Abstract
Biofluorescence, the absorption of high-energy light and its reemission at lower energy wavelengths, is widespread across vertebrate and invertebrate lineages, especially fishes. New observations over the past decade have significantly increased our understanding of the diversity and multifunctionality of fluorescence in fish lineages. In this study, we present a comprehensive account of all known biofluorescent teleosts and estimate the timing and frequency of the evolution of biofluorescence across this diverse group. We show that biofluorescence evolved numerous times in marine teleosts and is estimated to date back ~112 mya in Anguilliformes (true eels). Of the 459 known biofluorescent teleosts reported in this study, the majority are associated with coral reefs. We find that reef-associated species evolve biofluorescence at 10x the rate of non-reef species. Our results suggest that the chromatic and biotic conditions of coral reefs could have provided an ideal environment to facilitate the evolution and diversification of biofluorescence in teleost fishes.
Similar content being viewed by others
Introduction
Biofluorescence results from the absorption of higher-energy light and its reemission at longer, lower-energy wavelengths by living organisms1. It is a widespread phenomenon across the tree of life and occurs in most major clades of vertebrates1,2,3,4,5,6,7,8,9. Biofluorescence is found in both terrestrial and marine organisms, although these environments have vastly different lighting conditions. Terrestrial ecosystems are brightly lit by sunlight, which contains a wide spectrum of wavelengths spanning the visible light spectrum. Alternatively, vast portions of the photic ocean are characterized by a relatively monochromatic, blue-shifted environment10. As sunlight hits and enters oceanic waters, longer wavelengths (yellow, orange, red) are rapidly absorbed, resulting in a limited bandwidth of blue light (470–480 nm) by around 150 m depth (frequently much shallower depending on water clarity)10. Thus, the ability to absorb shorter wavelength ambient blue light at depth and reemit it as longer wavelengths through the use of fluorescent compounds may be advantageous to marine organisms to increase visibility and contrast amidst the more monochromatic blue environment of the ocean1.
Biofluorescence is phylogenetically pervasive, yet it remains unknown how much of this potential visual signal is biologically relevant and serves a functional role, versus an anatomical byproduct (e.g., enamel)4,11. For example, carnivorous pitcher plants (Nepenthaceae and Sarraceniaceae) fluoresce along the lip of the pitcher, which attracts insect prey2,12. Sexual dimorphism in green biofluorescence and ultraviolet (UV) reflectance is thought to aid in the mating rituals of jumping spiders (Salticidae)3. In marine fishes, where fluorescent emissions mainly occur in the green to red portions of the visible spectrum, biofluorescence has been implicated in camouflage, communication, species identification, mating, and prey attraction1,13,14,15. The Pacific spiny lumpsucker (Eumicrotremus orbis) exhibits sexually dichromatic fluorescent emission colors from the body that may enhance mate identification, whereas fluorescence of the pelvic disc in both males and females is thought to be utilized for signaling13. Biofluorescence is also notably prevalent in coral reef ecosystems. Scleractinian corals exhibit red and green fluorescence, which may increase contrast at depth, provide photoprotection for symbionts, and provide visual cues for other reef organisms16,17,18,19. Some reef fishes may utilize biofluorescent corals and marine algae for camouflage. Scorpionfishes (Scorpaenidae) and threadfin breams (Nemipteridae) have been observed residing on or near backgrounds with similar fluorescent emission wavelengths to their bodies1. Other reef fishes may be using biofluorescence for intraspecific signaling, including closely related species of reef lizardfishes (Synodontidae) that appear nearly identical under white light, but exhibit significant variation in fluorescent patterning1. The potential multifunctional roles of biofluorescence may be linked to the increased rates of diversification of coral reef fishes20,21,22.
These potential visual functions of biofluorescence all require that fluorescent emissions lie within the spectral sensitivity of relevant signal-receivers: conspecifics, predators, and/or prey1. Shallow water reef fishes often have relatively good color vision with two or three visual pigments, allowing them to navigate the chromatically complex ecosystem of coral reefs23. Although most reef fishes are sensitive to shorter wavelengths (blue), some species (e.g., Pomacentridae and Labridae) exhibit long-wavelength sensitivity as high as 600 nm (red)24. In addition, members of many families of marine fishes have been reported to possess yellow intraocular lenses that function as long-pass filters and can facilitate the visualization of longer fluorescent wavelengths1,25. Behavioral experiments support fluorescent recognition for sex identification in the fairy wrasse (Cirrhilabrus solorensis)26. Whereas, direct testing of the visual system in the swell shark (Cephaloscyllium ventriosum) and chain catshark (Scyliorhinus rotifer) revealed that fluorescence functions to increase luminosity contrast with the background environment and between skin patches at depth14. However, our knowledge of the visual capabilities of biofluorescent fishes is extremely limited23, as is our understanding of fluorescent molecules. Green fluorescent proteins (GFP), similar to the GFP that was first isolated from the hydrozoan Aequorea victoria27, have only been isolated and characterized in three species of Anguilliformes (true eels)28,29,30. Smaller fluorescent metabolites were found to be responsible for the green fluorescent emissions in elasmobranchs14, whereas no red fluorescent molecules have yet been isolated from fishes despite the prevalence of red fluorescence across Teleostei.
Given the various proposed functions of biofluorescence and the visual capabilities of marine fishes, further research into its evolution and diversification is crucial. Previous investigations of the phylogenetic distribution of biofluorescence in teleosts found that the phenomenon is both phylogenetically widespread and phenotypically variable across the assemblage1,31,32. However, no recent work has accounted for and incorporated information from the huge increase in biofluorescent teleost diversity that has been documented within the past decade.
Herein we investigate the early evolution of biofluorescence in Teleostei, determine when the phenomenon first evolved in this group, reconstruct the evolutionary patterns of fluorescent emission colors in various lineages, and report on the number of times the phenomenon is known to have independently evolved across this huge and remarkably diverse (>30,000 species) vertebrate assemblage. We also investigate whether the rise and global expansion of modern coral reefs contributed to the incredible diversity of biofluorescence we observe across modern teleost lineages.
