Abstract
KRAS is the most common mutated oncogenes in colorectal cancer (CRC), yet effective therapeutic strategies for targeting multiple KRAS mutations remained challenging. The prolonged protein stability of KRAS mutants contribute to their robust tumor-promoting effects, but the underlying mechanism is elusive. Herein by screening deubiquitinases (DUBs) siRNA library, we identify Josephin domain containing 2 (JOSD2) functions as a potent DUB that regulates the protein stability of KRAS mutants. Mechanistically, JOSD2 directly interacts with and stabilizes KRAS variants across different mutants, by reverting their proteolytic ubiquitination; while KRAS mutants reciprocally inhibit the catalytic activity of CHIP, a bona fide E3 ubiquitin ligase for JOSD2, thus forming a JOSD2/KRAS positive feedback circuit that significantly accelerates KRAS-mutant CRC growth. Inhibition of JOSD2 by RNA interference or its pharmacological inhibitor promotes the polyubiquitination and proteasomal degradation of KRAS mutants, and preferentially impede the growth of KRAS-mutant CRC including patient-derived cells/xenografts/organoids (PDCs/PDXs/PDOs) over that harboring wild-type KRAS. Collectively, this study not only reveals the crucial roles of JOSD2/KRAS positive feedback circuit in KRAS-mutant CRC, but also provides a rationale to target JOSD2 as the promising pan-KRAS-mutation-targeting strategy for the treatment of a broad population of CRC patients with KRAS variant across different mutant types.
Similar content being viewed by others
Introduction
Colorectal cancer (CRC) ranks as the third most common malignancy and the second leading cause of cancer mortality worldwide1,2. CRC is characterized by the heterogeneous mutational events involving APC, KRAS, and TP53 etc., among which KRAS is the most prominent oncogenic mutation and poses a major challenge for the management of CRC patients3. Those KRAS mutations-harboring patients (~53% of the total CRC population) show low response toward current therapeutic regimens, including chemotherapeutic agents and/or targeted therapy agents4,5. Therefore, novel therapies for KRAS-mutant CRC are urgently needed.
KRAS is the most frequently mutated cancer driver gene; substantial efforts have been devoted to targeting KRAS mutants over the past decades4,6,7. The GTP hydrolysis function of KRAS appears to be fundamental for the oncogenic activation. Nevertheless, due to the high affinity of KRAS for GTP (pM affinity) as well as the lack of an inhibitor binding pocket on the surface of the KRAS protein, KRAS mutants have been unamenable to pharmacological inhibition4. Until the recent success with the approval of the KRASG12C covalent inhibitor Sotorasib for those non-small-cell lung cancer (NSCLC) patients with G12C mutation8,9. However, the other types of KRAS mutants such as G12D and G12V cannot benefit from Sotorasib and the other G12C covalent inhibitors, while more than 90% of KRAS-mutant CRC patients harboring these non-G12C mutations; together with the fact that ~50% patients rapidly developed resistance against Sotorasib treatment10,11, there is still an unmet need for CRC patients harboring KRAS variants across all mutation.
Accumulating evidence shows that the RAS protein abundance and activities are regulated by ubiquitination and proteasomal degradation. Leucine zipper-like transcriptional regulator 1 (LZTR1), an adapter of the Cullin3 (CUL3)-based E3 ubiquitin ligase complex, was found to facilitate KRAS ubiquitination, and negatively regulate KRAS protein levels and MAPK pathway12,13,14. Similarly, WD40-repeat protein 76 (WDR76), a linker protein for E3 ubiquitin ligase, also interacted with RAS and functioned as a tumor suppressor to negatively regulate its protein abundance15. KRAS is a highly stable oncoprotein (half-life >12 h) and overexpression of its bona fide E3 ubiquitin ligases LZTR1 and WDR76 significantly decrease KRAS mutants’ stability and inhibit KRAS-mutant tumor growth13,15, these studies collectively underline that the high protein stability of KRAS mutants is critical for its robust tumor-promoting function, and the protein stability regulation may offer a potential strategy to intervene with the oncogenic function of KRAS variants across different mutation types. Compared to activating those tumor-suppressive E3 ubiquitin ligases, the inhibition of the deubiquitinases (DUBs) that are responsible for positively regulating the RAS protein stability by removing ubiquitin chains is considered a more ideal strategy to target KRAS mutants, since DUBs are intrinsically attractive as potential drug targets with well-defined catalytic clefts16. Therefore, the exploration of bona fide DUB(s) for KRAS mutants is expected to provide novel targets for CRC with differential KRAS mutants.
Josephin domain containing 2 (JOSD2) belongs to the Machado-Josephin Domain-containing proteases (MJDs) subfamily of DUBs, which consists of four family members: JOSD1, JOSD2, Ataxin-3, and Ataxin-3L17. Several lines of evidence have revealed the roles of JOSD2 in several cancer types, including nonsmall cell lung cancer (NSCLC)18, hepatocellular carcinoma (HCC)19, and cholangiocarcinoma (CCA)20. A recent study found that JOSD2 promoted glucose metabolism by deubiquitinating and stabilizing the metabolic enzyme complex, namely phosphoglycerate dehydrogenase (PHGDH), phosphofructokinase-1 (PFK-1), and Aldolase A18. Another study revealed that JOSD2 promoted cholangiocarcinoma proliferation by deubiquitinating and stabilizing Yes-associated protein (YAP) and transcriptional co-activator with PDZ-binding motif (TAZ), two essential transcriptional co-activators of the Hippo pathway20. These findings collectively suggest that JOSD2 is essential for the malignant progression of several cancer types. However, the roles of JOSD2 in CRC, particularly those harboring KRAS-mutant variants, remain unknown. Moreover, the underlying mechanism of JOSD2 hyperactivated in tumors is elusive.
In this study, we perform a functional RNA interference screen to globally profile the contribution of DUBs to KRAS mutants, and identify JOSD2 functions as a key deubiquitinase that positively regulates the stability of KRAS mutants. Reciprocally, the stabilized KRAS mutants inhibit the ubiquitination of JOSD2 by suppressing its bona fide E3 ubiquitin ligase CHIP, thus forming a JOSD2/KRAS positive feedback circuit that robustly accelerates proliferation in KRAS-mutant CRC cells. Conversely, JOSD2 inhibition by depleting or using its pharmacological inhibitor selectively impedes the growth of CRC models across different KRAS mutation types both in vitro and in vivo, while imposing less effect on KRAS wild-type CRC models. Taken together, our findings not only provide evidence for the vital importance of the JOSD2/KRAS positive feedback circuit in CRC, but also emphasize that inhibiting JOSD2 could serve as a promising therapeutic approach for CRC patients harboring various KRAS mutations.
Results
DUBs siRNA library screening identified JOSD2 as a crucial DUB regulating KRAS protein stability and activity
Multiple studies have revealed that the high protein stability of KRAS mutants is critical for its robust tumor-promoting function13,15, which is further verified in our study that the protein stability of KRAS mutants is significantly prolonged compared to that of KRAS wild-type (Supplementary Fig. 1a). Therefore, promoting the degradation of KRAS mutants may offer a potential strategy to disrupt the oncogenic activity of KRAS variants across different mutation types. The ubiquitination of KRAS induced by several known E3 ligases mediates the proteasomal degradation of KRAS. However, instead of activating these E3 ligases, a potentially more effective approach could involve inhibiting the potential DUB(s) responsible for deubiquitinating KRAS mutants. This is because the catalytic inhibition of DUBs has shown promising therapeutic potential for cancer treatment.
We conducted a comprehensive analysis to determine the impact of all 96 DUBs on direct regulation of KRAS, which remained an essential component. For this purpose, we established a KRASG12V-luciferase reporter system (Fig. 1a) that enabled the assessment of KRASG12V abundance based on luciferase signal. By screening the DUBs siRNA library (each DUB pool consists of four nonoverlapping siRNA oligos, Supplementary Data 1), we found JOSD2 siRNA pools significantly minimized the luciferase activity in a concentration-dependent manner, but JOSD1, another highly homologous member of MJDs, failed to pose a similar effect (Fig. 1b, c).
a The schematic diagram of screening the candidate DUBs of regulating KRAS stability by using KRASG12V-luciferase reporter system and DUBs siRNA library (The schematic diagram was created in BioRender. https://BioRender.com/w35e177). The KRASG12V-luciferase reporter system was constructed by cloning the KRASG12V coding sequences in front of the firefly-luciferase sequence, thus forming a fusion gene of KRASG12V-Luciferase. CDS coding sequences, DUB deubiquitinases. b JOSD2 siRNA pools significantly minimized the luciferase activity of the KRASG12V-luciferase reporter system. c JOSD2 depletion inhibited the luciferase activity of the KRASG12V-luciferase reporter system in a dose-dependent manner. (means ± SD, n = 3; three independent experiments were performed). d, e JOSD2, but not JOSD1, positively regulated KRAS mutants’ protein levels. f JOSD2 depletion dramatically decreased KRAS mutants’ stability. g JOSD2 depletion downregulated KRAS levels through the ubiquitin-proteasome systems but not the autophagy-lysosome system. h, i JOSD2, but not JOSD1, positively regulated MAPK signaling pathways. WT, wild-type; Mut, mutation. The significance analysis of (c) was conducted by One-way ANOVA (Bonferroni method was utilized to correct for multiple comparisons). The samples derived from the same experiment and processed on different gels in parallel are shown below: d SW480(JOSD2): one gel for GAPDH/KRAS, another for JOSD2; e SW480(shJOSD2) & SW620(shJOSD2): one gel for GAPDH/JOSD2, another for KRAS. f Different gels for GAPDH and Myc(KRAS); g one gel for GAPDH/JOSD2, another for KRAS; h SW480(shJOSD1): one gel for p-MEK/GAPDH/MEK, another for p-ERK/JOSD1/ERK; SW620(shJOSD1): one gel for p-ERK/MEK, another for p-MEK/ERK, another for GAPDH/JOSD1; SW480(shJOSD2): one gel for p-MEK/JOSD2/GAPDH, another three for ERK, p-ERK and MEK, respectively; SW620(shJOSD2): one gel for p-MEK/GAPDH, another for p-ERK/JOSD2, another two for ERK and MEK, respectively; i SW480(JOSD1): one gel for p-MEK/KRAS/GAPDH/MEK, another for p-ERK/JOSD1, another for ERK; SW620(JOSD1): one gel for p-ERK/KRAS/p-MEK/GAPDH, another for MEK/JOSD1, another for ERK; SW480(JOSD2): one gel for ERK/KRAS, another for p-MEK/GAPDH, another for p-ERK/JOSD2, another for MEK; SW620(JOSD2): one for ERK/KRAS, another for p-MEK/GAPDH, another for p-ERK/JOSD2, another for MEK. Source data are provided as a Source Data file.
The inhibitory effects of JOSD2 siRNA on the KRASG12V-luciferase reporter system led us to explore the roles of JOSD2 in the regulation of KRAS turnover and activity. We found overexpression of JOSD2, but not JOSD1, significantly upregulated KRAS levels (Fig. 1d). Correspondingly, depletion of JOSD2, but not JOSD1, significantly downregulated KRAS levels (Fig. 1e and Supplementary Fig. 1b, c), indicating JOSD2 positively regulates KRAS abundance. To further illustrate whether JOSD2 regulates KRAS at transcription or post-translational modification, we then examined the effects of JOSD2 on KRAS mRNA and protein stability. We found that JOSD2 posed little effect on KRAS transcription levels (Supplementary Fig. 1d), but depletion of JOSD2 dramatically shortened the half-life of KRAS (Fig. 1f and Supplementary Fig. 1e). Moreover, the inhibitory effect of JOSD2 knockdown on KRAS levels can be revered by the proteasome inhibitor MG-132, but not autophagy-lysosome inhibitor chloroquine (CQ), indicating JOSD2 regulates KRAS turnover via the ubiquitin-proteasome pathway (Fig. 1g). Furthermore, JOSD2, not JOSD1, positively regulates MAPK pathway, a classical downstream signaling pathway that reflects KRAS activity, in an enzyme activity-dependent manner (Fig. 1h, i and Supplementary Fig. 1f).
