 confocal laser scanning microscope (CLSM). Light microscope images were captured on a Nikon ECLIPSE Si biomicroscope. Cellular ROS detection was assessed by an EXFLOW-108 flow cytometer from Dakewe Biotech Co., Ltd. Scanning electron microscopy (SEM) images were obtained using a Phenom XL, Thermo Fisher Scientific, at an operation voltage of 10 kV.
Cells
Chlorella zofingiensis (ATCC 30412, denoted as CZ) was kindly provided by Shenzhen Key Laboratory of Marine Microbiome Engineering at Shenzhen University. These microalgae were cultured at 23 °C with agitation at 150 rpm and a continuous illumination of 50 µmol photons m−2 s−1 in a shaker incubator. BG11 medium was used for cultivating CZ66.
Preparation and 3D bioprinting of MHN
To prepare MHN, 10 mL of 80 mg mL−1 PEGDA (8%, w/v) was configured with 20 mg of LAP (0.2%, w/v). Then, 0.6 g SA (6%, w/v) was added to the mixture to obtain a clear, viscous solution. Next, microalgal cells (OD680 = 1, 10 mL) in the exponential growth phase were collected by centrifuge (4000×g, 5 min) and washed three times with deionized water to remove the residual medium. Washed microalgal cells were added to the above mixture. The prepared bioink was transferred into a 10 mL syringe of 3D bioprinter (Bio-Architect® SR, Regenovo, Hangzhou, China). The bioink was pneumatically dispensed through a nozzle with a pressure of 0.3 MPa and an extrusion flow speed of 5.0 mm s−1, and cured by a light-curing system during the printing process. Different customized 3D structures were designed by the printer’s software and the line diameters were adjusted by different nozzles. At the end of the printing, MHN was continued to cure under UV light for 2 min and crosslinked with 5 wt% CaCl2 solution for 5 min. Different customized 3D structures were designed by the printer’s software.
Preparation of MHN@TA and MHN@TA3
For the construction of the semipermeable coatings, the MHN@TA was obtained by immersing the as-prepared MHN in a 10 mg mL−1 TA solution for 2 min. MHN@TA3 was prepared by repeating the coating process three times.
Mechanical performance testing
The tensile, compressive tests, and cyclic tensile tests were performed on a universal tensile machine (CMT6202, MTS systems Co., Ltd., China) with a load cell capacity of 150 N. Dumbbell-shaped samples with ~2 mm in thickness, ~30 mm in length, and ~10 mm in width were prepared for tensile tests. The tensile speed was set at 10 mm min−1. For the compression tests, cylindrical samples with ~14 mm in diameter and ~10 mm in height were prepared at a strain rate of 10 mm min−1 compressed by the upper plate. Three samples were prepared for each mechanical performance experiment. The strain of the hydrogel sample was calculated by the length change related to the initial length of the samples, and the stress was estimated by dividing the force by the initial cross-sectional area of the samples.
Determination of cell viability
For live/dead cell staining assay, MHN and MHN@TA were incubated with 5 µM SYTOX Green Nucleic Acid Stain (Tianjin Alpha Biotechnology Co., Ltd.) in the dark at room temperature for 20 min. Then, MHN and MHN@TA were examined under a fluorescent inverted microscope. Live cells were identified by their red autofluorescence of chlorophyll, while dead cells were identified by their green fluorescence, as stained by SYTOX Green. 3D fluorescence microscope images (z-stack) were collected using a confocal laser scanning microscope.
Detection of microalgae and pigment yields
The microalgal cells was evaluated by measuring the absorbance of cell suspension at a wavelength of 680 nm (OD680) with a UV-vis spectrometer.
MHN and MHN@TA were lyophilized, ground into powder, and then pigments were extracted with methanol in the dark until the pellets were colorless. After centrifugation (4000×g, 10 min), the chlorophyll content was determined by measuring the absorbance of the supernatant at 435 nm. The absorbance of the supernatant was analysed at 665 and 652 nm67. The amounts of Chlorophyll a and Chlorophyll b were calculated as follows:
Antibiotic degradation
For a typical antibiotic degradation process, MHN@TA was immersed into the as-prepared TC solutions (100–400 mg L−1), and degradation was carried out under an environment maintained at 23 °C, with a continuous illumination of 50 µmol photons m−2 s−1. After degradation, the residual antibiotic was measured by HPLC. For SM system, microalgal cells in exponential growth phase were collected by centrifuge (4000×g, 5 min) and then re-suspended in different concentrations of TC solutions. The degradation was carried out in the same conditions as described aforementioned. For reuse experiments, MHN@TA was immersed in 100 mg L−1 TC solution for 24 h and the process was repeated five times. The degradation process of AMX (50–300 mg L−1), PG (25–200 mg L−1), and CIP (2–25 mg L−1) were conducted in the same protocol.