Results
Species-level survey of biofluorescence
We report 48 previously unreported biofluorescent teleost species based on observations of recently collected specimens (Supplementary Data 2), exhibiting red (11 spp.), green (32 spp.), and both red and green (5 spp.) fluorescent emissions. We found 413 currently known biofluorescent teleost species across peer reviewed literature1,13,15,26,28,31,32,33,34,35,36,37,38,39,40,41,42,43,44. Combined with our new records, this results in a total of 459 biofluorescent teleost species spanning 87 families and 34 orders (Supplementary Data 1). Of these, fluorescent emissions are red only in 261 species, green only for 150 species, and both red and green for 48 species. We also recorded a total of 469 species where fluorescence was not observed (Supplementary Data 1).
Ancestral state reconstructions of biofluorescence
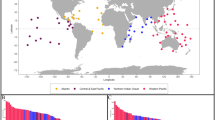
Biofluorescence was present in 267 of the 613 total species in the trimmed Rabosky et al.45 time-calibrated phylogeny, spanning 79 families (Fig. 1). The best-fit Mk models were both the equal-rates and the all-rates-different models, hence they were model-averaged during stochastic character mapping proportional to their Akaike model weights (67.9% and 32.1%, respectively). The root node of the tree (192.8 mya) likely exhibited an absence of fluorescence and had a posterior probability of 33.6% for biofluorescence presence. The oldest node with fluorescence (66.8% posterior probability) was the ancestor of all Anguilliformes (~112 mya) (Fig. 1A). The next oldest nodes where biofluorescence is present are ~104 mya in Syngnathiformes and ~87 mya in Perciformes45, which had posterior probabilities of 79.3% and 82.5% for biofluorescence presence, respectively (Fig. 1C, D). From the root, a mean of 178.9 changes occurred between states. Of these, ~101 were from absence to presence of biofluorescence, and ~78 were from presence to absence. These changes were almost proportional to the mean total time spent in each state: 55% (12,921.24 my) and 45% (10,571.26 my) for presence and absence, respectively.
Major clades where biofluorescence is widespread (A–F) and the oldest reconstructed node where biofluorescence is present (A) are labeled and indicated with an additional outline circle: A) Anguilliformes, B) Labriformes, C) Perciformes, D) Syngnathiformes, E) Pleuronectiformes, and F) Blenniiformes. Photographs: NOAA, Vincent Hyland/Wild Derrynane, John Sparks, Mason Thurman. We have obtained permission to use these images in a commercially published journal.
Ancestral state reconstructions of biofluorescent emission color
Of the 267 biofluorescent teleost species in the Rabosky et al. trimmed phylogeny, fluorescent color was red only in 142 species, green only in 99 species, and both red and green in 26 species (Supplementary Data 3, Fig. 2). The evolution of red and green fluorescence was best described by a corHMM model with two rate classes, both with an all-rates-different model and without dual transitions. Green first evolved in the ancestor of Anguilliformes (Fig. 2A), with node likelihoods of 62% for green fluorescence only, 14.6% for red fluorescence only, and 22.2% for both red and green fluorescence. The most recent common ancestor of Synodus (Aulopiformes: Synodontidae; Fig. 2B) exhibited a majority node likelihood for both red and green fluorescence (54.1%), followed by red only (40.3%) and green only (5.1%). The most recent common ancestor of Pseudocheilinus + Cirrhilabrus (Labriformes: Labridae; Fig. 2C) had node likelihoods of 83.4% for red fluorescence only and 15.4% for no fluorescence. However, the most recent common ancestor of Cheilinus (Labriformes: Labridae; Fig. 2D) had a majority node likelihood of 92.0% for only green fluorescence only. The node likelihoods were 94.3% for red fluorescence in Antennariidae (Lophiiformes; Fig. 2E) and 91.2% for green fluorescence in Nemipteridae (Spariformes; Fig. 2F).
Major clades where a certain biofluorescent color is widespread (A–O) are shown and indicated with an additional outline circle: A) Anguilliformes, B) Synodus (Aulopiformes: Synodontidae), C) Pseudocheilinus + Cirrhilabrus (Labriformes: Labridae), D) Cheilinus (Labriformes: Labridae), E) Antennariidae (Lophiiformes), F) Nemipteridae (Spariformes), G) Perciformes, H) Cyclopteridae + Liparidae (Perciformes), I) Rhamphocottidae + Agonidae + Psychrolutidae (Perciformes), J) Syngnathiformes, K) Hippocampus (Syngnathiformes), L) Pleurosicya + Gobiodon + Bryaninops + Paragobiodon + Eviota (Gobiiformes), M) Gobiodon (Gobiiformes), N) Pleuronectiformes, and O) Blenniiformes.
In Perciformes45, the ancestor of the entire order (Fig. 2G) exhibited node likelihoods of 68.9% for red fluorescence only and 28.9% for no fluorescence. However, two clades of Perciformes had different majority character states. Node likelihoods for Cyclopteridae + Liparidae (Perciformes; Fig. 2H) were 56.8% for both red and green fluorescence, 39.5% for red fluorescence only, and 3.4% for green fluorescence only, while sculpins (Rhamphocottidae + Agonidae + Psychrolutidae; Fig. 2I) had a 90.9% likelihood for green fluorescence only and 5.2% for no fluorescence. The ancestor of Syngnathiformes (Fig. 2J) had node likelihoods of 57.2% for red fluorescence only, 39.3% for no fluorescence, and 3.4% for green fluorescence only. However, the likelihoods for the most recent common ancestor of Hippocampus (Syngnathiformes; Fig. 2K) was 54.8% for both red and green fluorescence and 45.1% for red fluorescence only. Two clades of Gobiiformes had majority likelihoods for the evolution of red fluorescence only. The node likelihoods were 95.3% for red fluorescence only and 4.6% for no fluorescence in one clade of gobiid genera (Pleurosicya + Gobiodon + Bryaninops + Paragobiodon + Eviota; Fig. 2L), and 68.6% for red fluorescence only and 31.1% for both red and green fluorescence in another clade of Gobiidae (Gobiodon, Fig. 2M). The ancestor of Pleuronectiformes (Fig. 2N) had node likelihoods of 65.5% for red fluorescence only, 22.9% for both red and green fluorescence, 8.7% for no fluorescence, and 2.8% for green fluorescence only.