KRAS activates downstream pathways by binding to its classical downstream effector RAF1, whether the ubiquitination modification affects their binding. As shown in Supplementary Fig. 1g, h, we found ubiquitin overexpression inhibits KRAS activity and its binding with RAF1. Subsequently, we also introduced epidermal growth factor (EGF), a classical stimulator of KRAS, and found that EGF stimulation indeed significantly downregulated KRAS ubiquitination levels and promoted KRAS activity and its binding to RAF1 (Supplementary Fig. 1i, j). Moreover, we found that JOSD2, but not JOSD1, significantly promoted the binding between KRAS and RAF1 (Supplementary Fig. 1k), suggesting the ubiquitination modification of KRAS may also affect their binding with effectors. We hypothesized that steric hindrance might be generated when KRAS is modified by polyubiquitin chains, potentially affecting the binding between KRAS and its downstream effector RAF1. Specifically, when JOSD2 removes the polyubiquitin chains from KRAS, the steric hindrance is reduced, thereby promoting the KRAS-RAF1 interaction, which will be further investigated in our future study.
Taken together, these data confirmed that JOSD2 positively regulated KRAS abundance and activity in an enzymatic activity-dependent manner.
JOSD2 interacts with KRAS
To gain further insight into how JOSD2 regulated KRAS turnover and activity through its DUB activity, we first examined the exogenous interaction between JOSD2 and KRAS. We found both KRAS wild-type and mutants interacted with JOSD2 (Fig. 2a and Supplementary Fig. 2a), moreover, JOSD2 shRNA interrupted their interaction, further verifying the binding specificity (Supplementary Fig. 2b). More importantly, the levels of co-immunoprecipitation between KRAS mutants and JOSD2 were significantly higher than with KRAS wild-type (Fig. 2a). Considering JOSD2 regulated KRAS in an enzyme activity-dependent manner, we next asked whether JOSD2 binding to KRAS relying on its catalytic activity. As shown in Fig. 2b, both wild-type JOSD2 and its catalytic mutants interact with KRAS, suggesting that the catalytic activity of JOSD2 is not necessary for KRAS binding to JOSD2.
a The ability of KRASG12C/G12D/G12V to bind to JOSD2 was stronger than that of KRASWT. WT, wild-type; IP, immunoprecipitation. b The interaction between JOSD2 and KRAS was independent of the enzyme activity of JOSD2. CA&HY, JOSD2-C24A&H125Y. c–e JOSD2, not JOSD1, interacted with KRAS at semi-endogenous levels and endogenous levels. f In vitro GST pull-down assays to examine the interaction between JOSD2 and KRASWT/G12C/G12D/G12V. 293T cells were transfected with the indicated expression plasmids for 24 h. Cell lysates were incubated with the Streptavidin magnetic beads conjugated with bacterial-expressed recombinant GST or GST-JOSD2 protein. Proteins retained on Streptavidin magnetic beads were subjected to WB analysis. g, h JOSD2, not JOSD1, removed the polyubiquitination chains of KRASWT/G12C/G12D/G12V in an enzyme activity-dependent manner. i JOSD2 depletion promoted the polyubiquitination of KRASWT/G12C/G12D/G12V. j JOSD2 removed the polyubiquitination chains of KRASWT/G12C/G12D/G12V in vitro (The schematic diagram was created in BioRender. https://BioRender.com/w35e177). 293T cells were co-transfected with indicated expression plasmids, and cell lysates were incubated with anti-Flag affinity gel overnight. Then, proteins retained on anti-Flag affinity gel were incubated with bacterial-expressed recombinant human JOSD2 (rhJOSD2) protein at 37 °C for 3 h, followed by WB analysis. k JOSD2, not JOSD1, removed the polyubiquitination chains of KRASG12V promoted by LZTR1. The samples derived from the same experiment and processed on different gels in parallel were shown below: d Input: one gel for JOSD1/JOSD2/GAPDH, another for Flag(G12V), IP: one gel for JOSD1/Flag(G12V), another for JOSD2; e SW620: Input: one gel for JOSD1/KRAS/GAPDH, another for JOSD2, IP: one gel for JOSD1/KRAS, another for JOSD2; HCT-116: Input: one gel for JOSD2/GAPDH, another for JOSD1/KRAS, IP: different gels for JOSD1, KRAS and JOSD2; f one gel for KRAS/GST-JOSD2, another for GST; g Input of KRAS-WT/G12C: one gel for HA(ub)/GAPDH/ Flag(KRAS), another for JOSD1&2, Input of KRAS-G12D/G12V: one gel for HA(ub)/GAPDH/JOSD1&2, another for Flag(KRAS); i Input of KRAS-WT/G12C/G12D/G12V: one gel for HA(ub)/GAPDH/JOSD2, another for Flag(KRAS); k Input: one gel for HA(ub)/GAPDH, another for Flag(KRAS), another for Myc(LZTR1)/JOSD1&JOSD2. Source data are provided as a Source Data file.
In addition, we also observed an interaction between exogenous JOSD2 and endogenous KRAS as well as exogenous KRAS and endogenous JOSD2 (Fig. 2c, d). Moreover, the protein-protein interaction between endogenous JOSD2 and endogenous KRAS in SW620 and HCT-116 cells was further demonstrated in our co-immunoprecipitation assays (Fig. 2e). However, JOSD1 failed to bind to KRAS neither semi-endogenous nor endogenous levels (Fig. 2d, e). Subsequently, we purified recombinant protein GST-JOSD2/JOSD1 and His-KRAS (WT, G12C, G12D, G12V) by using Escherichia coli BL21, and the purity levels are above 90% (Supplementary Fig. 2c). Subsequently, we examined the biological activity of GST-JOSD2/JOSD1 by introducing their co-reported substrate proteins YAP20,21, as shown in Supplementary Fig. 2d, both GST-JOSD2 and GST-JOSD1 efficiently removing the polyubiquitin chains of YAP, confirming that our purified recombinant protein are biologically active. Subsequent in vitro GST pull-down assays further verify that JOSD2 is a bona fide KRAS binding protein, and the binding amount between KRAS mutants and JOSD2 was significantly stronger than that of KRAS wild-type (Fig. 2f and Supplementary Fig. 2e); however, whether the increased binding is physiologically relevant requires further investigation in future studies. To further demonstrate that the interaction between KRAS and JOSD2 is influenced by physiological or pathological conditions, we then introduced EGF and found that EGF stimulation indeed significantly promoted the MAPK signaling pathway (Supplementary Fig. 2f), which is consistent with the previous study22. Moreover, we found that the interaction between JOSD2 and KRAS was greatly increased by EGF, suggesting that this interaction might be influenced by KRAS activity stimulators. (Supplementary Fig. 2g). Taken together with the finding that KRAS mutants show significantly higher co-immunoprecipitation levels with JOSD2 compared to wild-type KRAS, these data collectively suggest that the active conformation of KRAS may influence its binding to JOSD2, which needs further investigation in future studies.
Collectively, these data demonstrated that JOSD2 is a bona fide KRAS binding partner at exogenous and endogenous levels.
JOSD2 deubiquitinates and stabilizes KRAS in an enzyme activity-dependent manner
Aforementioned data suggested that JOSD2 interacts with KRAS and positively regulates KRAS turnover and activity in an enzymatic activity-dependent manner. Considering JOSD2 functions as a DUB, we next examined whether JOSD2 removes the ubiquitin chains of KRAS. As shown in Fig. 2g, h and Supplementary Fig. 2h, i, JOSD2 wild-type significantly removed the polyubiquitin chains of KRAS wild-type and mutants (G12C, G12D, G12V, G12A, G13C, G13D, G13V, G13R) at exogenous/endogenous levels, while JOSD2 enzymatic mutants failed to posed a similar effect, indicating that JOSD2 deubiquitinates KRAS wild-type and mutants depending on its DUB enzymatic activity. However, neither JOSD1 wild-type nor its enzymatic mutant failed to decrease KRAS polyubiquitination levels (Fig. 2g). Subsequently, we also tested the effect of JOSD2 knockdown on KRAS ubiquitination and found that depletion of JOSD2 remarkably promoted the polyubiquitination levels of KRAS wild-type and mutants (Fig. 2i). More importantly, we performed in vitro deubiquitination assays using bacterial-expressed recombinant rhJOSD2 and found rhJOSD2 effectively removing the polyubiquitin chains of KRAS wild-type and mutants, while GST-JOSD1 failed to pose similar effects (Fig. 2j and Supplementary Fig. 2j).
Multiple studies reported that LZTR1 is a vital E3 ligase regulating KRAS12,13,14, so we next examined whether JOSD2 reversed the effect of LZTR1 on KRAS. As shown in Fig. 2k and Supplementary Fig. 2k, l, overexpression of LZTR1 significantly promotes the polyubiquitination of KRAS and downregulates KRAS levels. On this basis, overexpression of JOSD2, not JOSD1, dramatically decreased the polyubiquitin chains of KRAS promoted by LZTR1 and restored the LZTR1-mediated KRAS levels downregulation (Fig. 2k and Supplementary Fig. 2k, l), indicating that JOSD2 reversed the effect of LZTR1-mediated KRAS ubiquitination degradation. Subsequently, we further investigated the ubiquitin chain types of JOSD2 regulating KRAS. Given that JOSD2 has been shown to reverse the polyubiquitin chains of KRAS mediated by LZTR1, which is reported to promote K48, K63, and K33-linked ubiquitin chains as part of its function as a “RAS killer protein” and JOSD2 also can deubiquitinate K6-linked ubiquitination of LKB1 and affecting its kinase activity13,23, so we next tested the effects of GST-JOSD2 on K6-, K33-, K48-, K63- and K48R-linked ubiquitin chains of KRAS. As shown in Supplementary Fig. 2m, GST-JOSD2 efficiently removes K48-linked ubiquitin chains but failed to cleave the K6-, K33-, K63- and K48R-linked ubiquitin chains of KRAS. These data conclusively demonstrate the specificity of JOSD2 in regulating KRAS through K48-linked ubiquitin chains, reinforcing the unique role of this modification.
Taken together, JOSD2 directly deubiquitinated and stabilized KRAS wild-type and mutants in an enzyme-dependent manner in vivo and in vitro, further demonstrating that JOSD2 functions as a crucial DUB regulating KRAS.
Depletion of JOSD2 remarkably inhibits KRAS-mutant CRC growth in vitro/in vivo
Many studies have demonstrated that KRAS mutants function as a powerful oncogene and promote CRC cell proliferation24,25. Our above data demonstrated that JOSD2 functions as a vital DUB that regulates KRAS turnover and activity, so we next asked whether JOSD2 influenced the proliferation of KRAS-mutant CRC cells. We first examined the effects of JOSD2 knockdown on KRAS-mutant CRC cell growth and found depletion of JOSD2 dramatically inhibited the proliferation and colony formation of KRAS-mutant CRC cells SW480, SW620, and HCT-116 in vitro (Supplementary Fig. 3a–d). To further demonstrate that JOSD2 is involved in KRAS-mutant CRC cell growth, we also performed in vivo xenografts. As shown in Supplementary Fig. 3e–g and Supplementary Table 1, depletion of JOSD2 remarkably suppressed the tumor growth of two KRAS-mutant CRC cell-derived xenografts (CDXs), with an inhibition ratio of 68.70% (SW620 xenografts) and 55.48% (SW480 xenografts), but posed little effects on the body weight of mice (Supplementary Fig. 3h). To further clarify that JOSD2 influences KRAS-mutant CRC cells growth by regulating KRAS, we also examined the intratumor related protein levels. As shown in Supplementary Fig. 3i, depletion of JOSD2 significantly downregulated intratumor KRAS levels, indicating JOSD2 inhibition is capable of suppressing KRAS turnover in vitro and in vivo.