Antibiotic detection and degradation product analysis
Antibiotic concentrations were determined using a high-performance liquid chromatography (HPLC, CORUI, China) equipped with a COSMOSIL C18 column (4.6 × 150 mm, 5 μm). For TC, the mobile phases were ultra-pure water and acetonitrile with trifluoroacetic acid (0.1%, v/v), which was used for elution with a gradient changing from 85 to 20% (v/v). Other detection conditions were as follows. Column temperature: 25 °C. Flow rate: 2 mL min−1. Injection volume: 20 μL. Wavelength of the ultraviolet (UV) detector: 350 nm. For AMX, the mobile phases were 1% acetonitrile and 99% potassium dihydrogen phosphate (0.05 mol L−1, pH = 5). Other detection conditions were as follows. Column temperature: 30 °C. Flow rate: 2 mL min−1. Injection volume: 20 μL. The wavelength of the UV detector: 254 nm. For PG, the mobile phases were 30% acetonitrile and 70% potassium dihydrogen phosphate (0.1 mol L−1). Other detection conditions were as follows. Column temperature: 30 °C. Flow rate: 1.5 mL min−1. Injection volume: 20 μL. The wavelength of the UV detector: 225 nm. For CIP, the mobile phases were 15% acetonitrile and 85% formic acid water (0.1%, v/v). Other detection conditions were as follows. Column temperature: 30 °C. Flow rate: 2 mL min−1. Injection volume: 20 μL. Wavelength of the UV detector: 270 nm.
TC intermediates were analyzed using an UPLC-Q-TOF system (Agilent, 1290infinity -6545, USA) equipped with an Agilent C18 column (2.1 × 100 mm, 2.6 μm). Samples were filtered through 0.22- μm microporous membrane (Jinteng, China) filters prior to analysis.
The total inorganic carbon (TIC) was measured by TOC/TNb analyzer vario TOC cube (Elementar, German) with liquid mode. Samples were filtered through 0.22-μm microporous membrane (Jinteng, China) prior to analysis. Other detection conditions were as follows. Carrier gas: high-purity oxygen. Detecting temperature: 850 °C. Pressure: 950–1000 mbar. Flow: 200 mL min−1.
Toxicity analysis of degradation products
The toxicology experiment was performed by mixing 15 mL of bacterial suspension (S. aureus, OD600 = 0.6) with 5 mL of BG11 medium (blank), TC solution, and degradation solution, respectively. The growth of S. aureus was monitored by measuring the absorbance of cell suspension at a wavelength of 600 nm with a UV-vis spectrometer. After incubation for 24 h at 37 °C, the absorbance of the suspension was measured.
For the inhibition zone experiment, 100 μL of diluted S. aureus was dripped on the LB bacterial culture plates and scribbled evenly. The standard paper disks with a diameter of 6 mm were immersed in BG11 medium (blank), TC solution, and degradation solution for 0.5 h. Afterward, the treated paper disks were put onto the center of the plates. After incubation for 24 h at 37 °C, the inhibition zones were characterized.
Transcriptomics analysis
Microalgal cells were collected by centrifuge (4000×g, 5 min) after treating with antibiotic (20 mg L−1) for 24 h. The TRIzol® Reagent (Invitrogen, Carlsbad, CA, USA) was used to extract total RNA. The RNA purity and concentration were measured using a NanoPhotometer® spectrophotometer (IMPLEN, CA, USA), while integrity was assessed with the Agilent 2100 Bioanalyzer (Agilent, CA, USA). The double-stranded cDNA was purified by using AMPure XP beads. The final cDNA library was obtained by PCR enrichment with NEBNext® Ultra™ RNA Library Prep Kit for Illumina®. The library preparation and transcriptome sequencing were performed on an Illumina NovaSeq platform. The differentially expressed genes (DEGs) were analysed with the DEGSeq R package. Gene Ontology (GO) term enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses were performed using the GOseq R package to investigate the functional significance of the DEGs.