Diversification analysis of biofluorescence and reef association
The best fitting SSE models were character-independent models, indicating that factors besides fluorescence and reef-association influence lineage diversification. The best fitting models were the MuCID model with seven and eight hidden states. Average net diversification rates, informed by the ancestral state reconstructions, were similar (r = 0.029–0.031) with overlapping standard deviations across all character state combinations (Fig. 3B). However, based on transition rates, the rate of change between character states, teleost fishes on reefs evolved fluorescence at a rate 10x faster than species off reefs (Fig. 3A, bolded arrows; Fig. 4; Supplementary Fig. 3). However, reef fishes also lost fluorescence at a rate 1.38× faster than non-reef fishes. Additionally, fishes with fluorescence transitioned to living off reefs at a rate 2.6× faster than they transitioned on reefs (Fig. 3A).
A Q-matrix of transition rates (rate of change between character states), with dashed arrows showing that teleost fishes on reefs evolved fluorescence at a rate 10x faster than species off reefs. B Net diversification including standard deviation error bars for each state: non-reef and nonfluorescent (n = 63), reef-associated and non-fluorescent (n = 283), non-reef and fluorescent (n = 71), and reef-associated and fluorescent (n = 196).
Fluorescence evolves at a faster rate in reef-associated teleosts compared to non-reef species, which is correlated with the rise of scleractinian coral species diversity in the early Eocene (~50 mya). The dashed line marks the K-Pg boundary21. The number of scleractinian coral species was based on data from the fossil record53. The number of reef-associated and non-reef biofluorescent lineages was based on the number of nodes over time, as determined by node ages of the teleost phylogeny and majority node likelihood of each character state.
Discussion
We examined the evolutionary patterns of biofluorescence across Teleostei, a massive clade of fishes accounting for ~50% of all extant vertebrate species45. We show that biofluorescence is phylogenetically widespread across teleosts, occurring in at least 34 orders, 87 families, and nearly 460 species45. This significantly expands the known diversity of biofluorescent teleosts, as previous studies have reported only 180–272 fluorescent teleost species, 49–50 families, and 12–16 orders1,31. However, to date, we have only sampled a small percentage of the total diversity of teleosts, and these numbers almost certainly severely underestimate the true phylogenetic diversity of biofluorescent taxa. We find that biofluorescence is not restricted to any specific portion of the topology but is phylogenetically widespread across Teleostei. However, fluorescence is highly prevalent and exceptionally phenotypically variable in emission color in certain diverse assemblages (Figs. 1 and 2). These groups contain representatives across broad oceanic depth profiles and habitats, from intertidal zones (e.g., Perciformes: Liparis florae; Fig. 1C, Supplementary Data 1) to shallow water reefs (e.g., some Anguilliform families; Fig. 1A) to the deep sea (Aulopiformes: Chlorophthalmidae; Fig. 2B).
Despite this broad range of environments and habitats, we find biofluorescence to be most prevalent in species residing on or near coral reefs, such as Gobiiformes, Blenniiformes, and Syngnathiformes (Fig. 1D, F, O). Of the 267 biofluorescent species within the tree, 73% are reef associated46 (Fig. 4; Supplementary Data 3). These reef-associated fluorescent species account for 196 of the total 479 reef-associated species. Reefs may provide an ideal environment for biofluorescent emissions, as many coral species live in clear shallow water and require greater exposure to sunlight16. Thus, many reef fishes inhabit areas with high intensity light that can better facilitate the excitation and reemission of longer wavelength biofluorescent light. Biofluorescence is most conspicuous in bright, monochromatic blue water (~5 to 100 m), although it is likely overwhelmed by full-spectrum sunlight in the upper few meters of the water column, limiting its contribution as a visual signal23,47. Reef-building corals (Scleractinia) also exhibit fluorescence, which can increase contrast at depth16. Fluorescence in cryptic reef fishes may better camouflage them against fluorescent coral or algal backgrounds, as hypothesized in some scorpaenids (Scorpaenopsis) and nemipterids (Scolopsis)1. Further, complex structures like reef ledges and coral heads cast shadows, altering the light regime under and around these structures48. When fish occupy these shadowed spaces, their fluorescence may be accentuated due to the reduction of ambient light “noise”14. Biofluorescence has also been hypothesized to aid in mate identification through variation in fluorescent patterning (e.g., intraspecific identification in reef lizardfish, Synodus)25,37,45. Further, fluorescent pigments are prevalent in the fins of reef fishes, such as Eviota (Gobiidae), Enneapterygius (Tripterygiidae), and Corythoichthys (Syngnathidae), structures that these groups use for intraspecific communication32. Thus, biofluorescence may be advantageous for reef fishes, given its numerous potential functions in camouflage, mate identification, and intraspecific communication1,13,26,32,34. We note, however, that intraspecific communication, including mate recognition, could also be facilitated by fluorescence in non-reef habitats with lower visibility, such as turbid water.