To further investigate the contribution of JOSD2 in KRAS-mutant CRC growth in preclinical settings, we conducted KRAS-mutant CRC patient-derived cells (PDCs) and patient-derived xenografts (PDX) models (Fig. 3a). We found depletion of JOSD2 significantly downregulated KRAS, p-MEK and p-ERK levels in PDC cells (Supplementary Fig. 3j) and suppressed PDC cells growth in vitro, as measured by cell proliferation and colony formation assays (Supplementary Fig. 3k–l). Subsequent in vivo xenografts demonstrated that JOSD2 inhibition dramatically suppressed KRAS-mutant CRC PDXs growth and downregulated intratumor KRAS, p-MEK and p-ERK levels, with inhibition ratio as 73.24% (PDX-1 xenografts) and 81.12% (PDX-2 xenografts) (Fig. 3b–e, Supplementary Fig. 3m and Supplementary Table 2), which is consistent with the results of SW480 and SW620 xenografts.
a The schematic diagram of PDCs and PDXs models (The schematic diagram was created in BioRender. https://BioRender.com/w35e177). CRC, colorectal cancer; PDC, patient-derived cells; PDX, patient-derived xenografts. b–e Knocking down JOSD2 significantly inhibited the tumor growth of PDXs in vivo. The tumors were intratumorally injected with lentivirus-encoded scramble shRNA (Ctrl) or JOSD2 shRNA (shJOSD2) every two days (means ± S.E.M., n = 6 mice/group for PDX-1 xenografts; n = 7 mice/group for PDX-2 xenografts). b The bearing-tumor mouse and tumor images. c The relative tumor volume of indicated groups. Tumors were measured every two days. d The tumor weight of the indicated groups. e Knocking down JOSD2 downregulated intratumor KRAS levels and inhibited MAPK signaling pathways of PDXs. The protein isolated from the tumor was subjected to WB analysis (means ± S.E.M., n = 6 mice/group). The significance analysis of c–e was conducted by Two-tailed unpaired Student’s t-tests. The samples derived from the same experiment and processed on different gels in parallel are shown below: e one gel for p-MEK/GAPDH/JOSD2/MEK, another for p-ERK/ERK/KRAS. Source data are provided as a Source Data file.
These results collectively demonstrated that JOSD2 plays a crucial role in KRAS-mutant CRC and that targeting JOSD2 significantly suppressed KRAS-mutant CRC growth in vitro and in vivo.
KRAS mutants significantly upregulate JOSD2 by inhibiting the E3 ligase activity of CHIP
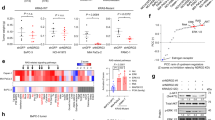
Considering that the binding amount of KRAS mutants to JOSD2 was significantly stronger than that of KRAS wild-type, we speculated whether JOSD2 plays different roles in KRAS wild-type or mutant CRC. To exclude the differences of cells genetic background, we conducted a HT-29 cells model harbored KRAS wild-type/mutants by separately infecting lentivirus that stably overexpressing KRASWT/G12C/G12D/G12V and performed a series of comparative studies (Supplementary Fig. 4a). We found overexpression of KRAS mutants promoted cellular malignant phenotype and cell proliferation compared to KRAS wild-type cells (Supplementary Fig. 4b, c). Moreover, JOSD2 displayed higher levels in HT-29-KRASG12C/G12D/G12V compared to HT-29-KRAS parent/WT (Supplementary Fig. 4d), prompting KRAS-mutant CRC may harbor higher JOSD2 levels. To demonstrate this assumption, we collected KRAS wild-type/mutant CRC tissues from the clinic and performed immunochemistry (IHC) staining to evaluate the intratumor JOSD2 protein levels. As shown in Fig. 4a, KRAS-mutant CRC patients displayed higher JOSD2 levels compared to KRAS wild-type patients, indicating that KRAS mutants may function as a driver promoting JOSD2 abnormally high expression in KRAS-mutant CRC.
a The intratumor JOSD2 levels of KRAS-mutant CRC patients were significantly higher than those of KRAS wild-type CRC patients. IHC, immunochemistry; WT, wild-type; Mut, mutation. (means ± SD, n = 6 patients/group; Scale bar, 100 μm). b, c KRAS mutants, not KRAS wild-type, positively regulated JOSD2 levels. d KRAS mutants posed little effect on JOSD2 mRNA levels (means ± SD, n = 3; three independent experiments were performed). e Overexpression of KRAS mutants, not KRAS wild-type, increased JOSD2 stability. CHX, cycloheximide. f KRAS mutants decreased the polyubiquitination levels of JOSD2. g There was no significant difference in the ubiquitination levels of JOSD2-WT and C24A when immunoprecipitated with KRAS-G12V. WB, western blot. h K48, K142, and K181 are three main ubiquitination sites on JOSD2 (The schematic diagram was created in BioRender. https://BioRender.com/w35e177). i The stability of JOSD2-3KR was significantly higher than JOSD2-WT. j KRAS mutants significantly increased the protein levels and stability of JOSD2-WT, but not that of JOSD2-3KR. DOX, doxycycline. k, l JOSD2 interacted with CHIP in vivo/in vitro. m, n CHIP positively regulated the polyubiquitination levels of JOSD2. o CHIP negatively regulated JOSD2 levels. p Overexpression of CHIP significantly decreased the protein levels of JOSD2-WT, but not of JOSD2-3KR. q Depletion of CHIP prevented the further upregulation of JOSD2 levels induced by KRASG12V. (means ± SD, n = 3; three independent experiments were performed). The significance analysis of (a) was conducted by Two-tailed unpaired Student’s t-tests, and (d, q) were conducted by One-way ANOVA (Bonferroni method was utilized to correct for multiple comparisons). The samples derived from the same experiment and processed on different gels in parallel were shown below: b one gel for Flag(KRAS)/GAPDH, another for JOSD2; f Input: one gel for HA(ub)/GAPDH/Flag(JOSD2), another for Myc(KRAS); k Input: different gels for GAPDH and Flag(CHIP); IP: different gels for JOSD2 and Flag(CHIP); l different gels for GST and Myc(CHIP); q one gel for CHIP/Flag(G12V), another for GAPDH/JOSD2. Source data are provided as a Source Data file.
Subsequently, we examined the effects of KRAS wild-type/mutants on JOSD2 and found that KRAS mutants significantly upregulated JOSD2 protein levels, while KRAS wild-type failed to pose similar effect (Fig. 4b and Supplementary Fig. 4e), and the effect of KRAS mutants mediated JOSD2 levels upregulation could be blocked by PD98059 and SB203580, the inhibitors of MEK and MAPK pathway (Supplementary Fig. 4f). Correspondingly, depletion of KRAS mutants dramatically downregulated JOSD2 levels (Fig. 4c), indicating KRAS mutants positively regulated JOSD2 levels. To further illustrate whether KRAS mutants regulate JOSD2 at the levels of transcription or post-translational modification, we then examined the effects of KRAS mutants on JOSD2 mRNA levels. We found that neither KRAS-mutant nor wild-type posed a little effect on JOSD2 transcription levels (Fig. 4d), but KRAS mutants dramatically prolonged the half-life of JOSD2 (Fig. 4e), indicating that KRAS mutants regulated JOSD2 at the levels of post-translational modification.
Ubiquitination modification is the key process controlling protein levels and stability, so we asked whether KRAS mutants regulate the ubiquitination modification of JOSD2. As shown in Fig. 4f and Supplementary Fig. 4g, KRAS mutants, but not KRAS wild-type, significantly decreased the polyubiquitination levels of JOSD2, similar results were also observed on JOSD2 enzymatic mutant C24A (Supplementary Fig. 4h). Moreover, EGF stimulation induced KRAS activation also posed a similar effect (Supplementary Fig. 4i), demonstrating the regulation of KRAS on JOSD2 ubiquitination depending on KRAS activity. Subsequently, we further compared JOSD2-WT/C24A ubiquitinylation levels in IPs with KRAS-G12V and found that the ubiquitinylation levels of JOSD2-WT and C24A in IPs with KRAS-G12V didn’t display a significant difference, indicated that the ubiquitinylation of the recovered JOSD2 in KRAS-G12V IPs is independent on JOSD2’s catalytic activity (Fig. 4g). We next attempted to identify the ubiquitin sites of JOSD2. We first analyzed the amino acid sequence of JOSD2 and identified four potential ubiquitin modification sites, namely lysine residues 48 (K48), 142 (K142), 176 (K176), and 181 (K181) (Fig. 4h). Subsequent sequences comparison revealed these potential sites are conserved in JOSD2 orthologs across different species (Supplementary Fig. 4j). To verify potential ubiquitin sites, we constructed a series of mutants only carrying single K and mutation another three K-to-R. By in vivo ubiquitination assays, we found K48, K142, and K181 displayed obvious ubiquitination, while no obvious ubiquitination was observed at K176 (Fig. 4h), indicating K48, K142, and K181 are the three main ubiquitination sites on JOSD2. Subsequently, we compared the protein stability of JOSD2 wild-type (JOSD2-WT) and its ubiquitin sites mutant (JOSD2-3KR) and found the half-life of JOSD2-3KR is significantly longer than that of JOSD2-WT (Fig. 4i). Moreover, KRAS mutants remarkably upregulated the protein levels and prolonged the half-life of JOSD2-WT, while posed little effect on JOSD2-3KR, demonstrating KRAS mutants regulating JOSD2 levels and stability mainly through K48, K142, and K181 (Fig. 4j).
Considering that E3 ligases are key proteins regulating the ubiquitination levels of substrates, we guessed whether KRAS mutants regulate JOSD2 ubiquitination through their E3 ligases. To screen the E3 ligase regulating JOSD2, we performed affinity purification mass spectrometry (AP-MS) analysis (Supplementary Data 2), and several E3 ligases, including CHIP, WWP1, RNF169, PPIL2, RNF138, and CBLL1, were identified. To verify whether these E3 ligases directly regulate JOSD2, we assessed their interactions in SW480 cells, reflective of our AP-MS conditions. As shown in Fig. 4k and Supplementary Fig. 4k, although all tested E3 ligases interact with JOSD2, the binding affinity with CHIP was significantly stronger than that with the others. Subsequent in vitro GST pull-down assays further verify that CHIP is a bona fide JOSD2 binding protein (Fig. 4l). Moreover, CHIP positively regulated the K48-linked polyubiquitination levels of JOSD2 (Fig. 4m, n and Supplementary Fig. 4l–n), indicating CHIP binds to JOSD2 and regulates the latter’s ubiquitination levels.
To further demonstrate that CHIP functions as an E3 ligase regulating JOSD2, we next examined the effect of CHIP on JOSD2 protein levels. As displayed in Fig. 4o and Supplementary Fig. 4o, overexpression of CHIP substantially reduced JOSD2 levels in SW480 cells, whereas the effects of WWP1, RNF169, PPIL2, RNF138 and CBLL1 on JOSD2 levels were minimal. Moreover, overexpression of CHIP did not alter JOSD2-3KR levels (Fig. 4p), indicating that CHIP negatively regulated JOSD2 levels through the ubiquitin sites of K48, K142, and K181 on JOSD2. Moreover, CHIP overexpression significantly increased the ubiquitination levels of KRAS, leading to a downregulation of KRAS protein levels. Conversely, the depletion of CHIP resulted in the opposite effect (Supplementary Fig. 4p, q). More importantly, KRAS mutants have been reported to suppress E3 enzymatic activity of CHIP26,27, we next attempted to demonstrate whether KRAS mutants upregulating JOSD2 levels depend on CHIP. As shown in Fig. 4q, KRAS mutants significantly upregulated JOSD2 levels, but failed to pose similar effects in CHIP-depleted cells, indicating that KRAS mutants upregulated JOSD2 relying on CHIP.