Photosynthetic performance measurements
The photosynthetic performance was measured using an Os30p (Opti-sciences Inc, USA). Specifically, samples were kept in complete darkness for 30 min, and then the steady-state chlorophyll fluorescence (F0) and maximum chlorophyll fluorescence (Fm) were obtained by using a modulated light and a brief pulse of saturating light, respectively. The ratio of variable fluorescence to maximum fluorescence (Fv/Fm) was calculated as:
Numerical analysis of antibiotics diffusion
We simulated the diffusion of antibiotics in MHN@TA and SM systems by solving a two-dimensional time-dependent diffusion model in COMSOL Multiphysics. In the MHN@TA system, MHN@TA has a thickness of 2.50 mm. The diffusion coefficient of antibiotic molecules in hydrogel is 8.51 × 10−12 m² s−1. Two TA layers (10.00 µm in thickness) are attached to the upper and lower surfaces of the hydrogel, respectively. The diffusion coefficient of the antibiotic molecules within the TA is 4.00 × 10−12 m² s−1. For comparison, in the SM system, the medium (water) also has a thickness of 2.50 mm, but the antibiotic diffusion coefficient in it is significantly higher, at 5.00 × 10−10 m² s−1. It should be noted that the degradation rate of the microalgae is neglected in the current numerical analysis, which may result in an overestimated concentration of antibiotics in the MHN@TA system.
Large-scale preparation of MHN@TA
For potential industrial wastewater treatment, we developed a custom 3D printer to prepare large MHN@TA membranes. Specifically, we used Blender software to construct a 3D model of the MHN film (70 cm × 40 cm × 0.5 cm). The model was then sliced using Slic3r software. The printing parameters were set as follows: a fill rate of 60%, a printing speed of 7000 mm min−1, a layer height of 1 mm, and an inner needle diameter of 0.84 mm. Then, the prepared MHN was coated with TA.
Statistics and reproducibility
All studies were performed with three independent replications. All samples used in the study were randomly allocated into different experimental groups. The average and standard deviation were calculated in Prism9.5 software (GraphPad Software, USA). All data were analyzed using SPSS software (version 17.0; IBM, USA) and expressed as the mean ± SD. To determine the statistical significance in the comparison of the two groups, a two-tailed t-test was performed. To determine the statistical significance in the comparison of multiple groups, a one-way analysis of variance (ANOVA) was performed with Tukey’s test for post hoc analysis. Results are considered significant at p < 0.05.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All relevant data of this study are available in the paper and its supplementary materials. The RNA-seq data under this study are available in the NCBI database under accession code PRJNA1235111 and PRJNA1234825. Source data are provided with this paper.
References
Sutherland, M. E. Antibiotic use across the globe. Nat. Hum. Behav. 2, 373 (2018).
Årdal, C. et al. Antibiotic development-economic, regulatory and societal challenges. Nat. Rev. Microbiol. 18, 267–274 (2020).
MacFadden, D. R., McGough, S. F., Fisman, D., Santillana, M. & Brownstein, J. S. Antibiotic resistance increases with local temperature. Nat. Clim. Change 8, 510–514 (2018).
Larsson, D. G. J. & Flach, C. F. Antibiotic resistance in the environment. Nat. Rev. Microbiol. 20, 257–269 (2022).
Xu, L. Y. et al. Occurrence, fate, and risk assessment of typical tetracycline antibiotics in the aquatic environment: a review. Sci. Total. Environ. 753, 141975 (2021).
Khare, A. Achilles’ heel of antibiotic resistance. Nat. Microbiol. 6, 1339–1340 (2021).
Li, F. et al. Efficient removal of antibiotic resistance genes through 4f-2p-3d gradient orbital coupling mediated Fenton-like redox processes. Angew. Chem. Int. Ed. 62, e202313298 (2023).
Jansen, K., Knirsch, C. & Anderson, A. The role of vaccines in preventing bacterial antimicrobial resistance. Nat. Med. 24, 10–19 (2018).
Zhang, S., Zheng, H. & Tratnyek, P. G. Advanced redox processes for sustainable water treatment. Nat. Water 1, 666–681 (2023).
Xu, J. et al. Organic wastewater treatment by a single-atom catalyst and electrolytically produced H2O2. Nat. Sustain 4, 233–241 (2021).
Alsbaiee, A. et al. Rapid removal of organic micropollutants from water by a porous β-cyclodextrin polymer. Nature 529, 190–194 (2016).