Due to its high prevalence on shallow reefs, fluorescence in teleosts could be linked to the diversification of coral reef-associated lineages. Reef-associated teleosts were previously found to exhibit higher diversification rates compared to non-reef associated groups due to many factors, including numerous microhabitats in reefs, increased niche partitioning, and protection from past extinction events20. Cowman and Bellwood20 investigated this trend in Labridae, Chaetodontidae, Pomacentridae, and Apogonidae, all of which have biofluorescent representatives (Figs. 1 and 2; Supplementary Data 1). Through a diversification analysis, we find that reef-associated teleosts evolve fluorescence at a much faster rate (10x transition rate) than non-reef species (Figs. 3A and 4). This indicates that a lineage already occupying a reef environment is much more likely to evolve fluorescence than a lineage that is not reef-associated. However, while fluorescence in teleosts is more likely to evolve on reefs, it is not restricted to reef environments, as fluorescent fish are 2.6x more likely to transition to living off reefs (Fig. 3A). We also find that reef-associated lineages have similar transition rates for the loss (q = 0.008) and gain (q = 0.01) of fluorescence (Fig. 3A). This could imply that fluorescence is a labile trait on reefs. Further, we find a distinct increase in the number of fluorescent teleost lineages following the K-Pg boundary (Fig. 4, dashed line). This trend coincides with the rise of modern coral dominated reefs and the rapid colonization of reefs by fishes, which occurred primarily during the early Cenozoic following a significant loss of coral diversity in the K-Pg extinction (Fig. 4)49,50,51,52,53,54,55. These correlations may suggest that the emergence of modern coral reefs could have facilitated the diversification of fluorescence in reef-associated teleosts20 (Fig. 4). However, due to the character-independent models having the best fit, our results imply that other factors besides reef-association or biofluorescence is likely driving global diversification across the entire tree. Thus, biofluorescence in reef fishes could be a byproduct of their increased diversification rate in these habitats, as coral reefs could have provided the chromatic and biotic conditions to facilitate the evolution of ecologically relevant fluorescence in fishes.
Although some biofluorescent teleost lineages evolved in the early Cenozoic, biofluorescence first evolved much earlier and independently in numerous groups. We find that fluorescence first evolved in Anguilliformes, the true eels, approximately 112 mya (Fig. 1A and Table 1). Subsequently, fluorescence independently evolved somewhat later in Syngnathiformes (~104 mya; Fig. 1D) and Perciformes45 (~87 mya; Fig. 1C). Our analyses imply that biofluorescence was independently gained ~101 times and lost ~78 times (Fig. 1). However, numerous species and even entire families have yet to be investigated for biofluorescence, so these numbers are most certainly underestimates. The numerous independent evolutions of biofluorescence across Teleostei may point towards similar selective pressures or its potential multifunctionality in different clades, such as camouflage, communication, and species identification1,13,14,15. However, we find that multiple transitions between absence and presence of biofluorescence even occur within teleost orders (e.g., Labriformes; Fig. 1B), which could indicate that fluorescence is more evolutionarily labile in some clades14,28,29,30.
Our results suggest that biofluorescence is not only phylogenetically widespread, but phenotypically diverse in emission color. Both red and green emissions are present within 48 species spanning 20 families (Supplementary Data 1), specifically within Aulopiformes (Fig. 2B), Cyclopteridae and Liparidae (Perciformes; Fig. 2H), and some Syngnathiformes (Hippocampus; Fig. 2K). However, it is important to note that a node with both red and green fluorescence could mean either individuals within a species have different emission colors (e.g., sexual dimorphism in Labridae26), or a species exhibits simultaneous dichromatic biofluorescence, where an individual has both red and green fluorescent emissions. Simultaneous dichromatic biofluorescence is a rarer phenomenon than singular color emission, and had previously been observed in only sixteen families1,13,35. However, through our observations, we also find simultaneous dichromatic biofluorescence in members of Antennariidae (Antennarius) and Blenniidae (Ecsenius; Fig. 4).
There are also many groups exhibiting only a single fluorescent emission color. We find that green fluorescence evolved first in Anguilliformes (Fig. 2A) and is present in three larger clades: a clade of sculpins (Rhamphocottidae + Agonidae + Psychrolutidae; Fig. 2I), Nemipteridae (Spariformes; Fig. 2F), and Cheilinus (Labriformes: Labridae; Fig. 2D). While red fluorescence evolved later, initially in Syngnathiformes (Fig. 2J), it is far more widespread than green fluorescence across the teleost phylogeny (Fig. 2C, G, J, L, N, O), notably evolving in the ancestor of all Perciformes45 (Fig. 2G). This is opposite the trend reported by Sparks et al.1, who found green biofluorescence to be more common. This was likely a result of reduced sampling effort for red fluorescence, which often requires additional long-pass filters to visualize, blocking out brighter green emissions in lineages where the entire body fluoresces in both green and red (e.g., Synodontidae, reef lizardfishes; Chlopsidae, false moray eels)1. Interestingly, in Labriformes red and green fluorescence evolved independently in two sister groups of Labridae: the clade comprising Pseudocheilinus + Cirrhilabrus (red; Fig. 2C), and Cheilinus (green; Fig. 2D). Whereas green wavelengths are within the spectral range of more reef-associated teleosts than red23, many coral reef fishes possess opsins with red-shifted long wavelength sensitivity (e.g., Pomacentridae)24. Red light also attenuates rapidly in seawater and may be beneficial for shorter distance communication than green fluorescence. However, our current understanding regarding the spectral sensitivities in fishes is limited and more taxonomically comprehensive studies are warranted14,47,56,57,58,59. There is also evidence that red emission wavelengths may vary both between and within lineages of reef fishes (e.g., intraspecific variation within a species), or even within a particular individual31. This variation in red emissions may increase the possibility of spectral tuning, especially when fluorescence is associated with conspecific signaling23,32,47.
Biofluorescence is a phylogenetically widespread and phenotypically diverse phenomenon that is found at deep ancestral nodes in teleost fishes. In this study, we recover ~101 independent evolutions of biofluorescence across Teleostei, suggesting that it may be an evolutionarily important phenomenon in this massive assemblage that comprises over 50% of all living vertebrate species and ~95% of all extant fishes. We estimate that fluorescence first evolved 112 mya in Anguilliformes. We also find that while green biofluorescence evolved first (Anguilliformes), red biofluorescence is much more prevalent, particularly in crown Perciformes. We show that reef-associated species evolve fluorescence at 10x the rate of non-reef species and, therefore, that the rise of modern coral reefs in the early Eocene could have provided the abiotic and biotic conditions necessary to facilitate the evolution and diversification of biofluorescence in marine teleosts.