Given that JOSD2 functions as a DUB, we further investigate whether JOSD2 exhibits auto-DUB activity. As shown in Supplementary Fig. 5a–c, we found that although JOSD2 does not stably interact with itself, it can remove its own polyubiquitination chains in an enzyme activity-dependent manner, and depletion of endogenous JOSD2 by using shRNA targeting 3′UTR significantly downregulated exogenously over-expressed JOSD2 exclusively in KRAS-mutant cells, these results further underscore that the regulatory effects of JOSD2 on itself depend on the JOSD2/KRAS positive feedback loop.
Collectively, these data demonstrated that KRAS mutants promoted JOSD2 abnormally high expression in CRC. Mechanically, KRAS mutants positively regulated JOSD2 protein levels and stability in a CHIP-dependent manner and then restrained JOSD2 proteasomal degradation at K48, K142, and K181.
Targeting JOSD2 poses more potent anti-tumor effects on KRAS-mutant CRC than that of KRAS wild-type
To further demonstrate the roles of JOSD2 differ in KRAS wild-type or mutant CRC, we next conducted a parallel comparative study in HT-29-KRASWT/Mut models to investigate the effects of JOSD2 knockdown on KRAS wild-type/mutant CRC growth. As shown in Fig. 5a–d, depletion of JOSD2 inhibited the growth of HT-29-KRASMut cells significantly stronger than that of HT-29-KRASWT, as measured by cell proliferation and colony formation assays. Subsequently, we also performed in vivo xenografts and found knocking down JOSD2 posed more potent anti-tumor effects on HT-29-KRASG12V xenografts, with the inhibition ratio as 88.34% (HT-29-KRASG12V xenografts) and 33.60% (HT-29-KRASWT xenografts) (Fig. 5e–g and Supplementary Table 3). Further intratumor protein analysis shown that depletion of JOSD2 significantly downregulated intratumor KRAS levels, and the effects on KRAS mutants is significantly stronger than KRAS wild-type (Fig. 5h and Supplementary Fig. 6a). Given that JOSD2 has been reported to regulate YAP/TAZ, two essential transcriptional co-activator of Hippo pathway, to further demonstrate that the growth-inhibitory effects of JOSD2 inactivation are predominantly due to the enhanced degradation of mutant KRAS, rather than YAP/TAZ, we introduced phosphorylation mutants YAP-5SA/TAZ-4SA that lack key serine residues. These mutants predominantly localize in the nucleus, circumventing ubiquitination and subsequent proteasomal degradation driven by Hippo pathway kinases LATS1/2 and the E3 ligase β-TRCP28,29,30. As shown in Supplementary Fig. 6b–f, overexpression of YAP-5SA/TAZ-4SA, leading to hyperactivation of YAP/TAZ, modestly accelerates tumor growth in both wild-type and mutant KRAS xenografts. However, despite the activated state of YAP/TAZ, depletion of JOSD2 still shows more robust anti-tumor effects in KRAS-mutant xenografts compared to the wild-type counterparts, further demonstrating that the growth-inhibitory effect of JOSD2 knockdown depends, at least partially, on the degradation of mutant KRAS.
a, b JOSD2 depletion inhibits the cell proliferation of KRAS-mutant cells strongly than KRAS wild-type cells in vitro. HT-29KRAS-WT/Mut cells were infected with indicated lentiviruses for 96 h, then these cells were seeded with 2000 cells per well in 96-well/6-well plates, followed by SRB staining. Ctrl, control. a The representative cells proliferation images of the indicated group; Scale bar, 6 mm. b The quantitative analyses of cells proliferation, P = 0.0965, WT-Ctrl vs. WT-shJOSD2; P = 4E-8, G12C-Ctrl vs. G12C-shJOSD2; P = 6E-8, G12D-Ctrl vs. G12D-shJOSD2; P = 1E-6, G12V-Ctrl vs. G12D-shJOSD2. (means ± SD, n = 3; three independent experiments were performed). c, d JOSD2 depletion inhibits the colony formation of KRAS-mutant cells strongly than KRAS wild-type cells in vitro (means ± SD, n = 3; three independent experiments were performed). c The representative colony formation images of the indicated group; Scale bar, 5 mm. d The quantitative analyses of colony formation. e–h JOSD2 depletion posed a more potent anti-tumor effect on KRAS-mutant xenografts compared to KRAS wild-type in vivo (means ± S.E.M., n = 7 mice/group). e The tumor images of the indicated groups. f The tumor volume of the indicated groups. Tumors were measured every two days. g The tumor weight of the indicated groups. h Knocking down JOSD2 downregulated intratumor KRAS levels of KRAS-mutant xenografts are stronger than those of KRAS wild-type xenografts. The proteins isolated from the tumor were subjected to WB analysis. The significance analysis of (b, d, f, g) was conducted by One-way ANOVA (Bonferroni method was utilized to correct for multiple comparisons). The samples derived from the same experiment and processed on different gels in parallel are shown below: h one gel for JOSD2/GAPDH and another for KRAS. Source data are provided as a Source Data file.
Given that KRAS mutants upregulated JOSD2 levels relying on CHIP, whether CHIP overexpression plays a similar role as JOSD2 shRNA? To demonstrate this hypothesis, we further investigated the impact of CHIP on CRC growth. As demonstrated in Supplementary Fig. 7a, b, CHIP overexpression markedly inhibited cell proliferation and colony formation in KRAS-mutant CRC cells. Importantly, the inhibitory effects of CHIP overexpression on KRAS-mutant CRC were stronger compared to KRAS-WT CRC (Supplementary Fig. 7c, d). Similar results were obtained in in vivo xenografts (Supplementary Fig. 7e–g).
To further demonstrate that JOSD2 is a potential clinically viable target, we performed in vivo xenografts to examine the combined effect of JOSD2 depletion and the MEK inhibitor trametinib on the growth of KRAS-mutant CRC xenografts. As shown in Supplementary Fig. 8a, b, silencing of JOSD2 notably suppressed KRAS-mutant GP-2d xenografts, yielding more pronounced anti-tumor effects than trametinib alone. Importantly, the combination of JOSD2 knockdown and trametinib resulted in significantly enhanced growth inhibition, with an inhibition ratio of 96.5% for the combination therapy compared to 64.5% for trametinib and 88.1% for shJOSD2 alone, further supporting the potential of JOSD2 as a viable therapeutic target for KRAS-mutant CRC, particularly in combination with MEK inhibition.
In addition, to investigate whether our findings also exist in other KRAS-mutant tumor types, we further tested this regulatory mechanism of JOSD2/KRAS positive feedback circuit in nonsmall cell lung cancer (NSCLC). As shown in Supplementary Fig. 8c–f, KRAS mutants, not KRAS wild-type, escalated JOSD2 expression in NSCLC cells, and KRAS-mutant NSCLC patients displayed higher JOSD2 levels compared to KRAS wild-type patients. Moreover, depletion of JOSD2 notably inhibited KRAS-mutant NSCLC cell proliferation, which is consistent with the results observed in KRAS-mutant CRC. These data fortify our findings and extend the applicability of our study beyond colorectal cancer to other KRAS-mutant tumors, thus providing a more comprehensive insight into the role of JOSD2 in cancer.
Collectively, these data demonstrated that JOSD2 inhibition or CHIP overexpression poses a more potent anti-tumor effect on KRAS-mutant CRC in vitro and in vivo.
JOSD2 catalytic inhibitor HY041004 preferentially impedes KRAS-mutant CRC growth in vitro/in vivo
A previous study has identified HY041004 as a potent JOSD2 inhibitor23, so we next investigated the anti-tumor activity of HY041004 on KRAS-mutant CRC. We first conducted an in vivo deubiquitination assay and found HY041004 effectively suppressing the ability of JOSD2 to remove KRAS mutants’ polyubiquitin chains (Fig. 6a). Moreover, HY041004 downregulated KRAS mutants’ levels in a concentration-dependent manner (Fig. 6b), indicating that HY041004 promoted KRAS mutants’ ubiquitination degradation by restraining JOSD2 deubiquitinating activity. Subsequently, we assessed the in vitro anti-tumor activity of HY041004 on KRAS-mutant CRC cells. As shown in Fig. 6c, d, HY041004 dramatically inhibited the growth of KRAS-mutant CRC cells in a concentration-dependent manner, as measured by cell proliferation and colony formation assays.
a HY041004 reversed the effects of JOSD2 deubiquitinating KRAS mutants. b HY041004 downregulated KRAS mutants’ protein levels in a concentration-dependent manner. c HY041004 inhibited the cell proliferation of KRAS-mutant CRC cells in vitro. WT, wild-type; Mut, mutation. (means ± SD, n = 3; three independent experiments were performed). d HY041004 inhibited the colony formation of KRAS-mutant CRC cells in a concentration-dependent manner in vitro. SW480/SW620 cells were treated with the indicated concentration of HY041004 for 72 h (SW480: 0, 0.03, 0.06, 0.12 μM; SW620: 0, 0.06, 0.09, 0.12 μM), then replaced with fresh medium and cultured for another 14 days followed by colony formation assays; Scale bar, 5 mm. e The inhibitory effects of HY041004 on the cells proliferation of KRAS-mutant cells are stronger than KRAS wild-type cells in vitro. P, vs. KRAS-WT. (means ± SD, n = 3; three independent experiments were performed). f The inhibitory effects of HY041004 on the colony formation of KRAS-mutant cells are stronger than KRAS wild-type cells in vitro; Scale bar, 5 mm. g–i HY041004 posed more potent anti-tumor effects on KRAS-mutant xenografts compared to KRAS wild-type in vivo (means ± S.E.M., n = 7 mice/group). g The tumor volume of the indicated groups. h The tumor weight of the indicated groups. i HY041004 downregulated intratumor KRAS levels of KRAS-mutant xenografts stronger than those of KRAS wild-type xenografts. j The represent images of CRC PDO. Scale bar, 200 μm. k HY041004 significantly downregulated KRAS levels in PDOs (patient-derived organoids). CRC, colorectal cancer; Scale bar, 100 μm. l HY041004 posed more potent anti-tumor activity on KRAS-mutant CRC PDO in vitro. (means ± SD, n = 3; three independent experiments were performed). The significance analysis of (e, g, h) was conducted by One-way ANOVA (Bonferroni method was utilized to correct for multiple comparisons), and (i, l) was conducted by Two-tailed unpaired Student’s t-tests. Source data are provided as a Source Data file.
Furthermore, we also conducted a series of parallel comparative studies in HT-29-KRASWT/Mut models to investigate the effects of HY041004 on KRAS wild-type/mutant CRC growth. As shown in Fig. 6e, f and Supplementary Fig. 9a, b, HY041004 remarkably suppressed the cell proliferation and colony formation of HT-29-KRASMut in vitro, and the effect of HY041004 on HT-29-KRASMut is significantly stronger than that on HT-29-KRASWT, whereas KRASG12C covalent inhibitors MRTX-849 only selectively inhibited KRASG12C CRC cells (Supplementary Fig. 9b), suggesting the potential of JOSD2 inhibition as the promising pan-KRAS-mutation-targeting strategy. Meanwhile, we also assessed the anti-tumor effect of HY041004 on HT-29-KRASWT/Mut xenografts in parallel. As shown in Fig. 6g, h, Supplementary Fig. 9c, and Supplementary Table 4, HY041004 posed a more potent anti-tumor effect on HT-29-KRASG12V xenografts. Subsequent intratumor protein analysis showed that HY041004 posed more potent effects on downregulating KRAS mutants (Fig. 6i), demonstrating the robust growth-inhibitory effects of HY041004 on KRAS-mutant CRC in vitro and in vivo.
To further compare the anti-tumor activity of HY041004 on KRAS wild-type and mutant CRC in preclinical settings, we investigated the anti-tumor effects of HY041004 on KRAS wild-type and mutant CRC patient-derived organoids (PDOs). As shown Fig. 6j–l, we found HY041004 significantly downregulated KRAS levels in PDOs, more importantly, HY041004 posed more potent anti-tumor effects on KRAS-mutant PDOs compared to that on KRAS wild-type PDOs, with the IC50 values as 1.06 μM and 3.43 μM, respectively, further demonstrating that JOSD2 manipulation prominently degrades KRAS mutants and exhibits robust anti-cancer activities against KRAS-mutant CRC PDO models.