Zhang, L. L. et al. Ultra-rapid and highly efficient enrichment of organic pollutants via magnetic mesoporous nanosponge for ultrasensitive nanosensors. Nat. Commun. 12, 6849 (2021).
Zhang, R. et al. Antifouling membranes for sustainable water purification: strategies and mechanisms. Chem. Soc. Rev. 45, 5888–5924 (2016).
Giorno, L. Membranes that filter and destroy pollutants. Nat. Nanotechnol. 17, 334–335 (2022).
Harms, H., Schlosser, D. & Wick, L. Untapped potential: exploiting fungi in bioremediation of hazardous chemicals. Nat. Rev. Microbiol. 9, 177–192 (2011).
Atashgahi, S. et al. Prospects for harnessing biocide resistance for bioremediation and detoxification. Science 360, 743–746 (2018).
Croce, R. & van Amerongen, H. Natural strategies for photosynthetic light harvesting. Nat. Chem. Biol. 10, 492–501 (2014).
Burlacot, A. et al. Alternative photosynthesis pathways drive the algal CO2-concentrating mechanism. Nature 605, 366–371 (2022).
Shahid, M. K., Kashif, A., Fuwad, A. & Choi, Y. Current advances in treatment technologies for removal of emerging contaminants from water-a critical review. Coordin. Chem. Rev. 442, 213993 (2021).
Liao, J., Mi, L., Pontrelli, S. & Lou, S. Fuelling the future: microbial engineering for the production of sustainable biofuels. Nat. Rev. Microbiol. 14, 288–304 (2016).
Lim, H. R. et al. Smart microalgae farming with internet-of-things for sustainable agriculture. Biotechnol. Adv. 57, 107931 (2022).
Nagarajan, D., Varjani, S., Lee, D. J. & Chang, J. S. Sustainable aquaculture and animal feed from microalgae–nutritive value and techno-functional components. Renew. Sust. Energ. Rev. 150, 111549 (2021).
Service, R. F. Algae’s second try. Science 333, 1238 (2011).
Cao, M. et al. Nanoparticles and antibiotics stress proliferated antibiotic resistance genes in microalgae-bacteria symbiotic systems. J. Hazard. Mater 443, 130201 (2023).
Zhang, D. et al. Microalgae-based oral microcarriers for gut microbiota homeostasis and intestinal protection in cancer radiotherapy. Nat. Commun. 13, 1413 (2022).
Wang, L. H. et al. A bioinspired scaffold for rapid oxygenation of cell encapsulation systems. Nat. Commun. 12, 5846 (2021).
Bose, S. et al. A retrievable implant for the long-term encapsulation and survival of therapeutic xenogeneic cells. Nat. Biomed. Eng. 4, 814–826 (2020).
Zhong, D. et al. Microalgae-based hydrogel for inflammatory bowel disease and its associated anxiety and depression. Adv. Mater 36, 2312275 (2024).
Hull, S. M., Brunel, L. G. & Heilshorn, S. C. 3D bioprinting of cell-laden hydrogels for improved biological functionality. Adv. Mater 2, 2103691 (2022).
Ren, C. et al. Bioinspired pH-responsive microalgal hydrogels for oral insulin delivery with both hypoglycemic and insulin sensitizing effects. ACS Nano 17, 14161–14175 (2023).
An, B. et al. Engineered living materials for sustainability. Chem. Rev 123, 2349–2419 (2022).
Liu, G. W. et al. Drinkable in situ-forming tough hydrogels for gastrointestinal therapeutics. Nat. Mater 23, 1292–1299 (2024).
Kang, Y. et al. Programmed microalgae-gel promotes chronic wound healing in diabetes. Nat. Commun. 15, 1042 (2024).
Datta, D. et al. Phenotypically complex living materials containing engineered cyanobacteria. Nat. Commun. 14, 4742 (2023).
Huang, K. X. et al. Enhancing the removal of sulfamethoxazole and microalgal lipid production through microalgae-biochar hybrids. Bioresour. Technol 413, 131510 (2024).
Liu, H. et al. Space-efficient 3D microalgae farming with optimized resource utilization for regenerative food. Adv. Mater 36, 2401172 (2024).
Lee, Y. W. et al. A tissue adhesion-controllable and biocompatible small-scale hydrogel adhesive robot. Adv. Mater 34, 2109325 (2022).