Methods
Species-level survey of biofluorescence
Research, collecting, and export permits were obtained from the government of the Bahamas, from the Ministry of Fisheries and Ministry of Environment, Honiara, Solomon Islands, and from the Department of Environment, Cayman Islands Government. Additional permits include Florida Atlantic University’s Institutional Animal Care and Use Committee (IACUC, protocols A16‐34 and A19-36), Florida Fish and Wildlife Conservation Commission Special Activity Licenses (SAL-18-1889-SRP and SAL-18-1785A-SRP), and Florida Department of Environmental Protection permit 09101815. All data supporting the findings of this study are available within the paper and its supplementary files.
We compiled a comprehensive list of all known fluorescent teleosts via a literature search from peer-reviewed descriptions of biofluorescence (Supplementary Data 1) in addition to our own observations of recently collected material (Supplementary Data 2). Live and frozen specimens used for examination and imaging (n = 107) were collected in the Solomons Islands in 2012, 2013, and 2019, Greenland in 2019, the Bahamas (Exumas) in 2011 and 2012, the Cayman Islands in 2016, Southern California, USA (San Diego) in 2015 and 2016, near-shore locations in Washington, USA in 2022, Florida, USA in 2022, Thailand (Gulf of Thailand and Andaman Sea) in 2024, and obtained through the aquarium trade (Supplementary Data 2). Additional fluorescence imaging was conducted at the Mystic Aquarium, Mystic, CT, USA in 2011 and 2012, and at the Birch Aquarium, Scripps Institution of Oceanography, La Jolla, CA, USA in 2015 and 2016.
For each fluorescent species, we recorded the color of fluorescent emission: either red (590-750 nm), green (515–570 nm), or both. For specimens that we examined directly, fluorescent emission colors were first judged by eye using a variety of blue excitation lights and scientific-grade long-pass filters. Specimens collected in Washington were examined for fluorescence using a NIGHTSEA Stereo Microscope Fluorescence illuminator with a Royal Blue (RB) head (440–460 nm; Supplementary Fig. 1) and a scientific grade long-pass filter (500 nm). Specimens in Florida were examined for fluorescence using a SeaLife Sea Dragon Mini Fluoro light (440–457 nm; Moorestown, NJ, USA; Supplementary Fig. 1) and a Semrock scientific grade 514 nm long-pass filter. For all other specimens, emission spectra were elicited via illumination with Royal Blue LED lights collimated to ensure perpendicular incidence on the scientific grade 450–470 nm interference filter surface (Omega Optical, Inc., Brattleboro, VT, USA) and fluorescence was confirmed via use of either 514 nm or 561 nm long-pass filters (Semrock, Rochester, NY). An Ocean Optics USB2000+ portable spectrophotometer (Dunedin, FL, USA) equipped with a hand-held fiber optic probe (Ocean Optics ZFQ-12135) was then used to record fluorescence emission spectra. Emission spectra were elicited via illumination with Royal Blue LED lights collimated to ensure perpendicular incidence on the scientific grade 450-470 nm interference filter surface (Omega Optical, Inc., Brattleboro, VT, USA; Semrock, Inc., Rochester, NY, USA). Emission spectra were recorded by placing the fiber optic probe near specific anatomical parts of the individual fish specimen exhibiting fluorescence. This was repeated several times for each specimen and each anatomical region to ensure the accuracy of measurements. Peak emissions (lambda-max) were used to confirm fluorescent emission colors (Supplementary Data 2).
Fluorescent macro photography
For specimen imaging, we placed individual fish in a narrow photographic tank and gently held them flat against the thin glass front. We imaged fluorescent emissions in a dark room using a Nikon D800 or D4 DSLR camera outfitted with a Nikon 60 or 105 mm macro lens, or a Sony A7SII camera outfitted with a Sony 90 mm macro lens. We covered the flashes (Nikon SB910) with blue interference bandpass excitation filters (Omega Optical, Inc.) and attached a long-pass (LP) emission filter (Semrock, Inc.) to the front of the camera lens to record any emitted fluorescence. To best capture the fluorescent emissions we tested multiple LP filter pairs. For example, a 514 nm LP filter was used to capture green fluorescence, whereas a 561 nm LP filter was used to image longer-wavelength fluorescence (orange and red) and to block any emitted green fluorescence in species with both fluorescent colors (i.e., overlapping fluorescent pigments).
Phylogenetic comparative methods
To assess the number of times fluorescence has evolved across Teleostei, we used the time-calibrated Rabosky et al.45 phylogeny. This tree includes 32 of the 34 orders of known biofluorescent teleosts45 (Supplementary Data 1). To avoid gaps in fluorescent data, we trimmed this dated phylogeny to only include species that have been investigated for biofluorescence (Supplementary Data 1). The resulting trimmed tree contained 613 species, including 85% of the 87 known biofluorescent teleost families and 58% of the 461 currently known biofluorescent species (Supplementary Data 1, 3; Supplementary Fig. 2).
Ancestral state reconstructions of biofluorescence
The evolution of fluorescence (presence/absence) was first examined in R60 by fitting four extended Mk models of evolution: equal-rates, all-rates-different, and two irreversible trait models (presence to absence only; absence to presence only) using the fitMk function in phytools 2.061 (Supplementary Code 1). Ancestral state reconstructions were then performed with stochastic character mapping (nsim = 1000) under the best-fit Mk model (determined by weighted AIC of the maximum likelihood estimates) using the simmap function61. The reconstructions were summarized to determine transition rates and total time spent in each state using phytools 2.061. We considered an ancestral node to likely be fluorescent if the posterior probability for presence was >60%.