To demonstrate the anti-tumor effects of HY041004 on KRAS-mutant CRC are mainly ascribe to JOSD2 inhibition, we next investigated the specificity of HY041004 toward JOSD2 by examining the inhibitory effects of HY041004 on other DUBs including all the 3 MJDs members (JOSD1, ATXN3 and ATXN3L) and some other DUBs subfamily members, as well as the key kinases of RAS-related pathways including EGFR, AKT1, PI3KCA, BRAF, MEK1, ERK, etc. As shown in Supplementary Fig. 9d, HY041004 displayed pronounced inhibitory effects on JOSD2 and JOSD1, while showing minimal effects on other tested DUBs and the key kinases of RAS-related pathways. Considering that JOSD2 and JOSD1 share a high similarity in amino acid sequence, it is reasonable that the inhibitory activities against JOSD1 of HY041004 are close to those on JOSD2. Moreover, our data have demonstrated that JOSD1 does not interact with or deubiquitinate KRAS and its depletion has minimal effects on KRAS-mutant CRC cell growth; the anti-tumor effects of HY041004 on KRAS-mutant CRC are primarily attributed to JOSD2 inhibition. Moreover, to validate whether the in vivo concentration of HY041004 falls within a range that effectively targets JOSD2 with specificity, we measured the intratumor concentration at different time points after the last HY041004 administration. The peak level reached was 1.38 nmol/g (~1.38 μM) at ~1 h (Supplementary Fig. 9e), correlating closely with the in vitro concentration range exhibiting selective anti-cancer activity against KRAS wild-type/mutant CRC PDO (the IC50 of HY041004 on KRAS-mutant/ wild-type PDO are 1.06 μM and 3.43 μM, respectively). This provides a strong indication that the observed anti-tumor effects of HY041004 in vivo are indeed mainly through JOSD2 inhibition.
Taken together, these data collectively demonstrated depletion of JOSD2 or pharmacological inhibiting JOSD2 significantly suppressed the growth of CRC in vitro and in vivo, especially KRAS-mutant CRC.
JOSD2 is elevated in KRAS-mutant CRC tissues and positively correlated with KRAS expression
To further investigate the clinical significance of JOSD2/KRAS positive feedback circuit in KRAS-mutant CRC tissues, we collected 36 KRAS wild-type CRC tissues and 36 KRAS-mutant CRC tissues from the clinic, then performed IHC staining to evaluate the expression of JOSD2 and KRAS. As shown in Fig. 7a–c, by scoring the expression levels of JOSD2 and KRAS in the tumor specimens, we found a positive correlation between JOSD2 and KRAS expression levels in CRC tissues (r = 0.6147, p < 0.001; n = 72). In CRC samples with high (JOSD2 +++) and medium (JOSD2 ++) JOSD2 expression, the percentage of samples with high (KRAS +++) and medium (KRAS ++) KRAS expression was approximately 90%. Conversely, in CRC patients exhibiting negative or low levels of JOSD2 (JOSD2- and JOSD2 +), this population decreased to around 45% (Fig. 7d). These observations suggest a significant clinical relevance between JOSD2 and KRAS expression. Moreover, JOSD2 expression levels are significantly higher in KRAS-mutant CRC tissues than KRAS wild-type tissues (Fig. 7e and Supplementary Fig. 9f), further consolidating our hypothesis that KRAS mutations are implicated in the abnormal overexpression of JOSD2 in KRAS-mutant CRC.
a, b KRAS-mutant (Mut) and wild-type (WT) colorectal cancer (CRC) tissues were performed IHC (immunohistochemistry) assays and stained with KRAS and JOSD2. Representative images of IHC staining of KRAS and JOSD2 in KRAS-Mut and WT CRC tissues (n = 36 patients/group) were shown; Scale bar, 1.5 mm. c The correlation analysis between JOSD2 and KRAS was performed, and the r value was calculated by the two-tailed Spearman correlation analysis via GraphPad Prism (P < 0.001, r = 0.6147, 95% confidence interval 0.4410~0.7440). IHC score was evaluated by multiplication of positive staining proportions (1 score, <25%; 2 score, 25%–50%; 3 score, 50%-75%; 4 score, 75%–100%) and intensity of protein expressions (1 score, weak staining; 2 score, moderate staining; 3 score, high staining). –, negative expression (1–3 score); +, low expression (4–6 score); ++, medium expression (7–9 score); +++, high positive expression (10–12 score). d JOSD2-high expression CRC displayed higher KRAS levels compared to JOSD2-low expression tissues. e KRAS-mutant CRC tissues displayed higher JOSD2 levels compared to KRAS wild-type CRC tissues. f The schematic diagram of JOSD2/KRAS positive feedback loop promoting the proliferation of KRAS-mutant CRC (The schematic diagram was created in BioRender. https://BioRender.com/w35e177). Source data are provided as a Source Data file.
To further confirm the clinical significance of JOSD2 in CRC, we next analyzed the correlation between JOSD2 levels and the prognosis of CRC patients via the SurvExpress database. As shown in Supplementary Fig. 9g, JOSD2 levels are associated with risk factors in CRC patients. Moreover, we also performed Kaplan–Meier Plotter database analysis and found that JOSD2 level is inversely correlated with the relapse-free survival of CRC patients (Supplementary Fig. 9h)31, highlighting that JOSD2 is closely associated with the poor prognosis of CRC patients.
Collectively, these data not only validated our hypothesis that JOSD2 and KRAS form a positive feedback circuit, synergistically promoting the growth of KRAS-mutant CRC, but also suggested that JOSD2 could be an underlying therapeutic target for CRC patients with a variety of KRAS mutants.
Discussion
Targeting KRAS, the most frequently and lethal mutated genes in CRC and other cancer types, has been one of the central goals in the last decades. Despite the success of the allele-specific KRASG12C inhibitors, a therapeutic strategy that comprehensively targets a broad range of KRAS mutations is urgently needed to address a large patient population32. Cumulative evidence has revealed that the increased protein stability of KRAS mutants is essential for its robust tumor-promoting function, therefore, the exploration of potential regulator(s) dictating the protein stability of KRAS mutants emerges as an attractive approach for the development of pan-KRAS mutations targeting strategies. In the present study, we performed global profiling of DUBs siRNA library on KRAS mutants’ protein stability and identified JOSD2 as a regulator that deubiquitinates and stabilizes three most common KRAS mutants, namely G12C, G12D, G12V, G12A, G12S, G13C, G13D, G13V, G13R, that covering ~70% of KRAS-mutated CRC patients4,32. Reciprocally, KRAS mutations prevented CHIP-mediated degradation of JOSD2, thus forming a JOSD2/KRAS positive feedback circuit to fuel malignant proliferation of KRAS-mutant CRC cells, and rendering these cells vulnerable to JOSD2 inhibition (Fig. 7f). Our study highlights the promise and merits of targeting JOSD2 to treat KRAS-mutated CRC harboring KRAS variant across different mutant types.
JOSD2 belongs to the MJDs family, the minimal subfamily of DUBs, which also includes three other members: JOSD1, Ataxin-3, and Ataxin-3L17. Among the four of them, JOSD1 and JOSD2 are highly similar in amino acid sequences, yet different in their modes of action and protein substrates. JOSD1 is endowed with catalytic functions only after it is monoubiquitinated, whereas JOSD2 could directly deubiquitinate substrates without such modification17. JOSD1 deubiquitinates and stabilizes myeloid cell leukemia 1 (MCL1)33 and JAK2-V617F34 to promote the malignancy in gynecological tumors and acute myeloid leukemia, respectively; while JOSD2 is found to regulate the glucose metabolic enzyme complex18, β-catenin19 in lung cancer and hepatocellular carcinoma. The tumor-promoting effects were also revealed by another study that JOSD2 deubiquitinates and stabilizes YAP/TAZ by cleaving their proteolytic ubiquitination and inhibiting the latter degradation20 in cholangiocarcinoma. However, the roles and the potential substrates of JOSD2/JOSD1 in CRC are still unclear. The current study not only uncovered a crucial DUB directly regulating KRAS mutants, but also demonstrated that it was JOSD2, but not JOSD1, that could physically interact with KRAS variants and cleave the proteolytic polyubiquitination chains to stabilize these tumor-promoting mutants, therefore robustly promoting cell proliferation in KRAS-mutated CRC. The diverse influence of JOSD2 vs. JOSD1 on KRAS mutants is probably due to their differential catalytic modes and the distinct reciprocal regulation imposed by KRAS mutants.
While the pro-tumor functions of JOSD2 have been identified, the precise mechanisms responsible for its elevation or activation in tumor regions remain unknown. By comparing the clinical tumor samples from KRAS-mutant/wild-type CRC patients, in addition to integrating oncogenic mutations (G12C, G12D, G12V, G12A, G12S, G13C, G13D, G13V) into KRAS wild-type cells, our study substantiated the hypothesis that the mutational activation of KRAS robustly augments JOSD2 expression. This may elucidate the distinct anti-cancer effects attained by JOSD2 depletion using either shRNA or a catalytic inhibitor, which exhibited a substantial inhibitory impact on KRAS-driven CRC both in vitro and in vivo models while sparing wild-type KRAS-harboring CRCs that were less influenced. The preferential regulation of oncogenic mutated substrates by DUBs, as previously reported, where USP10 and JOSD1 selectively deubiquitinate FLT-mutants and JAK2-V617F, respectively34,35, is also mirrored in the current study. Therefore, our data not only suggest that JOSD2 is a lynchpin for oncogenic KRAS dependence in CRC, but also implicate that KRAS mutants function as an inducer for JOSD2 overexpression. Our mechanistic work revealed that KRAS-modulated E3 ligase CHIP is essential for JOSD2 protein stability, therefore playing a crucial role in the mechanism underlying this specific regulation of KRAS/JOSD2 toward mutant KRAS. The regulation of CHIP activity by KRAS mutants has been reported previously26,27. In KRAS-mutant cancers, mutant KRAS promotes the activation of the MEK1/2-ERK1/2 cascade and inhibits the MEK5-ERK5 cascade. In contrast, inhibition of KRAS can lead to the activation of the MEK5-ERK5 cascade26. More importantly, ERK5 has been reported to function as a key regulator of CHIP activity, and ERK5 inhibition suppresses the E3 ligase activity of CHIP. Based on this understanding, we propose that KRAS mutants specifically upregulate JOSD2 levels by inhibiting the MEK5-ERK5 cascade, which mediates the promotion of CHIP activity27. Similar regulatory differences mediated by KRAS wild-type and mutants have been reported in previous studies. For example, Li et al. demonstrated that oncogenic KRAS mutants, but not wild-type KRAS, inhibit the proteolytic degradation of Branched-Chain Amino Acid Transaminase 2 (BCAT2) by disrupting its interaction with the E3 ubiquitin ligase Tripartite-motif-containing protein 21 (TRIM21)36. Yet our study provides clear evidence that DUB(s) could be upregulated by KRAS mutants, which further underlies the critical roles of JOSD2 in the cellular response to oncogenic KRAS-induced stress, therefore highlighting the promising application of JOSD2-based therapy as a pan-KRAS-mutation-targeting approach for KRAS-mutated cancers.