Ge, J. C. et al. Endogenous zinc-ion-triggered in situ gelation enables Zn capture to reprogram benign hyperplastic prostate microenvironment and shrink prostate. Adv. Mater 11, 2307796 (2023).
Wang, L. S. et al. Bioinspired polyacrylic acid-based dressing: wet adhesive, self-healing, and multi-biofunctional coacervate hydrogel accelerates wound healing. Adv. Sci 16, 2207352 (2023).
Wang, Z. et al. Facile biomimetic self-coacervation of tannic acid and polycation: tough and wide pH range of underwater adhesives. Chem. Eng. J. 404, 127069 (2021).
Zheng, Y. T. et al. Hemostatic patch with ultra-strengthened mechanical properties for efficient adhesion to wet surfaces. Biomaterials 301, 122240 (2023).
Yun, G. et al. Synthesis of metal nanoparticles in metal-phenolic networks: catalytic and antimicrobial applications of coated textiles. Adv. Healthc. Mater 7, 1700934 (2018).
Gorgich, M., Martins, A. A., Mata, T. M. & Caetano, N. S. Composition, cultivation and potential applications of Chlorella zofingiensis-a comprehensive review. Algal Res. 60, 102508 (2021).
Wang, Q., Unno, M. & Liu, H. Dual-function near-infrared emitting aerogel-based device for detection and sunlight-driven photodegradation of antibiotics: realizing the processability of silsesquioxane-based fluorescent porous materials. Adv. Funct. Mater 33, 2214875 (2023).
Svensson Grape, E. et al. Removal of pharmaceutical pollutants from effluent by a plant-based metal–organic framework. Nat. Water 1, 433–442 (2023).
Peydayesh, M. et al. Amyloid fibrils aerogel for sustainable removal of organic contaminants from water. Adv. Mater 12, 1907932 (2020).
Shaik, S., Kumar, D., de Visser, S. P., Altun, A. & Thiel, W. Theoretical perspective on the structure and mechanism of cytochrome P450 enzymes. Chem. Rev 105, 2279–2328 (2005).
Zhou, L. J. et al. Biotransformation of lincomycin and fluoroquinolone antibiotics by the ammonia oxidizers AOA, AOB and comammox: a comparison of removal, pathways, and mechanisms. Water Res. 196, 117003 (2021).
Xiong, J. Q., Qi, X. & Qin, J. Y. Transcriptomics unveiled metabolic perturbations in Desmodesmus quadricauda by sulfacetamide: key functional genes involved in the tolerance and biodegradation process. Sci. Total Environ. 826, 154436 (2022).
Zeng, J. L. et al. Magnetic field facilitated electrocatalytic degradation of tetracycline in wastewater by magnetic porous carbonized phthalonitrile resin. Appl. Catal. B Environ. Energy 340, 123225 (2024).
Gao, K. X., Hou, L. A., An, X. Q., Huang, D. D. & Yang, Y. BiOBr/MXene/gC3N4 Z-scheme heterostructure photocatalysts mediated by oxygen vacancies and MXene quantum dots for tetracycline degradation: process, mechanism and toxicity analysis. Appl. Catal. B Environ. Energy 323, 122150 (2023).
Mohsinul Reza, A. H. M., Zhu, X., Qin, J. & Tang, Y. Microalgae-derived health supplements to therapeutic shifts: redox-based study opportunities with AIE-based technologies. Adv. Healthc. Mater 24, 2101223 (2021).
Fan, Y. et al. Metagenomic and transcriptome elucidating the sulfamethoxazole degradation and metabolism pathways in fermentative bacteria and microalgae coupling system for mariculture wastewater treatment. Chem. Eng. J 474, 145560 (2023).
Jia, X., Shen, Z., Han, Q. & Bi, H. Rod-like Bi4O5I2/Bi4O5Br2 step-scheme heterostructure with oxygen vacancies synthesized by calcining the solid solution containing organic group. Chin. J. Catal. 2, 288–302 (2022).
Xie, D. et al. Tree-inspired efficient solar evaporation and simultaneous in-situ purification of ultra-highly concentrated mixed volatile organic wastewater. Nano Energy 93, 106802 (2022).
Chu, Y. et al. New insight into the concentration-dependent removal of multiple antibiotics by Chlorella sorokiniana. Bioresour. Technol. 385, 129409 (2023).