Ancestral state reconstructions of biofluorescent emission color
We also reconstructed the evolutionary history of red and green fluorescence across teleosts. We used the corHMM R package version 2.162 to incorporate hidden rate classes by leveraging hidden Markov models63 (Supplementary Code 1). These models allow for variation in transition rates throughout the tree and are commonly used for analyses of large clades spanning long evolutionary timeframes63. We fit equal-rate, symmetric, and all-rates-different transition rate models with and without a hidden rate category. We also fit hybrid hidden rates models where one rate class was an equal-rates model and the other was an all-rates different model. We fit all models with and without dual transitions for a total of 14 models. Weighted AICc values were used to determine the best-fitting model and calculate the model-averaged ancestral state likelihood values. The maximum likelihood estimates of some transition rates sat on a likelihood ridge and led to the estimation of unrealistically high transition rates. However, although high transition rates negatively influence stochastic character mapping, they have little effect on marginal ancestral state reconstruction64; as a result, the marginal ancestral state reconstructions were used.
Diversification analysis of biofluorescence and reef association
To investigate whether biofluorescence on coral reefs promotes increased diversification rates in marine teleosts, we implemented several Multicharacter Hidden State Speciation and Extinction (MuHiSSE) models using the hisse R package version 2.1.1165 (Supplementary Code 1). All species with biofluorescence presence/absence data were also coded as being reef associated or not using the classifications from Larouche et al.66, resulting in a total of 479 reef-associated species and 134 non-reef species (Supplementary Data 3). We fit a total of 10 models to the dataset. First, we ran a null MuSSE model with the same turnover parameter for all state combinations. Then we ran a “true” MuSSE model, where the turnover rate parameters differed between each fluorescence-habitat state combination. We also ran one character-dependent (MuHiSSE) and seven character-independent (MuCID) models that incorporated hidden states. Hidden states are used to account for unknown factors besides fluorescence and reef-association that may affect transition rates and lineage diversification. The MuHiSSE model included one hidden state, turnover rates were allowed to vary across the different states, and transition rate parameters were allowed to vary across hidden states. We ran MuCID models for two through eight hidden states, where the turnover rates were unlinked from the observed states and the transition rate parameters were constrained to be identical across hidden states. Note, for all models we used a single extinction fraction parameter, which was kept constant across all observed and hidden states. To account for incomplete phylogenetic sampling, we estimated sampling fractions for each fluorescence-habitat state (non-fluorescent/non-reef, fluorescent/non-reef, non-fluorescent/reef-associated, fluorescent/reef-associated). We first used the proportion of fluorescent fishes in the dataset reported in this study (Supplementary Data 1) to estimate the number of fluorescent and nonfluorescent species in the Larouche et al.66 dataset, which assigns a character state of reef-associated or non-reef for 3339 marine species. We then estimated sampling fractions based on the proportion of species sampled in the trimmed Rabosky et al.45 phylogeny with each fluorescence-habitat state out of the full Larouche et al.66 dataset. We also constrained the root state to be non-reef associated based on the findings of previous studies that marine teleostean fishes originated in non-reef habitats21,66.
We identified the best-fitting models using weighted AIC values, which estimated the transition rates between character states. We then used the transition rates to conduct marginal ancestral state reconstructions with the hisse R package. The results of the ancestral state reconstructions were summarized by calculating the tip-associated net diversification rates by model-averaging parameters from all models that contributed >5% of the AICc weight. Transition rates measure how frequently a trait is gained or lost (i.e., the rate of change between character states). A higher transition rate towards the loss of a character state, for example, indicates that a lineage is more likely to lose that character over evolutionary time. Diversification rates indicate the rate that new species evolve. If diversification rates are higher in lineages with a certain character state, that character state is likely contributing to the proliferation of species over time. The net diversification rates were visualized using the gghisse package version 0.1.1 (https://github.com/discindo/gghisse). We plotted the number of biofluorescent lineages through time in R60 by assigning character states (reef-associated vs non-reef) based on majority node probabilities.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all data supporting the findings of this study are available within the paper and its supplementary information files. The full Rabosky et al.45 phylogeny can be found at https://fishtreeoflife.org/.
Code availability
The authors declare that all code is available in the supplementary information files. All figures can be generated with Supplementary Code 1.
References
Sparks, J. S. et al. The covert world of fish biofluorescence: a phylogenetically widespread and phenotypically variable phenomenon. PLoS ONE 9, e83259 (2014).
Kurup, R. et al. Fluorescent prey traps in carnivorous plants. Plant Biol. 15, 611–615 (2013).
Lim, M. L. M., Land, M. F. & Li, D. Sex-specific UV and fluorescence signals in jumping spiders. Science 315, 481–481 (2007).
Marshall, J. & Johnsen, S. Fluorescence as a means of colour signal enhancement. Philos. Trans. R. Soc. B Biol. Sci. 372, 20160335 (2017).
Mazel, C. H. & Fuchs, E. Contribution of fluorescence to the spectral signature and perceived color of corals. Limnol. Oceanogr. 48, 390–401 (2003).
Welch, V. L., Hooijdonk, E. V., Intrater, N. & Vigneron, J.-P. Fluorescence in insects. In The Nature of Light: Light in Nature IV, vol. 8480 25–39 (SPIE, 2012).
Cox, J. L. & Fitzpatrick, B. M. Biofluorescent sexual dimorphism revealed in a southern Appalachian endemic salamander, Plethodon metcalfi. Sci. Rep. 13, 3588 (2023).
Travouillon, K. J., Cooper, C., Bouzin, J. T., Umbrello, L. S. & Lewis, S. W. All-a-glow: spectral characteristics confirm widespread fluorescence for mammals. R. Soc. Open Sci. 10, 230325 (2023).
Martin, R. P., Carr, E. M. & Sparks, J. S. Does biofluorescence enhance visual signals in birds-of-paradise? R. Soc. Open Sci. 12, 241905 (2025).
Jerlov, N. G. Optical Oceanography (Elsevier Publishing Company, Amsterdam, 1968).
Jeng, M.-L. Biofluorescence in terrestrial animals, with emphasis on fireflies: a review and field observation. In Bioluminescence - Analytical Applications and Basic Biology (ed Suzuki, H.) (IntechOpen, 2019).
Carney, C. L. et al. Fatal attraction: what is the role of visual cues in attracting prey to carnivorous plants? BIOS 90, 79–86 (2019).