To date, several E3 ligases, including LZTR112, Beta-transducing repeat-containing protein (β-TRCP)37 and WDR7615, have been reported to play a role in regulating RAS homeostasis. However, the specific DUBs that directly regulate KRAS mutants remain unidentified. Although previous studies have found that OTU domain-containing ubiquitin aldehyde-binding protein 1 (OTUB1)38 and ubiquitin-specific peptidase 18 (USP18)39 are involved in regulating KRAS, neither of these DUBs directly regulates KRAS. OTUB1 has been reported to enhance membrane localization and promote the activation of wild-type KRAS—but not KRAS mutants—by suppressing the enzymatic activity of E2 enzymes ubiquitin-conjugating enzyme 13 (UBC13) and ubiquitin-conjugating enzyme E2 D3 (UBE2D3), indicating that OTUB1 regulates wild-type KRAS in an indirect manner38. Mustachio et al. discovered a potential role of USP18 in regulating KRAS levels. However, they did not provide direct evidence of USP18 binding to and deubiquitinating KRAS, which are crucial for establishing USP18’s direct involvement in KRAS regulation. Consequently, it is reasonable to speculate that USP18 might indirectly influence KRAS levels through the regulation of other substrates39. To elucidate whether USP18 functions as a DUB directly regulating KRAS, we performed co-immunoprecipitation assays and found USP18 failed to interact with KRAS (Supplementary Fig. 9i). Moreover, in our DUBs siRNA library screening result, siUSPI8 and siOTUB1 only reduce KRAS levels from 1 to 0.622 and 0.953, respectively. These reductions are much weaker than the effect of siJOSD2, which brought KRAS levels down to 0.362, further implicating the indirect regulation of USP18 and OTUB1 toward KRAS. In our study, we demonstrated the interaction between JOSD2 and KRAS by exogenous/endogenous immunoprecipitation assays. Meanwhile, we also performed in vivo/in vitro deubiquitination assays to demonstrate the deubiquitination activity of JOSD2 toward KRAS. These data collectively demonstrated that JOSD2 directly binds to and deubiquitinates KRAS. In addition, given the high homology of KRAS, NRAS, and HRAS, whether JOSD2 is also involved in the deubiquitinating regulation toward NRAS and HRAS needs to be further studied.
In summary, we define a KRAS-driven circuitry that renders the KRAS-mutant CRC cells dependent on JOSD2, a DUB responsible for the deubiquitination and stabilization of a broad range of KRAS mutants. The pro-tumor effects are further reinforced by the reciprocal JOSD2 upregulation by activated KRAS, therefore forming a “JOSD2/KRAS positive feedback circuit” in CRC tumors. Targeting JOSD2 by knockdown or its catalytic inhibitor profoundly impedes the growth of KRAS-mutant CRC in vitro and in vivo. Such interplay between JOSD2 and KRAS not only reveals a unique mutual regulatory mode but also offers potential therapeutic strategies for KRAS-mutant CRC patients, particularly for those with undruggable KRAS oncoproteins.
Methods
Ethical statement
Our research complies with all relevant ethical regulations. Female BALB/c-Nude and NSG mice (4–5 weeks) were purchased from GemPharmatech Co., Ltd (Jiangsu, China) and maintained in a pathogen-free animal facility (temperatures between 21 and 26 °C and humidity levels ranging from 50% to 70%). All animal experiments were performed in accordance with the regulations of the Institutional Animal Care and Use Committee (IACUC). The protocols for the animal study were approved by the Animal Research Committee at Zhejiang University (approval number IACUC-s20-004) and Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University (approval numbers DW202410251219, DW202410221239, and DW202410251240). The humane endpoint for animals set by our ethics committee is when the weight of a single tumor exceeds 10% of the animal’s body weight, or the average diameter of a tumor in adult mice exceeds 20 mm. In our study, the maximal tumor burden did not exceed the humane endpoint. The patient-derived xenografts study obtained informed consent from all participants and was approved by the Ethics Committee of the Hangzhou First People’s Hospital (Ethics Approval License: KY-20220105-0011-01).
Study design
This study was aimed at identifying the key DUBs regulating KRAS mutants’. JOSD2 was identified as the key DUB regulating multiple KRAS mutants by directly interacting with and reverting their proteolytic ubiquitination. Moreover, we verified that KRAS mutants, not wild-type KRAS, reciprocally upregulated JOSD2, thus forming a JOSD2/KRAS positive feedback circuit that significantly accelerated cell proliferation. Mass molecular, cellular, and animal xenografts experiments, as well as clinical CRC samples, including patient-derived cells/xenografts/organoids (PDCs/PDXs/PDOs) were utilized to demonstrate our notion. Numbers of biological replicates in different experiments are included in the legend of each Figure and Supplementary Figure. No statistical methods were used to predetermine sample sizes, but our sample sizes are similar to those generally employed in this field. No sex or gender analysis was carried out. All experiments were randomized and blinded where possible, and no data were excluded. All experiments were repeated at least three times in our study, and the experiment results were presented as the mean ± SD or mean ± SEM.
Cell culture
All cell lines were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China) and Cobioer Biosciences Co., LTD (Nanjing, China). All cells were maintained at 37 °C in a humidified atmosphere containing 5% CO2. HT-29 and SW620 cells were maintained in RPMI 1640 medium (Gibico). SW480, HCT-116, 293T, and 293FT cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM) (Gibico). All culture mediums were supplemented with 10% (v/v) fetal bovine serum (FBS, Gibco). All cell lines were routinely tested for mycoplasma. Authentication of the cell lines was confirmed by short tandem repeat (STR) profiling every year according to the standard of the International Cell Line Authentication Committee (ICLAC).
Antibodies and reagents
The antibodies against p-MEK (#9154, dilution ratio: 1:1000), MEK (#4694, dilution ratio: 1:1000), p-ERK (#4370, dilution ratio: 1:1000), ERK (#4695, 1:1000) and CHIP (#2080, dilution ratio: 1:1000) were purchased from Cell Signaling Technology; anti-GAPDH (#db106, dilution ratio: 1:7500) and anti-HA (#db2603, dilution ratio: 1:5000) antibodies were purchased from Diagbio Biosciences; anti-RAF1 (#A19638, dilution ratio: 1:1000) and anti-KRAS (#A12704, dilution ratio: 1:2000) antibodies was purchased from ABclonal; anti-Myc (#A00172, dilution ratio: 1:5000) and anti-Flag (#A00187, dilution ratio: 1:5000) antibodies was purchased from Genscript; anti-JOSD2 (#SAB2103354, dilution ratio: 1:1000) antibodies was purchased from Sigma-Aldrich; anti-JOSD1 (#OTI3B11, dilution ratio: 1:1000) antibodies was purchased from Novus and anti-KRAS (#Sc30, dilution ratio: 1:1000) antibodies used for immunofluorescence was purchased from Santa Cruz Biotechnology. Proteasome inhibitor MG-132 (HY-13259), protein synthesis inhibitor cycloheximide (CHX, HY-12320), and MAPK inhibitors PD98059 (HY-12028) and SB203580 (HY-10256) were obtained from MedChemExpress.
Gene transfection and lentivirus infection
Cells were transfected with Jet PRIME (Polyplus, Strasbourg, France; #114-15) according to the manufacturer’s recommendation. The gene-specific shRNAs were cloned into the lentiviral vector pLKO.1/U6 (Addgene), and the CDS sequences of the targeting gene were cloned into pCDH-EF1-Puro constructs (System Biosciences). All these lentivirus-relevant plasmids were transfected into 293FT cells for packaging, and the relevant shRNA sequences (synthesized by TSINGKE Biological Technology, Hangzhou, China) were listed as follows:
shRNA-JOSD1#1: TTGCGAGGAAAGAACTGTGAA;
shRNA-JOSD1#2: CCTCGACTCCAAACTCAAGAT;
shRNA-JOSD2#1: CCTCGACTCCAAACTCAAGAT;
shRNA-JOSD2#2: CCAGGTGGACGGTGTCTACTA;
shRNA-KRAS#1: CCTATGGTCCTAGTAGGAAAT;
shRNA-KRAS#2: GAGGGCTTTCTTTGTGTATTT;
shRNA-CHIP#1: CCCAAGTTCTGCTGTTGGACT;
shRNA-CHIP#2: GAAGAGGAAGAAGCGAGACAT.
Real-time PCR
Cells were harvested in TRIzol (Invitrogen) for total RNA extraction, and cDNA was obtained by using TransScript One-Step gDNA Removal and cDNA Synthesis SuperMix Kit (TransGen Biotech) to perform reverse transcription of RNA. SYBR Premix Ex TaqTMα (TaKaRa) was utilized to perform real-time qPCR analysis with QuantStudio 6 Flex and QuantStudioTM Real-time PCR software. The related primer sequences (synthesized by TSINGKE Biological Technology, Hangzhou, China) were listed as follows:
KRAS, forward primer: ACAGAGAGTGGAGGATGCTTT;
KRAS, reverse primer: TTTCACACAGCCAGGAGTCTT;
JOSD2, forward primer: CCCACCGTGTACCACGAAC;
JOSD2, reverse primer: CTCCTGGCTAAAGAGCTGCTG;
GAPDH, forward primer: GGAGCGAGATCCCTCCAAAAT;
GAPDH, reverse primer: GGCTGTTGTCATACTTCTCATGG.
Cell proliferation and colony formation assay
CRC cells infected indicated lentivirus were seeded at 2000 cells/well in 96-well dishes, and cell proliferation was measured by sulforhodamine B (SRB) assay. For colony formation assay, the cells infected indicated lentivirus were seeded with 3000 cells /well in 6-well dishes and cultured for another 14 days, followed by SRB staining, and counted.
DUBs siRNA library screening
Hela cells were seeded 5000 cells/well in 96-well dishes and were transfected with individual pools of DUB siRNAs (each gene containing 4 different siRNA sequences) and the plasmids of KRASG12V-luciferase (constructed by cloning the KRASG12V CDS coding sequences in front of the firefly-luciferase sequence, thus forming a fusion gene of KRASG12V-Luciferase) and renilla-luciferase (utilized as internal control) for 24 h. The fluorescence signal was detected by dual-luciferase reporter assays following the manufacturer’s protocol (Promega, #E1910). The ratio of firefly/Renilla was normalized to the empty vector control.
Immunoblotting analysis and immunoprecipitation assays
For immunoblotting analysis, cells were lysed in RIPA lysis buffer (50 mM Tris-Base, 150 mM NaCl, 5 mM EDTA, 1% NP40, 0.1% SDS, pH 7.4), and protein quantification was measured by BCA Protein Quantification Kit (YEASEN, 20201ES86). Cell lysates were boiled for 30 min at 95 °C and separated using SDS-PAGE. For immunoprecipitation (IP) assay, cells were lysed in RIPA lysis buffer, and an equal amount of cell lysate was incubated with anti-Flag/Myc/HA sepharose overnight at 4 °C. The immunocomplexes were washed 6 times with RIPA lysis buffer and then subjected to immunoblotting analysis. For JOSD2-WT/C24A ubiquitinylation levels in IPs with KRAS-G12V, 293T cells transfected with Myc-KRAS-G12V, HA-ub, and JOSD2-WT-Flag or JOSD2-C24A-Flag were lysed in 1% NP40 lysis buffer, and an equal amount of cell lysate was incubated with anti-Myc magnetic beads overnight at 4 °C. The immunocomplexes of Myc-KRAS-G12V were washed 6 times with 1% NP40 lysis buffer and then lysed in 4% SDS lysis buffer, followed by ultrasonic cracking, to disrupt the interaction between KRAS-G12V and JOSD2; then the lysate (anti-Myc magnetic beads had been removed) was incubated with anti-Flag sepharose overnight at 4 °C. The immunocomplexes of JOSD2-WT/C24A-Flag were washed 6 times with 1% NP40 lysis buffer and then subjected to immunoblotting analysis. The software Quantity One was applied to perform density measurements. When quantitation of ubiquitin is performed, the entire length of the smears from each lane is analyzed and then normalized to the control group.