Yu, C. et al. Seawater Chlorella sp. biofilm for mariculture effluent polishing under environmental combined antibiotics exposure and ecological risk evaluation based on parent antibiotics and transformation products. Sci. Total. Environ. 939, 173643 (2024).
Pan, M. et al. Mitigating antibiotic pollution using cyanobacteria: removal efficiency, pathways and metabolism. Water Res. 190, 116735 (2021).
Qi, W. et al. Fe3+ enhanced degradation of oxytetracycline in water by pseudomonas. Water Res. 160, 361–370 (2019).
Chen, X. et al. Enrichment of tetracycline-degrading bacterial consortia: microbial community succession and degradation characteristics and mechanism. J. Hazard. Mater 448, 130984 (2023).
Zhao, Z., Zhang, G., Zhang, Y., Dou, M. & Li, Y. Fe3O4 accelerates tetracycline degradation during anaerobic digestion: synergistic role of adsorption and microbial metabolism. Water Res. 185, 116225 (2020).
Xu, J. et al. Enhanced removal of tetracycline from synthetic wastewater using an optimal ratio of co-culture of Desmodesmus sp. and Klebsiella pneumoniae. Bioresour. Technol. 351, 127056 (2022).
Aydin, S., Ünlü, İ. D., Arabacı, D., N. & Duru, Ö. A. Evaluating the effect of microalga Haematococcus pluvialis bioaugmentation on aerobic membrane bioreactor in terms of performance, membrane fouling and microbial community structure. Sci. Total. Environ. 807, 149908 (2022).
He, Z., Wei, Z., Zhao, Y., Zhang, D. & Pan, X. Enhanced performance of tetracycline treatment in wastewater using aerobic granular sludge with in-situ generated biogenic manganese oxides. Sci. Total. Environ. 735, 139533 (2020).
Guo, G. et al. Removal of antibiotics by four microalgae-based systems for swine wastewater treatment under different phytohormone treatment. Bioresour. Technol. 400, 130668 (2024).
Wang, Z., Hartline, C. J., Zhang, F. & He, Z. Enhanced microalgae cultivation using wastewater nutrients extracted by a microbial electrochemical system. Water Res. 206, 117722 (2021).
Li, X. et al. Cytoprotective metal–phenolic network sporulation to modulate microalgal mobility and division. Adv. Sci. 11, 2308026 (2024).
Acknowledgements
We acknowledge the Shenzhen Key Laboratory of Marine Microbiome Engineering for providing microalgae. The authors thank Prof. Qiang Hu for his guidance in microalgae cultivation. The authors thank Yu Du for his assistance with 3D bioprinting. This work was supported by the Opening Project of the Key Laboratory of Leather Chemistry and Engineering (Sichuan University), Ministry of Education. Jiajing Zhou acknowledges the Fundamental Research Funds for the Central Universities, and Natural Science Foundation of Sichuan Province (2024NSFSC0242). Jianfei Zhou and Jiajing Zhou acknowledge the funding from the Institutes of Leather and Footwear Industry of Wenzhou (2022WZSLCQ01, 202303GT01). Jin Liu acknowledges the Double First-Class Talent Team Leading Program of Nanchang University, and the Double Thousand Plan of Jiangxi Province. Yuan Ma acknowledges financial support from the Hong Kong Research Grants Council (ECS 25228722).
Author information
Authors and Affiliations
Contributions
Jiajing Zhou, J.L., and M.J. conceived and designed the experiments. M.J. performed the major experimental work and wrote the manuscript. M.J., J.Z., and H.L. performed the wastewater treatment. Jiajing Zhou and J.L. offered guidance and assistance for 3D printing and microalgae cultivation. Y.T. and Y.M. conducted the diffusion dynamics simulation and analyzed the results. Y.Y., Jianfei Zhou, and W.L. helped the analysis of the results. All authors discussed the results and contributed to manuscript writing and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jiang, M., Zheng, J., Tang, Y. et al. Retrievable hydrogel networks with confined microalgae for efficient antibiotic degradation and enhanced stress tolerance. Nat Commun 16, 3160 (2025). https://doi.org/10.1038/s41467-025-58415-z
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58415-z
This article is cited by
-
Revolutionizing Bioremediation: Synergistic Strains Tackle Ofloxacin Contamination
Water, Air, & Soil Pollution (2026)
-
Role of microalgae in reducing antibiotic-resistant bacteria in synthetic wastewater
Environmental Monitoring and Assessment (2025)