Cohen, K. E. & Summers, A. P. Dimorphic fluorescence in the pacific spiny lumpsucker. Ichthyol. Herpetol. 110, 350–353 (2022).
Gruber, D. F. et al. Biofluorescence in Catsharks (Scyliorhinidae): Fundamental description and relevance for elasmobranch visual ecology. Sci. Rep. 6, 24751 (2016).
Ludt, W. B. & Clardy, T. R. First detection of biofluorescence in a deep-sea anglerfish. J. Fish. Biol. 100, 843–846 (2022).
Gruber, D. F., Kao, H.-T., Janoschka, S., Tsai, J. & Pieribone, V. A. Patterns of fluorescent protein expression in scleractinian corals. Biol. Bull. 215, 143–154 (2008).
Bollati, E. et al. Green fluorescent protein-like pigments optimise the internal light environment in symbiotic reef-building corals. Elife 11, e73521 (2022).
Ben-Zvi, O., Wangpraseurt, D., Bronstein, O., Eyal, G. & Loya, Y. Photosynthesis and bio-optical properties of fluorescent mesophotic corals. Front. Mar. Sci. 8, 651601 (2021).
Palmer, C. V., Modi, C. K. & Mydlarz, L. D. Coral fluorescent proteins as antioxidants. PLOS ONE 4, e7298 (2009).
Cowman, P. F. & Bellwood, D. R. Coral reefs as drivers of cladogenesis: expanding coral reefs, cryptic extinction events, and the development of biodiversity hotspots. J. Evol. Biol. 24, 2543–2562 (2011).
Bellwood, D. R., Goatley, C. H., Cowman, P. F., Bellwood, O. & Mora, C. The evolution of fishes on coral reefs: fossils, phylogenies and functions. Ecol. Fishes Coral Reefs 6, 55–63 (2015).
Bellwood, D. R., Goatley, C. H. R., Brandl, S. J. & Bellwood, O. Fifty million years of herbivory on coral reefs: fossils, fish and functional innovations. Proc. R. Soc. B Biol. Sci. 281, 20133046 (2014).
Marshall, N. J., Cortesi, F., de Busserolles, F., Siebeck, U. E. & Cheney, K. L. Colours and colour vision in reef fishes: Past, present and future research directions. J. Fish. Biol. 95, 5–38 (2019).
Stieb, S. M. et al. Why UV vision and red vision are important for damselfish (Pomacentridae): structural and expression variation in opsin genes. Mol. Ecol. 26, 1323–1342 (2017).
Heinermann, P. H. Yellow intraocular filters in fishes. Exp. Biol. 43, 127–147 (1984).
Gerlach, T., Sprenger, D. & Michiels, N. K. Fairy wrasses perceive and respond to their deep red fluorescent coloration. Proc. R. Soc. B Biol. Sci. 281, 20140787 (2014).
Shimomura, O., Johnson, F. H. & Saiga, Y. Extraction, purification and properties of aequorin, a bioluminescent protein from the luminous hydromedusan. Aequorea. J. Cell. Comp. Physiol. 59, 223–239 (1962).
Hayashi, S. & Toda, Y. A novel fluorescent protein purified from eel muscle. Fish. Sci. 75, 1461–1469 (2009).
Gruber, D. F. et al. Adaptive evolution of eel fluorescent proteins from fatty acid binding proteins produces bright fluorescence in the marine environment. PLoS ONE 10, e0140972 (2015).
Guarnaccia, A. M., Krivoshik, S. R., Sparks, J. S., Gruber, D. F. & Gaffney, J. P. Discovery and characterization of a bilirubin inducible green fluorescent protein from the moray eel Gymnothorax zonipectis. Front. Mar. Sci. 8, 678571 (2021).
Anthes, N., Theobald, J., Gerlach, T., Meadows, M. G. & Michiels, N. K. Diversity and ecological correlates of red fluorescence in marine fishes. Front. Ecol. Evol. 4, 126 (2016).
Michiels, N. K. et al. Red fluorescence in reef fish: a novel signalling mechanism? BMC Ecol. 8, 16 (2008).
Gerlach, T., Theobald, J., Hart, N. S., Collin, S. P. & Michiels, N. K. Fluorescence characterisation and visual ecology of pseudocheilinid wrasses. Front. Zool. 13, 13 (2016).
De Brauwer, M. et al. Biofluorescence as a survey tool for cryptic marine species. Conserv. Biol. 32, 706–715 (2018).
Gruber, D. F. & Sparks, J. S. First report of biofluorescence in arctic snailfishes and rare occurrence of multiple fluorescent colors in a single species. Am. Mus. Novit. 2020, 1–12 (2021).
Juhasz-Dora, T., Teague, J., Doyle, T. K. & Maguire, J. First record of biofluorescence in lumpfish (Cyclopterus lumpus), a commercially farmed cleaner fish. J. Fish. Biol. 101, 1058–1062 (2022).
Miyamoto, K., Mcmahan, C. D. & Kaneko, A. Parascolopsis akatamae, a new species of dwarf monocle bream (Perciformes: Nemipteridae) from the Indo-West Pacific, with redescription of closely related species P. eriomma. Zootaxa 4881, 91–103 (2020).
Schliewen, U. K., Kovačić, M., Cerwenka, A. F., Svensen, R. & Ordines, F. Lebetus patzneri (Teleostei: Gobiidae), a new goby species from the Balearic Islands, western Mediterranean, with first records of Lebetus guilleti (Le Danois, 1913) from this area and Norway, and with notes on its biology. Zootaxa 4706, 231–254 (2019).
Vaccani, A. C., Freret-Meurer, N. V., Bertoncini, Á. A. & Santos, L. N. Shining in the dark: First record of biofluorescence in the seahorse Hippocampus reidi. PLoS ONE 14, e0220561 (2019).
Huie, J. M., Wainwright, D. K., Summers, A. P. & Cohen, K. E. Sticky, stickier and stickiest – a comparison of adhesive performance in clingfish, lumpsuckers and snailfish. J. Exp. Biol. 225, jeb244821 (2022).