Immunohistochemistry
Microarray tissues or tissue slides were first de-paraffinized and immersed in PBS. The slides were heated in a microwave oven for 15 min in Citrate Antigen Retrieval buffer. After cooling to room temperature, the slides were incubated with 3% H2O2 to block endogenous peroxidase activity and then incubated with 10% goat serum to block nonspecific staining for 30 min, respectively. Subsequently, the slides were incubated with primary antibodies at 4 °C overnight and then with secondary antibodies at room temperature for 1 h. After washing with PBS for 15 min, the slides were then exposed for 2–3 min to 3,3′-diaminobenzidine tetrahydrochloride (DAB) and rinsed off in deionized water to terminate the DAB reaction. The evaluation of the IHC staining was performed by a pathologist who is blind to the gene information of these patients. IHC score was evaluated by multiplication of positive staining proportions (1 score, <25%; 2 score, 25%–50%; 3 score, 50%–75%; 4 score, 75%–100%) and intensity of protein expressions (1 score, weak staining; 2 score, moderate staining; 3 score, high staining). –, negative expression (1–3 score); +, low expression (4–6 score); ++, medium expression (7–9 score); +++, high positive expression (10–12 score).
Recombinant protein purification
The His-tagged proteins were inserted into the plasmid of pET-28a, and the GST fusion proteins were inserted into the plasmid of pGEX-4T-1. The recombinant proteins were inducibly expressed in Escherichia coli BL21 (DE3) by using 0.4~2 mM isopropyl-β-D-thiogalactopyranoside (IPTG) (16 °C, 18~20 h) once the OD600nm value hit 0.6. Subsequently, the bacteria were collected and lysed in lysis buffer (20 mM Tris-HCl, pH 7.5, 500 mM NaCl, 10% glycerol, and 1 mM DTT) by using the high-pressure homogenizer. The bacterial lysate was centrifuged (4 °C, 15,000 × g, 30 min), and the supernatants were incubated with either GST or Ni-NTA agarose beads for affinity chromatography, following the manufacturer’s instructions. BCA assays were performed to measure the protein concentrations, and Coomassie brilliant blue staining was used to evaluate the protein purity.
GST pull-down assay
GST fusion proteins were bound to GSH Sepharose (Sangon Biotech, C600031) at 4 °C for 2 h. The GSH Sepharose was washed with RIPA lysis buffer three times and incubated with recombinant His-KRAS-WT/G12C/G12D/G12V or indicated cell lysates at 4 °C for another 2 h. After washing with RIPA lysis buffer six times, the bound proteins were subjected to immunoblotting with the indicated antibodies.
In vitro deubiquitination assay
293T cells were transfected with wild-type or mutant HA-tagged ubiquitin and Flag-tagged KRAS for 24 h, and the cell lysates were incubated with anti-Flag sepharose at 4 °C overnight, and the ubiquitinated KRAS was purified. The ubiquitinated KRAS was incubated with bacterial-expressed recombinant human JOSD2/JOSD1 (rhJOSD2/rhJOSD1) protein at 37 °C for 3 h in vitro, followed by immunoblotting analysis.
Protein stability assay
JOSD2-WT/3KR-HA and Myc-KRAS-G12V were cloned into pLenti-TRE-gene-CBH-Tet-On plasmid to construct doxycycline (DOX)-inducible Tet-On system. Cell stably expressed Tet-On-Myc-KRAS-G12V or Tet-On-JOSD2-WT/3KR-HA were infected indicating lentivirus, then these cells were treated with 0.4 μg/ml DOX for 24 h and replaced with fresh medium, cultured for another 48 h, and then the cell lysates at the indicated times were subjected to Western Blot (WB) analysis.
Affinity purification mass spectrometry (AP-MS) analysis
SW480 cells stably overexpressing Vector/JOSD2-SBP (Streptavidin binding peptide) were collected and lysed in the presence of 1% NP40 lysis buffer. An equal amount of cell lysate from the two groups was then incubated with Streptavidin Magnetic Beads (MedChemExpress, HY-K0208) for 6 h at 4 °C, and then the beads were washed with RIPA lysis buffer 3 times and 1×PBS 3 times. The immunoprecipitated JOSD2-SBP complexes were subjected to trypsin digestion and MS analysis. The AP-MS analysis was performed by Jingjie PTM BioLab (Hangzhou). Co. Inc. and the operation procedures were carried out according to the instructions provided by the company. To exclude the nonspecific interacting proteins, we used the blank group that SW480 cells were transfected with Vector, but not JOSD2-SBP, as a negative control and excluded the proteins identified in both two groups, moreover, those proteins identified by only one peptide were also excluded.
The specificity study of HY041004
In vitro ubiquitin-AMC assays were utilized to assess the specificity of HY041004 toward DUBs, 1 μM HY041004 was incubated with purified recombinant DUBs in reaction buffer (50 mM Tris-HCl, 5 mM MgCl2, 2 mM DTT, 2 mM ATP, pH 7.5) at 37 °C for 30 min, then the reactions were initiated by adding 800 nM ubiquitin-AMC (Boston Biochem, #U-550). Fluorescence levels were measured continuously at 25 °C using a Tecan Spark Microplate Reader at an excitation wavelength of 345 nm and an emission wavelength of 445 nm. The specificity of HY041004 toward the key kinases of RAS-related pathways was performed by ICE Bioscience Inc (Beijing), and the operation procedures were carried out according to the instructions provided by the company.
SW480 and SW620 xenografts studies
SW480 and SW620 cells were subcutaneously transplanted in BALB/c-Nude mice at a density of 5 × 106. When tumor volumes reached approximately 600 mm3, the tumor tissues were divided into 1 mm3 tumor mass and then were subcutaneously transplanted in new BALB/c-Nude mice. When tumor volumes reached approximately 60 mm3, the mice were randomly divided into control or shJOSD2 groups. The control group was intratumorally injected with lentivirus-encoded scramble shRNA (Ctrl), while the shJOSD2 group was intratumorally injected with lentivirus-encoded JOSD2 shRNA (shJOSD2) every two days. Tumor volumes and body weight of each mouse were periodically monitored.
GP-2d xenografts studies
GP-2d cells were infected with lentiviruses encoded scramble shRNA (Ctrl) or JOSD2 shRNA (shJOSD2) for 96 h, then these cells were subcutaneously transplanted in NSG mice at a density of 1.8 × 107. When the tumor grown up, the mice were randomly divided into control or trametinib groups. The mice were intragastrically administered with 3 mg/kg trametinib or the same volume of saline once every two days. Tumor volumes and body weight of mice were periodically monitored.
HT-29KRAS-WT/Mut xenografts studies
For the anti-tumor effect of shJOSD2 on HT-29KRAS-WT/G12V xenografts, HT-29KRAS-WT/G12V cells were infected with lentiviruses encoding scramble shRNA (Ctrl) or JOSD2 shRNA (shJOSD2) for 96 h. After screening for puromycin, these cells were subcutaneously transplanted in NSG mice at a density of 5 × 106. For the anti-tumor effect of CHIP overexpression on HT-29KRAS-WT/G12V xenografts, HT-29KRAS-WT/G12V cells were infected with lentiviruses encoding vector and CHIP for 96 h. After screening for puromycin, these cells were subcutaneously transplanted in NSG mice at a density of 2 × 106. For the anti-tumor effect of shJOSD2 on HT-29KRAS-WT/G12D xenografts, HT-29KRAS-WT/G12D cells infected with vector or YAP-5SA/TAZ-4SA were infected with lentiviruses encoded scramble shRNA (Ctrl) or JOSD2 shRNA (shJOSD2) for 96 h. After screening for puromycin, these cells were subcutaneously transplanted in NSG mice at a density of 1 × 106. For the anti-tumor activity of HY041004 on HT-29KRAS-WT/G12V xenografts, HT-29KRAS-WT/G12V cells were subcutaneously transplanted in NSG mice at a density of 5 × 106. When tumor volumes reached approximately 600 mm3, the tumor tissues were divided into 1 mm3 tumor mass and then were subcutaneously transplanted in new NSG mice. Once tumor volumes reached approximately 60 mm3, the mice were randomly divided into control or HY041004 groups. The mice were intragastrically administered with 100 mg/kg HY041004 or the same volume of saline twice a day for a week, and then the doses were increased to 200 mg/kg. Tumor volumes and body weight of mice were periodically monitored.
Detection of intratumor HY041004 concentration
The tumor tissues were collected at different time points (0 min, 5 min, 15 min, 30 min, 1 h, 1.5 h, 2 h, 4 h, 6 h, 8 h, 24 h) after the last HY041004 administration and the intratumor concentrations were measured by Liquid chromatography-tandem mass spectrometry (LC-MS/MS) as bellow:
Instruments and LC-MS/MS settings
HY041004 and its internal standard (IS) were chromatographically separated from tissue homogenate samples using an LC-40 high-performance liquid chromatographic system (Shimadzu, Japan). Analytical detection was carried out with a SCIEX Triple Quad 7500+ mass spectrometer (AB Sciex, Canada), which was equipped with an electrospray ionization (ESI) source. Data acquisition and processing were managed by OS (version 3.1.6.44) from AB Sciex. For the analysis, an ACQUITY BEH C18 column (50 × 2.1 mm, 1.7 μm) (Waters, USA) was employed, maintained at a temperature of 40 °C. The mobile phases for gradient elution consisted of (A) water with 0.1% formic acid and (B) methanol with 0.1% formic acid.
The gradient elution for tumor samples analysis commenced at 40% B for 0.4 min, then increased linearly to 70% B over 2.6 min, followed by a linear increase to 90% B over 0.2 min. The elution was held at 90% B for 0.8 min, then returned to 40% B in 0.1 min, and maintained at this level for 0.9 min. The eluent flow rate was set at 0.4 mL/min, and the injection volume was 1.0 μL.
The analytes were ionized in the ESI (+) mode with an ion spray voltage of 1500 V. The nitrogen settings for the nebulizer (GAS 1), heater (GAS 2), and curtain gas (CUR) were 50 psi, 50 psi, and 40 psi, respectively. The source temperature was adjusted to 500 °C, and the collision gas pressure was set at 9 psi. Multiple reaction monitoring mode was employed to detect HY041004 and IS, using ion transitions from m/z 343.2 to 143.1 and from m/z 271.1 to 155.0, respectively. The optimized collision energies (CE) were 33 V and 15 V, the Entry Potential (EP) was 11 V for both HY041004 and IS, and the Collision Cell Exit Potential (CXP) was 10 V for both.
Preparation of stock solutions, working solutions, calibration standards, and quality control (QC) samples
The stock solutions of HY041004 and the IS were prepared in DMSO at a concentration of 1.00 mg/mL. The working solutions were obtained through serial dilution with a Methanol-Water (50:50, v/v) mixture, followed by a further 20-fold dilution with blank tumor homogenate to yield concentrations of 1.00, 2.00, 5.00, 10.0, 100, 200, 500, 1000, and 5000 ng/mL for calibration standards, and 3.00, 80.0, 800, and 4000 ng/mL for QC samples. The IS working solution was prepared by diluting the IS stock solution with acetonitrile to a concentration of 200 ng/mL.
Sample preparation
Tumor tissues were thawed at room temperature and cut into pieces using surgical scissors. An appropriate amount of each sample was accurately weighed in homogenization tubes, followed by the addition of methanol at a volume twice the weight of the tissue. The mixture was homogenized using a Tiss-48 automatic sample fast grinder (Jingxin Industry, China) at 60 Hz for 2 min and then sonicated for 5 min to obtain tumor homogenate samples. To a 20.0 μL aliquot of tumor homogenate sample in a 96-well plate, 100 μL of acetonitrile (containing 200 ng/mL IS) was added. The plate was sealed, vortexed at 1000 rpm for 15 min, and then centrifuged at 2465 rcf for 15 min. Subsequently, 40 µL of the supernatant was transferred to a 96-well plate and mixed with 100 µL of water containing 0.1% formic acid. Finally, 1.0 μL aliquots of the supernatant were injected into the LC-MS/MS system.
Patient-derived cells (PDCs) studies
The tumor tissues from CRC patients harboring KRAS mutations were washed with PBS six times and were divided into 1 mm3 tumor mass, and the tumor mass was maintained in DMEM/F12 medium with 20% (v/v) fetal bovine serum (FBS, Gibco, Grand Island, NY, USA). The fresh culture medium was exchanged every two days, and cell passage cultivation was performed when cell confluence reached ~80%.
Patient-derived organoids (PDOs) studies
The anti-tumor activity assays of HY041004 on KRAS wild-type and mutant PDOs were entrusted to Accurate International Biotechnology Limited Company (Guangzhou, China). To prepare PDOs for drug response analyses, PDOs were dissociated into small clusters and resuspended in 2% Matrigel/organoid culture medium (200–1000 clusters per milliliter), then dispensed into 384-well plates in triplicate. At 48 h after plating, a 9-point, 2-fold dilution series of HY041004 was dispensed. Drug concentrations ranged from 20 µmol/L to 0.078 µmol/L. After 96 h, cell viability was assayed using a CellTiter-Glo 3D Cell Viability Assay (Promega, Madison, WI) according to the manufacturer’s instructions, and the results were normalized to those of vehicle controls.
Patient-derived xenografts (PDX) studies
The tumor tissues from CRC patients harboring KRAS mutations were divided into 1 mm3 tumor masses and then were subcutaneously transplanted in BALB/c-Nude mice. When tumor volumes reached approximately 60 mm3, the mice were randomly divided into control or shJOSD2 groups. The control group was intratumorally injected with lentivirus-encoded scramble shRNA (Ctrl), while the shJOSD2 group was intratumorally injected with lentivirus-encoded JOSD2 shRNA (shJOSD2) every two days. The tumor volumes and body weight of each mouse were periodically monitored. The calculation was carried out as follows: tumor volume (TV) = (length × width2) / 2, relative tumor volume (RTV) = TVday n/TVday 1, relative tumor growth rate (T/C) = RTVtreatment/RTVcontrol × 100%, inhibition ratio = Tumor weighttreatment/Tumor weightcontrol × 100%.
Statistics and reproducibility
Data were analyzed using GraphPad Prism software Version 8.0.1, and quantification of western blotting was done by Quantity One Version 4.6.2. Two-tailed unpaired Student’s t-tests were used to analyze the significant differences between the two groups, and One-way ANOVA for the differences among multiple groups (Bonferroni method was utilized to correct for multiple comparisons). P < 0.05 was considered statistically significant (P < 1E-13 labeled as P = 0.0; P > 0.9999 labeled as P = 1.0). In this study, all experiments were repeated three times independently with similar results, and all statistical tests were derived from at least three independent experiments/samples. The investigators were blinded to group allocation during data collection and analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The mass spectrometry proteomics data generated in this study have been deposited to the ProteomeXchange Consortium via the Proteomics Identification Database (PRIDE) (https://www.ebi.ac.uk/pride/archive?keyword=PXD054525). The survival analysis of colorectal cancer patients was performed by the Kaplan–Meier Plotter database (https://kmplot.com/analysis/index.php?p=service&cancer=colon)40, the relevance analysis between JOSD2 expression and risk factors in colorectal cancer patients was performed via the SurvExpress database (http://victortrevino.bioinformatics.mx:8080/Biomatec/SurvivaX.jsp)41. Source data are provided with this paper.
References
Siegel, R. L. et al. Colorectal cancer statistics, 2020. CA Cancer J. Clin. 70, 145–164 (2020).
Biller, L. H. & Schrag, D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA 325, 669–685 (2021).
Li, J., Ma, X., Chakravarti, D., Shalapour, S. & DePinho, R. A. Genetic and biological hallmarks of colorectal cancer. Genes Dev. 35, 787–820 (2021).
Cox, A. D., Fesik, S. W., Kimmelman, A. C., Luo, J. & Der, C. J. Drugging the undruggable RAS: mission possible? Nat. Rev. Drug Discov. 13, 828–851 (2014).
Wang, W. H. et al. Post-translational modification of KRAS: potential targets for cancer therapy. Acta Pharm. Sin. 42, 1201–1211 (2021).
Moore, A. R., Rosenberg, S. C., McCormick, F. & Malek, S. RAS-targeted therapies: is the undruggable drugged? Nat. Rev. Drug Discov. 19, 533–552 (2020).
Papke, B. & Der, C. J. Drugging RAS: know the enemy. Science 355, 1158–1163 (2017).
Hong, D. S. et al. KRAS(G12C) inhibition with sotorasib in advanced solid tumors. N. Engl. J. Med. 383, 1207–1217 (2020).
Skoulidis, F. et al. Sotorasib for lung cancers with KRAS p.G12C mutation. N. Engl. J. Med. 384, 2371–2381 (2021).
Awad, M. M. et al. Acquired resistance to KRAS(G12C) inhibition in cancer. N. Engl. J. Med. 384, 2382–2393 (2021).
Xue, J. Y. et al. Rapid non-uniform adaptation to conformation-specific KRAS(G12C) inhibition. Nature 577, 421–425 (2020).
Bigenzahn, J. W. et al. LZTR1 is a regulator of RAS ubiquitination and signaling. Science 362, 1171–1177 (2018).
Abe, T. et al. LZTR1 facilitates polyubiquitination and degradation of RAS-GTPases. Cell Death Differ. 27, 1023–1035 (2020).
Steklov, M. et al. Mutations in LZTR1 drive human disease by dysregulating RAS ubiquitination. Science 362, 1177–1182 (2018).
Jeong, W. J. et al. WDR76 is a RAS binding protein that functions as a tumor suppressor via RAS degradation. Nat. Commun. 10, 295–305 (2019).
Harrigan, J. A., Jacq, X., Martin, N. M. & Jackson, S. P. Deubiquitylating enzymes and drug discovery: emerging opportunities. Nat. Rev. Drug Discov. 17, 57–78 (2018).
Zeng, C. et al. Machado-Joseph deubiquitinases: from cellular functions to potential therapy targets. Front. Pharmacol. 11, 1311–1318 (2020).
Krassikova, L. et al. The deubiquitinase JOSD2 is a positive regulator of glucose metabolism. Cell Death Differ. 28, 1091–1109 (2021).
Huang, Y. et al. Deubiquitinating enzyme JOSD2 promotes hepatocellular carcinoma progression through interacting with and inhibiting CTNNB1 degradation. Cell Biol. Int. 46, 1089–1097 (2022).
Qian, M. et al. Deubiquitinase JOSD2 stabilizes YAP/TAZ to promote cholangiocarcinoma progression. Acta Pharm. Sin. B 11, 4008–4019 (2021).
Sun, Y. et al. Regulation of Hippo/YAP axis in colon cancer progression by the deubiquitinase JOSD1. Cell Death Discov. 10, 365 (2024).
Kang, D. Y. et al. The Inhibitory mechanisms of tumor PD-L1 expression by natural bioactive gallic acid in non-small-cell lung cancer (NSCLC) cells. Cancers 12, 727 (2020).
Yuan, T. et al. Josephin domain containing 2 (JOSD2) promotes lung cancer by inhibiting LKB1 (Liver kinase B1) activity. Signal Transduct. Target. Ther. 9, 11–26 (2024).
Chu, P. C. et al. Mutant KRAS promotes liver metastasis of colorectal cancer, in part, by upregulating the MEK-Sp1-DNMT1-miR-137-YB-1-IGF-IR signaling pathway. Oncogene 37, 3440–3455 (2018).
Yoo, B. H. et al. Oncogenic RAS-induced downregulation of ATG12 is required for survival of malignant intestinal epithelial cells. Autophagy 14, 134–151 (2018).
Vaseva, A. V. et al. KRAS suppression-induced degradation of MYC is antagonized by a MEK5-ERK5 compensatory mechanism. Cancer Cell 34, 807–822 (2018).
Le, N. T. et al. p90RSK targets the ERK5-CHIP ubiquitin E3 ligase activity in diabetic hearts and promotes cardiac apoptosis and dysfunction. Circ. Res. 110, 536–550 (2012).
Shimomura, T. et al. The PDZ-binding motif of Yes-associated protein is required for its co-activation of TEAD-mediated CTGF transcription and oncogenic cell transforming activity. Biochem. Biophys. Res. Commun. 443, 917–923 (2014).
Moroishi, T. et al. The hippo pathway kinases LATS1/2 suppress cancer immunity. Cell 167, 1525–1539.e1517 (2016).
Li, H. et al. YAP/TAZ drives cell proliferation and tumour growth via a polyamine-eIF5A hypusination-LSD1 axis. Nat. Cell Biol. 24, 373–383 (2022).
Kovács, S. A., Fekete, J. T. & Győrffy, B. Predictive biomarkers of immunotherapy response with pharmacological applications in solid tumors. Acta Pharm. Sin. 44, 1879–1889 (2023).
Hofmann, M. H., Gerlach, D., Misale, S., Petronczki, M. & Kraut, N. Expanding the reach of precision oncology by drugging all KRAS mutants. Cancer Discov. 12, 924–937 (2022).
Wu, X. et al. JOSD1 inhibits mitochondrial apoptotic signalling to drive acquired chemoresistance in gynaecological cancer by stabilizing MCL1. Cell Death Differ. 27, 55–70 (2020).
Yang, J. et al. Small molecule inhibition of deubiquitinating enzyme JOSD1 as a novel targeted therapy for leukemias with mutant JAK2. Leukemia 36, 210–220 (2022).
Weisberg, E. L. et al. Inhibition of USP10 induces degradation of oncogenic FLT3. Nat. Chem. Biol. 13, 1207–1215 (2017).
Li, J. T. et al. BCAT2-mediated BCAA catabolism is critical for development of pancreatic ductal adenocarcinoma. Nat. Cell Biol. 22, 167–174 (2020).
Kim, S. E. et al. H-Ras is degraded by Wnt/beta-catenin signaling via beta-TrCP-mediated polyubiquitylation. J. Cell Sci. 122, 842–848 (2009).
Baietti, M. F. et al. OTUB1 triggers lung cancer development by inhibiting RAS monoubiquitination. EMBO Mol. Med. 8, 288–303 (2016).
Mustachio, L. M. et al. Deubiquitinase USP18 loss mislocalizes and destabilizes KRAS in lung cancer. Mol. Cancer Res. 15, 905–914 (2017).
Győrffy, B. Integrated analysis of public datasets for the discovery and validation of survival-associated genes in solid tumors. Innovation 5, 100625 (2024).
Aguirre-Gamboa, R. et al. SurvExpress: an online biomarker validation tool and database for cancer gene expression data using survival analysis. PLoS ONE 8, e74250 (2013).
Acknowledgements
We sincerely appreciate Dr. Wencheng Kong of Hangzhou First People’s Hospital for clinical sample collection, and Jiamin Du, Jiale Shen of the Innovation Institute for Artificial Intelligence in Medicine of Zhejiang University for their technical support. This work was supported by the Natural Science Foundation of China (U21A20420 for B.Y., 82173836 for H.Z., 82304517 for T.Y.), Zhejiang Provincial Natural Science Foundation (LQ23H310009 for T.Y.).
Author information
Authors and Affiliations
Contributions
Conceptualization: T.Y., Q.J.H., H.Z., and B.Y. Methodology: T.Y., Q.J.H., H.Z., and B.Y. Investigation: T.Y., Y.L., R.L.W., M.J.Q., W.H.W., Y.H.L., H.D.Z., J.E.W., F.J.G., C.M.Z., Q.J.H., H.Z., and B.Y. Visualization: T.Y., Q.J.H., H.Z., and B.Y. Funding acquisition: T.Y., H.Z., and B.Y. Project administration: X.Y.D., R.G.H., T.H.Z., Q.J.H., H.Z., and B.Y. Supervision: X.Y.D., R.G.H, and T.H.Z. Writing – original draft: T.Y., Q.J.H., H.Z., and B.Y. Writing – review & editing: T.Y., Q.J.H., H.Z., and B.Y.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Livio Trusolino and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yuan, T., Liu, Y., Wu, R. et al. Josephin Domain Containing 2 (JOSD2) inhibition as Pan-KRAS-mutation-targeting strategy for colorectal cancer. Nat Commun 16, 3623 (2025). https://doi.org/10.1038/s41467-025-58923-y
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-58923-y