De Brauwer, M. & Hobbs, J.-P. A. Stars and stripes: biofluorescent lures in the striated frogfish indicate role in aggressive mimicry. Coral Reefs 35, 1171 (2016).
Smith, W. L., Everman, E. & Richardson, C. Phylogeny and taxonomy of flatheads, scorpionfishes, sea robins, and stonefishes (Percomorpha: Scorpaeniformes) and the evolution of the Lachrymal Saber. Copeia 106, 94–119 (2018).
Tea, Y.-K., Allen, G. R. & Dailami, M. Redescription of Cirrhilabrus solorensis Bleeker, with description of two new species of fairy wrasses (Teleostei: Labridae: Cirrhilabrus). Ichthyol. Herpetol. 109, 669–684 (2021).
Poding, L. H. et al. New observations of fluorescent organisms in the Banda Sea and in the Red Sea. PLOS ONE 19, e0292476 (2024).
Rabosky, D. L. et al. An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395 (2018).
Price, S. A., Claverie, T., Near, T. J. & Wainwright, P. C. Phylogenetic insights into the history and diversification of fishes on reefs. Coral Reefs 34, 997–1009 (2015).
Bitton, P.-P. et al. Red fluorescence of the triplefin Tripterygion delaisi is increasingly visible against background light with increasing depth. R. Soc. Open Sci. 4, 161009 (2017).
Endler, J. A. The color of light in forests and its implications. Ecol. Monogr. 63, 2–27 (1993).
Cowman, P. F. Historical factors that have shaped the evolution of tropical reef fishes: a review of phylogenies, biogeography, and remaining questions. Front. Genet. 5, 394 (2014).
Price, S. A. et al. Two waves of colonization straddling the K–Pg boundary formed the modern reef fish fauna. Proc. R. Soc. B Biol. Sci. 281, 20140321 (2014).
Budd, A. F. Diversity and extinction in the Cenozoic history of Caribbean reefs. Coral Reefs 19, 25–35 (2000).
Lipps, J. H. & Stanley, G. D. Reefs through time: an evolutionary view. In Coral Reefs at the Crossroads (eds Hubbard, D. K. et al.) 175–196 (Springer Netherlands, Dordrecht, 2016).
Simpson, C., Kiessling, W., Mewis, H., Baron‐Szabo, R. C. & Müller, J. Evolutionary diversification of reef corals: a comparison of the molecular and fossil records. Evolution 65, 3274–3284 (2011).
Siqueira, A. C., Kiessling, W. & Bellwood, D. R. Fast-growing species shape the evolution of reef corals. Nat. Commun. 13, 2426 (2022).
Quattrini, A. M. et al. Palaeoclimate ocean conditions shaped the evolution of corals and their skeletons through deep time. Nat. Ecol. Evol. 4, 1531–1538 (2020).
Utne-Palm, A. C. & Bowmaker, J. K. Spectral sensitivity of the two-spotted goby Gobiusculus flavescens (Fabricius): a physiological and behavioural study. J. Exp. Biol. 209, 2034–2041 (2006).
Phillips, G. A. C., Carleton, K. L. & Marshall, N. J. Multiple genetic mechanisms contribute to visual sensitivity variation in the labridae. Mol. Biol. Evol. 33, 201–215 (2016).
Mosk, V. et al. Spectral sensitivities of the seahorses Hippocampus subelongatus and Hippocampus barbouri and the pipefish Stigmatopora argus. Vis. Neurosci. 24, 345–354 (2007).
Kondrashev, S. L. Long-wave sensitivity in the masked greenling (Hexagrammos octogrammus), a shallow-water marine fish. Vis. Res 48, 2269–2274 (2008).
R Core Team: R: A Language and Environment for Statistical Computing (R Foundation for Statistical Computing, Vienna, Austria, 2023).
Revell, L. J. phytools 2.0: an updated R ecosystem for phylogenetic comparative methods (and other things). PeerJ 12, e16505 (2024).
Beaulieu, J., O’Meara, B., Oliver, J. & Boyko, J. corHMM: Hidden Markov models of character evolution. R Package Version 2 (2021).
Boyko, J. D. & Beaulieu, J. M. Generalized hidden Markov models for phylogenetic comparative datasets. Methods Ecol. Evol. 12, 468–478 (2021).
Boyko, J. Automatic discovery of optimal discrete character models. Preprint at bioRxivhttps://doi.org/10.1101/2024.11.15.623760 (2024).
Beaulieu, J. M. & O’Meara, B. C. Detecting hidden diversification shifts in models of trait-dependent speciation and extinction. Syst. Biol. 65, 583–601 (2016).
Larouche, O. et al. Reef-associated fishes have more maneuverable body shapes at a macroevolutionary scale. Coral Reefs 39, 1427–1439 (2020).
Acknowledgements
We are grateful to D. Harrington and T. Romano at Mystic Aquarium and N. Hillgarth and R. Elkus at Birch Aquarium (UCSD) for access to their collections. We would also like to thank J. Boyko for helpful discussions about corHMM. Funding for this work was provided by the National Science Foundation (DEB 1257555 to J.S.S.; GRFP DGE-1938103 to E.M.C.; GRFP DGE-1746914 to J.M.H.), the American Museum of Natural History, and the Richard Gilder Graduate School (Gerstner Scholarship to R.P.M.).
Author information
Authors and Affiliations
Contributions
E.M.C. and J.S.S. conceived the project. E.M.C, J.S.S., D.F.G, M.A.T, K.E.C, and J.M.H collected the data. E.M.C, J.S.S, D.F.G., R.P.M, M.A.T, and J.M.H. analyzed the data. E.M.C., R.P.M, M.A.T, J.M.H., and J.S.S. wrote the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Nils Anthes, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.”
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Carr, E.M., Martin, R.P., Thurman, M.A. et al. Repeated and widespread evolution of biofluorescence in marine fishes. Nat Commun 16, 4826 (2025). https://doi.org/10.1038/s41467-025-59843-7
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1038/s41467-025-59843-7